ABSTRACT
Caldicellulosiruptor bescii, an anaerobic Gram-positive bacterium with an optimal growth temperature of 78°C, is the most thermophilic cellulose degrader known. It is of great biotechnological interest, as it efficiently deconstructs nonpretreated lignocellulosic plant biomass. Currently, its genetic manipulation relies on a mutant uracil auxotrophic background strain that contains a random deletion in the pyrF genome region. The pyrF gene serves as a genetic marker to select for uracil prototrophy, and it can also be counterselected for loss via resistance to the compound 5-fluoroorotic acid (5-FOA). To expand the C. bescii genetic tool kit, kanamycin resistance was developed as a selection for genetic manipulation. A codon-optimized version of the highly thermostable kanamycin resistance gene (named Cbhtk) allowed the use of kanamycin selection to obtain transformants of either replicating or integrating vector constructs in C. bescii. These strains showed resistance to kanamycin at concentrations >50 μg · ml−1, whereas wild-type C. bescii was sensitive to kanamycin at 10 μg · ml−1. In addition, placement of the Cbhtk marker between homologous recombination regions in an integrating vector allowed direct selection of a chromosomal mutation using both kanamycin and 5-FOA. Furthermore, the use of kanamycin selection enabled the targeted deletion of the pyrE gene in wild-type C. bescii, generating a uracil auxotrophic genetic background strain resistant to 5-FOA. The pyrE gene functioned as a counterselectable marker, like pyrF, and was used together with Cbhtk in the ΔpyrE background strain to delete genes encoding lactate dehydrogenase and the CbeI restriction enzyme.
IMPORTANCE Caldicellulosiruptor bescii is a thermophilic anaerobic bacterium with an optimal growth temperature of 78°C, and it has the ability to efficiently deconstruct nonpretreated lignocellulosic plant biomass. It is, therefore, of biotechnological interest for genetic engineering applications geared toward biofuel production. The current genetic system used with C. bescii is based upon only a single selection strategy, and this uses the gene involved in a primary biosynthetic pathway. There are many advantages with an additional genetic selection using an antibiotic. This presents a challenge for thermophilic microorganisms, as only a limited number of antibiotics are stable above 50°C, and a thermostable version of the enzyme conferring antibiotic resistance must be obtained. In this work, we have developed a selection system for C. bescii using the antibiotic kanamycin and have shown that, in combination with the biosynthetic gene marker, it can be used to efficiently delete genes in this organism.
INTRODUCTION
Caldicellulosiruptor bescii is a thermophilic anaerobic Gram-positive bacterium with an optimal growth temperature of 78°C, and it has the ability to efficiently deconstruct nonpretreated lignocellulosic plant biomass (1, 2). It is, therefore, of biotechnological interest for genetic engineering applications geared toward biofuel production (3, 4). Such organisms require a tractable genetic system for metabolic engineering applications, and such a system has been established for C. bescii in recent years (5, 6). A major step in developing a genetic system for C. bescii was the discovery that it has a restriction-modification system that severely limits its transformability (7). This barrier to transformation was overcome through the use of a methylated donor plasmid with a methylation pattern that protected the transformed DNA from restriction digestion (5). The system was further improved by deleting the gene encoding the restriction enzyme, CbeI, making it possible to use nonmethylated donor DNA in transformation (6).
The current C. bescii genetic system is based upon use of the uracil biosynthetic pathway gene pyrF as a selectable marker for uracil prototrophy in a uracil auxotrophic mutant strain (5, 6). The pyrF gene encodes orotidine 5′-monophosphate (OMP) decarboxylase, which is required for uracil biosynthesis. This enzyme is also able to convert the pyrimidine analog 5-fluoroorotic acid (5-FOA) to the toxic product fluorodeoxyuridine (8). Strains deficient in OMP decarboxylase activity are, therefore, uracil auxotrophs and resistant to 5-FOA. In the development of pyrF as a counterselectable marker for C. bescii, mutant uracil auxotrophic strains of C. bescii were isolated using 5-FOA to select for cells with random mutations in the pyrF gene. The first strain isolated contained a deletion within pyrB, pyrC, and pyrF and required complementation with all three of these genes (5). A second uracil auxotrophic mutant was isolated that had a random deletion within pyrF and pyrA, and this strain, designated JWCB005, required complementation with only the pyrF gene (9).
The JWCB005 strain has become the basis for all genetically modified strains of C. bescii to date and has led to the development of genetic strategies and engineering efforts in this and related organisms. For example, it has been used in developing a replicating shuttle vector for C. bescii and other Caldicellulosiruptor species (9), and this vector has been used for homologous overexpression of the genes celA, encoding a bifunctional glycoside hydrolase (10), and xor, encoding a tungsten-dependent oxidoreductase (11). Numerous gene deletions in addition to cbeI have also been constructed, including the ldh gene encoding lactate dehydrogenase (12), a gene cluster encoding pectin-degrading enzymes (13), the celA gene (14), and a gene cluster encoding [Ni-Fe] hydrogenase maturation enzymes (15). The JWCB005 genetic background has also been used in genetic engineering efforts to insert foreign gene expression constructs into the chromosome for production of ethanol (16, 17), for detoxification of furan aldehydes (18), and for enhanced capability to deconstruct crystalline cellulose (19) and xylan (20).
While the counterselectable pyrF marker is versatile and has been used in genetic systems developed for various thermophilic bacteria as well as for archaea (21, 22), there are certain advantages that can be gained with the additional option of genetic selection with an antibiotic. Antibiotic selection does not preclude the use of defined medium and can be used in conjunction with nutritional markers to increase selective pressure. For thermophilic organisms, the use of antibiotics presents somewhat of a challenge, as the number of antibiotics that have stability at temperatures >50°C is limited (23). Furthermore, to employ these antibiotics, there must also be a corresponding thermostable version of the enzyme conferring antibiotic resistance.
Once such antibiotic that has been demonstrated as sufficiently thermostable is kanamycin (23), and thermostable versions of the corresponding resistance marker are also available. The Staphylococcus aureus kanamycin resistance gene knt, encoding kanamycin nucleotidyltransferase (KNT), was passaged through an Escherichia coli mutator strain, and thermostable variants were selected in Bacillus stearothermophilus at temperatures up to 70°C (24). This KNT variant, which contained two amino acid substitutions, was further enhanced for thermostability at temperatures up to 80°C by directed evolution in Thermus thermophilus. The resulting protein contained 19 amino acid substitutions and was named HTK for highly thermostable kanamycin nucleotidyltransferase (25). These thermostable kanamycin resistance markers have been used in development of genetic systems for a number of thermophilic organisms, including Moorella thermoacetica at 55°C (26), Thermoanaerobacter tengcongensis at 60°C (27), Geobacillus thermoglucosidasius at 68°C (28), Thermus thermophilus at 70°C (29), and Thermotoga spp. at 77°C (30).
We show that kanamycin selection can also be applied in C. bescii, thereby expanding the genetic tool kit available for this organism. Cbhtk, a codon-optimized version of the htk gene, conferred kanamycin resistance to C. bescii and was used to construct an alternative genetic background strain based on a clean deletion of the pyrE gene in wild-type C. bescii. Deletion of pyrE resulted in a strain auxotrophic for uracil and resistant to 5-FOA. The pyrE gene was applied as a marker, in addition to Cbhtk, and together these were used to construct additional deletions in this strain using kanamycin for selection of plasmid integration and 5-FOA for counterselection of plasmid loss, establishing this system as another method for making iterative chromosomal modifications in C. bescii.
MATERIALS AND METHODS
Growth of C. bescii.
C. bescii was routinely grown under strict anaerobic conditions at 70 to 75°C. For genetic applications, C. bescii was cultured in low-osmolarity defined (LOD) medium (31) with argon used in the headspace. For growth of uracil auxotrophic strains, uracil was added at a concentration of 20 to 40 μM. When required, kanamycin was used at a concentration of 50 μg · ml−1, unless otherwise noted. Solid medium was prepared by mixing preheated 2× LOD medium with an equal volume of 6% (wt/vol) agar. A volume of 100 μl fresh culture or culture dilution was pipetted into an empty petri dish, and medium was poured into the plate while swirling to distribute the cells within the medium. Plates were allowed to solidify and cool 20 min, after which the plates were inverted, placed into anaerobic canisters flushed with argon, and incubated at 65 to 75°C for 4 days. Colonies were picked by stabbing through the agar with a toothpick and inoculated into 4 ml medium in screw-cap Hungate tubes. Plating and colony picking were done either on the benchtop (aerobically) or in an anaerobic Coy chamber (for increased cell viability).
Growth experiments were performed in modified DSMZ 516 medium having the following composition per liter: 1× salt solution, 1× vitamin solution, 1× trace element solution, 0.16 μM sodium tungstate, 0.25 mg resazurin, 5 g cellobiose, 0.5 g yeast extract, 1 g cysteine hydrochloride, 1 g sodium bicarbonate, and 1 mM potassium phosphate buffer (pH 7.2). The 50× stock salt solution contained the following per liter: 16.5 g NH4Cl, 16.5 g KCl, 16.5 g MgCl2·6H2O and 7 g CaCl2·2H2O. Stock solutions of 200× vitamins and 1,000× trace elements were prepared as previously described (2). This complex medium containing cellobiose as the carbon source is referred to as CC516. A defined version of this medium was prepared by excluding the yeast extract and supplementing with uracil when necessary, and this medium is referred to as DC516. Culture headspace used with CC516 and DC516 media was composed of 80% N2 and 20% CO2.
Vector construction.
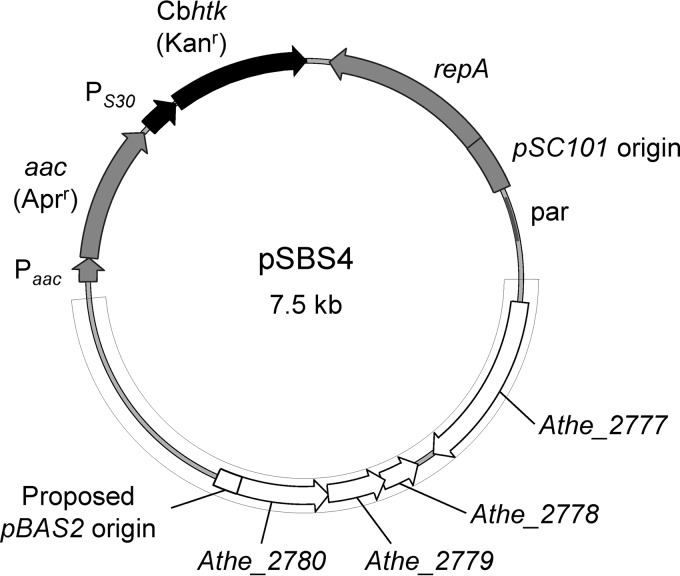
The htk gene sequence (from pUB110; GenBank accession no. AB121443) was codon optimized for use in C. bescii and is referred to as Cbhtk. Codons were changed to those that occur most frequently in C. bescii genes, thereby eliminating any potential rare codons (see Fig. S1 in the supplemental material). The codon-optimized Cbhtk gene was synthesized by GeneArt gene synthesis (Thermo Fisher Scientific, Inc.). The pSBS4 replicating shuttle vector employing the Cbhtk gene (Fig. 1) was constructed by replacing the pyrF gene with Cbhtk in pDCW89 (9) via Gibson assembly (32). The Cbhtk cassette, unless otherwise noted, employs the promoter region of Athe_2105 (6), a hypothetical conserved protein annotated as sigma 54 modulation protein/ribosomal protein S30EA; this promoter is referred to as PS30.
FIG 1.
Replicating shuttle vector with Cbhtk cassette for selection of transformants via kanamycin resistance. Plasmid map of pSBS4 with Cbhtk cassette (black), E. coli elements (gray), and C. bescii elements (white) from native plasmid pBAS2/pATHE02 (delineated by gray line).
The pJMC010 plasmid (see Fig. S2 in the supplemental material) for insertion of PslpCbhtk at the ΔcbeI locus was constructed via Gibson assembly (32) of the following PCR products: the plasmid backbone, including the pyrF cassette and E. coli elements (amplified from pDCW121 [12]), the 200-bp promoter region immediately upstream of the slp gene (Athe_2303), the Cbhtk gene, and ∼1-kb flanking regions to cbeI. The Cbhtk gene was placed under the control of the Pslp promoter, and the PslpCbhtk cassette was placed between the cbeI flanking regions. The plasmid was found to contain a mutation in the htk gene (L227I) which did not appear to affect the kanamycin resistance of the strain generated using this plasmid.
For deletion of pyrE (Athe_1382), pGL090 (see Fig. S2 in the supplemental material) was constructed via Gibson assembly (New England BioLabs) of the following PCR products: ∼1-kb upstream and downstream flanking regions to pyrE and the Cbhtk cassette and E. coli elements amplified from pSBS4. For deletion of cbeI (Athe_2438), pGL100 (see Fig. S2) was constructed via Gibson assembly (New England BioLabs) of the following PCR products: ∼0.5-kb upstream and downstream flanking regions to cbeI, the Aprr (apramycin resistance) and Kanr (kanamycin resistance) markers (amplified from pGL090), the E. coli ColE1 replication origin (amplified from pSET152), and the pyrE gene. The pyrE gene was placed immediately downstream from the Cbhtk gene in a synthetic operon, referred to as the Cbhtk-pyrE cassette. A 20-bp ribosomal binding site region (5′-aggaggtttggtgagtagtt), taken from immediately upstream of the slp gene (Athe_2303), was placed between Cbhtk and pyrE. A terminator region was placed at the end of the operon and consisted of 122 bp from the 3′ untranslated region of the slp gene, wherein lies a predicted rho-independent terminator (33). The general-purpose cloning plasmid, pGL104 (see Fig. S2), was constructed via Gibson assembly (New England BioLabs) of the following PCR products: the Cbhtk-pyrE cassette, a mini multiple cloning site containing restriction sites that are rare in C. bescii (SphI-AscI-PmeI-NotI), and E. coli plasmid elements (Aprr cassette and pSC101 origin). For deletion of ldh (Athe_1918), flanking regions of ∼1 kb on either side of the ldh gene were combined via splice-overlap extension PCR (34) and cloned into pGL104 via the SphI and AscI restriction sites to generate pGL108 (see Fig. S2). All plasmids were sequence verified.
Plasmid DNA methylation.
For transformation of C. bescii wild type (DSM 6725) and the strain MACB1018, plasmid DNA was methylated with recombinant CbeI methyltransferase (M.CbeI). The E. coli strain JW284, harboring the M.CbeI expression plasmid (pDCW73), was used for heterologous expression of tagged M.CbeI, as previously described (5). M.CbeI was purified from cell lysate using the His-Spin protein miniprep kit (Zymo Research). Plasmid DNA was subjected to methylation by M.CbeI followed by purification, essentially as previously described (5).
C. bescii transformation and strain construction.
For competent cell preparation and transformation, C. bescii strains were inoculated to a cell density of 6 × 105 to 8 × 105 cells · ml−1 into 500 ml LOD medium containing amino acids (LOD-AA) (31) and grown at 70°C to an optical density at 680 nm (OD680) of 0.06 to 0.07. Cultures were cooled for 10 min in a bath of running cool water. Cells from a 500-ml culture were harvested at 6,000 × g for 10 min. Cell pellets were washed twice consecutively in 250 ml of 10% (wt/vol) sucrose, with or without gentle suspension, and centrifugation was performed at 6,000 × g for 10 min each time. Cells were washed a third time with 50 ml of 10% (wt/vol) sucrose, after which cells were suspended in 1 ml of 10% (wt/vol) sucrose, transferred to a microcentrifuge tube, and centrifuged at 14,000 rpm for 30 s. Most of the supernatant was removed, and competent cells were gently suspended to a final volume of 100 to 125 μl in 10% (wt/vol) sucrose. Plasmid DNA (0.5 to 1 μg) was added in a volume of up to 5 μl to a 50-μl aliquot of competent cells, and this mixture was then transferred to a 1 mM gap electroporation cuvette. Cells were electroporated in a Bio-Rad gene pulser using the following conditions: 1.8 kV, 400 Ω, and 25 μF. Immediately after electroporation, cells were transferred to 20 ml of LOC medium (31) preheated to 70°C, and these recovery cultures were incubated at 70°C for 1 to 4 h. For selection of transformants, aliquots from the recovery cultures were periodically removed and subcultured into low-osmolarity complex (LOC) medium (31) containing 50 μg · ml−1 kanamycin, using a 10% inoculum. All centrifugation, wash, and electroporation steps were performed under aerobic conditions at room temperature. Transformants were colony purified at least once on solid LOD medium containing 50 μg · ml−1 kanamycin.
C. bescii strains used and constructed in this study are listed in Table 1. C. bescii JWCB018 (ΔpyrFA ΔcbeI) and the wild type (using methylated plasmid) were transformed with the replicating shuttle vector pSBS4 to generate strains resistant to kanamycin. The wild-type transformation isolate was purified once on solid LOD medium containing 50 μg · ml−1 kanamycin and designated MACB1015. The presence of the replicating pSBS4 plasmid was verified using total DNA isolated from MACB1015 to transform E. coli so that isolated plasmids could be restriction digested and compared to the original transformation plasmid. JWCB018 was transformed with pJMC010 for insertion of Cbhtk at the ΔcbeI locus. Counterselection for plasmid loss was performed on LOD solid medium containing 4 mM 5-FOA, 40 μM uracil, and 10 μg · ml−1 kanamycin. The isolate containing Cbhtk inserted at the ΔcbeI locus was purified twice on solid LOD medium containing 40 μM uracil and 25 μg · ml−1 kanamycin, and the strain was designated RKCB106. Wild-type C. bescii was transformed with methylated pGL090, which targeted integration at the pyrE locus. Counterselection on LOD solid medium containing 4 mM 5-FOA and 40 μM uracil resulted in plasmid loss and pyrE deletion. Isolates were further purified once on solid LOD medium containing 40 μM uracil. The deleted pyrE gene was confirmed by sequencing the locus in five isolates, and one isolate was designated MACB1018. The ΔpyrE strain MACB1018 was transformed with methylated pGL100 (for deletion of cbeI) or pGL108 (for deletion of ldh), which each contained the Cbhtk marker for transformation selection using kanamycin and the pyrE marker for counterselection using 5-FOA. Counterselection and purification were performed as described above for MACB1018. Strains containing deletions of cbeI and ldh were designated MACB1032 and MACB1034, respectively. For routine screening of cultures, genomic DNA was isolated using the ZymoBead genomic DNA kit (Zymo Research), with an added brief sonication step to improve cell lysis. All chromosomal mutations were verified by PCR screening and sequencing.
TABLE 1.
C. bescii strains used and constructed in this study
| Strain designation | Parent | Transforming plasmid | Description | Reference |
|---|---|---|---|---|
| DSM 6725 | Wild type | 2 | ||
| JWCB018 | JWCB005 | pDCW88 | ΔpyrFA ΔcbeI | 6 |
| MACB1015 | DSM 6725 | pSBS4 | pSBS4 | This study |
| RKCB106 | JWCB018 | pJMC010 | ΔpyrFA ΔcbeI::PslpCbhtk | This study |
| MACB1018 | DSM 6725 | pGL090 | ΔpyrE | This study |
| MACB1032 | MACB1018 | pGL100 | ΔpyrE ΔcbeI | This study |
| MACB1034 | MACB1018 | pGL108 | ΔpyrE Δldh | This study |
RESULTS AND DISCUSSION
Sensitivity of C. bescii to kanamycin.
The antibiotic kanamycin is widely used in bacterial genetic systems and is well suited for application in thermophilic bacteria due to its high stability over a range of temperatures and pH values (23). Thermostable versions of the S. aureus kanamycin nucleotidyltransferase gene (knt) have been developed (24, 25), and these have been used in a number of thermophilic bacteria, e.g., species of Thermoanaerobacter (27), Geobacillus (28), Thermus (29), and Thermotoga (30). To determine if kanamycin would be an option for selection in C. bescii, the sensitivity of C. bescii to kanamycin was determined. Wild-type and JWCB018 (ΔpyrFA ΔcbeI) (6) strains of C. bescii were inoculated into medium containing kanamycin at concentrations ranging from 10 to 200 μg · ml−1, and these cultures were incubated at 75°C to late stationary phase (20 h). Growth of both wild type and JWCB018 was inhibited by 10 μg · ml−1 kanamycin (see Fig. S3 in the supplemental material), making kanamycin selection a viable option for further development in C. bescii.
Use of the thermostable htk gene in transformation of C. bescii.
To test the possibility of employing kanamycin as a selection in C. bescii, the highly thermostable kanamycin nucleotidyltransferase (HTK), having increased thermostability up to 80°C (25), was chosen to test as a selectable marker. The sequence of the htk gene was optimized to use only the most frequently occurring codons in C. bescii (see Fig. S1 in the supplemental material). A replicating shuttle vector employing the Cbhtk gene was constructed by replacing the pyrF gene with Cbhtk in pDCW89 (9), thereby generating pSBS4 (Fig. 1). C. bescii contains a restriction modification system that severely affects its transformability, such that methylation of transforming DNA using the corresponding methylase is necessary (5). This barrier to transformation was overcome by deletion of the gene encoding the restriction enzyme CbeI, generating the JWCB018 strain (6). We selected this strain to use for transformation trials with pSBS4. As a positive control for transformation, the pDCW89 replicating shuttle vector (9) was used with uracil prototrophic selection. After electroporation, samples from the recovery cultures were inoculated into LOD medium either lacking uracil, for selection of pDCW89 transformants, or containing uracil and 50 μg · ml−1 kanamycin, for selection of pSBS4 transformants. Growth was routinely observed in these selective outgrowth cultures for both pDCW89 and pSBS4 electroporation trials, indicating that the Cbhtk gene was functional in conferring kanamycin resistance to C. bescii. Accordingly, wild-type C. bescii competent cells were prepared and electroporated with pSBS4 that had been methylated with heterologously expressed and purified M.CbeI (5), and similar results were observed. A purified pSBS4 transformation isolate was designated MACB1015. To confirm the presence of the replicating plasmid, total DNA from MACB1015 was transformed into E. coli, and 12 plasmid isolates were screened by restriction digestion. All isolates had restriction digestion patterns that were identical to pSBS4, indicating that the intact pSBS4 plasmid was present in the strain and that no plasmid rearrangement had taken place (see Fig. S4 in the supplemental material).
A major advantage of antibiotic selection versus uracil prototrophic selection is that there is no concern about nutrient carryover from the postelectroporation, rich recovery medium into defined selective medium. For uracil prototrophic selection, only ∼1 to 2% inoculum can be used from the recovery culture into defined medium lacking uracil to prevent the carryover of too much uracil that would negate the selection. For kanamycin selection, up to 10% inoculum can be used, allowing 100% of the recovery culture to be subcultured into kanamycin-containing liquid medium without the experiment becoming too cumbersome.
Use of Cbhtk for direct selection of chromosomal mutations.
Due to the low efficiency of transformation and recombination, introduction of a chromosomal mutation in C. bescii requires the use of a circular plasmid as opposed to linear DNA. A selection is used for plasmid integration into the chromosome, and a counterselection is used for resolving a second crossover event to achieve the desired mutation. Once integration of a plasmid containing the pyrF marker is obtained, counterselection for loss of pyrF using 5-FOA can yield the desired second crossover event or can result in reversion to the original state.
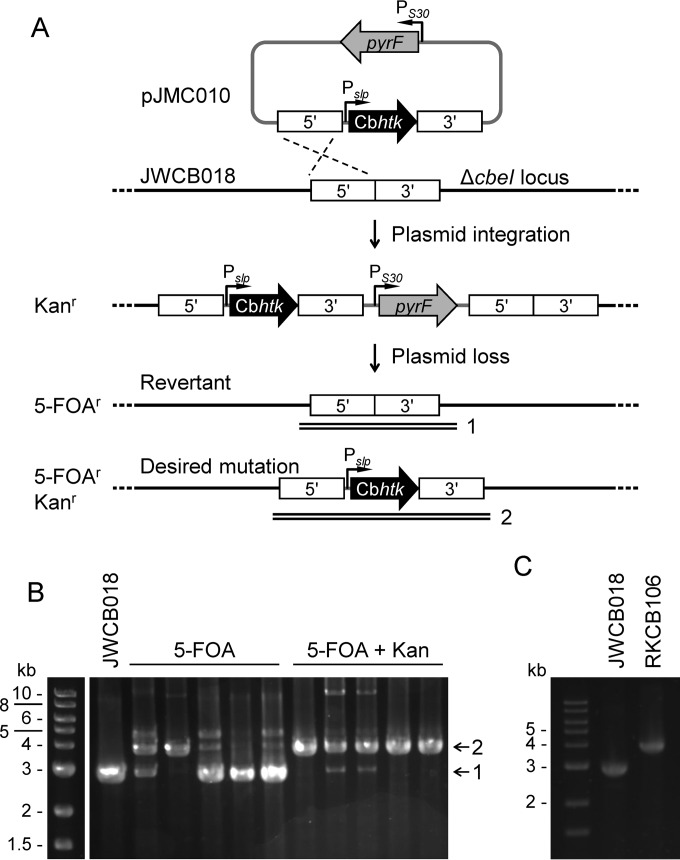
Placement of the Cbhtk marker between the flanking regions for homologous recombination allows for direct selection of only the desired second crossover event, resulting in gene insertion and/or deletion. This direct selection method was demonstrated by using pJMC010 to insert Cbhtk at the ΔcbeI locus (Athe_2438) in C. bescii strain JWCB018 (ΔpyrFA ΔcbeI). The pJMC010 plasmid contained the PslpCbhtk cassette between flanking regions to the cbeI locus and also contained the pyrF cassette on the plasmid backbone. Chromosomal integration of the plasmid was selected via kanamycin resistance, and the second crossover event was resolved by using both 5-FOA and kanamycin to select for loss of the pyrF marker while retaining the Cbhtk marker (Fig. 2A). When counterselection for plasmid loss was performed in the presence of 5-FOA and kanamycin, all five screened isolates contained the desired mutation, although two also contained faint bands for the full plasmid as well as the wild-type allele. When selection with only 5-FOA was performed, four out of five screened isolates had predominant bands indicating revertants (Fig. 2B). One ΔcbeI::PslpCbhtk isolate was further colony purified on medium containing kanamycin, and this strain was designated RKCB106 (Fig. 2C). For C. bescii, it is not uncommon for the initial screening of transformants or 5-FOA isolates to display bands indicating the presence of two alleles (the wild type and the desired allele) or the integrated plasmid. Typically, the purest isolates are chosen for moving forward, and colony purification with routine PCR screening results in final strains that are pure.
FIG 2.
Direct selection of chromosomal mutations using Cbhtk. (A) Scheme showing selection method for targeted insertion of the PslpCbhtk expression cassette (black) by homologous recombination (dashed lines) into the ΔcbeI locus in C. bescii strain JWCB018 (ΔpyrFA). Kanamycin selected for plasmid integration. To resolve the second crossover event, 5-FOA selected only for plasmid loss, and 5-FOA with kanamycin selected for plasmid loss while maintaining the inserted Cbhtk cassette. Numbered double lines indicate the locations of confirmation PCR products shown in B. (B) PCR screening of isolates counterselected for a second crossover event using solid LOD medium containing 4 mM 5-FOA with or without 10 μg · ml−1 kanamycin. Colonies that resulted from 5-FOA selection were grown in LOD medium with 40 μM uracil, while colonies that resulted from 5-FOAr and Kanr selection were grown in LOD medium with 40 μM uracil and 25 μg · ml−1 kanamycin. The Cbhtk knock-in had an expected amplicon size of 3.9 kb (2) compared to 2.9 kb (1), which was the expected size for the JWCB018 strain. (C) PCR confirmation of PslpCbhtk insertion at the ΔcbeI locus in RKCB106 compared to parent strain JWCB018. PCR products shown in B and C were amplified with primers outside the cbeI flanking regions used for homologous recombination.
Kanamycin resistance of Cbhtk-containing strains.
Since C. bescii is highly sensitive to kanamycin, it was of interest to test the kanamycin resistance level in strains containing the Cbhtk marker. Two strains were used: the MACB1015 strain containing the Cbhtk cassette expressed on a replicating shuttle vector and the RKCB106 strain containing a genome-integrated copy of the PslpCbhtk cassette. When cultivated in medium containing various concentrations of kanamycin up to 800 μg · ml−1 for 20 h, the RKCB106 strain reached the same cell density as the control culture lacking kanamycin. The MACB1015 strain showed decreased cell density at concentrations ≥200 μg · ml−1 kanamycin (see Fig. S5 in the supplemental material). The difference in the levels of kanamycin resistance between the two strains may be due to the location of Cbhtk expression (replicating plasmid versus chromosome). The copy number of the pSBS4 shuttle vector is 1:1 with the chromosome, assuming it is the same as that previously published for pDCW89, upon which it is based (9); however, little is known about the replication origin of this plasmid and how plasmid partitioning to daughter cells is controlled. Differing expression levels of the Cbhtk marker (PS30 versus Pslp) is also a possible contributing factor. The native genes downstream of both promoters are expressed at or above the expression level of the gene encoding the glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase, with the native gene for Pslp being expressed about 2-fold higher than that of PS30 (see Fig. S6 in the supplemental material).
Use of kanamycin selection for deletion of pyrE.
Uracil auxotrophic strains can be generated by deletion of a number of genes encoding enzymes within the uracil biosynthetic pathway. Two of these can also serve as counterselectable markers: the pyrF and pyrE genes, encoding OMP decarboxylase and orotate phosphoribosyltransferase, respectively. It has been shown in Saccharomyces cerevisiae and in various bacteria and archaea that deletion of the genes encoding these enzymes can confer resistance to 5-FOA, an analog of a uracil biosynthetic intermediate (8, 21, 22, 35).
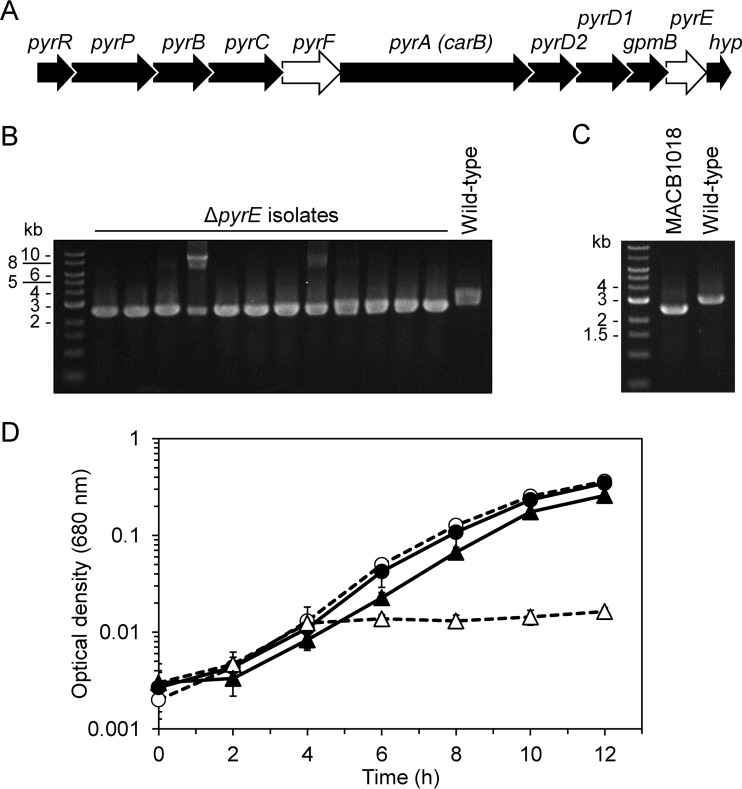
In C. bescii, the genes encoding enzymes in the uracil biosynthetic pathway are arranged in an 11-gene putative operon (Fig. 3A). The pyrF gene lies in the middle of the putative operon, while the pyrE gene is located near the end of the operon, with only one gene encoding a small hypothetical protein lying immediately downstream from it. We therefore selected pyrE as a target for deletion due to the lower possibility of detrimental polar effects arising during strain construction (due to integration of the entire deletion plasmid into the middle of the operon). The Cbhtk cassette was used in a vector designed with ∼1-kb flanking regions to the pyrE gene (see Fig. S2 in the supplemental material). Wild-type C. bescii was transformed with methylated plasmid DNA. After electroporation, cells were transferred to recovery medium and incubated at 72°C for 2 h, during which time samples were withdrawn and inoculated into medium containing 50 μg · ml−1 kanamycin for outgrowth. Genomic DNA was extracted from cultures that were able to grow in kanamycin-containing medium, and PCR screening confirmed the presence of the Cbhtk construct at the pyrE locus. These single-crossover isolates were counterselected for plasmid loss and pyrE deletion using solid medium containing 5-FOA and uracil. Colonies were subcultured into liquid medium containing uracil, and all 12 screened isolates contained a deletion of pyrE (Fig. 3B), indicating that loss of pyrE in C. bescii confers resistance to 5-FOA. After further colony purification and sequence verification, one ΔpyrE isolate was designated MACB1018 (Fig. 3C). Growth of wild-type C. bescii and MACB1018 in defined medium with and without supplemental uracil showed that MACB1018 exhibits uracil auxotrophy, which can be attributed to loss of the pyrE gene (Fig. 3D). The initial minimal growth of MACB1018 (to an OD680 of 0.016) can be attributed to uracil carryover from the inoculum culture.
FIG 3.
Deletion of pyrE in C. bescii confers uracil auxotrophy. (A) Uracil biosynthetic operon in C. bescii (Athe_1373 to Athe_1383). Gene symbols stand for the following enzymes: pyrR, uracil phosphoribosyltransferase; pyrP, uracil-xanthine permease; pyrB, aspartate carbamoyltransferase catalytic subunit; pyrC, dihydroorotase; pyrF, OMP decarboxylase; pyrA (also known as carB), carbamoyl phosphate synthase large subunit; pyrD2, dihydroorotate dehydrogenase electron transfer subunit; pyrD1, dihydroorotate dehydrogenase (NAD+) catalytic subunit; gpmB, phosphoglycerate mutase (glycolysis); pyrE, orotate phosphoribosyltransferase; and hyp, hypothetical protein. Targets for uracil auxotrophic selection and 5-FOA counterselection are indicated in white. (B) PCR screen of the pyrE locus in 5-FOA-resistant isolates, confirming successful pyrE deletion (2.5 kb) compared to wild type (3.0 kb). In two isolates, there are bands indicating that a single crossover of the pGL090 pyrE deletion plasmid is still present (8.9 kb). (C) PCR confirmation of pyrE gene deletion in MACB1018 (2.5 kb) versus wild type (3.0 kb). PCR products shown in B and C were amplified with primers outside the pyrE flanking regions used for homologous recombination. (D) Growth curve of wild-type strain (circles) and ΔpyrE strain MACB1018 (triangles) on DC516 medium either containing (closed symbols, solid lines) or lacking (open symbols, broken lines) 20 μM uracil. Error bars represent standard deviation; n = 3.
Use of pyrE as a selectable marker in a ΔpyrE background strain.
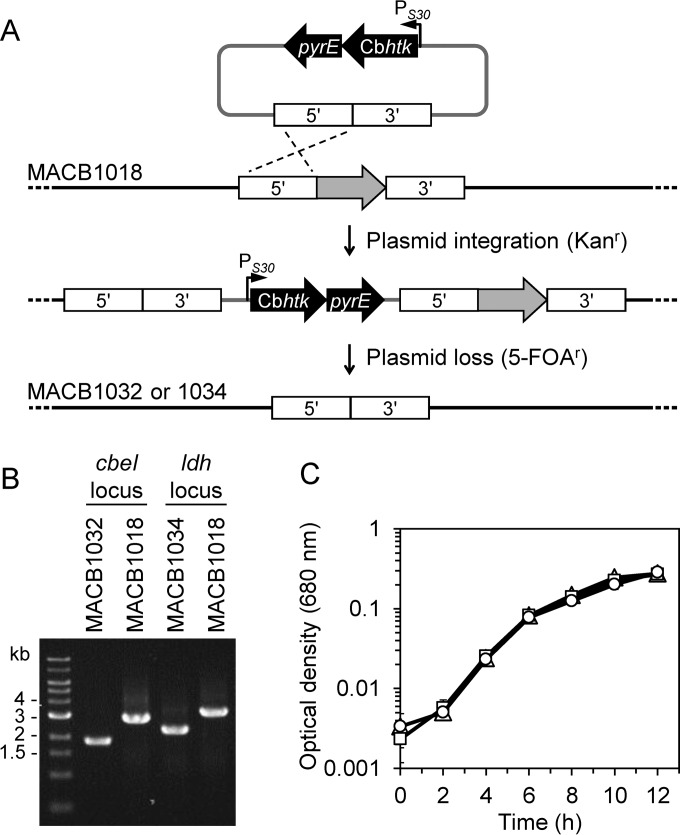
In order to demonstrate that the pyrE gene can be used as a selectable marker in a pyrE-deficient background strain, we chose two gene targets for deletion, cbeI (Athe_2438) and ldh (Athe_1918), both of which have previously been successfully deleted in the JWCB005 background (6, 12). Although pyrE should function in uracil prototrophic selection similar to pyrF, we chose to use Cbhtk in conjunction with kanamycin as the selection for transformation. This selection is more robust, since the use of kanamycin eliminates issues with uracil carryover from the electroporation recovery medium when transformants are selected via uracil prototrophy. The pyrE marker was used for 5-FOA counterselection of plasmid loss (Fig. 4A). Therefore, the cbeI and ldh deletion plasmids contained both markers combined into a synthetic operon, the Cbhtk-pyrE cassette. This cassette utilized the PS30 promoter, a ribosomal binding site between the genes, and a rho-independent terminator sequence at the end of the operon.
FIG 4.
Use of Cbhtk and pyrE markers for genetic selection of mutations in ΔpyrE strain MACB1018. (A) Scheme showing selection method for targeted deletion of ldh or cbeI (gray) using the Cbhtk-pyrE cassette (black) by homologous recombination (dashed lines) into the chromosome of C. bescii strain MACB1018 (ΔpyrE). Kanamycin selected for plasmid integration. To resolve the second crossover event, 5-FOA selected for plasmid loss, resulting in gene deletion or reversion to wild type (not shown). (B) PCR confirmation of cbeI deletion (1.9 kb) and ldh deletion (2.3 kb) in MACB1032 and MACB1034, respectively, compared to the expected sizes for wild-type cbeI (2.8 kb) and ldh (3.2 kb) in the MACB1018 background strain. PCR products were amplified with primers outside the flanking regions used to delete the genes. (C) Growth curve of ΔpyrE strain MACB1018 (triangles), ΔpyrE ΔcbeI strain MACB1032 (squares), and ΔpyrE Δldh strain MACB1034 (circles) in DC516 medium containing 20 μM uracil. Error bars represent standard deviation; n = 3.
The cbeI and ldh deletion plasmids were methylated and transformed into the ΔpyrE background strain MACB1018 using kanamycin to select for transformants. Postelectroporation selective outgrowth cultures were screened by PCR and verified to contain integration of the pGL100 and pGL108 plasmids at the cbeI and ldh genome regions, respectively. After colony purification, these isolates were counterselected for plasmid loss using solid medium containing uracil and 5-FOA. In this case, 5-FOA does not directly select for deletion of the genes but selects only for plasmid loss, which can lead to either gene deletion or reversion to the parent strain; therefore, resulting colonies were screened by PCR to identify ΔcbeI and Δldh isolates. These isolates were further colony purified, sequence verified, and designated MACB1032 (ΔpyrE ΔcbeI) and MACB1034 (ΔpyrE Δldh) (Fig. 4B). Growth characteristics of these strains on complex medium were similar to the parent MACB1018 (Fig. 4C). These strains will be useful in future genetic engineering efforts, since loss of CbeI function allows plasmid transformation without prior methylation of the DNA (6), and deletion of lactate dehydrogenase abolishes lactate production and can increase ethanol yield in engineered strains (12, 16).
We have demonstrated that kanamycin selection can be used in wild-type C. bescii to construct a uracil prototrophic strain based on deletion of the pyrE gene, and this strain can then be used as a genetic background to make other mutations (ΔcbeI and Δldh). Use of Cbhtk with kanamycin has proven to be a more robust method for selecting transformants in C. bescii than uracil prototrophic selection, since there are no issues with uracil contamination which can lead to background growth and false positives. When used in combination with a counterselectable marker, such as pyrF or pyrE, Cbhtk can be used iteratively in the same strain to make multiple mutations at multiple genome locations. Furthermore, kanamycin selection can be performed in a wild-type strain using a complex medium. It is therefore an ideal selectable marker for the initial development of nutritional auxotrophs in wild-type strains, allowing for targeted gene deletion as opposed to selection of random auxotrophic mutants that can potentially also contain mutations elsewhere in the genome. The Cbhtk marker should aid in the development of genetic systems in other members of the Caldicellulosiruptor genus as well as in related species.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daehwan Chung and Janet Westpheling for many helpful discussions as well as for supplying Caldicellulosiruptor bescii strain JWCB018, Escherichia coli strain JW284, and plasmids pDCW73, pDCW89, and pDCW121.
Funding Statement
This research was supported by a grant (DE-PS02-06ER64304) from the Bioenergy Science Center (BESC), Oak Ridge National Laboratory, a U.S. Department of Energy (DOE) Bioenergy Research Center supported by the Office of Biological and Environmental Research in the DOE Office of Science.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00570-16.
REFERENCES
- 1.Yang SJ, Kataeva I, Hamilton-Brehm SD, Engle NL, Tschaplinski TJ, Doeppke C, Davis M, Westpheling J, Adams MW. 2009. Efficient degradation of lignocellulosic plant biomass, without pretreatment, by the thermophilic anaerobe “Anaerocellum thermophilum” DSM 6725. Appl Environ Microbiol 75:4762–4769. doi: 10.1128/AEM.00236-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang SJ, Kataeva I, Wiegel J, Yin Y, Dam P, Xu Y, Westpheling J, Adams MW. 2010. Classification of “Anaerocellum thermophilum” strain DSM 6725 as Caldicellulosiruptor bescii sp. nov. Int J Syst Evol Microbiol 60:2011–2015. doi: 10.1099/ijs.0.017731-0. [DOI] [PubMed] [Google Scholar]
- 3.Zeldes BM, Keller MW, Loder AJ, Straub CT, Adams MW, Kelly RM. 2015. Extremely thermophilic microorganisms as metabolic engineering platforms for production of fuels and industrial chemicals. Front Microbiol 6:1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumer-Schuette SE, Brown SD, Sander KB, Bayer EA, Kataeva I, Zurawski JV, Conway JM, Adams MW, Kelly RM. 2014. Thermophilic lignocellulose deconstruction. FEMS Microbiol Rev 38:393–448. doi: 10.1111/1574-6976.12044. [DOI] [PubMed] [Google Scholar]
- 5.Chung D, Farkas J, Huddleston JR, Olivar E, Westpheling J. 2012. Methylation by a unique alpha-class N4-cytosine methyltransferase is required for DNA transformation of Caldicellulosiruptor bescii DSM6725. PLoS One 7:e43844. doi: 10.1371/journal.pone.0043844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung D, Farkas J, Westpheling J. 2013. Overcoming restriction as a barrier to DNA transformation in Caldicellulosiruptor species results in efficient marker replacement. Biotechnol Biofuels 6:82. doi: 10.1186/1754-6834-6-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung DH, Huddleston JR, Farkas J, Westpheling J. 2011. Identification and characterization of CbeI, a novel thermostable restriction enzyme from Caldicellulosiruptor bescii DSM 6725 and a member of a new subfamily of HaeIII-like enzymes. J Ind Microbiol Biotechnol 38:1867–1877. doi: 10.1007/s10295-011-0976-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeke JD, LaCroute F, Fink GR. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet 197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 9.Chung D, Cha M, Farkas J, Westpheling J. 2013. Construction of a stable replicating shuttle vector for Caldicellulosiruptor species: use for extending genetic methodologies to other members of this genus. PLoS One 8:e62881. doi: 10.1371/journal.pone.0062881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung D, Young J, Bomble YJ, Vander Wall TA, Groom J, Himmel ME, Westpheling J. 2015. Homologous expression of the Caldicellulosiruptor bescii CelA reveals that the extracellular protein is glycosylated. PLoS One 10:e0119508. doi: 10.1371/journal.pone.0119508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott IM, Rubinstein GM, Lipscomb GL, Basen M, Schut GJ, Rhaesa AM, Lancaster WA, Poole FL II, Kelly RM, Adams MW. 2015. A new class of tungsten-containing oxidoreductase in Caldicellulosiruptor, a genus of plant biomass-degrading thermophilic bacteria. Appl Environ Microbiol 81:7339–7347. doi: 10.1128/AEM.01634-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cha M, Chung D, Elkins JG, Guss AM, Westpheling J. 2013. Metabolic engineering of Caldicellulosiruptor bescii yields increased hydrogen production from lignocellulosic biomass. Biotechnol Biofuels 6:85. doi: 10.1186/1754-6834-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung D, Pattathil S, Biswal AK, Hahn MG, Mohnen D, Westpheling J. 2014. Deletion of a gene cluster encoding pectin degrading enzymes in Caldicellulosiruptor bescii reveals an important role for pectin in plant biomass recalcitrance. Biotechnol Biofuels 7:147. doi: 10.1186/s13068-014-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young J, Chung D, Bomble YJ, Himmel ME, Westpheling J. 2014. Deletion of Caldicellulosiruptor bescii CelA reveals its crucial role in the deconstruction of lignocellulosic biomass. Biotechnol Biofuels 7:142. doi: 10.1186/s13068-014-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha M, Chung D, Westpheling J. 2016. Deletion of a gene cluster for [Ni-Fe] hydrogenase maturation in the anaerobic hyperthermophilic bacterium Caldicellulosiruptor bescii identifies its role in hydrogen metabolism. Appl Microbiol Biotechnol 100:1823–1831. doi: 10.1007/s00253-015-7025-z. [DOI] [PubMed] [Google Scholar]
- 16.Chung D, Cha M, Guss AM, Westpheling J. 2014. Direct conversion of plant biomass to ethanol by engineered Caldicellulosiruptor bescii. Proc Natl Acad Sci U S A 111:8931–8936. doi: 10.1073/pnas.1402210111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung D, Cha M, Snyder EN, Elkins JG, Guss AM, Westpheling J. 2015. Cellulosic ethanol production via consolidated bioprocessing at 75°C by engineered Caldicellulosiruptor bescii. Biotechnol Biofuels 8:163. doi: 10.1186/s13068-015-0346-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung D, Verbeke TJ, Cross KL, Westpheling J, Elkins JG. 2015. Expression of a heat-stable NADPH-dependent alcohol dehydrogenase in Caldicellulosiruptor bescii results in furan aldehyde detoxification. Biotechnol Biofuels 8:102. doi: 10.1186/s13068-015-0287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chung D, Young J, Cha M, Brunecky R, Bomble YJ, Himmel ME, Westpheling J. 2015. Expression of the Acidothermus cellulolyticus E1 endoglucanase in Caldicellulosiruptor bescii enhances its ability to deconstruct crystalline cellulose. Biotechnol Biofuels 8:113. doi: 10.1186/s13068-015-0296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Conway JM, Pierce WS, Le JH, Harper GW, Wright JH, Tucker AL, Zurawski JV, Lee LL, Blumer-Schuette SE, Kelly RM. 2016. Multidomain, surface layer associated glycoside hydrolases contribute to plant polysaccharide degradation by Caldicellulosiruptor species. J Biol Chem 291:6732–6747. doi: 10.1074/jbc.M115.707810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor MP, van Zyl L, Tuffin IM, Leak DJ, Cowan DA. 2011. Genetic tool development underpins recent advances in thermophilic whole-cell biocatalysts. Microb Biotechnol 4:438–448. doi: 10.1111/j.1751-7915.2010.00246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Farkas JA, Picking JW, Santangelo TJ. 2013. Genetic techniques for the Archaea. Annu Rev Genet 47:539–561. doi: 10.1146/annurev-genet-111212-133225. [DOI] [PubMed] [Google Scholar]
- 23.Peteranderl R, Shotts EB Jr, Wiegel J. 1990. Stability of antibiotics under growth conditions for thermophilic anaerobes. Appl Environ Microbiol 56:1981–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao H, McKenzie T, Hageman R. 1986. Isolation of a thermostable enzyme variant by cloning and selection in a thermophile. Proc Natl Acad Sci U S A 83:576–580. doi: 10.1073/pnas.83.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoseki J, Yano T, Koyama Y, Kuramitsu S, Kagamiyama H. 1999. Directed evolution of thermostable kanamycin-resistance gene: a convenient selection marker for Thermus thermophilus. J Biochem 126:951–956. doi: 10.1093/oxfordjournals.jbchem.a022539. [DOI] [PubMed] [Google Scholar]
- 26.Iwasaki Y, Kita A, Sakai S, Takaoka K, Yano S, Tajima T, Kato J, Nishio N, Murakami K, Nakashimada Y. 2013. Engineering of a functional thermostable kanamycin resistance marker for use in Moorella thermoacetica ATCC 39073. FEMS Microbiol Lett 343:8–12. doi: 10.1111/1574-6968.12113. [DOI] [PubMed] [Google Scholar]
- 27.Liu B, Wang C, Yang H, Tan H. 2012. Establishment of a genetic transformation system and its application in Thermoanaerobacter tengcongensis. J Genet Genomics 39:561–570. doi: 10.1016/j.jgg.2012.07.003. [DOI] [PubMed] [Google Scholar]
- 28.Taylor MP, Esteban CD, Leak DJ. 2008. Development of a versatile shuttle vector for gene expression in Geobacillus spp. Plasmid 60:45–52. doi: 10.1016/j.plasmid.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto Y, Yano T, Kuramitsu S, Kagamiyama H. 2001. Disruption of Thermus thermophilus genes by homologous recombination using a thermostable kanamycin-resistant marker. FEBS Lett 506:231–234. doi: 10.1016/S0014-5793(01)02926-X. [DOI] [PubMed] [Google Scholar]
- 30.Han D, Norris SM, Xu Z. 2012. Construction and transformation of a Thermotoga-E. coli shuttle vector. BMC Biotechnol 12:2. doi: 10.1186/1472-6750-12-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farkas J, Chung D, Cha M, Copeland J, Grayeski P, Westpheling J. 2013. Improved growth media and culture techniques for genetic analysis and assessment of biomass utilization by Caldicellulosiruptor bescii. J Ind Microbiol Biotechnol 40:41–49. doi: 10.1007/s10295-012-1202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments. Methods Enzymol 498:349–361. doi: 10.1016/B978-0-12-385120-8.00015-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. 2011. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol 8:11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 34.Horton RM, Cai ZL, Ho SN, Pease LR. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques 8:528–535. [PubMed] [Google Scholar]
- 35.de Montigny J, Kern L, Hubert JC, Lacroute F. 1990. Cloning and sequencing of URA10, a second gene encoding orotate phosphoribosyl transferase in Saccharomyces cerevisiae. Curr Genet 17:105–111. doi: 10.1007/BF00312853. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.