ABSTRACT
Deep Lake in the Vestfold Hills is hypersaline and the coldest system in Antarctica known to support microbial growth (temperatures as low as −20°C). It represents a strong experimental model because the lake supports a low-complexity community of haloarchaea, with the three most abundant species totaling ∼72%. Moreover, the dominant haloarchaea are cultivatable, and their genomes are sequenced. Here we use metaproteomics linked to metagenome data and the genome sequences of the isolates to characterize the main pathways, trophic strategies, and interactions associated with resource utilization. The dominance of the most abundant member, Halohasta litchfieldiae, appears to be predicated on competitive utilization of substrates (e.g., starch, glycerol, and dihydroxyacetone) produced by Dunaliella, the lake's primary producer, while also possessing diverse mechanisms for acquiring nitrogen and phosphorus. The second most abundant member, strain DL31, is proficient in degrading complex proteinaceous matter. Hht. litchfieldiae and DL31 are inferred to release labile substrates that are utilized by Halorubrum lacusprofundi, the third most abundant haloarchaeon in Deep Lake. The study also linked genome variation to specific protein variants or distinct genetic capacities, thereby identifying strain-level variation indicative of specialization. Overall, metaproteomics revealed that rather than functional differences occurring at different lake depths or through size partitioning, the main lake genera possess major trophic distinctions, and phylotypes (e.g., strains of Hht. litchfieldiae) exhibit a more subtle level of specialization. This study highlights the extent to which the lake supports a relatively uniform distribution of taxa that collectively possess the genetic capacity to effectively exploit available nutrients throughout the lake.
IMPORTANCE Life on Earth has evolved to colonize a broad range of temperatures, but most of the biosphere (∼85%) exists at low temperatures (≤5°C). By performing unique roles in biogeochemical cycles, environmental microorganisms perform functions that are critical for the rest of life on Earth to survive. Cold environments therefore make a particularly important contribution to maintaining healthy, stable ecosystems. Here we describe the main physiological traits of the dominant microorganisms that inhabit Deep Lake in Antarctica, the coldest aquatic environment known to support life. The hypersaline system enables the growth of halophilic members of the Archaea: haloarchaea. By analyzing proteins of samples collected from the water column, we determined the functions that the haloarchaea were likely to perform. This study showed that the dominant haloarchaea possessed distinct lifestyles yet formed a uniform community throughout the lake that was collectively adept at using available light energy and diverse organic substrates for growth.
INTRODUCTION
Antarctica supports a rich diversity of life (1), which, like most ecosystems on Earth, is predicated on microorganisms forming the base of the food web (2–4). In Antarctic aquatic (e.g., lakes and ice) and lithic (soil and rock) environments, microorganisms dominate, and higher trophic organisms are rare (2–4). In the past decade, “omic”-based studies have generated many discoveries about Antarctic microbial ecology, including identifying previously unknown key taxa, microbial processes, gene exchange events, and virus-host interactions (reviewed in references 2–4). Metaproteomics has proven particularly useful for translating metagenomic community potential into knowledge of functionality for Southern Ocean (5, 6) and Antarctic lake (7–10) communities.
The Vestfold Hills are located on the coastal fringe of East Antarctica, covering an area of ∼411 km2, and contain numerous lakes that were isolated from the ocean ∼3,000 to 7,000 years ago during the isostatic rebound of the continent (4, 11). Deep Lake (68°33′36.8S, 78°11′48.7E) sits ∼50 m below sea level, and the salt has concentrated to the highest level (∼10 times marine) for any lake in the Vestfold Hills (4, 12, 13). The 36-m-deep, 0.58-km2 (surface area), monomictic system remains ice-free even during winter, when the air temperature drops to around −40°C (12, 13). The largest physical changes that occur in Deep Lake are elevated levels of UV and temperatures (≤10°C) in surface waters during summer compared to no UV and low temperatures (≥−20°C) in winter and input from snow melt during summer that consists primarily of freshwater and inorganic material (12–14).
The unicellular green alga Dunaliella (Chlorophyta) is regarded as the major primary producer in Deep Lake (15, 16), where it represents a small proportion of the community (Dunaliella 18S rRNA, ∼1.5% of the total small-subunit [SSU] rRNA [17]). Summer sunlight not only stimulates phototrophic growth and kinetically accelerates growth in Antarctic aquatic systems but also can be so intense that it causes algal photoinhibition (15). Since there is very little organic input (especially colloidal material) into Deep Lake, and minimal biological activity and no microbial blooms or mats, light penetrates to the bottom (36 m) of the lake (14).
Throughout the lake, photoheterotrophic haloarchaea dominate, with the indigenous species differing from typical species in lower-latitude hypersaline systems and with the three most abundant members being distinct genera: Halohasta litchfieldiae strain tADL (∼44% of the community), strain DL31 (unknown genus closely related to Halolamina) (∼18%), and Halorubrum lacusprofundi (∼10%) (17). All three members belong to the family Halobacteriaceae of the order Halobacteriales. Other saline (e.g., Ace Lake) or hypersaline (e.g., Organic Lake) lakes in the Vestfold Hills have significantly more complex communities, and the physical differences in these lakes contribute to major shifts in community composition and function associated with distinct layers (8, 18). Reflecting the monomictic limnology of Deep Lake, throughout its depth (assessed at 5, 13, 24, and 36 m), the community is essentially homogeneous, with only a small decrease in the relative abundance of Hht. litchfieldiae and an increase in the abundance of minor species in the deepest waters (17). The community composition also exhibits little change by size fraction (20 to 3.0 μm, 3.0 to 0.8 μm, and 0.8 to 0.1 μm), with the main difference being a relatively even distribution of Hht. litchfieldiae across all size fractions compared to the higher partitioning of DL31 and Hrr. lacusprofundi on 0.8- and 3.0-μm filters (17). Again, this is in contrast to other lakes such as Ace Lake, where size partitioning is very apparent (8).
The main physiological traits of the three most abundant Deep Lake genera, plus an additional strain, Halobacterium sp. strain DL1, which represents a minor fraction (∼0.3%) of the lake community, were predicted from their genome sequences (19). Hht. litchfieldiae was inferred to possess swimming motility with a saccharolytic metabolism that included a preference for glycerol utilization, DL31 was inferred to target mainly peptides and amino acids, and Hrr. lacusprofundi was inferred to be less specialized in the nutrients that it targets (19). Strain-level phylotypes were also identified by using fragment recruitment of metagenome data to the closed genomes of the isolates, single-nucleotide polymorphism (SNP) analyses, and GC/read-depth profiling of the de novo assembly of metagenome data (17). The latter revealed a cluster of 52 large contigs (>15 kb) totaling 1.89 Mb, which had the highest identity (∼85%) to Hht. litchfieldiae tADL. Differing significantly from the Hht. litchfieldiae tADL genome, the partially reconstructed genome was referred to as the “tADL-related fifth genome” (17), and here we designate this phylotype of Hht. litchfieldiae strain “tADL-II.” Overall, the community structure data for Deep Lake in association with its specific limnology indicate that ecological niches occur based on trophic preference (not physical partitioning) that manifests in genetic differences between distinct genera and between phylotypes/strains of the same species.
Recently, metaproteomics was developed for Deep Lake to identify viruses and host responses involved in viral infection, evasion, and defense (10). Viruses play a particularly prominent role in the microbial food web in Antarctica because higher trophic predators (e.g., protists) are uncommon (4, 20). As reservoirs of biodiversity providing unique genetic potential and mediating gene transfer, viruses control ecosystem function by lysing their hosts and causing the recycling of nutrients, thereby making a unique contribution to global biogeochemical cycles (21–23). Viruses are therefore predicted to play the key role in controlling community composition and microbial processes ranging from primary production to nutrient recycling (4). Lytic viruses in Deep Lake are predicted to cause nutrient turnover and make an important contribution to ecology, as the lake is dominated by heterotrophs (4, 10). A feature of the Deep Lake metaproteome study was the use of high-coverage metagenome data to generate contigs suitable for identifying proteins that were derived from strain variation (10). In particular, “variants” of cell surface proteins (e.g., main S-layer protein) were linked to providing a mechanism of infection evasion by preventing virus adsorption (10). With metaproteomic methods developed for this Antarctic lake, the first for any hypersaline environment (10), in this study, we sought to describe proteins that pertain to pathways and cellular processes present in these haloarchaea, test the validity of the previous genome-based predictions (17, 19), and establish how the main genera and strains coexist and utilize available natural resources. By utilizing biomass captured on 3.0-, 0.8-, and 0.1-μm filters from 0-, 5-, 13-, 24-, and 36-m depths of the lake, this study was able to test the hypothesis that functional distinctions in the community occurred primarily between genera and between strains and not between communities or individuals separated by lake depth or filter size.
MATERIALS AND METHODS
Metaproteomics was performed as described previously, using biomass collected by sequential size fractionation from Deep Lake water (25 to 50 liters) that was collected from 30 November to 5 December 2008 from depths of 0, 5, 13, 24, and 36 m (10). The mass spectrometry data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository under project name Metaproteome, Deep Lake, Antarctica, with project accession number PXD001436 (10). Analytical approaches were based on methods described previously for studying Antarctic lake ecosystems (7–10, 17–19). Briefly, a composite database comprised of assemblies of Deep Lake metagenome data and the genome sequences of the Deep Lake haloarchaea Halohasta litchfieldiae, undescribed genus strain DL31, Halorubrum lacusprofundi, and Halobacterium sp. DL1 were used for generating peptides to match to mass spectra to make protein identifications (10). Proteins were manually verified to contain at least one unique peptide, and any proteins sharing the same set of detected peptides were grouped into protein families. Proteins were assigned as variant proteins if they had <100% amino acid sequence identity to sequences from the genomes of the Deep Lake haloarchaea and at least one unique peptide mapped to a region of sequence variation within a metagenome contig (10). Percent identity for a variant (the lower the identity, the higher the extent of variation) indicates the extent of protein sequence identity relative to the best-matching sequence in the genome of one of the four isolates. Contigs assigned to tADL-II typically had multiple open reading frames (ORFs) that were syntenic and had 70 to 99% amino acid identity to tADL sequences. Contigs assigned as variants of tADL had ORFs with 100% identity to tADL plus some ORFs (typically one) with 97 to 99% identity. In a small number of cases, the gene context of the contig enabled assignments only to Hht. litchfieldiae (see the supplemental material for more details). Proteins were assigned to functional categories based on their manual annotations. Patterns of coabundance of specific proteins, taxonomic categories, or functional categories across the 15 samples (representing both depth and filter size) were evaluated by performing correlation statistics using normalized total spectrum counts (see Tables S1 and S2 in the supplemental material). Nonmetric multidimensional scaling (NMDS) analysis and analysis of similarity (ANOSIM) were performed to test for statistically significant functional differences in the metaproteome data between lake depths or filter size fractions using the PRIMER 6 software package (24). Epifluorescence microscopy was performed on filtered lake water to examine the presence of cells associated with particulate matter on large-pore-size filters. Growth studies were performed to evaluate inferences about substrate utilization. Hht. litchfieldiae was grown in batch cultures containing potential carbon, nitrogen, or phosphorus sources (dihydroxyacetone [DHA], starch, 2-aminoethylphosphonic acid, and DNA). Additional details are provided in the supplemental material.
RESULTS AND DISCUSSION
The metaproteomic data were combined with data from growth assays with potential substrates to determine specific metabolic pathways utilized and functional cellular processes performed by the Deep Lake haloarchaea. Statistical analyses (ANOSIM and NMDS) were performed to assess the relationship of taxon and cellular function with lake depth and size fraction. Below, we present findings that verify or refute previous inferences (17, 19) and advance our overall understanding of Deep Lake ecology. Additional results are provided in the supplemental material.
Relationship between organism function, lake depth, and size fraction.
A total of 1,109 distinct proteins (see Table S3 in the supplemental material) were identified from biomass collected by sequential size fractionation on 3.0-, 0.8-, and 0.1-μm filters from the five depths of Deep Lake (total of 15 samples). Fewer proteins were detected with the 0.1-μm filters than with the 0.8- or 3.0-μm filters, reflecting a lower level of biomass in the 0.1- to 0.8-μm size range (see Fig. S1 and Table S3 in the supplemental material). An increase in the amount of particulate matter with an increase in filter pore size and cell association with particulate matter were observed by epifluorescence microscopy (see Fig. S2 in the supplemental material).
A total of 902 (81%) proteins were assigned to the three most abundant haloarchaea, Hht. litchfieldiae, DL31, and Hrr. lacusprofundi (see Fig. S3 and Table S3 in the supplemental material). Consistent with its known abundance in the lake (17), Hht. litchfieldiae recruited the highest number of proteins (655 proteins accounting for 59% of all detected proteins and 63% of protein spectra) in the metaproteome. The proteins matched 513 distinct genes and accounted for ∼15% of the 3,465 protein-encoding genes in the Hht. litchfieldiae genome (Fig. 1). DL31 is the second most abundant species in Deep Lake and accounted for ∼14% (154 proteins) of the total number of protein identifications. A total of 93 proteins (∼9% of the metaproteome) were detected in Hrr. lacusprofundi.
FIG 1.
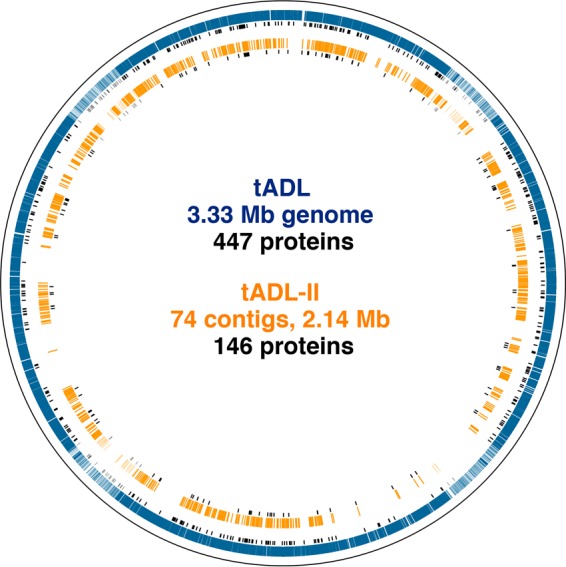
Circular plot of Hht. litchfieldiae tADL and tADL-II genomes. The outer blue annulus shows the coding sequences of the tADL genome, the inner orange annulus shows the tADL-II metagenome contigs mapped onto the tADL genome, and the short black bars are genes corresponding to proteins detected in the metaproteome.
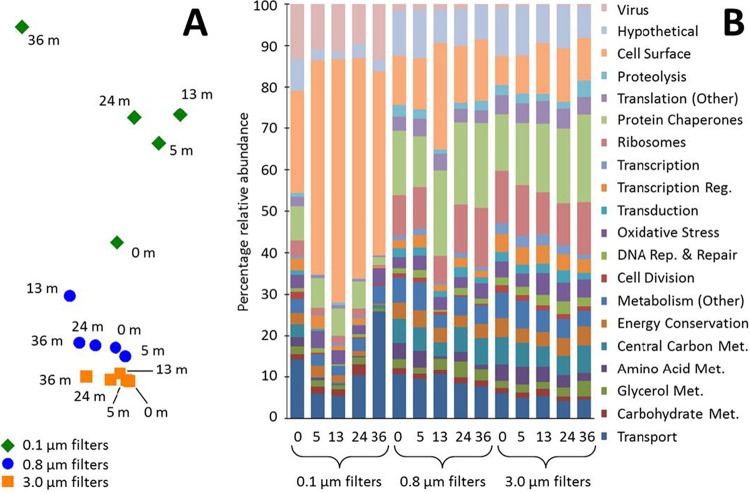
To test if there were functional distinctions between communities or individuals separated by lake depth or filter size, ANOSIM was performed with all identified proteins grouped into functional categories, and a significant difference (P < 0.01) was found between filter sizes but not between depths. The NMDS plot (Fig. 2A) showed that all the communities from the 3.0-μm and 0.8-μm filters cluster closely together, separated from the communities from the 0.1-μm filters, which were not as tightly clustered. Relatively high proportions of viral proteins and cell surface proteins were present on the 0.1-μm filters compared to the 0.8- and 3.0-μm filters (Fig. 2B).
FIG 2.
Relationship between function, depth, and size fraction. Proteins detected from each sample (5 depths and 3 filter sizes) were grouped into functional categories, and their relative abundance was calculated based on the normalized total spectrum count. (A) Ordination of samples in an NMDS plot (two-dimensional stress, 0.02). (B) Bar chart showing relative abundances of functional categories in each sample. Numbers on the x axis (0, 5, 13, 24, and 36) stipulate the lake depth, in meters, from which the samples were obtained.
To test the hypothesis that functional distinctions in the community occurred between genera and strains, the proteins identified from each depth and filter size were pooled to generate a single metaproteome for the lake, and the proteins assigned to each of the three main haloarchaea were used to compare their functional properties. Pairwise correlations of protein abundances (spectrum counts) by depth and filter size were also used to compare the abundances of single proteins or proteins from specific functional categories. By identifying covarying proteins or functional processes, inferences were drawn about the ecophysiological properties of the organisms (described below).
Transport proteins reveal distinctions in nutrient preferences.
The Deep Lake metaproteome included 64 transport proteins from the three main species (Fig. 3; see also Table S4 in the supplemental material). A relatively high abundance of transport proteins was identified for DL31 and Hrr. lacusprofundi (see Fig. S4 in the supplemental material), which may be partially explained by the lower abundance of these species than of Hht. litchfieldiae. As a consequence of nutrient kinetics in aquatic microorganisms, processing of imported substrates requires relatively lower expression levels of cytoplasmic enzymes than of extracytoplasmic proteins for substrate capture (25), making detection of expressed cytoplasmic enzymes more difficult for less abundant species (26). The most abundant transport proteins were the solute-binding components of ATP-binding cassette (ABC) transporters. In metaproteomic data sets, solute-binding components of active transporters tend to be overrepresented compared to membrane-associated components (26–28). This can be attributed to a high cellular abundance of solute-binding proteins in order to increase the frequency of solute capture (26–28). The relative abundance of the different types of transport proteins can therefore provide insight into substrate preference.
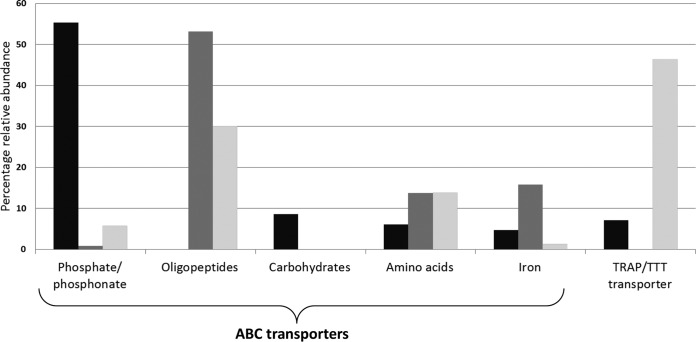
FIG 3.
Relative abundance of transport proteins in the metaproteome. Black, Hht. litchfieldiae; dark gray, DL31; light gray, Hrr. lacusprofundi. Abundance was calculated relative to the sum of the normalized total spectrum count for transport proteins across all 15 samples. In addition to the relatively abundant ABC and TRAP/TTT transporter proteins shown, other transporter proteins were identified, representing 18, 17, and 3% of the transport spectra for Hht. litchfieldiae, DL31, and Hrr. lacusprofundi, respectively (see Table S4 in the supplemental material).
The relative abundances of transporter proteins targeting different substrates varied greatly among the three main species (Fig. 3). Phosphate-binding transporter lipoproteins accounted for ∼55% of spectra for all the transport proteins of Hht. litchfieldiae, compared to only ∼1% and 6% for DL31 and Hrr. lacusprofundi, respectively. Most abundant were the ABC transporter phosphate-binding PstS proteins, but the ATPase PstB and multiple PhoU phosphate uptake regulator proteins were also detected. A high abundance of phosphate-binding ABC transporters indicates high phosphate demand by Hht. litchfieldiae and may be linked to a requirement for the phosphorylation of carbohydrates (including glycerol and simple sugars) as part of intracellular utilization, consistent with a highly saccharolytic metabolism (see “Hht. litchfieldiae carbohydrate metabolism and dependency on Dunaliella,” below). In Deep Lake, dissolved reactive phosphorus concentrations are low throughout the lake (0.52 to 2.34 nM) (14, 19). Therefore, the abundance of phosphate transport proteins likely reflects a shortage of bioavailable phosphate for Hht. litchfieldiae, as was described previously for the phosphorus-depleted Sargasso Sea (27). One ABC transporter lipoprotein targeting phosphonates was detected for Hht. litchfieldiae, the only Deep Lake haloarchaeon known to have the genomic potential to break down phosphonates (19). Phosphonates are components of phosphonolipids and phosphonoglycans (29) and can serve as sources of phosphate when the environmental concentration of bioavailable phosphate is low (30). In support of this, Hht. litchfieldiae can grow using 2-aminoethylphosphonate (a ubiquitous, naturally occurring phosphonate) as a phosphorus source (see Fig. S5A in the supplemental material).
DL31 recruited a relatively large number of abundant (∼25% of all DL31 spectra) transport proteins (16 in total). Most prominent were ABC transporter oligopeptide-binding lipoproteins (8 proteins; 53% of all transport spectra) (Fig. 3; see also Table S4 in the supplemental material), which is consistent with its predicted preference for proteinaceous matter (19) (see also “Diverse strategies of nitrogen acquisition,” below). Protein abundances of DL31 ABC transporter lipoproteins for oligopeptides, amino acids, and iron uptake were positively correlated across the 15 samples (see Table S1 in the supplemental material), indicating that these substrates might be derived from the same nutrient and mineral source(s).
The Hrr. lacusprofundi ABC transporter lipoproteins represented a wide range of substrates: oligopeptides, amino acids, nucleotides, phosphate, and iron (Fig. 3; see also Table S4 in the supplemental material). However, the most abundant transporter category was tripartite ATP-independent periplasmic transporter (TRAP)/tripartite tricarboxylate transporter (TTT), accounting for ∼46% of all Hrr. lacusprofundi transporter spectra, compared to only ∼7% in Hht. litchfieldiae and none for DL31 (TRAP/TTT transporters are not encoded in the DL31 genome). Hrr. lacusprofundi TRAP (TAXI family) transporter and amino acid and oligopeptide ABC transporter proteins were positively correlated with DL31 oligopeptide and amino acid ABC transporter proteins (see Table S1 in the supplemental material), suggesting that both species derived substrates from similar nutrient sources in Deep Lake. Hrr. lacusprofundi has minimal genomic capacity for extracytoplasmic proteolysis (19). The metaproteome data are consistent with a lifestyle by which Hrr. lacusprofundi benefits from labile substrates released by Hht. litchfieldiae and DL31 during the degradation of complex proteinaceous matter.
Hht. litchfieldiae carbohydrate metabolism and dependency on Dunaliella.
Fifty Hht. litchfieldiae proteins involved in carbohydrate uptake and metabolism were detected (see Table S5 in the supplemental material), compared to only five each for DL31 and Hrr. lacusprofundi (see Table S3 in the supplemental material). Five proteins were carbohydrate ABC transporter proteins for the uptake of sugars (four solute-binding lipoproteins), and one was for an ATPase. These data support previous inferences that Hht. litchfieldiae is highly saccharolytic (19).
Nine Hht. litchfieldiae enzymes involved in the catabolism of glycerol were detected, in comparison to only one low-abundance glycerol kinase for Hrr. lacusprofundi and none for DL31. Mass spectra for proteins from Hht. litchfieldiae and Dunaliella chloroplasts are positively correlated across the individual filter fractions throughout the lake (see Table S1 in the supplemental material). Glycerol is produced by Dunaliella sp. (31) and is regarded as a major carbon and energy source for haloarchaea in hypersaline habitats (16, 31, 32). The Hht. litchfieldiae genome encodes two pathways for the conversion of glycerol into DHA phosphate (19). Evidence for the first glycerol catabolic pathway included the detection of two distinct glycerol kinases and glycerol-3-phosphate dehydrogenase (see Table S5 and further discussion in the supplemental material). Hht. litchfieldiae, which was previously shown to grow on glycerol (19), can also grow on DHA (see Fig. S5B in the supplemental material). The detection of DHA kinase subunits L and K is consistent with glycerol catabolism by the second pathway but alternatively raises the possibility of DHA being directly obtained from the environment and catabolized. DHA is exuded as a by-product of the breakdown of surplus glycerol in Dunaliella (33, 34) and has been hypothesized to be an important growth substrate for haloarchaea in hypersaline lakes (34).
Three glycosidases inferred to be involved in starch degradation (glucoamylase, α-amylase, and α-4-glucanotransferase) were detected. Dunaliella produces large (up to ∼1-μm) internal starch granules as a carbohydrate storage product (35). Hht. litchfieldiae grew weakly with starch, strongly with pyruvate, and the best with pyruvate plus starch as the sole defined carbon sources (see Fig. S5C in the supplemental material). A similar growth pattern was observed with sucrose (19), suggesting that both sucrose and starch are most readily utilized in the presence of pyruvate. Enzymes for both the Emden-Meyerhof pathway for glycolysis and the modified (semiphosphorylative) part of the Entner-Doudoroff pathway involving glucose oxidation to gluconate (36, 37) were detected (see Table S5 in the supplemental material), indicating that the starch breakdown products and other simple sugars were catabolized by these pathways in Hht. litchfieldiae.
Carbohydrate ABC transporters detected for Hht. litchfieldiae could be used to import linear oligosaccharides or simple sugars generated by extracytoplasmic polysaccharide degradation. The spectral count of the most abundant α-amylase (also see “Variation within the Hht. litchfieldiae population of Deep Lake,” below) was positively correlated with Hht. litchfieldiae archaellins (see Table S1 in the supplemental material), which is suggestive of a link between motility and targeting starch granules. There was a negative correlation between Hht. litchfieldiae archaellins and proteins associated with central carbon metabolism, suggesting that the expression of archaella might occur in response to less favorable nutrient conditions, as observed for Sulfolobus solfataricus (38, 39). As a result, we hypothesize that motile Hht. litchfieldiae cells primarily express polysaccharide-degrading enzymes while actively searching for nutrients.
Overall, the metaproteome data indicate the Hht. litchfieldiae has a strong dependency on substrates produced by Dunaliella (starch, glycerol, and DHA) and is more competitive in utilizing them than the other haloarchaeal species, thereby contributing to its dominance in the lake.
Diverse strategies of nitrogen acquisition.
Hht. litchfieldiae appears the most versatile in acquiring nitrogen from the environment, with evidence for utilization of proteins, ammonium, amino acids, and urea (see Table S6 in the supplemental material for proteins of the three species involved in nitrogen acquisition/metabolism). Some nitrogen sources overlap those targeted by Hrr. lacusprofundi (ammonium and amino acids) and DL31 (proteins and amino acids). Unlike DL31 and Hrr. lacusprofundi, Hht. litchfieldiae apparently lacks the capacity to import oligopeptides (19); however, it encodes and we detected amino acid ABC transporter lipoproteins, including those that target amino acids that result from the cleavage of proteins or oligopeptides. Two putatively secreted, proteolytic enzymes were detected for Hht. litchfieldiae: a serine protease (halolysin) and an aminopeptidase. These enzymes likely perform extracytoplasmic digestion of proteinaceous substrates, degrading them into smaller oligopeptides and amino acids. The detection of these secreted proteins might indicate that they remain associated with the S-layer of cells (rather than diffuse into the extracellular milieu), as was reported previously for exoenzymes of certain bacteria (40).
The detection of a urea ABC transporter lipoprotein and a urease subunit (UreB) indicates that urea was utilized by Hht. litchfieldiae. Hht. litchfieldiae is the only Deep Lake haloarchaeon known to possess urease, although it cannot utilize urea as a carbon source (19), and urease genes are rarely found in Archaea (41–43). The bioenergetic costs of urease synthesis and urea uptake by ABC transport are high, so the utilization of urea by Hht. litchfieldiae may be advantageous only when the availability of ammonium is low or intermittent (43).
An ammonium transporter, glutamine synthetase (GS), and glutamate synthase (GOGAT) were detected for Hht. litchfieldiae, and GS was detected for Hrr. lacusprofundi. Operation of the GS-GOGAT cycle for ammonium assimilation in Hht. litchfieldiae and Hrr. lacusprofundi would be consistent with nitrogen limitation relative to carbon (28, 44). DL31 is the second most abundant organism in Deep Lake, and GS was not detected, which may indicate that DL31 was not nitrogen limited. Eight distinct DL31 oligopeptide-binding ABC transporter lipoproteins (see “Transport proteins reveal distinctions in nutrient preferences,” above; see also Table S4 in the supplemental material) and two secreted proteolytic enzymes (halolysin and aminopeptidase) were detected. Glutamate dehydrogenase was also detected, which likely functions in glutamate catabolism (deamination of glutamate released from oligopeptides), as posited for Bacteroidetes that degrade oligopeptides (6, 45). The data are consistent with predictions that DL31 is proteolytic (19), with the metaproteome data indicating that nitrogen is sourced exclusively from proteins, oligopeptides, and amino acids.
Other enzymes involved in the biosynthesis of proteinogenic amino acids were detected for Hht. litchfieldiae, including several for the biosynthesis of branched-chain amino acids (BCAAs) and aromatic amino acids. In total, the metaproteome contains 49 Hht. litchfieldiae proteins that are involved in the metabolism of amino acids, compared to only 4 and 2 for DL31 and Hrr. lacusprofundi, respectively (see Table S6 in the supplemental material). It is therefore possible that sugars imported by Hht. litchfieldiae are used to biosynthesize amino acids, whereas DL31 and Hrr. lacusprofundi are more reliant on amino acids derived from the active uptake of oligopeptides and free amino acids.
Hht. litchfieldiae motility.
Archaella are filaments made up of archaellin subunits that allow cells to swim by ATP-driven rotation (38). In the metaproteome, 10 archaellin homologs that were derived from six genes present in three Hht. litchfieldiae genomic loci were detected (see Table S7 in the supplemental material). The archaellin proteins were some of the most abundant in the metaproteome (e.g., 1st-, 3rd-, 7th-, and 12th-highest spectrum counts) and were most abundant on the 0.1-μm filters, and individual archaellin proteins were significantly positively correlated with one another. In contrast, archaellin proteins were negatively correlated with central carbon metabolism proteins and ribosomes, which were enriched on the 0.8-μm and 3.0-μm filters (see Tables S1 and S3 in the supplemental material), suggesting that cells engaged in swimming motility undergo less metabolic activity and biosynthesis than attached cells.
Hht. litchfieldiae bacteriorhodopsin and multiple methyl-accepting chemotaxis proteins (MCPs) were detected, including MCPs indicative of chemo-, photo-, and aerotaxis (see text and Table S7 in the supplemental material). Also detected were CheW, CheY, and a PBS lyase HEAT-like repeat protein that functions in Halobacterium salinarum to link the taxis signal transduction system to the archaellar apparatus and is essential for chemotaxis and phototaxis (46). Hht. litchfieldiae may therefore use light to generate ATP and exhibits a capacity to swim toward or away from specific environmental stimuli (see Table S7 in the supplemental material).
Haloarchaeal responses to Antarctic solar irradiation.
Numerous haloarchaeal proteins implicated in protection against or repair of damage due to oxidative stress or UV irradiation were detected in the Deep Lake metaproteome (see Table S8 in the supplemental material). It is likely that the very high levels of UV (both intensity and duration) that occur during the austral summer in Antarctica (UVA, 4.6 × 105 J m−2; UVB, 3.2 × 103 J m−2 [47]) enhance DNA and protein damage in Deep Lake haloarchaea through the production of reactive oxygen species. Compounding the problem, dissolved oxygen concentrations in Deep Lake are close to saturation, with levels increasing with decreasing temperature (14), and low temperature causes increased solubility of molecular oxygen and increased stability of reactive oxygen species. Laboratory studies of the response of Halobacterium sp. strain NRC-1 to UV irradiation revealed that RadA, RecJ exonuclease, ribonucleotide reductase, and topoisomerase VI subunits were upregulated (48, 49), and all these proteins were detected in Hht. litchfieldiae (see Table S8 in the supplemental material). The metaproteome data represent the first step in learning about the molecular responses of haloarchaea to high summer sunlight irradiation in Antarctica.
Variation within the Hht. litchfieldiae population in Deep Lake.
Most variants in the metaproteome were assigned to Hht. litchfieldiae (∼97%), and a total of 146 were assigned to tADL-II (Fig. 1; see also Table S9 in the supplemental material for details about protein assignments). For 122 of these variants, distinct peptides with a 100% match to tADL were also detected. The tADL proteins had an ∼3.5-fold-higher median abundance than the equivalent tADL-II proteins. The total number (146 proteins) and abundance (∼20% of the spectra assigned to Hht. litchfieldiae) of tADL-II proteins are consistent with calculations from metagenome read depth coverage that tADL-II represents ∼15% of the Hht. litchfieldiae Deep Lake population, a proportion similar to those for Hrr. lacusprofundi and DL31 (17).
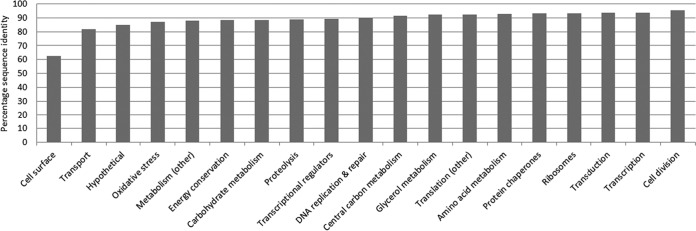
A high level of variation was observed between tADL and tADL-II sequences for cell surface proteins (63% average amino acid identity), in contrast to the substantially lower level of variation for typically conserved transcription and cell division proteins (≥94%) (Fig. 4). The divergence between tADL and tADL-II proteins is likely to result in phenotypic distinctions, as even single amino acid changes can confer functional differences (e.g., changes in the active site, substrate-binding site, site of interaction for effector molecules, and protein-protein interactions). Proteomic distinctions between tADL and tADL-II included proteins encoded by genes present on tADL-II contigs that were absent in the tADL genome. The detection of a nitrate/sulfonate/bicarbonate ABC transporter solute-binding lipoprotein (unique to tADL-II) may confer the ability to target distinct nutrient sources (see Table S9 in the supplemental material). A small number of tADL-II proteins (12 in total) also had higher abundances than the equivalent tADL proteins (see Table S9 in the supplemental material). One of these was an ABC transporter BCAA-binding lipoprotein with ∼10-fold-higher levels (when normalized to strain abundance), which may indicate that tADL-II has a greater preference for amino acids than does tADL. Other metabolic variants included ABC transporter phosphate-binding lipoproteins, glycerol kinases, and α-amylases (see Tables S10 and S11 in the supplemental material), which may allow the population to make use of a wider range of substrates (50, 51). The extent of variation pertaining to metabolic processes provides support for the existence of Hht. litchfieldiae ecotypes or a population occupying a broad environmental niche (52, 53).
FIG 4.
Extent of sequence variation between proteins in different functional categories from Hht. litchfieldiae tADL and tADL-II. Shown are average sequence identities for all proteins within a functional category for tADL-II proteins compared to tADL proteins. Categories are ranked based on their average sequence identity, highlighting the extent of variation within cell surface proteins.
Conclusion.
The metaproteomic data, combined with data from growth assays for certain potential substrates, have provided a strong understanding about specific metabolic pathways, functional cellular processes, and overall lake ecology (Fig. 5). The number of protein identifications (1,109 distinct proteins) provided reasonable coverage of abundant lake species and compares well to data from other Antarctic metaproteome studies (6–8).
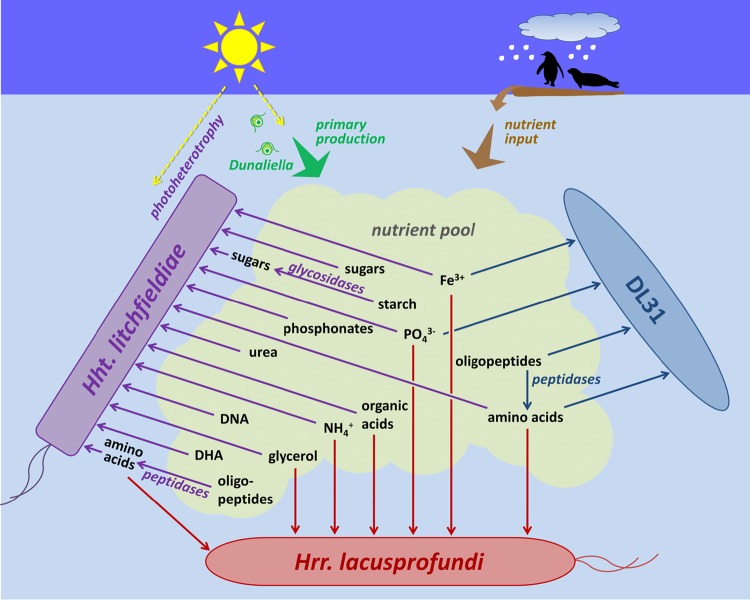
FIG 5.
Proposed substrate preferences and ecophysiological interactions of the three dominant haloarchaea. Shown is a cartoon derived from the interpretation of metaproteome data, highlighting the distinct functional properties of Hht. litchfieldiae, DL31, and Hrr. lacusprofundi and their inferred interactions that occur in Deep Lake. DHA, dihydroxyacetone. Scale is not indicative of organism size or relative contribution to the system. (Animal silhouettes courtesy of PhyloPic [http://phylopic.org/].)
Every protein identified in this study represents one of significance to organism function in the natural environment. Thus, rather than being only a confirmation of data from genomic analyses (19), this study revealed cellular processes relevant to growth and survival in the Antarctic, including processes that can be difficult to evaluate in the laboratory. For example, environmental microorganisms often lose motility when cultivated in the laboratory (54), but this study revealed the expression of proteins enabling Hht. litchfieldiae to swim and perform taxis in the lake, and we hypothesize that when actively searching for nutrients, motile cells express polysaccharide-degrading enzymes. The data also indicate that Hht. litchfieldiae not only mounts a protective response to UV light but also uses light to generate ATP for photoheterotrophic growth (Fig. 5). Such findings provide solid foundations for follow-up hypothesis-driven laboratory and field studies.
Distinctions in transport and metabolic proteins revealed nutrient preferences, with Hht. litchfieldiae being predicted to benefit from carbohydrates (e.g., starch, glycerol, and DHA) produced by Dunaliella, including for the biosynthesis of amino acids. In contrast, DL31 and Hrr. lacusprofundi are predicted to be much more reliant on free amino acids and peptides, with Hrr. lacusprofundi being inferred to benefit from the degradation of complex proteinaceous matter by Hht. litchfieldiae and DL31 (Fig. 5). The data provide a variety of evidence for protein variation for a range of functions, most notably metabolism related, and such variation should provide the population with enhanced flexibility to exploit available nutrients within Deep Lake. Our view is that the lake supports a relatively uniform distribution of taxa that have evolved a collective genetic capacity to effectively exploit light energy and diverse organic substrates throughout the lake.
Supplementary Material
ACKNOWLEDGMENTS
Mass spectrometric results were obtained at the Bioanalytical Mass Spectrometry Facility within the Analytical Centre of The University of New South Wales. This work was undertaken using infrastructure provided by NSW Government coinvestment in the National Collaborative Research Infrastructure Scheme. We gratefully acknowledge subsidized access to this facility. We thank Susanne Erdmann and Yan Liao for assistance with growth of the haloarchaea and Sarah Payne and Alyce Hancock for collecting samples in Antarctica.
Funding Statement
This work was supported by the Australian Antarctic Science program (AAS projects 2899 and 4031). This work was also funded by the Department of Industry, Innovation, Science, Research and Tertiary Education, Australian Government | Australian Research Council (ARC) (DP140100582 [to R.C.] and DP150100244 [to R.C. and M.J.R.]). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00473-16.
REFERENCES
- 1.Chown SL, Clarke A, Fraser CI, Cary SC, Moon KL, McGeoch MA. 2015. The changing form of Antarctic biodiversity. Nature 522:431–438. doi: 10.1038/nature14505. [DOI] [PubMed] [Google Scholar]
- 2.Cary SC, McDonald IR, Barrett JE, Cowan DA. 2010. On the rocks: the microbiology of Antarctic Dry Valley soils. Nat Rev Microbiol 8:129–138. doi: 10.1038/nrmicro2281. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins D, Yau S, Williams TJ, Allen MA, Brown MV, DeMaere MZ, Lauro FM, Cavicchioli R. 2013. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol Rev 37:303–335. doi: 10.1111/1574-6976.12007. [DOI] [PubMed] [Google Scholar]
- 4.Cavicchioli R. 2015. Microbial ecology of Antarctic aquatic systems. Nat Rev Microbiol 13:691–706. doi: 10.1038/nrmicro3549. [DOI] [PubMed] [Google Scholar]
- 5.Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ, Ducklow H, Grzymski JJ, Murray AE, Cavicchioli R. 2012. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J 6:1883–1900. doi: 10.1038/ismej.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Williams TJ, Wilkins D, Long E, Evans F, DeMaere MZ, Raftery MJ, Cavicchioli R. 2013. The role of planktonic flavobacteria is processing algal organic matter in coastal East Antarctica revealed using metagenomics and metaproteomics. Environ Microbiol 15:1302–1317. doi: 10.1111/1462-2920.12017. [DOI] [PubMed] [Google Scholar]
- 7.Ng C, DeMaere MZ, Williams TJ, Lauro FM, Raftery M, Gibson JA, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Thomas T, Cavicchioli R. 2010. Metaproteogenomic analysis of a dominant green sulfur bacterium from Ace Lake, Antarctica. ISME J 4:1002–1019. doi: 10.1038/ismej.2010.28. [DOI] [PubMed] [Google Scholar]
- 8.Lauro FM, DeMaere MZ, Yau S, Brown M, Ng C, Wilkins D, Raftery MJ, Gibson JA, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Thomas T, Cavicchioli R. 2011. An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5:879–895. doi: 10.1038/ismej.2010.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yau S, Lauro FM, DeMaere MZ, Brown MV, Thomas T, Raftery MJ, Andrews-Pfannkoch C, Lewis M, Hoffman JM, Gibson JA, Cavicchioli R. 2011. Virophage control of Antarctic algal host-virus dynamics. Proc Natl Acad Sci U S A 108:6163–6168. doi: 10.1073/pnas.1018221108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tschitschko B, Williams TJ, Allen MA, Páez-Espino D, Kyrpides N, Zhong L, Raftery MJ, Cavicchioli R. 2015. Antarctic archaea-virus interactions: metaproteome-led analysis of invasion, evasion and adaptation. ISME J 9:2094–2107. doi: 10.1038/ismej.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson JAE. 1999. The meromictic lakes and stratified marine basins of the Vestfold Hills, East Antarctica. Antarct Sci 11:175–192. [Google Scholar]
- 12.Campbell PJ. 1978. Primary productivity of a hypersaline Antarctic lake. Aust J Mar Freshw Res 29:717–724. doi: 10.1071/MF9780717. [DOI] [Google Scholar]
- 13.Ferris JM, Burton HR. 1988. The annual cycle of heat content and mechanical stability of hypersaline Deep Lake, Vestfold Hills, Antarctica. Hydrobiologia 165:115–128. doi: 10.1007/BF00025579. [DOI] [Google Scholar]
- 14.Barker RJ. 1981. Physical and chemical parameters of Deep Lake, Vestfold Hills, Antarctica. Publication no. 130. Australian National Antarctic Research Expeditions series B(V) limnology. Antarctic Division, Department of External Affairs, Melbourne, Australia. [Google Scholar]
- 15.Wright SW, Burton HR. 1981. The biology of Antarctic saline lakes. Hydrobiologica 82:319–338. [Google Scholar]
- 16.Franzmann PD, Stackebrandt E, Sanderson K, Volkman JK, Cameron DE, Stevenson PL, McMeekin TA, Burton HR. 1988. Halobacterium lacusprofundi, sp. nov., a halophilic bacterium isolated from Deep Lake, Antarctica. Syst Appl Microbiol 11:20–27. doi: 10.1016/S0723-2020(88)80044-4. [DOI] [Google Scholar]
- 17.DeMaere MZ, Williams TJ, Allen MA, Brown MV, Gibson JAE, Rich J, Lauro FM, Dyall-Smith M, Davenport KW, Woyke T, Kyrpides NC, Tringe SG, Cavicchioli R. 2013. High level of intergenera gene exchange shapes the evolution of haloarchaea in an isolated Antarctic lake. Proc Natl Acad Sci U S A 110:16939–16944. doi: 10.1073/pnas.1307090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yau S, Lauro FM, Williams TJ, DeMaere MZ, Brown MV, Rich J, Gibson JA, Cavicchioli R. 2013. Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J 7:1944–1961. doi: 10.1038/ismej.2013.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams TJ, Allen MA, DeMaere MZ, Kyrpides NC, Tringe SG, Woyke T, Cavicchioli R. 2014. Microbial ecology of an Antarctic hypersaline lake: genomic assessment of ecophysiology among dominant haloarchaea. ISME J 8:1645–1658. doi: 10.1038/ismej.2014.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brum JR, Hurwitz BL, Schofield O, Ducklow HW, Sullivan MB. 2016. Seasonal time bombs: dominant temperate viruses affect Southern Ocean microbial dynamics. ISME J 10:437–449. doi: 10.1038/ismej.2015.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohwer F, Prangishvili D, Lindell D. 2009. Roles of viruses in the environment. Environ Microbiol 11:2771–2774. doi: 10.1111/j.1462-2920.2009.02101.x. [DOI] [PubMed] [Google Scholar]
- 22.Suttle CA. 2013. Viruses: unlocking the greatest biodiversity on Earth. Genome 56:542–544. doi: 10.1139/gen-2013-0152. [DOI] [PubMed] [Google Scholar]
- 23.Brum JR, Sullivan MB. 2015. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat Rev Microbiol 13:147–159. doi: 10.1038/nrmicro3404. [DOI] [PubMed] [Google Scholar]
- 24.Clarke KR, Gorley RN. 2006. PRIMER v6: user manual/tutorial. Primer-E, Plymouth, United Kingdom. [Google Scholar]
- 25.Button DK, Robertson B. 2000. Effect of nutrient kinetics and cytoarchitecture on bacterioplankter size. Limnol Oceanogr 45:499–505. doi: 10.4319/lo.2000.45.2.0499. [DOI] [Google Scholar]
- 26.Williams TJ, Cavicchioli R. 2014. Marine metaproteomics: deciphering the microbial metabolic food web. Trends Microbiol 22:248–260. doi: 10.1016/j.tim.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ. 2009. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3:93–105. doi: 10.1038/ismej.2008.83. [DOI] [PubMed] [Google Scholar]
- 28.Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF, Giovannoni SJ. 2011. Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5:856–865. doi: 10.1038/ismej.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu X, Doroghazi JR, Janga SC, Zhang JK, Circello B, Griffin BM, Labeda DP, Metcalf WW. 2013. Diversity and abundance of phosphonate biosynthetic genes in nature. Proc Natl Acad Sci U S A 110:20759–20764. doi: 10.1073/pnas.1315107110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolowith L, Ingall ED, Benner R. 2001. Composition and cycling of marine organic phosphorus. Limnol Oceanogr 46:309–320. doi: 10.4319/lo.2001.46.2.0309. [DOI] [Google Scholar]
- 31.Elevi Bardavid R, Khristo P, Oren A. 2008. Interrelationships between Dunaliella and halophilic prokaryotes in saltern crystallizer ponds. Extremophiles 12:5–14. doi: 10.1007/s00792-006-0053-y. [DOI] [PubMed] [Google Scholar]
- 32.Sherwood KE, Cano DJ, Maupin-Furlow JA. 2009. Glycerol-mediated repression of glucose metabolism and glycerol kinase as the sole route of glycerol catabolism in the haloarchaeon Haloferax volcanii. J Bacteriol 191:4307–4315. doi: 10.1128/JB.00131-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elevi Bardavid R, Oren A. 2008. Dihydroxyacetone metabolism in Salinibacter ruber and in Haloquadratum walsbyi. Extremophiles 12:125–131. doi: 10.1007/s00792-007-0114-x. [DOI] [PubMed] [Google Scholar]
- 34.Ouellette M, Makkay AM, Papke RT. 2013. Dihydroxyacetone metabolism in Haloferax volcanii. Front Microbiol 4:376. doi: 10.3389/fmicb.2013.00376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gimmler H, Möller EM. 1981. Salinity-dependent regulation of starch and glycerol metabolism in Dunaliella parva. Plant Cell Environ 4:367–375. doi: 10.1111/j.1365-3040.1981.tb02114.x. [DOI] [Google Scholar]
- 36.Danson MJ, Lamble HJ, Hough DW. 2007. Central metabolism, p 260–287. In Cavicchioli R. (ed), Archaea. Molecular and cellular biology. ASM Press, Washington, DC. [Google Scholar]
- 37.Falb M, Müller K, Königsmaier L, Oberwinkler T, Horn P, von Gronau S, Gonzales O, Pfeiffer F, Bornberg-Bauer E, Oesterhelt D. 2008. Metabolism of halophilic archaea. Extremophiles 12:177–196. doi: 10.1007/s00792-008-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albers SV, Jarrell KF. 2015. The archaellum: how Archaea swim. Front Microbiol 6:23. doi: 10.3389/fmicb.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szabó Z, Sani M, Groeneveld M, Zolghadr B, Schelert J, Albers SV, Blum P, Boekema EJ, Driessen AJ. 2007. Flagellar motility and structure in the hyperthermoacidophilic archaeon Sulfolobus solfataricus. J Bacteriol 189:4305–4309. doi: 10.1128/JB.00042-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sára M, Sleytr UB. 2000. S-layer proteins. J Bacteriol 182:859–868. doi: 10.1128/JB.182.4.859-868.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solomon C, Collier J, Berg G, Glibert P. 2010. Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat Microb Ecol 59:67–88. doi: 10.3354/ame01390. [DOI] [Google Scholar]
- 42.Shi Y, Tyson GW, Eppley JM, DeLong EF. 2011. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alonso-Sáez L, Waller AS, Mende DR, Bakker K, Farnelid H, Yager PL, Lovejoy C, Tremblay JÉ, Potvin M, Heinrich F, Estrada M, Riemann L, Bork P, Pedrós-Alió C, Bertilsson S. 2012. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci U S A 109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoch MP, Jeffrey WH, Snyder RA, Dillon KS, Coffin RB. 2006. Expression of glutamine synthetase and glutamate dehydrogenase by marine bacterioplankton: assay optimization and efficacy for assessing nitrogen to carbon metabolic balance in situ. Limnol Oceanogr 4:308–328. doi: 10.4319/lom.2006.4.308. [DOI] [Google Scholar]
- 45.Takahashi N, Sato T, Yamada T. 2000. Metabolic pathways for cytotoxic end product formation from glutamate- and aspartate-containing peptides by Porphyromonas gingivalis. J Bacteriol 182:4704–4710. doi: 10.1128/JB.182.17.4704-4710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlesner M, Miller A, Streif S, Staudinger WF, Müller J, Scheffer B, Siedler F, Oesterhelt D. 2009. Identification of Archaea-specific chemotaxis proteins which interact with the flagellar apparatus. BMC Microbiol 9:56. doi: 10.1186/1471-2180-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson AT, Marchant HJ, de la Mare WK. 1996. Natural UVB exposure changes the species composition of Antarctic phytoplankton in mixed culture. Aquat Microb Ecol 10:299–305. doi: 10.3354/ame010299. [DOI] [Google Scholar]
- 48.McCready S, Müller JA, Boubriak I, Berquist BR, Ng WL, DasSarma S. 2005. UV irradiation induces homologous recombination genes in the model archaeon, Halobacterium sp. NRC-1. Saline Systems 1:3. doi: 10.1186/1746-1448-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boubriak I, Ng WL, DasSarma P, DasSarma S, Crowley DJ, McCready SJ. 2008. Transcriptional responses to biologically relevant doses of UV-B radiation in the model archaeon, Halobacterium sp. NRC-1. Saline Systems 4:13. doi: 10.1186/1746-1448-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhaya D, Grossman AR, Steunou AS, Khuri N, Cohan FM, Hamamura N, Melendrez MC, Bateson MM, Ward DM, Heidelberg JF. 2007. Population level functional diversity in a microbial community revealed by comparative genomic and metagenomic analyses. ISME J 1:703–713. doi: 10.1038/ismej.2007.46. [DOI] [PubMed] [Google Scholar]
- 51.Cuadros-Orellana S, Martin-Cuadrado AB, Legault B, D'Auria G, Zhaxybayeva O, Papke RT, Rodriguez-Valera F. 2007. Genomic plasticity in prokaryotes: the case of the square haloarchaeon. ISME J 1:235–245. doi: 10.1038/ismej.2007.35. [DOI] [PubMed] [Google Scholar]
- 52.Krause DJ, Whitaker RJ. 2015. Inferring speciation processes from patterns of natural variation in microbial genomes. Syst Biol 64:926–935. doi: 10.1093/sysbio/syv050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosen MJ, Davison M, Bhaya D, Fisher DS. 2015. Fine-scale diversity and extensive recombination in a quasisexual bacterial population occupying a broad niche. Science 348:1019–1023. doi: 10.1126/science.aaa4456. [DOI] [PubMed] [Google Scholar]
- 54.Kearns DB. 2010. A field guide to bacterial swarming motility. Nat Rev Microbiol 8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.