ABSTRACT
The phloem-feeding Southern chinch bug, Blissus insularis, harbors a high density of the exocellular bacterial symbiont Burkholderia in the lumen of specialized midgut crypts. Here we developed an organ culture method that initially involved incubating the B. insularis crypts in osmotically balanced insect cell culture medium. This approach enabled the crypt-inhabiting Burkholderia spp. to make a transition to an in vitro environment and to be subsequently cultured in standard bacteriological media. Examinations using ribotyping and BOX-PCR fingerprinting techniques demonstrated that most in vitro-produced bacterial cultures were identical to their crypt-inhabiting Burkholderia counterparts. Genomic and physiological analyses of gut-symbiotic Burkholderia spp. that were isolated individually from two separate B. insularis laboratory colonies revealed that the majority of individual insects harbored a single Burkholderia ribotype in their midgut crypts, resulting in a diverse Burkholderia community within each colony. The diversity was also exhibited by the phenotypic and genotypic characteristics of these Burkholderia cultures. Access to cultures of crypt-inhabiting bacteria provides an opportunity to investigate the interaction between symbiotic Burkholderia spp. and the B. insularis host. Furthermore, the culturing method provides an alternative strategy for establishing in vitro cultures of other fastidious insect-associated bacterial symbionts.
IMPORTANCE An organ culture method was developed to establish in vitro cultures of a fastidious Burkholderia symbiont associated with the midgut crypts of the Southern chinch bug, Blissus insularis. The identities of the resulting cultures were confirmed using the genomic and physiological features of Burkholderia cultures isolated from B. insularis crypts, showing that host insects maintained the diversity of Burkholderia spp. over multiple generations. The availability of characterized gut-symbiotic Burkholderia cultures provides a resource for genetic manipulation of these bacteria and for examination of the mechanisms underlying insect-bacterium symbiosis.
INTRODUCTION
Many plant-sap-feeding insects harbor exocellular bacterial symbionts in their digestive tracts (1, 2). These symbionts usually colonize specialized regions of the midgut, are transmitted to offspring by egg smearing, symbiont-containing capsules, or coprophagy, and play a role in host insect fitness (3–7). Exocellular symbionts can provide nutrients to supplement the unbalanced diets of their host insects (1, 8–10)m thus conferring fitness advantages (e.g., body size, coloration, development, and growth) (11–13). In addition, these symbionts have been reported to elicit symbiont-mediated plant specialization (14), to modulate plant virus transmission (15, 16), and/or to induce protection against pathogenic gut bacteria (17), protozoan parasites (18), and synthetic insecticides (19).
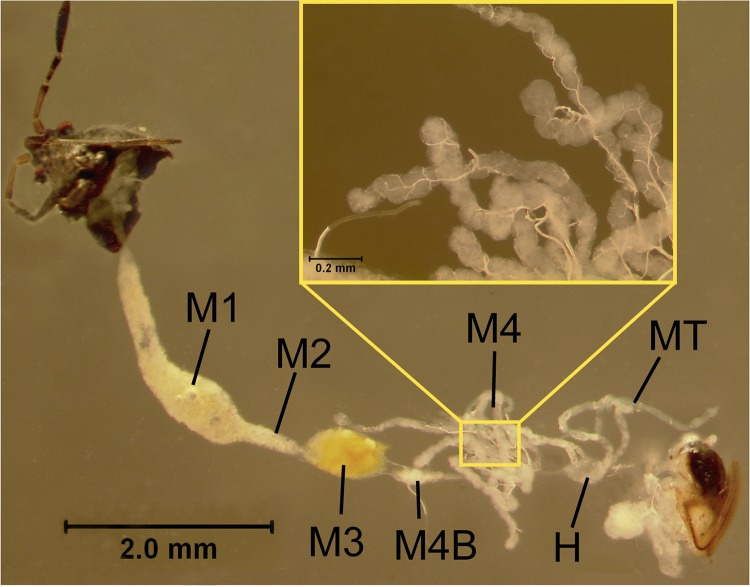
Recently, Burkholderia spp. have been detected in the lumens of specialized crypts, at the fourth region of the midgut (M4) (Fig. 1), in the Southern chinch bug, Blissus insularis Barber (Hemiptera: Lygaeoidea: Blissidae) (12). This insect, a primary pest of St. Augustinegrass, Stenotaphrum secundatum (Walter) Kuntze (20, 21), feeds on the grass phloem, resulting in diminished grass growth, yellowing and brown blade color, and the eventual death of grass patches (22, 23). Investigations of multiple B. insularis field populations demonstrated that complex Burkholderia ribotypes were present within and among the populations (12). The copy number of the Burkholderia 16S rRNA gene increased with the age of B. insularis insects, and antibiotic treatment reduced Burkholderia numbers and slowed B. insularis development, pointing to a mutualistic Blissus-Burkholderia association (12). Another chinch bug species, Cavelerius saccharivorus Okajima (Heteroptera: Lygaeoidea: Blissidae), also harbors Burkholderia spp. in the lumens of midgut crypts (24). Preliminary evidence suggests that Burkholderia spp. in both chinch bug species may be vertically transmitted from bacteria deposited on the egg surface. Specifically, Burkholderia 16S rRNA gene amplicons were detected in DNA preparations from both B. insularis (12) and C. saccharivorus (24) eggs, and sterilization of egg surfaces eliminated the Burkholderia 16S rRNA gene amplicons of the DNA extracted from C. saccharivorus hatchlings (24). Even though the biological function of gut symbionts in chinch bugs is not well understood, the consistent occurrence of Burkholderia spp. in these insects implies that these gut microorganisms play an important role in host insect fitness.
FIG 1.
Micrographs of dissected digestive and reproductive tracts of a female Blissus insularis bug. Abbreviations: M1, M2, and M3, first, second, and third sections of the midgut, respectively; M4, fourth section of the midgut with crypts; M4B, M4 bulb; H, hindgut; MT, Malpighian tubules. The labeling corresponds to that used for Cavelerius saccharivorus (24).
Previous attempts to culture the exocellular gut-symbiotic Burkholderia spp. from B. insularis have failed (12). The inability to culture these bacteria has hindered studies to address the functional role(s) of gut symbionts in B. insularis. In the current study, a novel culturing method was developed to produce Burkholderia cultures from the midgut of B. insularis. This strategy involved an initial organ culture step that probably allowed these symbionts to transit from a crypt-associated to a free-living lifestyle. The resulting bacterial cultures were transferred to bacteriological media and were characterized, providing phenotypic and molecular data on various Burkholderia ribotypes derived from B. insularis.
MATERIALS AND METHODS
Rearing and genetic background of insects.
Two field populations of B. insularis from two St. Augustinegrass lawns that were 480 km apart (in Alachua County and Santa Rosa County) in Florida were sampled. Insects from each site, reared as separate laboratory colonies (BiR [insecticide resistant] and BiS [insecticide susceptible], respectively), were provisioned with cut St. Augustinegrass and fresh, surface-sterilized yellow corn cobs (25). The genetic backgrounds of insects from the BiR and BiS colonies were examined by comparing the partial sequences of the mitochondrial cytochrome c oxidase subunit I (COI) gene (26). Specifically, four B. insularis females from each colony were surface sterilized by sequential immersion for 3 min in 70% ethanol (EtOH), 5% bleach, and 70% EtOH. Intact midgut crypts were dissected individually from the digestive tracts, rinsed in sterile H2O, and subjected to DNA extraction using the MasterPure yeast DNA purification kit (Epicentre, Madison, WI). The COI gene was amplified by PCR using the crypt genomic DNA of eight B. insularis individuals (see Table S1 in the supplemental material for primer sequences). Positive PCR amplicons were purified using a PCR purification kit (Agencourt AMPure XP; Beckman Coulter, Beverly, MA) and were sequentially subjected to unidirectional Sanger sequencing (ICBR Sequencing Core, University of Florida). The sequences of the COI gene were trimmed manually to 519 bp and were aligned with each other using MUSCLE, version 3.7 (27), in order to examine the sequence similarity between these two colonies.
Culturing of crypt-associated bacteria.
Conventional culturing of crypt-associated bacteria was tested initially by plating homogenates of dissected midgut crypts. Intact crypts were dissected individually from surface-sterilized B. insularis adults and were rinsed in sterile H2O, 1× phosphate-buffered saline (PBS, comprising 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4 [pH 7.4]), or Grace's insect cell culture medium (ICM) amended with l-glutamine (Orbigen, San Diego, CA). The rinsed crypts were homogenized in 50 μl of sterile H2O, PBS, or ICM, respectively, and were then streak-plated onto nutrient agar (0.3% beef extract, 0.5% peptone, 1.5% agar; Becton, Dickinson and Company, NJ) plates and were incubated at 28°C for 14 days. Plates were examined daily for bacterial growth.
The second culturing approach involved initial culturing of the dissected crypts in ICM. Intact crypts were dissected individually from surface-sterilized adult and fifth-instar B. insularis individuals that had been maintained for 2 to 3 generations, 4 to 5 generations, or 8 to 9 generations in BiR or BiS colonies. Half of the crypts dissected from each insect were rinsed in sterile H2O and were subjected to DNA extraction using the MasterPure yeast DNA purification kit. The remaining dissected crypt halves were rinsed three times in ICM, placed in 24-well cell culture plates (Corning Incorporated, Corning, NY) containing 500 μl of ICM, and incubated at 28°C. Organ cultures were examined daily under a dissecting microscope to monitor crypt morphology and under an inverted compound microscope fitted with Hoffman modulation contrast optics (Modulation Optics, Inc., Greenvale, NY) to assess bacterial growth over a 14-day period. Subsequently, preparations were inoculated onto nutrient agar plates and were incubated at 28°C. Duplicate bacterial colonies were selected from each plate, homogenized individually in 25 μl of sterile H2O, boiled for 3 min, and subjected to diagnostic PCR amplification using Burkholderia-specific 16S rRNA gene primers (see Table S1 in the supplemental material), as described below. Colonies producing positive PCR amplicons by use of Burkholderia-specific 16S rRNA gene primers were considered to be Burkholderia colonies; they were then inoculated into nutrient broth medium (0.3% beef extract, 0.5% peptone) and were shaken at 200 rpm and 28°C overnight. The MasterPure yeast DNA purification kit was used to extract bacterial DNA from the cultures for subsequent sequencing and analyses. The Burkholderia culture stocks were prepared in 60% sterile glycerol and were stored at −80°C.
Sequencing and analyses of symbiotic bacteria.
The 16S rRNA gene sequences of bacteria inhabiting 30 dissected crypts and those of bacteria derived from 20 cultured crypts were generated by PCR amplification with universal primers (10F and 1507R) (see Table S1 in the supplemental material) (28). Subsequently, the corresponding 1.5-kb PCR products were purified using a PCR purification kit (Agencourt AMPure XP) and were subjected to bidirectional Sanger sequencing. In addition to the 16S rRNA gene, three Burkholderia cepacia complex multilocus sequence typing (MLST) genes (atpD, encoding the ATP synthase beta chain; lepA, encoding a GTP binding protein; and recA, encoding recombinase A) (29) were amplified by PCR and sequenced in order to further genotype the Burkholderia spp. associated with B. insularis (see Table S1). DNA preparations derived from both dissected crypt preparations and cultured bacterial isolates originating from BiR and BiS insects were tested.
Trace chromatograms were examined in order to determine the complexity of bacteria associated with various DNA preparations. Universal 16S rRNA gene sequences generated from 26 dissected crypt preparations and 20 cultured bacterial isolates that produced clean chromatograms were trimmed to ∼1.4 kb and were uploaded to the Ribosomal Database Project (RDP; release 11) website to be aligned and to be compared with the database sequences, using the Sequence Match online analysis tool (30). The published database sequences with high similarity (≥0.98) and seqmatch (≥0.94) scores, and the Burkholderia 16S rRNA gene sequences obtained from B. insularis field populations (12), were included as reference sequences in subsequent phylogenetic analyses. Pandoraea norimbergensis (GenBank accession no. AF139171) served as the outgroup. Phylogenetic analyses of aligned nucleotides were conducted using the maximum likelihood method, as described by Dereeper et al. (31). A bootstrap test with 100 replicates was performed to generate the likelihood bootstrap values. The final version of the phylogenetic tree was edited using TreeGraph 2 software (32). Additionally, the 16S rRNA gene sequences of the same dissected B. insularis crypts were aligned pairwise with those of their cultured bacterial counterparts by use of MUSCLE in order to determine whether or not their ribotypes were identical.
For the MLST genes, the sequences were trimmed manually to the same length (443 bp for atpD, 397 bp for lepA, 393 bp for recA) as those of other B. cepacia complex MLST sequences available in the database (33). The concatenated MLST gene sequences (1,233 bp) generated from four dissected crypts and eight cultured bacterial isolates were subjected to phylogenetic analyses as conducted for the 16S rRNA gene. The phylogenetic trees of MLST gene sequences and the corresponding 16S rRNA gene sequences were edited by TreeGraph 2 and were constructed side by side for comparison. Burkholderia cepacia (strain ATCC 49709) served as the outgroup.
BOX-PCR fingerprinting.
In addition to 16S rRNA ribotyping and MLST analysis, BOX-PCR fingerprinting was conducted to examine the similarity of the bacteria inhabiting the dissected crypts with their cultured counterparts. The DNA preparations from 18 dissected-crypt DNA preparations and from their respective cultured bacterial isolates were subjected to PCR amplification using a BOX-A1R primer (5′-CTACGGCAAGGCGACGCTGACG-3′) (34). PCR products were electrophoresed in a 1.5% SynerGel–agarose gel (Diversified Biotech, Boston, MA) that was dissolved in 0.5× Tris-borate-EDTA buffer for 10 h at 40 V and were stained with ethidium bromide. Gel images were digitized using a ChemiDoc XRS system and were analyzed using Quantity One software (Bio-Rad, Hercules, CA). Lane-based background subtraction was applied to remove background from lanes, and a similarity matrix of lane-based samples was generated to determine the similarities (expressed as percentages) between dissected-crypt preparations and their respective cultured counterparts.
PFGE.
Twenty cultured Burkholderia isolates derived from crypts of 10 BiR and 10 BiS B. insularis individuals were subjected to pulsed-field gel electrophoresis (PFGE) for genomic typing using the CHEF-DR II pulsed-field electrophoresis system (Bio-Rad). Mid-log-phase cultures of Burkholderia isolates were mixed with 180 μg ml−1 of chloramphenicol (Bioline, Taunton, MA) and were incubated for an additional hour to terminate chromosomal replication. An estimated 5 × 108 Burkholderia cells were harvested and were suspended in 10 mM Tris–20 mM NaCl–50 mM EDTA (pH 7.2). The cell suspension was mixed (1:1) with 1.6% pulsed-field-certified agarose (Bio-Rad) at 50°C, and the mixture was loaded into plug molds. Solidified sample plugs were incubated in lysozyme buffer (10 mM Tris, 50 mM NaCl, 0.2% sodium deoxycholate, 0.5% sodium lauryl sarcosine, 1 mg ml−1 lysozyme [pH 7.2]) at 37°C for 1 h, incubated in proteinase K reaction buffer (100 mM EDTA, 0.2% sodium deoxycholate, 1% sodium lauryl sarcosine, 1 mg ml−1 proteinase K [pH 8.0]) at 50°C overnight, and then rinsed four times with 1× wash buffer (20 mM Tris, 50 mM EDTA [pH 8.0]). Sample plugs initially were run with the Hansenula wingei standard (1.05 to 3.13 Mb; strain YB-4662-VIA; Bio-Rad) in a 0.8% pulsed-field-certified agarose gel in 1× Tris-acetate-EDTA buffer recirculated at 14°C. The samples that had DNA fragments larger than those of H. wingei (>3.13 Mb) were run subsequently with the Schizosaccharomyces pombe standard (3.5 to 5.7 Mb; strain 972 h-; Bio-Rad). The run time for H. wingei was 50 h at 3 V cm−1 with a 250- to 900-s switch time ramp, whereas the run time for S. pombe was 70 h at 2 V cm−1 with a 1,200- to 1,800-s switch time ramp. Gels were stained with 1× SYBR gold nucleic acid gel stain (Molecular Probes, Eugene, OR), inspected visually under UV light, and imaged. DNA fragment sizes were estimated based on standard molecular weight using Quantity One software (Bio-Rad). Sample plug preparation and gel electrophoresis were repeated at least twice for each Burkholderia isolate. The numbers of replicons and estimated genomic sizes of Burkholderia isolates clustered from clades were analyzed separately using the two-sample t test (PROC TTEST, SAS 9.3).
In vitro growth of Burkholderia isolates.
The growth rates of 20 cultured Burkholderia isolates derived from 10 BiR and 10 BiS B. insularis individuals were measured using a modified 96-well microtiter plate assay (35). Initially, Burkholderia cells were harvested at the mid-log phase, diluted serially in nutrient broth medium, and plated on nutrient agar plates for determination of the CFU counts per milliliter in the starting culture inoculum. A 200-μl volume of each starting culture inoculum (105 CFU ml−1) was loaded in triplicate into the wells of a sterile 96-well microtiter plate and was placed in an incubator shaker at 150 rpm and 28°C for 36 h. Absorbance at 600 nm was measured hourly using the microtiter plate reader (BioTek Instruments, Inc.). The growth rate, expressed as the doubling time (hours per generation), was calculated during the exponential phase. The growth kinetics of each isolate were examined at least twice using freshly prepared inocula. The growth rates of BiR Burkholderia isolates were compared with those of BiS isolates using the two-sample t test (PROC TTEST, SAS 9.3). Comparisons among isolates from three clades were performed by analysis of variance (ANOVA) (PROC ANOVA, SAS 9.3).
Biofilm formation.
Culturing of the dissected crypts in liquid ICM resulted in the production of a bacterial biofilm on the ICM surface. To confirm that the biofilm was produced by the crypt-associated bacteria rather than by contaminants, the cultured bacterial isolates that produced positive Burkholderia-specific 16S rRNA gene amplicons by diagnostic PCR (see “Sequencing and analyses of symbiotic bacteria” above) were inoculated into fresh liquid ICM and were incubated at 28°C for 48 h. The resulting metallic biofilms were washed at least three times with sterile H2O using an injection syringe, transferred to microscope cover glasses, examined by differential interference microscopy at ×1,600 magnification using a Leica DMRB microscope (Leica Microsystems Inc., Buffalo Grove, IL), and then imaged with a SPOT Insight QE camera system (SPOT Imaging Solutions, Sterling Heights, MI).
Antibiotic sensitivities of Burkholderia isolates.
Twenty cultured Burkholderia isolates were subjected to disc diffusion sensitivity assays with six antibiotics, including kanamycin (Bioline), oxytetracycline hydrochloride (Sigma-Aldrich, St. Louis, MO), trimethoprim (Teknova, Hollister, CA), ampicillin (Sigma-Aldrich), penicillin G (Sigma-Aldrich), and chloramphenicol (Bioline). At the mid-log phase, 1 ml of Burkholderia culture grown in nutrient broth (approximately 5 × 108 cells ml−1) was overlaid onto a nutrient agar plate (95 by 15 mm; Fisher Scientific) and was incubated at room temperature for 15 min. Excess inoculum was removed, and plates were allowed to dry in a laminar flow hood. Sterile blank paper discs (diameter, 6 mm; BBL; Becton, Dickinson, and Company, Sparks, MD) were each inoculated with 25 μl of an antibiotic solution, air dried, and placed on the nutrient agar plate covered by Burkholderia cells. The solvent for chloramphenicol and trimethoprim was EtOH. Based on preliminary assays (unpublished results), a concentration of 1 mM antibiotic solution was selected to be assayed in triplicate against Burkholderia isolates. After 24 to 48 h of incubation at 28°C, the inhibition zone of each antibiotic was measured (diameter in millimeters) in order to determine the relative sensitivities of cultured Burkholderia isolates to antibiotics. Comparisons between Burkholderia isolates from the BiR and BiS populations were analyzed by the two-sample t test (PROC TTEST, SAS 9.3).
Nucleotide sequence accession numbers.
All DNA sequences obtained from this study were deposited in the GenBank nucleotide sequence database with the following accession numbers: KP683095 to KP683096, KP683112 to KP683113, KU242589 to KU242608, and KU244285 to KU244310 (16S rRNA gene sequences), KU242609 to KU242610 (COI gene sequences), KU242611 to KU242622 (atpD gene sequences), KU247540 to KU247551 (recA gene sequences), and KU247552 to KU247563 (lepA gene sequences).
RESULTS
Sequencing of the COI gene for B. insularis.
Eight crypt genomic DNA preparations from four BiR and four BiS B. insularis individuals produced chromatograms of COI gene amplicons free of mixed reads within the target sequence. The 519-bp sequences derived from seven B. insularis individuals were 100% identical to each other (GenBank accession no. KU242609), whereas the sequence of one individual from the BiS colony had 99% similarity to the others, with five single-nucleotide polymorphisms (SNPs) (GenBank accession no. KU242610).
Culturing of crypt-associated bacteria.
Attempts to directly culture bacteria from 15 B. insularis midgut crypts, homogenized in sterile H2O, PBS, or ICM, on bacteriological media failed to produce detectable colonies on the nutrient agar plate after 14 days. Among 30 dissected crypts from 13 BiR and 17 BiS B. insularis individuals that were inoculated in ICM, 22 crypt preparations (73%) became increasingly swollen (Fig. 2), with short, rod-shaped bacteria detected in ICM within 7 days, and produced lawns of identical colony phenotypes on the nutrient agar plate after being streak-plated and incubated for 48 h. Bacterial colonies from these 22 preparations were identified as Burkholderia by diagnostic PCR amplification using Burkholderia-specific 16S rRNA gene primers. The other eight crypt preparations (Bi15_R, Bi26_R, Bi13_S, Bi17_S, Bi18_S, Bi23_S, Bi24_S, and Bi25_S) either did not produce any detectable bacteria in ICM after 14 days or were unculturable upon transfer to nutrient agar plates. Based on the ribotypes produced by the genomic DNA derived from their dissected-crypt counterparts, six of these eight preparations were grouped into different clades (the stinkbug-associated beneficial and environmental [SBE] and Burkholderia cepacia complex [BCC] clades) identified in the phylogenetic tree (Fig. 3). The remaining two dissected-crypt preparations (Bi15_R_vivo and Bi18_S_vivo) produced mixed 16S rRNA sequences and therefore were excluded from the phylogenetic analysis.
FIG 2.
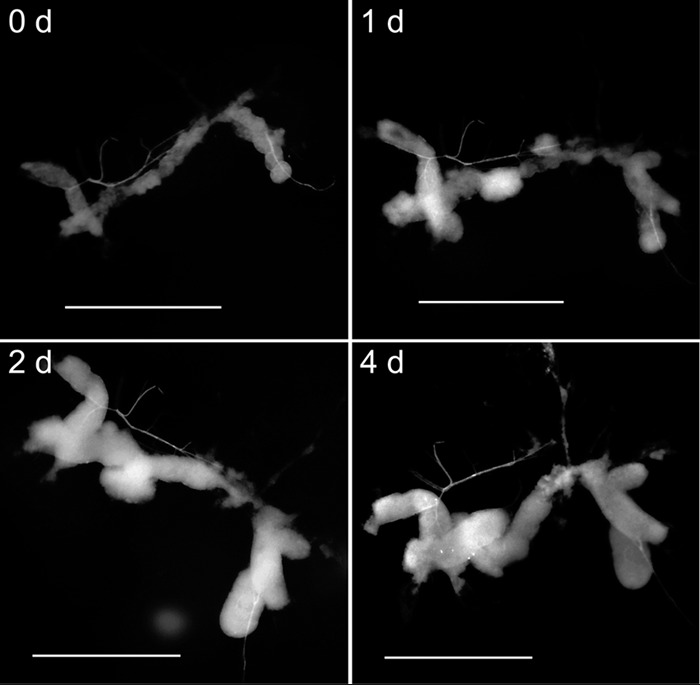
Morphological features of the Blissus insularis midgut crypts inoculated into insect culture medium on days 0, 1, 2, and 4 postdissection. Bars, 0.6 mm.
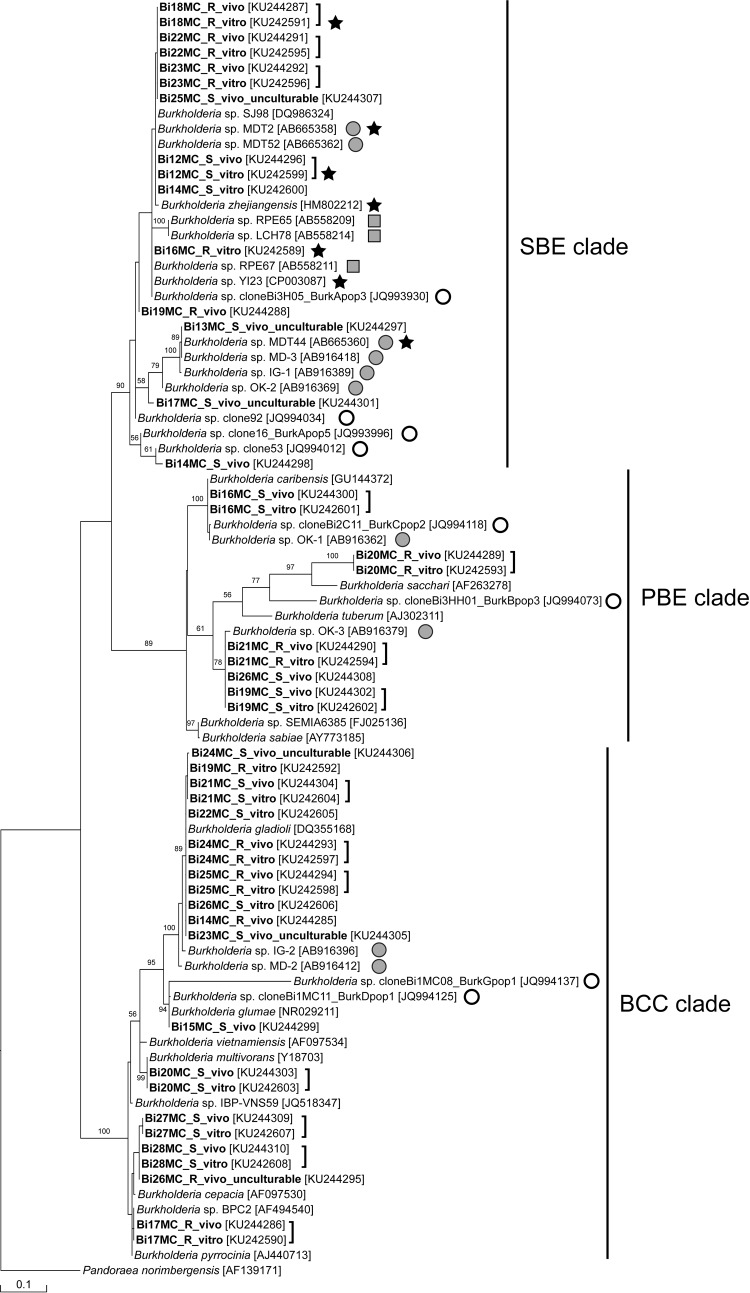
FIG 3.
Phylogenetic relationships of crypt-associated bacteria (labeled “vivo”) and their cultured counterparts (labeled “vitro”) obtained from Blissus insularis midgut crypts (designated Bi12MC to Bi28MC) on the basis of universal 16S rRNA gene sequences (∼1.4 kb). R and S indicate sequences obtained from the BiR and BiS B. insularis colonies, respectively. The sequences detected in the present study are shown in boldface. Numbers at the tree nodes represent the maximum likelihood bootstrap values obtained after 100 repetitions; only values of >50% are shown. GenBank nucleotide sequence accession numbers are given in brackets. Open and shaded circles indicate the Burkholderia spp. detected in the B. insularis field populations (12) and in Cavelerius saccharivorus (24), respectively; squares indicate the Burkholderia spp. detected in other heteropteran hosts; stars indicate the pesticide-degrading strains. Clades SBE, PBE, and BCC correspond to those described in references 24, 36, and 37, respectively.
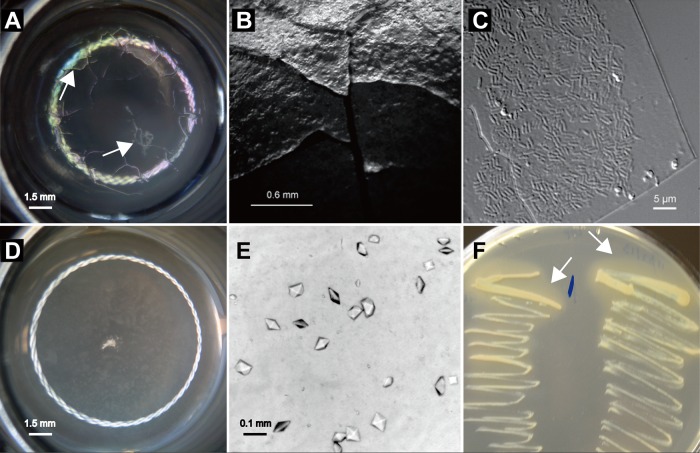
Among the 22 pure cultures generated from inoculated crypt preparations shown to be Burkholderia spp. by diagnostic PCR, 12 preparations (7 from BiR and 5 from BiS B. insularis individuals) produced ridged metallic biofilms on the surface of ICM (Fig. 4A and B). Microscopy revealed numerous short, rod-shaped bacteria, measuring approximately 0.5 by 2.0 μm, accumulating on the undersides of these biofilms (Fig. 4C). Six cultures (three from BiR and three from BiS B. insularis individuals) produced crystal deposits without detectable biofilms (Fig. 4D and E), whereas four cultures (one BiR and three BiS cultures) produced neither biofilm nor crystals (Table 1). Moreover, the cultures that produced crystals in ICM formed halos around the bacterial colonies on the nutrient agar plate (Fig. 4F). The cultured Burkholderia counterparts were reinoculated into fresh ICM and produced the same phenotypic characteristics as the original inoculated crypt preparations. These findings indicated that at least two distinct phenotypes (biofilm formation and crystal production) of crypt-associated Burkholderia spp. were present after the isolates were cultured in ICM, regardless of their source (BiR or BiS B. insularis) (Table 1).
FIG 4.
Microscopy of two typical phenotypic characteristics of culturable Burkholderia isolates recovered from Blissus insularis midgut crypts. (A to C) Biofilm formation. (A) The metallic biofilm established on the surface of ICM with the inoculated midgut crypts. Arrows indicate the metallic biofilm. (B) A close look at the metallic biofilm formation. (C) The short rod-shaped bacteria underneath the metallic biofilm were examined with a light microscope at a magnification of ×1,600. (D to F) Precipitation of salt crystals. (D) Salt crystal precipitation produced by the inoculated crypts in ICM. (E) A close look at the precipitated salt crystals. (F) The Burkholderia isolates that precipitated salt crystals in ICM formed halos (indicated by arrows) around the bacterial colonies after being plated on the nutrient agar plate.
TABLE 1.
Phenotypes, growth rates, and antibiotic sensitivities of cultured Burkholderia isolates recovered from Blissus insularis crypts
| Cladea | Isolate designation | Gender of host | Phenotypeb | Halo formationc | Mean (SE) doubling timed | Mean (SE) diam of inhibition zone (mm)e with: |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kan | Oxy | Tri | Amp | Pen | Chl | ||||||
| SBE | Bi16MC_R_vitro | Female | MB | − | 4.7 (0) | 24 (1) | 18 (2) | 25 (3) | 0 | 0 | 18 (1) |
| Bi18MC_R_vitro | Female | MB | − | 8.1 (0.1) | 23 (1) | 18 (2) | 25 (3) | 0 | 0 | 14 (1) | |
| Bi22MC_R_vitro | Male | MB | − | 3.4 (0.6) | 21 (1) | 22 (1) | 26 (2) | 0 | 0 | 20 (1) | |
| Bi23MC_R_vitro | Female | MB | − | 8.7 (1.4) | 31 (3) | 26 (1) | 24 (4) | 0 | 0 | 22 (2) | |
| Bi12MC_S_vitro | Female | MB | − | 4.5 (0.3) | 24 (1) | 18 (2) | 21 (2) | 0 | 0 | 17 (1) | |
| Bi14MC_S_vitro | Female | MB | − | 5.6 (0.9) | 23 (1) | 18 (2) | 18 (3) | 0 | 0 | 16 (1) | |
| PBE | Bi20MC_R_vitro | Female | MB | − | 11.1 (0.8) | 21 (2) | 22 (0) | 32 (2) | 26 (1) | 26 (1) | 17 (0) |
| Bi21MC_R_vitro | Male | MB | − | 5.4 (0.2) | 20 (2) | 22 (0) | 23 (5) | 27 (1) | 28 (2) | 15 (2) | |
| Bi16MC_S_vitro | Femalef | MB | − | 8.7 (0.6) | 22 (1) | 25 (1) | 28 (0) | 19 (2) | 18 (2) | 16 (1) | |
| Bi19MC_S_vitro | Femalef | MB | − | 5.2 (0.5) | 27 (2) | 22 (0) | 34 (2) | 26 (2) | 24 (1) | 15 (0) | |
| BCC | Bi14MC_R_vitro | Female | SC | + | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| Bi17MC_R_vitro | Female | MB | − | 5.4 (0.3) | 17 (1) | 12 (1) | 24 (1) | 0 | 0 | 14 (2) | |
| Bi19MC_R_vitro | Female | N/A | + | 4.6 (0.4) | 14 (1) | 23 (1) | 21 (4) | 34 (1) | 34 (2) | 9 (1) | |
| Bi24MC_R_vitro | Female | SC | + | 4.6 (0.2) | 22 (2) | 14 (1) | 22 (1) | 0 | 0 | 0 | |
| Bi25MC_R_vitro | Male | SC | + | 4.3 (0.5) | 21 (1) | 13 (1) | 21 (2) | 0 | 0 | 0 | |
| Bi15MC_S_vitro | Female | SC | + | N/A | N/A | N/A | N/A | N/A | N/A | N/A | |
| Bi20MC_S_vitro | Female | MB | − | 7.5 (0.6) | 21 (0) | 15 (1) | 23 (1) | 0 | 0 | 14 (2) | |
| Bi21MC_S_vitro | Female | SC | + | 4.8 (0.1) | 24 (1) | 11 (1) | 31 (3) | 0 | 0 | 0 | |
| Bi22MC_S_vitro | Female | SC | + | 6.3 (0.1) | 16 (0) | 11 (1) | 30 (4) | 0 | 0 | 0 | |
| Bi26MC_S_vitro | Male | N/A | + | 6.4 (0.6) | 19 (0) | 16 (1) | 34 (1) | 0 | 0 | 13 (1) | |
| Bi27MC_S_vitro | Male | N/A | − | 5.8 (0.4) | 15 (1) | 12 (2) | 10 (1) | 0 | 0 | 11 (1) | |
| Bi28MC_S_vitro | Female | N/A | − | 6.7 (0.5) | 22 (3) | 13 (3) | 27 (2) | 0 | 0 | 14 (1) | |
Defined by the universal 16S rRNA gene sequences (see Fig. 3 for details).
The phenotypic characteristics examined were biofilm formation and crystal precipitation (see Fig. 4). MB, metallic biofilm; SC, salt crystal precipitation; N/A, not available (no biofilm formed and no crystal precipitated).
Halo zone formed around the bacterial colonies on nutrient agar plates (see Fig. 4). +, halo formed; −, no halo formed.
Hours per generation. A minimum of two replicates were conducted using freshly prepared initial culture inocula. N/A, the isolate was not available for assay.
Including the disc diameter of 6 mm. Antibiotics were tested at 1 mM concentrations. Abbreviations: Kan, kanamycin; Oxy, oxytetracycline; Tri, trimethoprim; Amp, ampicillin; Pen, penicillin; Chl, chloramphenicol. N/A, the isolate was not available for assay.
The fifth instar of the B. insularis individual was examined.
Sequencing of the16S rRNA and MLST genes.
Twenty-six of the 30 dissected-crypt-associated 16S rRNA amplicons (∼1.4 kb) examined produced chromatograms free of mixed reads within the target sequence. All 26 reads were identified as belonging to the genus Burkholderia. The remaining four reads (Bi15_R_vivo, Bi16_R_vivo, Bi18_S_vivo, and Bi22_S_vivo) were mixed and therefore were excluded from the phylogenetic analysis. Among the 22 pure cultures of inoculated crypt preparations, 2 (Bi14_R_vitro and Bi15_S_vitro) were contaminated during isolation and were discarded before 16S rRNA gene sequencing. The remaining 20 16S rRNA amplicons (∼1.4 kb) generated from the cultured bacteria produced clean chromatograms; all sequences were identified as Burkholderia isolates. Pairwise alignments of 16S rRNA gene sequences from 9 BiR and 9 BiS B. insularis individuals revealed that 15 ribotypes (8 BiR and 7 BiS individuals) derived from crypt DNA preparations were identical to the ribotypes derived from their cultured counterparts (see Table S2 in the supplemental material). The 16S rRNA amplicons from the other three crypt DNA preparations had 1 to 4% nucleotide differences from the amplicons generated from their respective cultured preparations (see Table S2). The SNPs identified in these pairwise alignments were localized in the hypervariable V1-to-V8 regions (see Fig. S1 in the supplemental material).
Phylogenetic analyses placed the 16S rRNA gene sequences of crypt genomic DNA preparations and those of their cultured bacterial counterparts within three major clades: the SBE clade (24), the plant-associated beneficial and environmental (PBE) clade (36), and the BCC clade (37) (Fig. 3). Forty-six sequences obtained from the BiR and BiS colonies failed to form distinct clades and were distributed throughout the phylogenetic tree. Fifteen of these 46 sequences (33%) were clustered in clade SBE (bootstrap value, 90%) with the environmental and insect gut-symbiotic Burkholderia isolates, some of which were capable of degrading pesticides. Nine sequences (20%) were grouped in clade PBE and were related to various Burkholderia plant-associated species—Burkholderia caribensis, Burkholderia tuberum, Burkholderia sabiae, and Burkholderia sacchari—as well to the gut-symbiotic Burkholderia sp. detected in another chinch bug species, C. saccharivorus (bootstrap value, 89%). The other 22 sequences (47.8%) were grouped in clade BCC with the pathogenic species Burkholderia gladioli and Burkholderia glumae, species in the B. cepacia complex, and the C. saccharivorus-associated symbiotic Burkholderia sp. (bootstrap value, 100%) (Fig. 3).
MLST gene (atpD, recA, lepA) amplicons from four dissected-crypt preparations (two from BiR and two from BiS) and eight Burkholderia isolates (four from BiR and four from BiS) produced chromatograms free of mixed reads and were identified as belonging to the genus Burkholderia. Generally, the phylogenetic relationships of the concatenated 1,233-bp MLST gene sequences agreed with the associations observed using the 16S rRNA gene sequences (see Fig. S2 in the supplemental material). The MLST gene sequences obtained from BiR and BiS isolates did not form distinct clades but were distributed throughout the phylogenetic tree.
BOX-PCR fingerprinting.
BOX-PCR gels exhibited similar patterns with DNA from the cultures and their crypt counterparts (see Fig. S3 in the supplemental material). The similarity analyses of BOX-PCR patterns revealed that among the 18 examined B. insularis individuals, the majority (83.3%) had 71 to 98% similarity between dissected crypts and their cultured Burkholderia counterparts, whereas the other individuals (16.7%) had 12 to 28% similarity (see Table S2 in the supplemental material). The BOX-PCR data agreed with the 16S rRNA gene sequence analysis. Specifically, the 15 individuals, exhibiting >70% similar BOX-PCR patterns between in vivo bacteria and in vitro cultures had 16S rRNA gene sequences (∼1.4 kb) of dissected crypts that were identical to those of their cultured counterparts. Similarly, the three dissected-crypt preparations that did not produce BOX-PCR patterns similar to those of their respective cultured isolates had 1 to 4% nucleotide differences in their 16S rRNA gene sequences (see Table S2).
PFGE.
Sixteen cultured Burkholderia isolates (eight BiR and eight BiS isolates) that were typed by PFGE yielded various patterns containing two to six bands, demonstrating the presence of multiple replicons in the B. insularis-associated Burkholderia isolates (see Fig. S4 in the supplemental material). Their cumulative genome sizes ranged from 6.6 to 8.7 Mb (Table 2). Four additional isolates (two BiR and two BiS isolates) produced smear patterns on PFGE gels. The addition of 50 μM thiourea to the gel electrophoresis buffer did not prevent the degradation pattern obtained with these four isolates (see Fig. S5 in the supplemental material). Three of the isolates producing smears were grouped in clade PBE with plant-associated beneficial and environmental Burkholderia spp. by ribotyping, whereas the other was in clade BCC (Table 2). In general, the six isolates with ribotypes that clustered in clade SBE had significantly more replicons (mean ± standard error [SE], 5.2 ± 0.3; t = 5.07; df = 13; P = 0.0002), but smaller genomes (7.1 ± 0.2 Mb; t = −2.20; df = 13; P = 0.0469), than the nine isolates with ribotypes that clustered in clade BCC (2.8 ± 0.3 replicons; genome size, 7.8 ± 0.2 Mb). There was no correlation between the PFGE profile and the source of the culture (BiR or BiS insects).
TABLE 2.
Estimated genome sizes of Blissus insularis-associated symbiont Burkholderia isolates detected by PFGE and of Burkholderia reference strains
| Cladea | Strain designation | No. of replicatesb | Mean (SE) size (Mb)c of: |
Total (SE) size (Mb) | |||||
|---|---|---|---|---|---|---|---|---|---|
| Replicon 1 | Replicon 2 | Replicon 3 | Replicon 4 | Replicon 5 | Replicon 6 | ||||
| SBE | Bi16MC_R_vitro | 2 | 3.4 (0) | 1.8 (0) | 1.6 (0) | 0.5 (0) | 7.3 (0) | ||
| Bi18MC_R_vitro | 2 | 3.0 (0.1) | 1.3 (0.1) | 1.2 (0) | 1.1 (0) | 0.5 (0) | 0.5 (0) | 7.5 (0.1) | |
| Bi22MC_R_vitro | 2 | 3.1 (0) | 1.5 (0) | 1.2 (0) | 1.0 (0) | 0.5 (0) | 0.5 (0) | 7.7 (0) | |
| Bi23MC_R_vitro | 2 | 3.0 (0) | 1.4 (0) | 1.0 (0) | 0.8 (0) | 0.5 (0) | 6.8 (0) | ||
| Bi12MC_S_vitro | 2 | 3.0 (0.1) | 1.4 (0.1) | 1.0 (0) | 0.7 (0) | 0.6 (0) | 6.7 (0.2) | ||
| Bi14MC_S_vitro | 2 | 3.1 (0) | 1.4 (0) | 1.0 (0) | 0.8 (0) | 0.6 (0) | 6.8 (0) | ||
| Burkholderia sp. strain RPE67 | 8.7d | ||||||||
| Burkholderia sp. strain YI23 | 8.9e | ||||||||
| PBE | Bi20MC_R_vitro | 3 | 3.5 (0) | 2.0 (0) | 1.3 (0) | 1.2 (0) | 8.0 (0) | ||
| Bi21MC_R_vitro | 5 | N/A | N/A | ||||||
| Bi16MC_S_vitro | 2 | N/A | N/A | ||||||
| Bi19MC_S_vitro | 3 | N/A | N/A | ||||||
| Burkholderia caribensis | 9.0f | ||||||||
| BCC | Bi17MC_R_vitro | 2 | 3.4 (0) | 2.8 (0) | 1.0 (0) | 7.2 (0) | |||
| Bi19MC_R_vitro | 3 | N/A | N/A | ||||||
| Bi24MC_R_vitro | 2 | 4.4 (0) | 3.6 (0.1) | 0.7 (0) | 8.7 (0.1) | ||||
| Bi25MC_R_vitro | 2 | 4.5 (0) | 3.8 (0) | 8.3 (0) | |||||
| Bi20MC_S_vitro | 2 | 3.7 (0.1) | 2.3 (0) | 0.7 (0) | 6.6 (0.1) | ||||
| Bi21MC_S_vitro | 2 | 4.4 (0.1) | 3.8 (0) | 8.2 (0.1) | |||||
| Bi22MC_S_vitro | 2 | 4.4 (0) | 3.7 (0) | 8.1 (0) | |||||
| Bi26MC_S_vitro | 2 | 4.4 (0) | 3.7 (0) | 8.1 (0) | |||||
| Bi27MC_S_vitro | 3 | 3.6 (0) | 2.6 (0) | 0.9 (0) | 0.4 (0) | 0.4 (0) | 7.9 (0) | ||
| Bi28MC_S_vitro | 2 | 3.6 (0) | 2.6 (0.1) | 1.0 (0) | 7.2 (0.1) | ||||
| Burkholderia gladioli | 9.1g | ||||||||
| Burkholderia cepacia | 6.4h | ||||||||
Defined by the universal 16S rRNA gene sequences (see Fig. 3 for details).
Number of replicates conducted using freshly prepared sample plugs on separate PFGE gels.
Replicons were separated on a 0.8% pulsed-field-certified agarose without restriction digestion. N/A, not available due to DNA degradation (a smear pattern) during PFGE (see Fig. S5 in the supplemental material).
Burkholderia sp. strain RPE67 has three chromosomes (3.1, 1.8, and 1.7 Mb) and three plasmids (1.4, 0.5, and 0.2 Mb). See reference 61.
Burkholderia sp. strain YI23 has three chromosomes (3.1, 1.8, and 1.6 Mb) and three plasmids (1.9, 0.4, and 0.1 Mb). See reference 65.
Burkholderia caribensis has two chromosomes (3.7 and 2.9 Mb) and two plasmids (2.0 and 0.4 Mb). See reference 66.
Burkholderia gladioli has two chromosomes (4.4 and 3.7 Mb) and four plasmids (0.4, 0.3, 0.1, and 0.1 Mb). See reference 67.
Data from the draft genome sequence of Burkholderia cepacia (68).
In vitro growth of Burkholderia isolates.
The calculated doubling time of 20 Burkholderia isolates ranged from 3.4 to 11.1 h per generation (Table 1). Although isolates assigned to clade PBE required approximately 1.3-fold-longer doubling times than isolates in clades SBE and BCC, no statistically significant difference was detected among isolates from the three clades (F = 1.73; df = 2; P = 0.2076). The growth rates of 10 BiR Burkholderia isolates (doubling time, 6.0 ± 0.8 h) were not significantly different (t = 0.14; df = 18; P = 0.8906) from those of 10 BiS Burkholderia isolates (6.1 ± 0.4 h), suggesting that there was no correlation between the doubling time and the source of the culture (BiR or BiS insects).
Antibiotic sensitivities of Burkholderia isolates.
Twenty cultured Burkholderia isolates were susceptible to kanamycin, oxytetracycline, and trimethoprim, with variable responses. These three antibiotics produced inhibition zones of 14 to 31 mm, 11 to 26 mm, and 10 to 34 mm, respectively (Table 1). Sixteen isolates also were inhibited by chloramphenicol (inhibition zones, 9 to 22 mm). The four isolates that were not inhibited by chloramphenicol belonged to clade BCC, based on the 16S rRNA sequence analysis. Overall, five isolates were susceptible to both ampicillin (inhibition zones, 19 to 34 mm) and penicillin (inhibition zones, 18 to 34 mm). Among these five, the ribotypes of four isolates clustered in clade PBE; the ribotype of the other isolate was in clade BCC. When the susceptibilities of cultured Burkholderia isolates to kanamycin (t = 0.09; df = 18; P = 0.9263), oxytetracycline (t = 1.47; df = 18; P = 0.1593), trimethoprim (t = 0.52; df = 12; P = 0.6122), and chloramphenicol (t = 1.47; df = 13; P = 0.0968) were compared, no significant difference was found between cultured Burkholderia isolates from BiR and BiS insects.
DISCUSSION
Although exocellular gut-symbiotic Burkholderia spp. have been isolated and cultured in vitro from true bugs in the superfamilies Lygaeoidea and Coreoidea (6, 11, 38), previous attempts to isolate Burkholderia spp. from B. insularis insects using both midgut crypt homogenates and intact crypts in various axenic media have failed (12). Similarly, with the conventional plating cultivation method, only 10% of the attempts made to culture the crypt-inhabiting Burkholderia sp. from another chinch bug species, Cavelerius saccharivorus, have been successful (10/100 dissected crypts [Y. Kikuchi, personal communication]). These findings suggest that the ability to culture insect-associated Burkholderia spp. differs depending on the host species. It should be noted that both B. insularis and C. saccharivorus are phloem-feeding insects in the family Blissidae and possess tubular crypts that harbor closely related Burkholderia spp., as determined on the basis of ribotyping (12, 24). Therefore, the difficulty of culturing Burkholderia spp. from both blissid species may be related to the in vivo environment within the crypts as a specialized symbiotic organ and/or to the physiological adaptations of these symbiotic bacteria to this specialized microhabitat.
In the current study, the crypt-inhabiting Burkholderia spp. were cultured successfully from 73% of B. insularis individuals (n = 30) by incubating the symbiont organ (midgut crypts) in insect cell culture medium (ICM). Importantly, ICM maintained the viability of crypts, as witnessed by peristaltic movement during the initial 72 h postdissection. During this period, crypt-inhabiting bacteria probably adapted, switching from a symbiotic to a free-living phenotype. Subsequent bacterial propagation led to detectable swelling and, eventually, lysis of the crypts. In addition to the laboratory-reared B. insularis insects examined here, 20 field-collected adults were subjected to the organ culture method; the majority of these attempts produced Burkholderia cultures (unpublished data). Both ribotyping and BOX-PCR fingerprinting confirmed that the majority of culturable isolates were identical to the predominant Burkholderia strains inhabiting crypts. Crypt preparations that did not produce cultures had 16S rRNA gene sequences that clustered with those of culturable Burkholderia isolates. These results implied that the bacteria inhabiting these crypt preparations did not have distinct ribotypes, suggesting that there is room for improvement of the culture method.
Previously, insect cell lines have been reported as substrates for the propagation of bacterial symbionts. For example, selected strains of the highly fastidious endocellular symbiont Wolbachia can be propagated in combination with insect cells (39, 40); however, when transferred to a cell-free medium, the bacteria remain viable but do not divide (41). Cultures of the endocellular symbiont Sodalis glossinidius were propagated by inoculating tsetse fly hemolymph into the insect cell line; bacteria adapted to the insect cell line were subsequently grown in an agar-based medium under a microaerobic atmosphere (42, 43). The secondary symbiont “Candidatus Arsenophonus arthropodicus” of the hippoboscid louse fly Pseudolynchia canariensis was propagated using this biphasic approach (44). The availability of in vitro cultures has provided a platform for examination of the mechanisms underlying insect-bacterium symbiosis (44–46).
In agreement with the previous study on B. insularis field populations (12), the 16S rRNA gene sequences of crypt-associated bacteria and their in vitro-produced counterparts isolated from two separate B. insularis laboratory colonies (BiR and BiS) revealed the consistent presence of multiple Burkholderia ribotypes, which clustered in three distinct clades (Fig. 3). It should be emphasized that within both laboratory colonies, an array of gut symbiont ribotypes existed and persisted over a multigenerational time frame. Significantly, 87% of B. insularis individuals examined harbored a single Burkholderia ribotype in their respective midgut crypts, suggesting a clonal association between Burkholderia spp. and B. insularis. The mechanism by which these insects acquire and maintain these exocellular symbionts in the crypt lumen is unknown.
Like B. insularis, C. saccharivorus also contains Burkholderia ribotypes that belong to the SBE, PBE, and BCC clades (24). The symbiotic Burkholderia spp. inhabiting various true bug species are represented by ribotypes belonging to the SBE clade (6, 19). The presence of this less-diverse ribotype complex may be associated with the nature of the symbiotic organ, the feeding preference, and/or the habitat of host insects. Specifically, in a legume seed-sucking bug species, Riptortus pedestris (Hemiptera: Coreoidea: Alydidae), symbiotic Burkholderia spp. populate the lumens of two-row midgut crypts (M4) (6, 19). In contrast, the chinch bugs, B. insularis and C. saccharivorus, have tubular crypts (12, 24) and are primarily phloem-sucking insects that feed on the monocots St. Augustinegrass (20) and sugarcane, Saccharum officinarum L. (47), respectively. In sugarcane soils, a diversity of sequences of the Burkholderia complex, including ribotypes within the SBE, PBE, and BCC clades, has been revealed using Illumina sequencing (48). Moreover, the abundance of soil-derived Burkholderia sequences (58%) in the SBE clade (48) correlates with the predominance of SBE ribotypes (68%) detected in C. saccharivorus crypts (24). These findings suggest that the gut symbionts of C. saccharivorus are acquired from the environment (i.e., soils and host plants) (24). Like C. saccharivorus (47), B. insularis feeds and oviposits on plant blade sheaths and basal shoots (22). Currently, no published information on the microbiome complex in St. Augustinegrass field soils is available, but preliminary examinations using diagnostic PCR amplification and 16S rRNA gene sequencing of St. Augustinegrass tissues have revealed the presence of Burkholderia spp. (unpublished data), suggesting their potential acquisition from plants by the phloem-feeding B. insularis.
In addition to the ribotype, biofilm production, phenotypes, PFGE profiles, growth rates, and antibiotic sensitivities for cultured Burkholderia spp. isolated from B. insularis crypts varied among isolates and did not correlate with the colony source (BiR or BiS). Examination of 20 cultured Burkholderia isolates revealed a correlation between the ribotype, phenotype, and physiological characteristics. Many (55%) of the dissected crypts and their respective Burkholderia cultures inoculated into ICM produced metallic biofilms. Based on ribotyping, these biofilm-producing Burkholderia isolates were distributed throughout the phylogenetic tree, but the majority of cultures (83%) were in the SBE and PBE clades. Biofilm formation has been reported in other Burkholderia species (Burkholderia pseudomallei and B. cepacia complex species), and it may be associated with many factors, such as nutrient content, osmolarity, oxygen, pH, and temperature (49–51). In selected microbe-invertebrate symbioses, biofilm formation contributes to the establishment and persistence of symbiotic bacteria in host symbiont organs (52–55). Biofilms produced by crypt-associated exocellular Burkholderia spp. are detected in the crypt lumens of R. pedestris as an exocellular matrix with polysaccharides (52), and they are associated with host insect fitness (56). However, the nature of the biofilm, and the potential role it plays, in the Burkholderia-Blissus interaction is unknown.
The genus Burkholderia contains diverse species that can be free-living in various environmental niches and/or can inhabit specific hosts (i.e., plants, fungi, or animals) (36, 57). One of the notable features of Burkholderia species is the presence of multiple chromosomes that contribute to the adaptability of these bacteria to different microhabitats (57–59). Based on PFGE assays, the genomes of crypt-inhabiting Burkholderia spp. ranged from 6.6 to 8.7 Mb, located in two to six replicons (Table 2). Four Burkholderia isolates (two from BiR and two from BiS) run with 50 μM thiourea (60) produced unresolvable DNA patterns. Of these four isolates, three belonged to the PBE clade. The large genomes with multiple replicons in the B. insularis-associated Burkholderia spp. were consistent with the genome sizes of other Burkholderia species (6 to 11 Mb) reported in the current genome database online (http://www.ncbi.nlm.nih.gov/genome/browse). Two symbiotic Burkholderia strains from R. pedestris contained a 7.0- to 8.7-Mb genome with three circular chromosomes and two to three plasmids (61, 62). Ribotypes of these two strains (RPE64 and RPE67) were grouped into a single clade, SBE (19), which contained six B. insularis-associated Burkholderia isolates (four from BiR and two from BiS) that had 6.7- to 7.7-Mb genomes with four to six replicons. In contrast to those in clade SBE, the Burkholderia isolates in clade BCC contained a minimum of two large replicons and had an average genome size of 7.8 Mb. Based on ribotyping, these isolates were closely related to pathogenic B. gladioli and B. cepacia complex species, which have genomes ranging from 6 to 9 Mb with two to three chromosomes (63). Overall, the estimated genome sizes and the numbers of replicons of gut-symbiotic Burkholderia isolates appeared to be correlated with the ribotype rather than with the colony (BiR or BiS) of the B. insularis host.
Independently of the host source (BiR or BiS), all of the Burkholderia isolates tested that were assigned to clade SBE and most isolates in clade BCC were resistant to ampicillin and penicillin, whereas the isolates in clade PBE were susceptible to both antibiotics. However, regardless of the ribotype and host source, gut-symbiotic Burkholderia spp. in B. insularis were susceptible to kanamycin, oxytetracycline, and trimethoprim. These antibiotics may allow us to produce axenic insects and to examine the function of Burkholderia in B. insularis in future studies. In vitro Burkholderia spp. isolated from B. insularis had doubling times (3.4 to 11.1 h) considerably longer than those of B. cepacia complex species (1.2 to 2.9 h) measured in a previous study (64). This difference may be due to differences in the nutritional content of media, temperature, and/or other components of the culturing technique. No correlation was found between the doubling times of the gut-symbiotic Burkholderia isolates tested in the current study and their ribotypes.
In summary, the organ culture method established in vitro cultures of a fastidious Burkholderia symbiont associated with the midgut crypts of B. insularis. The identities of the resulting Burkholderia cultures were confirmed using their genomic and physiological properties. The results demonstrated that B. insularis insects maintained, over multiple generations, a diversity of gut-symbiotic Burkholderia strains by harboring a single ribotype within an individual insect. Access to cultures of these crypt-inhabiting bacteria provides an opportunity to investigate the interaction between symbiotic Burkholderia spp. and the B. insularis host. Furthermore, the culturing method outlined here provides an alternative strategy for establishing in vitro cultures of other fastidious insect-associated bacterial symbionts.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the technical support provided by S. Shanker at the ICBR Sequencing Core (University of Florida). We also thank C. Verhoeven, J. McDaniel, N. Kaur, S. Sharma, and T. Henson (University of Florida) for technical assistance. D. Dye (Florida Pest Control and Chemical Company, FL) and J. McDaniel (SummerGlen Golf Club Community, FL) provided the sampling sites used in this research. Special thanks to A. Tartar (Nova Southeastern University, FL) for providing helpful insight on phylogenetic analyses.
This study was partially supported by the Insecticide Resistance Action Committee and the Steinmetz endowment (University of Florida).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00367-16.
REFERENCES
- 1.Engel P, Moran NA. 2013. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev 37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 2.Buchner P. 1965. Endosymbiosis of animals with plant microorganisms. John Wiley & Sons, New York, NY. [Google Scholar]
- 3.Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. 2010. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Appl Environ Microbiol 76:3486–3494. doi: 10.1128/AEM.00421-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kikuchi Y, Meng XY, Fukatsu T. 2005. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae). Appl Environ Microbiol 71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi Y, Hosokawa T, Nikoh N, Meng XY, Kamagata Y, Fukatsu T. 2009. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol 7:2. doi: 10.1186/1741-7007-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J 5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kikuchi Y. 2009. Endosymbiotic bacteria in insects: their diversity and culturability. Microbes Environ 24:195–204. doi: 10.1264/jsme2.ME09140S. [DOI] [PubMed] [Google Scholar]
- 8.Baines S. 1956. The role of the symbiotic bacteria in the nutrition of Rhodnius prolixus (Hemiptera). J Exp Biol 33:533–541. [Google Scholar]
- 9.Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol 43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 10.Salem H, Bauer E, Strauss AS, Vogel H, Marz M, Kaltenpoth M. 2014. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc Biol Sci 281:20141838. doi: 10.1098/rspb.2014.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kikuchi Y, Hosokawa T, Fukatsu T. 2007. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol 73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boucias DG, Garcia-Maruniak A, Cherry R, Lu H, Maruniak JE, Lietze VU. 2012. Detection and characterization of bacterial symbionts in the heteropteran, Blissus insularis. FEMS Microbiol Ecol 82:629–641. doi: 10.1111/j.1574-6941.2012.01433.x. [DOI] [PubMed] [Google Scholar]
- 13.Hosokawa T, Hironaka M, Inadomi K, Mukai H, Nikoh N, Fukatsu T. 2013. Diverse strategies for vertical symbiont transmission among subsocial stinkbugs. PLoS One 8:e65081. doi: 10.1371/journal.pone.0065081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosokawa T, Kikuchi Y, Shimada M, Fukatsu T. 2007. Obligate symbiont involved in pest status of host insect. Proc Biol Sci 274:1979–1984. doi: 10.1098/rspb.2007.0620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinheiro PV, Kliot A, Ghanim M, Cilia M. 2015. Is there a role for symbiotic bacteria in plant virus transmission by insects? Curr Opin Insect Sci 8:69–78. doi: 10.1016/j.cois.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Kliot A, Cilia M, Czosnek H, Ghanim M. 2014. Implication of the bacterial endosymbiont Rickettsia spp. in interactions of the whitefly Bemisia tabaci with tomato yellow leaf curl virus. J Virol 88:5652–5660. doi: 10.1128/JVI.00071-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dillon RJ, Vennard CT, Buckling A, Charnley AK. 2005. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett 8:1291–1298. doi: 10.1111/j.1461-0248.2005.00828.x. [DOI] [Google Scholar]
- 18.Koch H, Schmid-Hempel P. 2011. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. Proc Natl Acad Sci U S A 108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 2012. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci U S A 109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerr SH. 1966. Biology of the lawn chinch bug, Blissus insularis. Fla Entomol 49:9–18. doi: 10.2307/3493308. [DOI] [Google Scholar]
- 21.Reinert JA. 1972. Control of the Southern chinch bug, Blissus insularis, in South Florida. Fla Entomol 55:231–235. doi: 10.2307/3493371. [DOI] [Google Scholar]
- 22.Reinert JA, Kerr SH. 1973. Bionomics and control of lawn chinch bugs. Bull Entomol Soc Am 19:91–92. [Google Scholar]
- 23.Rangasamy M, Rathinasabapathi B, McAuslane HJ, Cherry RH, Nagata RT. 2009. Role of leaf sheath lignification and anatomy in resistance against Southern chinch bug (Hemiptera: Blissidae) in St. Augustinegrass. J Econ Entomol 102:432–439. doi: 10.1603/029.102.0156. [DOI] [PubMed] [Google Scholar]
- 24.Itoh H, Aita M, Nagayama A, Meng XY, Kamagata Y, Navarro R, Hori T, Ohgiya S, Kikuchi Y. 2014. Evidence of environmental and vertical transmission of Burkholderia symbionts in the oriental chinch bug, Cavelerius saccharivorus (Heteroptera: Blissidae). Appl Environ Microbiol 80:5974–5983. doi: 10.1128/AEM.01087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vázquez JC, Hoy MA, Royalty RN, Buss EA. 2010. A synchronous rearing method for Blissus insularis (Hemiptera: Blissidae). J Econ Entomol 103:726–734. doi: 10.1603/EC09254. [DOI] [PubMed] [Google Scholar]
- 26.Rand D, Heath A, Suderman T, Pierce N. 2000. Phylogeny and life history evolution of the genus Chrysoritis within the Aphnaeini (Lepidoptera: Lycaenidae), inferred from mitochondrial cytochrome oxidase I sequences. Mol Phylogenet Evol 17:85–96. doi: 10.1006/mpev.2000.0820. [DOI] [PubMed] [Google Scholar]
- 27.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sandström JP, Russell JA, White JP, Moran NA. 2001. Independent origins and horizontal transfer of bacterial symbionts of aphids. Mol Ecol 10:217–228. doi: 10.1046/j.1365-294X.2001.01189.x. [DOI] [PubMed] [Google Scholar]
- 29.Spilker T, Baldwin A, Bumford A, Dowson CG, Mahenthiralingam E, LiPuma JJ. 2009. Expanded multilocus sequence typing for Burkholderia species. J Clin Microbiol 47:2607–2610. doi: 10.1128/JCM.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, Dufayard JF, Guindon S, Lefort V, Lescot M, Claverie JM, Gascuel O. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stöver BC, Müller KF. 2010. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics 11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baldwin A, Mahenthiralingam E, Thickette KM, Honeybourne D, Maiden MCJ, Govan JR, Speert DP, LiPuma JJ, Vandamme P, Dowson CG. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J Clin Microbiol 43:4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koeuth T, Versalovic J, Lupski JR. 1995. Differential subsequence conservation of interspersed repetitive Streptococcus pneumoniae BOX elements in diverse bacteria. Genome Res 5:408–418. doi: 10.1101/gr.5.4.408. [DOI] [PubMed] [Google Scholar]
- 35.Hall BG, Acar H, Nandipati A, Barlow M. 2014. Growth rates made easy. Mol Biol Evol 31:232–238. doi: 10.1093/molbev/mst187. [DOI] [PubMed] [Google Scholar]
- 36.Suárez-Moreno ZR, Caballero-Mellado J, Coutinho BG, Mendonça-Previato L, James EK, Venturi V. 2012. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb Ecol 63:249–266. doi: 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- 37.Coenye T, Vandamme P, Govan JRW, Lipuma JJ. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J Clin Microbiol 39:3427–3436. doi: 10.1128/JCM.39.10.3427-3436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia JR, Laughton AM, Malik Z, Parker BJ, Trincot C, Chiang SSL, Chung E, Gerardo NM. 2014. Partner associations across sympatric broad-headed bug species and their environmentally acquired bacterial symbionts. Mol Ecol 23:1333–1347. doi: 10.1111/mec.12655. [DOI] [PubMed] [Google Scholar]
- 39.O'Neill SL, Pettigrew MM, Sinkins SP, Braig HR, Andreadis TG, Tesh RB. 1997. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol Biol 6:33–39. doi: 10.1046/j.1365-2583.1997.00157.x. [DOI] [PubMed] [Google Scholar]
- 40.Dobson SL, Marsland EJ, Veneti Z, Bourtzis K, O'Neill SL. 2002. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl Environ Microbiol 68:656–660. doi: 10.1128/AEM.68.2.656-660.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasgon JL, Gamston CE, Ren X. 2006. Survival of Wolbachia pipientis in cell-free medium. Appl Environ Microbiol 72:6934–6937. doi: 10.1128/AEM.01673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dale C, Maudlin I. 1999. Sodalis gen. nov. and Sodalis glossinidius sp. nov., a microaerophilic secondary endosymbiont of the tsetse fly Glossina morsitans morsitans. Int J Syst Bacteriol 49:267–275. doi: 10.1099/00207713-49-1-267. [DOI] [PubMed] [Google Scholar]
- 43.Welburn SC, Maudlin I. 1987. In vitro cultivation of rickettsia-like-organisms from Glossina spp. Ann Trop Med Parasitol 81:331–335. [DOI] [PubMed] [Google Scholar]
- 44.Dale C, Beeton M, Harbison C, Jones T, Pontes M. 2006. Isolation, pure culture, and characterization of “Candidatus Arsenophonus arthropodicus,” an intracellular secondary endosymbiont from the hippoboscid louse fly Pseudolynchia canariensis. Appl Environ Microbiol 72:2997–3004. doi: 10.1128/AEM.72.4.2997-3004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kikuchi Y, Fukatsu T. 2014. Live imaging of symbiosis: spatiotemporal infection dynamics of a GFP-labelled Burkholderia symbiont in the bean bug Riptortus pedestris. Mol Ecol 23:1445–1456. doi: 10.1111/mec.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohbayashi T, Takeshita K, Kitagawa W, Nikoh N, Koga R, Meng X-Y, Tago K, Hori T, Hayatsu M, Asano K, Kamagata Y, Lee BL, Fukatsu T, Kikuchi Y. 2015. Insect's intestinal organ for symbiont sorting. Proc Natl Acad Sci U S A 112:E5179–E5188. doi: 10.1073/pnas.1511454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murai M. 1975. Population studies of Cavelerius saccharivorus Okajima (Heteroptera: Lygaeidae): a few findings on population interchange. Res Popul Ecol 17:51–63. doi: 10.1007/BF02510877. [DOI] [Google Scholar]
- 48.Tago K, Itoh H, Kikuchi Y, Hori T, Sato Y, Nagayama A, Okubo T, Navarro R, Aoyagi T, Hayashi K, Hayatsu M. 2014. A fine-scale phylogenetic analysis of free-living Burkholderia species in sugarcane field soil. Microbes Environ 29:434–437. doi: 10.1264/jsme2.ME14122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Conway BAD, Venu V, Speert DP. 2002. Biofilm formation and acyl homoserine lactone production in the Burkholderia cepacia complex. J Bacteriol 184:5678–5685. doi: 10.1128/JB.184.20.5678-5685.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramli NSK, Eng Guan C, Nathan S, Vadivelu J. 2012. The effect of environmental conditions on biofilm formation of Burkholderia pseudomallei clinical isolates. PLoS One 7:e44104. doi: 10.1371/journal.pone.0044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.O'Toole GO, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu Rev Microbiol 54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 52.Kim JK, Kwon JY, Kim SK, Han SH, Won YJ, Lee JH, Kim CH, Fukatsu T, Lee BL. 2014. Purine biosynthesis, biofilm formation, and persistence of an insect-microbe gut symbiosis. Appl Environ Microbiol 80:4374–4382. doi: 10.1128/AEM.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator RscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–1600. doi: 10.1111/j.1365-2958.2006.05475.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ciche TA, Kim K, Kaufmann-Daszczuk B, Nguyen KCQ, Hall DH. 2008. Cell invasion and matricide during Photorhabdus luminescens transmission by Heterorhabditis bacteriophora nematodes. Appl Environ Microbiol 74:2275–2287. doi: 10.1128/AEM.02646-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.An R, Grewal PS. 2011. purL gene expression affects biofilm formation and symbiotic persistence of Photorhabdus temperata in the nematode Heterorhabditis bacteriophora. Microbiology 157:2595–2603. doi: 10.1099/mic.0.048959-0. [DOI] [PubMed] [Google Scholar]
- 56.Kim JK, Jang HA, Won YJ, Kikuchi Y, Han SH, Kim CH, Nikoh N, Fukatsu T, Lee BL. 2014. Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J 8:552–563. doi: 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Compant S, Nowak J, Coenye T, Clément C, Ait Barka E. 2008. Diversity and occurrence of Burkholderia spp. in the natural environment. FEMS Microbiol Rev 32:607–626. doi: 10.1111/j.1574-6976.2008.00113.x. [DOI] [PubMed] [Google Scholar]
- 58.Lessie TG, Hendrickson W, Manning BD, Devereux R. 1996. Genomic complexity and plasticity of Burkholderia cepacia. FEMS Microbiol Lett 144:117–128. doi: 10.1111/j.1574-6968.1996.tb08517.x. [DOI] [PubMed] [Google Scholar]
- 59.Kim HS, Schell MA, Yu Y, Ulrich RL, Sarria SH, Nierman WC, DeShazer D. 2005. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics 6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Römling U, Tümmler B. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J Clin Microbiol 38:464–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeshita K, Shibata TF, Nikoh N, Nishiyama T, Hasebe M, Fukatsu T, Shigenobu S, Kikuchi Y. 2014. Whole-genome sequence of Burkholderia sp. strain RPE67, a bacterial gut symbiont of the bean bug Riptortus pedestris. Genome Announc 2(3):e00556-14. doi: 10.1128/genomeA.00556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shibata TF, Maeda T, Nikoh N, Yamaguchi K, Oshima K, Hattori M, Nishiyama T, Hasebe M, Fukatsu T, Kikuchi Y, Shigenobu S. 2013. Complete genome sequence of Burkholderia sp. strain RPE64, bacterial symbiont of the bean bug Riptortus pedestris. Genome Announc 1(4):e00441-13. doi: 10.1128/genomeA.00441-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahenthiralingam E, Baldwin A, Dowson CG. 2008. Burkholderia cepacia complex bacteria: opportunistic pathogens with important natural biology. J Appl Microbiol 104:1539–1551. doi: 10.1111/j.1365-2672.2007.03706.x. [DOI] [PubMed] [Google Scholar]
- 64.Caraher E, Duff C, Mullen T, Mc Keon S, Murphy P, Callaghan M, McClean S. 2007. Invasion and biofilm formation of Burkholderia dolosa is comparable with Burkholderia cenocepacia and Burkholderia multivorans. J Cyst Fibros 6:49–56. doi: 10.1016/j.jcf.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 65.Lim JS, Choi BS, Choi AY, Kim KD, Kim DI, Choi IY, Ka JO. 2012. Complete genome sequence of the fenitrothion-degrading Burkholderia sp. strain YI23. J Bacteriol 194:896. doi: 10.1128/JB.06479-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan Y, Kong KF, Tsang JSH. 2016. Complete genome sequence of the exopolysaccharide-producing Burkholderia caribensis type strain MWAP64. Genome Announc 4(1):e01636-15. doi: 10.1128/genomeA.01636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Seo YS, Lim J, Choi BS, Kim H, Goo E, Lee B, Lim JS, Choi IY, Moon JS, Kim J, Hwang I. 2011. Complete genome sequence of Burkholderia gladioli BSR3. J Bacteriol 193:3149. doi: 10.1128/JB.00420-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Belcaid M, Kang Y, Tuanyok A, Hoang TT. 2015. Complete genome sequence of Burkholderia cepacia strain LO6. Genome Announc 3(3):e00587-15. doi: 10.1128/genomeA.00587-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.