ABSTRACT
Epoxyalkane:coenzyme M transferase (EaCoMT) plays a critical role in the aerobic biodegradation and assimilation of alkenes, including ethene, propene, and the toxic chloroethene vinyl chloride (VC). To improve our understanding of the diversity and distribution of EaCoMT genes in the environment, novel EaCoMT-specific terminal-restriction fragment length polymorphism (T-RFLP) and nested-PCR methods were developed and applied to groundwater samples from six different contaminated sites. T-RFLP analysis revealed 192 different EaCoMT T-RFs. Using clone libraries, we retrieved 139 EaCoMT gene sequences from these samples. Phylogenetic analysis revealed that a majority of the sequences (78.4%) grouped with EaCoMT genes found in VC- and ethene-assimilating Mycobacterium strains and Nocardioides sp. strain JS614. The four most-abundant T-RFs were also matched with EaCoMT clone sequences related to Mycobacterium and Nocardioides strains. The remaining EaCoMT sequences clustered within two emergent EaCoMT gene subgroups represented by sequences found in propene-assimilating Gordonia rubripertincta strain B-276 and Xanthobacter autotrophicus strain Py2. EaCoMT gene abundance was positively correlated with VC and ethene concentrations at the sites studied.
IMPORTANCE The EaCoMT gene plays a critical role in assimilation of short-chain alkenes, such as ethene, VC, and propene. An improved understanding of EaCoMT gene diversity and distribution is significant to the field of bioremediation in several ways. The expansion of the EaCoMT gene database and identification of incorrectly annotated EaCoMT genes currently in the database will facilitate improved design of environmental molecular diagnostic tools and high-throughput sequencing approaches for future bioremediation studies. Our results further suggest that potentially significant aerobic VC degraders in the environment are not well represented in pure culture. Future research should aim to isolate and characterize aerobic VC-degrading bacteria from these underrepresented groups.
INTRODUCTION
Short-chain alkenes (e.g., ethene, propene, and butenes) are common hydrocarbons in the environment, primarily encountered as fossil fuel components or products of living organisms or generated by the chemical industry (1). For instance, ethene is generated by both plants (2) and bacteria (3).
Chlorinated alkenes (e.g., vinyl chloride [VC]) are also naturally occurring, albeit at very low levels (4). VC is produced industrially as a monomer for polyvinyl chloride plastics. However, most environmental VC is generated by incomplete anaerobic dechlorination of the widely used solvents tetrachloroethene (PCE) and trichloroethene (TCE) in groundwater, where ethene can also be generated as a complete dechlorination product (1). PCE, TCE, and VC are common groundwater contaminants (5), and sites contaminated with chloroethenes are widely distributed across the United States (6) and elsewhere (1, 7). VC is of particular concern as a known human carcinogen (8).
In aerobic bacteria that utilize short-chain and chlorinated alkenes as carbon and energy sources, a monooxygenase enzyme typically catalyzes the initial attack (1, 9), forming aliphatic epoxides. Aliphatic epoxides are highly reactive molecules that covalently bind proteins and nucleic acids, leading to toxic and mutagenic effects (10, 11) in most organisms. Certain aerobic alkene-oxidizing bacteria metabolize and/or detoxify these epoxides by conjugation to coenzyme M (CoM) with the enzyme epoxyalkane:coenzyme M transferase (EaCoMT; encoded by the gene designated etnE in some organisms) (9, 12, 13).
EaCoMT has been implicated in aerobic assimilation of propene (12, 14), ethene, and VC (1, 13, 15). EaCoMT belongs to a subset of the alkyltransferase family, in which zinc catalyzes thiol activation for nucleophilic attack (14, 16). A functionally analogous transferase enzyme is the cobalamin-independent methionine synthase MetE, which transfers a methyl group to homocysteine during methionine synthesis (17). Both EaCoMT and MetE contain a conserved His-X-Cys-X-Cys zinc binding motif (13, 18, 19), which is important in thiol group transfer. However, with the exception of the EaCoMT from Xanthobacter sp. strain Py2 (Xanthobacter Py2) (9), MetE and EaCoMT do not share significant nucleotide sequence identity.
Bacteria expressing EaCoMT during growth on VC and/or ethene include strains of Mycobacterium (20–23), Nocardioides (15, 21), Pseudomonas (24), and Ochrobactrum (24). Propene-assimilating Gordonia rubripertincta strain B-276 (previously identified as Nocardia corallina and Rhodococcus rhodochrous strain B-276) (25, 26) and Xanthobacter Py2 (27) express homologous EaCoMT genes during growth on propene but can also grow on ethene. EaCoMT genes are known to be carried on linear plasmids in several VC-, ethene-assimilating bacteria as well as the propene-assimilating Gordonia sp. strain B-276 (Gordonia B-276) and Xanthobacter Py2 (15, 22, 28–30). A recently isolated ethene-assimilating Haliea strain (31) also contains a putative EaCoMT gene.
Some ethene-assimilating bacteria degrade VC fortuitously (i.e., via cometabolism [32, 33]), while others can use VC as a carbon and energy source (VC assimilators). Several ethene-assimilating isolates have successfully transitioned from cometabolic VC degradation to growth-coupled VC metabolism after an extended incubation period with VC (34, 35). Mutations in the EaCoMT gene were implicated in the evolution of an ethene-assimilating bacterium into a VC-assimilating bacterium in laboratory experiments (23).
The EaCoMT gene (often designated etnE) could be a useful biomarker for aerobic VC biodegradation in the field. Quantitative PCR (qPCR) assays for etnE have been developed (36–38) and applied (38–41) in an effort to understand VC biodegradation in microcosm studies as well as directly from environmental samples. However, because etnE sequences in ethene- and VC-assimilating bacteria are very similar, the EaCoMT gene qPCR assay cannot distinguish VC-assimilating bacteria from ethene-assimilating bacteria that fortuitously degrade VC.
Despite the importance of EaCoMT in global carbon and halogen cycling and as a diagnostic biomarker for VC bioremediation, very little is known about its distribution and diversity in the environment. Therefore, the primary goals of this study were to retrieve EaCoMT genes from environmental samples and expand the available database of EaCoMT gene sequences, examine EaCoMT diversity and distribution patterns at contaminated sites, and investigate relationships between EaCoMT gene abundance and diversity and VC concentrations and attenuation rates at contaminated sites. Because of the burgeoning use of the EaCoMT gene as a biomarker for VC biodegradation in the environment, we focused our efforts on geographically diverse chloroethene-contaminated groundwater samples.
MATERIALS AND METHODS
Site information, environmental sample collection, and DNA extraction.
Groundwater samples from six sites featuring various VC concentrations (Table 1) were collected in collaboration with consulting firms and U.S. agencies. The Kotzebue, AK, and Fairbanks, AK, sites were contaminated with TCE and cis-dichloroethene (i.e., potential VC precursors), but VC was not detected. The Carver, MA, Soldotna, AK, and Oceana Naval Air Station, VA, sites contained relatively dilute groundwater VC plumes (i.e., less than 100 μg/liter VC) at the time of sampling. Finally, the basalt site in Melbourne, Australia, contained relatively high concentrations of VC (up to 72 mg/liter).
TABLE 1.
Summary of groundwater sample information in this studya
| Location (abbreviation) | Well ID no. | Sample designation | Sampling date (mo/day/yr) | DNA concn (ng/μl) | VC concn (μg/liter) | Ethene concn (μg/liter) | DO concn (mg/liter) | kpoint (yr−1)b | kbulk (yr−1)b | EaCoMT gene abundance (genes/liter)c | Diversity (H′ and 1/D) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Carver, MA (CARV) |
RB46D | CARV46 | 9/15/2010 | 3.25 | 1.5 | NA | 0.65 | 0.365 (P < 0.001) | 0.159 (P = 0.001) | 1.2 × 105 | NA |
| RB63I | CARV63 | 9/15/2010 | 3.95 | 1.3 | NA | 0.29 | 0.183 (P < 0.001) | 0.159 (P = 0.001) | 1.9 × 104 | 0.73 ± 0.01 1.55 ± 0.02 | |
| RB46D | CARV46-1 | 9/29/2009 | NA | 1.8 | 3.9 | 0.31 | 0.365 (P < 0.001) | 0.119 (P = 0.001) | NA | NA | |
| RB64I | CARV64 | 9/29/2009 | NA | <0.46 | <0.1 | 0.45 | 0.1095 (P < 0.001) | 0.119 (P = 0.001) | NA | NA | |
| Soldotna, AK (SOLD) | MW6 | SOLD6 | 05/12/2009 | 1.00 | 9.6 | 50 | 0.53 | 0.621 (P < 0.001) | 0.992 (P = 0.050) | 6.3 × 105 | 1.05 ± 0.50 2.28 ± 0.94 |
| MW40 | SOLD40 | 09/22/2008 | 2.61 | 20.7 | 90 | 1.29 | 0.730 (P < 0.001) | 2.093 (P = 0.059) | 1.6 × 105 | 1.53 ± 0.15 2.64 ± 0.38 | |
| Oceana, VA (OCEA) | MW18 | OCEA18 | 08/06/2009 | 0.14 | 0.8 | <1 | 1.89 | 0.730 (P < 0.001) | 0.008 (P = 0.524) | 4.3 × 103 | 1.29 ± 0.22 3.48 ± 0.64 |
| MW25 | OCEA25 | 11/21/2008 | 8.96 | 19 | <1 | 1.20 | 0.256 (P = 0.132) | 0.003 (P = 0.379) | 2.4 × 104 | 1.54 ± 0.14 3.73 ± 0.38 | |
| Melbourne, Australia (AUS) | 039IJ-1 | AUS39-1 | 10/10/2011 | 1.24 | 72,000 | 780 | 0.07 | NA | 0.368 (P = 0.034) | 8.6 × 106 | 1.47 ± 0.00 3.93 ± 0.01 |
| 039IJ-3 | AUS39-3 | 10/10/2011 | 0.32 | 4,400 | NA | 0.28 | NA | 0.368 (P = 0.034) | 4.7 × 104 | 0.98 ± 0.07 2.12 ± 0.04 | |
| 039IJ-6 | AUS39-6 | 10/17/2011 | 1.00 | 53,000 | 230 | 1.07 | NA | 0.368 (P = 0.034) | 3.0 × 106 | 1.17 ± 0.03 2.45 ± 0.04 | |
| 039IJ-7 | AUS39-7 | 10/17/2011 | 0.78 | 15,000 | 40 | 2.2 | NA | 0.368 (P = 0.034) | 3.0 × 105 | 1.09 ± 0.03 2.41 ± 0.05 | |
| 039IJ-8 | AUS39-8 | 10/18/2011 | 2.28 | 24,000 | NA | 0.65 | NA | 0.368 (P = 0.034) | 1.3 × 106 | 1.08 ± 0.07 2.36 ± 0.08 | |
| Kotzebue, AK (KOTZ) | MW10-01 | KOTZ01 | 10/22/2013 | 10.4 | <0.62 | NA | 1.46 | NA | NA | 6.6 × 105 | 0.95 ± 0.09 2.11 ± 0.33 |
| MW10-03 | KOTZ03 | 10/22/2013 | 1.31 | <0.62 | NA | 8.70 | NA | NA | 9.8 × 104 | 0.91 ± 0.08 2.15 ± 0.05 | |
| Fairbanks, AK (FAIR) | MW-4 M | FAIR4 | 03/27/2014 | 15.7 | <0.4 | <0.06 | 1.36 | NA | NA | 7.0 × 103 | 1.39 ± 0.42 2.87 ± 1.13 |
| MW-13 M | FAIR13 | 03/27/2014 | 14.3 | <0.4 | <0.06 | 0.88 | NA | NA | 3.9 × 104 | 1.68 ± 0.15 3.54 ± 0.45 |
VC, ethene, and geochemical data were provided by personal communication and/or publicly available reports as follows: CARV, James Begley of MT Environmental Restoration and Sam Fogel of Bioremediation Consulting, Inc.; SOLD, May 2009 Groundwater Monitoring Report, River Terrace RV Park, Soldotna, AK, Alaska Department of Environmental Conservation via Tim McDougall of Oasis Environmental and James Fish of Alaska Department of Environmental Quality; OCEA, Long-term Monitoring Report (2009) for SMWUs 2B, 2C, and 2E, Oceana NAS, Virginia Beach, VA, via Laura Cook of CH2MHill; AUS, Dora Ogles from Microbial Insights, Inc.; KOTZ and FAIR, James Fish of Alaska Department of Environmental Conservation. NA, not analyzed.
kpoint and kbulk are estimated values of the point VC decay rate and bulk VC attenuation rate, respectively. Statistically significant k values are based on P values of <0.1.
Quantification of etnE genes at CARV, SOLD, and OCEA were published previously (36) and are provided here for reference and used in correlation analyses.
These VC-contaminated sites also feature conditions favorable for VC oxidation processes. Molecular oxygen and ethene were injected into the VC plume at the Carver site to promote VC oxidation, with apparent success (42). Reverse transcription-qPCR (RT-qPCR) evidence of VC oxidizer activity in Carver site groundwater has also been reported (38). Biostimulation of the VC plume with an oxygen-releasing compound was conducted at the Oceana site, where VC concentrations have been decreasing since 2004 (43). VC oxidation in aerobic microcosms constructed with sediments from the Soldotna site has been reported (44). Finally, evidence of aerobic VC degradation in the vadose zone was observed at the Australia site (40). Groundwater geochemical parameters collected at these sites (i.e., dissolved oxygen [DO], pH, temperature, and oxidation reduction potential [ORP]) as well as VC and ethene concentrations in monitoring wells at the time of sampling are provided (Table 1; see also Table S1 in the supplemental material).
Biomass for DNA extraction was collected by passing groundwater (1 to 3 liters) through Sterivex-GP 0.22-μm membrane filter cartridges (Millipore Corporation, Billerica, MA) in the field as described previously (36). Filters (with the exception of the Australia samples) were shipped overnight to the University of Iowa and stored at −80°C until extraction. Sterivex filter samples from Australia were handled by Microbial Insights, Inc. (Knoxville, TN).
Australia, Carver, Oceana, and Soldotna samples were extracted using the MoBio PowerSoil DNA isolation kit (MoBio, Carlsbad, CA) as described previously (36), while the Kotzebue and Fairbanks samples were extracted using the MoBio PowerWater Sterivex DNA isolation kit. Elution buffer volumes were 100 μl (Australia, Carver, Soldotna, Kotzebue), 40 μl (Oceana), or 50 μl (Fairbanks). DNA concentrations were estimated with a Qubit fluorometer (Invitrogen, Waltham, MA) using the Quant-iT double-stranded DNA (dsDNA) HS assay kit (Table 1).
Quantitative PCR.
Reaction mixtures for qPCR (25 μl) contained 12.5 μl of Power SYBR green PCR master mix (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA), 750 nM RTE primers (see Table S2 in the supplemental material), and 2 μl of DNA extract. Bovine serum albumin (0.5 μg) was added to alleviate possible PCR inhibition (45). All qPCRs were performed in at least triplicate for all samples with the ABI 7000 Sequence detection system and analyzed by ABI 7000 System SDS software (Applied Biosystems) using the “auto baseline” and “auto CT” (where CT is threshold cycle) functions. Standards were prepared using Nocardioides sp. strain JS614 genomic DNA as the template as described previously (36), except that a 0.2 μM concentration of each of CoMF1L and CoMR2E primers (see Table S2 in the supplemental material) was used to minimize the formation of primer dimers. Other detailed qPCR information (e.g., fluorescent threshold and efficiency) is provided (see Table S3 in the supplemental material) in accordance with MIQE guidelines (46). In dissociation curve analysis, etnE amplicons generated with RTE primers displayed melting temperatures of 84.7 to 85.3°C (JS614 standards), 82.8 to 84.1°C (Australia samples), 82.6 to 84.4°C (Kotzebue samples), and 84.7 to 85.6°C (Fairbanks samples).
PCR amplification of EaCoMT genes.
In general, conventional PCR amplification of EaCoMT genes from environmental DNA extracts did not yield visible PCR products on agarose gels for every site. Therefore, we investigated successive rounds of conventional PCR, touchdown (TD) PCR, and nested-PCR approaches (see Table S4 in the supplemental material). The nested-PCR modification, which effectively amplified EaCoMT genes from all environmental samples, was performed as follows. The first round of PCR utilized the CoMF1L and CoMR2E primer set (0.2 μM each; see Table S2 in the supplemental material) as described previously (36) and 1 μl of DNA extract (containing 0.14 to 15.7 ng template) (Table 1). A subsequent round of nested PCR was performed with F131 and R562 primers (0.2 μM each; see Table S2 in the supplemental material) (47) and 2 μl of the initial reaction mixture. The nested-PCR thermocycler program consisted of an amplification phase (30 cycles of 94°C for 20 s, 60°C for 45 s, 72°C for 30 s) and a final extension (72°C for 15 min). Negative controls for the nested PCR, which used 2 μl of negative-control reaction mixtures used in the first round of PCR, showed no amplification.
Amplification of EaCoMT genes from Carver DNA extracts required a touchdown phase during the first round of PCR. The thermocycling program consisted of an initial denaturation step (94°C, 5 min) followed by a touchdown phase (20 cycles of 94°C for 30 s, 65°C for 45 s [0.5°C decrease of each cycle], and 72°C for 1 min), a general amplification phase (10 cycles of 94°C for 30 s, 55°C for 45 s, and 72°C for 1 min), and a final extension (72°C for 15 min). Nested PCR was then performed as described above.
The potential that nested and touchdown PCR could introduce lower apparent gene diversity was investigated by constructing a pooled sample of DNA (referred to as AUS39) extracted from five different monitoring wells at the Australia site (i.e., samples 39-1, 39-3, 39-6, 39-7, and 39-8). DNA from AUS39 was amplified with three methods: conventional PCR, nested PCR, and a combined touchdown PCR-nested PCR. The amplicons generated were subjected to T-RFLP analysis, and diversity indices were calculated to compare these three methods.
Only nested-PCR results were used for comparative analyses between sites. However, when constructing clone libraries, both conventional-PCR and nested-PCR amplicons from Australian samples were used. We also performed combined touchdown-nested PCR on three additional samples (Soldotna MW6, Oceana MW18, and Oceana MW25) (see Table S4 in the supplemental material) for PCR bias comparisons in clone libraries.
Terminal restriction fragment length polymorphism (T-RFLP) analysis.
The restriction enzyme AcoI (EaeI) (New England BioLabs, Inc., Ipswich, MA) was selected for EaCoMT T-RFLP analysis using the default settings of the software program REPK (48). This restriction enzyme maximized differentiation between EaCoMT sequences, based on analysis of a database populated with full-length EaCoMT sequences from VC- and ethene-assimilating isolates deposited in GenBank (see Table S5 in the supplemental material).
All samples used in T-RFLP analysis were subjected to nested PCR with fluorescently labeled 6-carboxyfluorescein (6-FAM) F131 and unlabeled R562 primers. Digestions were performed in duplicate. PCR products (12 μl) were digested with AcoI (EaeI) and precipitated with glycogen, sodium acetate, and ethanol in accordance with the manufacturer's protocol. The resulting terminal restriction fragments (T-RFs) were analyzed with an Applied Biosystems 3730 DNA analyzer with GeneScan 500 LIZ size standards at the Iowa Institute of Human Genetics at the University of Iowa. T-RF sizes were estimated using Peak Scanner software (Applied Biosystems).
T-RFLP data analysis.
T-RFLP data were further processed with T-REX software (49) using peak area to evaluate T-RF abundance, filter background noise, and round fragment sizes to the nearest whole number. Fragment sizes of <48 bp and >453 bp were excluded from further analysis. The final data matrix contained 28 samples (rows, duplicated for each well) from 14 groundwater sampling wells at six sites and 192 unique EaCoMT T-RFs (columns).
A T-RFLP profile clustered heatmap was generated using the gplots, vegan, and RColorBrewer packages in R (50). Briefly, this was achieved by calculating Bray-Curtis dissimilarities and using these values to execute the “-hclust” command in R, which performs the average-linkage hierarchical clustering method. T-RFs with relative abundance of <1% in at least one sample were removed from the heatmap to better visualize the results.
T-RFLP profiles were also analyzed by nonmetric multidimensional scaling (NMDS) in R with the vegan package (51), standardized with Wisconsin smoothing. Bray-Curtis dissimilarities were calculated with a random starting configuration, and a two-dimensional solution was reached. The final stress was 0.1402. T-RFLP profile composition differences between all samples were evaluated by the multiresponse permutation procedure (MRPP) using the vegan package in R, based on Euclidean dissimilarity and 999 permutations as the default. The chance-corrected within-group (i.e., each contaminated site) agreement was 0.5109, indicating that samples were homogenous within groups. The P value was 0.001, indicating that the differences in EaCoMT gene T-RFLP profiles among groups were statistically significant.
The Shannon-Wiener index (H′) and inverse Simpson index (1/D) (52) were calculated with the vegan package in R for each sample using the T-RFLP results. The relative abundance of each T-RF was treated as the “number of species” in the analysis. Australian composite sample T-RFs amplified with conventional PCR and combined touchdown-nested PCR were also analyzed with diversity indices, with fragment sizes of <48 bp and >453 bp excluded.
VC attenuation rate estimation and correlation analysis.
Groundwater VC attenuation rates (i.e., the point decay rate, kpoint, for each well and the bulk attenuation rate, kbulk, over a plume transect) were calculated in accordance with a U.S. EPA protocol (54), which is used under the assumption that VC attenuation follows a pseudo-first-order rate law (54) (for examples, see Fig. S3 in the supplemental material). Linear regressions of ln VC concentration versus time and ln VC concentration versus distance were performed in order to estimate the attenuation rate. The estimated rates were not corrected for dilution, dispersion, or sorption. Attenuation rates with a P value of <0.1 (two tailed) were considered statistically significant and used for further analysis. Groundwater flow rates, VC concentrations, and site maps used for the calculation were obtained as described in Table 1.
Linear regressions and Spearman's correlations were used to analyze the relationship between the abundance and diversity of EaCoMT genes and groundwater parameters (i.e., VC and ethene concentration, DO, temperature, pH, and ORP) as well as estimated VC attenuation rates (kpoint and kbulk) (see Table 3). The potential significance of each relationship was established based on a P value of <0.05 (two tailed). Wells or sites without adequate information for rate estimation and correlation analysis were excluded from this analysis.
TABLE 3.
Correlation analysis between geochemical parameters with EaCoMT gene abundance and diversity
| Parameter | Correlation with EaCoMT gene abundance (no. of genes/liter of GW) |
Correlation with etnE diversity |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Linear regression |
Spearman |
n | Index | Linear regression |
Spearman |
|||||||
| Slope | R2 | P value | Rho | P value | Slope | R2 | P value | Rho | P value | ||||
| Vinyl chloride concn (μg/liter) | 15 | 9.36 × 101 | 0.851 | <0.001 | 0.699 | 0.005 | 14 | H′ | 1.69 × 10−6 | 0.019 | 0.641 | 0.107 | 0.715 |
| 1/D | 9.19 × 10−6 | 0.086 | 0.309 | 0.189 | 0.514 | ||||||||
| Ethene concn (μg/liter) | 11 | 1.12 × 104 | 0.987 | <0.001 | 0.859 | 0.001 | 9 | H′ | 6.60 × 10−5 | 0.006 | 0.850 | −0.319 | 0.386 |
| 1/D | 9.00 × 10−4 | 0.130 | 0.341 | −0.151 | 0.682 | ||||||||
| Bulk VC attenuation rate (yr−1) | 11 | −2.57 × 105 | 0.004 | 0.853 | 0.636 | 0.039 | 10 | H′ | 9.59 × 10−2 | 0.058 | 0.503 | −0.110 | 0.636 |
| 1/D | −2.32 × 10−1 | 0.040 | 0.580 | −0.200 | 0.465 | ||||||||
| Point VC decay rate (yr−1) | 6 | 3.33 × 105 | 0.134 | 0.475 | 0.203 | 0.700 | 5 | H′ | 5.06 × 10−1 | 0.152 | 0.516 | 0.359 | 0.567 |
| 1/D | 8.17 × 10−2 | 0.059 | 0.693 | 0.359 | 0.567 | ||||||||
| DO concn (mg/liter) | 14 | −1.46 × 106 | 0.158 | 0.160 | −0.257 | 0.374 | 13 | H′ | 7.20 × 10−2 | 0.027 | 0.590 | 0.170 | 0.579 |
| 1/D | 1.24 × 10−1 | 0.012 | 0.721 | 0.104 | 0.737 | ||||||||
| ORP (mV) | 9 | −8.55 × 102 | 0.087 | 0.441 | −0.350 | 0.359 | 8 | H′ | −2.10 × 10−3 | 0.167 | 0.315 | −0.262 | 0.536 |
| 1/D | −5.90 × 10−3 | 0.250 | 0.207 | −0.286 | 0.501 | ||||||||
| pH | 9 | −4.98 × 104 | 0.024 | 0.693 | −0.250 | 0.521 | 8 | H′ | 8.73 × 10−2 | 0.044 | 0.619 | 0.238 | 0.582 |
| 1/D | 3.94 × 10−1 | 0.163 | 0.321 | 0.452 | 0.268 | ||||||||
| Temp (°C) | 9 | −2.11 × 104 | 0.367 | 0.084 | −0.683 | 0.050 | 8 | H′ | 3.25 × 10−3 | 0.006 | 0.852 | 0.167 | 0.703 |
| 1/D | 4.04 × 10−2 | 0.177 | 0.300 | 0.405 | 0.327 | ||||||||
a Bold values indicate relationships that are significant (two-tailed P value < 0.05); n represents the number of wells available for the analysis. GW, groundwater.
Cloning and sequencing.
Purifed PCR products amplified from environmental samples with both conventional PCR and nested PCR were ligated overnight at 4°C into the pCR2.1 vector (1:1 molar insert to vector ratio) using the Original TA Cloning kit (Invitrogen). Ligations were transformed into One Shot TOP10 chemically competent Escherichia coli. Transformants were analyzed according to the cloning kit instructions. Plasmids were extracted using the QIAprep Spin Miniprep kit. Clones were PCR screened with M13F and M13R primers (see Table S2 in the supplemental material), and those with the appropriately sized inserts were Sanger sequenced at the Iowa Institute of Human Genetics with the M13F and/or M13R primers.
Sequence analyses.
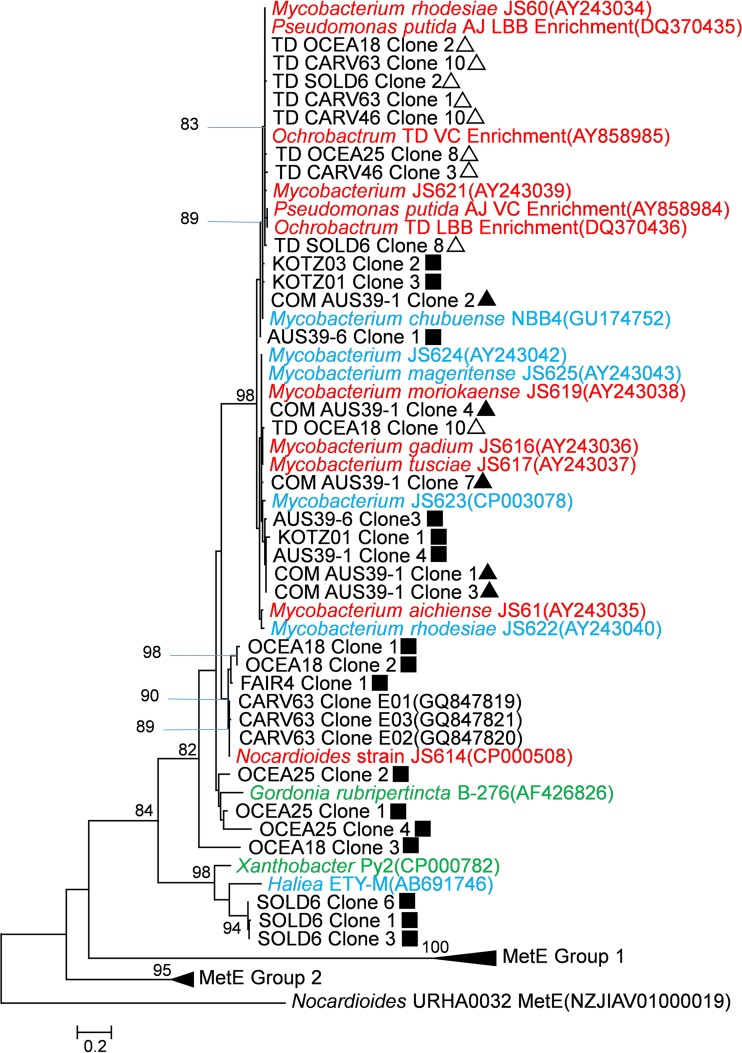
Sequences with <99% identity to each other from the same sample were included in an amino acid sequence analysis as representatives. Deduced EtnE amino acid sequences (adjusted with ORF finder [55]) from partially sequenced clones were aligned with deduced EtnE (see Table S6 in the supplemental material) and MetE (see Table S7 in the supplemental material) sequences from GenBank using ClustalW (56) and trimmed to 166 amino acids (aa) (including gaps). MetE genes were included in the phylogenetic analysis to account for potential homology with EtnE. Phylogenetic trees were generated with MEGA5 using the maximum likelihood method (57). The Nocardioides sp. strain URHA0032 MetE gene (GenBank accession no. WP_028637114) was used as the outgroup. A nucleotide phylogenetic tree was also constructed with all 139 clone sequences obtained in this study plus existing EaCoMT gene sequences from GenBank (see Fig. S1 in the supplemental material). Nucleotide sequences were processed as described above for amino acid analysis, except that Pseudomonas putida metE (58) was used as the outgroup. The results were visualized with EvolView (59).
Nucleotide sequence accession numbers.
Representative unique sequences included in the phylogenetic trees (see Fig. 2; see also Fig. S1 in the supplemental material) were deposited in GenBank under accession numbers KR936138 to KR936167 (see Table S8).
FIG 2.
A phylogenetic tree depicting the relationship of deduced EtnE amino acid sequences from environmental samples, enrichment cultures, and isolates (see Table S6 in the supplemental material), along with MetE sequences from related strains (see Table S7 in the supplemental material). Isolates are color coded as follows: red, VC assimilators; blue, etheneotrophs; green, propene oxidizers. Carver sequences from GenBank were from ethene enrichment cultures (60). Environmental samples are identified as described in Table 1. The symbols refer to the PCR amplification method used: ▲, conventional PCR; ■, nested PCR; △, nested PCR with a touchdown modification. Amino acid sequences were deduced from partial EaCoMT sequences (excluding gaps). An alignment of 166 aa (including gaps) was generated in ClustalW (56), and the tree was constructed and visualized in MEGA5 using the maximum likelihood method (57) with the Nocardioides sp. strain URHA0032 MetE gene as the outgroup. The bar represents a 20% sequence difference. For environmental samples, only sequences of <99% identity with other sequences from the same samples were included on the tree. Refer to Fig. S1 in the supplemental material for the complete nucleotide phylogenetic tree generated with all 139 sequences obtained in this study. See Table S8 in the supplemental material for GenBank accession numbers of clone sequences presented in this study.
RESULTS
T-RFLP analysis of EaCoMT gene diversity in environmental samples.
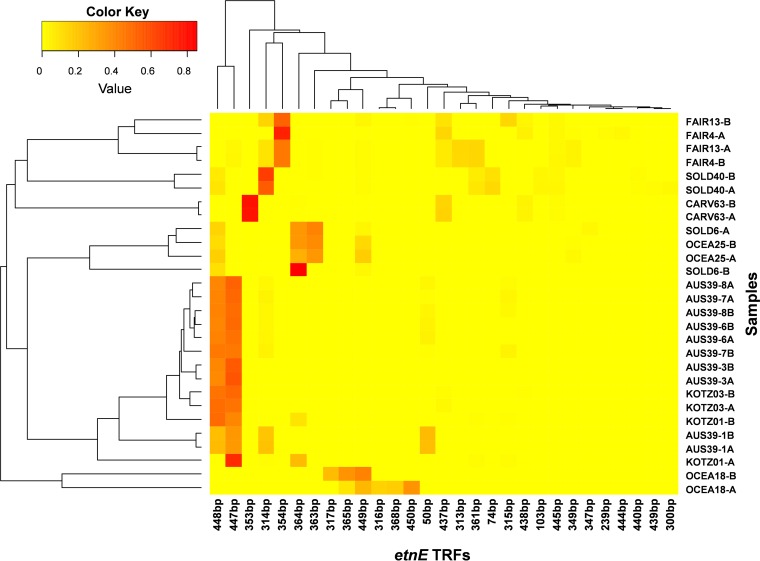
A heatmap (Fig. 1) displays EaCoMT gene T-RFLP profile patterns and clustering among different sites and monitoring wells. The heatmap reveals that several of the longer T-RFs were prominent among all groundwater samples, and some of them (e.g., the 314-bp, 354-bp, 364-bp, and 448-bp T-RFs) matched T-RFs of EaCoMT clone sequences recovered from environmental samples (Table 2). However, none of these abundant T-RFs matched predicted T-RFs from in silico digestion of EaCoMT gene sequences previously deposited in GenBank (see Table S5 in the supplemental material).
FIG 1.
Clustered heatmap of T-RFLP profiles generated by AcoI (EaeI) restriction-digested partial EaCoMT genes. For clarity, T-RFs with relative abundance of <1% were excluded from the graph. The higher the relative abundance of a particular T-RF in the sample, the warmer the coloring. Sample identifiers are formatted by site (using the first four characters of the name of each site), well number, and replicate (A or B). Please see Table 1 for specific site names.
TABLE 2.
In vitro and in silico AcoI (EaeI) digestion of selected EaCoMT gene clones (those having <99% identity with other clone sequences retrieved from the same sample) matched with similar EaCoMT gene sequences from isolated strainsa
| Site | Well ID no. | Clone ID(s) | Observed T-RF (bp) | Predicted T-RF (bp) | Organism with closest BLAST hit (% identity) |
|---|---|---|---|---|---|
| Oceana NAS, VA | MW18 | TD OCEA18 clone 10 | ND | 24 | Mycobacterium tusciae strain JS617 (99) |
| Australia | 039IJ-1 | COM AUS39-1 clones 4 and 7 | 50 | 48 | Mycobacterium gadium strain JS616 (95) |
| Australia | 039IJ-6 | AUS39-6 clone 1 | 50 | 48 | Mycobacterium chubuense NBB4 (92) |
| Oceana NAS, VA | MW25 | OCEA25 clone 2 | 73 | 73 | Nocardioides sp. JS614 (83) |
| Oceana NAS, VA | MW25 | OCEA25 clone 4 | 104 | 103 | Nocardioides sp. JS614 (83) |
| Oceana NAS, VA | MW18 | OCEA18 clones 1 and 2 | 104 | 103 | Nocardioides sp. JS614 (87) |
| Oceana NAS, VA | MW25 | OCEA25 clone 1 | 313 | 312 | Nocardioides sp. JS614 (85) |
| Oceana NAS, VA | MW18 | OCEA18 clone 3 | 314 | 318 | Nocardioides sp. JS614 (76) |
| Kotzebue, AK | 10-01 | KOTZ01 clone 1 | 314 | 312 | Mycobacterium sp. JS624 (92) |
| Australia | 039IJ-1 | COM AUS39-1 clones 1 and 3 | 314 | 312 | Mycobacterium smegmatis JS623 (94) |
| Australia | 039IJ-1 | AUS39-1 clone 4, AUS39-6 clone 3 | 314 | 312 | Mycobacterium sp. JS624 (93–94) |
| Fairbanks, AK | MW-4 M | FAIR4 clone 2 | 354 | 356 | Nocardioides sp. JS614 (98–99) |
| Fairbanks, AK | MW-13 M | FAIR13 clone 3 | |||
| Carver, MA | RB46D | TD CARV46 clones 3 and 10 | 364 | 363 | Mycobacterium rhodesiae strain JS60 (99–100) |
| RB63I | TD CARV 63 clones 1 and 10 | ||||
| Oceana NAS, VA | MW18 | OCEA18 clone 2 | |||
| MW25 | OCEA25 clone 8 | ||||
| Soldotna, AK | MW6 | SOLD6 clones 2 and 8 | |||
| Australia | 039IJ-1 | COM AUS39-1 clone 2 | 448 | 453b | Mycobacterium chubuense NBB4 (99) |
| Kotzebue, AK | 10-01 | KOTZ01 clone 3 | |||
| 10-03 | KOTZ03 clone 2 | ||||
| Soldotna, AK | MW6 | SOLD6 clones 1, 3, and 6 | 440 | 447b | Haliea sp. ETY-M (76–79) |
| MW40 | SOLD40 clone 1 |
The maximum T-RF size is 453 bp, as PCR was performed with F131/R562 primers. ND, not determined.
No predicted AcoI (EaeI) restriction site.
An NMDS analysis was also performed on the T-RFLP profiles (see Fig. S2 in the supplemental material). The NMDS analysis showed that EaCoMT genes from geographically distinct areas in some cases were similar (e.g., Australia and Kotzebue samples) but that EaCoMT genes recovered from geographically close locations (e.g., Alaska sites) did not necessarily cluster together. However, EaCoMT genes from Soldotna, AK, well MW40 did group closely with EaCoMT genes from Fairbanks, AK.
Phylogenetic analysis of EaCoMT gene diversity in environmental samples.
A total of 139 sequences, 121 of which were unique (i.e., contained at least a 1-bp difference from other sequences), were retrieved from clone libraries (see Table S4 in the supplemental material). The nucleotide sequence identity of each clone to previously documented EaCoMT gene sequences found in isolates varied from 76% to 99%. A nucleotide phylogenetic tree depicting the complete current EaCoMT gene database (see Fig. S1 in the supplemental material) revealed four potential EaCoMT gene subgroups (named according to the genus name of the first cultured representative in that group): Mycobacterium, Nocardioides, Gordonia, and Xanthobacter. Of the 121 unique EaCoMT sequences, 82 were related to EaCoMT genes found in isolated VC- and ethene-assimilating Mycobacterium strains (i.e., the Mycobacterium group). Notably, 11 of 12 EaCoMT sequences retrieved from the Fairbanks samples contained a 7-bp deletion at the 318-bp location of the 447-bp F131/R562 PCR product. These Fairbanks sequences were 98 to 99% identical to the etnE allele in Nocardioides sp. strain JS614 designated etnE1 (15). The remaining EaCoMT sequence from Fairbanks was 95% identical to that of Nocardioides sp. strain JS614 etnE. All EaCoMT gene sequences from Soldotna MW40 contained a 1-bp deletion at the 12-bp location of the 447-bp PCR product. These Soldotna MW40 sequences were 76 to 78% identical to the putative EaCoMT gene sequence from the ethene-assimilating Hailea sp. ETY-M.
The maximum likelihood amino acid phylogenetic tree (Fig. 2), constructed with 30 representative EaCoMT gene sequences (<99% identity with other clones from the same well) that did not contain frameshift deletions or internal stop codons and 19 EaCoMT sequences from isolated strains, was consistent with the nucleotide phylogenetic tree. Of these 30 sequences, 40% formed a phylogenetic subgroup with etnE found in Mycobacterium strains and 6% grouped with the etnE gene from Nocardioides sp. strain JS614.
Relationships between EaCoMT gene diversity, EaCoMT gene abundance, and contaminated-site conditions.
Using the T-RFLP data, we quantified EaCoMT gene diversity in each groundwater sample by calculating Shannon-Wiener (H′) and inverse Simpson (1/D) diversity indices (Table 1). When considering T-RFs amplified with nested PCR only, the Soldotna, Carver, and Kotzebue samples showed less EaCoMT gene diversity than did other sites, with Carver well RB63I displaying the lowest EaCoMT gene diversity (H′ = 0.73 ± 0.01; 1/D = 1.55 ± 0.02). Interestingly, the highest diversity was observed in Australia well 39-1 (H′ = 1.47 ± 0.00; 1/D = 3.93 ± 0.01), which also contained the highest VC concentration (72 mg/liter) of all the wells investigated in this study.
Quantification of EaCoMT gene abundance with qPCR confirmed the presence of EaCoMT genes in all samples included in this study (Table 1), ranging from 103 to 106 genes/liter of groundwater. The highest EaCoMT gene abundances (4.7 × 104 to 8.7 × 106 genes/liter of groundwater) were observed in the Australian samples, which were collected from a groundwater plume with high VC concentrations.
Correlation analysis (Table 3) showed little evidence that the EaCoMT gene diversity has any significant relationship with VC attenuation rates, dissolved oxygen (DO), or other geochemical parameters. However, there were significant positive associations between VC concentration and EaCoMT gene abundance (see Fig. S4 in the supplemental material), both quantitatively (linear correlation, P < 0.001) and qualitatively (Spearman's rank correlation, P < 0.005). There was also a significant linear correlation between ethene concentration and EaCoMT gene abundance (P < 0.001) (see Fig. S4 in the supplemental material). Although there was no significant linear relationship between the bulk VC attenuation rate and EaCoMT gene abundance, we did notice a significant rank correlation between these two variables (Spearman's correlation, P = 0.039).
PCR modifications affect EaCoMT gene diversity estimates.
Because we could amplify EaCoMT genes from the Australia samples with conventional PCR, we used the composite Australia DNA sample (AUS39) to assess potential bias introduced by nested-PCR and touchdown PCR modifications. The AUS39 sample, when amplified directly with COM primers, showed greater EaCoMT gene diversity (H′ = 2.04 ± 0.00; 1/D = 5.36 ± 0.04) than when it was amplified by nested PCR (H′ = 1.21 ± 0.41; 1/D = 2.70 ± 1.02) (see Fig. S5 in the supplemental material). The touchdown-nested-PCR-amplified AUS39 sample showed less EaCoMT gene diversity (H′ = 0.47 ± 0.09; 1/D = 1.41 ± 0.13) (see Fig. S5 in the supplemental material) than did the sample amplified by nested PCR alone.
To further assess the potential bias introduced by touchdown PCR, clone libraries were constructed with the touchdown-nested-PCR-amplified SOLD6, OCEA18, and OCEA25 samples (see Table S4 in the supplemental material). A total of 19 clones were sequenced from these clone libraries, and representative sequences were included in the phylogenetic analysis (Fig. 2; see also Fig. S1 in the supplemental material). All EaCoMT sequences retrieved by the touchdown modification grouped with those found in Mycobacterium isolates.
DISCUSSION
This study has expanded our view of EaCoMT gene diversity. Previously, our understanding of EaCoMT gene diversity was based on 16 sequences from VC-, ethene-, and propene-assimilating isolates and a few EaCoMT genes sequenced from VC and ethene enrichment cultures (41, 60). T-RFLP analysis has now revealed 192 different EaCoMT T-RFs. The 139 partial EaCoMT genes sequenced from groundwater samples were 76 to 99% identical to EaCoMT sequences found in isolates. These observations suggest that we have uncovered several novel EaCoMT sequences.
EaCoMT genes were present at all six sites surveyed in this study, even at sites where VC and ethene were not detected (Kotzebue and Fairbanks). The frequent occurrence of EaCoMT sequences (i.e., found in 112 metagenomes from 37 separate and geographically diverse sites; see Table S9 in the supplemental material) in the MG-RAST metagenomics database (61) further supports the notion that EaCoMT genes are widespread and globally distributed among a variety of environments. Most of the EaCoMT sequences in these metagenomes were similar to those of EaCoMT genes found in Mycobacterium and Nocardioides strains (see Table S9 in the supplemental material).
VC- and ethene-assimilating strains isolated to date are primarily members of the genus Mycobacterium (21), and the majority of environmental EaCoMT sequences recovered so far are similar to those found in Mycobacterium strains. This suggests that Mycobacterium strains are significant contributors to EaCoMT gene diversity in the environment. However, as the EaCoMT genes found in mycobacteria are known to be plasmid borne, it is possible that they are transferred to or have originated from other bacteria. Observing EaCoMT genes in VC-assimilating Pseudomonas and Ochrobactrum strains grouping with those in mycobacteria supports this hypothesis (Fig. 2).
The emerging clade of Nocardioides sp. JS614-like EaCoMT genes in the environment is notable. Although Nocardioides sp. JS614 is the sole isolated VC- and ethene-assimilating representative of the Nocardioides genus (21), a related Nocardioides strain was implicated as a dominant microbial community member of a VC-degrading enrichment culture likely using VC as a carbon and energy source (41). Taken together, these data suggest that Nocardioides sp. could play a significant, yet currently underappreciated, role in VC and ethene assimilation in the environment.
Interestingly, 11 EaCoMT sequences obtained from the Fairbanks site, which contained a 7-bp deletion, grouped closely with the etnE1 allele in Nocardioides sp. strain JS614 (see Fig. S1 in the supplemental material). The JS614 etnE1 allele also contains a 7-bp deletion that would yield a frameshift mutation in the gene product and an expected loss of activity (15, 62). The JS614 etnE1 allele appears to have resulted from a gene duplication event involving the functional EaCoMT gene found on the plasmid harbored by JS614 (62). Inspection of Fig. 1 reveals that additional EaCoMT sequences, presumably without deletions, were amplified from Fairbanks samples. It is possible that a related plasmid containing two EaCoMT alleles was present in Fairbanks groundwater and the PCR primers used (COM-F1L/COM-R2E) preferentially amplify the EaCoMT allele with the 7-bp deletion. Further work is required to confirm this hypothesis.
Another emerging EaCoMT clade contains the 2-hydroxylpropyl CoM lyase from propene-assimilating Xanthobacter Py2. Until recently, strain Py2 was the only cultured representative harboring an EaCoMT gene from this group. However, the recently isolated marine ethene-assimilating Haliea sp. strain ETY-M (Haliea ETY-M) (31) also contains a putative EaCoMT. The newly discovered phylogenetic relationship between EaCoMT genes from Xanthobacter Py2 and Haliea ETY-M suggests that this group of EaCoMT genes could participate in ethene and VC biodegradation in contaminated groundwater, contrary to what has been proposed previously (63).
The environmental conditions that facilitate the apparently widespread occurrence of EaCoMT genes in environmental samples are not yet understood. The only known function for EaCoMT is the transfer of CoM onto epoxides of VC, ethene, and propene (12, 13). The presence of VC and ethene in groundwater at relatively high concentrations in groundwater (e.g., VC at >1 mg/liter and/or ethene at >230 μg/liter at the Australia site) should promote plasmid-encoded EaCoMT gene maintenance as well as elevated EaCoMT gene abundance. This is supported by the strong positive correlations between alkene concentration (VC, ethene) and EaCoMT gene abundance in this study (Table 3). The presence of EaCoMT genes in groundwater could also facilitate the natural attenuation of VC as long as other parameters are not limiting (e.g., dissolved oxygen). Spearman's correlation between EaCoMT gene abundance and the VC bulk attenuation rate supports this hypothesis (Table 3). Although there is a positive correlation between ethene and VC concentration and EaCoMT gene abundance in the various monitoring wells included in this study, EaCoMT genes were found at the Fairbanks and Kotzebue sites (where VC and ethene were not detected). Additional research is required to determine why EaCoMT genes are maintained at sites such as these.
Alkene oxidation is an obligately aerobic process; however, we observed no significant relationship between EaCoMT gene abundance and dissolved oxygen. Aerobic VC oxidation can occur at very low DO levels (below 0.02 mg/liter) (64), and VC-assimilating bacteria have been isolated from anaerobic groundwater (65). The fact that we detected EaCoMT genes in low-DO groundwater indicates that molecular oxygen either continuously enters the system or has been present in the groundwater in the past.
We did not observe any relationship between VC or ethene concentrations and EaCoMT gene diversity indices. This suggests that EaCoMT gene diversity patterns are currently not useful in site assessment for VC bioremediation. It is possible that the concentration of VC and/or ethene at a contaminated site could facilitate changes in EaCoMT gene diversity over time, as microorganisms use these compounds as carbon and energy sources. In addition, we currently cannot differentiate between a VC assimilator and a VC cometabolizer by analyzing EaCoMT sequences in environmental samples. Furthermore, the presence of EaCoMT genes in the environment that cannot produce an active EaCoMT (i.e., they contain 7-bp deletions or internal stop codons) complicates the interpretation of EaCoMT sequence data from the environment. These and many other variables (e.g., CoM availability) could affect EaCoMT gene patterns in the environment. Future studies that address the activities of different types of EaCoMT genes in the environment with respect to VC biodegradation rates could shed new light on this issue.
The PCR modifications employed in this study (nested and touchdown PCR) successfully amplified EaCoMT genes from environmental samples, but the results should be interpreted carefully with respect to PCR bias. The F131/R562 primers used in nested PCR were developed with etnE sequences from VC-assimilating Mycobacterium isolates (47) and are less degenerate than the CoMF1L/R2E primers, which were based on EaCoMT sequences from Mycobacterium sp. strain JS60, Gordonia B-276, and Xanthobacter Py2. This was confirmed by diversity analysis, which showed lower EaCoMT diversity indices in the AUS39 composite sample T-RFLP data generated with nested PCR. However, since the nested-PCR method was used on all the samples for T-RFLP analysis, the results allow a logical comparison between sites.
Touchdown (TD) PCR targets genes that bind primers more specifically (66). TD PCR with F131/R562 primers appeared to amplify more Mycobacterium-like etnE, as reflected by clone library analysis. TD PCR-amplified etnE sequences also did not group with sequences obtained by nested PCR from the same samples (Fig. 2), an observation that further supports the bias introduced by TD PCR. Our T-RFLP comparison experiment also showed that using TD PCR will underestimate gene diversity (see Fig. S5 in the supplemental material). However, a merit of TD PCR lies in its potential to reveal underrepresented genes in a sample.
Although the F131/R562 primers were successful when performing nested PCR, these primers do not amplify some important regions of EaCoMT genes. These include the region where missense mutations (W243R, R257L) associated with adaptation to VC as a carbon and energy source observed in Mycobacterium sp. strain JS623 (23) and the His-X-Cys-Xn-Cys zinc binding motif found in EaCoMTs and homologs (18, 19, 21).
Because EaCoMT and cobalamin (vitamin B12)-independent methionine synthase (MetE) are members of the same alkyl transferase family, many EaCoMT genes found in VC- and ethene-assimilating bacterial genome sequences (notably Nocardioides sp. strain JS614 [67]) are incorrectly annotated as MetE genes in GenBank. Examples of inaccurate annotation are compiled and presented in Table S10 in the supplemental material. MetE shares functional homology but does not share significant nucleotide or amino acid with most EaCoMT sequences. In our phylogenetic analysis (Fig. 2), the MetE sequences (see Table S7 in the supplemental material) clearly grouped separately from translated EaCoMT sequences (bootstrap support, 62%). The analysis also indicates that the misannotated MetE sequences clearly group with other EaCoMT sequences.
Conclusion.
The EaCoMT gene plays a critical role in the assimilation of short-chain alkenes, such as ethene, VC, and propene. This is the first study that reports amplification, diversity analysis, correlation analysis, and sequencing of EaCoMT genes from environmental samples. The EaCoMT gene database was significantly expanded, and potentially novel EaCoMT genes were discovered. Incorrectly annotated EaCoMT genes currently deposited in the database were also noted. These new sequences and insights will be useful in further developing environmental molecular diagnostic tools such as qPCR primers and probes and will aid in the development and application of high-throughput sequencing approaches in future bioremediation studies.
The presence of VC and ethene in groundwater could help sustain EaCoMT gene pools in the environment. Mycobacterium and Nocardioides-like EaCoMT sequences were the most widely distributed among the six sites investigated. The expanded clades of Nocardioides-like EaCoMT sequences and the discovery of Haliea-like EaCoMT sequences at the Soldotna site further suggest that potentially significant aerobic VC degraders in the environment are not well represented in pure culture. Future research should aim to isolate and characterize aerobic VC-degrading bacteria from these underrepresented groups.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Dora Ogles (Microbial Insights, Inc.) and James Fish (Alaska Department of Environmental Quality), who provided site information, facilitated groundwater sample collection, and/or donated DNA samples for this study. We thank the Iowa Institute of Human Genetics for providing qPCR, T-RFLP, and Sanger sequencing services. We also thank Yi Liang and Carly Lintner for technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00673-16.
REFERENCES
- 1.Mattes TE, Alexander AK, Coleman NV. 2010. Aerobic biodegradation of the chloroethenes: pathways, enzymes, ecology, and evolution. FEMS Microbiol Rev 34:445–475. doi: 10.1111/j.1574-6976.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang KLC, Li H, Ecker JR. 2002. Ethylene biosynthesis and signaling networks. Plant Cell 14:S131–S151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mansouri S, Bunch AW. 1989. Bacterial ethylene synthesis from 2-oxo-4-thiobutyric acid and from methionine. J Gen Microbiol 135:2819–2827. [DOI] [PubMed] [Google Scholar]
- 4.Keppler F, Borchars R, Pracht J, Rheinberger S, Scholer H. 2002. Natural formation of vinyl chloride in the terrestrial environment. Environ Sci Technol 36:2479–2483. doi: 10.1021/es015611l. [DOI] [PubMed] [Google Scholar]
- 5.Squillace PJ, Moran MJ, Lapham WW, Price CV, Clawges RM, Zogorski JS. 1999. Volatile organic compounds in untreated ambient groundwater of the United States, 1985-1995. Environ Sci Technol 33:4176–4187. doi: 10.1021/es990234m. [DOI] [Google Scholar]
- 6.HazDat. 2006. HazDat database: ATSDR's hazardous substance release and health effects database. Agency for Toxic Substances and Disease Registry, Atlanta, GA. [Google Scholar]
- 7.Swartjes FA. 2011. Dealing with contaminated sites: from theory towards practical application. Springer Science & Business Media, New York, NY. [Google Scholar]
- 8.Bucher JR, Cooper G, Haseman JK, Jameson CW, Longnecker M, Kamel F, Maronpot R, Matthews HB, Melnick R, Newbold R, Tennant RW, Thompson C, Waalkes M. 2014. Report on carcinogens, 13th ed U.S. Department of Health and Human Services, Public Health Service, National Toxicology Program, Rockville, MD: http://ntp.niehs.nih.gov/pubhealth/roc/roc13/. [Google Scholar]
- 9.Ensign SA. 2001. Microbial metabolism of aliphatic alkenes. Biochemistry 40:5845–5853. doi: 10.1021/bi015523d. [DOI] [PubMed] [Google Scholar]
- 10.Barbin A, Bresil H, Croisy A, Jacquignon P, Malaveille C, Montesano R, Bartsch H. 1975. Liver microsome mediated formation of alkylating agents from vinyl bromide and vinyl chloride. Biochem Biophys Res Commun 67:596–603. doi: 10.1016/0006-291X(75)90854-2. [DOI] [PubMed] [Google Scholar]
- 11.Henschler D. 1994. Toxicity of chlorinated organic compounds—effects of the introduction of chlorine in organic molecules. Angew Chem Int Ed Engl 33:1920–1935. doi: 10.1002/anie.199419201. [DOI] [Google Scholar]
- 12.Allen JR, Clark DD, Krum JG, Ensign SA. 1999. A role for coenzyme M (2-mercaptoethanesulfonic acid) in a bacterial pathway of aliphatic epoxide carboxylation. Proc Natl Acad Sci U S A 96:8432–8437. doi: 10.1073/pnas.96.15.8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coleman NV, Spain JC. 2003. Epoxyalkane: coenzyme M transferase in the ethene and vinyl chloride biodegradation pathways of Mycobacterium strain JS60. J Bacteriol 185:5536–5545. doi: 10.1128/JB.185.18.5536-5545.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ensign SA, Allen JR. 2003. Aliphatic epoxide carboxylation. Annu Rev Biochem 72:55–76. doi: 10.1146/annurev.biochem.72.121801.161820. [DOI] [PubMed] [Google Scholar]
- 15.Mattes TE, Coleman NV, Spain JC, Gossett JM. 2005. Physiological and molecular genetic analyses of vinyl chloride and ethene biodegradation in Nocardioides sp strain JS614. Arch Microbiol 183:95–106. doi: 10.1007/s00203-004-0749-2. [DOI] [PubMed] [Google Scholar]
- 16.Matthews RG, Goulding CW. 1997. Enzyme-catalyzed methyl transfers to thiols: the role of zinc. Curr Opin Chem Biol 1:332–339. doi: 10.1016/S1367-5931(97)80070-1. [DOI] [PubMed] [Google Scholar]
- 17.Goulding CW, Matthews RG. 1997. Cobalamin-dependent methionine synthase from Escherichia coli: Involvement of zinc in homocysteine activation. Biochemistry 36:15749–15757. doi: 10.1021/bi971988l. [DOI] [PubMed] [Google Scholar]
- 18.Zhou ZHS, Peariso K, Penner-Hahn JE, Matthews RG. 1999. Identification of the zinc ligands in cobalamin-independent methionine synthase (MetE) from Escherichia coli. Biochemistry 38:15915–15926. doi: 10.1021/bi992062b. [DOI] [PubMed] [Google Scholar]
- 19.Krum JG, Ellsworth H, Sargeant RR, Rich G, Ensign SA. 2002. Kinetic and microcalorimetric analysis of substrate and cofactor interactions in epoxyalkane:CoM transferase, a zinc-dependent epoxidase. Biochemistry 41:5005–5014. doi: 10.1021/bi0255221. [DOI] [PubMed] [Google Scholar]
- 20.Hartmans S, Debont JAM. 1992. Aerobic vinyl-chloride metabolism in Mycobacterium aurum L1. Appl Environ Microbiol 58:1220–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman NV, Mattes TE, Gossett JM, Spain JC. 2002. Phylogenetic and kinetic diversity of aerobic vinyl chloride-assimilating bacteria from contaminated sites. Appl Environ Microbiol 68:6162–6171. doi: 10.1128/AEM.68.12.6162-6171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coleman NV, Spain JC. 2003. Distribution of the coenzyme M pathway of epoxide metabolism among ethene- and vinyl chloride-degrading Mycobacterium strains. Appl Environ Microbiol 69:6041–6046. doi: 10.1128/AEM.69.10.6041-6046.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jin YO, Cheung S, Coleman NV, Mattes TE. 2010. Association of missense mutations in epoxyalkane coenzyme M transferase with adaptation of Mycobacterium sp. strain JS623 to growth on vinyl chloride. Appl Environ Microbiol 76:3413–3419. doi: 10.1128/AEM.01320-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Danko AS, Saski CA, Tomkin Freedman S JPDL. 2006. Involvement of coenzyme M during aerobic biodegradation of vinyl chloride and ethene by Pseudomonas putida strain AJ and Ochrobactrum sp. strain TD. Appl Environ Microbiol 72:3756–3758. doi: 10.1128/AEM.72.5.3756-3758.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Furuhashi K, Taoka A, Uchida S, Karube I, Suzuki S. 1981. Production of 1,2-epoxyalkane from 1-alkenes by Nocardia corallina B-276. Appl Microbiol Biotechnol 12:39–45. doi: 10.1007/BF00508117. [DOI] [Google Scholar]
- 26.Clark DD, Ensign SA. 1999. Evidence for an inducible nucleotide-dependent acetone carboxylase in Rhodococcus rhodochrous B276. J Bacteriol 181:2752–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ensign SA. 1996. Aliphatic and chlorinated alkenes and epoxides as inducers of alkene monooxygenase and epoxidase activities in Xanthobacter strain Py2. Appl Environ Microbiol 62:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danko AS, Luo MZ, Bagwell CE, Brigmon RL, Freedman DL. 2004. Involvement of linear plasmids in aerobic biodegradation of vinyl chloride. Appl Environ Microbiol 70:6092–6097. doi: 10.1128/AEM.70.10.6092-6097.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saeki H, Akira M, Furuhashi K, Averhoff B, Gottschalk G. 1999. Degradation of trichloroethene by a linear-plasmid-encoded alkene monooxygenase in Rhodococcus corallinus (Nocardia corallina) B-276. Microbiology 145:1721–1730. doi: 10.1099/13500872-145-7-1721. [DOI] [PubMed] [Google Scholar]
- 30.Krum JG, Ensign SA. 2001. Evidence that a linear megaplasmid encodes enzymes of aliphatic alkene and epoxide metabolism and coenzyme M (2-mercaptoethanesulfonate) biosynthesis in Xanthobacter strain Py2. J Bacteriol 183:2172–2177. doi: 10.1128/JB.183.7.2172-2177.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suzuki T, Nakamura T, Fuse H. 2012. Isolation of two novel marine ethylene-assimilating bacteria, Haliea species ETY-M and ETY-NAG, containing particulate methane monooxygenase-like genes. Microbes Environ 27:54–60. doi: 10.1264/jsme2.ME11256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freedman DL, Herz SD. 1996. Use of ethylene and ethane as primary substrates for aerobic cometabolism of vinyl chloride. Wat Environ Res 68:320–328. doi: 10.2175/106143096X127767. [DOI] [Google Scholar]
- 33.Koziollek P, Bryniok D, Knackmuss HJ. 1999. Ethene as an auxiliary substrate for the cooxidation of cis-1,2-dichloroethene and vinyl chloride. Arch Microbiol 172:240–246. doi: 10.1007/s002030050766. [DOI] [PubMed] [Google Scholar]
- 34.Verce MF, Ulrich RL, Freedman DL. 2001. Transition from cometabolic to growth-linked biodegradation of vinyl chloride by a Pseudomonas sp. isolated on ethene. Environ Sci Technol 35:4242–4251. doi: 10.1021/es002064f. [DOI] [PubMed] [Google Scholar]
- 35.Jin YO, Mattes TE. 2008. Adaptation of aerobic, ethene-assimilating Mycobacterium strains to vinyl chloride as a growth substrate. Environ Sci Technol 42:4784–4789. doi: 10.1021/es8000536. [DOI] [PubMed] [Google Scholar]
- 36.Jin YO, Mattes TE. 2010. A quantitative PCR assay for aerobic, vinyl chloride- and ethene-assimilating microorganisms in groundwater. Environ Sci Technol 44:9036–9041. doi: 10.1021/es102232m. [DOI] [PubMed] [Google Scholar]
- 37.Jin YO, Mattes TE. 2011. Assessment and modification of degenerate qPCR primers that amplify functional genes from etheneotrophs and vinyl chloride-assimilators. Lett Appl Microbiol 53:576–580. doi: 10.1111/j.1472-765X.2011.03144.x. [DOI] [PubMed] [Google Scholar]
- 38.Mattes TE, Jin YO, Livermore J, Pearl M, Liu X. 2015. Abundance and activity of vinyl chloride (VC)-oxidizing bacteria in a dilute groundwater VC plume biostimulated with oxygen and ethene. Appl Microbiol Biotechnol 99:9267–9276. doi: 10.1007/s00253-015-6771-2. [DOI] [PubMed] [Google Scholar]
- 39.Atashgahi S, Maphosa F, Dogan E, Smidt H, Springael D, Dejonghe W. 2013. Small-scale oxygen distribution determines the vinyl chloride biodegradation pathway in surficial sediments of riverbed hyporheic zones. FEMS Microbiol Ecol 84:133–142. doi: 10.1111/1574-6941.12044. [DOI] [PubMed] [Google Scholar]
- 40.Patterson BM, Aravena R, Davis GB, Furness AJ, Bastow TP, Bouchard D. 2013. Multiple lines of evidence to demonstrate vinyl chloride aerobic biodegradation in the vadose zone, and factors controlling rates. J Contam Hydrol 153:69–77. doi: 10.1016/j.jconhyd.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 41.Paes F, Liu X, Mattes TE, Cupples AM. 2015. Elucidating carbon uptake from vinyl chloride using stable isotope probing and Illumina sequencing. Appl Microbiol Biotechnol 99:7735–7743. doi: 10.1007/s00253-015-6606-1. [DOI] [PubMed] [Google Scholar]
- 42.Begley JF, Czarnecki M, Kemen S, Verardo A, Robb AK, Fogel S, Begley GS. 2012. Oxygen and ethene biostimulation for a persistent dilute vinyl chloride plume. Ground Water Monit Remediat 32:99–105. doi: 10.1111/j.1745-6592.2011.01371.x. [DOI] [Google Scholar]
- 43.Cook LJ, Hickman G, Chang A, Landin P, Reisch T. 2006. Comparison of aerobic and anaerobic biotreatments of low-level vinyl chloride, abstr 351 Fifth Int Conf Remediat Chlorinated and Recalcitrant Compounds, Monterey, CA, 22 to 25 May 2006. Battelle Memorial Institute, Columbus, OH. [Google Scholar]
- 44.Bradley PM, Chapelle FH. 2004. Chloroethene biodegradation potential in the “lower” contaminant plume, River Terrace RV Park, Soldotna, Alaska. United States Geological Survey, Reston, VA. [Google Scholar]
- 45.Kreader CA. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol 62:1102–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. 2009. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem 55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 47.Begley JF, Hansen E, Wells AK, Fogel S, Begley GS. 2009. Assessment and monitoring tools for aerobic bioremediation of vinyl chloride in groundwater. Remediat J 20:107–117. doi: 10.1002/rem.20232. [DOI] [Google Scholar]
- 48.Collins RE, Rocap G. 2007. REPK: an analytical web server to select restriction endonucleases for terminal restriction fragment length polymorphism analysis. Nucleic Acids Res 35:W58–W62. doi: 10.1093/nar/gkm384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Culman SW, Bukowski R, Gauch HG, Cadillo-Quiroz H, Buckley DH. 2009. T-REX: software for the processing and analysis of T-RFLP data. BMC Bioinformatics 10:171. doi: 10.1186/1471-2105-10-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Development Core Team R. 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org. [Google Scholar]
- 51.da C Jesus E, Marsh TL, Tiedje JM, Moreira FMD. 2009. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J 3:1004–1011. doi: 10.1038/ismej.2009.47. [DOI] [PubMed] [Google Scholar]
- 52.Magurran AE. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, NJ. [Google Scholar]
- 53.Reference deleted.
- 54.Newell CJ, Rifai HS, Wilson JT, Connor JA, Aziz JA, Suarez MP. 2002. Calculation and use of first-order rate constants for monitored natural attenuation studies. U.S. Environmental Protection Agency, National Risk Management Research Laboratory, Ada, OK. [Google Scholar]
- 55.Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. Biotechniques 28:1102–1104. [DOI] [PubMed] [Google Scholar]
- 56.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. doi: 10.1002/0471250953.bi0203s00. [DOI] [PubMed] [Google Scholar]
- 57.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alaminos M, Ramos JL. 2001. The methionine biosynthetic pathway from homoserine in Pseudomonas putida involves the metW, metX, metZ, metH and metE gene products. Arch Microbiol 176:151–154. doi: 10.1007/s002030100293. [DOI] [PubMed] [Google Scholar]
- 59.Zhang HK, Gao SH, Lercher MJ, Hu SN, Chen WH. 2012. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chuang AS, Jin YO, Schmidt LS, Li YL, Fogel S, Smoler D, Mattes TE. 2010. Proteomic analysis of ethene-enriched groundwater microcosms from a vinyl chloride-contaminated site. Environ Sci Technol 44:1594–1601. doi: 10.1021/es903033r. [DOI] [PubMed] [Google Scholar]
- 61.Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386–386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chuang AS, Mattes TE. 2007. Identification of polypeptides expressed in response to vinyl chloride, ethene, and epoxyethane in Nocardioides sp. strain JS614 by using peptide mass fingerprinting. Appl Environ Microbiol 73:4368–4372. doi: 10.1128/AEM.00086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnakumar AM, Sliwa D, Endrizzi JA, Boyd ES, Ensign SA, Peters JW. 2008. Getting a handle on the role of coenzyme M in alkene metabolism. Microbiol Mol Biol Rev 72:445–456. doi: 10.1128/MMBR.00005-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gossett JM. 2010. Sustained aerobic oxidation of vinyl chloride at low oxygen concentrations. Environ Sci Technol 44:1405–1411. doi: 10.1021/es9033974. [DOI] [PubMed] [Google Scholar]
- 65.Fullerton H, Rogers R, Freedman D, Zinder S. 2014. Isolation of an aerobic vinyl chloride oxidizer from anaerobic groundwater. Biodegradation 25:893–901. doi: 10.1007/s10532-014-9708-z. [DOI] [PubMed] [Google Scholar]
- 66.Don RH, Cox PT, Wainwright BJ, Baker K, Mattick JS. 1991. Touchdown PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res 19:4008–4008. doi: 10.1093/nar/19.14.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coleman NV, Wilson NL, Barry K, Brettin TS, Bruce DC, Copeland A, Dalin E, Detter JC, Glavina del Rio T, Goodwin LA, Hammon NM, Han S, Hauser LJ, Israni S, Kim E, Kyrpides N, Land ML, Lapidus A, Larimer FW, Lucas S, Pitluck S, Richardson P, Schmutz J, Tapia R, Thompson S, Tice HN, Spain JC, Gossett JG, Mattes TE. 2011. Genome sequence of the ethene- and vinyl chloride-oxidizing actinomycete Nocardioides sp. strain JS614. J Bacteriol 193:3399–3400. doi: 10.1128/JB.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.