Abstract
Despite a wealth of knowledge about the significance of individual signal transducers and activators of transcription (STATs), essential functions of their upstream Janus kinases (JAKs) during postnatal development are less well defined. Using a novel mammary gland-specific JAK1 knockout model, we demonstrate here that this tyrosine kinase is essential for the activation of STAT1, STAT3, and STAT6 in the mammary epithelium. The loss of JAK1 uncouples interleukin-6-class ligands from their downstream effector, STAT3, which leads to the decreased expression of STAT3 target genes that are associated with the acute-phase response, inflammation, and wound healing. Consequently, JAK1-deficient mice exhibit impaired apoptosis and a significant delay in mammary gland remodeling. Using RNA sequencing, we identified several new JAK1 target genes that are upregulated during involution. These include Bmf and Bim, which are known regulators of programmed cell death. Using a BMF/BIM-double-knockout epithelial transplant model, we further validated that the synergistic action of these proapoptotic JAK1 targets is obligatory for the remodeling of the mammary epithelium. The collective results of this study suggest that JAK1 has nonredundant roles in the activation of particular STAT proteins and this tyrosine kinase is essential for coupling inflammatory cytokine signals to the cell death machinery in the differentiated mammary epithelium.
INTRODUCTION
The postnatal growth, functional differentiation, and postlactational remodeling of the epithelial compartment of the mammary gland are controlled by hormones and locally synthesized cytokines (1). While estrogen is primarily required for the elongation of mammary ducts after the onset of puberty (2, 3), progesterone and prolactin orchestrate the specification, proliferation, and differentiation of secretory alveolar cells during pregnancy (4–6). Following lactation and the weaning of the young, circulating levels of prolactin (PRL) decline and milk stasis induces the local production of interleukin-6 (IL-6)-class cytokines, in particular, IL-6, leukemia inhibitory factor (LIF), and oncostatin M (OSM). These IL-6-class cytokines trigger a cascade of intracellular events that orchestrate a process known as mammary gland remodeling, which entails the death and selective removal of terminally differentiated alveolar cells (7–9).
PRL and IL-6-class cytokines signal through their corresponding receptors and utilize Janus kinases (JAKs) to activate downstream signal transducers and activators of transcription (STATs) that subsequently alter the transcriptional profile, growth, and homeostasis of mammary epithelial cells. Five of the seven STAT proteins that are known in mammals (i.e., STAT1, STAT3, STAT5a, STAT5b, and STAT6) have been found to be sequentially activated at defined stages of mammary gland development (10, 11). The levels of phosphorylated STAT1 have been reported to be elevated in nulliparous females, but this particular transcription factor seems to be largely dispensable for normal mammogenesis (12). A pregnancy-associated surge in circulating PRL results in a significant increase in the activation of both STAT5 proteins (i.e., STAT5a and STAT5b) as well as their downstream transcriptional targets, such as genes encoding milk proteins (13–16). Although STAT5a is the predominant isoform expressed in the mammary epithelium, both STAT5 proteins are required for alveolar development and lactogenesis (17, 18). A recent report highlighted the importance of IL-4 and IL-13 in the activation of STAT6 in the mammary epithelium (19). Unlike STAT5-knockout mice, however, females deficient in STAT6 are able to lactate, and therefore, this particular STAT protein seems to play a subordinate role in alveologenesis during late pregnancy. The beginning of the postlactational remodeling period is characterized by a decrease in the levels of active STAT5 and a significant increase in the phosphorylation and nuclear accumulation of STAT3 (10). LIF is suggested to serve as the initial upstream ligand that activates STAT3 during the reversible phase of mammary gland involution. The subsequent, STAT3-mediated expression of OSM establishes an autocrine loop for a sustained activation of this signal transducer, which initiates the death of terminally differentiated epithelial cells and the remodeling of the mammary gland (9).
Despite a wealth of knowledge about the significance of individual STATs, the biologically relevant functions of their upstream Janus kinases during the development of the mammary gland are less defined. Among the four known members of the JAK family, higher levels of JAK3 and Tyk2 expression are mainly restricted to hematopoietic cells. In contrast, the biological roles of JAK1 and JAK2 are more pleiotropic due to their ubiquitous expression profile and their suggested coupling to multiple cytokine receptors in specific target cells (reviewed by Kisseleva et al. [20]). A deficiency in just one of these two JAKs results in embryonic or neonatal lethality (21–23). Using mammary gland-specific gene deletion models, we and others have demonstrated that JAK2 is essential for alveologenesis and the prolactin-mediated activation of STAT5 (24, 25). The conditional excision of the Jak2 gene from the mammary epithelium at defined stages of development revealed that this kinase is equally important for the specification and proliferation of alveolar progenitors and the survival of terminally differentiated epithelial cells (25). In contrast to JAK2, the biological significance of JAK1 during postnatal organogenesis and tissue homeostasis in adults has not been defined. Moreover, the specific contribution of JAK1 to the sequential activation of individual STAT proteins during normal mammary gland development is unknown.
In this work, we report for the first time the generation and analysis of a JAK1 conditional knockout model. We show here that JAK1 has a nonredundant role in the activation of STAT1, STAT3, and STAT6, and we demonstrate that this particular Janus kinase is essential for coupling extracellular cytokine signals to the cell death machinery in the functionally differentiated mammary epithelium.
MATERIALS AND METHODS
Mouse models and genotyping protocols.
For a comprehensive protocol about the use of bacterial artificial chromosome recombineering to generate targeting vectors and conditional knockout mice by homologous recombination, please refer to our recent publication (26). Technical details about the cloning of the Jak1 genomic locus, the construction of the targeting construct, and the production of genetically engineered mice with two Jak1 conditional knockout alleles (Jak1flox [Jak1fl] or Jak1tm1Kuw) or the derived null alleles (Jak1− or Jak1tm1.1Kuw) will be described elsewhere (K. Sakamoto et al., unpublished data). The presence of one or two Jak1 wild-type or conditional knockout (i.e., floxed) alleles was determined by PCR of genomic tail DNA using a primer set specific for a sequence spanning the 5′ loxP site within the intronic sequence that precedes the second coding exon (forward primer 2411 [5′-GAG ACA GGA TAC CTG GTG GCT TGG-3′] and reverse primer 2412 [5′-GTA GCA GTC CTG GAC ATT GAG TCC-3′]). The wild-type and floxed alleles are approximately 250 and 350 bp, respectively. A less than 390-bp recombined Jak1-null allele was identified by PCR using the same forward primer, primer 2411, described above in combination with a reverse primer specific for a sequence downstream of the 3′ loxP site within the intronic region that follows the second coding exon (reverse primer 2373 [5′-AGG TGC CAC TCC CAC TGT CCT TTC C-3′]). The PCR protocols for genotyping of mouse mammary tumor virus (MMTV)-Cre transgenic (line A) and WAP-Cre transgenic mice [Tg(MMTV-cre)1Mam and Tg(Wap-cre)11738Mam, respectively] as well as the CAG-LSL-green fluorescent protein (GFP) Cre/loxP reporter strain can be found elsewhere (27–29). The generation of conventional BMF- and BIM-knockout mice as well as BMF/BIM-double-knockout mice was described earlier (30–32). All animals used in this study were treated humanely and in accordance with institutional guidelines and federal regulations. This study was carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Research Council (33). The protocol was approved by the Institutional Animal Care and Use Committee of the University of Nebraska Medical Center.
Mammary gland transplantation.
Athymic nude mice (NCr strain) were used for the transplantation studies. The surgical procedures for clearing the fat pad of 3-week-old female mice and the method of implanting tissue fragments and cell suspensions have been described previously (25). Viably frozen fragments of mammary epithelium from BMF/BIM-double-knockout mice (32) of about 1 mm3 were implanted into the cleared fat pad of 3-week-old recipients. The recipients were kept as nulliparous virgins for 12 weeks to provide sufficient time for the transplanted epithelium to form a ductal tree. The transplant-carrying females were then bred, and mammary glands were taken from the recipients immediately after delivering the young (postpartum) or 3 to 5 days later (involution). The mammary glands were prepared as whole mounts and stained as described below.
Administration of LIF and OSM.
Nulliparous JAK1-deficient mice and their wild-type controls were injected intraperitoneally with LIF (500 U/g body weight) or OSM (12.5 ng/g body weight). The inguinal mammary glands were collected 30 min later and processed for histology and immunostaining of tyrosine-phosphorylated STAT3 (pY-STAT3).
Cell culture.
Mouse embryonic fibroblasts (MEFs) were isolated from 12.5-day-old Jak1+/+, Jak1+/−, and Jak1−/− embryos as described previously (34). Cells were starved in serum-free Dulbecco modified Eagle medium for 16 h and then treated with ovine growth hormone (GH; 20 nM), recombinant mouse IL-4 (50 ng/ml; BD Pharmingen), recombinant mouse OSM (25 ng/ml; R&D Systems), or LIF (103 U/ml; Millipore) for 15 min at 37°C. Ovine GH was kindly provided by A. F. Parlow (National Hormone and Peptide Program) under the sponsorship of the National Hormone and Pituitary Program. Cells were harvested, frozen on dry ice, and stored at −80°C before immunoblotting was performed.
IP and Western blot analysis.
Detailed experimental procedures for immunoprecipitation (IP) and Western blot analysis were described elsewhere (35). The following antibodies were used for immunoblotting: anti-β-actin (catalog number I-19), anti-JAK1 (catalog number sc-7228), anti-STAT5 (catalog number sc-836), anti-STAT1 (catalog number sc-592), and anti-STAT6 (catalog number sc-981) from Santa Cruz Biotechnology; anti-phospho-Tyr (catalog number 05-321), anti-STAT3 (catalog number 9139S), anti-phospho-STAT3 (anti-pSTAT3; Tyr705) (catalog number 9145S), and anti-pSTAT5 (Tyr694/699; catalog number 9351S) from Cell Signaling, Inc.; anti-pSTAT1 (phosphorylated on S727; catalog number ab109461), anti-pSTAT6 (catalog number ab54461), anti-BIM (catalog number ab7888), and anti-c-FOS (catalog number ab7963) from Abcam; anti-RUNX1 (catalog number TA307515) and anti-pSTAT1 (catalog number TA309955) from Origene; anti-pSTAT5 (Tyr694/699; catalog number AX1) from Advantex BioReagents; WFDC5 (catalog number LS-C297015) from LifeSpan BioSciences, Inc.; and anti-α-tubulin (catalog number 1878-1) from Epitomics. The anti-BMF mouse monoclonal antibody (clone 17C2) was from Enzo Life Sciences.
Preparation of mammary gland whole mounts and H&E staining and immunostaining of histological sections.
Whole mounts of the 4th inguinal mammary glands were prepared and stained in carmine alum as described previously (36). For histological examination, mammary tissues were fixed in 10% buffered formalin (Fisher Scientific Company), paraffin embedded, and sectioned. Slides were stained with hematoxylin and eosin (H&E) or processed for immunostaining. Detailed protocols for immunohistochemistry and immunofluorescent staining of histological sections can be found elsewhere (36). The sources for the following primary antibodies were listed above: anti-pSTAT3, anti-pSTAT5, anti-RUNX1, and anti-c-FOS. The anti-E-cadherin antibody was purchased from BD Transduction Laboratories. Corresponding secondary antibodies conjugated to Alexa Fluor dyes 488 and 594 (Invitrogen) were used for the detection of the specific targets. Slides were counterstained briefly with DAPI (4′,6-diamidino-2-phenylindole) or hematoxylin to visualize the nuclei. Alternatively, the staining of biotinylated secondary antibodies was performed using Vectastain Elite ABC kits from Vector Laboratories. Fluorescence and bright-field images of the histological sections on the slides were taken on an Axio Imager microscope (Carl Zeiss) equipped with a Spot Flex camera (Diagnostic Instruments, Inc.).
Transcriptome sequencing (RNA-Seq) analysis.
Total RNA was extracted from flash-frozen mammary gland tissues of seven conditional knockout females (three day 7 lactating mice and four day 2 involuting females) and six wild-type control mice (three day 7 lactating mice and three day 2 involuting females) using an RNeasy minikit (Qiagen). A SuperScript II kit from Invitrogen with oligo(dT) primers was used to perform the first-strand synthesis according to the manufacturer's protocol. Following quality control using a Bioanalyzer 2100 instrument, RNA samples were processed using a TruSeq RNA sample kit and sequenced using a HiSeq2000 sequencer (Illumina). The quality of the sequenced reads was determined using the FastQC program (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and contaminated adaptor portions were trimmed using the Cutadapt program (https://github.com/marcelm/cutadapt). The trimmed single-end reads were mapped to the mouse reference genome mm9 using STAR aligner software (37). Transcript abundance was estimated using the Tuxedo tools (Cufflinks, Cuffmerge, Cuffquant, Cuffnorm, and Cuffdiff) as described earlier (38). Differential transcript expression between JAK1-knockout and control mice during the two developmental stages was determined by estimation of the number of fragments per kilobase of transcript per million mapped reads (FPKM). To identify candidate target genes that are upregulated in a JAK1-dependent manner during involution, we focused our main efforts on comparison of the gene expression profiles between the tissues of the six lactating and involuting wild-type control mice and comparison of the tissues of the seven involuting JAK1-knockout and wild-type control mice. Genes showing more than 2-fold up- or downregulation between these samples were selected. A gene set enrichment analysis (GSEA) was performed to identify groups of genes that are controlled by JAK1 according to their cellular functions and pathways, and the Broad Institute's Integrative Genomics Viewer (IGV) was used to visualize the expression of individual genes and their exons.
ChIP assay and quantitative real-time PCR.
The chromatin immunoprecipitation (ChIP) assay was performed as described previously (39) using antibodies directed against STAT5 and STAT3 (see above) or an isotype-matched control IgG. Input and bound chromatin was detected by quantitative PCR using primer sets surrounding STAT binding sites (TTCN3GAA) in the murine Bmf and Bcl2l11 loci. The sequences of the primer sets specific for STAT binding sites will be provided upon request. The assay background was detected using PCR primers directed against a nonpromoter site and was used for normalization, in addition to comparison against a serial dilution of genomic DNA, as described previously (39).
ChIP-Seq analysis.
Snap-frozen mammary tissues were homogenized, and cross-linking of proteins to DNA was done with 1% formaldehyde. Following the isolation of cell nuclei in Farnham lysis buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 8.0, 85 mM KCl, 0.5% NP-40}, samples were sonicated, centrifuged, and resuspended in 1× radioimmunoprecipitation assay buffer. For ChIP, 600 to 1,000 μg of cell lysate was incubated overnight with Dynabead-antibody complexes. We used only validated or ChIP-certified commercial antibodies against STAT5a and STAT3 (catalog numbers sc-1081 and sc-482, respectively; Santa Cruz). Following IP, reverse cross-linking, and purification of DNA, indexed libraries were generated using a TruSeq ChIP sequencing (ChIP-Seq) sample kit from Illumina. The libraries were sequenced using a HiSeq2000 sequencer. The single-end reads were mapped to the mouse reference genome mm9 using the Bowtie aligner. To visualize peaks within promoters or intragenic regions that may control alternative transcripts, we used the HOMER or Galaxy program to convert BAM files into the BedGraph or bigWig format, and the files were viewed with the IGV browser.
Statistical analysis.
All graphic illustrations were prepared and statistical analyses were performed with Prism (version 6) software (GraphPad Software, Inc., La Jolla, CA). Data are expressed as the mean ± standard deviation (SD) unless otherwise indicated and were compared using an unpaired Student t test. A P value of less than 0.05 was considered significant.
Nucleotide sequence accession number.
The RNA-Seq data sets have been deposited in NCBI's Gene Expression Omnibus (GEO) database and are accessible through GEO series accession number GSE79452.
RESULTS
Expression levels of JAK1 correlate with the activation of STAT1 and STAT6 in the mammary glands of virgin and pregnant females as well as that of STAT3 during involution.
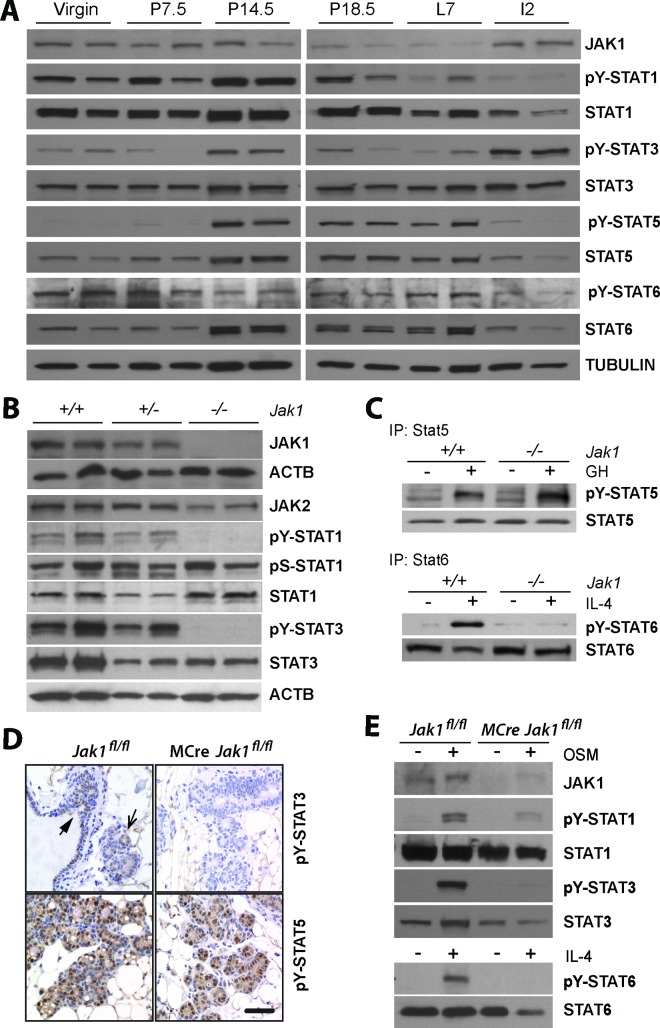
Previous studies have investigated the biological significance of five of the seven STAT proteins (STAT1, STAT3, STAT5a, STAT5b, and STAT6) during mammary gland development. To correlate their activation with changes in the expression of JAK1, we performed an inaugural analysis of the phosphorylation levels of these five STAT proteins in mammary gland tissues of virgin, pregnant, and lactating females as well as during the initial phase of mammary gland involution (i.e., day 2 postweaning). STAT1 was predominantly tyrosine phosphorylated in virgin and pregnant females (Fig. 1A). Its activation declined during lactation, and the expression and activation of STAT1 remained lower during the onset of involution. The highest levels of STAT3 phosphorylation were observed during pregnancy and involution, and active STAT5 was predominantly present in the mammary glands of pregnant and lactating mice. Active STAT6 was observed at all stages of mammary gland development, but its expression declined somewhat during the onset of involution. JAK1 levels were high in the glands of virgin and pregnant females. The expression of this Janus kinase was significantly lower during late pregnancy and lactation, but we observed a swift increase in the expression of JAK1 at the onset of involution. The collective results of this inaugural analysis confirm that STATs are sequentially activated during mammary gland development. However, all of these signal transducers were phosphorylated during midpregnancy. More importantly, higher levels of JAK1 expression coincided with the activation of STAT1 and STAT6 in the mammary glands of nulliparous and pregnant mice and more prominently with the phosphorylation of STAT3 during involution.
FIG 1.
JAK1 is essential for the activation of STAT1, STAT3, and STAT6 in embryonic fibroblasts and in mammary epithelial cells. (A) Immunoblot analysis to assess the expression of JAK1 and tyrosine phosphorylation of STAT1, STAT3, STAT5, and STAT6 during mammary gland development (P numbers, day of pregnancy; L7, lactation day 7; I2, involution day 2). The individual panels represent immunoblots of tissues from different mice. (B) Expression of JAK1 and JAK2 as well as steady-state activation of STAT1 and STAT3 in primary mouse embryonic fibroblasts (MEFs) from heterozygous and homozygous JAK1-knockout embryos that were generated through germ line deletion of the floxed locus (Jak1+/− [+/−] and Jak1−/− [−/−], respectively) as well as MEFs from wild-type controls (Jak1+/+ [+/+]). (C) IP/Western blot analysis of STAT5 and STAT6 in GH- and IL-4-treated wild-type and JAK1-knockout MEFs. (D) Immunohistochemical staining of active STAT5 and STAT3 on mammary tissue sections from a JAK1 conditional knockout mouse (MMTV-Cre Jak1flox/flox [MCre Jak1fl/fl] and a littermate wild-type control (Jak1flox/flox [Jak1fl/fl]) at day 14.5 of gestation. The slides were counterstained with hematoxylin. Bar, 50 μm. (E) Immunoblot analysis of phosphorylated STAT1, STAT3, and STAT6 in explanted mammary epithelial organoids from mice with mammary gland-specific JAK1 knockout and their wild-type controls with and without treatment with OSM and IL-4.
JAK1 is essential for the cytokine-induced activation of STAT1, STAT3, and STAT6 but not STAT5 in embryonic fibroblasts and in the mammary gland epithelium.
To study the biologically significant functions of JAK1 during postnatal mammary gland development, we performed gene targeting in embryonic stem cells and generated a Cre/lox-based conditional knockout mouse model. A description of the construction of the targeting vector and the phenotypic consequences of the deletion of JAK1 in the germ line will be described elsewhere (Sakamoto et al., unpublished). The insertion of loxP sites flanking the second coding exon of the Jak1 gene and the presence of the neomycin selection marker in intron 3 did not cause any phenotypic abnormalities when the targeted allele was crossed into homozygous mice. Jak1fl/fl mice were phenotypically indistinguishable from the wild-type controls (Jak1+/+). In an inaugural experiment, we induced a Cre-mediated recombination of the Jak1 floxed allele (Jak1fl) in the germ line of mice to validate that the conditional deletion of the second coding exon resulted in a true null allele. Similar to the phenotype for mice with the conventional knockout (21), the germ line from which the Jak1fl allele was deleted (Jak1−) causes postnatal lethality when crossed into homozygosity. Newborn Jak1−/− pups were visibly smaller, exhibited signs of apnea, and died within hours after birth (Sakamoto et al., unpublished). Next, we derived mouse embryonic fibroblasts from homozygous JAK1-knockout embryos at embryonic day 12.5 and their heterozygous and wild-type littermate controls to assess the activation of STAT proteins. As expected, the deletion of the second coding exon, which causes a frameshift of almost the entire coding sequence, led to a complete absence of the JAK1 protein (Fig. 1B). A deficiency in JAK1 did not result in a compensatory upregulation of JAK2, but we observed a complete absence of STAT1 and STAT3 tyrosine phosphorylation at steady state when cells were grown in the presence of serum (Fig. 1B). Interestingly, STAT1 was still phosphorylated on serine 727, despite its lack of tyrosine phosphorylation, supporting the previously postulated notion that both phosphorylation events may occur independently of each other (40). Similar to the findings for STAT1 and STAT3, the loss of JAK1 also abolished the phosphorylation of STAT6 in response to IL-4 signaling (Fig. 1C), but the growth hormone-induced activation of STAT5 was not affected in JAK1-deficient fibroblasts.
To determine the importance of JAK1 for the activation of STATs in the mammary gland, we generated conditional knockout mice that carried the MMTV-Cre transgene in a Jak1-floxed homozygous background. STAT activation in the mammary glands of MMTV-Cre Jak1fl/fl females and their littermate controls lacking Cre recombinase was analyzed at about gestation day 14.5, which represents a developmental window when all five STAT proteins exhibit elevated levels of activation (Fig. 1A). JAK1 deficiency did not seem to have any noticeable effect on the pregnancy-associated development of alveoli (Fig. 1D). The immunohistochemical analysis revealed that a deficiency in JAK1 led to a complete absence of tyrosine-phosphorylated STAT3 (pY-STAT3) in the epithelial compartment. Active STAT3 was predominantly localized in the ductal epithelium and was significantly less abundant in developing alveoli. In contrast to STAT3, a stronger nuclear accumulation of pY-STAT5 was observed in alveolar cells, and the activation of STAT5 was not affected by the lack of JAK1. To assess the activation of STAT1, STAT3, and STAT6, we freshly prepared mammary epithelial organoids and treated them for 20 min with OSM or IL-4 (Fig. 1E). Similar to the findings for the fibroblasts, the conditional deletion of JAK1 led to an uncoupling of these ligand-receptor signaling complexes from their downstream mediators, STAT1, STAT3, and STAT6. Given the similarities between mammary epithelial cells and fibroblasts, it is evident that essential functions of JAK1 for the activation of these three STAT proteins in response to OSM and IL-4 signaling are not restricted to one particular cell type or organ.
JAK1 deficiency causes impaired postlactational mammary gland remodeling.
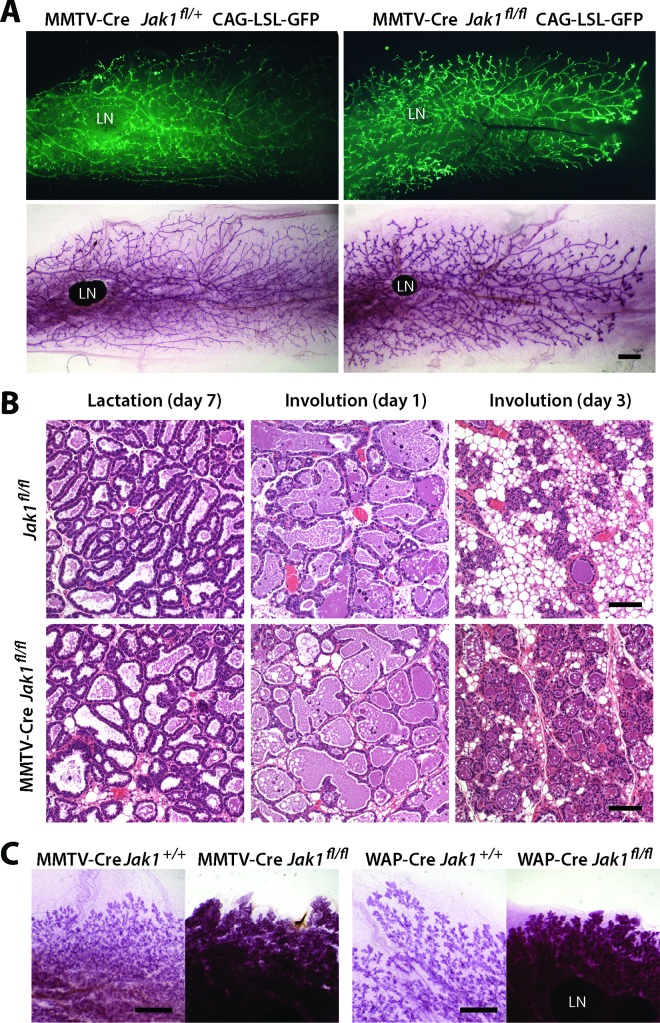
To assess the essential biological functions of JAK1 during mammogenesis, we next examined the morphology of mammary gland tissues from JAK1 conditional knockout females and controls during a normal gestation cycle. The inclusion of a GFP-based reporter transgene for Cre recombinase in the conditional knockout model showed that the MMTV-Cre-mediated deletion of the Jak1 gene occurred throughout the mammary ductal system of virgin females (Fig. 2A, top), suggesting that there was no negative selection against JAK1-deficient epithelial cells prior to pregnancy. The lack of JAK1 also did not have any discernible effect on ductal elongation or branching morphogenesis (Fig. 2A, bottom). Although JAK1 deficiency disrupts the activation of STAT6, females lacking JAK1 specifically in the mammary epithelium did not exhibit any noticeable delay in alveologenesis during pregnancy. MMTV-Cre Jak1fl/fl females lactated normally and were capable of supporting their litters until weaning. The histological appearance of secretory alveoli during lactation in JAK1-deficient females was indistinguishable from that of the secretory alveoli in their controls lacking Cre recombinase (Fig. 2B, left), and there was also no apparent difference in the mammary gland architecture 24 h after weaning of the offspring (Fig. 2B, middle). However, JAK1 conditional knockout females exhibited a significant delay in postlactational mammary gland remodeling starting at day 3 postweaning (Fig. 2B, right), and these phenotypic abnormalities were still evident after 5 days of involution (Fig. 2C, left). The impairment of the involution process as a consequence of the ablation of JAK1 in the mammary epithelium was confirmed using a WAP-Cre-mediated, mammary gland-specific knockout of JAK1, where this kinase is deleted specifically in alveolar cells during late pregnancy and lactation (Fig. 2C, right).
FIG 2.
The biologically significant role of JAK1 is restricted to postlactational mammary gland remodeling. (A) (Top) Fluorescence images of unfixed mammary gland tissues to visualize expression of GFP in mammary ducts of a nulliparous JAK1 conditional knockout female (MMTV-Cre Jak1flox/flox CAG-LSL-GFP) and its littermate control expressing JAK1 (MMTV-Cre Jak1flox/+ CAG-LSL-GFP). (Bottom) The same glands from the top panels following carmine alum staining. Bar, 1 mm. (B) H&E-stained histological sections of mammary glands from females with mammary gland-specific JAK1 knockout and wild-type controls during lactation as well as at 1 and 3 days of involution. Bars, 100 μm. (C) Whole mounts of mammary glands from females with MMTV-Cre- and WAP-Cre-mediated mammary gland-specific JAK1 knockout and their JAK1-expressing controls at day 5 of involution. Bars, 2 mm. LN, lymph node.
Deletion of JAK1 uncouples gp130-mediated signaling through IL-6-class cytokine receptor complexes from its downstream effector, STAT3.
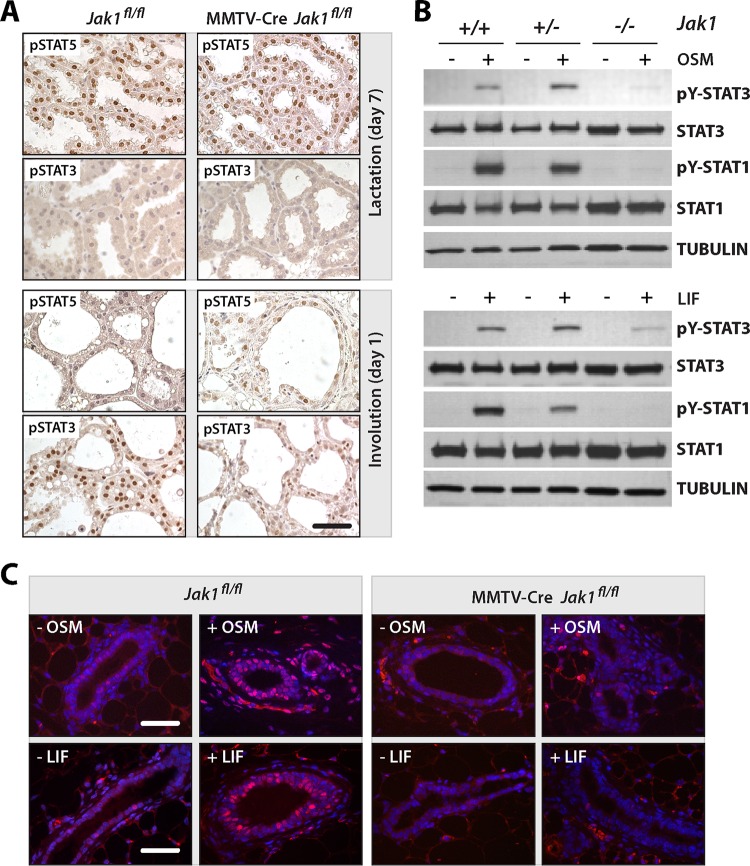
The initial analysis of the tyrosine phosphorylation of STATs during mammary gland development (Fig. 1A) showed that STAT5 and STAT3 exhibited the most dramatic switch in their activities at the onset of involution. Using immunostaining, we confirmed that the high levels of nuclear pSTAT5 that were present in the lactating glands of JAK1-knockout mice and their wild-type controls declined significantly within 24 h after weaning (Fig. 3A). Some postlactational glands of JAK1-deficient females still contained cells with a very weak but still noticeable staining of pSTAT5. In sharp contrast to the findings for STAT5, there was a significant reduction in the level of nuclear, tyrosine-phosphorylated STAT3 in JAK1-deficient females compared to the wild-type controls at day 1 of involution. This finding and our previous observation that the expression of JAK1 was swiftly upregulated following the weaning of the offspring (Fig. 1A) suggest that JAK1 plays a pivotal role in the activation of STAT3 in the involuting mammary gland.
FIG 3.
JAK1 is essential for the activation of STAT3 in response to OSM and LIF signaling in the mammary gland in vivo and cultured MEFs. (A) Immunohistochemical staining of active STAT5 and STAT3 on mammary tissue sections of JAK1 conditional knockout females (MMTV-Cre Jak1fl/fl) and wild-type controls (Jak1fl/fl) during lactation and the first day of involution. Bar, 50 μm. (B) Immunoblot analysis to assess the activation of STAT1 and STAT3 in heterozygous and homozygous JAK1-knockout MEFs (Jak1+/− and Jak1−/−, respectively) and wild-type controls (Jak1+/+) with and without treatment with OSM and LIF. (C) Staining of tyrosine-phosphorylated STAT3 on mammary tissue sections from nulliparous JAK1 conditional knockout mice and their controls with and without administration of exogenous OSM and LIF. Bars, 50 μm.
LIF and OSM are two IL-6-class inflammatory cytokines that have been shown to control mammary gland remodeling through activation of STAT3 (8, 9). These cytokines signal through specific ligand-receptor complexes that share the glycoprotein 130 (gp130) signal transduction subunit, which is essential for STAT3 activation in the involuting gland (41). To assess the importance of JAK1 as a mediator of signaling through gp130, we treated JAK1-deficient embryonic fibroblasts and their controls with LIF or OSM and examined the activation of STAT3 and STAT1. The immunoblotting results shown in Fig. 3B provide clear evidence that a deficiency in JAK1 uncouples IL-6-class cytokine signaling from these STAT proteins. To validate these findings in the mammary gland, we injected mice with both OSM and LIF and performed an immunofluorescent staining of pSTAT3 (Fig. 3C). While treatment with these IL-6-class ligands led to a swift nuclear accumulation of pSTAT3 in the mammary epithelium as well as surrounding stromal cells of control mice (Fig. 3C, left), active STAT3 was not detected in epithelial cells of JAK1 conditional knockout females (Fig. 3C, right). Nuclear STAT3 was still present in stromal cells of ligand-treated MMTV-Cre Jak1fl/fl females. The staining of these cells served as an internal positive control, since the mammary gland stroma does not express the MMTV-Cre transgene and is therefore wild type for JAK1. Collectively, the results from the analysis of JAK1-deficient fibroblast and mammary gland-specific knockout mice suggest that JAK1 is the major tyrosine kinase that transduces signals through gp130-receptor complexes downstream of IL-6-class cytokines.
Deletion of Jak1 leads to deregulated expression of downstream target genes associated with the acute-phase response, inflammation, wound healing, and programmed cell death.
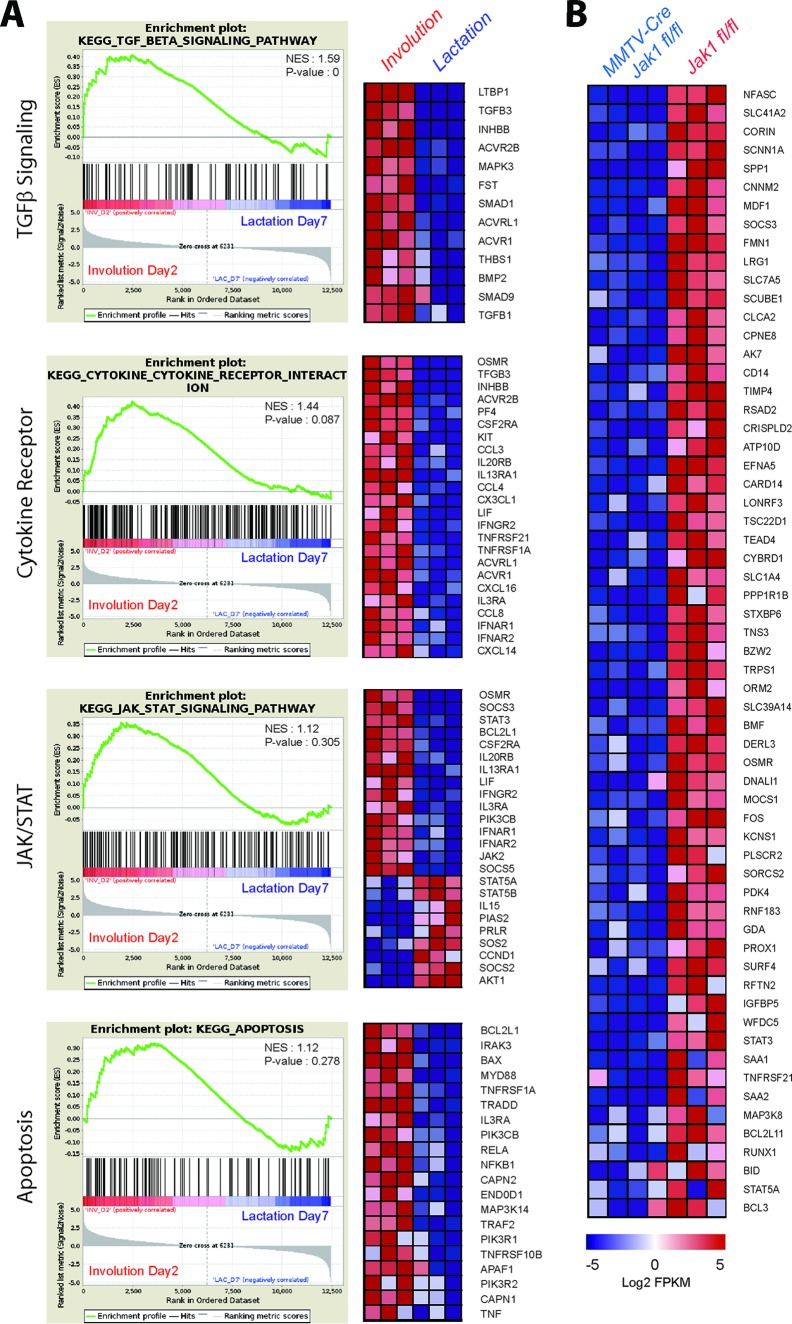
To gain insight into the underlying mechanism(s) by which the loss of JAK1 causes a delay in postlactational mammary gland remodeling, we performed a comprehensive, genome-wide RNA-Seq analysis of lactating and involuting mammary gland tissues of JAK1-knockout females and their wild-type controls. Given the canonical function of JAK1 as an activator of STAT transcription factors, we first focused on the identification of genes and pathways that were consistently upregulated during the cessation of lactation in the wild-type controls. We then concentrated our efforts on detecting a subset of genes that lack a normal increase in their expression during involution in the absence of JAK1. A comparison of the gene expression profiles of tissues from three lactating and three involuting wild-type mice revealed that 1,131 genes exhibited a 2-fold or higher upregulation during involution and 1,378 genes were downregulated. As anticipated, many known targets of PRL and JAK2/STAT5 signaling, such as Elf5, Socs2, Ccnd1, Akt1, and major milk protein genes (i.e., Wap, Csn1s1, and Csn2) were downregulated. The results of the gene set enrichment analyses (GSEAs) of the RNA-Seq data for the tissues from the six wild-type mice shown in Fig. 4A illustrate the synchronous switch in cytokine receptor expression and the utilization of distinctly different JAK/STAT pathways at the onset of involution. The reduced expression of the JAK2/STAT5 target genes coincided with significantly elevated levels of expression of genes that were activated in response to inflammatory cytokine receptor signaling. In addition to an increased expression of Lif, Osmr, and their known downstream effectors, Stat3 and Socs3, we detected an upregulation of genes associated with transforming growth factor β signaling and apoptosis during involution (Fig. 4A). We also observed a consistent deregulated expression of mediators of NF-κB signaling and an upregulation of several members of the tumor necrosis factor receptor superfamily, in particular, Tnfrsf1A, Tnfrsf10B, and Tnfrsf21.
FIG 4.
Deficiency in JAK1 inhibits the expression of downstream transcriptional target genes that are normally upregulated during mammary gland involution. (A) Gene set enrichment plots with heat maps of genes that are selectively upregulated during the second day of involution compared to their regulation during lactation. TGF-β, transforming growth factor β. (B) Heat maps of selected genes that are normally upregulated during involution but that exhibit a 2-fold or more downregulation in females with mammary gland-specific JAK1 knockout (MMTV-Cre Jak1fl/fl) compared to their regulation in the wild-type controls (Jak1fl/fl) on the second day of involution.
Next, we compared the gene expression profiles of mammary glands from four conditional knockout mice to those of mammary glands from the three wild-type controls at the second day of involution. A 2-fold or more downregulated expression was observed for only 135 genes, and 203 genes exhibited elevated levels of expression. Among the most consistently downregulated genes in the tissues from JAK1-knockout mice (Fig. 4B) were a number of previously identified direct or indirect transcriptional targets of STAT3. These included Socs3, Osmr, Bcl3, and the Stat3 gene itself. Similar to the findings for STAT3-knockout mice (42), we observed a reduced expression of genes associated with the acute-phase response and inflammation (i.e., Cd14, Saa1/2, Lrg1, and Orm2) as well as wound healing (Chi3L1). In addition to these previously reported genes, JAK1-deficient glands lacked expression of the transcription factors Runx1 and c-Fos, whose levels sharply increased during involution in wild-type tissues. We also identified a new WAP four-disulfide core domain protein gene (Wfdc5), which, unlike the milk protein WAP, was specifically upregulated during involution in a JAK1-dependent manner. Finally, we found several JAK1-induced target genes that control cell survival and programmed cell death, and their lack of expression in the JAK1 conditional knockout mice might be responsible for impaired mammary gland remodeling. These genes include the death receptor 6 gene (Tnfrsf21) and the tumor susceptibility gene Tpl2 (Map3k8). However, the most consistently downregulated proapoptotic factor in the JAK1-knockout glands was the gene encoding the BH3-only protein BMF. We also observed a reduced expression of the related genes Bim (Bcl2l11) and Bid in the absence of JAK1.
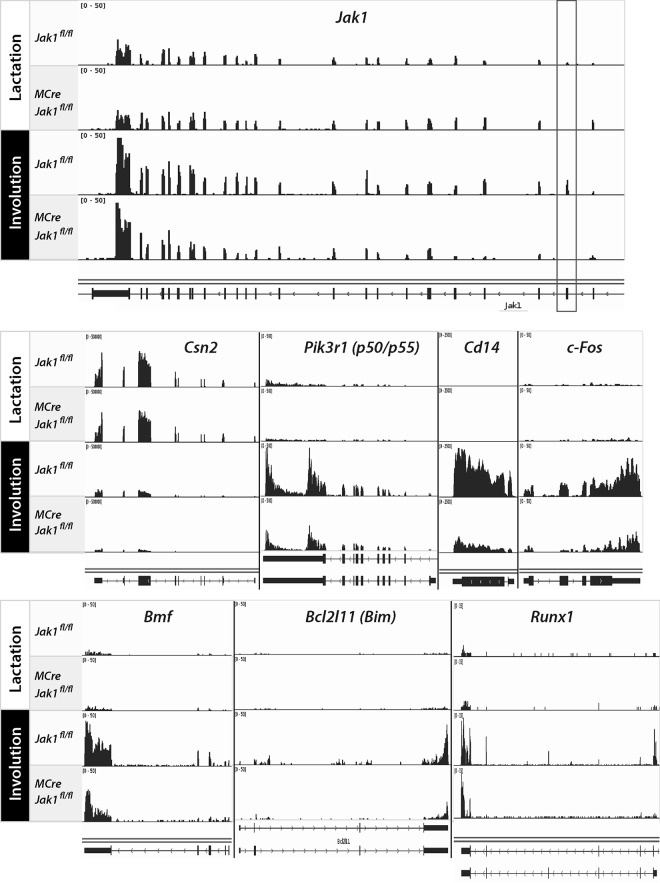
The inclusion of an additional three expression libraries from samples from lactating JAK1 conditional knockout females complemented the RNA-Seq analysis. Besides determining differences in the expression of genes, the remarkable fidelity of the RNA-Seq method also allowed us to confirm the precise excision of the floxed coding exon of Jak1 during lactation and involution (Fig. 5, top, boxed region). Jak1 was transcriptionally upregulated during the first phase of involution, which may explain the acute increase in JAK1 protein levels at this developmental stage (Fig. 1A). JAK1 deficiency had no noticeable effect on the expression of JAK2/STAT5-controlled genes during lactation, such as those encoding major milk proteins (e.g., Csn2) (Fig. 5, middle). The visual reexamination of selected JAK1 target genes (e.g., Cd14, c-Fos, Bmf, and Bim) confirmed their upregulation during involution and their dependence on JAK1, but we also observed unique patterns of promoter and exon usage of individual target genes. Similar to the previously reported upregulation of the gene for the p50/p55 isoforms of the phosphatidylinositol 3-kinase (PI3 kinase) regulatory subunit (Pik3r1) (42), we found that Runx1 was selectively expressed from an intronic promoter encoding the shorter isoform 3 in a JAK1-dependent manner (Fig. 5, bottom). While these examples highlight a superior fidelity of RNA-Seq, they also emphasize the importance of using only appropriate antibodies that recognize alternative isoforms for further validation on the level of the protein.
FIG 5.
JAK1 is required for the upregulation of isoform-specific STAT3 transcriptional targets and newly identified JAK1-responsive genes during mammary gland involution. The individual panels show the histograms of RNA-Seq data sets for selected genes, illustrating their expression levels and usage of exons in lactating and involuting mammary tissues from females with mammary gland-specific JAK1 knockout (MMTV-Cre Jak1fl/fl) and their wild-type controls (Jak1fl/fl). The box surrounding the second coding exon of Jak1 highlights the precise deletion of this exon in the conditional knockout mice. The beta-casein gene (Csn2) served as a control for a STAT5-responsive target gene that is downregulated during involution. Cd14 and Pik3r1 are examples of STAT3-responsive genes whose upregulation during involution is JAK1 dependent. c-Fos, Bmf, Bim, and Runx1 are newly identified targets of JAK1 signaling. Note the specific exon usage in Pik3r1, which encodes the alternative p50/p55 PI3 kinase regulatory subunit, and that expression of Runx1 during involution is driven by a shorter transcript known as isoform 3.
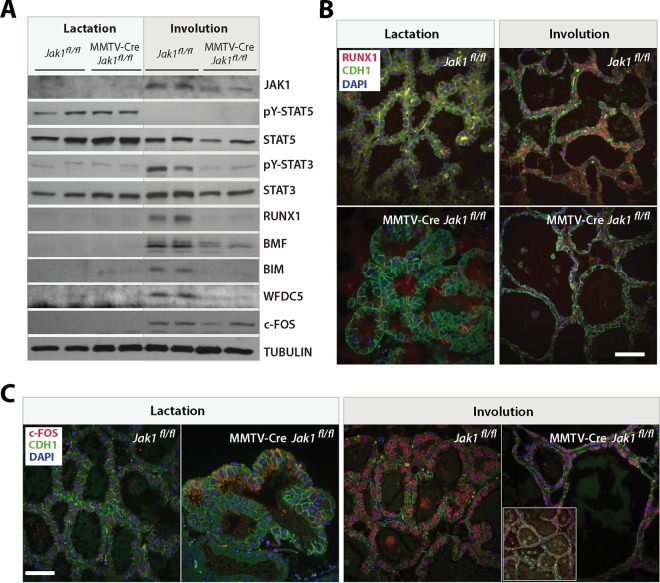
Next, we confirmed some of the newly identified downstream effectors of JAK1 signaling using Western blot analysis. Identical to the immunohistochemistry results shown in Fig. 3A, deletion of Jak1 had no effect on the activation of STAT5 during lactation, but the tyrosine phosphorylation of STAT3 was abrogated at the onset of involution (Fig. 6A). Along with an increase in the level of JAK1, we observed a significant upregulation of RUNX1, c-FOS, WDFC5, as well as BMF and BIM in the postlactational wild-type controls. With the exception of c-FOS, all other proteins exhibited a reduced expression in the JAK1-deficient glands. RUNX1 and c-FOS are known to be expressed in hematopoietic cells, but the immunostaining of these proteins on histological sections confirmed that these transcription factors were selectively upregulated in secretory epithelial cells during involution (Fig. 6B and C). RUNX1 was virtually absent from all epithelial cells in the JAK1 knockout, and even the nuclear staining of c-FOS was significantly lower or completely lacking in many areas of the involuting knockout glands. In summary, the Western blotting and immunostaining results confirmed the JAK1-dependent activation of STAT3 and the downregulated expression of selected JAK1 targets that were identified by RNA-Seq.
FIG 6.
A reduction in the corresponding protein levels of putative JAK1 target genes correlates with a decreased phosphorylation of STAT3 in postlactational mammary glands of JAK1 mutant females. (A) Immunoblot analysis of putative JAK1 target genes. These selected target genes exhibited a significant upregulation in their mRNA levels during the lactation/involution switch in wild-type females and a reduced mRNA expression in JAK1-deficient mice. (B and C) Immunofluorescence staining of RUNX1 (B) and c-FOS (C) on histologic sections of lactating (day 9) and involuting (day 2) mammary tissues from females with mammary gland-specific JAK1 knockout (MMTV-Cre Jak1fl/fl) and their wild-type controls (Jak1fl/fl). Slides were counterstained with E-cadherin (CDH1) and DAPI. Bars, 50 μm. (Inset in panel C) Example of areas that were completely devoid of c-FOS expression.
BMF and BIM are downstream targets of JAK1, and their combined deficiency phenocopies an impairment of mammary gland remodeling similar to that in JAK1-deficient mice.
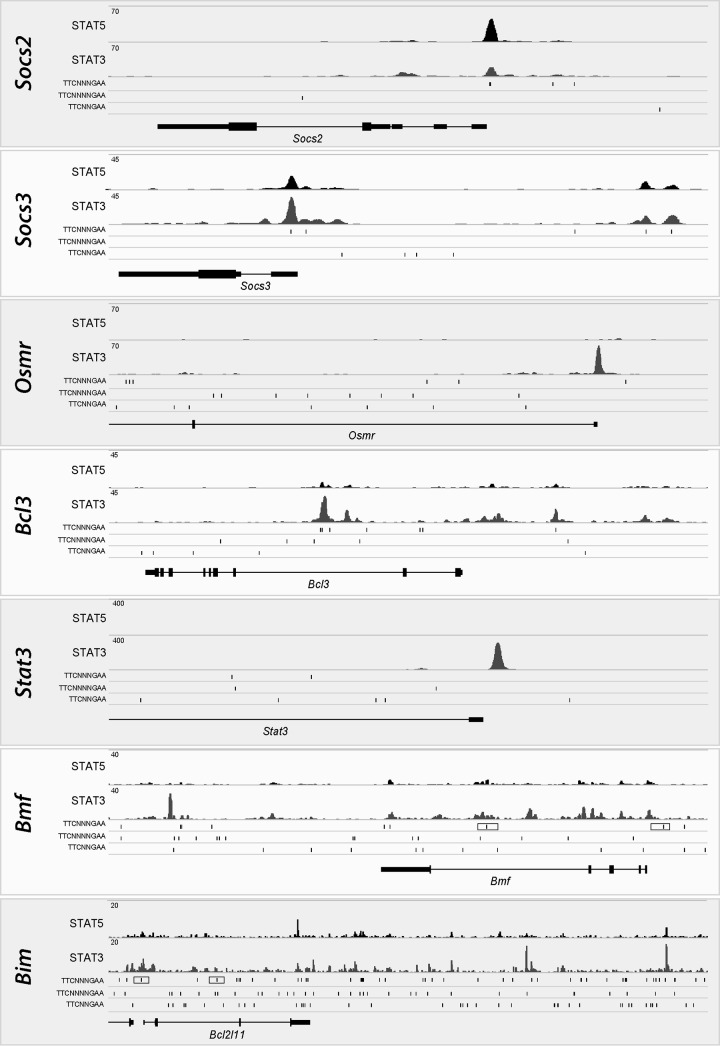
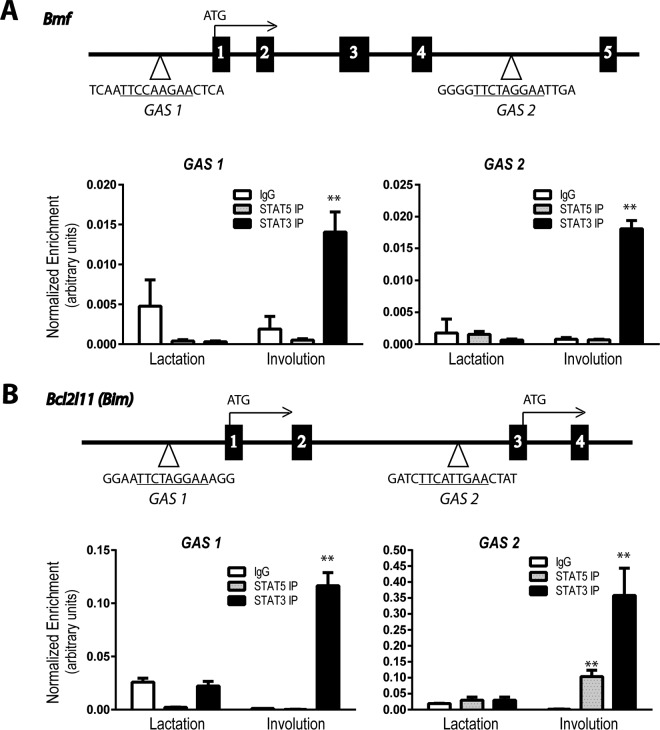
The comparative analysis of the mRNA profiles of lactating and involuting tissues revealed that several members of the Bcl2 gene family exhibited substantial changes in their transcriptional activation. We observed not only a more than 2-fold upregulation of Bax, Bcl-x (Bcl2l1), Bmf, Bim (Bcl2l11), and Bid (Fig. 4) but also a 3- and 7-fold upregulation of Bbc3 (Puma) and Bik, respectively, as well as a more than 2-fold downregulation of the prosurvival factor Bcl-w (Bcl2l2) during involution (not shown). The most significantly downregulated member of the Bcl2 family in the JAK1-knockout mice compared to wild-type controls was Bmf. The level of Bim was also consistently decreased, but its transcripts were about 4-fold less abundant than those of Bmf. To establish whether these two proapoptotic genes are directly regulated by STAT3 or STAT5, we performed a preliminary analysis of the ChIP-Seq data set for mammary tissues obtained from females during midpregnancy, a time that represents a developmental window when all STAT proteins are active in the gland in specific epithelial subtypes (Fig. 7). Socs2 and Socs3 served as controls for the predominant binding of STAT5 or STAT3. STAT3 was exclusively present within the Osmr, Bcl3, and Stat3 loci. More importantly, we observed a higher level of enrichment of STAT3 in particular regions in close proximity to Bmf and Bim. Next, we performed ChIP in combination with quantitative reverse transcription-PCR (qRT-PCR) to assess whether STAT3 binding to canonical gamma interferon activation sites (GAS) increases during involution, when this member of the STAT family exhibits its highest level of activity during development (Fig. 8). We found that STAT3 occupied GAS sequences that are located near the transcriptional start sites of Bmf and Bim, and there was a significant increase in STAT3 binding in postlactational mammary glands. Collectively, the ChIP-Seq data and qRT-PCR results support the notion that active STAT3 might be directly involved in the transcriptional activation of the proapoptotic genes Bmf and Bim during involution.
FIG 7.
The DNA binding of the transcription factors STAT3 and STAT5 varies among downstream target genes. The individual panels show the histograms of ChIP-Seq data sets for selected STAT3 and STAT5 target genes, illustrating the binding of these transcription factors to these loci in the mammary glands of mice during midpregnancy. Socs2 and Socs3 are controls that preferentially bind STAT5 or STAT3; also note that STAT3 binds exclusively to sites near Osmr, Bcl3, and Stat3, which are known targets of STAT3 signaling. Bmf and Bim are newly identified targets of JAK1 that exhibit an enrichment of STAT3 binding to regions in close proximity to these loci (boxed canonical STAT recognition sites [TTCN3GAA] refer to those shown in Fig. 8).
FIG 8.
The binding of STAT3 to STAT recognition sites located near the transcriptional start sites of Bmf and Bim increases during involution. Schematic outline of the location of canonical STAT binding sites (i.e., gamma interferon activation sites [GAS]; TTCN3GAA) near or within the genes encoding the BH3-only proteins BMF (A) and BIM (B). Boxes represent coding exons of the corresponding genes, Bmf and Bim; the numbers indicate the numbers of the exons. The bar graphs show the results from quantitative real-time PCR analyses of DNA isolated by ChIP from lactating (day 7) or involuting (day 2) mammary tissues with antibodies against STAT5, STAT3, or an IgG control. The specific primer sets that were used to amplify the regions surrounding the STAT-binding sites are specific for sequences located within the 5′ and intragenic regions of Bmf and Bcl2l11 (Bim). Values were normalized against those for background DNA by using primer sets specific for nonconsensus binding sites. Bars represent means and SDs. **, P < 0.01.
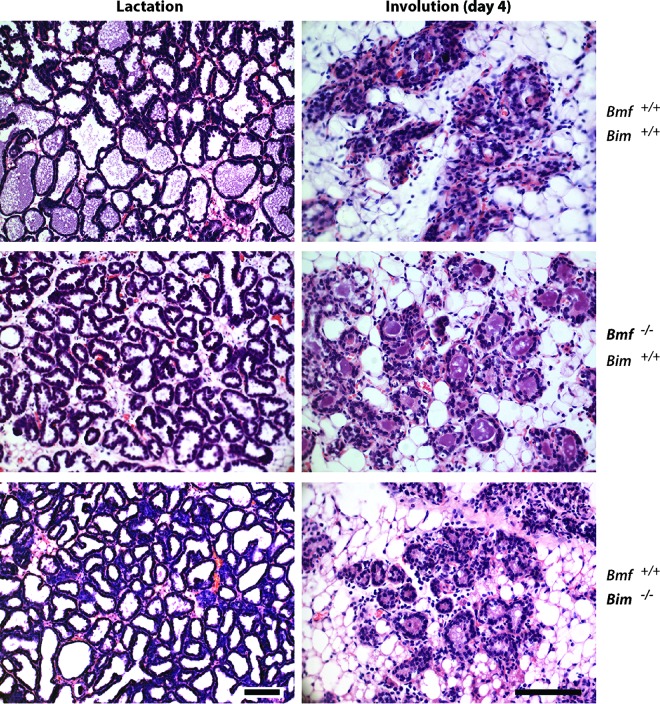
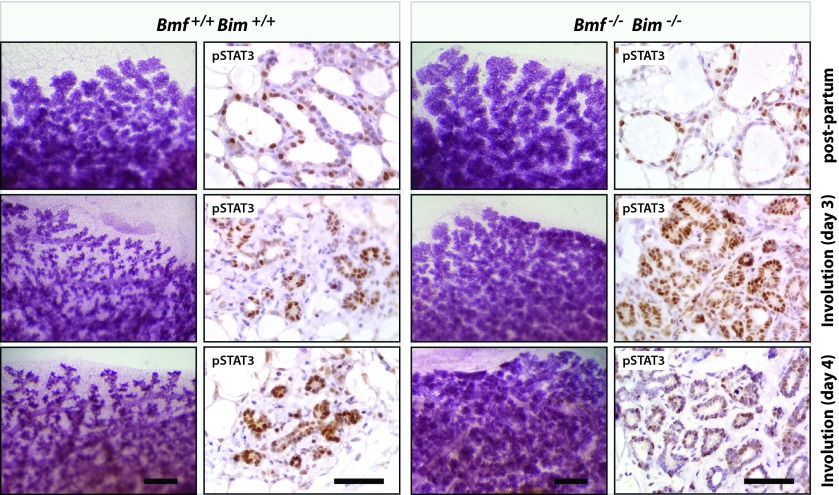
As a putative downstream effector of JAK1/STAT3 signaling, we anticipated that a genomic deletion of the Bmf gene alone or in combination with a loss of both alleles of Bim phenocopies the delay in postlactational remodeling that we observed in the JAK1 conditional knockout mice. To address this hypothesis, we first examined the histology of lactating and involuting mammary glands from BMF- and BIM-single-knockout mice. While the lactating mammary glands of both mutant mouse models were indistinguishable from the lactating mammary glands of the wild-type controls, we observed a delay in postlactational remodeling in the absence of either BMF or BIM (Fig. 9). Although the phenotypic abnormalities were slightly more pronounced in the BMF-deficient mice, these findings suggest that both members of the BH3-only family are regulators of the initiation of apoptosis of differentiated epithelial cells. Since both proteins were synchronously upregulated in a JAK1-dependent manner, we assumed that they might act synergistically during involution. To address this notion, we transplanted mammary epithelium from BMF/BIM-double-knockout mice into the cleared fat pad of wild-type recipient females. Following engraftment and ductal outgrowth, the recipient mice were bred at 12 weeks posttransplantation and their mammary glands were examined immediately following the birth of the offspring (Fig. 10, postpartum). We did not observe any morphological differences in pregnancy-induced alveologenesis between the double-knockout and wild-type control transplants. Since postpartum mammary transplants swiftly activate STAT3 and initiate the involution process due to milk stasis, we removed the newborn pups and analyzed the mammary glands at 2 to 5 days postweaning. We found that mammary gland remodeling was dramatically delayed in the absence of BMF and BIM compared to that in mice wild-type control transplants (Fig. 10, involution), and a 4- or 5-day involuting mammary gland of the double-knockout transplants was phenotypically similar to that in JAK1-deficient mice. The BMF- and BIM-single-knockout mice as well as the transplants that were doubly deficient in both BH3-only proteins exhibited a normal activation of STAT3 that was indistinguishable from that in the wild-type controls (Fig. 10). The collective findings obtained with each of the single-knockout mice and the double-knockout transplant model suggest that these proapoptotic factors genetically function downstream of JAK1 and STAT3. Since the mammary stroma and systemic environment in the recipient mice carrying the BMF/BIM-null epithelium are wild type, it is evident that these cell death-initiating proteins act in a cell-autonomous manner. The findings from the transplant model therefore correspond closely to the epithelium-specific conditional deletion of JAK1 or STAT3.
FIG 9.
Postlactational mammary gland remodeling is delayed in mice that lack BMF or BIM. H&E-stained histological sections of lactating and involuting mammary glands from BMF-knockout (Bmf−/− Bim+/+) and BIM-knockout (Bmf+/+ Bim−/−) females and their wild-type controls (Bmf+/+ Bim+/+) are shown. Bars, 100 μm.
FIG 10.
Mice with an epithelium-specific double knockout of BMF and BIM in the mammary gland exhibit a dramatic impairment in postlactational remodeling. Whole mounts of mammary glands and histological sections from wild-type recipient mice that were transplanted with mammary epithelium from BMF/BIM-double-knockout (Bmf−/− Bim−/−) or wild-type control (Bmf+/+ Bim+/+) donor females are shown. Tissues were collected within hours following the birth of the offspring (postpartum) as well as at 3 and 4 days after weaning (involution). (Left) Carmine alum-stained mammary gland whole mounts. Bars, 500 μm. (Right) Immunohistochemical staining of tyrosine-phosphorylated STAT3. Slides were counterstained with hematoxylin. Bars, 50 μm.
DISCUSSION
Biologically essential functions of JAK1 are restricted to postlactational mammary gland remodeling.
The molecular characterization of STAT activity in the JAK1 conditional knockout mice showed that this member of the Janus kinase family is essential for the cytokine-induced tyrosine phosphorylation of STAT1, STAT3, and STAT6 in cultured embryonic fibroblasts and mammary epithelial cells in vivo. Despite its critical role for the activation of three STAT proteins in the mammary gland, the biologically relevant function of JAK1 for the development of this organ is restricted to the remodeling of the epithelial compartment during postlactational involution. The phenotypic abnormalities of mice with mammary gland-specific JAK1 knockout closely resemble the defects in programmed cell death that were observed in STAT3-deficient females (43) and genetic models that lack gp130 or IL-6 (7, 41). The loss of the cytokine-mediated activation of STAT1 and STAT6 in the JAK1 conditional knockout mice seems to play a subordinate role that did not have any major impact on the development of the gland. It was previously reported that the expression and activation of STAT1 in the epithelium are dispensable for ductal elongation and alveologenesis (12), and recent work by Chen et al. (44) suggested that optimal ductal branching morphogenesis depends on the presence of active STAT1 in the stroma. In contrast to STAT1-deficient females, STAT6 conventional knockout mice exhibited a delay in the development of alveolar cells during early pregnancy (19). These phenotypic abnormalities were relatively subtle at the later stages of gestation, and STAT6-deficient females were able to nurse their offspring. A closer examination of the immunostaining results reported by Khaled et al. (19) showed that nuclear STAT6 was present in the epithelium as well as some adjacent stromal cells. It is therefore possible that activation of STAT6 in the stroma exerts paracrine effects that control the pregnancy-induced proliferation of mammary epithelial cells. This might explain why JAK1 conditional knockout mice that lack an IL-4-mediated activation of STAT6 in the epithelium were not phenotypically similar to females that systemically lack STAT6 expression.
JAK1 and JAK2 have nonredundant roles in the activation of particular STAT proteins and mammary gland development.
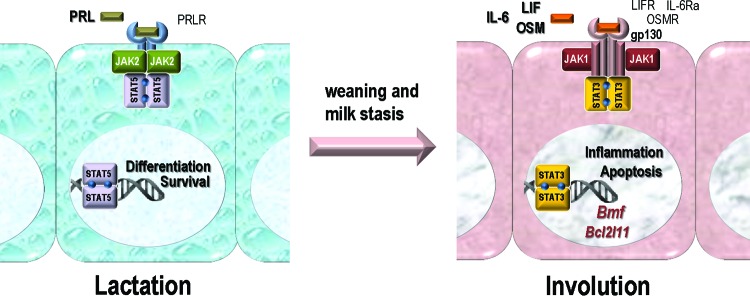
Our previous work on JAK2 conditional knockout mice revealed a remarkable specificity of this particular member of the Janus kinase family for the activation of STAT5 in response to PRL signaling (25). Despite the fact that the PRL-mediated activation of SRC and mitogen-activated protein kinases was not affected by the loss of JAK2 (35), neither these PRL-induced downstream mediators nor other Janus or receptor tyrosine kinases were able to compensate for the loss of JAK2 and to restore the development and function of milk-producing alveolar cells. The examination of JAK1 conditional knockout mice in this study demonstrated very clearly that the activation of STAT5 was not affected in the absence of JAK1. Although JAK2 was functional in the differentiated epithelium throughout lactation, it was unable to effectively compensate for the loss of JAK1 to activate STAT3 immediately after weaning. The collective results of our studies suggest that neither JAK1 nor JAK2 alone is sufficient to activate STAT5 or STAT3 in the same cell type in response to a swift change in ligand-receptor signaling complexes prior to and within hours of involution (Fig. 11). It is therefore evident that both Janus kinases have nonredundant roles in the activation of particular STAT proteins, and their biologically important functions do not overlap during normal mammary gland development. These are two new and important findings because previous studies have suggested that JAK1 and JAK2 are capable of activating STAT3 in response to inflammatory cytokines in diverse cell culture models (reviewed by Kisseleva et al. [20]). Although it was recognized nearly 2 decades ago that JAK1 plays a role in IL-6 signaling (21, 45, 46), more recent work has singled out JAK2 as the master regulator for STAT3 activation in breast cancer and other malignancies (47–49). This paradigm is likely the result of the fact that pharmacological agents labeled “JAK2 inhibitors” were being utilized in most of the recently published experiments. However, these drugs are known to block several JAKs, sometimes with identical efficacies (50). A striking argument against the role of JAK2 as the master regulator of STAT3 comes from the analysis of our JAK2 conditional knockout model. Specifically, the deletion of Jak2 after neoplastic transformation had no effect on the constitutive tyrosine phosphorylation of STAT3 in different mammary tumor models (51, 52). The activation of STAT3 in malignant cells has been shown to depend on an IL-6 autocrine signaling loop that is triggered by receptor tyrosine kinases (47–49, 53, 54). If that is correct, our findings that JAK1 and not JAK2 has a central role in gp130-mediated receptor signaling complexes may have significant implications beyond normal development for the treatment of breast cancer and other malignancies.
FIG 11.
JAK1 and JAK2 have nonredundant biological functions and discrete roles in the activation of STAT proteins in mammary epithelial cells during the switch from lactation to postlactational mammary gland involution. PRLR, PRL receptor; LIFR, LIF receptor; IL-6Ra, IL-6 receptor.
JAK1 is an essential link between IL-6-class inflammatory cytokines and the cell death machinery in functionally differentiated mammary epithelial cells.
The collective findings of this study demonstrate that the level of JAK1 increases at the cessation of lactation. The loss of JAK1 specifically in the mammary epithelium resulted in impaired involution, and this conditional knockout model closely phenocopies the major developmental abnormalities that were previously observed in STAT3-, gp130-, or IL-6-deficient mice (7, 41, 43). We further demonstrated that the deletion of JAK1 uncouples IL-6-class ligands from their downstream effector, STAT3. Consequently, this leads to the decreased expression of known STAT3 target genes that are associated with the acute-phase response, inflammation, and wound healing (42). The genome-wide expression analysis of JAK1-knockout mammary glands and control tissues revealed a number of novel target genes that were upregulated during involution in a JAK1-dependent manner. These included Runx1 and c-Fos, which was identified to be one of the first cytokine-induced proto-oncogenes (55) that can mediate tissue remodeling through expression of metalloproteinases (56, 57). Other potential targets of JAK1 that were previously linked to cell death were the death receptor 6 gene (Tnfrsf21) and the tumor susceptibility gene Tpl2 (Map3k8), whose biological functions in the mammary gland have not yet been determined. Among known factors that control canonical mitochondrion-associated apoptosis, we identified the genes encoding the BH3-only proteins BMF and BIM (Bcl2l11) to be targets of JAK1 signaling. The results of analysis of involuting mammary glands of single-knockout mice confirm the findings presented in a recent report that showed that both proapoptotic proteins contribute to the initiation of apoptosis (58). More importantly, using a BMF/BIM-double-knockout transplant model, we now provide clear evidence that both of these JAK1 targets act synergistically in an epithelium-specific manner. The combined loss of these BH3-only proteins closely resembles the phenotypic abnormalities that we observed in JAK1 conditional knockout females. According to the ChIP-Seq and ChIP/qRT-PCR results, STAT3 is bound to the chromatin in proximity to the Bmf and Bim loci, and its association with GAS sequences near transcriptional start sites was increased during involution. This finding suggests that STAT3 might be directly involved in the transcription of these genes, and it is therefore puzzling that STAT3-knockout mice did not show the same deregulated expression in Bmf and Bim that was observed in the JAK1-knockout mice (58). One possible explanation is that other transcription factors whose expression levels are controlled by JAK1 (e.g., Runx1) also contribute to the expression of Bmf and Bim. Whether STAT5 acts as a suppressor of Bim transcription, as suggested previously (58), warrants further validation using appropriate genetic in vivo models. The fact that the nuclear accumulation of pSTAT3 is not altered by the loss of BMF and BIM still supports the notion that these BH3-only proapoptotic factors function genetically downstream of the IL-6/JAK1/STAT3 axis. This seems to be likely, since an alternative model that may favor a major STAT3-independent function for BMF and BIM in mammary gland remodeling would require further clarification of how STAT3 might drive other cell death mechanisms, such as lysosome-mediated cell death (59), that are suggested to function without involvement of the classical apoptotic pathway. STAT3 was strongly nuclear in the recipients of BMF/BIM-double-knockout epithelial transplants postpartum, and it is therefore evident that the proposed alternative functions of active STAT3 were insufficient to override the inhibition of apoptosis that was caused by the impairment of the mitochondrion-associated cell death program. From the generation and analysis of JAK1 conditional knockout mice to the identification of downstream targets and their validation on the molecular as well as phenotypic level using a double-knockout transplant model, the collective results of this comprehensive study suggest that JAK1 is essential for IL-6-class inflammatory cytokine signaling and mammary gland involution. This member of the Janus kinase family is therefore essential for the coupling of extracellular cytokine signals to the cell death machinery in the functionally differentiated mammary epithelium.
ACKNOWLEDGMENTS
We thank Harold Smith, head of the NIDDK Genomics Core Facility, for his assistance with the next-generation sequencing (NGS) service. We are grateful to Karen K. Dulany for the preparation of histological sections and the members of the UNMC Mouse Genome Engineering Core Facility, in particular C. B. Gurumurthy, for assistance with the generation of JAK1-knockout mice.
This work was supported, in part, by Public Health Service grant CA117930 (to K.-U.W.). Additional financial support provided to K.S. and K.-U.W. by the Nebraska Cancer and Smoking Disease Research Program (NE DHHS LB506 2015-47 and 2016-54) was imperative to finance the maintenance of the JAK1 mutant mice. A.V. and F.S. were supported by the Austrian Science Fund, FWF (W1101), and the DOC-Fellowship program of the Austrian Academy of Science (ÖAW). The RNA-Seq and ChIP-Seq analyses were funded, in part, by the IRP of NIDDK, NIH (to L.H., K.H.Y., and H.Y.S.). B.L.W. received a graduate fellowship through the UNMC Cancer Research Training Program (CA009476). N.R. and B.L.W. were awarded a Program of Excellence graduate assistantship from the UNMC Graduate Studies Office.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
REFERENCES
- 1.Hennighausen L, Robinson GW. 2005. Information networks in the mammary gland. Nat Rev Mol Cell Biol 6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 2.Feng Y, Manka D, Wagner KU, Khan SA. 2007. Estrogen receptor-alpha expression in the mammary epithelium is required for ductal and alveolar morphogenesis in mice. Proc Natl Acad Sci U S A 104:14718–14723. doi: 10.1073/pnas.0706933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mallepell S, Krust A, Chambon P, Brisken C. 2006. Paracrine signaling through the epithelial estrogen receptor alpha is required for proliferation and morphogenesis in the mammary gland. Proc Natl Acad Sci U S A 103:2196–2201. doi: 10.1073/pnas.0510974103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Humphreys RC, Lydon J, O'Malley BW, Rosen JM. 1997. Mammary gland development is mediated by both stromal and epithelial progesterone receptors. Mol Endocrinol 11:801–811. doi: 10.1210/mend.11.6.9891. [DOI] [PubMed] [Google Scholar]
- 5.Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, Kelly PA. 1997. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev 11:167–178. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- 6.Horseman ND, Zhao W, Montecino-Rodriguez E, Tanaka M, Nakashima K, Engle SJ, Smith F, Markoff E, Dorshkind K. 1997. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. EMBO J 16:6926–6935. doi: 10.1093/emboj/16.23.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao L, Melenhorst JJ, Hennighausen L. 2002. Loss of interleukin 6 results in delayed mammary gland involution: a possible role for mitogen-activated protein kinase and not signal transducer and activator of transcription 3. Mol Endocrinol 16:2902–2912. doi: 10.1210/me.2001-0330. [DOI] [PubMed] [Google Scholar]
- 8.Kritikou EA, Sharkey A, Abell K, Came PJ, Anderson E, Clarkson RW, Watson CJ. 2003. A dual, non-redundant, role for LIF as a regulator of development and STAT3-mediated cell death in mammary gland. Development 130:3459–3468. doi: 10.1242/dev.00578. [DOI] [PubMed] [Google Scholar]
- 9.Tiffen PG, Omidvar N, Marquez-Almuina N, Croston D, Watson CJ, Clarkson RW. 2008. A dual role for oncostatin M signaling in the differentiation and death of mammary epithelial cells in vivo. Mol Endocrinol 22:2677–2688. doi: 10.1210/me.2008-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson CJ, Neoh K. 2008. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol 19:401–406. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Wagner KU, Schmidt JW. 2011. The two faces of Janus kinases and their respective STATs in mammary gland development and cancer. J Carcinog 10:32. doi: 10.4103/1477-3163.90677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klover PJ, Muller WJ, Robinson GW, Pfeiffer RM, Yamaji D, Hennighausen L. 2010. Loss of STAT1 from mouse mammary epithelium results in an increased Neu-induced tumor burden. Neoplasia 12:899–905. doi: 10.1593/neo.10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Robinson GW, Hennighausen L. 1996. Activation of Stat5a and Stat5b by tyrosine phosphorylation is tightly linked to mammary gland differentiation. Mol Endocrinol 10:1496–1506. [DOI] [PubMed] [Google Scholar]
- 14.Wakao H, Gouilleux F, Groner B. 1994. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J 13:2182–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burdon TG, Maitland KA, Clark AJ, Wallace R, Watson CJ. 1994. Regulation of the sheep beta-lactoglobulin gene by lactogenic hormones is mediated by a transcription factor that binds an interferon-gamma activation site-related element. Mol Endocrinol 8:1528–1536. doi: 10.1210/mend.8.11.7877621. [DOI] [PubMed] [Google Scholar]
- 16.Mukhopadhyay SS, Wyszomierski SL, Gronostajski RM, Rosen JM. 2001. Differential interactions of specific nuclear factor I isoforms with the glucocorticoid receptor and STAT5 in the cooperative regulation of WAP gene transcription. Mol Cell Biol 21:6859–6869. doi: 10.1128/MCB.21.20.6859-6869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu X, Gallego MI, Smith GH, Robinson GW, Hennighausen L. 1998. Functional release of Stat5a-null mammary tissue through the activation of compensating signals including Stat5b. Cell Growth Differ 9:795–803. [PubMed] [Google Scholar]
- 18.Cui Y, Riedlinger G, Miyoshi K, Tang W, Li C, Deng CX, Robinson GW, Hennighausen L. 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol 24:8037–8047. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khaled WT, Read EK, Nicholson SE, Baxter FO, Brennan AJ, Came PJ, Sprigg N, McKenzie AN, Watson CJ. 2007. The IL-4/IL-13/Stat6 signalling pathway promotes luminal mammary epithelial cell development. Development 134:2739–2750. doi: 10.1242/dev.003194. [DOI] [PubMed] [Google Scholar]
- 20.Kisseleva T, Bhattacharya S, Braunstein J, Schindler CW. 2002. Signaling through the JAK/STAT pathway, recent advances and future challenges. Gene 285:1–24. doi: 10.1016/S0378-1119(02)00398-0. [DOI] [PubMed] [Google Scholar]
- 21.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM Jr, Schreiber RD. 1998. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell 93:373–383. doi: 10.1016/S0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 22.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. 1998. Jak2 is essential for signaling through a variety of cytokine receptors. Cell 93:385–395. doi: 10.1016/S0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 23.Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. 1998. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell 93:397–409. doi: 10.1016/S0092-8674(00)81168-X. [DOI] [PubMed] [Google Scholar]
- 24.Shillingford JM, Miyoshi K, Robinson GW, Grimm SL, Rosen JM, Neubauer H, Pfeffer K, Hennighausen L. 2002. Jak2 is an essential tyrosine kinase involved in pregnancy-mediated development of mammary secretory epithelium. Mol Endocrinol 16:563–570. doi: 10.1210/mend.16.3.0805. [DOI] [PubMed] [Google Scholar]
- 25.Wagner KU, Krempler A, Triplett AA, Qi Y, George NM, Zhu J, Rui H. 2004. Impaired alveologenesis and maintenance of secretory mammary epithelial cells in Jak2 conditional knockout mice. Mol Cell Biol 24:5510–5520. doi: 10.1128/MCB.24.12.5510-5520.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakamoto K, Gurumurthy CB, Wagner KU. 2014. Generation of conditional knockout mice. Methods Mol Biol 1194:21–35. doi: 10.1007/978-1-4939-1215-5_2. [DOI] [PubMed] [Google Scholar]
- 27.Wagner KU, Wall RJ, St-Onge L, Gruss P, Wynshaw-Boris A, Garrett L, Li M, Furth PA, Hennighausen L. 1997. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res 25:4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagner KU, McAllister K, Ward T, Davis B, Wiseman R, Hennighausen L. 2001. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res 10:545–553. doi: 10.1023/A:1013063514007. [DOI] [PubMed] [Google Scholar]
- 29.Kawamoto S, Niwa H, Tashiro F, Sano S, Kondoh G, Takeda J, Tabayashi K, Miyazaki J. 2000. A novel reporter mouse strain that expresses enhanced green fluorescent protein upon Cre-mediated recombination. FEBS Lett 470:263–268. doi: 10.1016/S0014-5793(00)01338-7. [DOI] [PubMed] [Google Scholar]
- 30.Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O'Reilly L, Strasser A, Villunger A. 2008. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates gamma irradiation-induced thymic lymphoma development. J Exp Med 205:641–655. doi: 10.1084/jem.20071658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F, Adams JM, Strasser A. 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 32.Labi V, Woess C, Tuzlak S, Erlacher M, Bouillet P, Strasser A, Tzankov A, Villunger A. 2014. Deregulated cell death and lymphocyte homeostasis cause premature lethality in mice lacking the BH3-only proteins Bim and Bmf. Blood 123:2652–2662. doi: 10.1182/blood-2013-11-537217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 34.Krempler A, Qi Y, Triplett AA, Zhu J, Rui H, Wagner KU. 2004. Generation of a conditional knockout allele for the Janus kinase 2 (Jak2) gene in mice. Genesis 40:52–57. doi: 10.1002/gene.20063. [DOI] [PubMed] [Google Scholar]
- 35.Sakamoto K, Creamer BA, Triplett AA, Wagner KU. 2007. The Janus kinase 2 is required for expression and nuclear accumulation of cyclin D1 in proliferating mammary epithelial cells. Mol Endocrinol 21:1877–1892. doi: 10.1210/me.2006-0316. [DOI] [PubMed] [Google Scholar]
- 36.Wagner KU, Young WS, Liu X, Ginns EI, Li M, Furth PA, Hennighausen L. 1997. Oxytocin and milk removal are required for post-partum mammary-gland development. Genes Funct 1:233–244. doi: 10.1046/j.1365-4624.1997.00024.x. [DOI] [PubMed] [Google Scholar]
- 37.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamaji D, Kang K, Robinson GW, Hennighausen L. 2013. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res 41:1622–1636. doi: 10.1093/nar/gks1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schmidt JW, Wehde BL, Sakamoto K, Triplett AA, Anderson SM, Tsichlis PN, Leone G, Wagner KU. 2014. Stat5 regulates the phosphatidylinositol 3-kinase/Akt1 pathway during mammary gland development and tumorigenesis. Mol Cell Biol 34:1363–1377. doi: 10.1128/MCB.01220-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Decker T, Kovarik P. 2000. Serine phosphorylation of STATs. Oncogene 19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- 41.Zhao L, Hart S, Cheng J, Melenhorst JJ, Bierie B, Ernst M, Stewart C, Schaper F, Heinrich PC, Ullrich A, Robinson GW, Hennighausen L. 2004. Mammary gland remodeling depends on gp130 signaling through Stat3 and MAPK. J Biol Chem 279:44093–44100. doi: 10.1074/jbc.M313131200. [DOI] [PubMed] [Google Scholar]
- 42.Hughes K, Wickenden JA, Allen JE, Watson CJ. 2012. Conditional deletion of Stat3 in mammary epithelium impairs the acute phase response and modulates immune cell numbers during post-lactational regression. J Pathol 227:106–117. doi: 10.1002/path.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chapman RS, Lourenco PC, Tonner E, Flint DJ, Selbert S, Takeda K, Akira S, Clarke AR, Watson CJ. 1999. Suppression of epithelial apoptosis and delayed mammary gland involution in mice with a conditional knockout of Stat3. Genes Dev 13:2604–2616. doi: 10.1101/gad.13.19.2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen JQ, Mori H, Cardiff RD, Trott JF, Hovey RC, Hubbard NE, Engelberg JA, Tepper CG, Willis BJ, Khan IH, Ravindran RK, Chan SR, Schreiber RD, Borowsky AD. 2015. Abnormal mammary development in 129:STAT1-null mice is stroma-dependent. PLoS One 10:e0129895. doi: 10.1371/journal.pone.0129895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guschin D, Rogers N, Briscoe J, Witthuhn B, Watling D, Horn F, Pellegrini S, Yasukawa K, Heinrich P, Stark GR, Ihle GR, Kerr IM. 1995. A major role for the protein tyrosine kinase JAK1 in the JAK/STAT signal transduction pathway in response to interleukin-6. EMBO J 14:1421–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutticken C, Wegenka UM, Yuan J, Buschmann J, Schindler C, Ziemiecki A, Harpur AG, Wilks AF, Yasukawa K, Taga T, Kishimoto T. 1994. Association of transcription factor APRF and protein kinase Jak1 with the interleukin-6 signal transducer gp130. Science 263:89–92. doi: 10.1126/science.8272872. [DOI] [PubMed] [Google Scholar]
- 47.Corcoran RB, Contino G, Deshpande V, Tzatsos A, Conrad C, Benes CH, Levy DE, Settleman J, Engelman JA, Bardeesy N. 2011. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res 71:5020–5029. doi: 10.1158/0008-5472.CAN-11-0908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marotta LL, Almendro V, Marusyk A, Shipitsin M, Schemme J, Walker SR, Bloushtain-Qimron N, Kim JJ, Choudhury SA, Maruyama R, Wu Z, Gonen M, Mulvey LA, Bessarabova MO, Huh SJ, Silver SJ, Kim SY, Park SY, Lee HE, Anderson KS, Richardson AL, Nikolskaya T, Nikolsky Y, Liu XS, Root DE, Hahn WC, Frank DA, Polyak K. 2011. The JAK2/STAT3 signaling pathway is required for growth of CD44(+)CD24(−) stem cell-like breast cancer cells in human tumors. J Clin Invest 121:2723–2735. doi: 10.1172/JCI44745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abubaker K, Luwor RB, Zhu H, McNally O, Quinn MA, Burns CJ, Thompson EW, Findlay JK, Ahmed N. 2014. Inhibition of the JAK2/STAT3 pathway in ovarian cancer results in the loss of cancer stem cell-like characteristics and a reduced tumor burden. BMC Cancer 14:317. doi: 10.1186/1471-2407-14-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warsch W, Walz C, Sexl V. 2013. JAK of all trades: JAK2-STAT5 as novel therapeutic targets in BCR-ABL1+ chronic myeloid leukemia. Blood 122:2167–2175. doi: 10.1182/blood-2013-02-485573. [DOI] [PubMed] [Google Scholar]
- 51.Sakamoto K, Triplett AA, Schuler LA, Wagner KU. 2010. Janus kinase 2 is required for the initiation but not maintenance of prolactin-induced mammary cancer. Oncogene 29:5359–5369. doi: 10.1038/onc.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sakamoto K, Lin WC, Triplett AA, Wagner KU. 2009. Targeting Janus kinase 2 in Her2/neu-expressing mammary cancer: implications for cancer prevention and therapy. Cancer Res 69:6642–6650. doi: 10.1158/0008-5472.CAN-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berishaj M, Gao SP, Ahmed S, Leslie K, Al Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. 2007. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res 9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, Bromberg JF. 2007. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest 117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yang E, Lerner L, Besser D, Darnell JE Jr. 2003. Independent and cooperative activation of chromosomal c-fos promoter by STAT3. J Biol Chem 278:15794–15799. doi: 10.1074/jbc.M213073200. [DOI] [PubMed] [Google Scholar]
- 56.Han Z, Boyle DL, Chang L, Bennett B, Karin M, Yang L, Manning AM, Firestein GS. 2001. c-Jun N-terminal kinase is required for metalloproteinase expression and joint destruction in inflammatory arthritis. J Clin Invest 108:73–81. doi: 10.1172/JCI12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vincenti MP, Brinckerhoff CE. 2002. Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors. Arthritis Res 4:157–164. doi: 10.1186/ar401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schuler F, Baumgartner F, Klepsch V, Chamson M, Muller-Holzner E, Watson CJ, Oh S, Hennighausen L, Tymoszuk P, Doppler W, Villunger A. 2016. The BH3-only protein BIM contributes to late-stage involution in the mouse mammary gland. Cell Death Differ 23:41–51. doi: 10.1038/cdd.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kreuzaler PA, Staniszewska AD, Li W, Omidvar N, Kedjouar B, Turkson J, Poli V, Flavell RA, Clarkson RW, Watson CJ. 2011. Stat3 controls lysosomal-mediated cell death in vivo. Nat Cell Biol 13:303–309. doi: 10.1038/ncb2171. [DOI] [PubMed] [Google Scholar]