Abstract
Lysine selectivity is of critical importance during polyubiquitin chain formation because the identity of the lysine controls the biological outcome. Ubiquitins are covalently linked in polyubiquitin chains through one of seven lysine residues on its surface and the C terminus of adjacent protomers. Lys 48-linked polyubiquitin chains signal for protein degradation; however, the structural basis for Lys 48 selectivity remains largely unknown. The ubiquitin-conjugating enzyme Ube2R1/2 has exquisite specificity for Lys 48, and computational docking of Ube2R1/2 and ubiquitin predicts that Lys 48 is guided to the active site through a key electrostatic interaction between Arg 54 on ubiquitin and Asp 143 on Ube2R1/2. The validity of this interaction was confirmed through biochemical experiments. Since structural examples involving Arg 54 in protein-ubiquitin complexes are exceedingly rare, these results provide additional insight into how ubiquitin-protein complexes can be stabilized. We discuss how these findings relate to how other ubiquitin-conjugating enzymes direct the lysine specificity of polyubiquitin chains.
INTRODUCTION
The assembly of polyubiquitin (poly-Ub) chains onto proteins is a critical signaling process that is required for eukaryotic cellular homeostasis. Polyubiquitin chain synthesis is initiated when ubiquitin, a highly conserved 76-amino-acid protein, is activated by ubiquitin-activating enzyme (E1). In an ATP-dependent process, E1 forms a high-energy thioester bond, denoted with “∼,” to the C terminus of ubiquitin. Next, the ubiquitin is transferred from E1 to a ubiquitin-conjugating enzyme (E2). Finally, a ubiquitin ligase (E3) recruits both the E2∼ubiquitin complex and a protein substrate. E3 enzymes in the RING family (1) stimulate the transfer of ubiquitin from the E2 to a lysine residue on the substrate (2–9), resulting in the formation of an isopeptide bond between ubiquitin's C terminus and the amino group on lysine side chains.
During the process of RING class E3-catalyzed ubiquitylation, ubiquitin serves two major roles: it serves as a donor when an E2 transfers the ubiquitin from its active site to a protein substrate bound to an E3, and it serves as an acceptor when ubiquitin conjugated to substrate attacks an E2∼donor ubiquitin. Ubiquitin contains seven lysine residues located on its surface, and consecutive ubiquitins in a polyubiquitin chain are either covalently linked between one of these lysines or the N-terminal amino group on an acceptor ubiquitin and the C terminus of the donor. The identity of the site that tethers ubiquitins in the chain is of critical importance since different linkage types adopt unique conformations that signal alternative biological outcomes (10). For instance, a protein modified with Lys 48-specific polyubiquitin chains is targeted to the 26S proteasome for its degradation (11), whereas Lys 63-specific chains can promote intracellular trafficking to endocytic vesicles or protein-protein interactions (12, 13).
Lys 48-linked chains are the predominant chain type in human cells that promote protein degradation (14), and the human E2s Ube2K, Ube2G1/2, and Ube2R1/2 (which has historically been referred to as Cdc34) are known to assemble polyubiquitin chains onto protein substrates with Lys 48 specificity. Ubc1, the Saccharomyces cerevisiae ortholog of Ube2K, has three critical residues, Thr 84, Gln 122, and Ala 124, that were identified on the E2 surface that interact with an acceptor ubiquitin to direct Lys 48 to the Ubc1 active site (15). More recently, it was found that Ube2K forms an electrostatic interaction between Glu 51 on acceptor ubiquitin and a Lys residue on the E2 surface (16). Ube2G2 has been shown to form homodimers that may help direct Lys 48 to its active site (17). Ube2R1/2 is a critical E2 that functions with the cullin-RING ubiquitin ligases (1, 18), and, together, these enzymes may be responsible for 20% of all proteasome-dependent degradation in human cells (19). Ube2R1/2 contains an atypical insertion distal to its active site that contains several conserved acidic residues (20–25), and this acidic loop has also been shown to have an important role in Lys 48 selectivity (26, 27). More recent work has identified a loop on acceptor ubiquitin that contains residues that interact with the E2 in order to help place Lys 48 in the E2 active site (26). Despite these advances, a detailed molecular characterization showing how an E2 achieves Lys 48 specificity during polyubiquitin chain synthesis is lacking.
Here we show that human Ube2R1/2 forms a critical salt bridge interaction between a conserved aspartic acid residue on Ube2R1/2 and acceptor ubiquitin residue Arg 54 and that perturbation of this interaction leads to the severe loss of UbeR2 activity. Our results also provide new insight into how the Ube2R1/2 acidic loop may participate in catalysis, specifically through the formation of stabilizing interactions with the E3 as well as the donor ubiquitin.
MATERIALS AND METHODS
Molecular modeling. (i) Building initial models.
All of the molecular modeling steps were carried out using protocols in the Rosetta molecular modeling suite (28). To construct initial Ube2R1-Rbx1 models, Rbx1 (Protein Data Bank identifier [PDB ID] 2LGV [29]) and Ube2R1 (PDB ID 4MDK [30]) were aligned to the respective components from the Ube2G2/Rnf45 cocrystal structure (PDB ID 2LXP [8]) and energy was minimized with Rosetta algorithms relax (31) and fixbb (32). Next, an initial conformation for the acidic loop (residues 97 to 115) was built onto the top-scoring Ube2R1-Rbx1 model using the CCD algorithm with fragments derived from the Ube2R1 amino acid sequence (33). This model of the Ube2R1-Rbx1 complex was used to create initial models of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex using the UBQ_E2_thioester protocol (34), which allows the user to sequentially model the orientation of the thioesterified donor ubiquitin and the approach of an acceptor ubiquitin and to perform standard loop modeling on the acidic loop. To preorder the donor ubiquitin tail in a conformation consistent with previous E2/RING-E3 structures, the ubiquitin molecule was extracted from a cocrystal structure containing ubiquitin thioesterified to UbcH5b/Birc7 (PDB ID 4AUQ [4]). The acceptor ubiquitin was based on an apo structure (PDB ID 1UBQ [35]). To generate models consistent with the crystallized orientation of donor ubiquitin, a constraint was implemented between Leu 129 on Ube2R1 and both Ile 44 and Val 70 on the donor ubiquitin. Using this procedure, 4,000 theoretical models of Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 were generated yielding 286 low-scoring models. The executable protocol and flags for running the protocol can be found at www.rosetta.org. The subsequent models were clustered upon the acceptor ubiquitin position using a 4-Å cutoff value, yielding 14 clusters of acceptor ubiquitin approach.
(ii) Refining the acceptor ubiquitin.
To refine the acceptor ubiquitin position, the refinement option of the rosettadock protocol (36) was used with a constraint holding the ε-amino group on Lys 48 near the backbone carbonyl carbon atom of Gly 76 on the donor ubiquitin. Additionally, consistent with the experimental data, ambiguous constraints between Arg 54's three guanidinium nitrogen atoms in acceptor ubiquitin and Asp143's two carboxyl oxygen atoms in Ube2R1 were implemented. Using this procedure, 1,000 models were generated and the lowest scoring 1% were selected, yielding an ensemble of acceptor ubiquitin positions. The flags used to execute the Rosetta docking protocol can be found at www.rosetta.org.
(iii) Acidic-loop refinement.
During the modeling efforts, a structure of Rbx1 in complex with Ubc12 oxyesterified to Nedd8 was published (PDB ID 4P5O [7]), and Rbx1 from this cocrystal was therefore used to regenerate the Ube2R1-Rbx1 complex followed by more-extensive modeling of the Ube2R1 acidic loop. Based on the results from the charge-swapped mutations between Arg 91 on Rbx1 and Asp 102 on Ube2R1, ambiguous constraints were implemented between Arg 91's three guanidinium nitrogen atoms and Asp 102's two carboxyl oxygen atoms. To more effectively sample the conformational space, the “next-generation KIC” options (37) were used, allowing backbone flexibility for Ube2R1 residues 100 to 115. These results produced a variety of possible orientations, and low-scoring representative models were selected from the ensemble. The flags used to execute the next-generation KIC loop modeling protocol can be found at www.rosetta.org.
Protein expression and purification.
Human E2s and ubiquitin constructs were expressed in Rosetta (DE3) bacterial cells (EMD Millipore). Whereas Ube2R1 was used for structural modeling owing to the availability of X-ray structures, Ube2R2 protein was used for the subsequent biochemical analysis. Note that Ube2R1 and Ube2R2 share greater than 90% amino acid sequence identity within the catalytic domain and that the biochemical activities of these proteins are identical (G. Kleiger and R. Deshaies, unpublished data). E1 and the βTrCP-Skp1 complex were expressed and purified from Hi5 insect cells as previously described (38, 39). Cul1-Rbx1 was expressed and purified according to the “Split-n-Coexpress” method (40). Proteins were purified with N-terminal polyhistidine (His)6 or glutathione S-transferase (GST) tags on nickel-nitrilotriacetic acid (NTA)-agarose beads (Qiagen) or glutathione-Sepharose 4B beads (GE Healthcare), respectively. Acceptor ubiquitin and the ubiquitin constructs used in the Skp1-cullin-Fbox (SCF)-dependent ubiquitylation reactions had a noncleavable (His)6 affinity tag; otherwise, tags on the proteins were removed prior to gel filtration with tobacco etch virus (TEV) or thrombin protease. All ubiquitin and E2 proteins were subjected to gel filtration on a Superdex 75 10/300 column (GE Healthcare). Cul1-Rbx1 proteins were first purified by cation exchange chromatography before gel filtration was performed using a Superdex 200 10/300 column. All purified proteins were stored in a buffer containing 30 mM Tris-Cl (pH 7.5), 100 mM NaCl, 10% glycerol, and 1 mM dithiothreitol (DTT). Proteins were flash frozen in liquid nitrogen prior to storage at −80°C and were never subjected to multiple freeze-thaw cycles. All Cul1-Rbx1 proteins were subjected to neddylation and purified as previous described (39). β-Catenin peptide was purchased from New England Peptide and was identical to peptides used in previous studies, and monoubiquitylated peptides were also generated and purified using previously described methods (39).
Diubiquitin synthesis assay.
Diubiquitin synthesis was performed in a reaction buffer containing 30 mM Tris-Cl (pH 7.5), 100 mM NaCl, 5 mM MgCl2, 2 mM ATP, and 2 mM DTT. 32P-labeled K48R donor ubiquitin (5 μM) was first incubated with E1 enzyme (0.5 μM) for 2 min. Ube2R2 (2 μM) was then added, and the reaction mixture was incubated for a further 2 min. The reaction was initiated by the addition of acceptor ubiquitin (50 μM) that contained an additional Asp residue at its C terminus (this is referred to as D77 ubiquitin, which cannot be thioesterified to Ube2R2). Samples were collected at several time points and quenched by the addition of reducing SDS-PAGE loading buffer. Reaction mixtures were resolved on 4% to 20% SDS-PAGE gels (Lonza), dehydrated, and exposed to a phosphor screen. The reaction mixtures were imaged using a Typhoon 9410 scanner, and the amounts of substrate and product were quantified using ImageQuant software (GE Healthcare). Specifically, the amount of product was quantified by dividing the amount of diubiquitin by the total signal per lane and then the product was normalized by multiplying by the ratio of the concentrations of donor ubiquitin and Ube2R2. The rate of product formation was then determined by linear regression.
Multiturnover ubiquitylation assays.
Multiturnover ubiquitylation reactions were performed using β-catenin peptide as the substrate. Unlabeled ubiquitin (60 μM) was charged with E1 (1 μM) for 2 min in a reaction buffer that was identical to that used in the diubiquitin synthesis assay. E2 (10 μM) was then added and the reaction mixture incubated for 2 min, followed by the addition of neddylated SCFβ-TrCP (0.1 μM). 32P-labeled substrate (5 μM) was added to the reaction mixture, and samples were collected at several time points and quenched using reducing SDS-PAGE loading buffer. Reaction mixtures containing Ube2D3/UbcH5c were resolved on 15% SDS-PAGE gels, whereas all others were resolved on 4% to 20% gels.
The Km values of E2 constructs for SCF were determined as follows. Ubiquitylation reaction mixtures were assembled to contain a 2-fold dilution series of each E2 protein, while the remaining reaction components were identical to those described above. The highest concentration of a given E2 construct and the time of incubation for the ubiquitylation reactions were determined to ensure that the amounts of substrate conversion did not exceed 20%. The fractions of substrates converted to products were quantified by dividing all ubiquitylated products by the total signal, including substrate, and were normalized by multiplying by the ratio of the concentrations of β-catenin peptide and SCF. The reaction rates were then estimated by dividing the normalized fractions of substrates converted to products by the time of incubation. The reaction velocities were plotted as a function of E2 concentration and were fitted to the Michaelis-Menten equation using nonlinear curve fitting (GraphPad Prism software).
Single-encounter quench flow.
Single-encounter ubiquitylation assays were performed using a KinTek RQF-3 Rapid Quench-Flow instrument. Ube2R2 (20 μM) was charged in the presence of (His)6-tagged R54D ubiquitin (30 μM), E1 (1 μM), and reaction buffer, followed by the addition of unlabeled β-catenin peptide (200 μM). This reaction mixture was then mixed equally with SCF (1 μM) that had been preincubated with 32P-labeled Ub–β-catenin peptide (0.2 μM). Reaction mixtures were quenched in reducing SDS-PAGE buffer and substrate, and products were resolved on a 4% to 20% gradient SDS-PAGE gel. Reactions were run in duplicate, and each product band was quantified as a percentage of the signal from the total lane for each time point. The rates of ubiquitin transfer and substrate dissociation from SCF were determined using both KinTek Explorer global fitting software and nonlinear curve fitting to analytical closed-form solutions as described by Pierce et al. (41).
RESULTS
To explore the molecular basis for Lys 48 polyubiquitin chain synthesis, molecular models of Ube2R1 were generated in complex with a donor ubiquitin thioesterified to the Ube2R1 active site, an acceptor ubiquitin with Lys 48 constrained to the active site, and the Rbx1 subunit of the Skp1-cullin-Fbox (SCF) ubiquitin ligase. Ube2R1/2 was chosen for the following reasons: (i) it has critical importance for cellular homeostasis; (ii) it forms polyubiquitin chains with exquisite Lys 48 specificity (27); and (ii) previous modeling efforts elucidated the conformation of the Ube2R1-donor ubiquitin interface, which was subsequently confirmed by X-ray crystallography (30, 34). The orientation of the acidic loop has been previously characterized by molecular dynamics (24) and was also included during the modeling procedure.
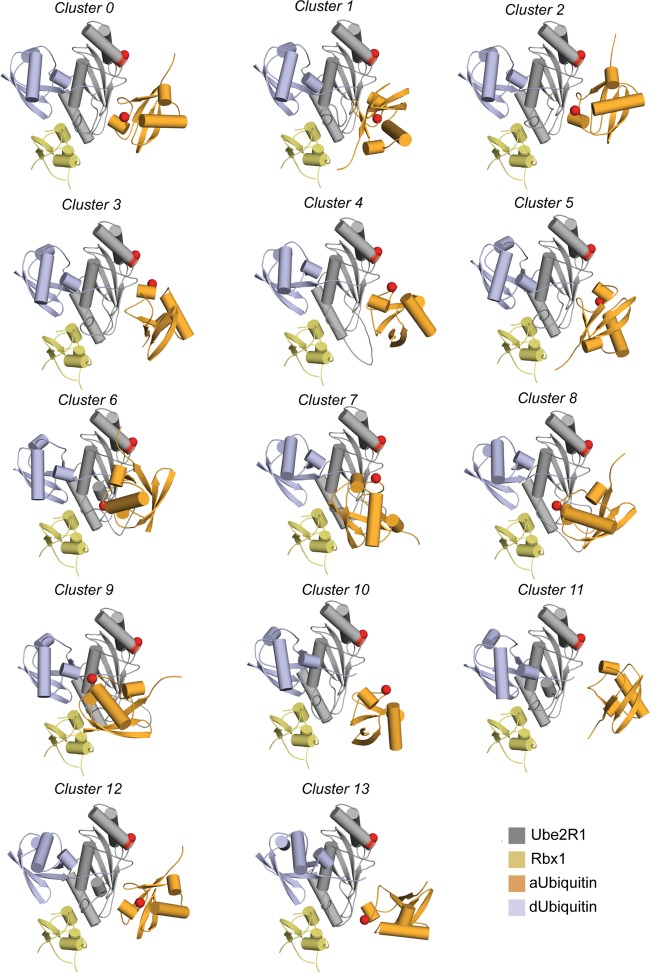
Molecular modeling of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex resulted in 14 distinct clusters of the Ube2R1-acceptor ubiquitin conformation (Fig. 1). To distinguish between the theoretical models, compensatory charge-swapped mutations were introduced into Ube2R2 and ubiquitin that individually should have disrupted the interface and resulted in decreased Ube2R2 activity; in combination, however, the mutant Ube2R2 and ubiquitin proteins should have restored the interface and product formation.
FIG 1.
Results of the clustering analysis of the acceptor ubiquitin position in the initial modeling effort with the UBQ_E2_thioester protocol. This analysis produced 14 clusters. The Cα atoms for Ube2R1 Asp 143 and acceptor ubiquitin Arg 54 are depicted as red spheres. Cluster 3 permits the closest approach of Arg 54 and Asp 143, and models from that cluster were used for refinement. aUbiquitin, acceptor ubiquitin; dUbiquitin, donor ubiquitin.
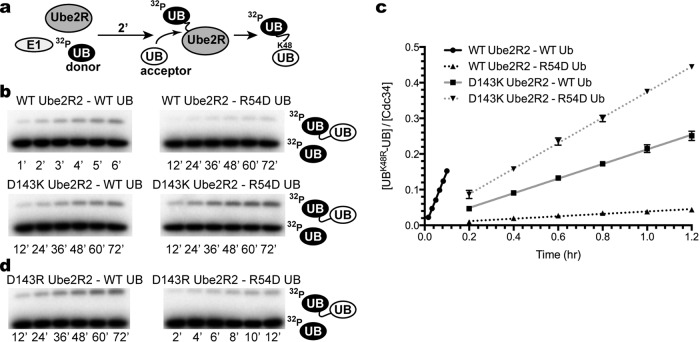
Ube2R2 activity was characterized using a previously established, simplified ubiquitylation assay in the absence of E3 (Fig. 2a). Ube2R2 was first thioesterified to radiolabeled donor ubiquitin, and the reaction was initiated by the addition of unlabeled acceptor ubiquitin, resulting in a diubiquitin product.
FIG 2.
The double mutant cycle of R54D ubiquitin and D143K or D143R Ube2R2 demonstrates that each mutant is individually defective but that the mutants complement each other in combination during Ube2R2 catalysis. (a) Schematic showing the components and order of addition for the diubiquitin synthesis assay. UB, ubiquitin. (b) Time course (in hours) for the diubiquitin synthesis assay and all four combinations of the R54D ubiquitin and D143K Ube2R2 double mutant cycle. Representative data are shown. (c) Graph plotting the quantitation of the amounts of diubiquitin product shown in panel b versus time. Ub, ubiquitin. (d) Same as in panel b, except D143R Ube2R2 activity has been assessed in the presence of either WT or R54D acceptor ubiquitin. Error bars represent the standard errors of measurements from duplicate data sets.
A previous study had shown that acceptor ubiquitin residue Arg 54 participated in Ube2R1-catalyzed Lys 48 polyubiquitin chain formation (26), and, consistent with this, the mutation of Arg 54 to an Asp residue in acceptor ubiquitin led to a 52-fold reduction in Ube2R2 activity compared with wild-type (WT) ubiquitin results (Fig. 2b and c and Table 1). Models from 1 of the 14 clusters were consistent with an interaction between Arg 54 and Ube2R1 residue Asp 143. To experimentally validate this interaction, D143K Ube2R2 protein was expressed and purified (note that lysine was chosen for the charge swap instead of arginine since the Rosetta algorithm identified this and R54D ubiquitin as the most energetically favorable compensatory mutant pair). Consistent with this, D143K Ube2R2 activity with WT acceptor ubiquitin was reduced by 7-fold in comparison with WT Ube2R2 results (Fig. 2b and c and Table 1). Impressively, the combination of D143K Ube2R2 and R54D acceptor ubiquitin resulted in only a 4-fold loss of Ube2R2 activity compared to the results seen with WT Ube2R2 and acceptor ubiquitin. Thus, the pairing of D143K Ube2R2 and R54D acceptor ubiquitin restored Ube2R2 activity to nearly WT levels despite the presence of the R54D mutation in ubiquitin. Additionally, to determine whether Asp 143 can also be charge swapped with arginine, D143R Ube2R2 was produced and showed a loss of activity nearly identical to that seen with D143K Ube2R2 as well as the ability to produce diubiquitin in the presence of R54D acceptor ubiquitin (Fig. 2d and Table 1).
TABLE 1.
Rates of diubiquitin formation (kobs) for mutant Ube2R2 and acceptor ubiquitin proteins and comparison with wild-type components
| Ube2R2 | Ubiquitin | kobs (h−1) | Fold changea |
|---|---|---|---|
| WT | WT | 1.56 ± 0.03 | |
| WT | R54D | 0.03 ± 0.00 | 52 |
| D143K | WT | 0.21 ± 0.01 | 7 |
| D143K | R54D | 0.36 ± 0.01 | 4 |
| D143R | WT | 0.24 ± 0.01 | 7 |
| D143R | R54D | 0.41 ± 0.02 | 4 |
| WT | I44A | 0.58 ± 0.04 | 3 |
| WT | I44R | 0.79 ± 0.04 | 2 |
| D91K | WT | 1.64 ± 0.04 | 1 |
| D91K | R54D | 0.04 ± 0.01 | 44 |
| WT | D58R | 0.02 ± 0.01 | 65 |
| D143K | D58R | NDb | |
| WT | R54A | 0.08 ± 0.01 | 20 |
| D143A | WT | 0.34 ± 0.02 | 5 |
Value of the rate for the reaction mixture containing WT Ube2R2 and WT ubiquitin divided by the rate of the reaction mixture containing mutant proteins.
ND, not determined since product levels were too low for quantitation.
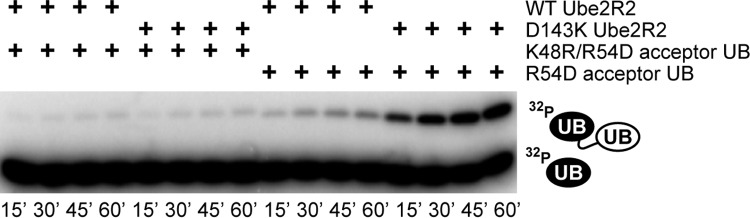
To determine whether the Lys 48 chain linkage specificity of the D143K Ube2R2 was compromised, SCF-independent Ube2R2 assays were conducted using a K48R ubiquitin variant that also contained the R54D mutation. Note that while Ube2R1/2 shows high fidelity for generating Lys 48-specific polyubiquitin chains, it is capable of generating non-Lys 48 chain linkages in the presence of K48R acceptor ubiquitin, albeit at a severely diminished rate (22, 27). The activities of both WT and D143K Ube2R2 with K48R/R54D acceptor ubiquitin were similar (Fig. 3), confirming that the Lys 48 specificity of D143K Ube2R2 remained intact.
FIG 3.
Diubiquitin synthesis assay demonstrating that ubiquitin chains generated by D143K Ube2R2 and R54D ubiquitin are predominantly linked through Lys 48. Time course data show that reaction mixtures containing K48R/R54D acceptor ubiquitin and WT or D143K Ube2R2 resulted in similar amounts of product formation, whereas the reaction mixture containing R54D acceptor ubiquitin and D143K Ube2R2 resulted in far greater product formation than the reaction mixture containing WT Ube2R2 and R54D ubiquitin. The acceptor ubiquitin concentration was 100 μM.
Based on these results, molecular models of the acceptor ubiquitin approach were refined using a constraint between Ube2R1 residue Asp 143 and acceptor ubiquitin residue Arg 54. This effort resulted in an ensemble of acceptor ubiquitin approaches (Fig. 4a and b) and allowed several important observations. First, the acceptor ubiquitin orientation permits the side chains of acceptor ubiquitin residue Arg 54 and Ube2R1 residue Asp 143 to adopt several rotamers capable of forming hydrogen bonds or salt bridges between side chain atoms while maintaining the Lys 48 side chain and the Ube2R1-donor ubiquitin thioester bond in proximity to each other. Second, the highly conserved Ube2R1/2 residues Tyr 87, His 98, and Ser 138, which all had previously been shown to have catalytic roles during polyubiquitin chain formation (22), were localized near Lys 48 on acceptor ubiquitin and the Ube2R1 active site (Fig. 4c). Lastly, Ile 44 on acceptor ubiquitin, a residue that typically is important in the stabilization of protein-protein interactions involving ubiquitin, was not participating in the Ube2R1-acceptor ubiquitin interface (Fig. 4c). Consistent with the latter observation, mutation of Ile 44 in only acceptor ubiquitin to either an Ala or Arg residue led to only a 3- or 2-fold reduction in Ube2R2 activity, respectively (Fig. 5a and Table 1). This result is all the more striking given that Ile 44 has been shown to stabilize the docking of donor ubiquitin to Ube2R1/2 (34).
FIG 4.
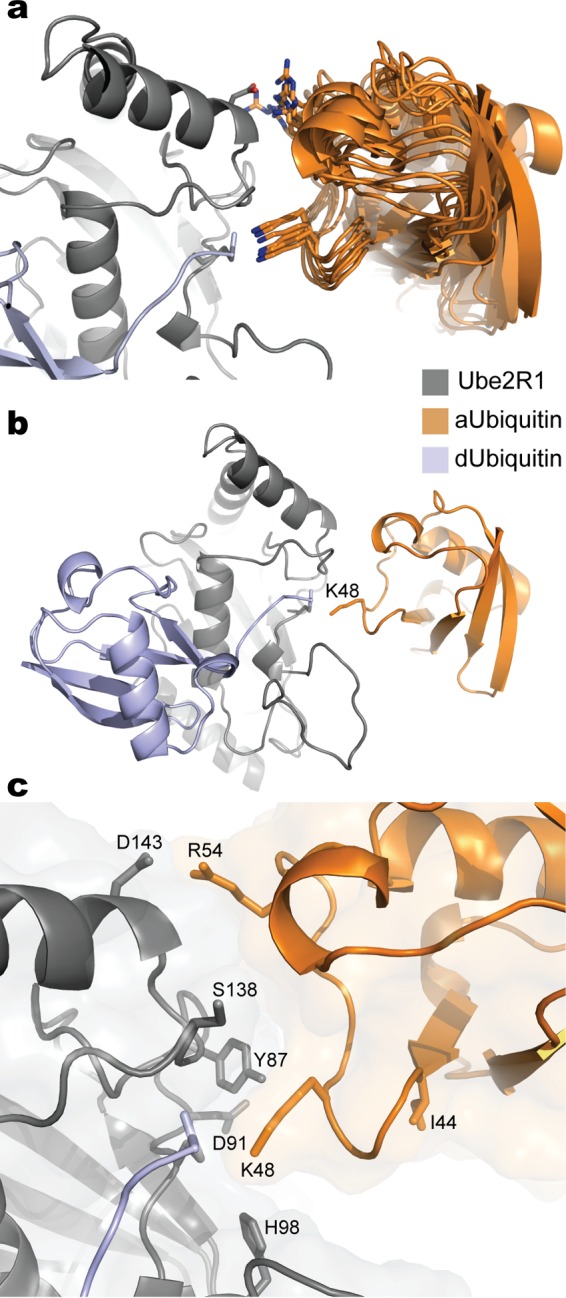
Molecular models of the Ube2R1-donor ubiquitin/acceptor ubiquitin complex after constraining Arg 54 in ubiquitin and Asp 143 in Ube2R1 to be in proximity. (a) Ribbon diagrams for 10 models randomly selected from an ensemble derived from the acceptor ubiquitin refinement procedure. Nitrogen atoms on Arg 54 and Lys 48 are blue; oxygen atoms on Asp 143 are red. (b) Ribbon diagram of a representative model from the ensemble showing the entire Ube2R1-donor ubiquitin/acceptor ubiquitin complex. (c) Closeup view of the Ube2R1-acceptor ubiquitin interface highlighting the locations of key residues that have previously been shown to participate in catalysis. Notice that Ube2R1 residue Asp 91 and acceptor ubiquitin residue Ile 44 are not located at the predicted Ube2R1-ubiquitin interface. Images were generated in Pymol.
FIG 5.
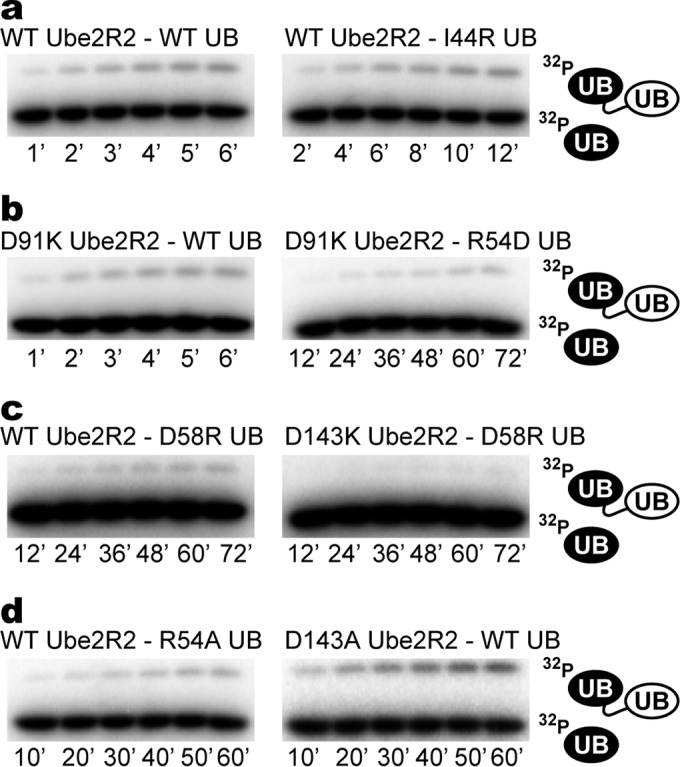
Experimental validation of the Ube2R1/2-acceptor ubiquitin complex confirming that Ile 44 on ubiquitin and Asp 91 on Ube2R2 do not help stabilize the interaction. (a) Time courses for diubiquitin synthesis comparing product formation for WT components with reaction mixtures containing WT Ube2R2 and I44R acceptor ubiquitin. (b) Same as in panel a, except D91K Ube2R2 activity has been assessed in the presence of WT or R54D acceptor ubiquitin. (c) Same as in panel a, except D58R acceptor ubiquitin activity has been assessed in the presence of either WT or D143K Ube2R2. (d) Same as in panel a, except R54A acceptor ubiquitin activity has been assessed in the presence of WT Ube2R2 or D143A Ube2R2 activity has been assessed in the presence of WT ubiquitin. Each experiment was performed in duplicate, and representative data are shown.
To further validate the Rosetta models, it was noticed that Ube2R1 residue Asp 91 was located on the periphery of the Ube2R1-acceptor ubiquitin interface and was not participating in stabilization of the complex (Fig. 4c), and thus a charge-swapped mutation at this position should not reduce Ube2R2 activity or restore activity in the presence of R54D acceptor ubiquitin. Consistent with the models, D91K Ube2R2 activity was both comparable to that of WT Ube2R2 in the presence of WT acceptor ubiquitin and similarly defective in the presence of R54D acceptor ubiquitin (Fig. 5b and Table 1). Next, a charge-swapped mutation to acceptor ubiquitin residue Asp 58, a residue in close proximity to Arg 54, led to a 65-fold decrease in WT Ube2R2 activity, and product formation was even more defective in the presence of D143K Ube2R2 (Fig. 5c and Table 1). These results collectively exclude the alternative hypothesis that the D143K mutation suppresses R54D ubiquitin by converting Ube2R2 into a promiscuous enzyme and support the ab initio Rosetta model of Ube2R1/2 Lys 48 ubiquitin chain formation.
To further explore the nature of the putative Asp 143 Ube2R1/2-Arg 54 ubiquitin interaction, D143A Ube2R2 and R54A acceptor ubiquitin proteins were expressed and purified. Specifically, if the interaction was deterministic in stabilizing the complex between Ube2R1/2 and acceptor ubiquitin, both D143A Ube2R2 and R54A acceptor ubiquitin should be defective in activity, whereas if the interaction was only permissive, these mutants should not lead to defects in activity. Product formation was defective in a reaction mixture containing WT Ube2R2 and R54A acceptor ubiquitin, though less so than in the reaction mixture containing charge-swapped R54D ubiquitin (Fig. 5d and Table 1). Similarly, D143A Ube2R2 activity was compromised in the presence of WT ubiquitin and also to a lesser extent than for D143K Ube2R2. Taken together, these results suggest that Asp 143 on Ube2R1/2 and Arg 54 on ubiquitin interact to stabilize the Ube2R1/2-acceptor ubiquitin complex.
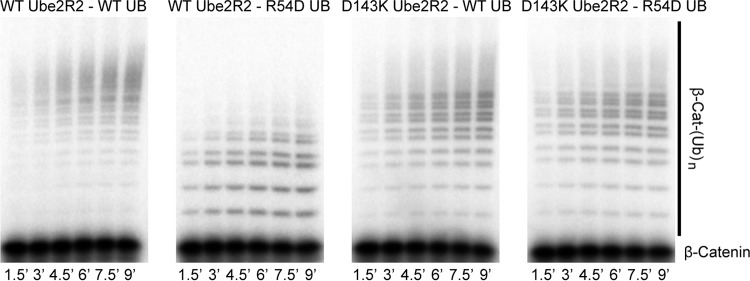
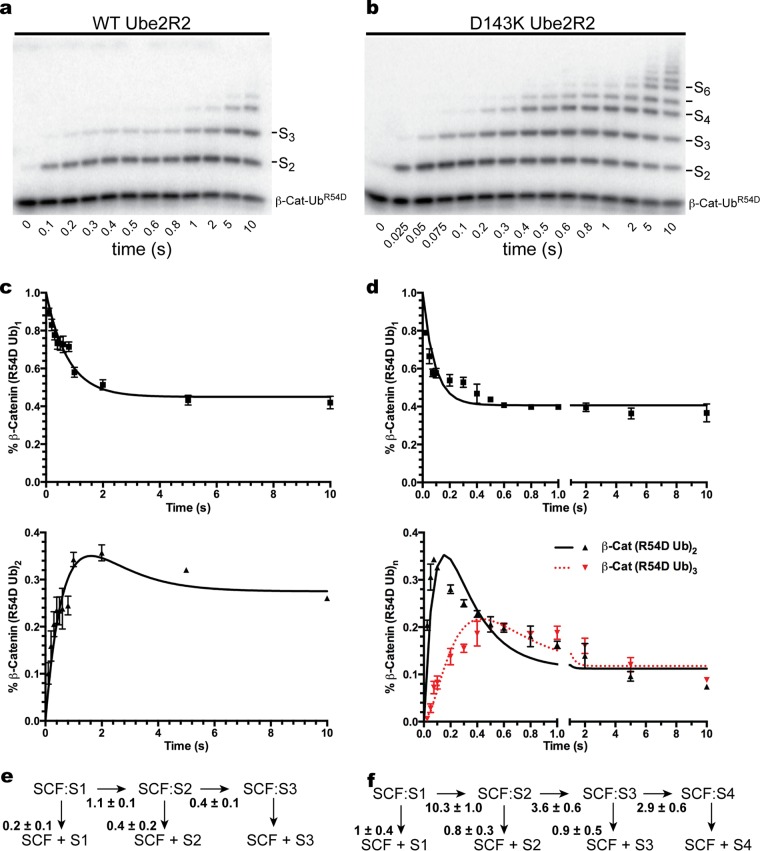
The significance of the Asp 143 Ube2R1/2-Arg 54 acceptor ubiquitin ion pair was examined next in the context of a fully reconstituted, ubiquitylation reaction mixture containing both ubiquitin ligase SCF and a physiologically relevant substrate bound to SCF. Multiturnover ubiquitylation reaction mixtures were assembled with combinations of WT or mutant Ube2R2 and ubiquitin and a 32P-labeled β-catenin peptide containing a single N-terminal lysine residue. The combination of WT Ube2R2 and ubiquitin in the SCF-dependent ubiquitylation reaction resulted in the formation of long polyubiquitin chains onto the β-catenin peptide substrate, and the combination of WT Ube2R2 with R54D ubiquitin led to a substantial reduction in the number of ubiquitins contained within the chains (Fig. 6). The combination of D143K Ube2R2 and WT ubiquitin also led to a reduction in the average lengths of the polyubiquitin chains compared to WT Ube2R2, although to a lesser extent than was seen with WT Ube2R2 and R54D ubiquitin. Finally, the combination of D143K Ube2R2 and R54D ubiquitin resulted in polyubiquitin chains attached to substrate that were longer than those seen with the reaction mixture containing WT Ube2R2 and R54D ubiquitin.
FIG 6.
Multiturnover ubiquitylation reactions with SCF demonstrate that the presence of R54D ubiquitin diminishes the lengths of the polyubiquitin chains on product compared with WT ubiquitin and that the introduction of D143K Ube2R2 with R54D ubiquitin substantially increases the lengths of the chains. Data represent time courses for the ubiquitylation of β-catenin (β-Cat) peptide for the entire double mutant cycle. Notice that some of the polyubiquitin chains in the reaction mixture containing WT Ube2R2 and ubiquitin migrated as high-molecular-weight smears, and while these were still present in the reaction mixtures containing D143K Ube2R2, the intensities had shifted significantly lower in the direction of the substrate band.
While polyubiquitin chain lengths were significantly reduced in reaction mixtures containing WT Ube2R2 and R54D ubiquitin in comparison with reaction mixtures containing WT ubiquitin, it was surprising that polyubiquitin chains with as many as 10 R54D ubiquitins were formed on the substrate, especially considering that, in the absence of SCF, WT Ube2R2 activity was reduced by 52-fold in the presence of R54D ubiquitin (Fig. 2 and Table 1). It would be ideal to estimate the individual rates of ubiquitin transfer from Ube2R2 to one or more ubiquitins on a SCF-bound substrate for comparison with the rates derived from the SCF-independent assay; however, this is not possible using the results from the multiturnover reactions. To address in detail how the R54D mutation affects ubiquitin's activity as an acceptor on a SCF-bound substrate, the pre-steady-state kinetics of the SCF-dependent ubiquitylation reaction were measured using a quenched-flow instrument in the presence of WT Ube2R2 and a monoubiquitylated β-catenin peptide substrate containing the R54D mutation.
Single-encounter ubiquitylation reactions were initiated in the presence of WT Ube2R2 and R54D ubiquitin, and the rates of ubiquitin transfer were estimated (Fig. 7a, c, and e). The rates for the first and second events of ubiquitin transfer to substrate-modified ubiquitins were 1.1 s−1 and 0.4 s−1, respectively, which were considerably lower than the previously measured rates of ubiquitin transfer for WT proteins (4 s−1 to 6 s−1) (41). Single-encounter ubiquitylation reactions using mixtures containing an identical substrate were then repeated in the presence of D143K Ube2R2 and R54D ubiquitin (Fig. 7b, d, and f). In this case, the rates for the first, second, and third transfers of ubiquitin to SCF-bound substrate (10.3 s−1, 3.6 s−1, and 2.9 s−1, respectively) were now comparable with the rates of ubiquitin transfer for WT components. Also note that in the presence of SCF and WT Ube2R2, the reduction in the average rate of ubiquitin transfer comparing WT and R54D ubiquitin was 8-fold; this is significantly less than the 52-fold reduction observed in the SCF-independent assay (Table 1). This key observation provides additional evidence supporting the notion that the R54D mutation in acceptor ubiquitin affects binding to Ube2R2; this point is elaborated on further in the Discussion.
FIG 7.
Pre-steady-state single-encounter ubiquitylation reactions allow the estimation of the individual rates of ubiquitin transfer from Ube2R2 to substrate-modified ubiquitins bound to SCF, indicating that the rate of R54D ubiquitin transfer is substantially higher in the presence of D143K Ube2R2 than in the presence of WT Ube2R2. (a) Time course showing the stepwise transfer of R54D ubiquitin to SCF-bound substrate in the presence of WT Ube2R2. S, substrate. (b) Same as in panel a, except with D143K Ube2R2. (c) Graph plotting either the disappearance of monoubiquitylated substrate (above) or the appearance of diubiquitylated product (below) over time. Error bars represent the standard errors of measurements. (d) Same as in panel c, except with D143K Ube2R2. (e) Schematic showing the rates of ubiquitin transfer to substrate as well as the rates of substrate and product dissociation from SCF with WT Ube2R2 and R54D acceptor ubiquitin. (f) Same as in panel e, except with D143K Ube2R2.
The Ube2R1/2 acidic loop participates in Lys 48-specific polyubiquitin chain formation by binding to SCF.
Ube2R1/2 contains a conserved 12-amino-acid insertion distal to the active site referred to as the acidic loop (owing to its four invariably conserved acidic residues), and previous studies have suggested an important role for the acidic loop in controlling Lys 48 specificity (22, 27). The conformation of the acidic loop is highly flexible, and loop residues are disordered in all X-ray structures of Ube2R1 analyzed to date (30, 42, 43). We reasoned that the presence of both donor and acceptor ubiquitins and Rbx1 may limit the conformational space afforded to the acidic loop, and the models of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex were analyzed to explore how the acidic loop participates in Lys 48 specificity.
Despite the presence of both ubiquitins and Rbx1, the modeling effort resulted in multiple conformations of the Ube2R1 acidic loop. Nevertheless, many of these models resulted in an interaction between Rbx1 residue Arg 91 and acidic-loop residue Asp 102 and/or Asp 103 (Fig. 8). Indeed, the previous structural characterization of the Ube2R1-Rbx1 interface by NMR had identified Arg 91 as a critical residue in promoting Ube2R1 interaction with SCF (29). To determine whether acidic-loop residues might contact Arg 91 in Rbx1, charge-swapped mutations were generated in Ube2R2 residues Asp 102 and Asp 103 as well as in Rbx1 residue Arg 91.
FIG 8.
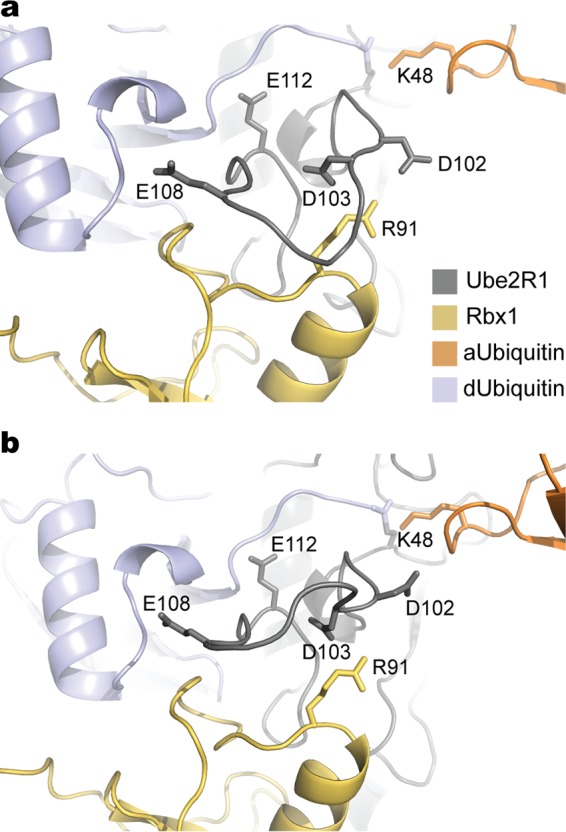
Molecular modeling suggests that the Ube2R1/2 acidic loop interacts with the Rbx1 subunit of SCF. (a) Ribbon diagram of a representative model of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex where Ube2R1 residue Asp 102 and Rbx1 residue Arg 91 are in close proximity. (b) Same as in panel a, except Ube2R1 residue Asp 103 and Rbx1 residue Arg 91 are predicted to be in close proximity.
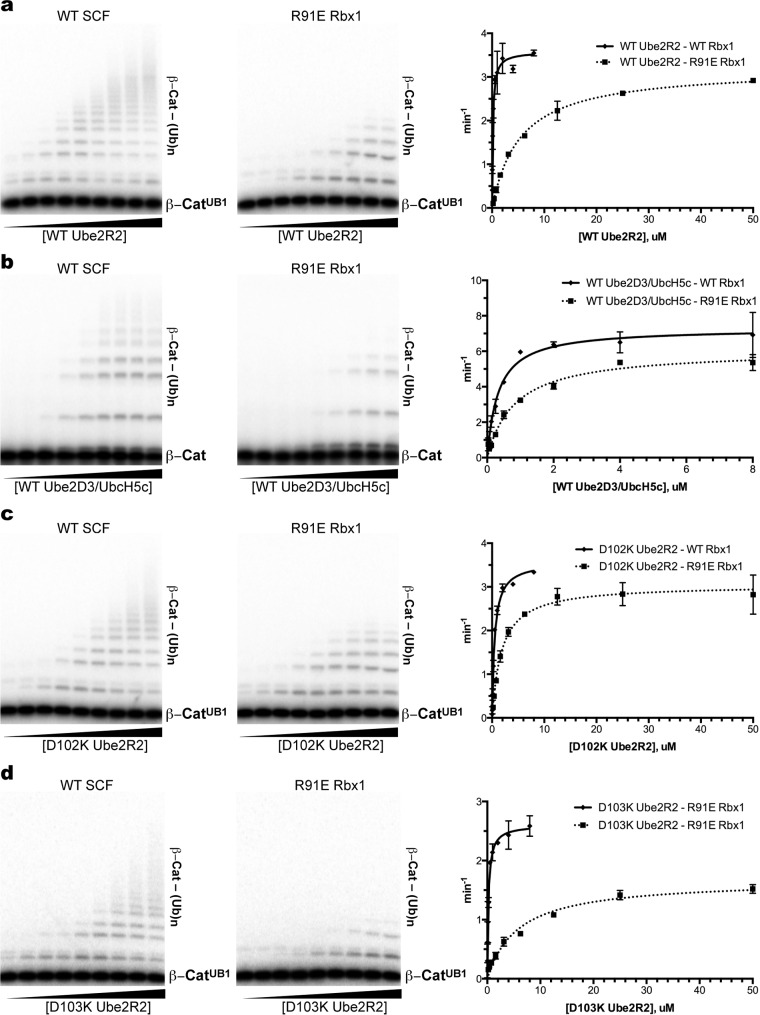
The affinities of Ube2R2 proteins for WT or mutant SCF were assessed in ubiquitylation assays by estimating the Km of Ube2R2 for SCF. The Km of WT Ube2R2 for WT SCF was 0.15 μM, whereas the Km of Ube2R2 for SCFR91E Rbx1 was 5.4 μM, indicating a large decrease in the affinity of Ube2R2 for mutant SCF (Fig. 9a and Table 2). To determine whether the defect in Ube2R2 binding by the R91E mutation in Rbx1 was specific to the acidic-loop region, the Km of WT Ube2D3/UbcH5c, an E2 that does not contain an acidic loop, was determined for WT SCF as well as SCFR91E Rbx1, and in this case only a 2-fold difference was observed (Fig. 9b and Table 2). Finally, the Km of D102K Ube2R2 for WT SCF was 0.49 μM, whereas the Km of D102K Ube2R2 for SCFR91E Rbx1 was 1.8 μM (Fig. 9c and Table 2). While the affinity of D102K Ube2R2 for SCFR91E Rbx1 is approximately 10-fold weaker than the affinity of WT Ube2R2 for SCF, the affinity of D102K Ube2R2 for SCFR91E Rbx1 is nevertheless 3-fold greater than that of WT Ube2R2 for mutant SCF, demonstrating a modest but significant restoration of D102K Ube2R2's affinity for SCFR91E Rbx1.
FIG 9.
The Ube2R1/2 acidic loop interacts with the Rbx1 subunit of SCF specifically through Asp 102 on Ube2R1/2 and Arg 91 on Rbx1. (a) Multiturnover ubiquitylation reactions comparing the activities of WT SCF and SCFR91E Rbx1 in the presence of increasing concentrations of WT Ube2R2. Each lane represents a single ubiquitylation reaction mixture that was incubated for 2 min prior to quenching. The graph plots the rates of substrate ubiquitylation as a function of WT Ube2R2 concentration. Error bars represent the standard errors of measurements from duplicate data sets. (b) Same as in panel a, except titrations of Ube2D3/UbcH5c were performed in the presence of either WT SCF or SCFR91E Rbx1. (c) Same as in panel a, except titrations of D102K Ube2R2 were performed in the presence of either WT SCF or SCFR91E Rbx1. (d) Same as in panel a, except titrations of D103K Ube2R2 were performed in the presence of either WT SCF or SCFR91E Rbx1.
TABLE 2.
Michaelis constants for wild-type and mutant E2-E3 pairs
| E2 | Rbx1 | Km (μM) |
|---|---|---|
| WT Ube2R2 | WT | 0.15 ± 0.03 |
| WT Ube2R2 | R91E | 5.40 ± 0.40 |
| WT Ube2D3/UbcH5c | WT | 0.33 ± 0.06 |
| WT Ube2D3/UbcH5c | R91E | 0.84 ± 0.12 |
| D102K Ube2R2 | WT | 0.49 ± 0.05 |
| D102K Ube2R2 | R91E | 1.83 ± 0.27 |
| D103K Ube2R2 | WT | 0.22 ± 0.04 |
| D103K Ube2R2 | R91E | 5.79 ± 0.83 |
The possibility of an electrostatic interaction between Ube2R2 residue Asp 103 and Rbx1 residue Arg 91 was also predicted by the modeling effort (Fig. 8b); however, the Km values of WT and D103K Ube2R2 for WT SCF and for SCFR91E Rbx1 were similar (Fig. 9d and Table 2). Taken together, these results suggest that Ube2R1/2 acidic-loop residue Asp 102 forms an interaction with Rbx1 residue Arg 91 during Ube2R1/2-SCF complex formation, although it remains likely that Arg 91 forms additional interactions with other residues in the acidic loop since only a partial restoration in the affinity of D102K Ube2R2 for SCFR91E Rbx1 was observed.
Ube2R1/2 acidic-loop residue Glu 108 or Glu 112 may participate in Lys 48-specific polyubiquitin chain formation by interacting with donor ubiquitin.
We next wanted to address the structural roles of the other 2 conserved acidic residues in the Ube2R1/2 acidic loop, Glu 108 and Glu 112, and to determine how they may affect Lys 48 specificity. Some 5,000 models of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex were generated in which Ube2R1 residue Asp 102 and Arg 91 in Rbx1 were constrained to be in proximity; however, the position of the acidic loop was variable despite the constraint (Fig. 10). The top 15% scoring structures were analyzed; only 8 of the 750 models predicted an interaction between Glu 108 and acceptor ubiquitin, and none of the models predicted an interaction between Glu 112 and acceptor ubiquitin. Instead of interacting with acceptor ubiquitin, approximately 50% of the top models predicted an interaction between either Glu 108 or Glu 112 and donor ubiquitin. Thus, molecular modeling suggests that Ube2R1/2 residues Glu 108 and Glu 112 may promote Ube2R1/2 function by interacting with the donor ubiquitin without directly influencing the conformation of the Ube2R1/2-acceptor ubiquitin complex.
FIG 10.
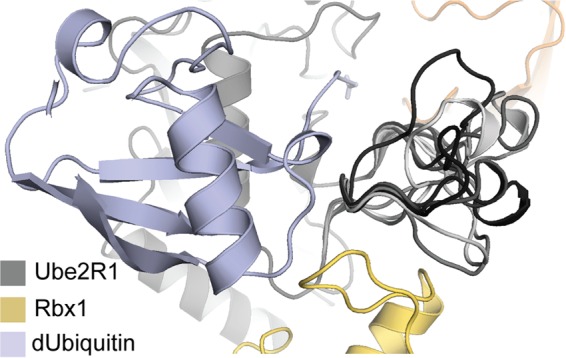
Lowest-scoring Ube2R1 acidic-loop orientations from 10 models of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex randomly selected from 5,000 trajectories. The acidic loops have been colored by a gray-scale gradient. These low-scoring structures result in a variety of possible conformations that can make contacts with both Rbx1 and the donor ubiquitin, but the acidic loop can contact the acceptor ubiquitin only rarely.
DISCUSSION
A combined molecular modeling and double mutant cycle analysis has been used to elucidate the mechanism of Lys 48 specificity during polyubiquitin chain formation by the Ube2R1/2 ubiquitin-conjugating enzyme. These results uncover a potential electrostatic interaction between Arg 54 on acceptor ubiquitin and Asp 143 on Ube2R1/2; however, we acknowledge that more-intricate models cannot be ruled out. For instance, Asp 143 and/or Arg 54 may form additional intermolecular interactions at the complex interface, which potentially explains why the double mutant combination does not fully restore Ube2R2 activity. It is not surprising that Asp 143 may have an important role in Ube2R1/2 function, as it is invariably conserved in Ube2R1/2 ortholog sequences. Furthermore, there is evidence that Arg 54 on ubiquitin is also important, since the replacement of endogenous ubiquitin with a R54D mutant in S. cerevisiae resulted in a mild slow-growth phenotype (44), and, when introduced into mammalian cells, expression of the double R54A/Y59A ubiquitin mutant led to cellular apoptosis (26). Taking those findings in combination with the results presented here, the importance of ubiquitin residue Arg 54 has been demonstrated both in vivo and in vitro.
Careful consideration of the rates of Ube2R2-catalyzed ubiquitin transfer in either the absence or the presence of SCF further supports the notion that acceptor ubiquitin residue Arg 54 participates in promoting ubiquitin binding to Ube2R1/2. Mutations in ubiquitin that negatively affect binding to Ube2R1/2 (and not catalysis) can in principle be compensated for by increasing the ubiquitin concentration to a level high enough to saturate Ube2R1/2. During analysis of the results of the SCF-dependent reactions, it was noted that the difference in the average rates of ubiquitin transfer in the presence of either WT or R54D ubiquitin was approximately 8-fold (Fig. 7), significantly less than the difference in the rates of ubiquitin transfer in the SCF-independent assay (52-fold) (Table 1). One critical difference between these two assays is the acceptor ubiquitin concentration: in the SCF-independent assay (Fig. 2a), the concentration of ubiquitin was 50 μM. In the SCF-dependent reaction, the simultaneous binding of both a substrate modified with an acceptor ubiquitin and Ube2R2 to SCF results in a substantial increase in the effective concentration of acceptor ubiquitin and Ube2R2 (previously estimated to be in the low millimolar range [27]). Considering that the Km of acceptor ubiquitin for Ube2R2 is approximately 1 mM (25), the increase in the effective concentration of acceptor ubiquitin for Ube2R2 most likely suppresses the reduced affinity of Ube2R2 for R54D ubiquitin in the SCF-dependent assay. Thus, these results help explain why substantial polyubiquitin chains are formed on a SCF-bound substrate in the presence of R54D ubiquitin during performance of the multiturnover reaction scheme (Fig. 6).
Does the presence of an acidic residue on E2s at structurally equivalent positions to Ube2R1/2 residue Asp 143 determine lysine specificity during catalysis? Approximately one-half of human E2 protein sequences contain either an aspartic or glutamic acid residue at positions that are aligned to Asp 143 in Ube2R1/2 in a multiple-sequence alignment of E2 paralogs. However, the presence of an acidic residue at this position does not necessarily determine the lysine specificity of polyubiquitin chains. For instance, Ube2S is an E2 that forms polyubiquitin chains with Lys 11 specificity, and though Glu 132 in Ube2S is structurally equivalent to Asp 143 in Ube2R1/2, a mutation in Glu 132 was not shown to disrupt Ube2S activity (45). Furthermore, the E2s that modify protein substrates with the ubiquitin-like proteins Nedd8 and SUMO also contain acidic residues at positions that are structurally equivalent to that of Ube2R1/2 residue Asp 143; however, these E2s do not assemble chains of Nedd8 or SUMO.
A survey of the Protein Data Bank for structures of proteins in complex with ubiquitin indicates that electrostatic interactions between a protein and ubiquitin residue Arg 54 are highly atypical (46). Indeed, only 3 examples were found, each involving the interaction of a deubiquitylating enzyme and ubiquitin (PDB ID 3IHP, 4DHJ, and 4BOZ) (47, 48). On the other hand, electrostatic interactions involving other charged residues have been shown to have important roles in determining the lysine specificity of polyubiquitin chains by also stabilizing protein-ubiquitin interactions. For instance, Ube2S forms polyubiquitin chains with Lys 11 specificity by stabilizing the Ube2S-ubiquitin interaction through a salt bridge between Lys 6 on ubiquitin and Glu 131 on Ube2S (45). Interestingly, the yeast E2 Ubc1, which, like Ube2R1/2, forms Lys 48-specific polyubiquitin chains, utilizes a polar cluster of residues near its active site that are not conserved in Ube2R1/2 (15), and the human ortholog Ube2K forms an electrostatic interaction between Glu 51 on ubiquitin and a Lys residue on Ube2K (16). Thus, it appears that Ube2R1/2 and Ubc1/Ube2K generate Lys 48-linked polyubiquitin chains through distinct mechanisms.
In addition to Lys 48- and Lys 11-specific polyubiquitin chains, the mechanism describing how Lys 63 specificity is achieved by an E2 has also been elucidated (2, 49). In this case, the E2 Ubc13 forms a heterodimer with its binding partner Mms2. Ubc13 contains the active site that participates in catalysis, while Mms2 has a ubiquitin binding site that places Lys 63 within close proximity of the Ubc13 active site. In summary, it is clear that, while only a small number of examples of how E2s determine lysine specificity during polyubiquitin chain formation are known, a diverse repertoire of mechanisms exists in nature.
We also used our modeling approach to help ascertain how the four highly conserved Ube2R1/2 acidic-loop residues participate in determining lysine specificity during polyubiquitin chain formation. Rather than forming direct interactions with the acceptor ubiquitin, it appears that the acidic-loop residues interact either with SCF or with the donor ubiquitin. In support of this, note that we had previously generated a human Ube2R2 mutant protein in which all four acidic residues were mutated to alanine, and while the activity of this mutant was severely defective, it was still capable of forming polyubiquitin chains on a SCF-bound substrate with Lys 48 specificity (25). Also, models of the Ube2R1-donor ubiquitin/acceptor ubiquitin-Rbx1 complex predicted an interaction between Ube2R1 residue Asp 102 and SCF subunit Rbx1 residue Arg 91 that was supported by functional binding experiments. Finally, acidic-loop residues Glu 108 and Glu 112 are not in proximity of acceptor ubiquitin in nearly all of the models that were generated. In summary, these results are most consistent with a model where SCF controls the conformation of the Ube2R1/2 acidic loop which may serve to optimize stabilizing interactions between residues Glu 108 and Glu 112 and the donor ubiquitin. One notable exception may be Asp 103, for which some of the models predicted an interaction between this residue and acceptor ubiquitin, a result that is consistent with previous protein cross-linking experiments (26).
While this work provides the most detailed molecular insight into the mechanism of Ube2R1/2 and SCF function to date, there are still many issues that need to be addressed. For instance, the modeling effort here suggests that the Ube2R1/2 acidic loop can adopt many different conformations, and it is possible that multiple conformations exist during Ube2R1/2 function (potentially explaining how both Glu 108 and Glu 112 may simultaneously participate in catalysis by interacting with donor ubiquitin). Another topic of interest involves an essential step during catalysis, the deprotonation of Lys 48 on acceptor ubiquitin. We had previously speculated that a conserved histidine (His 98 in Ube2R1/2) may participate in deprotonation (25), and while our models show that His 98 is in proximity to Lys 48 on acceptor ubiquitin and may therefore play some role in lysine activation, it is likely that the mechanism also involves Ube2R1/2 residue Ser 138, which is structurally equivalent to an aspartic acid residue present in most E2s that has been shown to be critical in promoting lysine deprotonation on the substrate (50).
Collectively, these data demonstrate the effectiveness of using computational tools to gain structural insights into transient protein complexes as well as into flexible regions within those structures. Using an iterative procedure of computational modeling and biochemical experiments that distinguish between theoretical models, the refinement of the initial models based on the experimental results provides a glimpse into protein function that may be inaccessible through more traditional structural biology methodologies. We believe that these procedures can be generally applied to a number of other protein complexes for which traditional methodologies have presented problems that have proved difficult or intractable.
ACKNOWLEDGMENTS
S.H. performed the ubiquitylation assays. J.S.H. performed all molecular modeling and helped with the experimental design. S.M.L. modified the Rosetta protocols. B.K. helped design and supervised the molecular modeling efforts. G.K. helped with the experimental design and wrote the manuscript with contributions from all of the other authors.
Funding Statement
J.S.H. was supported through the Lineberger Comprehensive Cancer Center postdoctoral fellowship (T32-CA009156). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Deshaies RJ, Joazeiro CA. 2009. RING domain E3 ubiquitin ligases. Annu Rev Biochem 78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Branigan E, Plechanovova A, Jaffray EG, Naismith JH, Hay RT. 2015. Structural basis for the RING-catalyzed synthesis of K63-linked ubiquitin chains. Nat Struct Mol Biol 22:597–602. doi: 10.1038/nsmb.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calabrese MF, Scott DC, Duda DM, Grace CR, Kurinov I, Kriwacki RW, Schulman BA. 2011. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat Struct Mol Biol 18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dou H, Buetow L, Sibbet GJ, Cameron K, Huang DT. 2012. BIRC7-E2 ubiquitin conjugate structure reveals the mechanism of ubiquitin transfer by a RING dimer. Nat Struct Mol Biol 19:876–883. doi: 10.1038/nsmb.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plechanovová A, Jaffray EG, Tatham MH, Naismith JH, Hay RT. 2012. Structure of a RING E3 ligase and ubiquitin-loaded E2 primed for catalysis. Nature 489:115–120. doi: 10.1038/nature11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruneda JN, Littlefield PJ, Soss SE, Nordquist KA, Chazin WJ, Brzovic PS, Klevit RE. 2012. Structure of an E3:E2∼Ub complex reveals an allosteric mechanism shared among RING/U-box ligases. Mol Cell 47:933–942. doi: 10.1016/j.molcel.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott DC, Sviderskiy VO, Monda JK, Lydeard JR, Cho SE, Harper JW, Schulman BA. 2014. Structure of a RING E3 trapped in action reveals ligation mechanism for the ubiquitin-like protein NEDD8. Cell 157:1671–1684. doi: 10.1016/j.cell.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das R, Liang YH, Mariano J, Li J, Huang T, King A, Tarasov SG, Weissman AM, Ji X, Byrd RA. 2013. Allosteric regulation of E2:E3 interactions promote a processive ubiquitination machine. EMBO J doi: 10.1038/emboj.2013.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vittal V, Stewart MD, Brzovic PS, Klevit RE. 2015. Regulating the regulators: recent revelations in the control of E3 ubiquitin ligases. J Biol Chem 290:21244–21251. doi: 10.1074/jbc.R115.675165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 11.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 12.Dikic I, Wakatsuki S, Walters KJ. 2009. Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol 10:659–671. doi: 10.1038/nrm2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hicke L, Schubert HL, Hill CP. 2005. Ubiquitin-binding domains. Nat Rev Mol Cell Biol 6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 14.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, Harper JW, Gygi SP. 2011. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell 44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodrigo-Brenni MC, Foster SA, Morgan DO. 2010. Catalysis of lysine 48-specific ubiquitin chain assembly by residues in E2 and ubiquitin. Mol Cell 39:548–559. doi: 10.1016/j.molcel.2010.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton AJ, Day CL. 2015. The molecular basis of lysine 48 ubiquitin chain synthesis by Ube2K. Sci Rep 5:16793. doi: 10.1038/srep16793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W, Shang Y, Zeng Y, Liu C, Li Y, Zhai L, Wang P, Lou J, Xu P, Ye Y, Li W. 2014. Dimeric Ube2g2 simultaneously engages donor and acceptor ubiquitins to form Lys48-linked ubiquitin chains. EMBO J 33:46–61. doi: 10.1002/embj.201385315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 19.Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 20.Arrigoni A, Bertini L, De Gioia L, Papaleo E. 2014. Inhibitors of the Cdc34 acidic loop: a computational investigation integrating molecular dynamics, virtual screening and docking approaches. FEBS Open Bio 4:473–484. doi: 10.1016/j.fob.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi YS, Lee YJ, Lee SY, Shi L, Ha JH, Cheong HK, Cheong C, Cohen RE, Ryu KS. 2015. Differential ubiquitin binding by the acidic loops of Ube2g1 and Ube2r1 enzymes distinguishes their Lys-48-ubiquitylation activities. J Biol Chem 290:2251–2263. doi: 10.1074/jbc.M114.624809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gazdoiu S, Yamoah K, Wu K, Pan Z-Q. 2007. Human Cdc34 employs distinct sites to coordinate attachment of ubiquitin to a substrate and assembly of polyubiquitin chains. Mol Cell Biol 27:7041–7052. doi: 10.1128/MCB.00812-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Papaleo E, Casiraghi N, Arrigoni A, Vanoni M, Coccetti P, De Gioia L. 2012. Loop 7 of E2 enzymes: an ancestral conserved functional motif involved in the E2-mediated steps of the ubiquitination cascade. PLoS One 7:e40786. doi: 10.1371/journal.pone.0040786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papaleo E, Ranzani V, Tripodi F, Vitriolo A, Cirulli C, Fantucci P, Alberghina L, Vanoni M, De Gioia L, Coccetti P. 2011. An acidic loop and cognate phosphorylation sites define a molecular switch that modulates ubiquitin charging activity in Cdc34-like enzymes. PLoS Comput Biol 7:e1002056. doi: 10.1371/journal.pcbi.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziemba A, Hill S, Sandoval D, Webb K, Bennett EJ, Kleiger G. 2013. Multimodal mechanism of action for the Cdc34 acidic loop: a case study for why ubiquitin-conjugating enzymes have loops and tails. J Biol Chem 288:34882–34896. doi: 10.1074/jbc.M113.509190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong RA, Wu K, Spratt DE, Yang Y, Lee C, Nayak J, Xu M, Elkholi R, Tappin I, Li J, Hurwitz J, Brown BD, Chipuk JE, Chen ZJ, Sanchez R, Shaw GS, Huang L, Pan ZQ. 2014. Pivotal role for the ubiquitin Y59-E51 loop in lysine 48 polyubiquitination. Proc Natl Acad Sci U S A 111:8434–8439. doi: 10.1073/pnas.1407849111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petroski MD, Deshaies RJ. 2005. Mechanism of lysine 48-linked ubiquitin-chain synthesis by the cullin-RING ubiquitin-ligase complex SCF-Cdc34. Cell 123:1107–1120. doi: 10.1016/j.cell.2005.09.033. [DOI] [PubMed] [Google Scholar]
- 28.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. 2011. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol 487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spratt DE, Wu K, Kovacev J, Pan ZQ, Shaw GS. 2012. Selective recruitment of an E2∼ubiquitin complex by an E3 ubiquitin ligase. J Biol Chem 287:17374–17385. doi: 10.1074/jbc.M112.353748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H, Ceccarelli DF, Orlicky S, St-Cyr DJ, Ziemba A, Garg P, Plamondon S, Auer M, Sidhu S, Marinier A, Kleiger G, Tyers M, Sicheri F. 2014. E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin. Nat Chem Biol 10:156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conway P, Tyka MD, DiMaio F, Konerding DE, Baker D. 2014. Relaxation of backbone bond geometry improves protein energy landscape modeling. Protein Sci 23:47–55. doi: 10.1002/pro.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhlman B, Baker D. 2000. Native protein sequences are close to optimal for their structures. Proc Natl Acad Sci U S A 97:10383–10388. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Canutescu AA, Dunbrack RL Jr. 2003. Cyclic coordinate descent: a robotics algorithm for protein loop closure. Protein Sci 12:963–972. doi: 10.1110/ps.0242703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha A, Lewis S, Kleiger G, Kuhlman B, Deshaies RJ. 2011. Essential role for ubiquitin-ubiquitin-conjugating enzyme interaction in ubiquitin discharge from Cdc34 to substrate. Mol Cell 42:75–83. doi: 10.1016/j.molcel.2011.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijay-Kumar S, Bugg CE, Cook WJ. 1987. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol 194:531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 36.Chaudhury S, Berrondo M, Weitzner BD, Muthu P, Bergman H, Gray JJ. 2011. Benchmarking and analysis of protein docking performance in Rosetta v3.2. PLoS One 6:e22477. doi: 10.1371/journal.pone.0022477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein A, Kortemme T. 2013. Improvements to robotics-inspired conformational sampling in Rosetta. PLoS One 8:e63090. doi: 10.1371/journal.pone.0063090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. 2009. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell 139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saha A, Deshaies RJ. 2008. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell 32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li T, Pavletich NP, Schulman BA, Zheng N. 2005. High-level expression and purification of recombinant SCF ubiquitin ligases. Methods Enzymol 398:125–142. doi: 10.1016/S0076-6879(05)98012-9. [DOI] [PubMed] [Google Scholar]
- 41.Pierce NW, Kleiger G, Shan SO, Deshaies RJ. 2009. Detection of sequential polyubiquitylation on a millisecond timescale. Nature 462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J, Durocher D, Meloche S, Webb DR, Tyers M, Sicheri F. 2011. An allosteric inhibitor of the human Cdc34 ubiquitin-conjugating enzyme. Cell 145:1075–1087. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]
- 43.Sheng Y, Hong JH, Doherty R, Srikumar T, Shloush J, Avvakumov GV, Walker JR, Xue S, Neculai D, Wan JW, Kim SK, Arrowsmith CH, Raught B, Dhe-Paganon S. 2012. A human ubiquitin conjugating enzyme (E2)-HECT E3 ligase structure-function screen. Mol Cell Proteomics 11:329–341. doi: 10.1074/mcp.O111.013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. 2013. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol 425:1363–1377. doi: 10.1016/j.jmb.2013.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickliffe KE, Lorenz S, Wemmer DE, Kuriyan J, Rape M. 2011. The mechanism of linkage-specific ubiquitin chain elongation by a single-subunit E2. Cell 144:769–781. doi: 10.1016/j.cell.2011.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison JS, Jacobs TM, Houlihan K, Van Doorslaer K, Kuhlman B. 2015. UbSRD: The ubiquitin structural relational database. J Mol Biol doi: 10.1016/j.jmb.2015.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiener R, DiBello AT, Lombardi PM, Guzzo CM, Zhang X, Matunis MJ, Wolberger C. 2013. E2 ubiquitin-conjugating enzymes regulate the deubiquitinating activity of OTUB1. Nat Struct Mol Biol 20:1033–1039. doi: 10.1038/nsmb.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, Freund SM, Ovaa H, Komander D. 2013. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eddins MJ, Carlile CM, Gomez KM, Pickart CM, Wolberger C. 2006. Mms2-Ubc13 covalently bound to ubiquitin reveals the structural basis of linkage-specific polyubiquitin chain formation. Nat Struct Mol Biol 13:915–920. doi: 10.1038/nsmb1148. [DOI] [PubMed] [Google Scholar]
- 50.Yunus AA, Lima CD. 2006. Lysine activation and functional analysis of E2-mediated conjugation in the SUMO pathway. Nat Struct Mol Biol 13:491–499. doi: 10.1038/nsmb1104. [DOI] [PubMed] [Google Scholar]