Abstract
Many bacteria form surface-attached communities known as biofilms. Due to the extreme resistance of these bacterial biofilms to antibiotics and mechanical stresses, biofilms are of growing interest not only in microbiology but also in medicine and industry. Previous studies have determined the extracellular polymeric substances present in the matrix of biofilms formed by Bacillus subtilis NCIB 3610. However, studies on the physical properties of biofilms formed by this strain are just emerging. In particular, quantitative data on the contributions of biofilm matrix biopolymers to these physical properties are lacking. Here, we quantitatively investigated three physical properties of B. subtilis NCIB 3610 biofilms: the surface roughness and stiffness and the bulk viscoelasticity of these biofilms. We show how specific biomolecules constituting the biofilm matrix formed by this strain contribute to those biofilm properties. In particular, we demonstrate that the surface roughness and surface elasticity of 1-day-old NCIB 3610 biofilms are strongly affected by the surface layer protein BslA. For a second strain, B. subtilis B-1, which forms biofilms containing mainly γ-polyglutamate, we found significantly different physical biofilm properties that are also differently affected by the commonly used antibacterial agent ethanol. We show that B-1 biofilms are protected from ethanol-induced changes in the biofilm's stiffness and that this protective effect can be transferred to NCIB 3610 biofilms by the sole addition of γ-polyglutamate to growing NCIB 3610 biofilms. Together, our results demonstrate the importance of specific biofilm matrix components for the distinct physical properties of B. subtilis biofilms.
INTRODUCTION
Bacteria embed themselves with secreted biopolymers (1–3), building a community that is referred to as a biofilm. Such biofilms can be formed by a variety of Gram-positive as well as Gram-negative bacteria (4). The composition of the biofilm matrix is dependent on the biofilm-forming bacterium and environmental conditions such as shear forces experienced, temperature, and nutrient availability (5); the matrix can consist of different extracellular polymeric substances (EPSs), such as polysaccharides, proteins, lipids, and nucleic acids (6). Biofilms can grow on various surfaces (7–9). While Bacillus subtilis biofilms are formed on solid nutrient surfaces or at liquid-air interfaces (10, 11), other bacteria can produce biofilms on surfaces under water-saturated conditions (in liquid) (12). Due to their high mechanical stability (13, 14) and their resistance to antibiotic or chemical treatment (15–18), such biofilms present significant problems in both industry and health care (19–22). Although the compositions of many biofilm matrices are known, the biomolecular reason for the outstanding resistance of bacterial biofilms is not well understood. Only a few studies have investigated the influence of specific matrix components on the mechanical properties of biofilms (23, 24). The majority of recent studies focuses on the analysis of the mechanical properties of wild-type biofilms (25–27). These studies comprise work on biofilm elasticity (16, 17, 28, 29), biofilm erosion stability (30), and adhesion properties (31–33), as well as theoretical investigations of mechanical biofilm characteristics (34, 35). Biofilm formation by the bacterium Bacillus subtilis has been studied intensively (36–43), but information about the mechanical properties of these biofilm-forming bacteria is just emerging (24, 30, 44). Hence, a detailed understanding of the mechanical characteristics of bacterial biofilms is urgently needed to fight the growth of bacterial biofilms on medical devices and other industrially used surfaces.
In this study, we investigated the physical properties of 1-day-old biofilms of B. subtilis NCIB 3610 under well-defined laboratory conditions, which allowed us to compare these results directly with data obtained for a second B. subtilis strain, B-1. Although both strains belong to the species B. subtilis, the strains are reported to differ in their biofilm matrix compositions. The biofilm matrix of B. subtilis NCIB 3610 (45) is composed mainly of an exopolysaccharide produced by the gene products of the epsA-O operon (i.e., the operon comprising epsA, epsO, and the genetic material between those genes) (46) and an amyloid-fiber-forming protein, TasA (47, 48). A second biofilm matrix protein, BslA, is a self-assembling hydrophobin on the surface of B. subtilis NCIB 3610 biofilms (49, 50). In contrast to that of strain NCIB 3610, the biofilm matrix of the wild-type strain B. subtilis B-1 is described as being composed mainly of γ-polyglutamate (γ-PGA) (51), a highly hydrophilic anionic polymer (52).
We present a gene expression analysis of B-1 biofilms (Fig. 1), revealing that genes encoding proteins known to be part of the NCIB 3610 biofilm matrix, in addition to the main B-1 biofilm matrix component γ-PGA, are transcribed in the B-1 biofilm variant. This indicates that proteins such as BlsA may also be present in the B-1 biofilm matrix, contributing to the material properties of this biofilm variant, although no direct conclusions on protein amounts present in the B-1 biofilm matrix can be drawn from mRNA expression levels. In contrast, γ-PGA is not produced in biofilms of strain NCIB 3610 (46, 51). Hence, we expect that biofilms formed by these two strains, B-1 and NCIB 3610, would differ in their physical properties. To investigate the physical properties of B-1 and NCIB 3610 biofilms, we used three different techniques. We performed surface indentation experiments using atomic force microscopy (AFM) to analyze the surface stiffness of the bacterial biofilms. To quantify the surface roughness of the biofilms, a profilometer study was performed. Finally, using macrorheology, we obtained quantitative information on the bulk stiffness of the 1-day-old B. subtilis biofilms. Detailed information on the techniques and the experimental procedures is given in Materials and Methods and in the supplemental material. We find that even such closely related wild-type strains generate biofilms with significantly different material properties. Using a set of different mutant strains, we demonstrate that the surface properties (i.e., stiffness and roughness) of the NCIB 3610 biofilm can be attributed mainly to the surface layer protein BslA. Furthermore, we study the effects of the antibacterial agent ethanol on specific material properties of the wild-type biofilms, and we provide evidence that γ-polyglutamate protects B. subtilis B-1 biofilms from ethanol-induced changes in the biofilm bulk elasticity.
FIG 1.
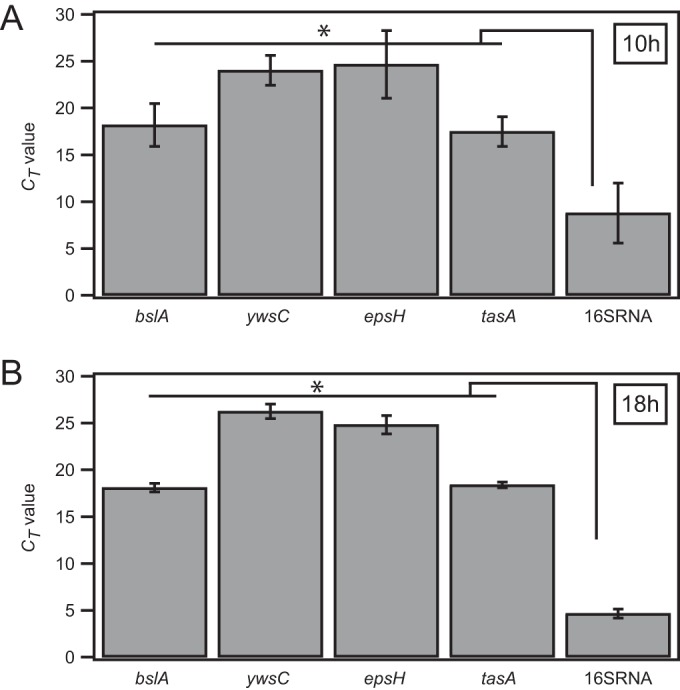
mRNA production of genes for biofilm matrix proteins in B. subtilis strain B-1. Gene expression analyses were performed for biofilms generated by wild-type strain B-1, at 10 h (A) and 18 h (B) of biofilm growth. The mRNA production of genes corresponding to matrix components described for wild-type strains NCIB 3610 and B-1, in comparison with the production of the control, 16S rRNA, was analyzed. CT values represent the mean values obtained from 3 biological and 3 technical triplicates. The error bars denote standard deviations. P values from pairwise t tests for these data are given in Table S1 in the supplemental material. Additionally, an ensemble significance test (one-way ANOVA and Tukey's honestly significant difference test; MATLAB) was performed to confirm that the genes blsA, ywsC, epsH, and tasA were transcribed at significantly lower levels than the 16S rRNA control. *, ensemble significant differences.
MATERIALS AND METHODS
Strains and growth conditions.
The B. subtilis strains used in this study are presented in Table 1. LB medium (Luria-Miller; Carl Roth GmbH, Karlsruhe, Germany) served as a complex medium for all B. subtilis strains, with the corresponding antibiotic (Table 1). Bacteria were cultivated for 8 to 9 h in 5 ml medium at 37°C, with agitation at 300 rpm. Liquid cultures were subsequently plated confluently onto LB agar plates without antibiotics and were incubated overnight at 37°C, resulting in 1-day-old biofilms fully covering the agar plates. The three experimental techniques used in this study, i.e., atomic force microscopy (see Fig. S1 in the supplemental material), macrorheology (see Fig. S2 in the supplemental material), and profilometry, require LB agar plates of different sizes. To ensure identical starting conditions for all experimental approaches, the plating volume was adjusted to the size of the agar plates (for example, 30 μl for agar plates with a diameter of 3.9 cm, as used for AFM measurements). To investigate the effects of ethanol on the material properties of B. subtilis biofilms, immediately, before the start of these experiments, the 1-day-old biofilms were covered with ethanol (99% [or 80%, if indicated]; Sigma-Aldrich, St. Louis, MO, USA). We applied ethanol at such volumes that, even with ongoing ethanol evaporation, enough ethanol remained over the time course of the experiments that the biofilms were fully covered by ethanol. The volume of ethanol applied was also adjusted to the sizes of the agar plates. Images of 1-day-old biofilm colonies, as shown in Fig. 2, represent single biofilm colonies and were taken using a Nikon SMZ1000 stereomicroscope with a DS-Fi2 camera. It should be noted that the strains used in this study form proper biofilms (confluently grown biofilms as well as single biofilm colonies) within 12 h of growth on agar plates. No significant increase in biofilm height was observed after 24 h of growth.
TABLE 1.
B. subtilis strains used in this study
| Strain | Description | Remaining matrix composition | Antibiotic (concn [μg/ml]) | Reference |
|---|---|---|---|---|
| NCIB 3610 | Wild type | Proteins TasA and BslA, exopolysaccharide | None | 45 |
| CA017 | TasA::Kan | Protein BslA, exopolysaccharide | Kanamycin (50) | 48 |
| N24 | BslA::Cam | Protein TasA, exopolysaccharide | Chloramphenicol (5) | 49 |
| ZK3660 | EpsA-O::Tet | Proteins TasA and BslA | Tetracycline (12.5) | 42 |
| B-1 | Wild type | Mainly γ-polyglutamate | None | 51 |
FIG 2.
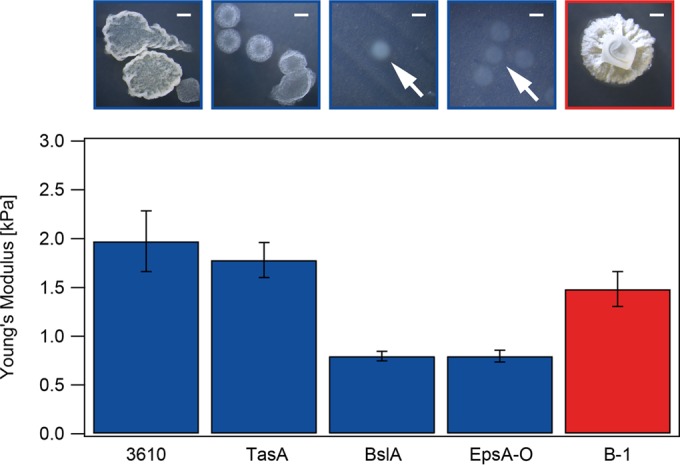
Surface stiffness of different B. subtilis biofilms. Young's modulus values were obtained for 1-day-old biofilms of the wild-type strains B. subtilis B-1 (red) and NCIB 3610 (blue) and three mutant strains of NCIB 3610 (blue). Error bars are Gaussian errors resulting from the error in the method. Photographs above the graph are images of single biofilm colonies. Scale bars, 1 mm. White arrows point to the faint single biofilm colonies of BslA and EpsA-O mutant strains. P values for these data are given in Table S2 in the supplemental material.
Gene expression analysis.
B-1 biofilms were grown and prepared as described above. RNA for gene expression analysis was extracted from biofilms that had been grown for 10 h or 18 h, by using the Qiagen RNeasy extraction kit and following the protocols provided with the kit. Gene expression analysis was performed by IMGM Laboratories, via quantitative real-time PCR (40 cycles) in custom TaqMan gene expression assays for the genes ywsC, blsA, epsH, and tasA. The conserved 16S rRNA housekeeping gene was used as a positive control. Negative controls did not contain genetic material or were performed with the corresponding RNA. Threshold cycle (CT) values given in Fig. 1 represent mean values obtained from 3 biological and 3 technical triplicates. For significance analysis, a two-sample t test was performed using MATLAB R2014a. P values obtained from this significance analysis are given in Table S1 in the supplemental material. Additionally, an “ensemble” significance test (one-way analysis of variance [ANOVA] and Tukey's honestly significant difference test; MATLAB) was performed to confirm that the genes blsA, ywsC, epsH, and tasA were transcribed at significantly lower levels than the 16S rRNA control. The ensemble significance results are indicated in Fig. 1 by asterisks.
AFM settings and cantilevers used in the study.
Force spectroscopy was performed using a JPK NanoWizard BioScience atomic force microscope (with a CellHesion module; JPK Instruments AG). For measurements of the elasticity of the biofilm surface, soft cantilevers with silicon dioxide (SiO2) beads (diameter, 6.62 μm) were used (sQube CP-PNPS-SiO-C-5; NanoAndMore GmbH). According to the manufacturer's data, the resonance frequencies of the cantilevers were ∼17 kHz and the cantilevers had a force constant of 0.08 N/m, as verified by thermal noise analysis (data from JPK Instruments [user manual]). For each individual cantilever, the sensitivity (in nanometers per volt) was calibrated by fitting the retraction part of a force curve measured on plastic under ethanol, thus setting a baseline. The individual spring constants of the cantilevers (in millinewtons per meter) were measured using the thermal noise method (as indicated by JPK Instruments). Experiments were performed in the contact mode using the following parameters of the control JPK Instruments software: sample rate, 6,000 Hz; z-length, 20 μm; set point, 5.0 nN; and speed, 5.0 μm/s.
Surface indentation measurements with AFM.
After the addition of ethanol, due to technical reasons, a minimum of 10 min elapsed before the first measurement could be performed. Each biofilm surface was measured at three different positions. Each of the measurements was composed of 64 single runs, measured at a distance of 1.25 μm from each other. For each strain, 10 experiments were performed on 9 different days. A schematic of the experimental procedure is given in Fig. S1 in the supplemental material. In order to compare the data obtained for the different strains, only those measurements that were obtained between 10 and 25 min after the addition of ethanol are presented in this study (Fig. S3). The obtained force curves (Fig. S1) were analyzed with JPK Instruments data-processing software (version spm-5.1.7). To obtain the Young's modulus, the Hertz model of elastic deformations was applied (53). A Poisson ratio of 0.5 was set, assuming a perfectly incompressible material that deforms elastically. Although this introduces a certain error to the absolute values obtained in this study, it does not interfere with the relative statements of this study. Further data analyses were performed using the software MATLAB R2014a and Igor Pro 4.06. A pairwise significance analysis was performed as described above. The obtained P values are given in Table S2.
Profilometric analysis.
The surface profiles of 1-day-old biofilms were obtained using a NanoFocus μsurf profilometer (NanoFocus AG, Oberhausen, Germany). For each sample, three randomly selected spots with an area of 320 by 300 μm were scanned using a 50× objective. Each scanned area was then evaluated with the software (μsoft version 6.0; NanoFocus AG, Oberhausen, Germany) to obtain the root mean squared roughness (Sq value). Profilometer images were obtained before and after 1 h of biofilm exposure to 2 ml of 99% (or 80%) ethanol. After ethanol exposure, the ethanol was discarded and the samples were kept at room temperature for 10 min, to allow evaporation of the remaining ethanol prior to profilometric analysis. A pairwise significance analysis was performed as described above. The obtained P values are given in Table S2 in the supplemental material.
Rheological characterization.
Rheological measurements were performed using a commercial shear rheometer (MCR 302; Anton Paar GmbH, Graz, Austria) with a 25-mm plate-plate geometry and 300-μm plate separation, in strain-controlled mode (see Fig. S2 in the supplemental material). One-day-old biofilms were harvested from agar plates by manual scraping and pooled. Even though the initial structure of the biofilm was changed due to the biofilm scraping, we saw no large differences in the elastic moduli from when biofilms were grown on the rheometer in situ (Fig. S4). For ethanol treatment, the biofilms were covered with 15 ml of 99% (or 80%) ethanol prior to harvesting and incubated for 10 or 60 min, respectively. After ethanol exposure, the ethanol was discarded and the samples were kept at room temperature for 10 min to allow evaporation of the remaining ethanol prior to rheological characterization. To study the protective effect of γ-polyglutamate on NCIB 3610 biofilms against ethanol-induced changes in biofilm elasticity, the NCIB 3610 biofilms were allowed to grow in the presence of γ-polyglutamate extracted from B-1 biofilms. In detail, γ-polyglutamate was added to liquid NCIB 3610 cultures at a concentration of 4.75 mg/ml. Then, 100 μl of this mixture was plated onto agar plates, from which 1-day-old biofilms were harvested and pooled as described above. γ-Polyglutamate was extracted from B-1 biofilms by following a protocol outlined previously (54); in a difference from that protocol, we used LB medium throughout the procedure. The dialysis step was performed using Spectra/Por dialysis tubes with a molecular mass cutoff value of 12 to 14 kDa (Spectrum Europe, Breda, The Netherlands). Frequency spectra from 0.1 Hz to 10 Hz were obtained at 21°C using small torques (∼1 μN·m) to guarantee a linear material response. A pairwise significance analysis was performed as described above. The P values obtained are given in Tables S2 and S3.
Determination of total and dried masses.
In order to determine the total mass of biofilms produced by wild-type strains NCIB 3610 and B-1, liquid cultures grown under the growth conditions described above were confluently plated onto agar plates (diameter, 9 cm) and cultivated for 18 h. Some of the plates were covered with 99% ethanol for 1 h, in the presence or absence of γ-polyglutamate. The ethanol was discarded, and the plates were dried for 60 min. For all charges of plates, treated or untreated, the biofilm was scraped from the agar and weighed in Eppendorf tubes. We visually checked that all biofilm material was removed and that the underlying agar surface was not damaged during the scraping process. All tubes were frozen in liquid nitrogen at −169°C and put in an Alpha 1-2 lyophilizer (Martin Christ Gefriertrocknungsanlagen GmbH, Osterode am Harz, Germany) at −55°C and 2.1 Pa, where they were kept for 8 h. The tubes were weighed again to determine the dried mass. A pairwise significance analysis was performed as described above. The obtained P values are given in Table S4 in the supplemental material.
RESULTS
One-day-old B-1 biofilms express the genes ywsC, blsA, tasA, and epsH.
The two Bacillus subtilis strains NCIB 3610 and B-1 are known to produce biofilms, and key elements that build up the biofilm matrices of these strains are known (46–51). Here, we performed a sequence comparison of genes involved in biofilm formation in the species B. subtilis (10) by using the B. subtilis strains B-1 and 168 (strain 168 is a well-studied laboratory strain that is genetically similar to the wild-type strain NCIB 3610 [55]) (Table 2). Among these genes are genes encoding key macromolecules forming the biofilm matrix of strain NCIB 3610: the genes for the surface layer protein BslA (49), the fiber-forming protein TasA (47), and the exopolysaccharide (the epsA-O operon is responsible for the production of the exopolysaccharide in strain NCIB 3610 [46]). The wild-type strain B-1 possesses all of these genes, and we found sequence homologies (BLASTx) to the laboratory strain B. subtilis 168 of 75% for tasA, 78% for the epsA-O operon, and 74% for bslA (Table 2). We also performed a gene expression analysis of 10-h-old and 18-h-old B-1 biofilms (see Materials and Methods) (Fig. 1; also see Table S1 in the supplemental material) to confirm that these genes, in addition to the ywsC gene, which is important for the production of γ-polyglutamate, are indeed transcribed in strain B-1. At both time points, mRNAs of all four genes—ywsC, bslA, tasA, and epsH—were produced at levels lower than that of a housekeeping gene, i.e., the 16S rRNA control. This indicates that, in addition to the main component (γ-polyglutamate) described by Morikawa et al. (51), proteins such as the surface layer protein BslA may be present in the B-1 biofilm matrix. The finding that genes such as bslA are transcribed in strain B-1 is somewhat unexpected, as Morikawa et al. (51) reported that the biofilm matrix of strain B-1 is composed predominantly of γ-polyglutamate. In the following sections, we address the question of how these different biofilm matrix components influence the material properties of the two B. subtilis biofilm variants.
TABLE 2.
Homology of genes involved in biofilm formation
| Gene(s) | Sequence position (bp)a |
Homology between strains 168 and B-1 (%) | Biofilm matrix component | Reference | |
|---|---|---|---|---|---|
| Strain B-1 | Strain 168 | ||||
| tasA | 3450573 | 2553866 | 75 | Matrix fiber | 48 |
| epsA-O | 2140149 | 3529911 | 78 | Polysaccharide | 42 |
| bslA | 2873830 | 3187503 | 74 | Surface layer protein | 49 |
| tapA | 3452069 | 2555247 | 72 | Lipoprotein (TasA assembly) | 61 |
| sipW | 489452 | 2554502 | 72 | Type I signal peptidase (surface adhesion) | 62 |
| pgsB or ywsC | 3360079 | 3700627 | 82 | γ-Polyglutamate | 51 |
| degU | 628676 | 3645296 | 86 | Two-component response regulator | 63 |
| abrB | 50582 | 45138 | 91 | Transcription factor | 64 |
| sinR | 3450190 | 2552653 | 97 | Master regulator for biofilm formation | 65 |
The positions show the start sites of the genes in the respective genomes. Comparisons between the genomes were performed with the NCBI online tools LAST and BLASTx. The recently sequenced B. subtilis strain B-1 (GenBank accession no. CP009684) was compared with the well-studied laboratory strain B. subtilis strain 168 (GenBank accession no. CP010052), which is genetically similar to the wild-type strain NCIB 3610 (55, 66).
The surface layer protein BslA contributes to the surface stiffness of NCIB 3610 biofilm.
As a first step, we investigated the surface stiffness of biofilms produced by the two B. subtilis strains NCIB 3610 and B-1 (Table 1). We performed AFM nanoindentation experiments with native 1-day-old biofilms formed by these two strains (see Materials and Methods; also see the supplemental material). At such an early phase of biofilm formation, it is expected that the mechanical properties of these biofilms can still be altered by chemical treatment and that further counteractions are possible. To allow for the mechanical analysis of soft materials, such as 1-day-old biofilms, we chose soft cantilevers with a silicon dioxide bead at the cantilever tip. Measurements of B. subtilis biofilms in air are not possible due to the strong electrostatic interactions between the biofilm and the cantilever; therefore, we covered our fully grown biofilms with a dielectric fluid (99% ethanol) to avoid this issue.
In a first experiment, we investigated the surface stiffness of biofilms generated by the well-studied B. subtilis wild-type strain NCIB 3610 (Fig. 2). As a quantitative measure of the biofilm surface stiffness, we calculated the Young's modulus, E, by fitting the obtained indentation data to the Hertz model (53), which yielded an average Young's modulus, ENCIB 3610, of 1.97 ± 0.31 kPa. In comparison, the average Young's modulus of the solid agar surface, Eagar, was 37.82 ± 5.87 kPa (see Fig. S3 in the supplemental material). The surface stiffness of wild-type B-1 biofilms was in a range similar to that for wild-type NCIB 3610 biofilm, and we measured EB-1 as 1.48 ± 0.18 kPa (Fig. 2). These values represent data obtained within 25 min after ethanol application to the biofilm surface (Fig. S3), as we did not observe any changes in the measured Young's modulus values for all studied strains during this time period. We did, however, detect strong day-to-day variations in the absolute values of the Young's moduli obtained for these wild-type strains; variations were less pronounced for mutant strains with smooth biofilm surfaces, as described below. As both wild-type strains showed very rough colony morphology (Fig. 2), we attribute the variations to differences in the biofilm surface structures. It is virtually impossible to perform AFM indentation measurements on two different biofilms in such a way that identical surface structures are probed, but such differences in biofilm morphology would certainly influence the results of indentation experiments.
In a next step, we investigated how different biofilm matrix components contribute to the observed biofilm surface stiffness. We repeated our AFM nanoindentation experiments with NCIB 3610 mutant strains lacking one particular matrix element (Table 1). Those mutant strain biofilms clearly differed in their macroscopic morphologies; whereas the TasA mutant formed rough biofilm colonies similar to those of the NCIB 3610 wild-type strain, single biofilm colonies of the BslA and EpsA-O mutants were smooth and had no defined colony edges (Fig. 2). We also tried to generate B-1 mutants with the same mutations, but so far, we have been unsuccessful in creating such mutants due to the low transformability of this strain. Biofilms formed by the NCIB 3610 TasA mutant strain (48) had an ETasA of 1.78 ± 0.18 kPa (Fig. 2), a Young's modulus similar to that of the NCIB 3610 wild-type strain. In contrast, the Young's moduli of the EpsA-O (46) and BslA mutant (49) biofilms were significantly decreased, with an EEpsA-O of 0.80 ± 0.05 kPa and an EBslA of 0.80 ± 0.06 kPa (see Table S2 in the supplemental material). These findings imply that the TasA fiber-forming protein does not contribute to the surface stiffness of NCIB 3610 biofilms. In contrast, the remaining two matrix components, i.e., the protein BslA and the exopolysaccharide, are important for this biofilm property. Kobayashi and Iwano (49) have shown that the production of the surface layer protein BslA of strain NCIB 3610 is dependent on the expression of the epsA-O operon, which is responsible for the production of the exopolysaccharide. Thus, the decrease in surface stiffness determined for the EpsA-O mutant may also be due to the additional absence of the BslA protein in this mutant strain, rather than to the absence of the exopolysaccharide itself. The equal decreases in biofilm surface stiffness observed for the EpsA-O and BslA deletion mutants support this hypothesis. Ostrowski et al. (56) showed that BslA is not involved in the synthesis, export, or polymerization of the TasA amyloid fibers or the exopolysaccharide. Hence, we attribute the decrease in the biofilm surface stiffness observed for the BslA mutant to the absence of the BslA protein.
The surface layer protein BslA increases the surface roughness of NCIB 3610 biofilms.
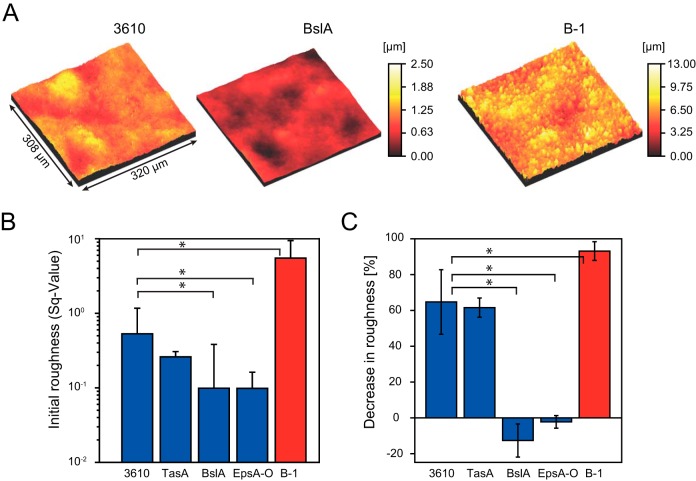
We had seen that the surface stiffness values for the two wild-type biofilms, those of NCIB 3610 and B-1, have similar ranges and that BslA contributes to the surface stiffness of NCIB 3610 biofilms. To further investigate the material properties of NCIB 3610 and B-1 biofilms, we performed a profilometric analysis of the surface roughness of these biofilms. Figure 3A displays the microscopic surface profiles obtained for biofilms formed by the strains NCIB 3610 and B-1. We found that the root mean squared roughness (Sq value) of the B-1 biofilm was ∼10-fold greater than that of the NCIB 3610 wild-type biofilm, with an SqB-1 of 5.5 ± 3.85 and an SqNCIB 3610 of 0.53 ± 0.63 (Fig. 3B; also see Table S2 in the supplemental material). Similar to the macroscopic roughness of single biofilm colonies (Fig. 2), the microscopic surface roughness of the biofilms was lowest for the BslA and EpsA-O mutants (Fig. 3A and B). This suggests that the surface layer protein BslA is mainly responsible for the roughness of NCIB 3610 biofilm surfaces.
FIG 3.
Surface roughness of different B. subtilis biofilms. The root mean squared surface roughness was determined for 1-day-old biofilms of wild-type B. subtilis strains B-1 (red) and NCIB 3610 (blue) and three mutant strains of NCIB 3610 (blue). (A) Profilometer images of wild-type strain NCIB 3610, BslA mutant NCIB 3610, and wild-type strain B-1 biofilms. The color code indicates the sample heights, according to the key. (B) Initial roughness of untreated B. subtilis biofilms. (C) Decreases in the roughness of B. subtilis biofilms after 60 min of exposure to ethanol. Error bars denote the standard deviations for 9 samples from 3 different growth batches. P values for these data are given in Table S2 in the supplemental material. P values below 0.05 indicate significant differences from the data obtained for strain NCIB 3610 and are indicated with an asterisk.
Our data show that biofilms formed by the two B. subtilis wild-type strains B-1 and NCIB 3610 significantly differ in surface roughness (P values are given in Table S3 in the supplemental material). We next addressed the question of whether the surface roughness of these two strains would be affected by exposure of the biofilm to ethanol, a commonly used antibacterial agent. We found that, after 60 min of exposure to 99% ethanol, the surface roughness values for the NCIB 3610 biofilm and the corresponding TasA mutant biofilm were decreased by about 60% (P values of 0.0431 and 0.0735, respectively), whereas the BlsA and EpsA-O mutant biofilms showed no significant changes in surface roughness upon ethanol exposure (P values of 0.8144 and 0.4110, respectively) (Fig. 3C). The latter finding reflects the fact that the biofilm surfaces of both BslA and EpsA-O mutant strains were found to be very smooth even before ethanol application (Fig. 3B). Wild-type B-1 biofilms possessed the greatest initial surface roughness, and this biofilm variant was also affected the most by ethanol exposure; we measured a decrease in the surface roughness of more than 90% (P = 0.0005). As typical ethanol concentrations used for disinfection are in the range of 70 to 80%, we repeated the profilometric experiments for strains NCIB 3610 and B-1 with a concentration of 80% ethanol. We found a similar decrease in roughness for NCIB 3610 but a smaller decrease of about 50% for B-1, compared to the finding when 99% ethanol was applied (see Fig. S5A in the supplemental material). In summary, these data demonstrate that disinfectant chemicals such as ethanol can affect the surface morphology of 1-day-old bacterial biofilms generated by B. subtilis strains B-1 and NCIB 3610.
γ-Polyglutamate protects B. subtilis biofilms from ethanol-induced changes in biofilm bulk elasticity.
So far, we had investigated the morphology and micromechanical properties of 1-day-old B. subtilis biofilm surfaces. Using macrorheology (see the supplemental material), we next studied the bulk viscoelasticity of biofilms formed by the two wild-type strains B-1 and NCIB 3610. For both biofilm variants, we found that the material responses were dominated by elasticity over the whole range of frequencies tested. The storage modulus G′(f), which quantifies the elastic properties of the biofilm, shows a pronounced plateau. Thus, we compared the elastic properties of the different biofilms at a fixed intermediate frequency of 1 Hz. In detail, we measured a G′NCIB 3610 (at 1 Hz) of 584 ± 132 Pa for wild-type NCIB 3610 biofilm and similar values for all three NCIB 3610 mutant strains (Fig. 4A, blue). In contrast, wild-type B-1 biofilms were considerably softer, as the storage moduli were ∼10-fold lower than those obtained for the NCIB 3610 biofilm variants (Fig. 4A, red; Table S2). Upon application of 99% ethanol (see Materials and Methods), we found that the elastic modulus of NCIB 3610 biofilms increased within 10 min, whereas the elastic modulus of B-1 biofilms remained unchanged. Further increases were observed after 60 min of ethanol exposure for all NCIB 3610 biofilm variants but not for the B-1 wild-type biofilm (Fig. 4A). When the biofilms were treated with 80% ethanol, the elastic properties of NCIB 3610 and B-1 biofilms were similar to those of the biofilms treated with 99% ethanol (Fig. S5B). One explanation for this finding may be that the application of ethanol leads to dehydration of the NCIB 3610 biofilm but not of the B-1 biofilm. Biofilms of B. subtilis B-1 are described as consisting mainly of strongly hydrophilic γ-polyglutamate (PGA) (51), a macromolecule that is highly hygroscopic when prepared as a powder. We thus speculate that PGA may prevent B-1 biofilms from ethanol-induced dehydration.
FIG 4.
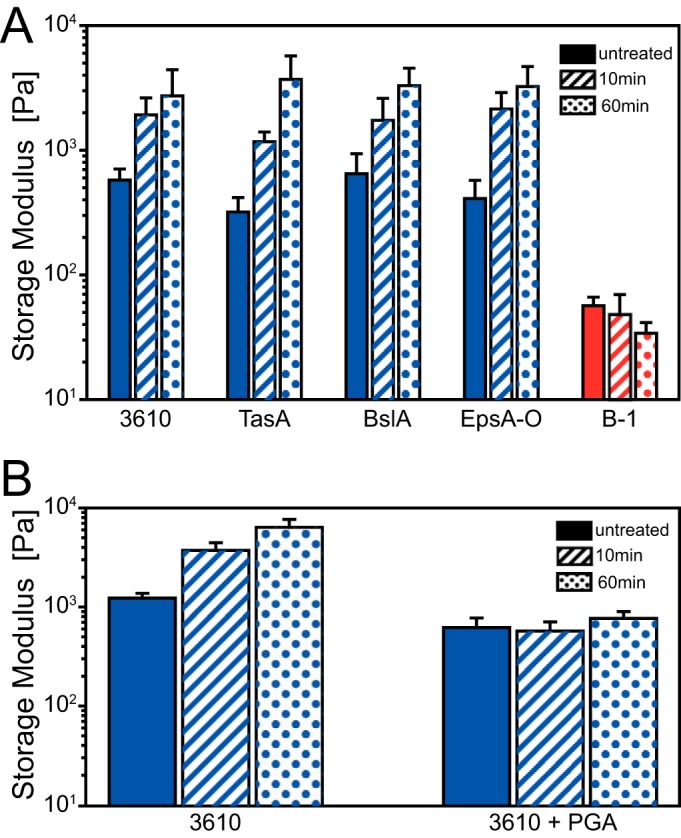
Bulk viscoelasticity of B. subtilis biofilms. (A) Storage modulus values for 1-day-old biofilms of wild-type strains B. subtilis B-1 (red) and NCIB 3610 (blue) and three mutant strains of NCIB 3610 (blue); (B) storage modulus values for 1-day-old biofilms of wild-type B. subtilis strain NCIB 3610 grown in the presence or absence of γ-polyglutamate (PGA) and after different periods of exposure to ethanol (see Materials and Methods). Biofilms were not treated or were exposed to ethanol for 10 or 60 min, as indicated. Error bars denote the standard deviations for 9 samples from 3 different growth batches. P values for these data are given in Tables S2 and S3 in the supplemental material.
To test this hypothesis, we grew NCIB 3610 biofilms in the presence of purified PGA and repeated our macrorheological experiments (see Materials and Methods). Indeed, such PGA-enriched NCIB 3610 biofilms were protected from the ethanol-induced stiffening (Fig. 4B; also see Table S4 in the supplemental material). This demonstrates the importance of γ-polyglutamate for the viscoelastic properties of B-1 biofilms and its potential to protect B. subtilis biofilms from chemically induced changes in their elastic properties.
Finally, we measured the total masses of NCIB 3610 and B-1 biofilms and compared them to the corresponding dried masses (Fig. 5), both with and without ethanol exposure (see Materials and Methods). On average, B-1 biofilms had an ∼4-fold greater biomass than did biofilms of strain NCIB 3610 (1,170 mg/agar plate and 303 mg/agar plate for B-1 and NCIB 3610 biofilms, respectively). After ethanol exposure, the total masses were decreased by ∼40% and ∼60% for biofilms generated by strains NCIB 3610 and B-1, respectively (Fig. 5; also see Table S4 in the supplemental material). However, this difference in the total mass decreases observed between the two biofilm variants was not statistically significant (P = 0.2231), indicating that the observed total mass decrease was approximately the same for B-1 and NCIB 3610 biofilms. In a next step, we repeated these experiments with both biofilm variants grown in the presence of γ-polyglutamate (see Materials and Methods). Interestingly, we obtained values for the total masses of biofilms grown in the presence of γ-polyglutamate after ethanol exposure that were similar to those of biofilms that had not been treated with ethanol (Fig. 5). This indicates that dehydration occurs for both types of B. subtilis biofilms and to comparable degrees, but this effect can be counteracted by artificially increasing the amounts of γ-polyglutamate in these biofilms. We conclude that, although PGA can prevent ethanol-induced stiffening of biofilms, this protective effect does not necessarily stem from a prevention of biofilm dehydration.
FIG 5.
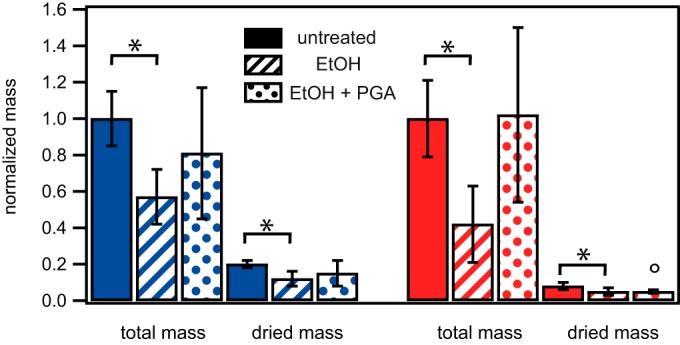
Comparison of normalized total and dry masses of biofilms formed by the wild-type strains NCIB 3610 and B-1. This bar plot shows the total produced masses of 1-day-old biofilms of the wild-type strains NCIB 3610 (blue) and B-1 (red), as well as the dry masses of biofilms of these two wild-type strains, as determined by lyophilization (see Materials and Methods). Biofilms were not treated, were treated with ethanol (EtOH) for 60 min, or were grown in the presence of γ-PGA and then treated with ethanol for 60 min, as indicated. The circle indicates that under these particular conditions, a subset of samples could not be successfully lyophilized, indicating that ethanol might be taken up into the B-1 biofilm. The data given here represent the dry weights of the samples for which lyophilization was successful. Error bars denote the standard deviations. P values for these data are given in Table S4 in the supplemental material. P values below 0.05 indicate that the data obtained in the presence of ethanol were significantly different from those obtained in the absence of ethanol for that strain and are indicated with an asterisk.
DISCUSSION
In this study, we demonstrate how specific biomolecules of the biofilm matrices of B. subtilis NCIB 3610 and B-1 contribute to the structural and mechanical characteristics of the corresponding biofilms. The key macromolecules described as forming the biofilm matrix of biofilms produced by strain NCIB 3610 are the proteins BslA and TasA (48, 49), as well as the exopolysaccharide expressed from the epsA-O operon (46). In contrast, B-1 biofilms are described as containing mainly γ-polyglutamate (51), which is expressed from the ywsC gene. However, to date, it is not known whether the key matrix components found in NCIB 3610 biofilms (e.g., the surface layer protein BslA) are also present in B-1 biofilms. Recently, the whole genome of the wild-type strain B-1 was sequenced (57), enabling a sequence comparison of strain B-1 with other B. subtilis genomes, such as B. subtilis 168, as performed in this study. A homology analysis of genes important for biofilm formation by the species B. subtilis (10), in combination with our data from the gene expression analysis, indicates that strain B-1 is in principle capable of expressing all of the biofilm matrix elements described as being part of the biofilm matrix of strain NCIB 3610. However, Morikawa et al. (51) showed that the final biofilm matrix of strain B-1 consists mainly of γ-polyglutamate, a biofilm matrix component whose expression is suppressed in NCIB 3610 biofilms (46, 51). As environmental conditions can affect the exact composition of the biofilm matrix (5), we focused on the above-mentioned key components of biofilms. We attribute the observed differences in the physical properties of strains B-1 and NCIB 3610 to their respective biofilm matrix compositions, particularly the presence of γ-polyglutamate in B-1 biofilms (51) and its absence in NCIB 3610 biofilms (46, 51).
Analysis of the surface elasticity of 1-day-old biofilms of strain NCIB 3610 revealed a Young's modulus of 2 kPa. In an earlier study, Asally et al. (24) determined a Young's modulus of 25 kPa for 7-day-old biofilms of wild-type strain NCIB 3610. This suggests that, for this bacterial strain, the biofilm becomes stiffer during maturation. This is in contrast to biofilms generated by Pseudomonas aeruginosa, for which both the elastic and viscous properties were found to be reduced during maturation (23). However, that study was performed with biofilms cultivated in flow cells, which might have influenced the experimental outcomes. This suggests that the development of the material properties of biofilms as a function of biofilm growth time may depend on both the particular bacterial species and the environmental conditions. Thus, detailed information on the material properties of biofilms in the different stages of biofilm formation is required to identify the ideal “window of opportunity” to fight bacterial biofilm formation on medical devices or industrial surfaces (19). This window of opportunity is most likely before the biofilm becomes established. Hence, future in situ investigations focusing on the transition from the two-dimensional (microcolony) growth to the three-dimensional growth of bacterial biofilms are needed, but such studies will probably require a combination of several experimental techniques to cover these two stages of biofilm formation. Information on the establishment of material properties is also helpful when biofilm growth is induced on purpose (58), e.g., for industrial applications, such as wastewater treatment.
Asally et al. observed that, for 7-day-old B. subtilis NCIB 3610 biofilms, general overproduction of the extracellular matrix (ECM) increased the biofilm surface stiffness by a factor of 2, whereas an SrfA deletion strain (SrfA regulates ECM production through cell-to-cell signaling) had a reduced surface stiffness of 8.1 ± 11 kPa (24). Here, we identified the surface layer protein BslA to be mainly responsible for the surface roughness and the surface stiffness of NCIB 3610 biofilms, whereas the contributions of the fiber-forming protein TasA and the exopolysaccharide were only minor.
Unlike with the surface properties of B. subtilis biofilms, the bulk elasticity values of NCIB 3610 biofilms were similar for all tested mutant strains. This is again in contrast to what occurs with P. aeruginosa biofilms; the exopolysaccharide Psl contributes significantly to the elasticity of P. aeruginosa biofilms (59). The bulk elasticity of the B-1 biofilms was lower than that of NCIB 3610 biofilms by a factor of 10, yet the surface stiffness values were comparable for the two biofilm variants. We found that the B-1 biofilm surface roughness was significantly greater than the roughness of NCIB 3610 biofilm surfaces. A high level of biofilm roughness can promote the adhesion of additional bacteria from the fluid and thus promote further biofilm growth (60).
Also, we found different mechanical characteristics for the biofilm variants (strains NCIB 3610 and B-1), which we attribute to the presence of γ-PGA in B-1 biofilms. γ-Polyglutamate seems to protect B-1 biofilms from ethanol-induced changes in biofilm bulk elasticity. This effect may be explained in part by the ability of PGA to trap water inside the biofilm matrix, thus reducing dehydration. Artificially PGA-enriched NCIB 3610 biofilms are slightly softer than their unmodified counterparts, which agrees with the notion that more water is trapped in those modified biofilms.
According to this hypothesis, considerable protection from ethanol-induced dehydration is reached only when the PGA content is extremely high. For biofilms formed by strain B-1, we observed bubble formation after the application of ethanol (Fig. 6; also see Movie S1 in the supplemental material), indicating that ethanol is taken up by B-1 biofilms and gas is released from the biofilm matrix. This penetration of ethanol deep into the biofilm matrix may induce chemical alterations that may stabilize the B-1 biofilms, thus preventing changes in the bulk mechanical properties of the material.
FIG 6.
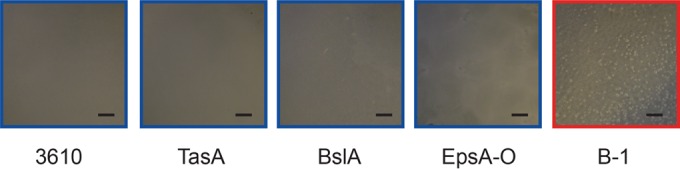
Images of plated B. subtilis biofilms immediately after the application of 99% ethanol. Images were obtained for the wild-type strains B. subtilis B-1 (red) and NCIB 3610 and three mutant strains of NCIB 3610 (blue). Scale bars, 1 mm.
Our findings demonstrate that strains of the same bacterial species, such as Bacillus subtilis strains NCIB 3610 and B-1, can produce biofilms with diverse physical properties.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Kolter for Bacillus subtilis strains NCIB 3610, CA017, and ZK3660. We thank K. Kobayashi for B. subtilis strain N24 and M. Morikawa for B. subtilis strain B-1. We thank H. Gaub, J. O. Rädler, and E. Frey for their support and fruitful discussions. For PGA purification, we thank K. Bidmon. For technical assistance, we thank M. Bennoit, F. Moormann, A. Mader, S. Schaffer, and M. Heinlein.
Funding Statement
Financial support by the Deutsche Forschungsgemeinschaft through SFB 863, project B11 (Mechanics of Bacterial Biofilms), is gratefully acknowledged. Additional support from the excellence cluster Nanosystems Initiative Munich and the Center for Nanoscience is acknowledged.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03957-15.
REFERENCES
- 1.Sutherland I. 2001. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology 147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 2.Marvasi M, Visscher PT, Casillas Martinez L. 2010. Exopolymeric substances (EPS) from Bacillus subtilis: polymers and genes encoding their synthesis. FEMS Microbiol Lett 313:1–9. doi: 10.1111/j.1574-6968.2010.02085.x. [DOI] [PubMed] [Google Scholar]
- 3.Wei Q, Ma LZ. 2013. Biofilm matrix and its regulation in Pseudomonas aeruginosa. Int J Mol Sci 14:20983–21005. doi: 10.3390/ijms141020983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hall-Stoodley L, Costerton JW, Stoodley P. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol 2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 5.Flemming H-C, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol 8:623–633. [DOI] [PubMed] [Google Scholar]
- 6.Sauer K, Rickard AH, Davies DG. 2007. Biofilms and biocomplexity. Microbe 2:347–353. [Google Scholar]
- 7.Zijnge V, van Leeuwen MBM, Degener JE, Abbas F, Thurnheer T, Gmür R, Harmsen HJM. 2010. Oral biofilm architecture on natural teeth. PLoS One 5:e9321. doi: 10.1371/journal.pone.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramey BE, Koutsoudis M, von Bodman SB, Fuqua C. 2004. Biofilm formation in plant-microbe associations. Curr Opin Microbiol 7:602–609. doi: 10.1016/j.mib.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 9.Danhorn T, Fuqua C. 2007. Biofilm formation by plant-associated bacteria. Annu Rev Microbiol 61:401–422. doi: 10.1146/annurev.micro.61.080706.093316. [DOI] [PubMed] [Google Scholar]
- 10.Vlamakis H, Chai Y, Beauregard P, Losick R, Kolter R. 2013. Sticking together: building a biofilm the Bacillus subtilis way. Nat Rev Microbiol 11:157–168. doi: 10.1038/nrmicro2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cairns LS, Hobley L, Stanley-Wall NR. 2014. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Mol Microbiol 93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drescher K, Shen Y, Bassler BL, Stone HA. 2013. Biofilm streamers cause catastrophic disruption of flow with consequences for environmental and medical systems. Proc Natl Acad Sci U S A 110:4345–4350. doi: 10.1073/pnas.1300321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simoes M, Cleto S, Pereira MO, Vieira MJ. 2007. Influence of biofilm composition on the resistance to detachment. Water Sci Technol 55:473–480. doi: 10.2166/wst.2007.293. [DOI] [PubMed] [Google Scholar]
- 14.Simoes M, Simoes LC, Vieira MJ. 2009. Species association increases biofilm resistance to chemical and mechanical treatments. Water Res 43:229–237. doi: 10.1016/j.watres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Davies D. 2003. Understanding biofilm resistance to antibacterial agents. Nat Rev Drug Discov 2:114–122. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- 16.Jones WL, Sutton MP, McKittrick L, Stewart PS. 2011. Chemical and antimicrobial treatments change the viscoelastic properties of bacterial biofilms. Biofouling 27:207–215. doi: 10.1080/08927014.2011.554977. [DOI] [PubMed] [Google Scholar]
- 17.Lieleg O, Caldara M, Baumgartel R, Ribbeck K. 2011. Mechanical robustness of Pseudomonas aeruginosa biofilms. Soft Matter 7:3307–3314. doi: 10.1039/c0sm01467b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zrelli K, Galy O, Latour-Lambert P, Kirwan L, Ghigo JM, Beloin C, Henry N. 2013. Bacterial biofilm mechanical properties persist upon antibiotic treatment and survive cell death. New J Physics 15:125026. doi: 10.1088/1367-2630/15/12/125026. [DOI] [Google Scholar]
- 19.Donlan RM. 2002. Biofilms: microbial life on surfaces. Emerg Infect Dis 8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costerton JW, Stewart PS. 2001. Battling biofilms. Sci Am 285:74–81. doi: 10.1038/scientificamerican0701-74. [DOI] [PubMed] [Google Scholar]
- 21.Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, Pollock DA, Cardo DM. 2007. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122:160–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penesyan A, Gillings M, Paulsen IT. 2015. Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20:5286–5298. doi: 10.3390/molecules20045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lau PC, Dutcher JR, Beveridge TJ, Lam JS. 2009. Absolute quantitation of bacterial biofilm adhesion and viscoelasticity by microbead force spectroscopy. Biophys J 96:2935–2948. doi: 10.1016/j.bpj.2008.12.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Asally M, Kittisopikul M, Rue P, Du Y, Hu Z, Cagatay T, Robinson AB, Lu H, Garcia-Ojalvo J, Suel GM. 2012. Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc Natl Acad Sci U S A 109:18891–18896. doi: 10.1073/pnas.1212429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bol M, Ehret AE, Bolea Albero A, Hellriegel J, Krull R. 2013. Recent advances in mechanical characterisation of biofilm and their significance for material modelling. Crit Rev Biotechnol 33:145–171. doi: 10.3109/07388551.2012.679250. [DOI] [PubMed] [Google Scholar]
- 26.Wilking JN, Angelini TE, Seminara A, Brenner MP, Weitz DA. 2011. Biofilms as complex fluids. MRS Bull 36:385–391. doi: 10.1557/mrs.2011.71. [DOI] [Google Scholar]
- 27.Wright CJ, Shah MK, Powell LC, Armstrong I. 2010. Application of AFM from microbial cell to biofilm. Scanning 32:134–149. doi: 10.1002/sca.20193. [DOI] [PubMed] [Google Scholar]
- 28.Pavlovsky L, Younger JG, Solomon MJ. 2013. In situ rheology of Staphylococcus epidermidis bacterial biofilms. Soft Matter 9:122–131. doi: 10.1039/C2SM27005F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rupp CJ, Fux CA, Stoodley P. 2005. Viscoelasticity of Staphylococcus aureus biofilms in response to fluid shear allows resistance to detachment and facilitates rolling migration. Appl Environ Microbiol 71:2175–2178. doi: 10.1128/AEM.71.4.2175-2178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grumbein S, Opitz M, Lieleg O. 2014. Selected metal ions protect Bacillus subtilis biofilms from erosion. Metallomics 6:1441–1450. doi: 10.1039/C4MT00049H. [DOI] [PubMed] [Google Scholar]
- 31.Kesel S, Mader A, Seeberger PH, Lieleg O, Opitz M. 2014. Carbohydrate coating reduces adhesion of biofilm-forming Bacillus subtilis to gold surfaces. Appl Environ Microbiol 80:5911–5917. doi: 10.1128/AEM.01600-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kannan A, Karumanchi SL, Krishna V, Thiruvengadam K, Ramalingam S, Gautam P. 2014. Nanoscale investigation on Pseudomonas aeruginosa biofilm formed on porous silicon using atomic force microscopy. Scanning 36:551–553. doi: 10.1002/sca.21148. [DOI] [PubMed] [Google Scholar]
- 33.Oh YJ, Lee NR, Jo W, Jung WK, Lim JS. 2009. Effects of substrates on biofilm formation observed by atomic force microscopy. Ultramicroscopy 109:874–880. doi: 10.1016/j.ultramic.2009.03.042. [DOI] [PubMed] [Google Scholar]
- 34.Picioreanu C, van Loosdrecht MC, Heijnen JJ. 1998. Mathematical modeling of biofilm structure with a hybrid differential-discrete cellular automaton approach. Biotechnol Bioeng 58:101–116. doi:. [DOI] [PubMed] [Google Scholar]
- 35.Ehret AE, Bol M. 2012. Modelling mechanical characteristics of microbial biofilms by network theory. J R Soc Interface doi: 10.1098/rsif.2012.0676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Epstein AK, Pokroy B, Seminara A, Aizenberg J. 2011. Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc Natl Acad Sci U S A 108:995–1000. doi: 10.1073/pnas.1011033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seminara A, Angelini TE, Wilking JN, Vlamakis H, Ebrahim S, Kolter R, Weitz DA, Brenner MP. 2012. Osmotic spreading of Bacillus subtilis biofilms driven by an extracellular matrix. Proc Natl Acad Sci U S A 109:1116–1121. doi: 10.1073/pnas.1109261108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beauregard PB, Chai Y, Vlamakis H, Losick R, Kolter R. 2013. Bacillus subtilis biofilm induction by plant polysaccharides. Proc Natl Acad Sci U S A 110:E1621–E1630. doi: 10.1073/pnas.1218984110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilking JN, Zaburdaev V, De Volder M, Losick R, Brenner MP, Weitz DA. 2013. Liquid transport facilitated by channels in Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 110:848–852. doi: 10.1073/pnas.1216376110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mielich-Suss B, Lopez D. 2015. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ Microbiol 17:555–565. doi: 10.1111/1462-2920.12527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bleich R, Watrous JD, Dorrestein PC, Bowers AA, Shank EA. 2015. Thiopeptide antibiotics stimulate biofilm formation in Bacillus subtilis. Proc Natl Acad Sci U S A 112:3086–3091. doi: 10.1073/pnas.1414272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chai Y, Beauregard PB, Vlamakis H, Losick R, Kolter R. 2012. Galactose metabolism plays a crucial role in biofilm formation by Bacillus subtilis. mBio 3:e00184-12. doi: 10.1128/mBio.00184-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aguilar C, Vlamakis H, Guzman A, Losick R, Kolter R. 2010. KinD is a checkpoint protein linking spore formation to extracellular-matrix production in Bacillus subtilis biofilms. mBio 1:e00035-10. doi: 10.1128/mBio.00035-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang W, Dai W, Tsai SM, Zehnder SM, Sarntinoranont M, Angelini TE. 2015. Surface indentation and fluid intake generated by the polymer matrix of Bacillus subtilis biofilms. Soft Matter 11:3612–3617. doi: 10.1039/C5SM00148J. [DOI] [PubMed] [Google Scholar]
- 45.Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. 2001. Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci U S A 98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Branda SS, Chu F, Kearns DB, Losick R, Kolter R. 2006. A major protein component of the Bacillus subtilis biofilm matrix. Mol Microbiol 59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- 47.Romero D, Aguilar C, Losick R, Kolter R. 2010. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A 107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vlamakis H, Aguilar C, Losick R, Kolter R. 2008. Control of cell fate by the formation of an architecturally complex bacterial community. Genes Dev 22:945–953. doi: 10.1101/gad.1645008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kobayashi K, Iwano M. 2012. BslA(YuaB) forms a hydrophobic layer on the surface of Bacillus subtilis biofilms. Mol Microbiol 85:51–66. doi: 10.1111/j.1365-2958.2012.08094.x. [DOI] [PubMed] [Google Scholar]
- 50.Hobley L, Ostrowski A, Rao FV, Bromley KM, Porter M, Prescott AR, MacPhee CE, van Aalten DM, Stanley-Wall NR. 2013. BslA is a self-assembling bacterial hydrophobin that coats the Bacillus subtilis biofilm. Proc Natl Acad Sci U S A 110:13600–13605. doi: 10.1073/pnas.1306390110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morikawa M, Kagihiro S, Haruki M, Takano K, Branda S, Kolter R, Kanaya S. 2006. Biofilm formation by a Bacillus subtilis strain that produces γ-polyglutamate. Microbiology 152:2801–2807. doi: 10.1099/mic.0.29060-0. [DOI] [PubMed] [Google Scholar]
- 52.Scoffone V, Dondi D, Biino G, Borghese G, Pasini D, Galizzi A, Calvio C. 2013. Knockout of pgdS and ggt genes improves γ-PGA yield in B. subtilis. Biotechnol Bioeng 110:2006–2012. doi: 10.1002/bit.24846. [DOI] [PubMed] [Google Scholar]
- 53.Kuznetsova TG, Starodubtseva MN, Yegorenkov NI, Chizhik SA, Zhdanov RI. 2007. Atomic force microscopy probing of cell elasticity. Micron 38:824–833. doi: 10.1016/j.micron.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Steinbüchel A, Oppermann-Sanio FB, Ewering C, Pötter M. 2013. Mikrobiologisches Praktikum Versuch und Theorie, vol 2 Springer Spektrum Verlag, Berlin, Germany. [Google Scholar]
- 55.McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. 2011. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol 193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. 2011. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J Bacteriol 193:4821–4831. doi: 10.1128/JB.00223-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kesel S, Moormann F, Gumperlein I, Mader A, Morikawa M, Lieleg O, Opitz M. 2014. Draft genome sequence of the biofilm-producing Bacillus subtilis strain B-1, isolated from an oil field. Genome Announc 2(6):e01163-14. doi: 10.1128/genomeA.01163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morikawa M. 2006. Beneficial biofilm formation by industrial bacteria Bacillus subtilis and related species. J Biosci Bioeng 101:1–8. doi: 10.1263/jbb.101.1. [DOI] [PubMed] [Google Scholar]
- 59.Chew SC, Kundukad B, Seviour T, van der Maarel JRC, Yang L, Rice SA, Doyle P, Kjelleberg S. 2014. Dynamic remodeling of microbial biofilms by functionally distinct exopolysaccharides. mBio 5:e01536-14. doi: 10.1128/mBio.01536-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y, Monroy GL, Derlon N, Janjaroen D, Huang C, Morgenroth E, Boppart SA, Ashbolt NJ, Liu WT, Nguyen TH. 2015. Role of biofilm roughness and hydrodynamic conditions in Legionella pneumophila adhesion to and detachment from simulated drinking water biofilms. Environ Sci Technol 49:4274–4282. doi: 10.1021/es505842v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Driks A. 2011. Tapping into the biofilm: insights into assembly and disassembly of a novel amyloid fibre in Bacillus subtilis. Mol Microbiol 80:1133–1136. doi: 10.1111/j.1365-2958.2011.07666.x. [DOI] [PubMed] [Google Scholar]
- 62.Terra R, Stanley-Wall NR, Cao G, Lazazzera BA. 2012. Identification of Bacillus subtilis SipW as a bifunctional signal peptidase that controls surface-adhered biofilm formation. J Bacteriol 194:2781–2790. doi: 10.1128/JB.06780-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hsueh YH, Cozy LM, Sham LT, Calvo RA, Gutu AD, Winkler ME, Kearns DB. 2011. DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis. Mol Microbiol 81:1092–1108. doi: 10.1111/j.1365-2958.2011.07755.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chumsakul O, Takahashi H, Oshima T, Hishimoto T, Kanaya S, Ogasawara N, Ishikawa S. 2011. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res 39:414–428. doi: 10.1093/nar/gkq780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Colledge VL, Fogg MJ, Levdikov VM, Leech A, Dodson EJ, Wilkinson AJ. 2011. Structure and organisation of SinR, the master regulator of biofilm formation in Bacillus subtilis. J Mol Biol 411:597–613. doi: 10.1016/j.jmb.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, Bai R, Wyss M, Perkins JB. 2008. The origins of 168, W23, and other Bacillus subtilis legacy strains. J Bacteriol 190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.