Abstract
Polycomb Group (PcG) proteins are transcriptional repressors that epigenetically modify chromatin and participate in the establishment and maintenance of cell fates. These proteins play important roles in both stem cell self-renewal and in cancer development. Our understanding of their mechanism of action has greatly advanced over the past 10 years, but many unanswered questions remain. In this review, we present the currently available experimental data that connect PcG protein function with some of the key processes which govern somatic stem cell activity. We also highlight recent studies suggesting that a delicate balance in PcG gene dosage is crucial for proper stem cell homeostasis and prevention of cancer stem cell development.
Stem Cells and Cancer
Throughout our lives, mature cells in adult tissues are continuously replenished through the activity of small populations of somatic stem cells. The defining feature of these cells is their capacity to undergo self-renewal division coupled with maintenance of multipotency. Somatic stem cells have been identified for most tissues (blood, brain, muscle, skin, gut, etc.), and harnessing their regenerative properties offers a great potential for future therapies. At the molecular level, much about self-renewal remains to be elucidated. However, we can envision this process as the fine orchestration of cell-cycle regulation with cell-fate decisions. To self-renew, stem cells must therefore enter the cell cycle and progress successfully through cell division. During this process, genome integrity has to be preserved through the coordinated regulation of cell-cycle checkpoints and DNA damage repair. While doing so, they must also ensure that at least one daughter cell restricts programs leading to differentiation, senescence, or apoptosis, thus retaining stemness.
Accumulating evidence indicates that a subpopulation of cancer cells within tumors possess stem cell-like properties. Bonnet and Dick (1997) showed that most leukemic blasts are limited in their proliferative capacity and must be constantly replenished by a rare self-renewing population of “leukemic stem cells.” Similar findings have been reported for cancers of the breast, brain, colon, ovary, pancreas, and prostate (Al-Hajj et al., 2003; Li et al., 2007; O’Brien et al., 2007; Singh et al., 2003; Zhang et al., 2008b). Thus, like normal tissues, tumors appear to be organized in a hierarchy that depends on the self-renewing and ever expanding “cancer stem cell,” which most likely retains remnants of the normal developmental program. In support of this model, cancer cells frequently exhibit stem cell-like gene expression and chromatin structure signatures (Ben-Porath et al., 2008; Widschwendter et al., 2007). This predicts similarities in the genes that determine self-renewal of normal and cancer stem cells and highlights the importance of identifying the key components regulating this function. As detailed below, the Polycomb Group (PcG) genes represent prime candidates for determining activity of normal and cancer stem cells. In this review, we discuss the proposed function of PcG proteins in stem cell activity with a particular focus on their role in cell-cycle regulation, differentiation, apoptosis, and senescence. We also briefly describe the growing importance of PcG genes in cancer development.
Polycomb Complexes
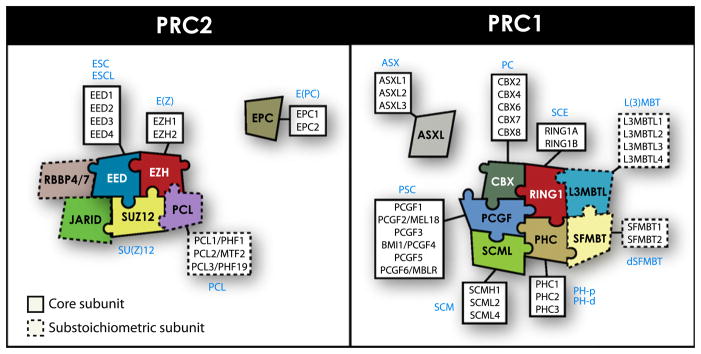
PcG genes were identified in Drosophila more than 30 years ago as regulators of anterior-posterior body patterning through the repression of Hox genes. They have since been recognized as global epigenetic transcriptional repressors and key regulators of cell fate (reviewed in Schwartz and Pirrotta, 2007). The Polycomb family comprises a structurally diverse set of proteins which assemble into chromatin-associated complexes. The composition of these complexes is variable and context-dependent (e.g., differentiation status). However, in mammals, two main families of PcG chromatin-modifying complexes, named Polycomb Repressive Complexes 1 and 2 (PRC1, PRC2) have been identified. The core of the PRC1 complex includes one subunit of the PCGF, CBX, PHC, SCML, and RING1 paralog groups (Figure 1, right panel) (Levine et al., 2002; Valk-Lingbeek et al., 2004). The RING1 protein has monoubiquitylation E3 ligase activity specific for the lysine 119 of H2A (H2AK119ub), a mark associated with repressive chromatin structure (de Napoles et al., 2004; Wang et al., 2004). Although less well characterized, L3MBTL and SFMBT proteins are also found in PRC1 complexes, whereas ASXL1 was recently identified in the new Polycomb Repressive H2A Deubiquitinase complex (Grimm et al., 2009; Ohtsubo et al., 2008; Peterson et al., 2004; Peterson et al., 1997; Sánchez et al., 2007; Scheuermann et al., 2010). The core of PRC2 complexes comprises SUZ12, one of the EED isoforms and the histone methyltransferase EZH1 or EZH2, which catalyze the trimethylation of lysine 27 of histone H3 (H3K27me3), another typical epigenetic silencing mark (Figure 1, left panel) (Cao et al., 2002; Cao and Zhang, 2004; Czermin et al., 2002; Kirmizis et al., 2004; Kuzmichev et al., 2002). The PRC2-associated PCL proteins are not essential for complex formation and stability but are required for enhancement of its methyltransferase activity (Nekrasov et al., 2007; Sarma et al., 2008). Similarly, RBBP4/7 and JARID proteins are cofactors that help to recruit and modulate the enzymatic activity of PRC2 (Cao et al., 2002; Kuzmichev et al., 2002; Landeira et al., 2010; Pasini et al., 2008; Peng et al., 2009; Shen et al., 2009). The incompletely characterized PcG protein EPC1, which was found in a repressive E2F6-EZH2 complex, can also be considered as an additional PRC2-associated protein (Attwooll et al., 2005). When targeted to specific loci, PcG complexes repress gene expression mainly through epigenetic modification of histone tails and compaction of chromatin. The current model of gene silencing by PcG complexes is summarized in Figure 2. Detailed information about the biochemical characterization and recruitment of PcG complexes to target genes can be found in other recent reviews (Surface et al., 2010; Bracken and Helin, 2009; Schuettengruber and Cavalli, 2009; Simon and Kingston, 2009).
Figure 1. Diversity of PRC1 and PRC2 Complexes Formed by Vertebrate PcG Proteins.
Subunits of the PRC1 (right panel) and PRC2 (left panel) complexes are indicated. The Drosophila homolog of each subunit is indicated in light blue. Multiple combinations of paralog subunits can generate a diversity of PRC1 and PRC2 complexes, which likely have specific and shared functions. Some subunits seem to be present in substoichiometric amounts and interact with the PcG complexes in a cell-context-dependent manner. The core subunits and the substoichiometric subunits are identified. The contacts illustrated in the diagrams are not intended to represent the actual interactions. Involvement of EPC and ASXL subunits with PRC2 or PRC1 complexes is still unclear and requires further investigations.
Figure 2. Coordinated Epigenetic Silencing Activity of PcG Complexes.
Following recruitment of the PRC2 complex to chromatin, the histone methyl-transferase EZH1/2 catalyzes the trimethylation of the lysine 27 of histone H3 (H3K27me3). Subsequent recruitment of the PRC1 complex occurs in part through affinity binding of the chromodomain of the CBX subunit to the H3K27me3 covalent mark. The PRC1 RING1 E3 ligase then monoubiquitylates the lysine 119 of histone H2A (H2AK119ub1), which was proposed to consolidate transcriptional repression by preventing access to chromatin remodelers, inhibiting RNA polymerase II-dependent transcriptional elongation and facilitating chromatin compaction (Francis et al., 2004; Zhou et al., 2008). PRC1 has also been reported to be targeted to chromatin independently of PRC2 (Boyer et al., 2006; Ku et al., 2008; Schoeftner et al., 2006). The ASXL subunit has recently been found to be involved in a H2A deubiquitinase complex required for PcG-mediated repression, but its precise role remains unclear.
During the evolution from invertebrates to vertebrates the number of PcG genes underwent significant expansion, rising from approximately 15 in Drosophila to 37 in Mouse and Humans. The apparent gene duplications that occurred, most notably in mammals, resulted in alternative paralogous subunits that diverged in their structural domains (Kerppola, 2009; Whitcomb et al., 2007). In some situations, this structural diversification confers to paralogs the ability to execute novel activities. For example, expression of the PRC1 protein BMI1 promotes cell proliferation and is essential for maintenance of cancer cells (Lessard and Sauvageau, 2003), while its paralog PCGF2/MEL18 (78% amino acid similarity; 97% similarity for RING domain) blocks cell proliferation and has tumor suppressive functions (Guo et al., 2007; Tetsu et al., 1998). Underlying the complexity of PcG biology, accumulating evidence indicates the existence of multiple PRC1 and PRC2 complexes whose specific function is defined by the activity of the distinct paralogs that participate in these complexes (Figure 1). A good characterization of these different PcG complexes and their specific or shared functions is greatly lacking, which impedes our ability to uncover the precise roles PcG play in stem cell activity and cancer development.
PcG and Regulation of Cell-Cycle Progression
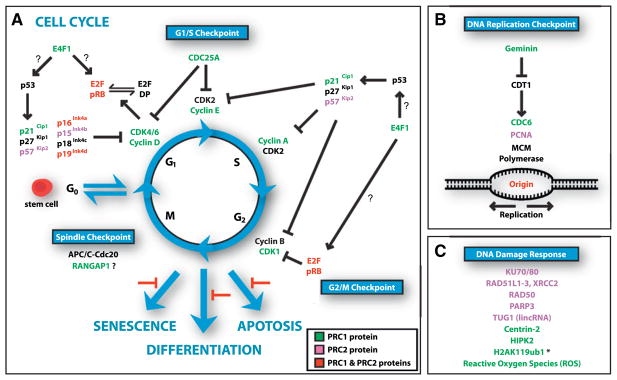
Accumulating evidence indicate that PcG proteins play important functions in multiple cell cycle checkpoints (Figure 3).
Figure 3. Regulation of Cell-Cycle Checkpoints and DNA Damage Repair Pathways by PcG.
(A) The essential role of PcG proteins in somatic stem cell self-renewal is multifaceted. It involves the regulation of several cell-cycle checkpoints, DNA damage repair pathways, control of differentiation, and prevention of senescence and apoptosis programs. Cell-cycle regulators directly interacting with or repressed by PRC1 and/or PRC2 proteins are indicated in green (PRC1), purple (PRC2), and red (PRC1 and PRC2). The question mark indicates that the role of PcG proteins in the process remains to be confirmed.
(B) PRC1 proteins directly interact with Geminin and mediate its degradation, thereby stabilizing the DNA replication licensing factor CDT1. PcG proteins also interact with CDC6, colocalize with PCNA, and associate with late DNA replication origins.
(C) PcG proteins play an important role in maintaining genome integrity by regulating and interacting with multiple proteins and long noncoding RNAs (lincRNA) involved in DNA damage repair pathways. They also appear to participate in reactive oxygen species metabolism. The asterisk indicates that the involvement of the PRC1 complex in DNA damage-induced monoubiquitylation of lysine 119 of histone H2A (H2AK119ub1) remains unclear. The color code in (B) and (C) is the same as in (A).
G0/G1Transition
Integration of diverse metabolic, environmental, and stress signals dictates whether cells remain quiescent (G0) or enter the cell cycle. No role has been ascribed yet to PcGs in G0/G1 transition. However, expression levels of the PRC2 proteins EZH2 and SUZ12 are greatly reduced in serum-starved nondividing fibroblasts and rapidly increase as they enter cell cycle, whereas EZH1 levels are not affected (Bracken et al., 2003; Pasini et al., 2004). It will be important to determine whether this differential regulation of PcG protein levels also characterizes somatic stem cells and what role these changes play in their G0/G1 transition.
G1/S Checkpoint
Regulation of the G1/S transition occurs mainly through the INK4A-pRB/E2F pathway. The PcG-mediated repression of the p16Ink4a/p19Arf locus is essential for G1/S progression (Figure 3A). The PRC1 proteins BMI1, PCGF1, PCGF2/MEL18, CBX2, CBX7, CBX8, RING1B, and the PRC2 proteins EED, SUZ12, and EZH2 have been shown in multiple cell types to bind the p16Ink4a/p19Arf promoter directly and repress this locus (Gil and Peters, 2006; Maertens et al., 2009). EZH2 also binds to and trimethylates H3K27 in the p15Ink4b promoter (Kheradmand Kia et al., 2009). By repressing these CDK inhibitors, the PcG proteins enable activation of the Cyclin D-CDK4/6 complex and consequently progression through the G1 phase of the cell cycle. Stress signals appear to displace PcG complexes from the promoters of these CDK inhibitors, allowing their expression, which subsequently blocks cells in G1.
The role of PcG proteins at the G1/S checkpoint is not restricted to repressing Ink4 CDK inhibitors. PcG complexes also require an intact pRB-E2F complex to repress a subset of cell-cycle genes (Blais et al., 2007). BMI1, RING1B, and SUZ12 bind the p16Ink4a locus in a pRB-dependent manner, and PRC2-dependent trimethylation of H3K27 at this locus requires an active pRB protein, indicating a regulatory feedback loop between p16INKA expression and pRB-E2F-PcG complexes (Kotake et al., 2007). Multiple PcG proteins directly interact with and modulate the activity of pRB-E2F. In fly and mammals, physical interaction between L3MBTL1 and pRB-E2F appears to be required for repression of target genes, notably cyclin E, an E2F2-pRB target gene crucial for S phase initiation (Lu et al., 2007; Trojer et al., 2007). Moreover, EZH2 and HDAC1 compete for interaction with pRB2/p130, indicating that PRC2 participates in the regulation of pRB/E2F activity (Tonini et al., 2004). Interestingly, E2Fs bind to and activate the Ezh2 and Eed promoters, indicating the presence of a regulatory feedback loop between PcG and pRB-E2F proteins (Bracken et al., 2003). Prohibitin (PHB), an antiproliferative protein that interacts with E2F-pRB and p53, represses E2F target genes by recruiting corepressors and RING1B/PRC1 complexes. This latter interaction is essential for the transcriptional repressive function of PHB, and knockdown of PHB results in reduction of RING1B levels and increase in p16Ink4a expression (Choi et al., 2008). Thus, RING1B and PHB directly interact with and inhibit E2F1’s transcriptional activity. Curiously, they also indirectly promote E2F1 activity by repressing p16Ink4a expression, which allows Cyclin D-CDK4/6-mediated phosphorylation of pRB and E2F1 release. This dual function could serve as a fine-tuning system for G1/S transition in response to mitogenic signals.
S Phase
The PRC1 complex has been implicated in regulation of S phase initiation. Within the PRC1 complex, SCMH1 directly interacts with Geminin, an inhibitor of the DNA replication licensing factor CDT1 (Figure 3B). The PRC1 complex, comprising PHC1, SCMH1, RING1B, and BMI1, exerts E3 ubiquitin ligase activity on Geminin. PRC1-induced ubiquitination of Geminin leads to its proteasome-mediated degradation and to initiation of S phase entry and/or release of stalled DNA replication forks (Luo et al., 2004; Ohtsubo et al., 2008). Accordingly, Phc1 deficiency impairs the ubiquitin-proteasome-mediated degradation of Geminin and blocks initiation of DNA replication. Interestingly, this regulatory role in S phase does not involve the standard chromatin-associated function of the PRC1 complex but, rather, a PRC1-mediated post-translational modification of a nonhistone protein.
As mentioned previously, L3MBTL1 represses expression of the cyclin E gene. In addition, L3MBTL1 physically interacts with the methyltransferase PRSET7 (Kalakonda et al., 2008). This latter protein interacts with PCNA, localizes to sites of DNA synthesis, and regulates the progression of replication forks. Knockdown of PRSET7 leads to loss of PRSET7-associated H4K20me1 marks, improper DNA replication, and massive occurrence of DNA double-strand breaks (Huen et al., 2008; Jørgensen et al., 2007; Tardat et al., 2007). Interestingly, L3MBTL1 levels increase during cell-cycle progression to peak in G1/S and S (Kalakonda et al., 2008). L3MBTL1 binds to the PRSET7-dependent H4K20me1 modification, and its accumulation mirrors the presence of this mark during S phase. Although it remains to be proven, it appears that L3MBTL1 and PRSET7 may cooperate in regulating the progression of DNA replication forks, and it would be important to find out whether, in this context, L3MBTL1 and PRESET7 act as components of the PRC1 complex.
Further evidence suggests that PRC1 and PRC2 complexes may control the timing of DNA replication. During S phase, PRC1 proteins are bound to chromatin and to newly replicated DNA. They also appear to inhibit DNA and chromatin replication in vitro (Francis et al., 2009). Interestingly, BMI1 interacts with CDC6, a protein essential for the assembly of the pre-replicative complex at origins of DNA replication. CDC6 represses p16Ink4a/p19Arf in a Bmi1-dependent manner (Agherbi et al., 2009). In addition, the PRC2 protein EZH2 colocalizes with PCNA at foci of ongoing DNA replication, and the PRC2-mediated H3K27me3 modifications positively correlate with late replication of large DNA segments (Birney et al., 2007; Hansen et al., 2008) (Figure 3B).
G2/M and Spindle Checkpoints
Triggering of M phase is under the control of the Cyclin B/CDK1 complex while control of chromosome segregation is regulated by the APC/CDC20 complex. Following stress signals, progression into mitosis can be blocked by association of pRB with E2F, CtBP, and the PRC1 proteins CBX4 and RING1, which together bind the promoters and repress the expression of the cyclin A and Cdk1 genes (Dahiya et al., 2001). Polycomb and pRB-E2F thus appear to play a dual role in cell-cycle regulation, first by enabling G1/S progression through repression of the p16Ink4a/p19Arf locus and then by arresting cells in G2/M through repression of cyclin A and Cdk1 (Figure 3A). Moreover, the p53-induced Cip/Kip CDK inhibitors, which act at both the G1/S and the G2/M transition, are also regulated by PcG proteins. EZH2 was reported to bind and repress the p57Kip2 locus in breast cancer cells (Yang et al., 2009), whereas p21Cip1 is repressed by BMI1 or its paralog PCFG1 (Fasano et al., 2007; Gong et al., 2006). Accordingly, knockdown of p21Cip1 partially rescues the proliferative defects and the decreased frequency and size of Bmi1-deficient neurospheres.
In primitive hematopoietic cells, BMI1 interacts with the atypical ubiquitin E3 ligase E4F1 and triggers its degradation (Chagraoui et al., 2006). E4F1 is a tumor suppressor that interacts with p53 and pRB and activates both pathways (Fajas et al., 2000; Le Cam et al., 2006) (Figure 3A). E4f1 null cells exhibit mitotic progression and chromosomal segregation defects, while ectopic expression of E4f1 results in inhibition of the G1/S transition and cytokinesis defects. Importantly, loss of E4F1 function is sufficient to rescue the clonogenic and repopulating ability of Bmi1 null hematopoietic stem cells (HSCs) in a p16Ink4a/p19Arf- and p53-independent manner. The mechanism of this rescue is still unknown, but it will be important to test whether this rescue is due to release of the G1/S or G2/M checkpoints or to some novel function(s) of E4F1.
Redistribution of PHC1, RING1B, BMI1, and PCGF6 from chromatin to the cytoplasm has been detected in human cells during mitosis (Akasaka et al., 2002; Miyagishima et al., 2003). Interestingly, both BMI1 and PCGF6 are specifically phosphorylated in G2/M, suggesting that post-translational modification of these proteins might regulate their functions and their association with chromatin (Akasaka et al., 2002; Voncken et al., 1999). PCGF6 can be phosphorylated in vitro by CDK7, a kinase required during G2 for Cyclin B/CDK1 complex assembly and M phase entry (Akasaka et al., 2002). However, if and how the cell-cycle phase-specific phosphorylations affect the PRC1 complex activity remains to be elucidated. Recent studies also revealed that heterozygous mutants of Ph-p, a Drosophila homolog of PHC, display mitotic chromosome segregation defects in fly embryos and impaired chromatin condensation in imaginal discs. PH-p does not localize to chromatin during M phase but is required for mitosis as suggested by formation of chromatin bridges and chromosome breakage observed in mutant flies (Beck et al., 2010; O’Dor et al., 2006). Interestingly, heterozygous mutants of other PRC1 genes (Pc, Psc, Asx) also display chromatid segregation defects, whereas impaired chromatin condensation and misalignment of chromosomes at the metaphase plate have been observed in E(z) mutants (O’Dor et al., 2006). In contrast to other PRC1 proteins, human L3MBTL1 associates with condensed mitotic chromosomes, and its overexpression leads to chromosome segregation defects, failure in cytokinesis, and the formation of multinucleated cells (Koga et al., 1999). PRSET7 also remains associated with chromosomes during mitosis and appears to be required for chromosome condensation (Oda et al., 2009; Rice et al., 2002). As mentioned previously, these two proteins interact together, suggesting they may function cooperatively during both S phase and mitosis.
Two recent studies provided potential novel functions for PCGF2/MEL18 in regulating the G2 and M phases. PCGF2/MEL18 inhibits sumoylation of the heat shock factor HSF2 by binding to and inhibiting the UBC9 E2 conjugating enzyme. HSF2 binds the promoter of other heat shock genes during mitosis. In M phase, PCGF2/MEL18 dissociates from HSF2, which renders it susceptible to sumoylation. This enables the interaction of HSF2 with the CAP-G subunit of the condensing complex during gene bookmarking, an epigenetic memory system that allows transmission of the marks that denote active genes or genes poised for activation to daughter cells (Zhang et al., 2008a). The anti-SUMO E3 activity of PCGF2/MEL18 also affects RANGAP1, a Ran GTPase-activating protein involved in mitotic spindle formation and postmitotic nuclear assembly. In contrast to HSF2, the PCGF2/MEL18-RANGAP1 interaction increases during mitosis, which prevents sumoylation of RANGAP1 (Zhang and Sarge, 2008). Interestingly, sumoylation of RANGAP1 is required for its association with RANBP2 and its localization to the spindle and kinetochores (Joseph et al., 2002). This pathway may thus provide a way to create a Ran-GTP gradient implicated in the assembly and orientation of the mitotic spindle (Clarke and Zhang, 2008). Disruption of the SUMO1-RANGAP1-RANBP2 complex leads to chromosome misalignments and segregation defects, a phenotype reminiscent of that observed in Drosophila PcG mutants (Beck et al., 2010; Joseph et al., 2002; O’Dor et al., 2006).
PcG AND DNA Damage Repair
Defects in the DNA damage response have been widely associated with aging, stem cell defects, and cancer (Ito et al., 2004; Nijnik et al., 2007; Rossi et al., 2007; Ruzankina et al., 2007). According to recent data, PcG proteins play a role in DNA damage repair (Figure 3C). The first observations were reported by Zeidler et al., who found that overexpression of EZH2 in human mammary epithelial cells leads to a drastic decrease in expression of multiple RAD51 paralogs required for homologous recombination double-strand break (DSB) repair. Overexpression of EZH2 leads to a significant decrease in the numbers of DNA repair foci, increased aneuploidy, and reduced survival rates of cells exposed to genotoxic stress (Zeidler et al., 2005). Recently, the PRC2-associated PCL1/PHF1 protein was reported to be rapidly recruited to DSBs in a KU70/KU80-dependent manner. Knockdown of PCL1/PHF1 increases the frequency of homologous recombination and sensitizes cells to irradiation. The direct physical interaction of PCL1/PHF1 with KU70/KU80 and RAD50 proteins strongly suggests that this PRC2 protein promotes the nonhomologous end joining (NHEJ) repair pathway (Hong et al., 2008). This possibility is further supported by studies showing that the PRC2 components EZH2 and SUZ12 interact with the poly (ADP-ribose) polymerase 3 (PARP3) protein, known to interact with components of the NHEJ repair pathway (Rouleau et al., 2007). In addition, 39 different long noncoding RNAs (lincRNA) were reported to be induced by DNA damage in a p53-dependent manner. Interestingly, one of them, named TUG1, recruits the PRC2 complex to repress genes that regulate mitosis, spindle formation, and cell-cycle phasing (Guttman et al., 2009; Khalil et al., 2009). Given that approximately 20% of the ~3,300 identified lincRNAs were estimated to bind to the PRC2 complex, it is likely that numerous other lincRNAs interact with PRC2 to mount an adequate DNA damage response. Interestingly, the PRC1-associated monoubiquitination of H2AK119 accumulates at DNA damage foci, but involvement of the PRC1 ubiquitin E3 ligase RING1A/B in this process remains to be clearly demonstrated (Bergink et al., 2006; Zhu et al., 2009).
The PRC1 protein CBX4, a SUMO E3 ligase, interacts with Centrin-2 and HIPK2, two proteins implicated in DNA damage repair pathways. Centrin-2 stimulates nucleotide excision repair (NER) by interacting with XPC, which recognizes DNA lesions. Sumoylation of Centrin-2 by CBX4 enhances its interaction with XPC and is essential for its nuclear localization and participation in NER (Klein and Nigg, 2009). HIPK2 is a kinase involved in transcriptional repression occurring early after the occurrence of DNA lesions and implicated in the induction of cell-cycle arrest and apoptosis. Interaction between CBX4 and the DNA damage-induced kinase HIPK2 generates an auto-regulatory loop where HIPK2 controls CBX4 intranuclear localization and phosphorylates it at threonine 495. This modification enhances the SUMO E3 ligase activity, which in turn increases sumoylation of HIPK2 and of CBX4 thereby promotes its DNA damage-induced transcriptional silencing function (Roscic et al., 2006).
Reactive oxygen species (ROS) induce double-strand breaks and base/nucleotide modifications through DNA oxidation. Accumulating evidence indicates that oxidative metabolism plays a crucial role in maintaining the self-renewal properties of somatic stem cells and that accumulation of high ROS levels leads to premature differentiation (Jang and Sharkis, 2007; Owusu-Ansah and Banerjee, 2009). Mice lacking genes involved in ROS response display genomic instability as well as defects in the maintenance of HSC quiescence and self-renewal ability (Ito et al., 2004; Zhang et al., 2007). Interestingly, antioxidant treatment of stem cells with high ROS levels restores their long term self-renewal potential (Ito et al., 2004; Jang and Sharkis, 2007; Zhang et al., 2007). To date, BMI1 is the only PcG protein that has been linked to oxidative metabolism. Chatoo et al. (2009) reported that BMI1 prevents accumulation of ROS in neurons through the repression of p53’s pro-oxidant activity. Bmi1 deficiency leads to impaired mitochondrial function and a marked increase in ROS levels, which in turn activate the ROS-mediated DNA damage response in a p16Ink4a/p19Arf-independent manner. Treatment with antioxidants or abrogation of the ATM-dependent DNA damage checkpoint extends the survival of Bmi1−/− mice and rescues the majority of their cellular defects. However, long-term repopulation potential of hematopoietic stem cells remains severely impaired (Liu et al., 2009). Although these observations argue against the possibility that defects of Bmi1−/− HSC result from ROS-induced activation of the DNA damage checkpoint, they do not exclude the possibility that repair and recovery of DNA lesions may be affected in the absence of BMI1. Together, these studies indicate that proteins from both PRC1 and PRC2 are involved in different DNA damage pathways and play important roles in mounting an adequate repair DNA damage response to maintain genome integrity.
PcG as Regulators of Cell Fate and Differentiation
The role of PcG proteins in stem cell fate is firmly established (reviewed in Surface et al., 2010; Sparmann and van Lohuizen, 2006). The genome-wide mapping of PcG target genes in mammalian and Drosophila cells provided a wealth of information about how PcG proteins control differentiation and cell identity. These studies revealed that PcG complexes bind to and repress numerous genes encoding key developmental regulators and signaling proteins in embryonic stem cells (ESCs). Interaction of PcG proteins with chromatin at these loci is associated with increased levels of H3K27me3 and H2AK119ub1. Importantly, these repressed PcG target genes are preferentially activated during ESC differentiation (Boyer et al., 2006; Bracken et al., 2006; Lee et al., 2006; Stock et al., 2007; Tolhuis et al., 2006). This correlates with the loss of PcG binding, loss of the H3K27me3 and H2AK119ub1 repressive marks, and increase in the active H3K4me3 modifications. Accordingly, knockout of the PRC2 and PRC1 genes Ezh2, Suz12, Eed, or Ring1b leads to a loss of H3K27me3 and H2AK119ub1 marks, a spontaneous increase in developmental gene expression, and consequently, differentiation defects of embryonic stem cells (Chamberlain et al., 2008; Pasini et al., 2007; Shen et al., 2008; van der Stoop et al., 2008). Similar observations have been reported for many somatic tissues (Ezhkova et al., 2009; Caretti et al., 2004; Hirabayashi et al., 2009; Zencak et al., 2005).
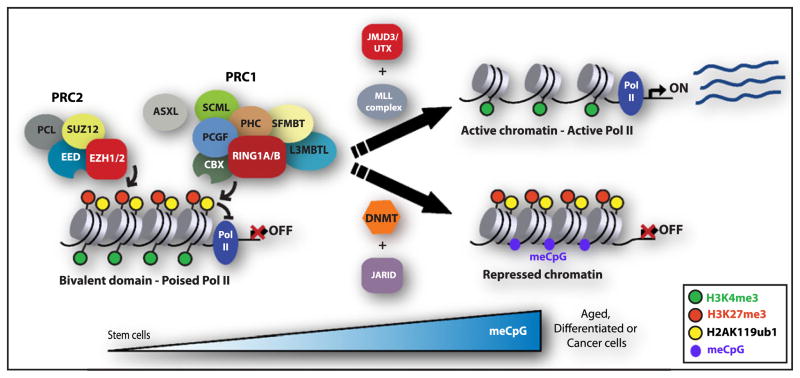
Genome-wide chromatin state maps in ESCs revealed that a large proportion of developmental and tissue specific genes simultaneously carry the PcG-repressive H3K27me3 and H2aK119ub marks overlapping with the H3K4me3 modifications associated with an active chromatin state (Bernstein et al., 2006a; Ku et al., 2008; Stock et al., 2007). These loci, termed bivalent, are silent or expressed at low levels in pluripotent ESCs. In fact, these loci appear to be in a poised state ready for activation since the RNA polymerase II phosphorylated at serine 5, which is associated with paused transcription, also localizes to these loci (Stock et al., 2007). The PRC1 and PRC2 complexes are bound to these bivalent domains (Ku et al., 2008). The PRC1-mediated H2AK119ub1 modification reinforces the poised RNA polymerase II configuration and deletion of Ring1a/b leads to the release of the poised RNA polymerase II, and premature expression of target genes (Stock et al., 2007; Zhou et al., 2008). In differentiating ESCs, these bivalent domains are resolved and retain chromatin modifications denoting either completely repressed (loss of H3K4me3) or activated (loss of H3K27me3 and H2AK119ub1) state (Figure 4). These bivalent domains are also found in hematopoietic cells, neural progenitors, and fibroblasts, indicating they are not restricted to cultured embryonic stem cells (Mikkelsen et al., 2007; Oguro et al., 2010; Pan et al., 2007; Roh et al., 2006). Deletion of Bmi1 in hematopoietic stem cells results in the resolution of bivalent domains at the Ebf1 and Pax5 B cell lineage-specific genes. Inappropriate expression of these genes in immature cells leads to premature lymphoid lineage specification associated with a loss of stem and progenitor cell populations (Oguro et al., 2010). More detailed evaluation of the mechanisms that either establish or resolve these bivalent domains in somatic stem cells will provide key information about the role PcG plays in the regulation of cell differentiation.
Figure 4. Resolution of Bivalent Chromatin Domains and DNA Methylation of PcG Target Genes.
In stem cells, a large proportion of developmental regulators carry simultaneously both PcG-mediated H3K27me3 and H2AK119ub1 marks and the H3K4me3 modifications associated with repressive and active chromatin states, respectively. While remaining silent, these “bivalent” loci are maintained in a poised state ready for activation, given that a nonprocessive form of RNA polymerase II phosphorylated at serine 5 localizes to these loci (poised RNA Pol II). Following differentiation, these domains are resolved and retain chromatin modifications denoting either completely repressed (loss of H3K4me3) or activated (loss of H3K27me3 and H2AK119ub1) state. Resolution of the “bivalent” domains appears to require the H3K4me3 JARID demethylases (for repression) or the H3K27me3 demethylases (JMJD3/UTX) and MLL complexes (for activation). PRC1 and PRC2 proteins control DNA methylation by directly interacting with DNA methyltransferases (DNMT).
PcG Proteins and Prevention of Senescence
Senescence is an irreversible phenomenon where cells that should normally be able to divide (i.e., have not undergone terminal differentiation) cease to do so. It can result from normal telomere attrition following multiple rounds of divisions (replicative senescence) or from unrepaired DNA damage (stress-induced senescence). Progressive reduction of the replicative potential of stem cells is believed to represent an important factor in the normal aging of tissues. Induction of a senescence program requires an intact pRB-p16INK4A pathway because deletion of p16Ink4a bypasses this process. As mentioned previously, PcG proteins participate in the transcriptional repressive function of pRB and directly inhibit expression of the p16Ink4a locus (Figure 3A). It is, therefore, not surprising that the majority of PcG genes prevent the onset of the senescent state. Expression of Bmi1, Cbx2, Cbx7, and Ezh2 is downregulated as cells approach or enter replicative senescence. This is accompanied by decreased binding of PcG proteins to the p16Ink4a/p19Arf promoter, loss of H3K27me3, and upregulation of p16INK4A and p19ARF expression. Similarly, knockout or downregulation of these PcG genes by RNAi increases the expression of p16INK4A/p19ARF and induces premature senescence (Gil and Peters, 2006). Homozygous deletion of Bmi1 in mice leads to a progeroid phenotype, with prematurely senescent cells and a drastic loss of hematopoietic and neural stem cells. Deletion of the p16Ink4a/p19Arf locus partially rescues the HSC and neural cell defects in these mice (Molofsky et al., 2005; Oguro et al., 2006). However, whether a combined p15Ink4b and p18Ink4c loss could provide an improved or even complete rescue of hematopoietic and neural defects in Bmi1 null mutant mice has not been investigated.
Overexpression of Bmi1, Cbx7, Ezh2, and Cbx8 extends the replicative lifespan of cells by bypassing senescence in a pRB-p16INK4A/p19ARF-dependent manner (Gil and Peters, 2006). Importantly, overexpression of Ezh2 prevents HSC exhaustion after replicative stress induced by serial transplantations (Kamminga et al., 2006). Prior to transformation, cells overexpressing the c-Myc oncogene go through a senescent phase. Direct upregulation of Bmi1 by c-MYC leads to repression of the p16Ink4a/p19Arf locus and bypasses the oncogene-induced senescence program, thus promoting tumorigenesis (Jacobs et al., 1999). Again, it would be critical to determine the contribution of the other Ink4 members to this important observation.
PcG and Apoptosis
The role of PcG proteins in the regulation of apoptosis has not been extensively examined. In Drosophila, overexpression of Ph in imaginal discs induces proliferation associated with increased JNK-dependent apoptosis (González et al., 2009). In mammalian cells, the absence of Bmi1 leads to an increase in apoptosis, whereas its overexpression promotes survival, protects cells from stress-induced death, and is accompanied by reduced caspase activity and Poly(ADP-ribose) polymerase (PARP) cleavage (Lee et al., 2008). In lymphoid cells, BMI1 directly represses the proapoptotic Noxa gene. Importantly, apoptosis of Bmi1−/− lymphoid cells can be rescued by reducing the expression of Noxa, but not p16Ink4a/p19Arf (Yamashita et al., 2008). Similarly, Ring1b−/− T cells, which are highly susceptible to apoptosis, express high levels of the proapoptotic gene Bim. The apoptosis-susceptible phenotype of these cells can be rescued by reducing the expression of Bim, but not other proapoptotic genes such as Perp, Noxa, or Bax, indicating that RING1B specifically inhibits BIM-mediated apoptosis (Suzuki et al., 2010). The fact that direct cleavage of RING1B by Caspases 3 and 9 inhibits its transcriptional repressive activity (Wong et al., 2007) suggests the presence of a positive feedback loop in apoptosis signaling pathways. E2F1 can also promote apoptosis by inducing the expression of Bim. Interestingly, EZH2, which interacts with E2F1, antagonizes the induction of E2F1-mediated apoptosis by directly inhibiting expression of Bim (Wu et al., 2010).
PcG and Cancer
The multifaceted functions of PcG proteins in the regulation of cell-cycle checkpoints, DNA damage repair, cell differentiation, senescence, and apoptosis makes them prime candidate cancer stem cell genes.
Deregulation of PcG Gene Expression in Cancer
One of the first indications that PcG proteins play a role in cancer was the identification of Bmi1 as a c-Myc-collaborating oncogene (Haupt et al., 1993; Jacobs et al., 1999). Multiple other PRC1 genes are aberrantly expressed in a broad spectrum of human cancers (summarized in Table 1 and Figure 5B, reviewed in Rajasekhar and Begemann, 2007; Sparmann and van Lohuizen, 2006). Bmi1 is overexpressed, and its locus amplified in several human leukemias and solid tumors. BMI1 activity is crucial for the maintenance of both normal and cancer stem cells, as the loss of its function leads to progressive depletion of these cell populations (Lessard and Sauvageau, 2003). Conversely, the Bmi1 paralog Pcgf2/Mel18 acts as a negative regulator of cell proliferation and possesses tumor suppressive functions. Overexpression of Pcgf2/Mel18 impairs proliferation and leads to G1/S arrest, whereas deletion of this gene promotes proliferation (Tetsu et al., 1998). In line with this, loss of Pcgf2/Mel18 expression is often found in different types of human cancers (Table 1).
Table 1.
Expression, Mutations, and Chromosomal Translocations of PcG Genes in Human Cancers
| Subunit | Aberrant Expression | |
|---|---|---|
| Expression | Cancer Type | |
| PRC2 Components | ||
| Ezh2 | overexpression and/or amplification | B cell non-Hodgkin lymphomas,35 bladder cancers,1,5,29,41 breast cancers,2,5,8,15,27 colon cancers,5,22 glioblastoma,5 Hodgkin lymphomas,28 oral cancers,5 liver cancers,33 lung cancers,5 lymphoma,5 mantle cell lymphomas,37 melanomas,2 prostate cancers,2,36 testicular cancers5 |
| Suz12 | overexpression and/or amplification | breast cancers,14 colon cancers,14,42 germinal cell-derived tumors,20 liver cancers,14 lung cancers,20 mantle cell lymphomas,20 melanomas,20 parathyroid and pituitary adenomas20 |
| Pcl3/Phf19 | overexpression | colon cancers,39 skin cancers,39 lung cancers,39 rectal cancers,39 cervical cancers,39 uterus cancers,39 liver cancers39 |
| PRC1 components | ||
| Cbx7 | overexpression | follicular lymphoma32 |
| loss | pancreatic cancers13 | |
| Phc1 | loss of heterozygosity | acute lymphoblastic leukemias34 |
| Phc3 | loss of heterozygosity | Osteosarcomas12,17 |
| Bmi1 | overexpression and/or amplification | acute myeloid leukemias,31 breast cancers,30 gastrointestinal tumors,30 lymphomas,3,30,35 medulloblastomas,18 neuroblastomas,26 lung cancers,38,30 parathyroid and pituitary adenomas30 |
| Pcgf2/Mel18 | loss | cutaneous squamous-cell carcinomas,30 gynecological tumors,30 prostate cancers30,40 |
| Ring1a | loss | clear-cell renal-cell carcinomas,30 testicular germ-cell tumors30 |
| Ring1b | overexpression | bladder cancers,30 breast cancers,30 colon tumors,30 gynecological tumors,30 kidney cancers,30 larynx cancers,30 livers cancers,30 lung cancers,30 lymphomas,30 melanomas,30 pancreas cancers,30 parathyroid and thyroid cancers,30 prostate cancers,30 skin cancers,30 thymus cancers,30 urinary tract cancers30 |
| Subunit | Mutations | |
| Type of Mutation | Cancer Type | |
| PRC2 Components | ||
| Ezh2 | missense | diffuse large B cell lymphomas,23 myelodysplasic/myeloproliferative syndromes10,25 |
| Epc1 | truncation | acute lymphoblastic leukemias24 |
| PRC1 Components | ||
| Phc3 | missense | osteosarcomas9,12 |
| Asxl1 | frameshift, truncation | acute myeloid leukemias,4,7 myelodysplasic/myeloproliferative syndromes4,6,11 |
| Subunit | Chromosomal Translocations | |
| Fusion Gene Partner | Cancer Type | |
| PRC2 Components | ||
| Suz12 | Jazf1 | endometrial stromal sarcomas16,19 |
| Pcl1/Phf1 | Jazf1, Epc1 | endometrial stromal sarcomas21 |
| Epc1 | Asxl2 | acute lymphoblastic leukemias24 |
| PRC1 Components | ||
| Asxl2 | Epc1 | acute lymphoblastic leukemias24 |
See Supplemental Information for detailed numerical references.
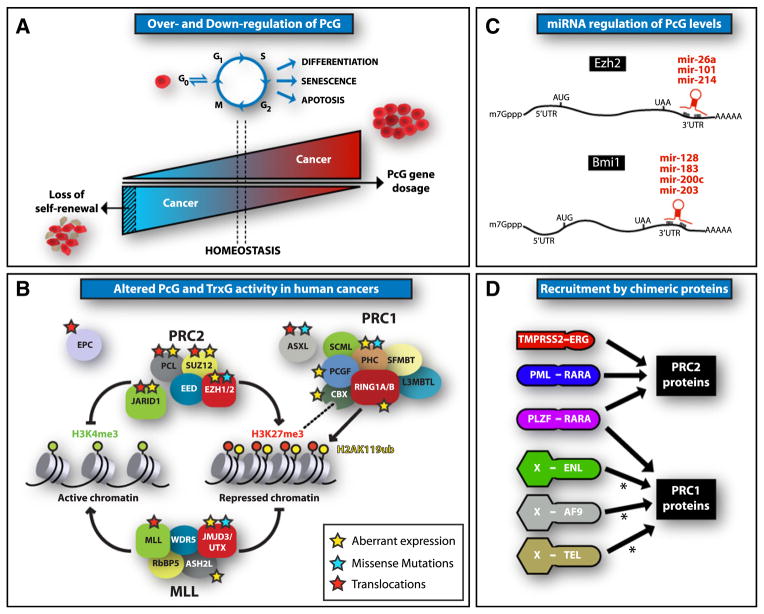
Figure 5. Deregulation of PcG Activity in Cancer.
(A) Proposed model for PcG gene dosage and its effect on stem cells and cancer development. Briefly, adequate PcG gene expression levels seem to be essential to maintain homeostasis and normal stem cell function. Gain or loss of PcG functions, such as deregulated expression or mutations, lead to increased tumor development, possibly by modifying the composition or perturbing the stoichiometry of the complexes. In turn, complete ablation of PcG activity appears to be detrimental and leads to impaired self-renewal and loss of stem cells.
(B) Schematic representation of the balance between PcG-mediated H3K27me3 and MLL complex-mediated H3K4me3 modifications in the maintenance of a transcriptional program. The function of many proteins from these complexes is altered by aberrant expression (yellow star), by missense mutations (blue star), or by chromosomal translocations (red star) in human cancers (Agger et al., 2009; Dalgliesh et al., 2010; Krivtsov and Armstrong, 2007; van Haaften et al., 2009; Wang et al., 2009; Xiang et al., 2007), likely leading to changes in transcriptional programs and cell states.
(C) Expression of Ezh2 and Bmi1 is regulated at the post-transcriptional level by miRNAs, targeting their 3′UTR, leading to transcript degradation.
(D) Multiple chimeric proteins resulting from chromosomal translocations found in human cancers interact with PRC1 and PRC2 complexes and appear to recruit them to target genes. AF9. ENL and TEL are found in multiple chromosomal translocation-derived fusion proteins in human cancers. The asterisk indicates that, although actual recruitment of PRC1 by the fusion proteins remains to be proven, the AF9, ENL, and TEL domains interacting with PRC1 proteins are present in the fusion proteins and are essential for their oncogenic activity (Boccuni et al., 2003; García-Cuéllar et al., 2001; Hemenway et al., 2001).
The PRC2 proteins EZH2, SUZ12 and PCL3/PHF19 are also overexpressed in a variety of different human cancers (summarized in Table 1 and Figure 5B, reviewed in Rajasekhar and Begemann, 2007; Sparmann and van Lohuizen, 2006). The Ezh2 gene is amplified in many human prostate cancer cell lines, in 27% of untreated prostate carcinomas, in 80% of xenografts, and is a good marker for aggressiveness and poor outcome in prostate and breast cancers (Kleer et al., 2003; Saramäki et al., 2006; Varambally et al., 2002). More than half of the hormone-refractory prostate cancers possess increased copies of Ezh2, which correlates with high expression levels of this gene. Ezh2 overexpression and amplification is rare in early stage prostate cancers, but much more common in late stages, highlighting its potential function in tumor progression (Saramäki et al., 2006).
Mutations and Chromosome Anomalies of PcG in Cancer
Recently, a number of PcG missense mutations and chromosomal translocations have been identified in different types of human cancers (also summarized in Table 1 and Figure 5B). Somatic Ezh2 mutations and deletions were found in 22% of germinal center diffuse large B cell lymphomas, 7% follicular lymphomas, and 12%–23% of patients with myelodysplasic and myeloproliferative disorders (Ernst et al., 2010; Morin et al., 2010; Nikoloski et al., 2010). Both monoallelic and biallelic mutations were found, and almost all are predicted to inactivate the methyltransferase activity of EZH2. This PcG enzyme, therefore, appears to have both oncogenic and tumor suppressive functions. Supporting this, heterozygous null or homozygous hypomorphic mutations of Eed in mice increase the incidence and reduce the latency of lymphoid tumors (Richie et al., 2002; Sauvageau et al., 2008), indicating a possible tumor suppressive role for this other PRC2 protein. Importantly, the methyltransferase activity of EZH2 depends on its interaction with EED, and mutations in EED have been reported to affect this interaction (Denisenko et al., 1998; Shumacher et al., 1996; van Lohuizen et al., 1998). Heterozygosity for a Suz12 allele carrying an inactivating point mutation was also reported to cause an increase in numbers of platelets, megakaryocytes, and hematopoietic progenitors. Moreover, HSCs harboring a Suz12 mutation are more competitive in repopulating the hematopoietic system than their wild-type counterparts, indicating that SUZ12 negatively regulates HSC/progenitor activity and suggesting a tumor suppressive role for SUZ12 (Majewski et al., 2008). Together, these studies indicate that a delicate balance of PRC2 gene dosage is crucial for homeostasis. Either upregulation or a decrease in PRC2 activity is sufficient to sensitize cells to transformation, whereas complete absence of PRC2 activity leads to stem cell extinction (Figure 5A).
The ASXL proteins have been implicated in cancer development by recent studies reporting Asxl1 mutations in 17%–25% of acute myeloid leukemias, 33%–43% of myelomonocytic leukemias, and 10%–18% of myelodysplasic syndromes and hyperproliferative disorders (Table 1). The majority of Asxl1 alleles are heterozygous frameshift mutations caused by duplications, insertions, substitutions, or deletions of nucleotides and are predicted to encode an ASXL1 protein lacking the C-terminal region comprising the nuclear receptor box and PHD domain (Boultwood et al., 2010; Carbuccia et al., 2009; Gelsi-Boyer et al., 2009). Interestingly, in acute myeloid leukemias, ASXL1 and NPM1 mutations appear to be mutually exclusive, ASXL1 mutations being present in 34% of non-NPM1-mutated cases (Carbuccia et al., 2010). Moreover, Asxl−/− mice have defects in lymphoid and myeloid progenitor cell frequency and differentiation, whereas multipotent progenitor and HSC populations appear to behave normally (Fisher et al., 2010). Missense mutations are also found in PHC3 in osteosarcomas (Deshpande et al., 2007; Iwata et al., 2010). More extensive analysis of PcG mutations in human cancers will likely be revealing.
Some PcG genes are involved in recurrent chromosomal rearrangements detected in human cancers (see Table 1). In addition, PcG proteins have been shown to be recruited to target genes by physically interacting with a number of chimeric fusion proteins involved in leukemia, such as PLZF-RARA and TMPRSS2-ERG (Figure 5D) (Boccuni et al., 2003; Boukarabila et al., 2009; García-Cuéllar et al., 2001; Hemenway et al., 2001; Villa et al., 2007; Yu et al., 2010).
PcG and Cancer Epigenetics
Aging is the strongest demographic risk factor for cancer, and several lines of evidence point to an association between aging and changes in DNA methylation at specific loci. Epigenetic silencing by DNA hypermethylation is prevalent in cancers and represents an efficient mechanism for inactivation of tumor suppressor genes. Interestingly, PcG target genes identified in ESCs are more likely to be hypermethlyated in aged somatic cells (Teschendorff et al., 2010). A large proportion of the PcG target methylation signature is present in ovarian cancer, follicular lymphoma, and glioblastoma multiforme (Martinez et al., 2009; O’Riain et al., 2009; Teschendorff et al., 2010). In fact, the PcG target genes identified in ESCs are 12 times more likely to be hypermethylated in cancers than non-PcG targets (Ohm et al., 2007; Widschwendter et al., 2007). A recent study showed that 50% of genes frequently hypermethylated in colon or prostate cancers are premarked by PcG proteins and by the PRC2-mediated H3K27me3 mark for de novo methylation in cancer (Schlesinger et al., 2007). The link between DNA methylation and polycomb target genes in cancer is further supported by the fact that PRC2 directly controls DNA methylation through interaction of EZH2 with the DNA methyltransferases DNMT1, DNMT3A, and DNMT3B. This interaction allows the recruitment of DNMTs to PRC2-repressed genes and their subsequent methylation (Viré et al., 2006). The PRC1 proteins CBX4 and CBX7 also interact with DNMT3B, whereas BMI1 forms a ternary complex with DNMT1 through direct interaction with the DNMT-associated protein DMAP1 (Kim et al., 2008; Mohammad et al., 2009; Negishi et al., 2007). It was estimated that approximately 47% of DNMT3B target genes are bound by PRC1 and PRC2 and that DNMT3B-PcG-bound genes in normal cells are good predictors of epigenetically silenced loci in colon cancer (Jin et al., 2009). Since most of the PcG targets are lineage and differentiation determinants, these studies suggests that gradual de novo DNA methylation of PcG targets locks cells in an undifferentiated state and predisposes them to malignant transformation (Figure 4).
Recently, many large intervening noncoding RNAs (lincRNA) in the Hox loci have been found to be deregulated during progression of breast cancer. The lincRNA HOTAIR, which recruits the PRC2 complex to silence the HoxD cluster, is overexpressed in primary breast tumors and metastases. In fact, high HOTAIR levels appear to be a good indicator of metastatic progression, and its knockdown decreases cancer invasiveness, especially in cells expressing high levels of PRC2 proteins (Gupta et al., 2010; Rinn et al., 2007). Conversely, ectopic expression of HOTAIR increases invasiveness and metastasic potential of epithelial cancer cells and induces relocalization of the PRC2 complex to bind target genes in a pattern similar to that observed in embryonic fibroblasts. The resulting differential gene expression pattern is likely responsible for the invasive phenotype because reduction of PRC2 activity in HOTAIR-overexpressing cells inhibits metastasis (Gupta et al., 2010). It is likely that numerous other lincRNAs are also deregulated in cancers, thus generating a new mechanism to redirect PRC2 complex activity and “reprogram” cells to an undifferentiated and/or malignant state. Interestingly, interaction between the lincRNA ANRIL and the PRC1 protein CBX7 in prostate cancer cells has recently been found to be crucial for repression of the p15Ink4b/p16Ink4a/p19Arf locus (Yap et al., 2010). Since all CBX proteins have sequence-specific RNA binding activity, they also likely interact with multiple lincRNAs and thereby target the PRC1 complex to specific loci (Bernstein et al., 2006b).
In recent years, small noncoding RNAs have been recognized as major players in regulatory pathways, and many of them are deregulated in cancers. Multiple miRNAs are repressed in ESCs and marked by H3K27me3 modification, indicating that PRC2 complexes regulate their expression (Marson et al., 2008). In addition, several miRNAs were found to bind PcG transcripts and inhibit their expression (e.g., mir-26a and mir-214 target Ezh2 transcript [Juan et al., 2009; Sander et al., 2008]) (Figure 5C). Interestingly, MYC decreases the expression of mir-26a and, thereby, contributes to tumorigenesis by allowing expression of Ezh2 (Sander et al., 2008). mir-101 targets the 3′UTR of Ezh2 mRNA and promotes its degradation. Expression of this miRNA is downregulated in bladder transitional cell carcinoma, hepatocellular carcinoma, and prostate tumors, which correlates with high Ezh2 levels (Friedman et al., 2009; Su et al., 2009; Varambally et al., 2008). Almost 40% of prostate cancers and 67% of metastatic stage tumors harbor loss of one or both mir-101 alleles. Overexpression of mir-101 inhibits proliferation and reduces the invasiveness potential and colony formation activity of cancer cells, a phenotype similar to that found with Ezh2 knockdown (Cao et al., 2010; Friedman et al., 2009; Varambally et al., 2008).
The PRC1 gene Bmi1 is also targeted by multiple miRNAs. Expression of Bmi1 is post-transcriptionally inhibited by mir-128, mir-183, mir-200c, and mir-203, which target the 3′UTR of the transcript. The mir-203 locus is silenced by DNA methylation in hepatocellular carcinoma and squamous cell carcinoma of the esophagus (Furuta et al., 2010; Kozaki et al., 2008). Similarly, mir-200 and mir-183 are often suppressed by DNA methylation in invasive cancers, providing a link between miRNAs and the deregulation of PcG in cancers. Human glioblastomas display reduced levels of mir-128, whereas ectopic expression of mir-128 inhibits glioma cell proliferation and xenograft growth. Bmi1 downregulation by mir-128 in glioma cells leads to a decrease in H3K27me3 levels, upregulation of p21Cip1, and a block in self-renewal (Godlewski et al., 2008). Normal mammary and breast cancer stem cell populations show reduced expression of mir-183 and mir-200 family members, resulting in increased Bmi1 levels. In addition to inhibiting the ability of normal mammary stem cells to form mammary ducts, expression of mir-200c suppresses breast cancer stem cell proliferation, clonogenicity, and tumorigenic potential (Shimono et al., 2009). Recently, a link between miRNA, Bmi1, and epithelial-mesenchymal transition (EMT) was also established. Carcinoma invasiveness and metastasic spread is promoted by the establishment of an embryonic EMT program. Expression of the ZEB1 protein activates epithelial-mesenchymal transition (EMT) and contributes to the maintenance of stem cell self-renewal. This protein also suppresses expression of mir-200c, mir-203, and mir-183, which results in high levels of BMI1. Highly invasive tumors express high levels of ZEB1, which appears to be essential for the expansion of primary and metastic pancreatic and colorectal tumors (Wellner et al., 2009).
Concluding Remarks
Through chromatin silencing and other non-histone functions, PcG complexes are involved in the regulation of multiple molecular programs essential for stem cell activity. Such programs involve cell cycle checkpoints and progression, DNA damage repair, differentiation, apoptosis and senescence. Aberrant expression, inactivating mutations, chromosomal translocations and recruitment of PcG proteins and complexes by chimeric proteins or lincRNAs are observed in human tumors. The frequency of these newly reported mutations/anomalies now suggests that deregulation of PcG activity is central to cancer development and cancer stem cell biology. A clear picture of the exact regulatory events involving PcG complexes in stem cell self-renewal and cancer is hindered by the multiplicity of PcG complexes and by the distinct functions of the different PcG proteins within these complexes. Future studies will have to address this issue.
Supplementary Material
Acknowledgments
The authors would like to thank J. Krosl, T. MacRae, and J. Chagraoui for critical comments and helpful discussions. This work was supported by a Grant from the Canadian Institutes of Health Research to G.S. and by a Terry Fox Foundation Research Studentship to M.S.
Footnotes
Supplemental Information includes Supplemental References and can be found with this article online at doi:10.1016/j.stem.2010.08.002.
References
- Agger K, Cloos PA, Rudkjaer L, Williams K, Andersen G, Christensen J, Helin K. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS ONE. 2009;4:e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka T, Takahashi N, Suzuki M, Koseki H, Bodmer R, Koga H. MBLR, a new RING finger protein resembling mammalian Polycomb gene products, is regulated by cell cycle-dependent phosphorylation. Genes Cells. 2002;7:835–850. doi: 10.1046/j.1365-2443.2002.00565.x. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, Wagener C, Sardet C, Moroni MC, Helin K. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem. 2005;280:1199–1208. doi: 10.1074/jbc.M412509200. [DOI] [PubMed] [Google Scholar]
- Beck SA, Falconer E, Catching A, Hodgson JW, Brock HW. Cell cycle defects in polyhomeotic mutants are caused by abrogation of the DNA damage checkpoint. Dev Biol. 2010;339:320–328. doi: 10.1016/j.ydbio.2009.12.031. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Salomons FA, Hoogstraten D, Groothuis TA, de Waard H, Wu J, Yuan L, Citterio E, Houtsmuller AB, Neefjes J, et al. DNA damage triggers nucleotide excision repair-dependent monoubiquitylation of histone H2A. Genes Dev. 2006;20:1343–1352. doi: 10.1101/gad.373706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006a;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Duncan EM, Masui O, Gil J, Heard E, Allis CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006b;26:2560–2569. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ENCODE Project ConsortiumNISC Comparative Sequencing Program-Baylor College of Medicine Human Genome Sequencing CenterWashington University Genome Sequencing CenterBroad InstituteChildren’s Hospital Oakland Research Institute. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blais A, van Oevelen CJ, Margueron R, Acosta-Alvear D, Dynlacht BD. Retinoblastoma tumor suppressor protein-dependent methylation of histone H3 lysine 27 is associated with irreversible cell cycle exit. J Cell Biol. 2007;179:1399–1412. doi: 10.1083/jcb.200705051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccuni P, MacGrogan D, Scandura JM, Nimer SD. The human L(3)MBT polycomb group protein is a transcriptional repressor and interacts physically and functionally with TEL (ETV6) J Biol Chem. 2003;278:15412–15420. doi: 10.1074/jbc.M300592200. [DOI] [PubMed] [Google Scholar]
- Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- Boukarabila H, Saurin AJ, Batsché E, Mossadegh N, van Lohuizen M, Otte AP, Pradel J, Muchardt C, Sieweke M, Duprez E. The PRC1 Polycomb group complex interacts with PLZF/RARA to mediate leukemic transformation. Genes Dev. 2009;23:1195–1206. doi: 10.1101/gad.512009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, Larrayoz MJ, Garcia-Delgado M, Giagounidis A, Malcovati L, et al. Frequent mutation of the poly-comb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24:1062–1065. doi: 10.1038/leu.2010.20. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer. 2009;9:773–784. doi: 10.1038/nrc2736. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22:5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Cao P, Deng Z, Wan M, Huang W, Cramer SD, Xu J, Lei M, Sui G. MicroRNA-101 negatively regulates Ezh2 and its expression is modulated by androgen receptor and HIF-1alpha/HIF-1beta. Mol Cancer. 2010;9:108. doi: 10.1186/1476-4598-9-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbuccia N, Murati A, Trouplin V, Brecqueville M, Adélaïde J, Rey J, Vainchenker W, Bernard OA, Chaffanet M, Vey N, et al. Mutations of ASXL1 gene in myeloproliferative neoplasms. Leukemia. 2009;23:2183–2186. doi: 10.1038/leu.2009.141. [DOI] [PubMed] [Google Scholar]
- Carbuccia N, Trouplin V, Gelsi-Boyer V, Murati A, Rocquain J, Adélaïde J, Olschwang S, Xerri L, Vey N, Chaffanet M, et al. Mutual exclusion of ASXL1 and NPM1 mutations in a series of acute myeloid leukemias. Leukemia. 2010;24:469–473. doi: 10.1038/leu.2009.218. [DOI] [PubMed] [Google Scholar]
- Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004;18:2627–2638. doi: 10.1101/gad.1241904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagraoui J, Niessen SL, Lessard J, Girard S, Coulombe P, Sauvageau M, Meloche S, Sauvageau G. E4F1: a novel candidate factor for mediating BMI1 function in primitive hematopoietic cells. Genes Dev. 2006;20:2110–2120. doi: 10.1101/gad.1453406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatoo W, Abdouh M, David J, Champagne MP, Ferreira J, Rodier F, Bernier G. The polycomb group gene Bmi1 regulates antioxidant defenses in neurons by repressing p53 pro-oxidant activity. J Neurosci. 2009;29:529–542. doi: 10.1523/JNEUROSCI.5303-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi D, Lee SJ, Hong S, Kim IH, Kang S. Prohibitin interacts with RNF2 and regulates E2F1 function via dual pathways. Oncogene. 2008;27:1716–1725. doi: 10.1038/sj.onc.1210806. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the Rb and polycomb pathways. Mol Cell. 2001;8:557–569. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Denisenko O, Shnyreva M, Suzuki H, Bomsztyk K. Point mutations in the WD40 domain of Eed block its interaction with Ezh2. Mol Cell Biol. 1998;18:5634–5642. doi: 10.1128/mcb.18.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshpande AM, Akunowicz JD, Reveles XT, Patel BB, Saria EA, Gorlick RG, Naylor SL, Leach RJ, Hansen MF. PHC3, a component of the hPRC-H complex, associates with E2F6 during G0 and is lost in osteosarcoma tumors. Oncogene. 2007;26:1714–1722. doi: 10.1038/sj.onc.1209988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, Waghorn K, Zoi K, Ross FM, Reiter A, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajas L, Paul C, Zugasti O, Le Cam L, Polanowska J, Fabbrizio E, Medema R, Vignais ML, Sardet C. pRB binds to and modulates the transrepressing activity of the E1A-regulated transcription factor p120E4F. Proc Natl Acad Sci USA. 2000;97:7738–7743. doi: 10.1073/pnas.130198397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Fisher CL, Pineault N, Brookes C, Helgason CD, Ohta H, Bodner C, Hess JL, Humphries RK, Brock HW. Loss-of-function Additional sex combs like 1 mutations disrupt hematopoiesis but do not cause severe myelodysplasia or leukemia. Blood. 2010;115:38–46. doi: 10.1182/blood-2009-07-230698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306:1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- Francis NJ, Follmer NE, Simon MD, Aghia G, Butler JD. Polycomb proteins remain bound to chromatin and DNA during DNA replication in vitro. Cell. 2009;137:110–122. doi: 10.1016/j.cell.2009.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, Zhou X, Jones PA. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive micro-RNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- García-Cuéllar MP, Zilles O, Schreiner SA, Birke M, Winkler TH, Slany RK. The ENL moiety of the childhood leukemia-associated MLL-ENL oncoprotein recruits human Polycomb 3. Oncogene. 2001;20:411–419. doi: 10.1038/sj.onc.1204108. [DOI] [PubMed] [Google Scholar]
- Gelsi-Boyer V, Trouplin V, Adélaïde J, Bonansea J, Cervera N, Carbuccia N, Lagarde A, Prebet T, Nezri M, Sainty D, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145:788–800. doi: 10.1111/j.1365-2141.2009.07697.x. [DOI] [PubMed] [Google Scholar]
- Gil J, Peters G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol. 2006;7:667–677. doi: 10.1038/nrm1987. [DOI] [PubMed] [Google Scholar]
- Godlewski J, Nowicki MO, Bronisz A, Williams S, Otsuki A, Nuovo G, Raychaudhury A, Newton HB, Chiocca EA, Lawler S. Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 2008;68:9125–9130. doi: 10.1158/0008-5472.CAN-08-2629. [DOI] [PubMed] [Google Scholar]
- Gong Y, Yue J, Wu X, Wang X, Wen J, Lu L, Peng X, Qiang B, Yuan J. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 2006;34:6158–6169. doi: 10.1093/nar/gkl834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González I, Simón R, Busturia A. The Polyhomeotic protein induces hyperplastic tissue overgrowth through the activation of the JAK/STAT pathway. Cell Cycle. 2009;8:4103–4111. doi: 10.4161/cc.8.24.10212. [DOI] [PubMed] [Google Scholar]
- Grimm C, Matos R, Ly-Hartig N, Steuerwald U, Lindner D, Rybin V, Müller J, Müller CW. Molecular recognition of histone lysine methylation by the Polycomb group repressor dSfmbt. EMBO J. 2009;28:1965–1977. doi: 10.1038/emboj.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo WJ, Zeng MS, Yadav A, Song LB, Guo BH, Band V, Dimri GP. Mel-18 acts as a tumor suppressor by repressing Bmi-1 expression and down-regulating Akt activity in breast cancer cells. Cancer Res. 2007;67:5083–5089. doi: 10.1158/0008-5472.CAN-06-4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KH, Bracken AP, Pasini D, Dietrich N, Gehani SS, Monrad A, Rappsilber J, Lerdrup M, Helin K. A model for transmission of the H3K27me3 epigenetic mark. Nat Cell Biol. 2008;10:1291–1300. doi: 10.1038/ncb1787. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Bath ML, Harris AW, Adams JM. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene. 1993;8:3161–3164. [PubMed] [Google Scholar]
- Hemenway CS, de Erkenez AC, Gould GC. The polycomb protein MPc3 interacts with AF9, an MLL fusion partner in t(9;11)(p22;q23) acute leukemias. Oncogene. 2001;20:3798–3805. doi: 10.1038/sj.onc.1204478. [DOI] [PubMed] [Google Scholar]
- Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal M, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [DOI] [PubMed] [Google Scholar]
- Hong Z, Jiang J, Lan L, Nakajima S, Kanno S, Koseki H, Yasui A. A polycomb group protein, PHF1, is involved in the response to DNA double-strand breaks in human cell. Nucleic Acids Res. 2008;36:2939–2947. doi: 10.1093/nar/gkn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huen MS, Sy SM, van Deursen JM, Chen J. Direct interaction between SET8 and proliferating cell nuclear antigen couples H4-K20 methylation with DNA replication. J Biol Chem. 2008;283:11073–11077. doi: 10.1074/jbc.C700242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Hirao A, Arai F, Matsuoka S, Takubo K, Hamaguchi I, Nomiyama K, Hosokawa K, Sakurada K, Nakagata N, et al. Regulation of oxidative stress by ATM is required for self-renewal of haematopoietic stem cells. Nature. 2004;431:997–1002. doi: 10.1038/nature02989. [DOI] [PubMed] [Google Scholar]
- Iwata S, Takenobu H, Kageyama H, Koseki H, Ishii T, Nakazawa A, Tatezaki S, Nakagawara A, Kamijo T. Polycomb group molecule PHC3 regulates polycomb complex composition and prognosis of osteosarcoma. Cancer Sci. 2010;101:1646–1652. doi: 10.1111/j.1349-7006.2010.01586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang YY, Sharkis SJ. A low level of reactive oxygen species selects for primitive hematopoietic stem cells that may reside in the low-oxygenic niche. Blood. 2007;110:3056–3063. doi: 10.1182/blood-2007-05-087759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B, Yao B, Li JL, Fields CR, Delmas AL, Liu C, Robertson KD. DNMT1 and DNMT3B modulate distinct polycomb-mediated histone modifications in colon cancer. Cancer Res. 2009;69:7412–7421. doi: 10.1158/0008-5472.CAN-09-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S, Elvers I, Trelle MB, Menzel T, Eskildsen M, Jensen ON, Helleday T, Helin K, Sørensen CS. The histone methyltransferase SET8 is required for S-phase progression. J Cell Biol. 2007;179:1337–1345. doi: 10.1083/jcb.200706150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M. SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol. 2002;156:595–602. doi: 10.1083/jcb.200110109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan AH, Kumar RM, Marx JG, Young RA, Sartorelli V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol Cell. 2009;36:61–74. doi: 10.1016/j.molcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK, et al. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293–4304. doi: 10.1038/onc.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Bystrykh LV, de Boer A, Houwer S, Douma J, Weersing E, Dontje B, de Haan G. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107:2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerppola TK. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheradmand Kia S, Solaimani Kartalaei P, Farahbakhshian E, Pourfarzad F, von Lindern M, Verrijzer CP. EZH2-dependent chromatin looping controls INK4a and INK4b, but not ARF, during human progenitor cell differentiation and cellular senescence. Epigenetics Chromatin. 2009;2:16. doi: 10.1186/1756-8935-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Park J, Choi MC, Park JH, Kim HP, Lee JH, Oh DY, Im SA, Bang YJ, Kim TY. DNA methyltransferase 3B acts as a co-repressor of the human polycomb protein hPc2 to repress fibroblast growth factor receptor 3 transcription. Int J Biochem Cell Biol. 2008;40:2462–2471. doi: 10.1016/j.biocel.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Kirmizis A, Bartley SM, Kuzmichev A, Margueron R, Reinberg D, Green R, Farnham PJ. Silencing of human polycomb target genes is associated with methylation of histone H3 Lys 27. Genes Dev. 2004;18:1592–1605. doi: 10.1101/gad.1200204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein UR, Nigg EA. SUMO-dependent regulation of centrin-2. J Cell Sci. 2009;122:3312–3321. doi: 10.1242/jcs.050245. [DOI] [PubMed] [Google Scholar]
- Koga H, Matsui S, Hirota T, Takebayashi S, Okumura K, Saya H. A human homolog of Drosophila lethal(3)malignant brain tumor (l(3)mbt) protein associates with condensed mitotic chromosomes. Oncogene. 1999;18:3799–3809. doi: 10.1038/sj.onc.1202732. [DOI] [PubMed] [Google Scholar]
- Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21:49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Krivtsov AV, Armstrong SA. MLL translocations, histone modifications and leukaemia stem-cell development. Nat Rev Cancer. 2007;7:823–833. doi: 10.1038/nrc2253. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nat Cell Biol. 2010;12:618–624. doi: 10.1038/ncb2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cam L, Linares LK, Paul C, Julien E, Lacroix M, Hatchi E, Triboulet R, Bossis G, Shmueli A, Rodriguez MS, et al. E4F1 is an atypical ubiquitin ligase that modulates p53 effector functions independently of degradation. Cell. 2006;127:775–788. doi: 10.1016/j.cell.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Adhikary G, Balasubramanian S, Gopalakrishnan R, McCormick T, Dimri GP, Eckert RL, Rorke EA. Expression of Bmi-1 in epidermis enhances cell survival by altering cell cycle regulatory protein expression and inhibiting apoptosis. J Invest Dermatol. 2008;128:9–17. doi: 10.1038/sj.jid.5700949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, Wicha M, Clarke MF, Simeone DM. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–1037. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- Liu J, Cao L, Chen J, Song S, Lee IH, Quijano C, Liu H, Keyvanfar K, Chen H, Cao LY, et al. Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature. 2009;459:387–392. doi: 10.1038/nature08040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Ruhf ML, Perrimon N, Leder P. A genome-wide RNA interference screen identifies putative chromatin regulators essential for E2F repression. Proc Natl Acad Sci USA. 2007;104:9381–9386. doi: 10.1073/pnas.0610279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Yang X, Takihara Y, Knoetgen H, Kessel M. The cell-cycle regulator geminin inhibits Hox function through direct and polycomb-mediated interactions. Nature. 2004;427:749–753. doi: 10.1038/nature02305. [DOI] [PubMed] [Google Scholar]
- Maertens GN, El Messaoudi-Aubert S, Racek T, Stock JK, Nicholls J, Rodriguez-Niedenführ M, Gil J, Peters G. Several distinct polycomb complexes regulate and co-localize on the INK4a tumor suppressor locus. PLoS ONE. 2009;4:e6380. doi: 10.1371/journal.pone.0006380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski IJ, Blewitt ME, de Graaf CA, McManus EJ, Bahlo M, Hilton AA, Hyland CD, Smyth GK, Corbin JE, Metcalf D, et al. Polycomb repressive complex 2 (PRC2) restricts hematopoietic stem cell activity. PLoS Biol. 2008;6:e93. doi: 10.1371/journal.pbio.0060093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez R, Martin-Subero JI, Rohde V, Kirsch M, Alaminos M, Fernandez AF, Ropero S, Schackert G, Esteller M. A microarray-based DNA methylation study of glioblastoma multiforme. Epigenetics. 2009;4:255–264. doi: 10.4161/epi.9130. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagishima H, Isono K, Fujimura Y, Iyo M, Takihara Y, Masumoto H, Vidal M, Koseki H. Dissociation of mammalian Polycomb-group proteins, Ring1B and Rae28/Ph1, from the chromatin correlates with configuration changes of the chromatin in mitotic and meiotic prophase. Histochem Cell Biol. 2003;120:111–119. doi: 10.1007/s00418-003-0551-2. [DOI] [PubMed] [Google Scholar]
- Mohammad HP, Cai Y, McGarvey KM, Easwaran H, Van Neste L, Ohm JE, O’Hagan HM, Baylin SB. Polycomb CBX7 promotes initiation of heritable repression of genes frequently silenced with cancer-specific DNA hypermethylation. Cancer Res. 2009;69:6322–6330. doi: 10.1158/0008-5472.CAN-09-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Bmi-1 promotes neural stem cell self-renewal and neural development but not mouse growth and survival by repressing the p16Ink4a and p19Arf senescence pathways. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, et al. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42:181–185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi M, Saraya A, Miyagi S, Nagao K, Inagaki Y, Nishikawa M, Tajima S, Koseki H, Tsuda H, Takasaki Y, et al. Bmi1 cooperates with Dnmt1-associated protein 1 in gene silencing. Biochem Biophys Res Commun. 2007;353:992–998. doi: 10.1016/j.bbrc.2006.12.166. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Köcher T, Cohen A, Stunnenberg HG, Wilm M, Müller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijnik A, Woodbine L, Marchetti C, Dawson S, Lambe T, Liu C, Rodrigues NP, Crockford TL, Cabuy E, Vindigni A, et al. DNA repair is limiting for haematopoietic stem cells during ageing. Nature. 2007;447:686–690. doi: 10.1038/nature05875. [DOI] [PubMed] [Google Scholar]
- Nikoloski G, Langemeijer SM, Kuiper RP, Knops R, Massop M, Tönnissen ER, van der Heijden A, Scheele TN, Vandenberghe P, de Witte T, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42:665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- O’Dor E, Beck SA, Brock HW. Polycomb group mutants exhibit mitotic defects in syncytial cell cycles of Drosophila embryos. Dev Biol. 2006;290:312–322. doi: 10.1016/j.ydbio.2005.11.015. [DOI] [PubMed] [Google Scholar]
- O’Riain C, O’Shea DM, Yang Y, Le Dieu R, Gribben JG, Summers K, Yeboah-Afari J, Bhaw-Rosun L, Fleischmann C, Mein CA, et al. Array-based DNA methylation profiling in follicular lymphoma. Leukemia. 2009;23:1858–1866. doi: 10.1038/leu.2009.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda H, Okamoto I, Murphy N, Chu J, Price SM, Shen MM, Torres-Padilla ME, Heard E, Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol Cell Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H, Iwama A, Morita Y, Kamijo T, van Lohuizen M, Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203:2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, Nakauchi H, Iwama A. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell. 2010;6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Ohm JE, McGarvey KM, Yu X, Cheng L, Schuebel KE, Cope L, Mohammad HP, Chen W, Daniel VC, Yu W, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hyper-methylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Yasunaga S, Ohno Y, Tsumura M, Okada S, Ishikawa N, Shirao K, Kikuchi A, Nishitani H, Kobayashi M, Takihara Y. Polycomb-group complex 1 acts as an E3 ubiquitin ligase for Geminin to sustain hematopoietic stem cell activity. Proc Natl Acad Sci USA. 2008;105:10396–10401. doi: 10.1073/pnas.0800672105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan G, Tian S, Nie J, Yang C, Ruotti V, Wei H, Jonsdottir GA, Stewart R, Thomson JA. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AJ, Kyba M, Bornemann D, Morgan K, Brock HW, Simon J. A domain shared by the Polycomb group proteins Scm and ph mediates heterotypic and homotypic interactions. Mol Cell Biol. 1997;17:6683–6692. doi: 10.1128/mcb.17.11.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson AJ, Mallin DR, Francis NJ, Ketel CS, Stamm J, Voeller RK, Kingston RE, Simon JA. Requirement for sex comb on midleg protein interactions in Drosophila polycomb group repression. Genetics. 2004;167:1225–1239. doi: 10.1534/genetics.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekhar VK, Begemann M. Concise review: roles of polycomb group proteins in development and disease: a stem cell perspective. Stem Cells. 2007;25:2498–2510. doi: 10.1634/stemcells.2006-0608. [DOI] [PubMed] [Google Scholar]
- Rice JC, Nishioka K, Sarma K, Steward R, Reinberg D, Allis CD. Mitotic-specific methylation of histone H4 Lys 20 follows increased PR-Set7 expression and its localization to mitotic chromosomes. Genes Dev. 2002;16:2225–2230. doi: 10.1101/gad.1014902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richie ER, Schumacher A, Angel JM, Holloway M, Rinchik EM, Magnuson T. The Polycomb-group gene eed regulates thymocyte differentiation and suppresses the development of carcinogen-induced T-cell lymphomas. Oncogene. 2002;21:299–306. doi: 10.1038/sj.onc.1205051. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscic A, Möller A, Calzado MA, Renner F, Wimmer VC, Gresko E, Lüdi KS, Schmitz ML. Phosphorylation-dependent control of Pc2 SUMO E3 ligase activity by its substrate protein HIPK2. Mol Cell. 2006;24:77–89. doi: 10.1016/j.molcel.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Seita J, Czechowicz A, Bhattacharya D, Bryder D, Weissman IL. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle. 2007;6:2371–2376. doi: 10.4161/cc.6.19.4759. [DOI] [PubMed] [Google Scholar]
- Rouleau M, McDonald D, Gagné P, Ouellet ME, Droit A, Hunter JM, Dutertre S, Prigent C, Hendzel MJ, Poirier GG. PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery. J Cell Biochem. 2007;100:385–401. doi: 10.1002/jcb.21051. [DOI] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez C, Sánchez I, Demmers JA, Rodriguez P, Strouboulis J, Vidal M. Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol Cell Proteomics. 2007;6:820–834. doi: 10.1074/mcp.M600275-MCP200. [DOI] [PubMed] [Google Scholar]
- Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TF, Möller P, Stilgenbauer S, Pollack JR, Wirth T. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202–4212. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- Saramäki OR, Tammela TL, Martikainen PM, Vessella RL, Visakorpi T. The gene for polycomb group protein enhancer of zeste homolog 2 (EZH2) is amplified in late-stage prostate cancer. Genes Chromosomes Cancer. 2006;45:639–645. doi: 10.1002/gcc.20327. [DOI] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Miller M, Lemieux S, Lessard J, Hébert J, Sauvageau G. Quantitative expression profiling guided by common retroviral insertion sites reveals novel and cell type specific cancer genes in leukemia. Blood. 2008;111:790–799. doi: 10.1182/blood-2007-07-098236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, de Ayala Alonso AG, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature. 2010;465:243–247. doi: 10.1038/nature08966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J. 2006;25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Cavalli G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development. 2009;136:3531–3542. doi: 10.1242/dev.033902. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Kim W, Fujiwara Y, Simon MD, Liu Y, Mysliwiec MR, Yuan GC, Lee Y, Orkin SH. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139:1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumacher A, Faust C, Magnuson T. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature. 1996;383:250–253. doi: 10.1038/383250a0. [DOI] [PubMed] [Google Scholar]
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol. 2007;9:1428–1435. doi: 10.1038/ncb1663. [DOI] [PubMed] [Google Scholar]
- Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y, Zhuang SM. MicroRNA-101, down-regulated in hepatocellular carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer Res. 2009;69:1135–1142. doi: 10.1158/0008-5472.CAN-08-2886. [DOI] [PubMed] [Google Scholar]
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–298. doi: 10.1016/j.stem.2010.08.004. this issue. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Iwamura C, Shinoda K, Tumes DJ, Kimura MY, Hosokawa H, Endo Y, Horiuchi S, Tokoyoda K, Koseki H, et al. Polycomb group gene product Ring1B regulates Th2-driven airway inflammation through the inhibition of Bim-mediated apoptosis of effector Th2 cells in the lung. J Immunol. 2010;184:4510–4520. doi: 10.4049/jimmunol.0903426. [DOI] [PubMed] [Google Scholar]
- Tardat M, Murr R, Herceg Z, Sardet C, Julien E. PR-Set7-dependent lysine methylation ensures genome replication and stability through S phase. J Cell Biol. 2007;179:1413–1426. doi: 10.1083/jcb.200706179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Menon U, Gentry-Maharaj A, Ramus SJ, Weisenberger DJ, Shen H, Campan M, Noushmehr H, Bell CG, Maxwell AP, et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome Res. 2010;20:440–446. doi: 10.1101/gr.103606.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, Ishihara H, Kanno R, Kamiyasu M, Inoue H, Tokuhisa T, Taniguchi M, Kanno M. mel-18 negatively regulates cell cycle progression upon B cell antigen receptor stimulation through a cascade leading to c-myc/cdc25. Immunity. 1998;9:439–448. doi: 10.1016/s1074-7613(00)80627-5. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Tonini T, Bagella L, D’Andrilli G, Claudio PP, Giordano A. Ezh2 reduces the ability of HDAC1-dependent pRb2/p130 transcriptional repression of cyclin. A Oncogene. 2004;23:4930–4937. doi: 10.1038/sj.onc.1207608. [DOI] [PubMed] [Google Scholar]
- Trojer P, Li G, Sims RJ, 3rd, Vaquero A, Kalakonda N, Boccuni P, Lee D, Erdjument-Bromage H, Tempst P, Nimer SD, et al. L3MBTL1, a histone-methylation-dependent chromatin lock. Cell. 2007;129:915–928. doi: 10.1016/j.cell.2007.03.048. [DOI] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS ONE. 2008;3:e2235. doi: 10.1371/journal.pone.0002235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O’Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, Laxman B, Cao X, Jing X, Ramnarayanan K, et al. Genomic loss of micro-RNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322:1695–1699. doi: 10.1126/science.1165395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa R, Pasini D, Gutierrez A, Morey L, Occhionorelli M, Viré E, Nomdedeu JF, Jenuwein T, Pelicci PG, Minucci S, et al. Role of the polycomb repressive complex 2 in acute promyelocytic leukemia. Cancer Cell. 2007;11:513–525. doi: 10.1016/j.ccr.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Viré E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, van Lohuizen M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112:4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, Waldvogel B, Vannier C, Darling D, zur Hausen A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11:1487–1495. doi: 10.1038/ncb1998. [DOI] [PubMed] [Google Scholar]
- Whitcomb SJ, Basu A, Allis CD, Bernstein E. Polycomb Group proteins: an evolutionary perspective. Trends Genet. 2007;23:494–502. doi: 10.1016/j.tig.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- Wong CK, Chen Z, So KL, Li D, Li P. Polycomb group protein RING1B is a direct substrate of Caspases-3 and -9. Biochim Biophys Acta. 2007;1773:844–852. doi: 10.1016/j.bbamcr.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Zheng SS, Li ZM, Qiao YY, Aau MY, Yu Q. Polycomb protein EZH2 regulates E2F1-dependent apoptosis through epigenetically modulating Bim expression. Cell Death Differ. 2010;17:801–810. doi: 10.1038/cdd.2009.162. [DOI] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase upregulated in prostate cancer. Proc Natl Acad Sci USA. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita M, Kuwahara M, Suzuki A, Hirahara K, Shinnaksu R, Hosokawa H, Hasegawa A, Motohashi S, Iwama A, Nakayama T. Bmi1 regulates memory CD4 T cell survival via repression of the Noxa gene. J Exp Med. 2008;205:1109–1120. doi: 10.1084/jem.20072000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Karuturi RK, Sun F, Aau M, Yu K, Shao R, Miller LD, Tan PB, Yu Q. CDKN1C (p57) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS ONE. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, et al. An integrated network of androgen receptor, polycomb, and TMPRSS2-ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17:443–454. doi: 10.1016/j.ccr.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Varambally S, Cao Q, Chinnaiyan AM, Ferguson DO, Merajver SD, Kleer CG. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia. 2005;7:1011–1019. doi: 10.1593/neo.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zencak D, Lingbeek M, Kostic C, Tekaya M, Tanger E, Hornfeld D, Jaquet M, Munier FL, Schorderet DF, van Lohuizen M, Arsenijevic Y. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci. 2005;25:5774–5783. doi: 10.1523/JNEUROSCI.3452-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sarge KD. Mel-18 interacts with RanGAP1 and inhibits its sumoylation. Biochem Biophys Res Commun. 2008;375:252–255. doi: 10.1016/j.bbrc.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sejas DP, Qiu Y, Williams DA, Pang Q. Inflammatory ROS promote and cooperate with the Fanconi anemia mutation for hematopoietic senescence. J Cell Sci. 2007;120:1572–1583. doi: 10.1242/jcs.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Goodson ML, Hong Y, Sarge KD. MEL-18 interacts with HSF2 and the SUMO E2 UBC9 to inhibit HSF2 sumoylation. J Biol Chem. 2008a;283:7464–7469. doi: 10.1074/jbc.M707122200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 2008b;68:4311–4320. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell. 2008;29:69–80. doi: 10.1016/j.molcel.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Wani G, Arab HH, El-Mahdy MA, Ray A, Wani AA. Chromatin restoration following nucleotide excision repair involves the incorporation of ubiquitinated H2A at damaged genomic sites. DNA Repair (Amst) 2009;8:262–273. doi: 10.1016/j.dnarep.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.