Significance
Cytochrome c oxidase is the last of the enzymes in the mitochondrial electron transport chain and represents a large integral membrane protein complex coupling conversion of molecular oxygen to water with proton translocation across the membrane. We used mass spectrometry to study cytochrome c oxidase extracted from bovine heart and provide the first evidence that both the monomer and the dimer are present in solution and are stabilized by annular and interfacial lipid interactions. We also identified posttranslational modifications and located them in the structure of cytochrome c oxidase; the location of the acetylated residues in particular suggests a role for this modification in modulating lipid interactions, and hence the interplay of the two monomeric subunits.
Keywords: cytochrome c oxidase, mass spectrometry, lipids
Abstract
Bovine cytochrome c oxidase is an integral membrane protein complex comprising 13 protein subunits and associated lipids. Dimerization of the complex has been proposed; however, definitive evidence for the dimer is lacking. We used advanced mass spectrometry methods to investigate the oligomeric state of cytochrome c oxidase and the potential role of lipids and posttranslational modifications in its subunit interfaces. Mass spectrometry of the intact protein complex revealed that both the monomer and the dimer are stabilized by large lipid entities. We identified these lipid species from the purified protein complex, thus implying that they interact specifically with the enzyme. We further identified phosphorylation and acetylation sites of cytochrome c oxidase, located in the peripheral subunits and in the dimer interface, respectively. Comparing our phosphorylation and acetylation sites with those found in previous studies of bovine, mouse, rat, and human cytochrome c oxidase, we found that whereas some acetylation sites within the dimer interface are conserved, suggesting a role for regulation and stabilization of the dimer, phosphorylation sites were less conserved and more transient. Our results therefore provide insights into the locations and interactions of lipids with acetylated residues within the dimer interface of this enzyme, and thereby contribute to a better understanding of its structure in the natural membrane. Moreover dimeric cytochrome c oxidase, comprising 20 transmembrane, six extramembrane subunits, and associated lipids, represents the largest integral membrane protein complex that has been transferred via electrospray intact into the gas phase of a mass spectrometer, representing a significant technological advance.
The electron transport chain in both bacteria and mitochondria consists of a series of membrane-embedded protein complexes that transfer electrons from donors to acceptors to establish a proton gradient across the inner membrane. This proton gradient is then used by ATP synthase to produce energy. Cytochrome c oxidase (CcO) is the last enzyme of the electron transport chain and receives electrons from cytochrome c molecules to catalyze the reduction of oxygen to water (1). CcO in bovine heart contains 13 different subunits (2) with various associated proteins with lower binding affinities (3). The three largest subunits (I–III) are encoded by mitochondrial genes (4) and form the core of the enzyme (5); subunits I and II contain metal binding sites, whereas subunit III stabilizes these catalytic centers and harbors the O2 transfer pathway (6). The remaining 10 subunits are thought to have a stabilizing function (5). CcO was found to be dimeric in the crystal lattice (7), making it one of the largest transmembrane protein complexes studied by X-ray crystallography.
A significant quantity of lipids (8) has been identified and located in crystal structures of both bacterial and bovine CcO (4, 9, 10). Two cardiolipins (CL), one phosphatidylcholine (PC), three phosphatidylethanolamines (PE), four phosphatidylglycerols (PG), and three triglycerides (TG) have been identified in bovine crystalline CcO (11). Ether phospholipid choline plasmalogens, typically present in mammalian cell membranes, were also identified but were found to be identical to the corresponding PCs and likely are derivatives thereof (11). Several lipids were also identified that bridge the two CcO monomers, suggesting that they may stabilize a putative dimer interface (11, 12), and a critical role for CL has been proposed following its removal and concomitant inactivation of the enzyme (13, 14).
CcO is regulated by expression of specific isoforms that are dependent on the tissue, species, or developmental stage, by allosteric effectors such as ATP/ADP, palmitate, or calcium and by reversible phosphorylation (3). To date, a total of 18 phosphorylation sites has been identified (3). One of these 18 sites (Thr11-VIa) was proposed in the X-ray structure of CcO although not confirmed at the resolution attained (15). Recently an interplay between allosteric ATP inhibition and reversible phosphorylation was proposed, implying inhibitory phosphorylation sites (16).
Dissociation of tightly bound CcO subunits is also thought to regulate the enzyme (3). Alkaline, urea, or n-dodecyl-β-d-maltoside (DDM) treatment leads to selective loss of protein subunits (17, 18) and is linked with inactivation of the enzyme (19). Thermal denaturation revealed a stabilizing role for tightly bound phospholipids in both monomeric and dimeric CcO (20). Subunit VIb has been proposed to increase the stability of dimeric CcO, as suggested by its position in the crystal structure (3). In addition to dimeric CcO, a mitochondrial supercomplex containing monomeric CcO has been detected (21). Because allosteric ATP inhibition requires cooperativity of the two monomers, however, a biological relevance for the monomeric form continues to be debated.
We address the question of how lipids and posttranslational modifications (PTMs) modulate dimerization of CcO by using mass spectrometry (MS). Transferring intact protein assemblies into the gas phase enabled us to investigate the oligomeric state of CcO in solution. Locating the phosphorylation sites in the atomic structure of CcO reveals multiple sites on the peripheral domains, exposed to the mitochondrial matrix. By contrast, the hotspots for acetylation occur primarily along the proposed dimer interfaces, in close contact with the “lipid plug,” suggesting the role of these acetyl groups in modulating the dimeric enzyme through changes in lipid interactions.
Results
Bovine CcO Contains 13 Individual Subunits.
CcO was isolated from bovine heart and we first used liquid chromatography-coupled tandem-MS (LC-MS/MS) of the peptides after protein hydrolysis to confirm the 13 subunits defined previously. Because CcO contains many integral membrane protein subunits we performed both tryptic and chymotryptic hydrolysis of the proteins to allow generation of sufficient peptides for confident protein and PTM identification. Subunits I–III, VIIb, and VIII are mostly embedded in the membrane and have only few tryptic cleavage sites; their sequence coverage after tryptic digestion was therefore low (6–40%). After chymotryptic hydrolysis these subunits could be identified with high sequence coverage (20–72%) (Table S1). Using this approach we confirmed the presence of all 13 subunits and also identified an associated protein, VDAC (UniProt ID P68002), with moderate sequence coverage (61%). VDAC was previously reported as associated with CcO but is not considered a constituent of the core complex (3, 22).
Table S1.
Identification of CcO protein subunits
| Trypsin 1 | Trypsin 2 | Chymotrypsin 1 | Chymotrypsin 2 | |||||||||||
| Subunit | UniProt ID | MW (Da) | No. spec | No. PS | SC (%) | No. spec | No. PS | SC (%) | No. spec | No. PS | SC (%) | No. spec | No. PS | SC (%) |
| I | P00396 | 57,032 (57,032) | 54 | 5 | 15 | 47 | 4 | 14 | 33 | 23 | 36 | 660 | 81 | 70 |
| II | P68530 | 26,021 (26,021) | 397 | 7 | 26 | 111 | 6 | 40 | 42 | 16 | 51 | 865 | 40 | 72 |
| III | P00415 | 29,933 (29,933) | 19 | 3 | 8 | 8 | 2 | 6 | 77 | 18 | 36 | |||
| IV | P00423 | 19,572 (17,150) | 1,265 | 37 | 86 | 323 | 26 | 84 | 48 | 14 | 64 | 344 | 33 | 76 |
| Va | P00426 | 16,735 (12,440) | 604 | 23 | 89 | 266 | 12 | 57 | 183 | 16 | 65 | 489 | 22 | 67 |
| Vb | P00428 | 13,834 (10,670) | 441 | 18 | 89 | 110 | 13 | 66 | 203 | 7 | 34 | |||
| VIa | P07471 | 10,800 (9,530) | 553 | 10 | 96 | 96 | 8 | 94 | 36 | 11 | 80 | 358 | 11 | 63 |
| VIb | P00429 | 10,156 (10,030) | 418 | 25 | 98 | 52 | 9 | 67 | 165 | 11 | 98 | |||
| VIc | P04038 | 8,610 (8,480) | 393 | 20 | 75 | 15 | 5 | 41 | 124 | 12 | 86 | |||
| VIIa | P07470 | 9,063 (6,670) | 128 | 7 | 41 | 46 | 6 | 85 | 3 | 3 | 38 | 116 | 7 | 68 |
| VIIb | P13183 | 9,065 (6,360) | 57 | 3 | 22 | 3 | 2 | 21 | 6 | 3 | 33 | 63 | 8 | 47 |
| VIIc | P00430 | 7,331 (5,440) | 241 | 7 | 84 | 10 | 3 | 31 | 169 | 10 | 55 | |||
| VIII | P10175 | 7,639 (4,960) | 6 | 2 | 20 | |||||||||
For each subunit the UniProt ID and the molecular weight (MW) is given. Masses in parentheses correspond to the proteins without transit peptide or initiator methionine. The proteins were hydrolyzed in two technical replicates with trypsin or chymotrypsin (Trypsin 1 and 2 or Chymotrypsin 1 and 2). The number of spectra (No. spec), the number of peptide sequences (No. PS), and the sequence coverage (SC) are listed for each replicate.
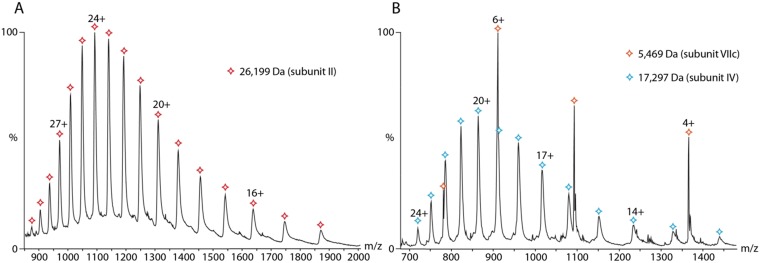
We used denaturing LC-MS to determine exact masses from the mass spectra of 10 of the 13 subunits (Fig. S1 and Table S2). We were not able to measure masses of subunits I, III, and VIIb, presumably because these membrane proteins are very hydrophobic and their separation by LC is impeded (also discussed above). Interestingly the proteins eluted over several minutes, in different chromatographic peaks, indicative of multiple forms and with higher masses than their theoretical values, suggesting that they contain PTMs.
Fig. S1.
Example spectra of denatured protein subunits. Exact masses could be determined from peak distributions. (A) CcO subunit II. (B) CcO subunits VIIc and IV.
Table S2.
Exact protein masses as determined by denaturing LC-MS
| Subunit | Theoretical mass (Da) | RT (min) | Experimental mass (Da) | Δ Mass (Da) | |
| With transit peptide | Without transit peptide | ||||
| I | 57,032 | 57,032 | / | / | / |
| II | 26,021 | 26,020 | 52.3-57.9 | 26,199 | 179 |
| III | 29,933 | 29,940 | / | / | / |
| IV | 19,572 | 17,150 | 35.8-37.8 | 17,297 | 147 |
| Va | 16,735 | 12,440 | 33.0-36.1 | 12,518 | 78 |
| Vb | 13,834 | 10,670 | 28.0-35.8 | 10,775 | 105 |
| VIa | 10,800 | 9,530 | 32.6-33.1 | 9,376 | -154 |
| VIb | 10,156 | 10,030 | 27.0-29.6 | 10,094 | 64 |
| VIc | 8,610 | 8,480 | 29.6-31.5 | 8,615 | 135 |
| VIIa | 9,063 | 6,670 | 31.3-31.9 | 6,713 | 43 |
| VIIb | 9,065 | 6,360 | / | / | / |
| VIIc | 7,331 | 5,440 | 33.0-37.8 | 5,469 | 29 |
| VIII | 7,639 | 4,960 | 33.0-35.8 | 4,978 | 18 |
| Σ | 225,791 | 204,722 | 205,366 | 644 | |
The theoretical masses obtained from the UniProt database, the retention time during LC-MS, the experimental masses, and the mass differences are given for each protein subunit. For proteins that contain a transit peptide, the masses with and without transit peptide are given. Subunits VIb and VIc include an initiator methionine, which is removed. Note that some proteins eluted over several minutes and multiple mass spectra were obtained. For these proteins the average masses were calculated. Subunits I, III, and VIIc were not identified by denaturing LC-MS.
PTMs Are Located on the Peripheral Subunits and in the Dimeric Interface of CcO.
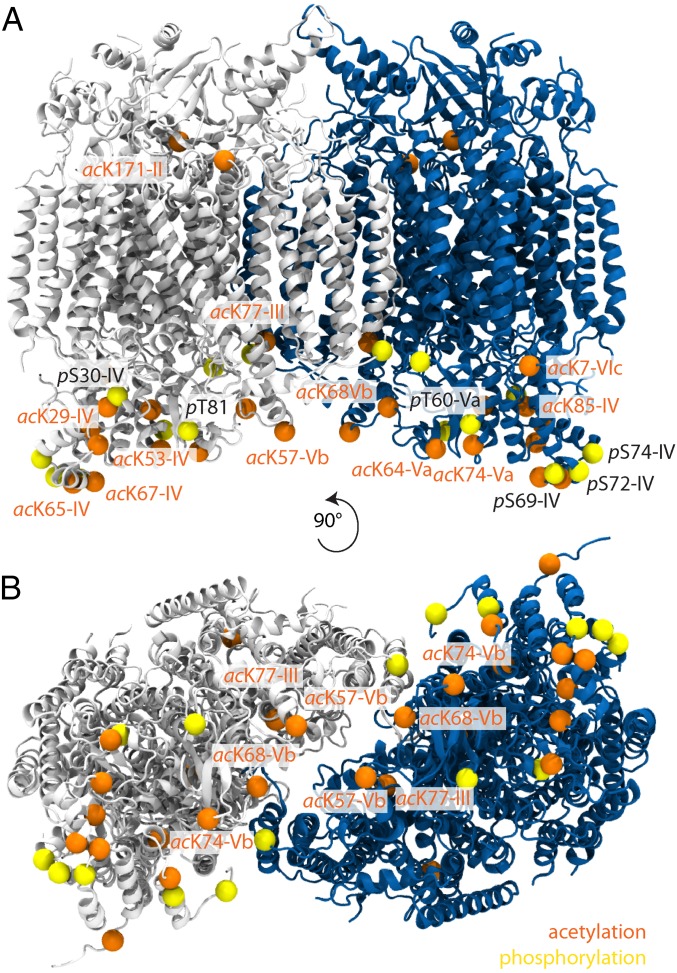
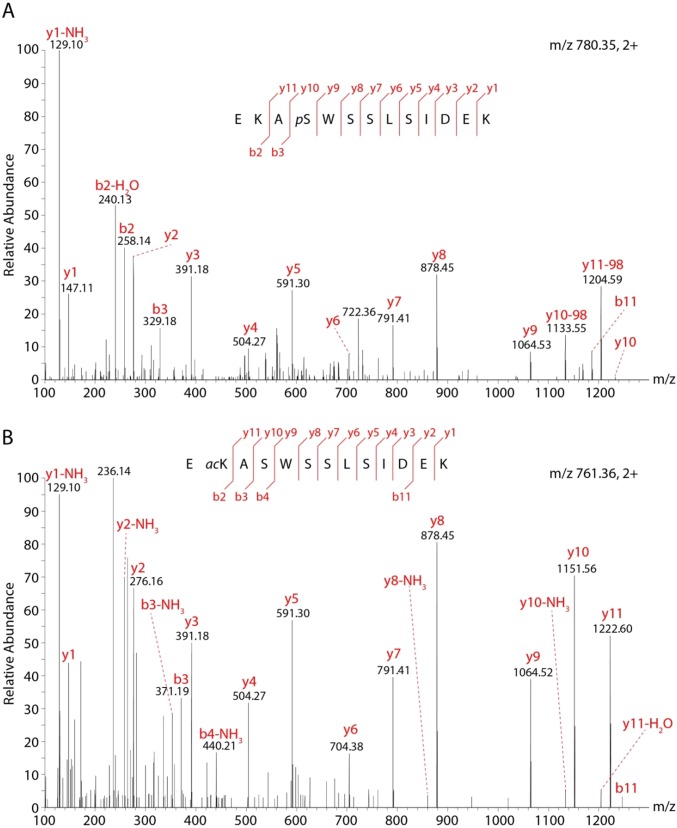
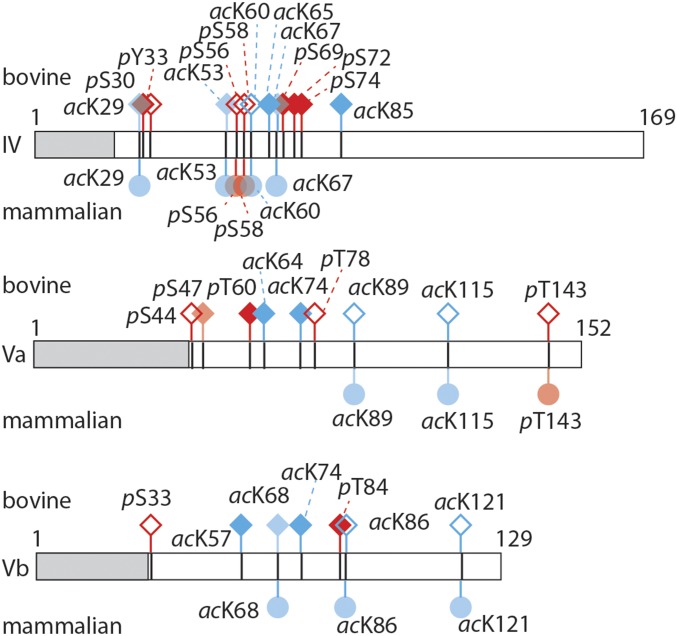
We carried out standard proteomics methods (Methods) and included potential phosphorylation of serine, threonine, and tyrosine residues as well as acetylation of lysine residues in database searches. In total, we identified 22 modified residues (8 phosphorylated and 14 acetylated) in 7 of the 13 CcO subunits (Fig. 1 and Table S3). Most of the modified residues are located in soluble domains of subunits IV, Va, and Vb, with at least four PTMs in each of these subunits. Interestingly, none of these subunits shows a preference for phosphorylation or acetylation; all three subunits are equally modified by both PTMs. However, all phosphorylated residues are serines or threonines, with no phosphorylated tyrosine detected. Representative spectra are shown for both phosphorylation and acetylation in Fig. S2. Comparing our results with previous studies in which 11 acetylation and 3 phosphorylated residues were identified in human, mouse, or rat (Table S4) we find that the majority of acetylated residues are conserved across species (Fig. 2 and Fig. S3). Locating the PTMs within the crystal structure of the CcO dimer (PDB ID code 2OCC) shows that phosphorylation occurs primarily on the matrix side of the soluble subunits (Fig. 1). By contrast, acetylated lysine residues are more widely distributed, often in proximity to phosphorylation sites.
Fig. 1.
PTMs are located in the peripheral subunits and in the dimer interface of CcO. Phosphorylated (yellow) and acetylated (orange) residues are highlighted on the crystal structure of CcO (PDB ID code 2OCC). Residue numbers and the type of PTM (p, phosphorylation or ac, acetylation) are given. Phosphosites are located mainly in peripheral subunits on the matrix side. Acetylated residues are located at protein interfaces and in the CcO dimer interface. (A) Side view of CcO. (B) Bottom view of CcO.
Table S3.
PTMs identified in CcO subunits
| Subunit | PTM | Modified residue | Peptide sequence | Maximum Mascot score | Known PTMs |
| II | Acetylation | acK171 | AVPSLGLKTDAIPGRL | 68 | pS115d, pY218a,e |
| III | Acetylation | acK77 | ESTFQGHHTPAVQKGLR | 64 | |
| IV | Acetylation | acK29 | AHGSVVKSEDYALPSYVDR | 58 | pY33c*, acK29a, acK53a, pS56a,b*, pS58a, acK60a, acK67a |
| IV | Phosphorylation | pS30 | AHGSVVKSEDYALPSYVDR | 86 | |
| IV | Acetylation | acK53 | DYPLPDVAHVKNLSASQK | 56 | |
| IV | Acetylation | acK65 | EKEKASWSSLSIDEK | 88 | |
| IV | Acetylation | acK67 | EKASWSSLSIDEK | 79 | |
| IV | Phosphorylation | pS769 | EKEKASWSSLSIDEK | 67 | |
| IV | Phosphorylation | pS72 | ASWSSLSIDEK | 66 | |
| IV | Phosphorylation | pS74 | EKASWSSLSIDEK | 56 | |
| IV | Acetylation | acK85 | RLKFKESFAEMNR | 53 | |
| Va | Phosphorylation | pS47 | SHGSHETDEEFDARWVTY | 75 | pS44e*, pS47b*, pT78b, acK89a, acK115a, pT143a |
| Va | Phosphorylation | pT60 | WVTYFNKPDIDAWELR | 89 | |
| Va | Acetylation | acK64 | WVTYFNKPDIDAWELR | 42 | |
| Va | Acetylation | acK74 | KGMNTLVGYDLVPEPK | 112 | |
| pS33c, acK68a, acK86a, acK121a | |||||
| Vb | Acetylation | acK57 | KGQDPYNILAPK | 78 | |
| Vb | Acetylation | acK68 | KGQDPYNILAPKATSGTK | 71 | |
| Vb | Acetylation | acK74 | ATSGTKEDPNLVPSITNK | 66 | |
| Vb | Phosphorylation | pT84 | AASGTKEDPNLVPSITNK | 71 | |
| VIa | Acetylation | acK70 | TKPFSWGDGNHTFFHNPR | 100 | pT23b* |
| VIb | acK62a | ||||
| VIc | Acetylation | acK7 | STALAKPQMR | 61 | |
| VIIc | pS17e, acK25a |
The protein subunit, type of modification, the modified residue number (ac, acetylation and p, phosphorylation), the peptide sequence of the modified peptides, and the maximum Mascot scores are listed for all identified residues. Known PTMs in CcO purified from bovine heart are listed for each protein subunit (a: UniProt; b: ref. 57; c: ref. 58 d: ref. 59; e: ref. 16). Known PTMs also identified in this study are highlighted in bold.
The numbering includes the mitochondrial transit peptide. Modified residues are underlined.
Fig. S2.
Example MS/MS spectra of phosphorylated (A) and acetylated (B) peptides. b and y ions are assigned and fragmentation of the peptides is indicated. The modified sites could be identified by fragmentation of the peptide.
Table S4.
PTMs identified in CcO subunits from human, mouse, or rat
| Subunit | Human | Mouse | Rat | |||
| UniProt ID | Acetylation sites | UniProt ID | Acetylation sites | UniProt ID | Acetylation sites | |
| I | P00395 | / | P00397 | / | Q06QA9 | / |
| II | P00403 | / | Q7JCZ1 | / | Q5UAJ6 | / |
| III | P00414 | / | Q7JCX7 | / | Q7H115 | / |
| IV | P13073 | acK29, acK53, pSer56, pSer58, acK60, acK67 | P19783 | acK29, pSer56, pSer58, acK60, acK67 | P10888 | acK29, acK53, pSer56, pSer58, acK60, acK67 |
| Va | P20674 | acK87, acK113, pThr141 | P12787 | acK83, acK109, pThr137 | P11240 | acK83, acK109, pThr137 |
| Vb | P10606 | acK68, acK86, acK121 | P19536 | acK67, acK85, acK120 | P12075 | acK68, acK86, acK121 |
| VIa | P12074 | / | P43024 | / | P10818 | / |
| VIb | P14854 | / | P56391 | acK62 | P80430 | / |
| VIc | P09669 | / | Q9CPQ1 | / | P11950 | / |
| VIIa | P24310 | / | P56392 | / | P35171 | acK33 |
| VIIb | P24311 | / | P56393 | / | P80431 | / |
| VIIc | P15954 | acK25 | P17665 | acK25 | P80432 | acK25 |
| VIII | P10176 | / | Q64445 | / | P80433 | / |
The subunit name, UniProt ID, and the modified residue numbers are given (ac, acetylation and p, phosphorylation). PTMs and modified sites were obtained from UniProt databases.
Fig. 2.
PTMs cluster in subunits IV, Va, and Vb. PTMs found in bovine CcO (above the bar) in this study (filled symbols) with those reported previously (open symbols) are indicated by squares. PTMs identified in both this and previous studies are transparent. Phosphorylation (red) and acetylation (blue) sites are shown. A comparison between the bovine sites (squares, on top) and those in other mammalia (rat/human/mouse, circles, below the bar) shows that mostly acetylation sites are conserved across the species (filled, transparent circles). The mitochondrial transit peptide is shown in gray.
Fig. S3.
Sequence alignment of bovine, human, mouse, and rat CcO subunits IV (A), Va (B), and Vb (C). PTMs identified in this study (red), previous studies (green), and identified in this and previous studies (blue) are highlighted. Sequence similarity between bovine subunits and human (H), mouse (M), and rat (R) are given for each subunit.
CcO Monomers and Dimers in Solution.
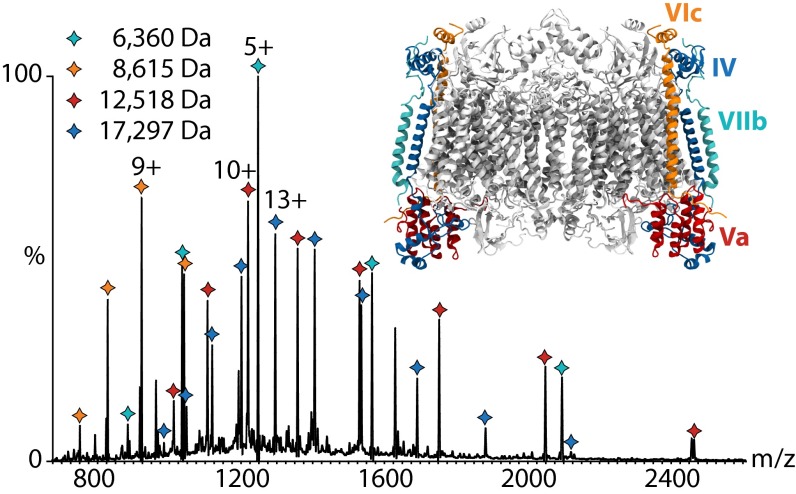
We next acquired mass spectra of CcO under high-energy conditions to strip the detergent micelle and release the intact protein complex in the gas phase (23, 24). The low m/z region of these spectra shows charge state series assigned to single protein subunits (Fig. 3). We identified these based upon their masses as subunits IV, Va, VIc, and VIIb and located them in the crystal structure. They correspond to peripheral membrane and soluble subunits of the complex.
Fig. 3.
MS of CcO reveals peripheral subunits. The low m/z region (<2,600 m/z) of the mass spectrum shows four peak series corresponding to peripheral subunits IV (blue), Va (red), VIc (orange), and VIIb (cyan). Charge states for the most intense peaks of each series are given. The peripheral subunits are highlighted on the crystal structure of CcO (PDB ID code 2OCC). Intact CcO was not observed in DM detergent, presumably due to the high activation energy required to release the proteins from the detergent micelle.
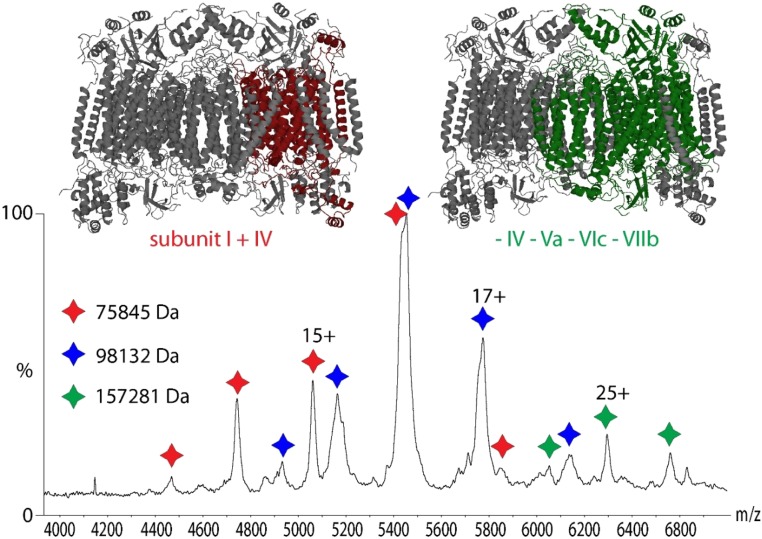
We also observed three peak series with a molecular weight of 76, 98, and 157 kDa corresponding to subcomplexes of CcO (Fig. S4). The mass of the smallest complex, together with the crystal structure, suggests that subunits I and IV form the core of these assemblies. Numerous assignments are possible for the 98-kDa complex; however, it is likely that it contains the core subunits I and IV as well as multiple peripheral subunits. The high mass complex corresponds to the intact CcO monomer that has lost several peripheral subunits. Interestingly, the mass of this subcomplex is in accord with the loss of the subunits IV, Va, VIc, and VIIb detected in the low m/z region of this mass spectrum (discussed above; see also Fig. 3), leaving a stable core containing subunits I, II, III, Vb, VIa, VIb, VIIa, VIIc, and VIII. Interestingly, CcO is not stable under the relatively harsh conditions required to release the protein complex from the n-decyl-β-d-maltopyranoside (DM) micelle. We therefore screened various detergents to identify one that requires lower activation energy to release the CcO complex intact from the detergent micelle. C8E4 is considered a “harsher” detergent (i.e., its ability to substitute for natural lipids is low compared with DM or DDM) but it requires lower activation energy to release proteins from micelles in the gas phase (25).
Fig. S4.
Subcomplexes of CcO. Three subcomplexes of CcO were observed by native MS in DM detergent. The complex of approximately 76 kDa (red) corresponds to the I–IV subcomplex. Due to similar masses of the protein subunits the 98-kDa complex (blue) could not be unambiguously assigned. A high-mass complex of 157 kDa (green) corresponds to the intact CcO monomer that lost several peripheral subunits.
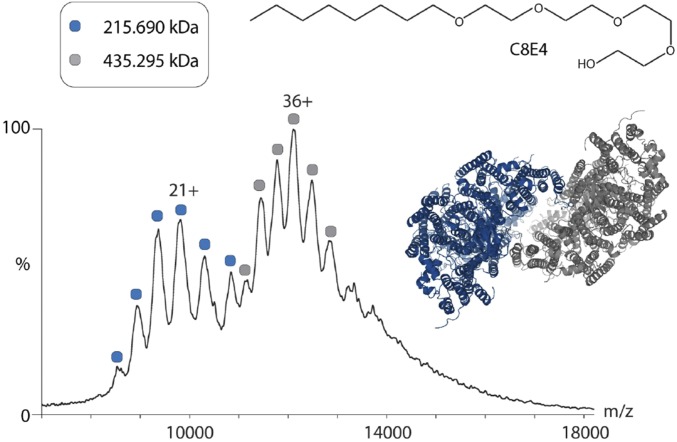
Indeed, after exchanging DM with C8E4 and recording a mass spectrum we observed two charge state series at high m/z (Fig. 4). Considering the experimental masses of the protein subunits the theoretical masses of the intact CcO monomer and dimer correspond to 205.4 and 410.8 kDa, respectively. The two peak series have masses of 215.7 and 435.3 kDa. The broad peaks observed in the mass spectrum are attributed to multiple incomplete PTMs and heterogeneous lipid binding. The difference between calculated and theoretical masses corresponds to an additional 10 and 25 kDa for the monomer and dimer, respectively. These masses suggest that the complexes are associated with a large lipid entity of ∼10 kDa per monomer, with a further 5 kDa of mass in dimeric CcO, ascribed to lipids located in the inner cavity within the complex.
Fig. 4.
MS of intact CcO reveals stabilization of monomeric/dimeric CcO by associated lipids. After exchanging DM detergent for C8E4 detergent (Upper) the CcO complex remains intact and the MS reveals peak series for monomeric (blue circles) and dimeric (gray circles) CcO. Charge states for the most intense peak of each series are given. Observed masses of the monomer (215 kDa) and dimer (435 kDa) are higher than the theoretical masses calculated from subunit masses, consistent with the association of lipids in monomeric and dimeric CcO. The broad peak width of the charge states is assigned to multiple lipid bound states and heterogeneous PTMs.
Lipids That Associate with CcO.
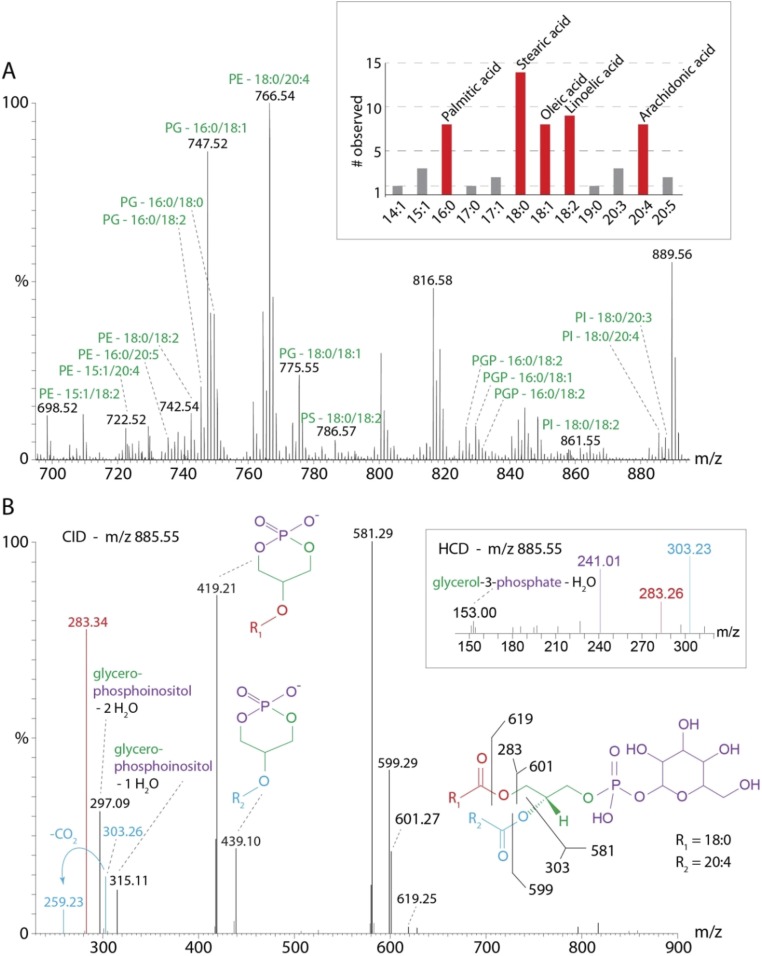
We analyzed the lipid content of CcO using an established protocol (26), digesting the proteins with trypsin and separating the resulting peptides from the associated lipids by LC. The lipids were eluted directly into a high-resolution mass spectrometer and analyzed by LC-MS/MS. We searched the precursor masses against a virtual lipid database containing TG, PC, phosphatidic acids, phosphatidylserines (PS), PE, PG, phosphatidylinositols (PI), PI phosphates, and CL with various acyl chain lengths (www.lipidmaps.org). This precursor ion search does not include the identification of the lipids by tandem MS. We therefore inspected the fragment spectra of abundant lipids by MS/MS data search (www.lipidmaps.org) to verify the lipids and identify the isomers. Following this workflow we identified four lipid species with different fatty acids including PG, PI, PS, and PE (Table S5). Manual inspection of the MS/MS spectra also revealed one species that was not included in the lipid database, phosphatidylglycerol phosphate (PGP), characterized by neutral loss of phosphate and coelution with the corresponding PG (Table S5). The presence of PGP in our analysis implies that the PG isomers that we identified originate from their related PGP by neutral loss of the phosphate group during activation in the mass spectrometer. Our lipid analysis further revealed preferences of fatty acids in these lipids, 16:0, 18:0, 18:1, 18:2, and 20:4 being clearly favored, consistent with the fatty acid composition of mammalian heart mitochondria with similar proportions (27).
Table S5.
Identified lipids in CcO
| Lipid species | m/z | RT (min) | XIC area | Side chains | |
| Phosphatidylserine | 786.53 | 17.06 | 5,176,354 | 18:1 | 18:1 |
| 788.55 | 18.65 | 5,434,349 | 18:0 | 18:1 | |
| 810.53 | 16.55 | 1,032,265 | 18:0 | 20:4 | |
| 812.55 | 17.42 | 3,990,216 | 20:3 | 18:0 | |
| Phosphatidylethanolamine | 698.52 | 16.10 | 86,942,694 | 18:2 | 15:1 |
| 708.50 | 16.38 | 2,500,554 | 20:4 | 14:1 | |
| 714.15 | 17.24 | 7,536,574 | 16:0 | 18:2 | |
| 720.50 | 16:38 | 3,252,866 | 20:5 | 15:1 | |
| 722.52 | 17.59 | 36,521,846 | 15:1 | 20:4 | |
| 726.51 | 19.93 | 108,115,079 | 18:2 | 17:1 | |
| 736.50 | 15.57 | 8,387,826 | 16:0 | 20:5 | |
| 740.53 | 17:59 | 27,057,547 | 18:1 | 18:2 | |
| 742.54 | 19.4 | 177,911,109 | 18:0 | 18:2 | |
| 750.55 | 19.57 | 55,999,319 | 20:4 | 17:1 | |
| 752.53 | 17.94 | 13,276,776 | 20:4 | 17:0 | |
| 766.54 | 20.03 | 351,505,712 | 20:4 | 18:0 | |
| 768.56 | 19.75 | 50,585,719 | 18:0 | 20:3 | |
| 780.52 | 15.18 | 2,266,030 | 20:4 | 19:0 | |
| Phosphpatidylinositol | 885.55 | 14.03 | 54,637,801 | 18:0 | 20:4 |
| 861.55 | 14.20 | 19,990,435 | 18:2 | 18:0 | |
| 863.55 | 16.52 | 3,662,266 | 18:1 | 18:0 | |
| 887.57 | 14.37 | 20,786,368 | 18:0 | 20:3 | |
| Phosphatidylglycerol | 747.52 | 14.53 | 998,032,532 | 16:0 | 18:1 |
| 749.54 | 15.35 | 428,464,867 | 16:0 | 18:0 | |
| 775.55 | 15.69 | 287,891,760 | 18:0 | 18:1 | |
| 745.51 | 14.03 | 240,267,363 | 18:2 | 16:0 | |
| 773.54 | 14.70 | 4,843,126 | 18:2 | 18:0 | |
| Phosphatidylglycerolphosphate | 829.52 | 14.53 | 114,242,798 | 16:0 | 18:1 |
| 831.54 | 15.35 | 30,210,395 | 16:0 | 18:0 | |
| 827.51 | 14.03 | 21,788,964 | 18:2 | 16:0 | |
The lipid class, the identified m/z and retention time (RT), the area obtained from extracted ion chromatograms (XIC), and the identified fatty acid side chains are given.
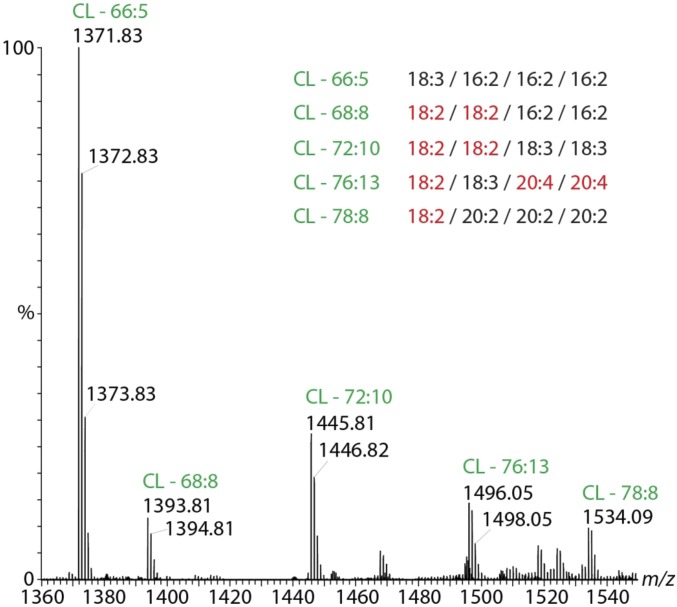
The most abundant lipid precursors are indicated in a combined mass spectrum (Fig. S5A), which also reveals singly charged species above m/z 1,300, most likely corresponding to CL isomers (Fig. S6). A precursor search revealed the numbers of carbon atoms and double bonds in the fatty acids, allowing us to conclude possible combinations from the favored side chains of other lipids (Fig. S5A). The most abundant CL isomer is assigned to 66:5 and is of significantly shorter chain length than the average mitochondrial CL (28). In total, we identified 35 lipid isomers from six lipid classes. Of these, PE is the most abundant species with 14 isomers and the highest precursor intensities (Fig. S5A and Table S5). Compared with previous studies we also identified lipid species that were not anticipated (e.g., PI and PGP) (Fig. S5B). PI and PS together represent minor constituents of the mitochondrial membrane (27). Interestingly, PGP has not been identified in the inner mitochondrial membrane previously.
Fig. S5.
Lipid identification. (A) The mass spectrum was obtained by combining all MS scans over the full LC elution time. Abundant lipid precursors are labeled with the lipid class and the number of C atoms and double bonds in the fatty acid side chains. PE is highly abundant in the CcO complex. The favored fatty acid side chains as identified here are shown (Inset). For each side chain the number of times it was observed is given. Fatty acids that were identified more than five times are highlighted (red). Those observed fewer than five times are depicted in gray. (B) Fragment spectrum of PI (precursor m/z 885.55). Specific fragments are labeled. HCD further confirmed the presence of PI by additional low mass fragments (Inset).
Fig. S6.
Cardiolipin identified by LC-MS. The combined mass spectrum over the elution time reveals several lipids at m/z > 1,300 assigned to cardiolipins. The numbers of carbon atoms and double bonds in the fatty acid side chains were identified by database searching. The fatty acids were assigned based on the most prevalent fatty acids identified for the other lipid species (Fig. S5A).
CcO Dimers Are Formed with Multiple Interfacial Lipids.
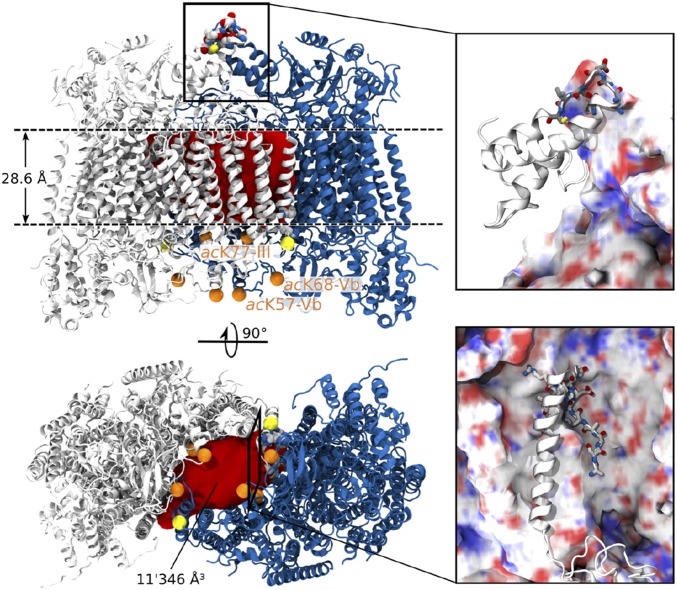
The high level of lipids associated with this complex implies not only that they are sufficient to form a protective shield but also that they may also reside within subunit interfaces. Indeed, the difference between the anticipated mass of the dimer and the measured mass equates to 25 kDa, consistent with over 30 phospholipids. The crystal structure of CcO (PDB ID code 2OCC) reveals a large cavity between the monomers with few protein–protein contacts. We calculated the volume of this cavity to define the number of lipids that could be accommodated (SI Methods) by aligning the crystal structure with the membrane and determining the hydrophobic region of the membrane-spanning subunits as 28.6 Å. The volume was then calculated by inserting the crystal structure into a mesh grid then removing all of the grid points that clash with protein atoms in the crystal structure. The volume of the cavity was thus defined by the volume of the remaining grid points within the hydrophobic region (Fig. 5). The volume for fatty acid side chains identified in our lipid analysis that could be accommodated in the central cavity equates to seven or eight lipids.
Fig. 5.
The lipid plug of CcO. Side view (Upper) and bottom view (Lower) of the CcO dimer (PDB ID code 2OCC). The membrane spanning hydrophobic region (28.6 Å) is indicated (Upper). The volume of the inner cavity (11′346 Å3, red) allows incorporation of seven or eight phospholipids or three or four CL, respectively. PTMs (phosphorylation, yellow and acetylation, orange) in the dimeric interface of CcO were located in the crystal structure. The bottom view of the complex reveals at least six acetylated lysine residues at the dimer interface, suggesting interactions with lipids. Protein–protein interactions in the dimer interface are shown as backbone and cartoon representation for the two monomers (magnifications). Amino acids in direct contact with other amino acids are shown as licorice, revealing a weak dimer interface stabilized by phospholipids.
We therefore propose that the “lipid shield” of monomeric and dimeric CcO accounts for 10 and 20 kDa, respectively, whereas the “lipid plug” within the dimeric interface corresponds to 5 kDa, consistent with the additional 25 kDa (Fig. 4). Highlighting the lipid plug in the crystal structure of CcO and locating the PTMs identified here reveals that these acetylated residues line the inner cavity and point toward several lipid molecules. (Fig. 5, Lower). Acetylation will change the charge properties of the amine groups and modulate electrostatic interactions with phosphoplipids, providing a plausible fine-tuning mechanism to modulate protein–lipid interactions.
SI Methods
Protocol for Purification of Bovine Heart CcO.
Minced bovine heart muscle (1.1 kg) obtained from one beef heart was homogenized in 20 mM sodium phosphate buffer, pH 7.4, followed by centrifugation for removal of the cell debris. The mitochondrial inner membrane fraction in the supernatant was collected by centrifugation after acidic pH treatment of the supernatant and suspended in 100 mM sodium phosphate buffer, pH 7.4. The suspension was treated with 3% (wt/vol) sodium cholate and ammonim sulfate at 33% saturation for solubilization of CcO. The solubilized fraction containing CcO was collected by centrifugation as the supernatant, followed by ammonium sulfate precipitation. The precipitant collected by centrifugation was dissolved in 100 mM sodium phosphate buffer, pH 7.4, containing 0.5% (wt/vol) sodium cholate and dialyzed against 40 mM sodium phosphate buffer, pH 7.4, for removal of sodium cholate to precipitate the CcO fraction. The precipitate containing CcO was dissolved in 100 mM sodium phosphate buffer, pH 7.4, containing 2–0.5% (wt/vol) sodium cholate and fractionated with ammonium sulfate three times. The resulting CcO-containing fraction was purified further by another ammonium sulfate fractionation in the presence of 0.2% decylmaltoside three times. The final preparation dissolved in 40 mM sodium phosphate buffer, pH 6.8, containing 0.2% (wt/vol) decylmaltoside was concentrated with an Amicon diaflow apparatus for microcrystallization. The microcrystals were dissolved in 60 mM sodium phosphate buffer, pH 6.8, containing 0.2% (wt/vol) decylmaltoside.
More detailed descriptions for the above purification procedure have been reported previously (51).The effect of detergent structure on the purity of the final preparation was reported previously (52).
Protein and PTM identification.
Proteins were separated by SDS/PAGE and digested as described (56). Peptides were separated by nano-LC using the EASY nLC 1000 system from Thermo Scientific [mobile phase A, 0.1% (vol/vol) formic acid (FA)/5% (vol/vol) DMSO; mobile phase B, 100% (vol/vol) acetonitrile (ACN)/0.1% (vol/vol) FA/5% (vol/vol) DMSO]. The peptides were loaded onto a trap column (5 mm, PepMap RSLC, C18, 300-μm i.d., particle size 3 μm; Thermo Scientific), separated with a flow rate of 200 nL/min and a gradient of 7–30% (vol/vol) mobile phase B over 30 min on an analytical C18 column (50 cm, PepMap RSLC, EASY-spray column, C18, 75-μm i.d., particle size 3 μm; Thermo Scientific) and directly eluted into a Q Exactive Orbitrap mass spectrometer (Thermo Scientific). MS conditions were as follows: spray voltage of 2.1 kV, capillary temperature of 320 °C, and operation in data-dependent mode. MS spectra were acquired in the orbitrap (m/z 350−1,500) with a resolution of 70,000 and an automatic gain control (AGC) target at 3 × 10e6. The 10 most intense ions were selected for HCD at an AGC target of 50,000.
Raw data were searched against UniProt database (541,762 sequences) using the Mascot v2.4.1 search engine (Matrix Science) with the following parameters: Mass accuracy filter: 20 ppm for precursor ions; 1.0 Da for MS/MS fragment ions; Enzyme: Trypsin or Chymotrypsin with maximal two missed cleavage sites; Carbamidomethylation of cysteine and oxidation of methionine as well as phosphorylation of serine, threonine, and tyrosine and acetylation of lysine as variable modifications.
Denaturing LC-MS.
The proteins were separated by nano-LC using a DionexUltiMate 3000 RSLC nano System from Thermo Scientific [mobile phase A: 0.05% (vol/vol) TFA; mobile phase B: 50% (vol/vol) acetonitrile/50% (vol/vol) isopropanol/0.04% (vol/vol) TFA]. The proteins were dissolved in 0.05% (vol/vol) TFA and loaded onto a monolithic PS-DVB column (100 μm × 25 cm; Thermo Scientific). Protein subunits were separated with a flow rate of 600 nL/min and a gradient from 8 to 98% solvent B over 30 min. The proteins were directly eluted into a QSTAR XL mass spectrometer (ABSciex). MS conditions were ion spray voltage 2,980 V, declustering potential 75 V, and collision energy 17 V.
MS of intact CcO.
Aliquots of CcO in DM-containing buffer were exchanged against 200 mM ammonium acetate using Micro Bio-spin 6 columns (Bio Rad). Spectra were acquired on a Q-ToF II or Synapt1 mass spectrometer (Waters) modified for high masses (48) using gold-coated glass capillaries prepared in-house (49). Optimized instrument parameters were as follows: capillary voltage 1.7 kV, cone voltage 190 V, extractor 5 V, source backing pressure 7–10 mbar, and a collision cell pressure of 10 psi. Collision cell energy was 100–200 V. Spectra were processed and assigned using MassLynx software.
Detergent exchange.
The detergent was exchanged as described previously (50). Briefly, DM detergent was exchanged against C8E4, LDAO, TX-100, β-OG, or FosCholine-16 in 130 mM NaCl, 50 mM Tris, pH 7.4, by size-exclusion chromatography using a Superdex 200 column (5 × 150 mm; GE Healthcare) with a flow rate of 0.2 mL/min at 4 °C. Before MS analysis the buffer was exchanged against 200 mM ammonium acetate, pH 7.4, supplemented with the respective detergents using the same chromatographic conditions.
Lipid identification.
CcO proteins were digested with Trypsin at 37 °C. The peptide/lipid mixture was lyophilized and redissolved in 90% (vol/vol) methanol/0.05% (vol/vol) ammonia solution. For LC-MS/MS analysis, the peptide/lipid mixture was separated by nano-LC (DionexUltiMate 3000 RSLCnano System; Thermo Scientific) [mobile phase A: 70% (vol/vol) methanol/25 mM ammonium acetate; mobile phase B:100% (vol/vol) methanol/25 mM ammonium acetate]. Peptides and lipids were loaded onto an analytical column (HPLC column Acclaim PepMap 100, C8, 100-μm i.d., particle size 5 μm; Thermo scientific) and separated with a flow rate of 300 nL/min and a gradient of 35–100% (vol/vol) mobile phase B over 21 min. Peptides and lipids directly eluted into an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Scientific).
MS conditions were spray voltage of 1.5 kV and capillary temperature of 160 °C. The LTQ-Orbitrap XL was operated in negative ion mode and in data-dependent mode. Survey full-scan MS spectra were acquired in the orbitrap (m/z 350−2,000) with a resolution of 60,000 at m/z 400 and an AGC target of 5·10e5. The three most intense ions were selected for collision-induced dissociation (CID) and high-energy collisional dissociation (HCD) MS/MS fragmentation in the linear ion trap or the orbitrap, respectively. Detection of previously selected precursors was excluded for 120 s. Ions with unrecognized charge state were excluded. CID fragmentation in the linear ion trap was performed at an AGC target of 3·10e4 and a normalized collision energy of 38% at an activation of q = 0.25 and an activation time of 30 ms. HCD fragmentation was performed in the orbitrap with a resolution of 7,500 at m/z 400 and a normalized collision energy of 50.0% at an activation time of 100 ms.
Computational Quantification of Lipids in the Lipid Plug.
The alignment of CcO crystal structure (PDB ID code 2OCC) against a lipid bilayer was determined using OPM server (53). As such, the protein was aligned along the z axis, the origin being the center of the predicted 28.6-Å-thick hydrophobic region. To calculate the volume of the central cavity in the z = [−14.3 Å,14.3 Å] region, we immersed it into a 1-Å-dense mesh grid. Every grid point at less than 3.5 Å from any protein atom was considered as clashing, and removed. The remaining grid point cloud featured few outliers, disconnected from the central cavity region. These were identified and removed using a DBSCAN algorithm using a 1-Å distance cutoff (55). The cavity volume was finally calculated by summing the volume occupied by every remaining grid point, and represented using the VMD volmap tool (56).
CH2, CH3, and HC=CH carbons forming an alkyl chain have a volume of 27.7, 53.3, and 44.4 Å3, respectively (54). We computed the volume occupied by the experimentally observed alkyl chains compositions 16:0, 18:0, 18:1, 18:2, and 20:4. We generated 100,000 random collections of chains, each trying to match the calculated cavity volume. We selected the 2,231 unique alkyl chains collections having a total volume within 5% of the target cavity volume. Supposing only lipids with two alkyl chains are present in the cavity, we selected only the 164 collections featuring an even number of chains. From these, mean and SD in number of represented lipids were calculated.
Discussion
We have characterized CcO purified from bovine heart by determining accurate subunit masses and identifying PTMs, lipids, and a protein associated with the intact complex. Transferring intact CcO into the gas phase of a mass spectrometer from C8E4 allowed us to release intact monomeric and dimeric CcO containing 13 and 26 subunits, respectively. The masses observed for these complexes revealed a large lipid entity associated with both oligomers, the CcO monomer binding an additional 10 kDa corresponding to ∼13 phospholipids when assuming an average mass of 775 Da (Table S5). The lipids associated with dimeric CcO constitute 25 kDa. Assuming that both individual monomers are stabilized by lipids, about six phospholipids likely stabilize the dimeric interface; specifically, CL is thought to stabilize the dimeric CcO interface (11, 14, 29, 30). Recent molecular dynamics simulations identified seven binding sites for CL in CcO (29), most of which are contradictory to sites identified from crystal structures (11, 15).
We identified five lipid classes, with several isomers, and a number of CL. Considering the number of isomers identified (Table S5) and the peak intensities recorded (Fig. S5A), PE is the most prevalent lipid. Given that PE and PC are most abundant in bovine heart tissue (27), their prevalence in the natural membrane suggests that they are often in contact with complexes and may constitute the annular lipids of the CcO complex. PC was not identified in this study but was identified previously (11). Our lipid analysis also revealed three lipid classes previously unidentified with bovine CcO: PI, PS, and PGP. Interestingly, these lipids are only minor constituents of the mitochondrial inner membrane (27) and are therefore likely sequestered from exchange with bulk lipids in the membrane (PE and PC) by virtue of their specific binding in the dimer interface. A similar phenomenon has been reported previously for the membrane-spanning ring in V- and F-type ATPases. In both cases the lipid plug is represented by low-abundant lipids in their respective membranes, sequestered from exchange with bulk lipids that make up the surrounding lipid environment (26, 31). We therefore propose that PI, PS, and PGP bind specifically, presumably to the inner cavity of the CcO dimer interface, to impart stability to dimeric CcO by binding in the inner cavity. We also observed CL, a lipid previously suggested to be in close contact to the proteins that form inner cavities (11, 13, 14). In high-resolution structures reported previously CL is located at the contact sites between the two monomeric complexes that make relatively few protein interactions (6) (Fig. 5). The chain length of the abundant CL isomer identified here (16:2) agrees well with the dimensions of the hydrophobic membrane-spanning domain, calculated for the lipid plug of CcO.
Molecular dynamics simulations of monomeric CcO revealed two high-affinity and five low-affinity CL binding sites only on the surface of the monomer, implying that binding of CL in the dimeric interface only occurs during dimer formation (29). Interestingly, the two high-affinity CL binding sites were located in close proximity to the proton channels and were associated with the regulation of the enzyme (32, 33), whereas the low-affinity binding sites were linked to stabilization of subunits VIa and VIb, important for dimerization of CcO (30, 34). A loss of CcO function in the absence of CL was shown recently through destabilization of quaternary structure following CL removal (13, 14). Specific association of CL, and location in the dimeric interface and/or at the regulatory sites of CcO, therefore represents an important issue to resolve to understand CcO function fully. We propose based on these results that CL likely mediates the equilibrium between monomeric and dimeric forms of CcO.
Comparing the previous X-ray structure of bovine heart CcO, wherein only the symmetric 72:8 CL was identified (11), with the lipids found here, which include 66:5, 68:8, 72:10, 76:13, and 78:8, a greater dispersity of chain length and a higher degree of unsaturation is revealed. This observation could be consistent with diverse locations for CL in different subunit interfaces requiring different chain lengths. We propose that with their two phosphatidic acid moieties CLs form a bridge between subunits, their tailored side chains acting as a “hydrophobic glue” to stabilize different subunit interactions as well as the CcO dimer interface.
To gain more insights into the lipid-mediated stabilization of subunit interfaces as well as possible regulatory mechanisms in CcO we also identified acetylation and phosphorylation sites. Interestingly, subunits IV, Va, and Vb were modified to the greatest extent. Locating these PTMs in the crystal structure of CcO, and taking into account the flexibility of their locations on unstructured loops, reveals that the phosphorylation sites are present predominantly in soluble domains, accessible to the matrix side of the inner membrane, accessible for kinases and phosphatases. Strikingly, phosphorylation was detected primarily on serine, and to a lower extent on threonine residues. In agreement with other studies (35) this indicates that these sites have a regulatory role rather than a stabilizing one, which is often ascribed to phosphorylation on tyrosine residues (35).
Comparing our PTMs with those reported in databases (www.uniprot.org) reveals that only five of our modified sites coincide with those identified previously (Table S3). This relatively low overlap might have several origins. Critically, phosphorylation and acetylation are highly dynamic and reversible, and as such they are often lost during purification or extraction either by dephosphorylation or deacetylation due to the presence of the respective enzymes (36). In addition, PTMs are often of low abundance in natural sources (<10%) due to their functional roles in signaling cascades (37). Low levels of PTMs have also been shown to regulate subunit interfaces (31, 38), with subtle differences in phosphorylation status having a major impact on complex stability (31). Even though developments of higher resolution and sensitivity have increased the numbers of PTMs identified by MS (39), often only subsets are captured and therefore do not necessarily correspond in different studies. Further modified sites have been proposed for CcO (4), in addition to the sites described here, implying that the number of PTMs in this complex is likely to increase with future MS developments and greater understanding of purification/extraction procedures. A comparison over different mammalian species presented here revealed that the acetylation sites identified, and those deposited in databases, are highly conserved over bovine, human, mouse, and rat in CcO subunits IV, Va, and Vb. Whereas only five PTMs were consistently identified in bovine CcO, nine acetylation sites and three phosphorylation sites are conserved in subunits IV, Va, and Vb over these four species. Aligning their amino acid sequences shows a high sequence homology (>82%), suggesting that the PTMs identified here, or previously, will likely be present in other mammalian species.
Although phosphorylation has been characterized extensively over the last decade, acetylation was initially only considered to play a role in histone modification. However, recent large-scale proteomic studies have revealed that acetylation represents a major PTM in eukaryotes (40) and prokaryotes (41). Interestingly, 63% of mitochondrial proteins contain acetylation sites, most of which have been assigned an inhibitory role (42). Interestingly, the acetylation sites identified here, and those reported previously, cluster with phosphorylation sites in close proximity to the dimeric interface (Fig. 5, Lower).
Given that there are very few protein–protein interactions in the CcO dimer interface, representing a rather weak interface, it is likely that lipids could play a role in stabilization (Fig. 5). Assuming that the low-abundance lipids PS and PI are sequestered in the inner cavity of CcO, lysine residues in this region might undergo electrostatic interactions with the headgroups of the lipids. Acetylation of these amino groups would thus modulate the stability of dimeric CcO, in line with previous reports of lysine acetylation changing lipid binding properties (43). PTM clusters at protein interfaces and in unstructured regions have been described previously; moreover, their importance for stabilizing and controlling protein interactions has been proposed (44). Interactions between PTMs of proteins and associated lipids is, however, a relatively new concept that is sure to have major implication for the organization of membrane–protein complexes.
In summary this protein–lipid assembly is the largest integral membrane complex analyzed intact by electrospray MS to date, to our knowledge. Although previous studies have demonstrated the survival of rotary ATPases (26, 31), these complexes have large soluble head regions facilitating their survival outside of the membrane. Moreover, early MS studies delivered only approximate subunit masses and lipid composition due to the low resolution available at that time (45). The fact that we could preserve the intact CcO dimer with an assembled lipid plug and transfer this from solution to gas phase, with both monomeric and dimeric species present, provides the first compelling evidence, to our knowledge, for the existence of this lipid-mediated dimer in solution. The functional role of this dimer is not yet clear, however. Similarly, other complexes of the electron transport chain, such as the bc1 complex, have been found to be dimeric (46). Although CcO and bc1 are functionally independent, an interplay between the monomeric complexes is often discussed (for example, see ref. 47). The role of lipids and PTMs in mediating interactions between CcO and bc1 is often debated but not yet defined. By distinguishing lipids that bind within an annular belt from those in the central lipid plug, and by locating PTMs within the structure, we propose that the interplay of the monomeric complexes is fine-tuned by multiple acetylation and tailored by selective lipid binding.
Methods
Further details of all methods are given in SI Methods.
CcO Purification.
CcO was purified as crystalline preparation in DM from bovine heart as described in SI Methods. Each sample was prepared from one heart.
Protein and PTM Identification.
Proteins were separated by SDS/PAGE and digested with trypsin. Peptides were separated by nano-LC (EASY nLC 1000 system; Thermo Scientific) and directly eluted into a mass spectrometer (Q Exactive Orbitrap; Thermo Scientific). Proteins and PTMs were identified by database searching.
Denaturing LC-MS.
Proteins were separated by nano-LC (DionexUltiMate 3000 RSLC; Thermo Scientific) and directly eluted into a QSTAR XL mass spectrometer (ABSciex).
MS of Intact CcO.
Mass spectra were acquired in ammonium acetate on a Q-ToF II or Synapt1 mass spectrometer (Waters) modified for high masses (48) using gold-coated glass capillaries prepared in-house (49).
Detergent Exchange.
The detergent was exchanged as described previously (50).
Lipid Identification.
CcO proteins were digested with trypsin and the peptide/lipid mixture was separated by nano-LC (DionexUltiMate 3000 RSLC; Thermo Scientific). Peptides and lipids directly eluted into an LTQ-Orbitrap XL mass spectrometer (Thermo Scientific).
Acknowledgments
We thank Emily Barlow for preliminary MS experiments and Kyoko Shinzawa-Ito for purification of bovine CcO. This work was funded by European Research Council Grant IMPRESS 268851 (to C.V.R.); Medical Research Council Grant 98101 (to C.V.R.); Wellcome Trust Grants WT008150 and WT099141 and an Investigator Award (to C.V.R.); and Swiss National Science Foundation Grant P2ELP3_155339 (to M.T.D.). S.Y. is funded by Grants-in-Aid for the Global Center of Excellence Program and for Scientific Research 2247012 and 26291033, by Core Research for Evolutionary Science and Technology, and by Targeted Proteins Research Program, each provided by the Japanese Ministry of Education, Culture, Sports, Science and Technology. C.V.R. is a Royal Society Professor. S.Y. is a Senior Visiting Scientist in the RIKEN Harima Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1600354113/-/DCSupplemental.
References
- 1.Babcock GT, Wikström M. Oxygen activation and the conservation of energy in cell respiration. Nature. 1992;356(6367):301–309. doi: 10.1038/356301a0. [DOI] [PubMed] [Google Scholar]
- 2.Kadenbach B, et al. The complexity of respiratory complexes. Trends Biochem Sci. 1983;8(11):398–400. [Google Scholar]
- 3.Kadenbach B, Hüttemann M. The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion. 2015;24:64–76. doi: 10.1016/j.mito.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Tsukihara T, et al. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272(5265):1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 5.Yoshikawa S, Muramoto K, Shinzawa-Itoh K. Proton-pumping mechanism of cytochrome C oxidase. Annu Rev Biophys. 2011;40:205–223. doi: 10.1146/annurev-biophys-042910-155341. [DOI] [PubMed] [Google Scholar]
- 6.Yoshikawa S, Muramoto K, Shinzawa-Itoh K, Mochizuki M. Structural studies on bovine heart cytochrome c oxidase. Biochim Biophys Acta. 2012;1817(4):579–589. doi: 10.1016/j.bbabio.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 7.Tsukihara T, et al. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A. Science. 1995;269(5227):1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 8.Robinson NC. Specificity and binding affinity of phospholipids to the high-affinity cardiolipin sites of beef heart cytochrome c oxidase. Biochemistry. 1982;21(1):184–188. doi: 10.1021/bi00530a031. [DOI] [PubMed] [Google Scholar]
- 9.Harrenga A, Michel H. The cytochrome c oxidase from Paracoccus denitrificans does not change the metal center ligation upon reduction. J Biol Chem. 1999;274(47):33296–33299. doi: 10.1074/jbc.274.47.33296. [DOI] [PubMed] [Google Scholar]
- 10.Svensson-Ek M, et al. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321(2):329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 11.Shinzawa-Itoh K, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26(6):1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshikawa S, Shimada A. Reaction mechanism of cytochrome c oxidase. Chem Rev. 2015;115(4):1936–1989. doi: 10.1021/cr500266a. [DOI] [PubMed] [Google Scholar]
- 13.Musatov A, Robinson NC. Bound cardiolipin is essential for cytochrome c oxidase proton translocation. Biochimie. 2014;105:159–164. doi: 10.1016/j.biochi.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sedlák E, Robinson NC. Destabilization of the quaternary structure of bovine heart cytochrome c oxidase upon removal of tightly bound cardiolipin. Biochemistry. 2015;54(36):5569–5577. doi: 10.1021/acs.biochem.5b00540. [DOI] [PubMed] [Google Scholar]
- 15.Tsukihara T, et al. The low-spin heme of cytochrome c oxidase as the driving element of the proton-pumping process. Proc Natl Acad Sci USA. 2003;100(26):15304–15309. doi: 10.1073/pnas.2635097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Helling S, et al. Multiple phosphorylations of cytochrome c oxidase and their functions. Proteomics. 2012;12(7):950–959. doi: 10.1002/pmic.201100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saraste M, Penttilä T, Wikström M. Quaternary structure of bovine cytochrome oxidase. Eur J Biochem. 1981;115(2):261–268. doi: 10.1111/j.1432-1033.1981.tb05232.x. [DOI] [PubMed] [Google Scholar]
- 18.Thompson DA, Gregory L, Ferguson-Miller S. Cytochrome c oxidase depleted of subunit III: Proton-pumping, respiratory control, and pH dependence of the midpoint potential of cytochrome a. J Inorg Biochem. 1985;23(3-4):357–364. doi: 10.1016/0162-0134(85)85046-7. [DOI] [PubMed] [Google Scholar]
- 19.Sedlák E, Robinson NC. Sequential dissociation of subunits from bovine heart cytochrome C oxidase by urea. Biochemistry. 2009;48(34):8143–8150. doi: 10.1021/bi900773r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sedlák E, Varhač R, Musatov A, Robinson NC. The kinetic stability of cytochrome C oxidase: Effect of bound phospholipid and dimerization. Biophys J. 2014;107(12):2941–2949. doi: 10.1016/j.bpj.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wittig I, Schägger H. Advantages and limitations of clear-native PAGE. Proteomics. 2005;5(17):4338–4346. doi: 10.1002/pmic.200500081. [DOI] [PubMed] [Google Scholar]
- 22.Roman I, Figys J, Steurs G, Zizi M. In vitro interactions between the two mitochondrial membrane proteins VDAC and cytochrome c oxidase. Biochemistry. 2005;44(39):13192–13201. doi: 10.1021/bi050674s. [DOI] [PubMed] [Google Scholar]
- 23.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321(5886):243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 24.Barrera NP, et al. Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat Methods. 2009;6(8):585–587. doi: 10.1038/nmeth.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reading E, et al. The role of the detergent micelle in preserving the structure of membrane proteins in the gas phase. Angew Chem Int Ed Engl. 2015;54(15):4577–4581. doi: 10.1002/anie.201411622. [DOI] [PubMed] [Google Scholar]
- 26.Zhou M, et al. Mass spectrometry of intact V-type ATPases reveals bound lipids and the effects of nucleotide binding. Science. 2011;334(6054):380–385. doi: 10.1126/science.1210148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daum G. Lipids of mitochondria. Biochim Biophys Acta. 1985;822(1):1–42. doi: 10.1016/0304-4157(85)90002-4. [DOI] [PubMed] [Google Scholar]
- 28.Schlame M, Brody S, Hostetler KY. Mitochondrial cardiolipin in diverse eukaryotes. Comparison of biosynthetic reactions and molecular acyl species. Eur J Biochem. 1993;212(3):727–735. doi: 10.1111/j.1432-1033.1993.tb17711.x. [DOI] [PubMed] [Google Scholar]
- 29.Arnarez C, Marrink SJ, Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci Rep. 2013;3:1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Musatov A, Robinson NC. Cholate-induced dimerization of detergent- or phospholipid-solubilized bovine cytochrome C oxidase. Biochemistry. 2002;41(13):4371–4376. doi: 10.1021/bi016080g. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt C, et al. Comparative cross-linking and mass spectrometry of an intact F-type ATPase suggest a role for phosphorylation. Nat Commun. 2013;4:1985. doi: 10.1038/ncomms2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robinson NC. Functional binding of cardiolipin to cytochrome c oxidase. J Bioenerg Biomembr. 1993;25(2):153–163. doi: 10.1007/BF00762857. [DOI] [PubMed] [Google Scholar]
- 33.Robinson NC, Strey F, Talbert L. Investigation of the essential boundary layer phospholipids of cytochrome c oxidase using Triton X-100 delipidation. Biochemistry. 1980;19(16):3656–3661. doi: 10.1021/bi00557a003. [DOI] [PubMed] [Google Scholar]
- 34.Sedlák E, Robinson NC. Phospholipase A(2) digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry. 1999;38(45):14966–14972. doi: 10.1021/bi9914053. [DOI] [PubMed] [Google Scholar]
- 35.Ly T, et al. A proteomic chronology of gene expression through the cell cycle in human myeloid leukemia cells. eLife. 2014;3:e01630. doi: 10.7554/eLife.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Jensen ON. Modification-specific proteomics: Strategies for characterization of post-translational modifications using enrichment techniques. Proteomics. 2009;9(20):4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baeza J, et al. Stoichiometry of site-specific lysine acetylation in an entire proteome. J Biol Chem. 2014;289(31):21326–21338. doi: 10.1074/jbc.M114.581843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morgner N, et al. Hsp70 forms antiparallel dimers stabilized by post-translational modifications to position clients for transfer to Hsp90. Cell Reports. 2015;11(5):759–769. doi: 10.1016/j.celrep.2015.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen JV, Mann M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol Cell Proteomics. 2013;12(12):3444–3452. doi: 10.1074/mcp.O113.034181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mischerikow N, Heck AJ. Targeted large-scale analysis of protein acetylation. Proteomics. 2011;11(4):571–589. doi: 10.1002/pmic.201000397. [DOI] [PubMed] [Google Scholar]
- 41.Soufi B, Soares NC, Ravikumar V, Macek B. Proteomics reveals evidence of cross-talk between protein modifications in bacteria: Focus on acetylation and phosphorylation. Curr Opin Microbiol. 2012;15(3):357–363. doi: 10.1016/j.mib.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 42.Baeza J, Smallegan MJ, Denu JM. Mechanisms and dynamics of protein acetylation in mitochondria. Trends Biochem Sci. 2016;41(3):231–244. doi: 10.1016/j.tibs.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maltsev AS, Ying J, Bax A. Impact of N-terminal acetylation of α-synuclein on its random coil and lipid binding properties. Biochemistry. 2012;51(25):5004–5013. doi: 10.1021/bi300642h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beilsten-Edmands V, et al. eIF2 interactions with initiator tRNA and eIF2B are regulated by post-translational modifications and conformational dynamics. Cell Discovery. 2015;1:15020. doi: 10.1038/celldisc.2015.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morgner N, Kleinschroth T, Barth HD, Ludwig B, Brutschy B. A novel approach to analyze membrane proteins by laser mass spectrometry: From protein subunits to the integral complex. J Am Soc Mass Spectrom. 2007;18(8):1429–1438. doi: 10.1016/j.jasms.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 46.Castellani M, et al. Direct demonstration of half-of-the-sites reactivity in the dimeric cytochrome bc1 complex: Enzyme with one inactive monomer is fully active but unable to activate the second ubiquinol oxidation site in response to ligand binding at the ubiquinone reduction site. J Biol Chem. 2010;285(1):502–510. doi: 10.1074/jbc.M109.072959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crofts AR, et al. The Q-cycle reviewed: How well does a monomeric mechanism of the bc(1) complex account for the function of a dimeric complex? Biochim Biophys Acta. 2008;1777(7-8):1001–1019. doi: 10.1016/j.bbabio.2008.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sobott F, Hernández H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74(6):1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 49.Hernández H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2(3):715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 50.Laganowsky A, Reading E, Hopper JT, Robinson CV. Mass spectrometry of intact membrane protein complexes. Nat Protoc. 2013;8(4):639–651. doi: 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoshikawa S, Choc MG, O’Toole MC, Caughey WS. An infrared study of CO binding to heart cytochrome c oxidase and hemoglobin A. Implications re O2 reactions. J Biol Chem. 1977;252(15):5498–5508. [PubMed] [Google Scholar]
- 52.Mochizuki M, et al. Quantitative reevaluation of the redox active sites of crystalline bovine heart cytochrome c oxidase. J Biol Chem. 1999;274(47):33403–33411. doi: 10.1074/jbc.274.47.33403. [DOI] [PubMed] [Google Scholar]
- 53.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of proteins in membranes database. Bioinformatics. 2006;22(5):623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 54.Ester M, Kriegel H-P, Sander J, Xu X. Proceedings of the Second International Conference on Knowledge Discovery and Data Mining. AAAI Press, Palo Alto; CA: 1996. A density-based algorithm for discovering clusters in large spatial databases with noise; pp. 226–231. [Google Scholar]
- 55.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph Model. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 56.Kucerka N, Tristram-Nagle S, Nagle JF. Structure of fully hydrated fluid phase lipid bilayers with monounsaturated chains. J Membr Biol. 2005;208(3):193–202. doi: 10.1007/s00232-005-7006-8. [DOI] [PubMed] [Google Scholar]
- 57.Helling S, et al. Phosphorylation and kinetics of mammalian cytochrome c oxidase. Mol Cell Proteomics. 2008;7(9):1714–1724. doi: 10.1074/mcp.M800137-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee I, et al. Isolation of regulatory-competent, phosphorylated cytochrome c oxidase. Methods Enzymol. 2009;457:193–210. doi: 10.1016/S0076-6879(09)05011-3. [DOI] [PubMed] [Google Scholar]
- 59.Huttemann M, et al. Regulation of mitochondrial respiration and apoptosis through cell signaling: Cytochrome c oxidase and cytochrome c in ischemia/reperfusion injury and inflammation. Biochim Biophys Acta. 2011;1817(4):598–609. doi: 10.1016/j.bbabio.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]