Abstract
Immune dysfunction is commonly associated with several neurological and mental disorders. Although the mechanisms by which peripheral immunity may influence neuronal function are largely unknown, recent findings implicate meningeal immunity influencing behavior, such as spatial learning and memory1. Here we show that meningeal immunity is also critical for social behavior; mice deficient in adaptive immunity exhibit social deficits and hyper-connectivity of fronto-cortical brain regions. Associations between rodent transcriptomes from brain and cellular transcriptomes in response to T cell–derived cytokines suggest a strong interaction between social behavior and interferon-gamma (IFN-γ) driven responses. Concordantly, we demonstrate that inhibitory neurons respond to IFN-γ and increase GABAergic currents in projection neurons, suggesting that IFN-γ is a molecular link between meningeal immunity and neural circuits recruited for social behavior. Meta-analysis on the transcriptomes of a range of organisms revealed that rodents, fish, and flies elevate IFN-γ/JAK-STAT–dependent gene signatures in a social context, suggesting that the IFN-γ signaling pathway could mediate a co-evolutionary link between social/aggregation behavior and an efficient anti-pathogen response. This study implicates adaptive immune dysfunction, in particular IFN-γ, in disorders characterized by social dysfunction and suggests a co-evolutionary link between social behavior and an anti-pathogen immune response driven by IFN-γ signaling.
Social behavior is beneficial for many processes critical to the survival of an organism, including foraging, protection, breeding, and for higher order species, mental health2,3. Social dysfunction manifests in several neurological and mental disorders such as autism spectrum disorder (ASD), frontotemporal dementia, and schizophrenia amongst others4. Likewise, imbalance of cytokines, a disparity of T cell subsets, and overall immune dysfunction is often associated with abovementioned disorders5–7. However, the fundamental mechanism(s) by which dysfunctional immunity may interfere with neural circuits and contribute to behavioral deficits remain unclear.
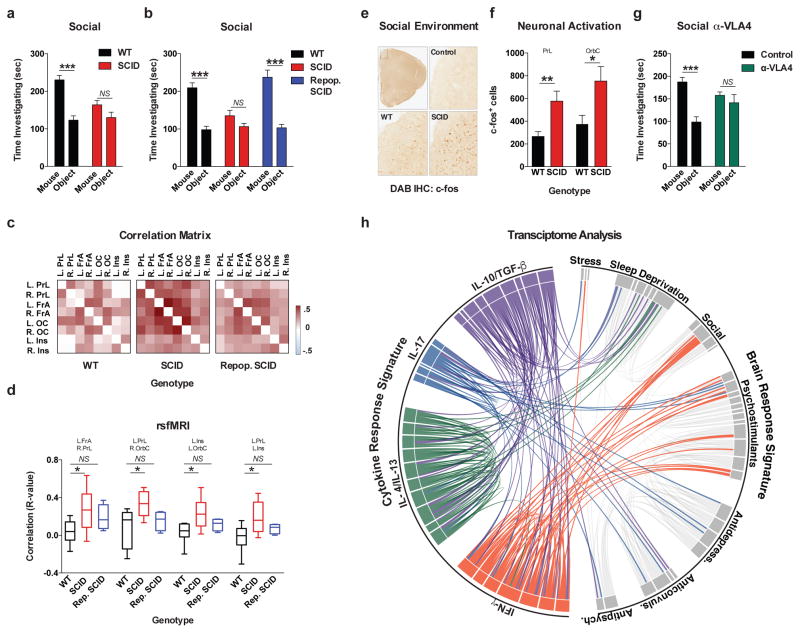
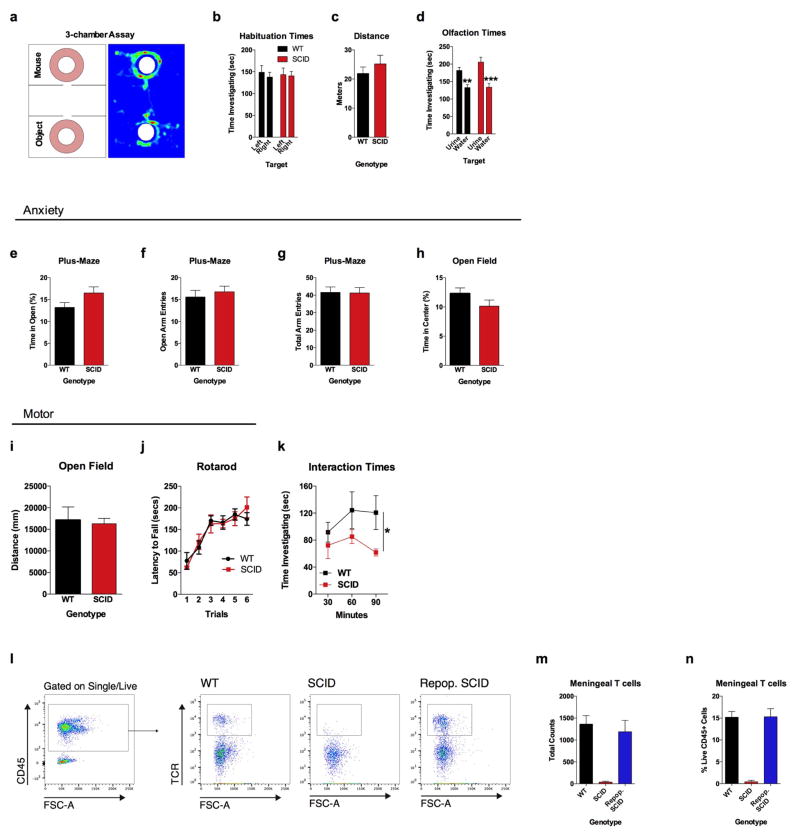
To test if adaptive immunity is necessary for normal social behavior, we tested SCID mice (deficient in adaptive immunity) using the 3-chamber sociability assay8 (Extended Data Fig. 1a). This assay quantifies the preference of a mouse for investigating a novel mouse versus object, and has been used to identify deficits in multiple mouse models of disorders that present with social dysfunction9. Unlike wild-type mice, SCID mice lacked social preference for a mouse over an object (Fig. 1a). Importantly, SCID mice did not show anxiety, motor, or olfactory deficits (Extended Data Fig. 1b–j). We confirmed that SCID mice have social deficits by analyzing social interactions in a home cage (Extended Data Fig. 1k). To test if social deficits were reversible, we repopulated 4-week-old SCID mice with wild-type lymphocytes (Extended Data Fig. 1l–n) and measured social behavior 4 weeks post transfer. SCID mice repopulated with lymphocytes, unlike those injected with the vehicle, showed social preference indistinguishable from wild-type mice (Fig. 1b).
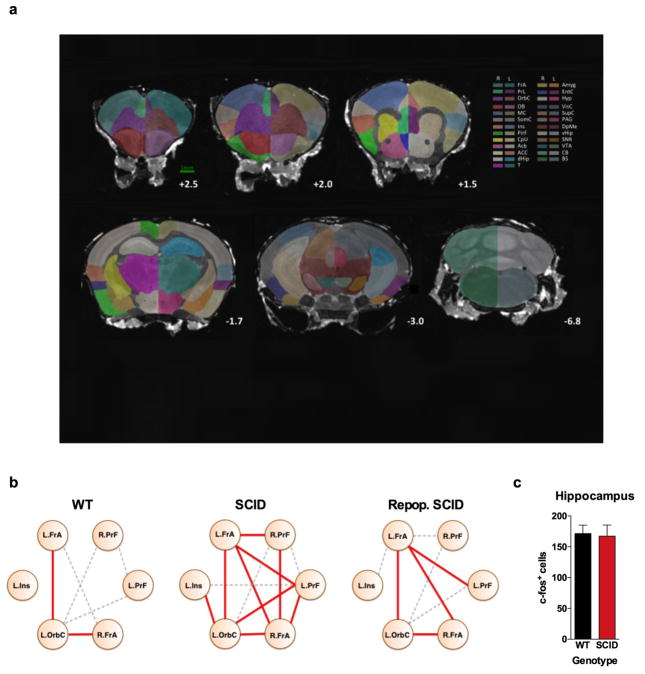
Figure 1. Meningeal T cell compartment is necessary for supporting neuronal connectivity and social behavior.
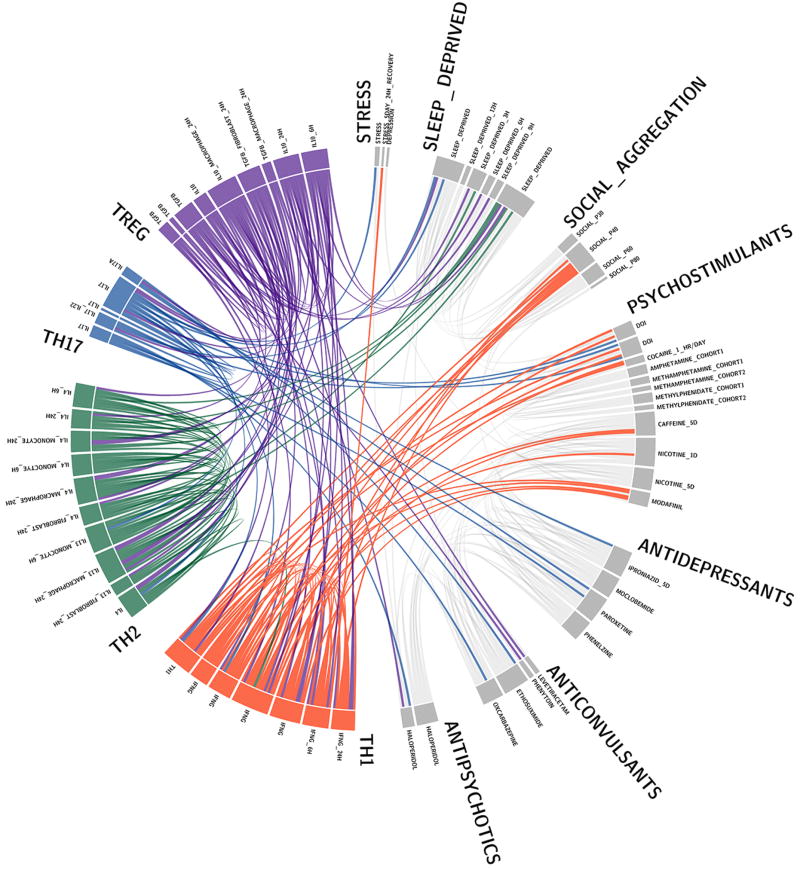
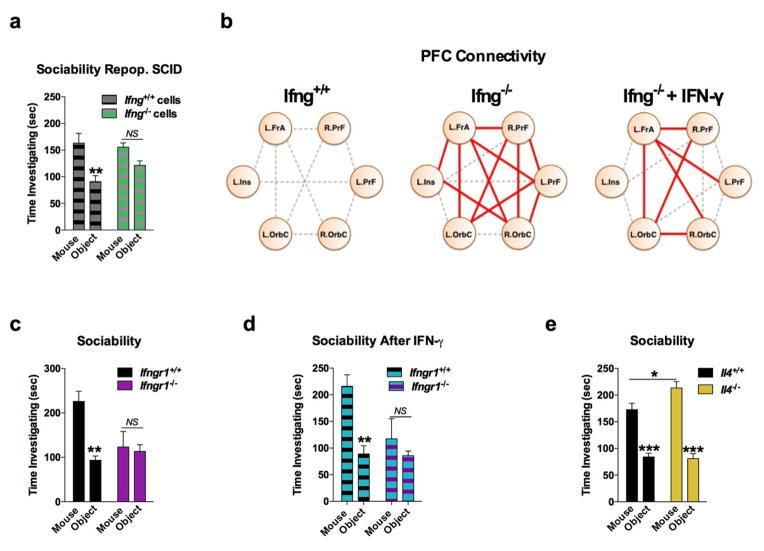
a, Wild-type mice exhibit social preference that is absent in SCID mice (ANOVA for genotype (F(1, 26)) = 6.370, P = 0.0181; n = 14 mice per group; **P < 0.01 Sidak’s post-hoc; pooled 2 independent experiments). b, Repopulating the adaptive immune system in SCID mice restored normal social behavior (n = 17;16;15 mice per group; ANOVA for genotype (F(2, 45)) = 8.282, P = 0.0009 and interaction (F(2, 45)) = 9.146, P = 0.0005; ***P < 0.001; **P < 0.01 Sidak’s post-hoc; pooled 3 independent experiments). c, Correlation matrices from wild-type, SCID, and repopulated (Repop.) SCID mice were generated by rsfMRI. Abbrev.: L=left; R=right; FrA=frontal association area; PrL=prelimbic cortex; Ins=insula; OrbC=orbital cortex. d, Correlation values from rsfMRI. The box and whisker plots extend to the 25th and 75th percentiles with the center-line showing the mean. The whiskers represent the min and max data points. (n = 8;9;4 mice per group; ANOVA < 0.05; *P < 0.05 Sidak’s post-hoc; pooled 2 independent experiments). e, Immunohistochemistry of c-fos in the PFC. f, Elevated c-fos+ cells in the prefrontal and orbital cortices of SCID compared to wild-type mice (n = 9;10 mice per group; **P < 0.01; *P < 0.05 t-test; single experiment). g, Acute partial depletion of meningeal T cells caused social deficits (n = 12;13 mice per group; ANOVA for interaction (F(1, 23)) = 7.900, P < 0.01; ***P < 0.001 Sidak’s post-hoc; pooled 2 independent experiments). h, Circos plot showing the connectivity map derived from the pairwise comparison of transcriptome datasets. IFN-γ signature genes are over-represented in cortex of animals exposed to social aggregation and psychostimulants. The representations of IFN-γ, IL-4/IL-13, IL-17, and IL-10/TGF-β dataset connectivity are shown in orange, green, blue, and purple, respectively. Each line represents a pairwise dataset overlap, which was determined using GSEA analysis and filtered by P < 0.05 and NES > 1.5. See Extended Data Fig. 5 for labels. Data from the 3-chamber test (a, b, g) were analyzed by applying a 2-way ANOVA for social behavior and genotype/treatment, followed by a post-hoc Sidak’s test. Bars represent average mean times investigating ± s.e.m.
Recent clinical findings indicate disturbed circuit homeostasis, resulting in hyper-connectivity, is a feature of children with ASD10. Imaging studies using task-free resting state fMRI (rsfMRI), revealed hyper-connectivity among frontal cortical nodes in ASD patients11. Disturbances in resting state connectivity are also observed in mice with social deficits12. rsfMRI is an unbiased technique used to assess synchrony between brain regions over time by comparing spontaneous fluctuations in blood oxygenation level dependent (BOLD) signals13. To assess the influence of adaptive immunity on functional connectivity, we analyzed resting-state BOLD signals from wild-type and SCID mice (Extended Data Fig. 2a). SCID mice exhibited hyper-connectivity between multiple frontal and insular regions (Fig. 1c, d; Extended Data Fig. 2b; Supplement Table 1) implicated in social behavior and ASD. Notably, repopulating SCID mice with lymphocytes rescued aberrant hyper-connectivity observed in vehicle-treated SCID controls (Fig. 1c, d; Extended Data Fig. 2b). Interestingly, other functionally connected regions, not directly implicated in social function, such as interhemispheric connectivity between motor and somatosensory cortex were not affected by a deficiency in adaptive immunity (Supplement Table 1). Using another approach to analyze neuronal activation in a task-based system, we demonstrated that SCID mice exposed to a social stimulus, exhibited hyper-responsiveness in the prefrontal cortex (PFC; increased number of c-fos+ cells in PFC; Fig. 1e, f) but not the hippocampus (Extended Data Fig. 2c).
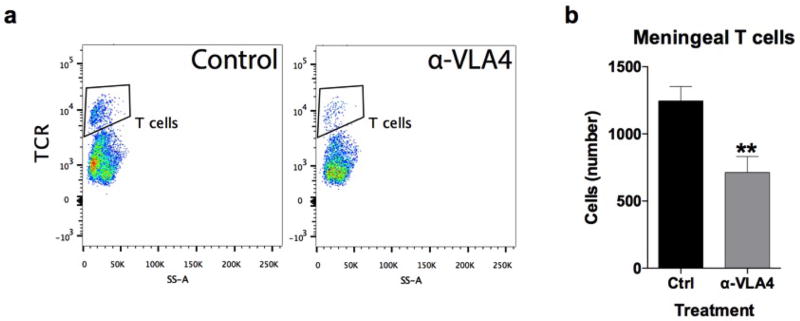
We previously demonstrated T cells influence learning behavior and exert their beneficial effects presumably from the meninges1,14. To address the role of meningeal T cells in social behavior, we decreased the extravasation of T cells into the meninges of wild-type mice using antibodies against VLA415, an integrin expressed on T cells (among other immune cells) required for CNS homing. Partial elimination of T cells from meninges (Extended Data Fig. 3) was sufficient to cause a loss in social preference (Fig. 1g). Despite their proximity to the brain, meningeal T cells do not enter the brain parenchyma, suggesting their effect is mediated by soluble factors. To identify which T cell–mediated pathways are involved in regulating social behavior, we used gene set enrichment analysis (GSEA) to search for T cell–mediated response signatures (IFN-γ, IL-4/IL-13, IL-17, Il-10, TGF-β) in 41 transcriptomes from mouse and rat brain cortices. GSEA assesses if the expression of a previously defined group of related genes is enriched in one biological state. In this case, we used GSEA to identify which cytokine induced response signatures were enriched in the transcriptomes of mice and rats exposed to different stimuli. The stimuli included: social aggregation, sleep deprivation, stress, psychostimulants, antidepressants, anticonvulsants, and antipsychotics. Transcriptomes from cortices of animals exposed to social aggregation and psychostimulants were enriched for IFN-γ regulated genes (Fig. 1h, Extended Data Fig. 4, and Supplementary Tables 2, 3).
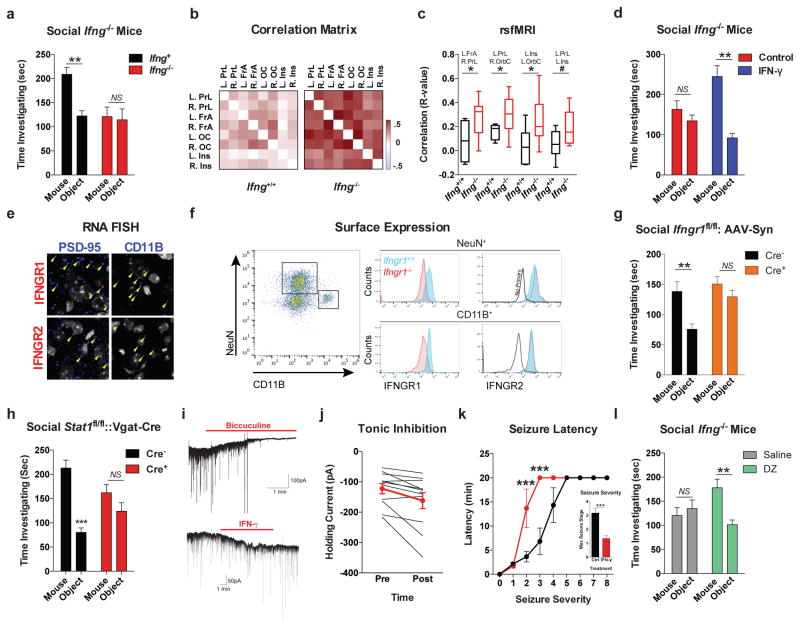
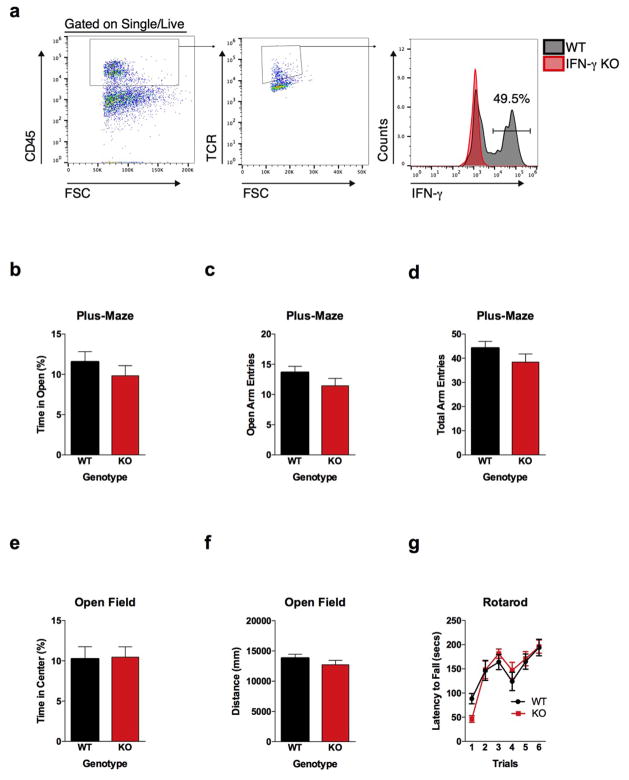
A substantial number of meningeal T cells are capable to express IFN-γ (41.95% ± 6.34 of TCR+ cells; Extended Data Fig. 5a) and recent work has proposed a role for IFN-γ in T cell trafficking into meningeal spaces16. To assess the potential role of IFN-γ in mediating the influence of T cells on social behavior, we first examined the social behavior of IFN-γ deficient mice and determined that they had social deficits (Fig. 2a). Importantly, IFN-γ deficient mice did not show anxiety or motor deficits (Extended Data Fig. 5b–g). Similar to SCID mice, IFN-γ deficient mice also exhibited aberrant hyper-connectivity in fronto-cortical/insular regions (Fig. 2b, c; Supplement Table 1). While repopulating SCID mice with lymphocytes from wild-type mice restored a social preference, repopulating SCID mice with lymphocytes from Ifng−/− mice did not have such an effect (Extended Data Fig. 6a). Remarkably, a single injection of recombinant IFN-γ into the cerebrospinal fluid (CSF) of Ifng−/− mice was sufficient to restore their social preference when tested after 24 hours post injection (Fig. 2d) and reduce overall hyper-connectivity in the PFC (Extended Date Fig. 6b). To further validate a role for IFN-γ signaling in social behavior, we tested mice deficient for the IFN-γ receptor (Ifngr1−/− mice) and found that they had a similar social deficit as Ifng−/− mice (Extended Data Fig. 6c), which, as expected, was not rescued by injecting recombinant IFN-γ into the CSF (Extended Data Fig. 6d). Based on our previous demonstration of a role for IL-4 produced by meningeal T cells in spatial learning behavior1, we assessed whether deficiency in IL-4 would also result in social deficits. IL-4 deficient mice did not demonstrate social deficits; in fact, they spent more time investigating a novel mouse than a novel object as compared to wild-type mice (Extended Data Fig. 6e).
Figure 2. IFN-γ supports proper neural connectivity and social behavior.
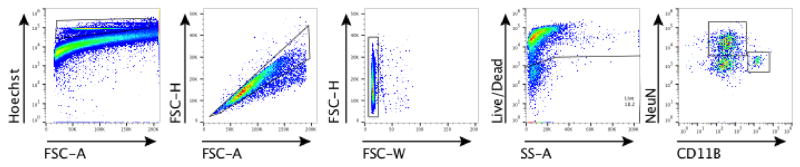
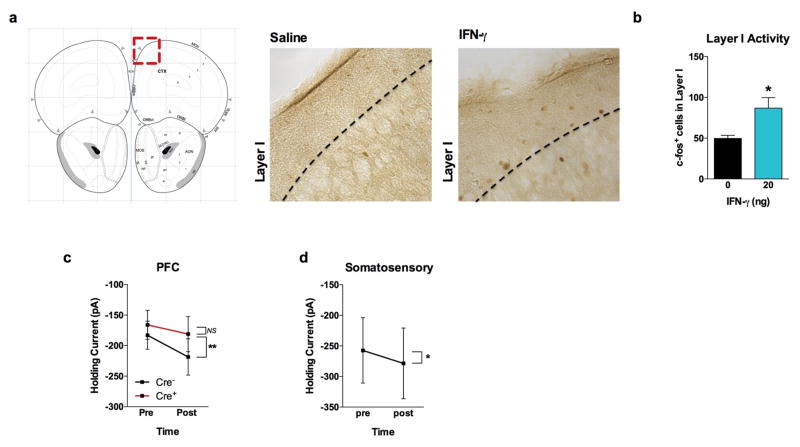
a, Ifng−/− mice exhibit social deficits (n = 16;12 mice per group; ANOVA for genotype (F(1, 52)) = 8.327, P < 0.01; **P < 0.01 Sidak’s post-hoc; pooled 2 independent experiments). b, Correlation matrix from Ifng+/+ and Ifng−/− mice. c, Box and whisker plots of correlation values (n = 8 mice per group; *P < 0.05; # P = 0.06 t-test; repeated 2 times). d, A single CSF injection of IFN-γ (20ng) was sufficient to rescue social deficits in Ifng−/− mice 24 hours post-injection (n = 14;11 mice per group; ANOVA for interaction (F(1, 46)) = 10.22 P < 0.01; ***P < 0.001 Sidak’s post-hoc; pooled 2 independent experiments). e, Expression of IFN-γ receptor subunit mRNA by fluorescent in situ hybridization in slices from mouse PFC. RNA probes and corresponding colors: left:psd95-blue (neurons); right: CD11B-blue (microglia); top: IFNGR1-red; bottom:IFGR2-red. Yellow arrowheads denote co-localization. f, Expression of IFN-γ receptor subunit protein by flow cytometry. Cells were gated on Hoechst+/live/single then neurons and microglia were gated on NeuN and CD11B, respectively. Ifngr1−/− mice and no primary antibody for IFNGR2 were included as negative controls. g, Deleting Ifngr1 from neurons in the PFC caused social deficits. Ifngr1fl/fl mice were injected with AAV-Syn-CRE-GFP into the PFC and tested for social behavior 3 weeks post injection (n = 11;12 mice per group; ANOVA for genotype (F(1, 21)) = 10.62, P < 0.01; *P < 0.05 Sidak’s post-hoc; pooled 3 independent experiments). h, VgatCre::Stat1fl/fl mice exhibit social deficits (n = 10;11 mice per group; ANOVA for interaction (F(1, 19)) = 10.30 < 0.01; ***P < 0.001 Sidak’s post-hoc; pooled 3 independent experiments). i, Layer 2/3 neurons in slices from wild-type mice are held under tonic GABAergic inhibition (top), which is blocked by the GABA-A receptor antagonist bicuculline. IFN-γ increases tonic GABAergic inhibitory current (n = 11 cells from 4 mice; bottom). j, Holding current pre and during IFN-γ (P = 0.01 t-test). k, IFN-γ increased latency to reach each seizure stage (n = 6 mice per group; ANOVA with repeated measures < 0.001; ***P < 0.001 Sidak post-hoc) and (inset) reduced the maximum severity of seizures (***P < 0.001 t-test; repeated 2 times). l, Diazepam rescued social deficits in Ifng−/− mice (n = 12 mice per group; ANOVA for interaction (F(1, 22)) = 9.204 < 0.01; **P < 0.01 Sidak’s post-hoc; repeated 2 times). Data from the 3-chamber test (a, d, g, h, i) were analyzed by applying a 2-way ANOVA for social behavior and genotype/treatment, followed by a Sidak’s post-hoc test. Bars represent average mean times investigating ± s.e.m. All experiments were repeated at least once.
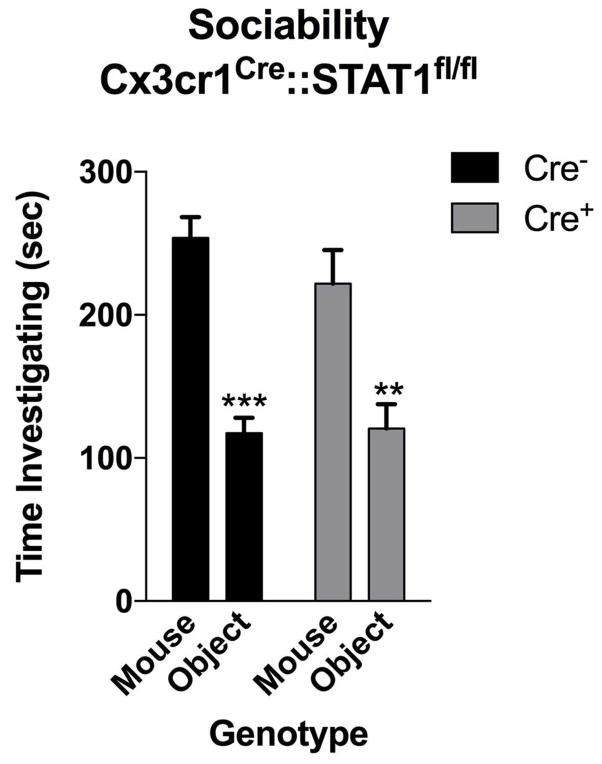
To determine which cell types in the brain respond to IFN-γ, we analyzed mouse PFC for the expression of IFN-γ receptor subunits 1 and 2 and found that both neurons and microglia express mRNA and protein for R1 and R2 subunits of the IFN-γ receptor (Fig. 2e, f; Extended Data Fig. 7). Microglia are CNS resident macrophages and are known to express the IFN-γ receptor17. However, genetically deleting STAT1, the signaling molecule downstream of the IFN-γ receptor, from microglia (and other cells of myeloid origin), did not disturb normal social preference (Extended Data Fig. 8). These results led us to focus on neuronal responses to IFN-γ as they relate to social behavior.
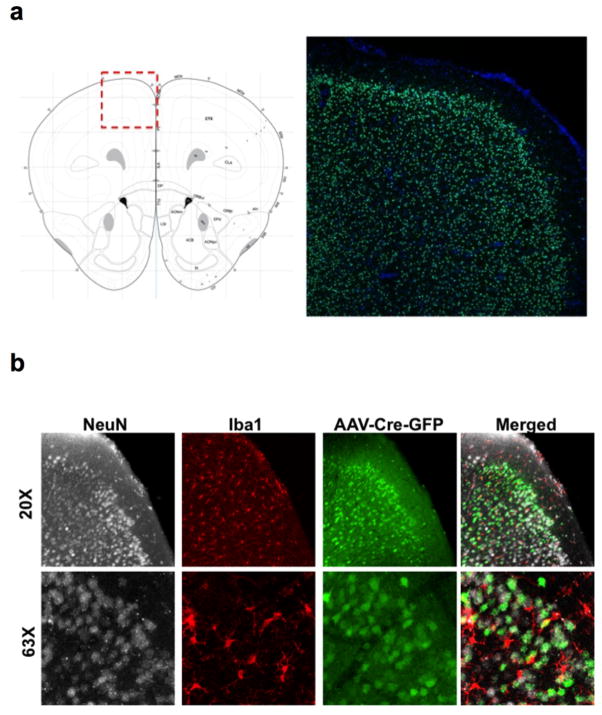
To assess a role for IFN-γ in neuronal signaling, we deleted Ifngr1 in PFC neurons via AAV delivery of Cre recombinase under the Synapsin I promoter (Extended Data Fig. 9). Attenuating IFN-γ signaling in PFC neurons was sufficient to alter mouse behavior in a 3-chamber social task and result in a lack of social preference (Fig. 2g), reinforcing the importance of IFN-γ signaling on neurons for social behavior. Injecting recombinant IFN-γ into the CSF, activated layer I neocortical neurons as assessed by c-fos immunoreactivity (Extended Data Fig. 10a, b). These neurons are almost entirely inhibitory18, suggesting that IFN-γ may drive regional inhibition of circuits by directly activating layer I inhibitory neurons located in close proximity to the brain surface and CSF. To investigate mechanisms downstream of the IFN-γ receptor, we used VgatCre::Stat1fl/fl mice. Deletion of STAT1 from GABAergic inhibitory neurons was sufficient to induce deficits in social behavior (Fig. 2h), suggesting that the IFN-γ may be signaling through inhibitory neurons.
To directly assess if IFN-γ can drive inhibitory tone in the PFC, we measured inhibitory currents in layer 2/3 pyramidal cells from acutely prepared brain slices from wild-type mice. In addition to receiving phasic inhibitory synaptic input, these cells are also held under a tonic GABAergic current that serves to hyperpolarize their resting membrane potential (Fig. 2i). Tonic GABAergic currents are extrasynaptic and can yield long-lasting network inhibition19. We observed that IFN-γ augmented tonic current (Fig. 2i, j, Extended Data Fig. 10c, d), suggestive of elevated levels of ambient GABA during application of IFN-γ. Deleting the IFN-γ receptor from inhibitory neurons (VgatCre::Ifngr1fl/fl mice) prevented IFN-γ from augmenting tonic inhibitory current (Extended Data Fig. 10c). Given that IFN-γ promotes inhibitory tone, we tested if IFN-γ could prevent aberrant neural discharges by injecting IFN-γ into the CSF and then chemically inducing seizures with the GABA type A receptor antagonist, pentylenetetrazole (PTZ). Mice injected with IFN-γ were less susceptible to PTZ induced seizures; IFN-γ delayed seizure onset and lowered seizure severity (Fig. 2k). Further, to test if overexcitation causes social deficits in IFN-γ deficit mice, we treated Ifng−/− mice with diazepam to augment GABAergic transmission20. Diazepam successfully rescued social behavior of Ifng−/− mice, similar to the effect observed with recombinant IFN-γ treatment (Fig. 2l), suggesting that social deficits, due to a deficiency in IFN-γ, may arise from inadequate control of GABAergic inhibition by IFN-γ.
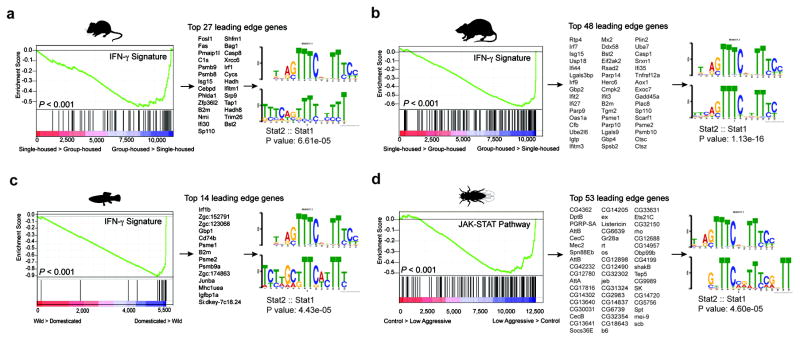
It is intriguing that IFN-γ, predominately thought of as an anti-pathogen cytokine, can play such a profound role in maintaining proper social function. Since social behavior is crucial for the survival of a species and aggregation increases the likeliness of spreading pathogens, we hypothesized that there was co-evolutionary pressure to increase an anti-pathogen response as sociability increased, and that the IFN-γ pathway may have influenced this co-evolution. To test this hypothesis, we analyzed metadata of publically available transcriptomes from multiple organisms including the rat, mouse, zebrafish, and fruit flies. Using GSEA, we determined transcripts from social rodents (acutely group-housed) are enriched for an IFN-γ responsive gene signature (Fig. 3a, b; Supplementary Tables 3–7). Conversely, rodents that experienced social isolation demonstrated a dramatic loss of the IFN-γ responsive gene signature (Supplementary Tables 3–7). Zebrafish and flies showed a similar association between anti-pathogen and social responses (Fig. 3c, d). We observed that immune response programs were highly enriched in the brain transcriptomes of flies selected for low aggressiveness traits (a physiological correlate for socially experienced flies21; Fig. 3d; Supplementary Tables 3–7). We next analyzed the promoters of these highly upregulated social genes and found them to be enriched for STAT1 transcription factor binding motifs (Fig. 3a–d). These data suggest, even in the absence of infection, an IFN-γ gene signature is upregulated in aggregated organisms. This is consistent with an interaction between social behavior and the anti-pathogen response, a dynamic that could be mediated by the IFN-γ pathway. Since low-aggressive flies upregulate genes in the JAK/STAT pathway (canonically downstream of IFN-γ receptors in higher species), yet lack IFN-γ or T cells, it is intriguing to speculate that T cell–derived IFN-γ may have evolved in higher species to more efficiently regulate an anti-pathogen response during increased aggregation of individuals.
Figure 3. Over-representation of IFN-γ transcriptional signature genes in social behavior-associated brain transcriptomes of rat, mouse, zebrafish, and drosophila.
GSEA plots demonstrate the over-representation of IFN-γ transcriptional signatures (derived from Molecular Signature Database C2, GSE33057, or Lopez-Munoz, A. et al30) in brain transcriptomes of (a) mice and (b) rats subjected to social or isolated housing and in brain transcriptomes of (c) domesticated zebrafish compared to a wild zebrafish strain. (d) Over-representation of JAK/STAT pathway transcriptional signature genes (derived from GSE2828) in head transcriptomes of flies selected for low-aggressive behavior (behavioral readout for social behavior in flies (see methods section “Meta-data analysis” for more details)). Genes are ranked into an ordered list according to their differential expression. The middle part of the plot is a bar code demonstrating the distribution of genes in the IFN-γ transcriptional signature gene set against the ranked list of genes. The list on the right shows the top genes in the leading edge subset. Promoter regions of these genes were scanned for transcription factor binding site (TFBS) using MEME suite. MEME output demonstrates significant STAT TFBS enrichment in cis-regulatory regions of leading edge genes up-regulated in a social context.
Our results reveal a novel role for meningeal immunity in regulating neural activity and social behavior through IFN-γ. The role of immune molecules has been previously shown to control brain development and function22–26 and a role for cytokines in influencing behavior has been proposed, primarily in the context of sickness behavior and pain27,28. These signaling paradigms, however, have predominately focused on peripheral nerves and non-neuronal targets. Here we show that CNS neurons directly respond to IFN-γ derived from meningeal T cells to elevate tonic GABAergic inhibition and prevent aberrant hyper-excitability in the PFC. These data suggest that social deficits in numerous neurological and psychiatric disorders may result from impaired circuitry homeostasis derived from dysfunctional immunity. Based on our findings, it is also plausible that subtle homeostatic changes in meningeal immunity may also contribute to modulating neuronal circuits that are responsible for our everyday behaviors and personality. Given this communication between immunity and neuronal circuits29, it is intriguing to hypothesize that these pathways may be vulnerable to manipulation by fast-evolving pathogens. Better comprehension of these pathways could improve our understanding of the etiology of neurodevelopmental and neuropsychiatric disorders associated with maternal infections and/or aberrant inflammation and may result in the development of new therapeutic targets.
Methods
Mice
All mice (C57BL/6) were either bred in-house or purchased from the Jackson Laboratory. For each individual experiment, the control mice were obtained from the same institution as test mice. In the case that mice were purchased, they were maintained for at least one week to habituate prior to manipulation/experimentation. When possible, mice used for experiments were littermates. These include all electrophysiology experiments, all cohorts using Stat1fl/fl mice, experiments analyzing induced seizures, experiments counting c-fos+ neurons, and experiments using Il4−/− mice. Experiments assessing the social behavior of SCID mice using the 3-chamber assay include mice breed in-house and mice purchased from the Jackson Laboratory. All other experiment used mice purchased from the Jackson Laboratory. Experimental groups were blinded and randomly assigned prior to the start of experimentation and remained blinded until all data were collected. Mice were housed under standard 12 hour light/dark cycle conditions in rooms equipped to control for temperature and humidity. Unless stated otherwise, male mice were tested at 8–10 weeks of age. Sample sizes were chosen based on a power analysis using estimates from previously published experiments. All experiments were approved by the Institutional Animal Care and Use Committee of the University of Virginia.
Behavior
For cohorts tested with multiple behavioral assay, the elevated plus-maze was performed first and then followed by the open field before any other assay. Prior to all experiments, mice were transported to the behavior room and given 1 hour to habituate. All behavioral testing was conducted during daylight hours.
Elevated Plus-Maze
Mice were placed into the center hub and allowed to explore the plus-maze for 10 minutes. Video tracking software (CleverSys) was used to quantify time spent in the open arms.
Open Field
Mice were placed into the open field (35cm × 35cm) and allowed to explore for 15 minutes. Total distance and time spent in the center (23cm × 23cm) were quantified using video tracking software (CleverSys).
Rotarod
Mice were placed on an accelerating rotarod (MedAssociates) that accelerated from 4.0–40 rpm over 5 minutes. Infrared beams were used to quantify the latency of a mouse to fall of the rod. Mice were given 6 trials with a 4-hour break between trial 3 and 4.
Three-chamber sociability assay
The three-chamber sociability test was conducted as previously described31. Briefly, mice were transported to the testing room and habituated for at least one hour. The room was maintained in dim light and a white noise generator used to mitigate any unforeseen noises. Test boxes were fabricated, in-house, by a machine shop. Test mice were placed in the center chamber with the two outer chambers containing empty wire cages (Spectrum Diversified Designs) and allowed to explore for ten minutes (habituation phase). After the habituation phase, mice were return to the center chamber. A novel mouse (an 8–10 week old male C57BL/6J ~ 18–22g) was placed under one cup and an object placed under the other. Prior to testing, the novel mouse was habituated to the cup by being placed under the cup for 10 minutes, 3 times a day for 5 days. Mice were allowed to explore for an additional ten minutes (social phase). A video tracking system (CleverSys) was used to quantify the time spent around each target. For 3-chamber experiments comparing social behavior between wild-type and SCID mice, data collected using males and females were combined because no significant effects were found between genders. When females were tested, juvenile C57BL/6J male (approximately 4 weeks old) mice were used as novel demonstrators.
Novel/Social environment
Mice were single housed and maintained un-manipulated for 5 days, then placed in a novel/social environment with a mouse (opposite sex and identical scid genotype) under video surveillance as previously described31. After 2 hours, mice were sacrificed and prepared for immunohistochemistry as described below. The time spent interacting was measured by a blinded observer for 5 minutes at the 30, 60, and 90 minute post-introduction time points.
Resting-state fMRI
The protocol for rsfMRI was adapted from Zhan et al.32. Mice were maintained under light anesthesia with (1–1.25%) isoflurane and images were acquired on a 7.0 Tesla MRI Clinscan system (Bruker, Etlingen, Germany) using a 30-mm inner-diameter mouse whole-body radiofrequency coil. High-resolution structural images were acquired by collecting 16 × 0.7mm thick coronal slices using TR/TE 5,500/65ms and an 180° flip angle. A BOLD rsfMRI time series of 16 × 0.7mm thick coronal slices were collected using TR/TE 4,000/17ms and a 60° flip angle. For analysis, a structural template33 was custom labeled using “The Mouse Brain” by Paxinos and Franklin as reference (Extended Data Fig. 3). The non-rigid transformation between the template and each individual mouse was estimated using the open source Advanced Normalization Tools (ANTs) package34 that was then used to propagate the regional labels to each subject. Cleaning of the bold fMRI data was performed using tools available in ANTsR35 – a statistical and visualization interface between ANTs and the R statistical project. fMRI preprocessing consisted of motion correction36, band pass filtering (freq. = [0.002, 0.1]), and CompCor estimation37. The correlation matrix was determined from the clean fMRI data using the regions labels. To determine if the functional connectivity between sample groups was significantly different, we tested for the equality of their corresponding correlation matrices. First, an aggregate correlation matrix was constructed for each group by calculating the median value for each connection, and then the aggregate matrices were compared using the Jennrich test38, as implemented in the cortest.jennrich function in the R psych package. To create a functional connectivity network for a sample group, a correlation threshold was applied to the connections between regions of interest. If a connection strength was above the threshold, it was kept as an edge in the network, otherwise it was discarded. When comparing networks from multiple sample groups, the threshold was determined by calculating the maximum threshold that leaves one of the networks connected (i.e., it is possible to reach any node in the network from any other node).
Immunohistochemistry
Mice were sacrificed under Euthasol then transcardially perfused with PBS with heparin. Brains were removed and drop fixed in 4% PFA for 48 hours. After fixation, brains were washed with PBS, cryoprotected with 30% sucrose, then frozen in O.C.T. compound (Sakura Finetek) and sliced (40μM) with a cryostat (Leica). Free-floating sections were maintained in PBS + Azide (0.02%) until further processing. Immunohistochemistry for c-fos (1:1000 dilution; Millipore) on free-floating sections was performed as previously described39.
Depletion of Meningeal T cells
A rat monoclonal antibody to murine VLA4 (clone PS/2) was affinity purified from hybridoma supernatants and was used with the kind permission of Dr. Klaus Ley (La Jolla Institute of Allergy and Immunology, San Diego, CA). Mice were given two separate injections (i.p.; 0.2mg in saline per mouse), four days apart, of either anti-VLA4 or rat anti-HRP for control (clone HRPN; BioXcell), then tested twenty-four hours post final injection.
Dissection of meninges and flow cytometry
Meninges were dissected as previously described40. Briefly, after sacrificing and perfusing, skulls caps were removed by making an incision along the parietal and squamosal bones. The meninges were removed from the internal side of the skull cap and gently pressed through a 70 μm nylon mesh cell strainer with sterile plastic plunger (BD Biosciences) to isolate a single cell suspension. Cells were then centrifuged at 300g at 4°C for 10 min, resuspended in cold FACS buffer (pH 7,4; 0.1 M PBS; 1 mM EDTA; 1% BSA), and stained for extracellular markers using the following antibodies at a 1:200 dilution: CD45 PerCP-Cyanine5.5 (eBioscience), TCR BV510 (BD Bioscience), CD4 FITC (eBioscience), L/D Zombie NIR (BioLegend). To measure intracellular IFN-γ, single cell isolates from meninges were maintained in T cell isolation buffer (RPMI + 2% FBS, 2mM L-glutamine, 1mM sodium pyruvate, 10mM HEPES, 1X non-essential amino acids, and 1X Antibiotic-Antimycotic (Thermo Fisher) and stimulated with PMA/ionomycin (Cell Stimulation Cocktail – eBioscience) + 10 μg/ml brefeldin A at 37°C prior to extracellular staining as stated above. Cells were then permeabilized with Cytofix/Cytoperm (BD Biosciences) and stained with IFN-γ APC (eBioscience; clone XMG1.2).
To measure expression of IFN-γ receptors, brains were removed and placed in Neurobasal media containing 10% fetal bovine serum. The meninges were removed from the brain and the frontal cortex was micro-dissected under a dissection microscope. Using a 2 mL dounce homogenizer, brains were homogenized in Neurobasal media with 50U/mL Dnase I. The homogenate was passed through a 70μm nylon filter and washed with cold FACS buffer. IFNGR were labeled with anti-IFNGR1 Biotin (BD Pharmingen; GR20) or rabbit anti-IFNGR2 (Santa Cruz; M-20). Cells were washed with FACS buffer and incubated with FITC conjugated streptavidin or 488 conjugated chicken anti-rabbit. Next, cells were washed again then incubated with CD11B PE-Cyanine7 (eBioscience), L/D Zombie NIR (BioLegend), and Hoechst for 45 minutes. Cells were washed, then permeabilized and fixed with Cytofix/Cytoperm (BD Biosciences). After another wash with permeabilization wash buffer (PBS with 10% fetal bovine serum, 1% sodium azide, and 1% saponin; pH 7.4), cells were incubated overnight with NeuN PE (Millipore). Cells underwent a final wash and again passed through a 70μm nylon filter. Samples were run on a flow cytometer Gallios (Beckman Coulter) then analyzed using FlowJo software (Treestar).
SCID repopulation
SCID mice were repopulated with cells from spleen and lymph nodes (axillary, brachial, cervical, inguinal, and lumbar). Spleen and lymph nodes were collected from a 3–4 week old donor and passed through a 70μm nylon mesh cell strainer with a sterile plastic plunger. ACK buffer was used to lyse red blood cells prior to washing with saline. Cells were counted on an automated cell counter (Nexcelom) and injected (i.v.) at 5×106 cells in 250μL of saline (control mice were injected with 250μL of saline only). For injections, the animal technician was blinded to the genotype of the mice and the content of the injection. Thus, all groups were handled identically. Mice aged 3–4 weeks were carefully placed into a tail vein injection platform. Their tails were briefly warmed using a heating pad and saline with cells or saline alone was slowly injected into the tail vein using a 28G needle. After the injection, mice were returned to their home cage.
IFN-γ injections
Mice were anaesthetized with a ketamine/xylazine [ketamine (100mg/kg) and xylazine (10mg/kg)] injection (i.p.) or isoflurane (2%), then placed into a stereotaxic frame with the head at an approximately 45° angle. The skin above cisterna magna was cleaned and sanitize before a 1 cm incision was made. The underlying muscles were separated with forceps, retracted, and a small Hamilton syringe (33G) was used to slowly inject 1uL of saline or IFN-γ (20ng; eBioscience) into the cisterna magna (i.c.m.). After injection the syringe was held in place for 5 minutes to avoid back-flow of CSF. After the syringe was removed, muscles were put back in place and skin was sutured. Mice were placed on a heating pad and given ketoprofen and baytril for recovery.
Induced seizures
IFN-γ was injected (i.c.m.) into the CSF through the cisterna magna (i.c.m.), as described above, 24 hours prior to inducing seizures. Control mice were injected with the same volume of saline. To induce seizures, mice were injected with PTZ (40mg/kg; i.p.). After injection, mice were placed into an empty housing cage and recorded for video analysis. Seizures were analyzed by a blinded observer using a behavior scoring system previously published41.
Diazepam treatment
Diazepam (1.25mg/kg) was delivered i.p. for 30 minutes prior to testing for social behavior.
Fluorescent in situ hybridization
Mice were euthanized then transcardially perused with PBS with heparin followed by 4% PFA. Brains were then removed and drop fixed in 4% PFA for 24 hours, frozen in OCT, and 12μM sections were cut on a cryostat. Fluorescent in situ hybridization was performed using RNA ISH tissue assay kits (Affymetrix) following the manufacture’s protocol. Tissues were treated with protease for 20 minutes at 40°C. 63× Images were acquired on a Leica TCS SP8 confocal system (Leica Microsystems) using the LAS AF Software.
AAV Delivery
AAV1.hSyn.HI.eGFP-Cre.WPRE.SV40 and AAV1.hSyn.eGFP.WPRE.bGH were purchased from Penn Vector Core. IFNGR1fl/fl mice were purchased from the Jackson lab. After 1 week of habituation, mice were anesthetized with 2% isoflurane and injected bilaterally with 2×1010 genome copies of AAV virus in 1 μL at stereotaxic coordinates: +2.5μM bregma A/P, 0.25μM lateral, 1.25μM deep.
Measuring inhibitory currents
Visualized whole-cell patch-clamp recordings were performed on layer 2/3 prefrontal cortical neurons prepared from acute brain slices (adult) using the protective recovery method42. Recordings were performed in 34°C artificial cerebrospinal fluid (ACSF) containing (in mM) 131.5 NaCl, 25 NaHCO3, 12 D-glucose, 2.5 KCl, 1.25 NaH2PO4, 2 CaCl2, and 1 MgCl2. ACSF also contained 3mM kynurenic acid to block synaptic excitation and 2.5mM NO-711 to enhance tonic inhibition43. Slices were incubated in this ACSF for 5–10 minutes prior to placement in the recording chamber. The patch pipette solution with elevated chloride contained (in mM) 140 CsCl, 4 NaCl, 1 MgCl2, 10 HEPES, 0.05 EGTA, 2 Mg-ATP, and 0.4 Mg-GTP43. Once recordings equilibrated, baseline holding current in ACSF was measured for 3.5 minutes, after which ACSF containing IFN-γ (20pg/mL) was applied for 8.5 minutes and then washed. Data presented show the mean holding current during the last minute of control (ACSF) and drug (ACSF+IFN-γ) conditions.
RNA isolation and sequencing
8 week old, male mice were purchased from The Jackson Lab and housed in standard housing boxes either 4 mice per cage or isolated for 6 days. Mice were euthanized as described above and the PFC was microdissected under a dissection microscope. RNA was isolated using RNAeasy mini kit (Qiagen) and a cDNA library was generated with a TruSeq Stranded mRNA Library Prep Kit (Illumina) with Agencourt AMPure XP beads for PCR cleanup. Samples were loaded onto a NextSeq 500 High-output 75 cycle cartridge and sequenced on a NextSeq 500 (Illumina).
Transcriptome analysis
Raw FASTQ sequencing reads were chastity filtered to remove clusters having outlying intensity corresponding to bases other than the called base. Filtered reads were assessed for quality using FastQC44. Reads were splice-aware aligned to the UCSC mm9 genome using STAR45, and reads overlapping UCSC mm9 gene regions were counted using featureCounts46. The DESeq2 Bioconductor package47 in the R statistical computing environment48 was used for normalizing count data, performing exploratory data analysis, estimating dispersion, and fitting a negative binomial model for each gene comparing the expression from the PFC of mice in a social environment versus isolation. After obtaining a list of differentially expressed genes, log fold changes, and P-values, Benjamini-Hochberg False Discovery Rate procedure was used to correct P-values for multiple testing. A Gene Set Enrichment Analysis (GSEA) algorithm49 was applied to identify the enrichment of transcriptional signatures and molecular pathways in PFC transcriptomes of mice exposed to group and isolation housing conditions. 4726 publicly available transcriptional signatures were obtained from the molecular Signature Database C2 version 4.0 and GSEA was used to examine the distribution of these curated gene sets in lists of genes ordered according to differential expression between group and isolation housing conditions. The statistical analysis was performed by evaluation of nominal P-value and normalized enrichment score (NES) based on 1,000 random sample permutations.
Meta-data analysis
The custom-made IFN-γ and pathogen-induced transcriptional signatures (Supplementary Table 2) were generated by retrieving genes upregulated ≥ 2 fold following IFN-γ stimulation or pathogen infection. All custom signatures were derived from publicly available transcriptomes downloaded from Gene Expression Omnibus (GEO). Specifically, mammalian IFN-γ transcriptional signatures were derived from transcriptomes: GSE33057, GSE19182, GSE36287, GSE9659, GSE1432 and GSE6353. Zebrafish IFN-γ transcriptional signature was used as described50. Drosophila pathogen-induced JAK/STAT-dependent transcriptional signatures were derived from transcriptomes: GSE54833 and GSE2828.
A GSEA algorithm was applied to identify the enrichment of custom-made mammalian IFN-γ transcriptional signatures in the publicly available brain cortex transcriptomes of mice and rats exposed to: social aggregation, sleep deprivation, stress, psychostimulants (DOI, cocaine, amphetamine, methamphetamine, methylphenidate, caffeine, nicotine and modafinil), antipsychotics (olanzapine, haloperidol and risperidone), anticonvulsants (levetiracetam, phenytoin, ethosuximide and oxcarbazepine), and antidepressants (iproniazid, moclobemide, paroxetine and phenelzine). In total, 41 transcriptomes were analyzed as indicated in Supplementary Table 2. The statistical analysis was performed by evaluation of nominal P-value and NES based on 1,000 random sample permutations. Zebrafish are social fish that aggregate into shoals51–54. Various Zebrafish strains differ in their preference for social interaction and novelty, resembling the phenotypic variation of inbred mouse strains55,56. Notably, domesticated zebrafish strains demonstrate higher social interaction and social novelty preference57–59 compared to wild zebrafish strains. Therefore, a GSEA algorithm was used to identify the enrichment of Zebrafish IFN-γ transcriptional signature in the publicly available whole-brain transcriptomic profiles of behaviorally distinct strains of domesticated and wild zebrafish (Supplementary Table 7). The brain transcriptomic profiles of domesticated (Scientific Hatcheries (SH) and Transgenic Mosaic 1 (TM1)) and wild (Nadia, Gaighata) zebrafish strains were derived from publicly available transcriptome data set: GSE38729. The statistical analysis was performed by evaluation of nominal P-value and NES based on 1,000 random gene set permutations.
Social interactions in Drosophila melanogaster flies play an important role in courtship, mating, egg-laying, circadian timing, food search and even life-span determination60–65. Notably, a number of studies have demonstrated that social isolation leads to an aggressive behavior in Drosophila flies, whereas group housing suppresses the aggressiveness66–68. Therefore, we employed a GSEA algorithm to identify the enrichment of JAK/STAT-dependent transcriptional signatures in the publicly available brain transcriptomes of Drosophila populations that were socially-induced or genetically-selected for low-aggressive (i.e. social) behavior (Supplementary Table 7). The transcriptomic profiles were derived from publicly available transcriptome data sets: GSE5404 and GSE6994. The statistical analysis was performed by evaluation of nominal P-value and NES based on 1,000 random gene set permutations.
Promoter Motif analysis
A GSEA algorithm was used to identify genes that are differentially expressed in brain transcriptomes of mice and rats exposed to social aggregation, domesticated Zebrafish strains and low-aggressive Drosophila melanogaster populations, compared to their control counterparts. High-scoring differentially expressed “leading-edge” social genes were selected based on their presence in the IFN-γ and pathogen-induced transcriptional signatures. Specifically, as shown in Figure 3, we have identified 31, 48, 14, 53 leading-edge genes in brain transcriptomes of mice and rats exposed to social aggregation, domesticated Zebrafish strain, and low-aggressive Drosophila melanogaster population, respectively. We next, extracted promoter sequences of 200bp up-stream of TSS of these “leading edge” genes using UCSC Genome Browser (https://genome.ucsc.edu/). The MEME suite was then used to discover overrepresented transcription factor binding motifs, as described69. MEME parameters used were any number of motif repetitions per sequence, with a minimum motif width of 5 bases and maximum motif width of 15 bases. The discovered MEME motifs were compared using Tomtom analysis. In this case, the Tomtom motif similarity analysis ranks the MEME motif most similar to the Vertebrates in vivo and in silico, canonical motif for STAT. The statistics was determined using Euclidean distance.
Circos plot
A GSEA algorithm was applied to identify the enrichment of IFN-γ, IL-4/IL-13, IL-17, and IL-10/TGF-β signaling pathways in the brain transcriptomes of rodent animals exposed to: social aggregation, stress, psychostimulants, antipsychotics, and antidepressants. All custom signatures were derived from publically available rodent transcriptomes downloaded from Gene Expression Omnibus. Statistical significance of GSEA results was assessed using 1,000 sample permutations. A NES greater than 1.5 and nominal P-value less than 0.05 was used to determined pairwise transcriptome connectivity. Circos graph was generated using circos package 0.68.1270.
Statistics
Data were analyzed using the statistical methods stated in each figure legend. For the 3-chamber assay, a 2-way ANOVA was performed using genotype/treatment and sociability as main effects, followed by applying a Sidak’s post-hoc comparison to assess if the group had a significant social preference. Prior to running an ANOVA, an equality of variance was determined by using a Brown-Forsythe test. Bars display the means, and error bars represent ranges of the standard error of the mean. For rsfMRI, data were analyzed using a 1-way ANOVA followed by a post-hoc Tukey’s test. The box and whisker plots extend to the 25th and 75th percentiles and the center line indicates the mean. The whiskers represent the min and max data points. Data for seizure latency was analyzed using a 2-way ANOVA with repeated measures followed by a post-hoc Sidak test. Additional details of statistical analysis are supplied in Supplementary Table 8.
Extended Data
Extended Data Figure 1. SCID mice have no observable anxiety, motor, or olfactory deficits.
a, The 3-chamber sociability assay was used to test social behavior. b, Neither wild-type nor SCID mice had a side bias in the habituation phase (empty cups) of the 3-chamber assay (n = 6). c, There was no effect of genotype on distance traveled in the 3-chamber assay during the habituation phase (n = 6). d, Both wild-type and SCID mice had an olfactory preference to urine, suggesting normal olfactory behavior (n = 8 mice per group; ANOVA for urine preference F (1, 28) = 31.01; P < 0.0001; *** P < 0.001, ** P < 0.01 Sidak’s post-hoc). e, Percent time spent in the open arms of plus-maze (n = 22 mice per group). f, Number of entries into the open arms of the plus-maze (n = 22 mice per group). g, Total arm entries of plus-maze (n = 22 mice per group). h, Percent time spent in the center of the open field (n = 22 mice per group). i, Total ambulatory distance in the open field (n = 22 mice per group). j, Latency to fall off the accelerating rotarod (n = 8 mice per group). k, SCID mice spent less time investigating each other than wild-type mice spent investigating each other when placed into a novel social environment (n = 5 mice per group; repeated-measures ANOVA for genotype F (1, 21) = 5.708 * P < 0.05). l, Repopulated SCID mice have similar numbers (m) and percentage (n) of meningeal T cells as wild-type mice (n = 4–5 mice per group). Cells were gated on singlets, live, CD45+, and TCR.
Extended Data Figure 2. Neuroanatomical Structures analyzed by rsfMRI.
a, Regions of interests (ROIs) were generated using “The Mouse Brain” by Paxinos and Franklin as a reference. Abbreviations are as follows: FrA=frontal association cortex; PrL=prelimbic cortex; OrbC=orbital cortex; OB=olfactory bulb; MC=motor cortex; SocC=somatosensory cortex; Ins=insula; PirF=piriform cortex; CpU= caudate putamen; Acb=accumbens; ACC=anterior cingulate cortex; dHip=dorsal hippocampus; T=thalamus; Amyg=amygdala; EntC=entorhinal cortex; Hyp=hypothalamus; VisC=visual cortex; SupC=superior colliculus; PAG=periductal grey; DpMe=deep mesencephalic nucleus; vHip=ventral hippocampus; SNR=substantia nigra; VTA=ventral tegmental area; CB=cerebellum; BS=brain stem. b, Connectivity of local PFC/Insular nodes. Correlation thresholds were applied to visualize the strength of the connection. Connections that pass a high threshold are shown in red; connections that pass a lower threshold are shown in dashed grey. SCID mice have aberrant hyper-connectivity in the PFC (n = 8–9 mice per group; P < 0.05 Jennrich test). c, c-fos+ cells in the hippocampus (n = 9–10 mice per group).
Extended Data Figure 3. Acute reduction of meningeal T cells with anti-VLA4.
a, Anti-VLA4 depletes meningeal T cells. Meninges were dissected and single cell suspensions were immune-stained. T cells were gated on live, single, CD45+, TCR+ events and counted by flow cytometry. b, Acute injection of anti-VLA4 reduced the amount of TCR+ T cells in the meninges (n = 4 mice per group; * P < 0.01).
Extended Data Figure 4. Circos plot showing the connectivity of Th1 response and social aggregation.
Here labels are shown for the datasets analyzed and presented in Fig. 1h.
Extended Data Figure 5. T cells in the meninges produce IFN-γ and IFN-γ deficient mice have normal levels of anxiety and motor behavior.
a, A substantial percentage of meningeal T cells produce IFN-γ. Cells were gated for live, singlets, CD45+, and TCR+. Ifng−/−mice were used to gate for IFN-γ staining. b, Percent time spent is open arms of the plus-maze (n = 20 mice per group). c, Entries into the open arms of plus-maze (n = 20 mice per group). d, Total entries into all arms of the plus-maze (n = 20 mice per group). e, Percent time spent in the center of the open field (n = 20 mice per group). f, Total ambulatory distance in the open field (n = 20 mice per group). g, Latency to fall off the accelerating rotarod (n = 8 mice per group).
Extended Data Figure 6. IFN-γ signaling is necessary for normal social behavior.
a, Repopulating SCID mice with wild-type lymphocytes rescued a social preference; repopulating with Ifng−/−lymphocytes did not rescue a social preference; AVONA for Social behavior F (1, 14) = 11.99; P = 0.0038 (** P < 0.01; n = 8 mice per group). b, Connectivity of local PFC/Insular nodes. Correlation thresholds were applied to visualize the strength of the connection. Connections that pass a high threshold are shown in red; connections that pass a lower threshold are shown in dashed grey. Ifng−/−mice have more connections than wild-type mice (Jennerich test; P = 0.0006). These connections were reduced by IFN-γ (Jennerich test; P = 0.02). c, Ifngr1−/−mice have social deficits (n = 6 mice per group; ANOVA for interaction P = 0.01; ** P < 0.01 Sidak’s post-hoc) that were not rescued by injecting IFN-γ into the CSF (d; n = 5–6 mice per group; ANOVA for interaction P = 0.01; ** P < 0.01 Sidak’s post-hoc). e, Il4−/− mice spend more time than wild-type mice investigating a novel mice; ANOVA for genotype F (1, 32) = 5.397; P = 0.0267 (* P < 0.05 Sidak’s post-hoc; n = 16–18 mice per group).
Extended Data Figure 7. Gating strategy for neurons and microglia.
Brain homogenates were stained and analyzed by flow cytometry. Cells were gated on nucleated, singlets, and live. Neurons were then gated on NeuN positive and microglia on CD11B positive cells.
Extended Data Figure 8. IFN-γ Signaling in Microglia is not Necessary for Normal Social Function.
Mice deficient for STAT1 in microglia have normal social preference (n = 9 mice per group; ANOVA for Cre F(1, 16) = 1.809 and sociability F(1, 16) = 30.10; P < 0.0001; **P < 0.01; *** P < 0.001 Sidak’s post-hoc).
Extended Data Figure 9. Deleting IFNGR1 by AAV transduction.
Mice were injected with AVVs expressing Cre and GFP under a synapsin promoter. a, GFP fluorescence in the PFC. Atlas image adapted from: c 2015 Allen Institute for Brain Science. Allen Brain Atlas [Internet]. Available from: http://www.brain-map.org. b, GFP fluorescence is only observed in NeuN+ neurons, not Iba+ microglia (top = 20×; bottom 63× Objective).
Extended Data Figure 10. IFN-γ increased the number of c-fos+ cells in Layer I of the PFC.
a, IFN-γ was injected into the CSF (i.c.m.) 2 hours prior to sacrificing and processing brains for immunohistochemistry. Slices were stained for c-fos. Atlas image adapted from: c 2015 Allen Institute for Brain Science. Allen Brain Atlas [Internet]. Available from: http://www.brain-map.org. b, Total c-fos+ cells in layer I of the PFC (n = 3 mice per group; * P < 0.05). Holding current pre and post IFN-γ application on acute slices from the PFC (c) and somatosensory cortex (d; n = 6 neurons from 3 mice). c, VgatCre::Ifngr1fl/fl mice. IFN-γ increased tonic inhibition in Cre- mice (n = 6–7 cells from 4 mice per group; ** P < 0.01 Sidak’s post-hoc test).
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (AG034113 and NS081026 to J.K), (T32-AI007496 to A.J.F.), and the Hartwell Foundation (to A.J.F.). We thank all the members of the Center for Brain Immunology and Glia (BIG) for their valuable comments during numerous discussions of this manuscript. We also thank Jack Roy for his expertise in MRI, Bonnie Tomlin and Nour Al Hamadani for animal care, as well as Stephen Rich, Suna Onengut-Gumuscu, and Emily Farber for sequencing the cDNA library.
Footnotes
Contributions
A.J.F and J.K. designed and performed experiments and wrote the manuscript; Y.X. and V.L., provided intellectual contributions and analyzed all transcriptome data. S.D.T, Z.W., S.N.P, H.C., analyzed transcriptome data. N.T., C.C.O analyzed BOLD data. R.L.M, W.B., I.S., S.P.G, M.M.S, M.P.B provided intellectual contributions and assisted with experimental procedures. M.P.B performed all electrophysiological experiments. K.S.L critically read the manuscript.
Author Information
RNA-seq data was deposited by chris.overall@virginia.edu into the Gene Expression Omnibus (GEO; GSE81783).
The authors do not have competing financial interests.
References
- 1.Derecki NC, et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J Exp Med. 2010;207:1067–1080. doi: 10.1084/jem.20091419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourke AF. Hamilton’s rule and the causes of social evolution. Philosophical transactions of the Royal Society of London. Series B, Biological sciences. 2014;369:20130362. doi: 10.1098/rstb.2013.0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cacioppo S, Capitanio JP, Cacioppo JT. Toward a neurology of loneliness. Psychol Bull. 2014;140:1464–1504. doi: 10.1037/a0037618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kennedy DP, Adolphs R. The social brain in psychiatric and neurological disorders. Trends Cogn Sci. 2012;16:559–572. doi: 10.1016/j.tics.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashwood P, et al. Altered T cell responses in children with autism. Brain Behav Immun. 2011;25:840–849. doi: 10.1016/j.bbi.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta S, Aggarwal S, Rashanravan B, Lee T. Th1- and Th2-like cytokines in CD4+ and CD8+ T cells in autism. J Neuroimmunol. 1998;85:106–109. doi: 10.1016/s0165-5728(98)00021-6. [DOI] [PubMed] [Google Scholar]
- 7.Waisman A, Liblau RS, Becher B. Innate and adaptive immune responses in the CNS. Lancet Neurol. 2015;14:945–955. doi: 10.1016/S1474-4422(15)00141-6. [DOI] [PubMed] [Google Scholar]
- 8.Moy SS, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 9.Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11:490–502. doi: 10.1038/nrn2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nelson SB, Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–698. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Supekar K, et al. Brain hyperconnectivity in children with autism and its links to social deficits. Cell Rep. 2013;5:738–747. doi: 10.1016/j.celrep.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan Y, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014;17:400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- 13.Shen HH. Core Concept: Resting-state connectivity. Proc Natl Acad Sci U S A. 2015;112:14115–14116. doi: 10.1073/pnas.1518785112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brynskikh A, Warren T, Zhu J, Kipnis J. Adaptive immunity affects learning behavior in mice. Brain Behav Immun. 2008;22:861–869. doi: 10.1016/j.bbi.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 15.Yednock TA, et al. Prevention of experimental autoimmune encephalomyelitis by antibodies against alpha 4 beta 1 integrin. Nature. 1992;356:63–66. doi: 10.1038/356063a0. [DOI] [PubMed] [Google Scholar]
- 16.Kunis G, et al. IFN-gamma-dependent activation of the brain’s choroid plexus for CNS immune surveillance and repair. Brain. 2013;136:3427–3440. doi: 10.1093/brain/awt259. [DOI] [PubMed] [Google Scholar]
- 17.Tsuda M, et al. IFN-gamma receptor signaling mediates spinal microglia activation driving neuropathic pain. Proc Natl Acad Sci U S A. 2009;106:8032–8037. doi: 10.1073/pnas.0810420106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prieto JJ, Peterson BA, Winer JA. Morphology and spatial distribution of GABAergic neurons in cat primary auditory cortex (AI) J Comp Neurol. 1994;344:349–382. doi: 10.1002/cne.903440304. [DOI] [PubMed] [Google Scholar]
- 19.Olah S, et al. Regulation of cortical microcircuits by unitary GABA-mediated volume transmission. Nature. 2009;461:1278–1281. doi: 10.1038/nature08503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han S, Tai C, Jones CJ, Scheuer T, Catterall WA. Enhancement of inhibitory neurotransmission by GABAA receptors having alpha2,3-subunits ameliorates behavioral deficits in a mouse model of autism. Neuron. 2014;81:1282–1289. doi: 10.1016/j.neuron.2014.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang L, Dankert H, Perona P, Anderson DJ. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci U S A. 2008;105:5657–5663. doi: 10.1073/pnas.0801327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Datwani A, et al. Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron. 2009;64:463–470. doi: 10.1016/j.neuron.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Djurisic M, et al. PirB regulates a structural substrate for cortical plasticity. Proc Natl Acad Sci U S A. 2013;110:20771–20776. doi: 10.1073/pnas.1321092110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinman L. Inflammatory cytokines at the summits of pathological signal cascades in brain diseases. Sci Signal. 2013;6:pe3. doi: 10.1126/scisignal.2003898. [DOI] [PubMed] [Google Scholar]
- 25.Smith LK, et al. beta2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat Med. 2015;21:932–937. doi: 10.1038/nm.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villeda SA, et al. The ageing systemic milieu negatively regulates neurogenesis and cognitive function. Nature. 2011;477:90–94. doi: 10.1038/nature10357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCusker RH, Kelley KW. Immune-neural connections: how the immune system’s response to infectious agents influences behavior. J Exp Biol. 2013;216:84–98. doi: 10.1242/jeb.073411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scholz J, Woolf CJ. The neuropathic pain triad: neurons, immune cells and glia. Nat Neurosci. 2007;10:1361–1368. doi: 10.1038/nn1992. [DOI] [PubMed] [Google Scholar]
- 29.Bhat R, et al. Inhibitory role for GABA in autoimmune inflammation. Proc Natl Acad Sci U S A. 2010;107:2580–2585. doi: 10.1073/pnas.0915139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopez-Munoz A, et al. Evolutionary conserved pro-inflammatory and antigen presentation functions of zebrafish IFNgamma revealed by transcriptomic and functional analysis. Mol Immunol. 2011;48:1073–1083. doi: 10.1016/j.molimm.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 31.Filiano AJ, et al. Dissociation of frontotemporal dementia-related deficits and neuroinflammation in progranulin haploinsufficient mice. J Neurosci. 2013;33:5352–5361. doi: 10.1523/JNEUROSCI.6103-11.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhan Y, et al. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat Neurosci. 2014;17:400–406. doi: 10.1038/nn.3641. [DOI] [PubMed] [Google Scholar]
- 33.Johnson GA, et al. Waxholm space: an image-based reference for coordinating mouse brain research. Neuroimage. 2010;53:365–372. doi: 10.1016/j.neuroimage.2010.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Avants BB, et al. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage. 2011;54:2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avants BB, et al. The pediatric template of brain perfusion. Sci Data. 2015;2:150003. doi: 10.1038/sdata.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59:2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Behzadi Y, Restom K, Liau J, Liu TT. A component based noise correction method (CompCor) for BOLD and perfusion based fMRI. Neuroimage. 2007;37:90–101. doi: 10.1016/j.neuroimage.2007.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jennrich RI. An Asymptotic χ2 Test for the Equality of Two Correlation Matrices. Journal of the American Statistical Association. 1970;65:904–912. [Google Scholar]
- 39.Palop JJ, Mucke L, Roberson ED. Quantifying biomarkers of cognitive dysfunction and neuronal network hyperexcitability in mouse models of Alzheimer’s disease: depletion of calcium-dependent proteins and inhibitory hippocampal remodeling. Methods Mol Biol. 2011;670:245–262. doi: 10.1007/978-1-60761-744-0_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derecki NC, et al. Regulation of learning and memory by meningeal immunity: a key role for IL-4. J Exp Med. 2010;207:1067–1080. doi: 10.1084/jem.20091419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Z, Hall AM, Kelinske M, Roberson ED. Seizure resistance without parkinsonism in aged mice after tau reduction. Neurobiol Aging. 2014;35:2617–2624. doi: 10.1016/j.neurobiolaging.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ting JT, Daigle TL, Chen Q, Feng G. Acute brain slice methods for adult and aging animals: application of targeted patch clamp analysis and optogenetics. Methods Mol Biol. 2014;1183:221–242. doi: 10.1007/978-1-4939-1096-0_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nusser Z, Mody I. Selective modulation of tonic and phasic inhibitions in dentate gyrus granule cells. J Neurophysiol. 2002;87:2624–2628. doi: 10.1152/jn.2002.87.5.2624. [DOI] [PubMed] [Google Scholar]
- 44.SA FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010 [Google Scholar]
- 45.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Team, R. C. D. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2010. http://www.R-project.org/ [Google Scholar]
- 49.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lopez-Munoz A, Roca FJ, Meseguer J, Mulero V. New insights into the evolution of IFNs: zebrafish group II IFNs induce a rapid and transient expression of IFN-dependent genes and display powerful antiviral activities. J Immunol. 2009;182:3440–3449. doi: 10.4049/jimmunol.0802528. [DOI] [PubMed] [Google Scholar]
- 51.Engeszer RE, Patterson LB, Rao AA, Parichy DM. Zebrafish in the wild: a review of natural history and new notes from the field. Zebrafish. 2007;4:21–40. doi: 10.1089/zeb.2006.9997. [DOI] [PubMed] [Google Scholar]
- 52.Krause J, Butlin RK, Peuhkuri N, Pritchard VL. The social organization of fish shoals: a test of the predictive power of laboratory experiments for the field. Biol Rev Camb Philos Soc. 2000;75:477–501. doi: 10.1111/j.1469-185x.2000.tb00052.x. [DOI] [PubMed] [Google Scholar]
- 53.Miller N, Gerlai R. Quantification of shoaling behaviour in zebrafish (Danio rerio) Behav Brain Res. 2007;184:157–166. doi: 10.1016/j.bbr.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Saverino C, Gerlai R. The social zebrafish: behavioral responses to conspecific, heterospecific, and computer animated fish. Behav Brain Res. 2008;191:77–87. doi: 10.1016/j.bbr.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barba-Escobedo PA, Gould GG. Visual social preferences of lone zebrafish in a novel environment: strain and anxiolytic effects. Genes Brain Behav. 2012;11:366–373. doi: 10.1111/j.1601-183X.2012.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moy SS, et al. Sociability and preference for social novelty in five inbred strains: an approach to assess autistic-like behavior in mice. Genes Brain Behav. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- 57.Moretz JA, Martins EP, Robison BD. Behavioral syndromes and the evolution of correlated behavior in zebrafish. Behavioral Ecology. 2007;18:556–562. doi: 10.1093/beheco/arm011. [DOI] [Google Scholar]
- 58.Wright D, Butlin RK, Carlborg O. Epistatic regulation of behavioural and morphological traits in the zebrafish (Danio rerio) Behav Genet. 2006;36:914–922. doi: 10.1007/s10519-006-9080-9. [DOI] [PubMed] [Google Scholar]
- 59.Zala SM, Määttänen I, Penn DJ. Different social-learning strategies in wild and domesticated zebrafish, Danio rerio. Animal Behaviour. 2012;83:1519–1525. http://dx.doi.org/10.1016/j.anbehav.2012.03.029. [Google Scholar]
- 60.Levine JD, Funes P, Dowse HB, Hall JC. Resetting the circadian clock by social experience in Drosophila melanogaster. Science. 2002;298:2010–2012. doi: 10.1126/science.1076008. [DOI] [PubMed] [Google Scholar]
- 61.Mery F, et al. Public versus personal information for mate copying in an invertebrate. Curr Biol. 2009;19:730–734. doi: 10.1016/j.cub.2009.02.064. [DOI] [PubMed] [Google Scholar]
- 62.Ruan H, Wu CF. Social interaction-mediated lifespan extension of Drosophila Cu/Zn superoxide dismutase mutants. Proc Natl Acad Sci U S A. 2008;105:7506–7510. doi: 10.1073/pnas.0711127105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sarin S, Dukas R. Social learning about egg-laying substrates in fruitflies. Proc Biol Sci. 2009;276:4323–4328. doi: 10.1098/rspb.2009.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sokolowski MB. Social interactions in “simple” model systems. Neuron. 2010;65:780–794. doi: 10.1016/j.neuron.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 65.Wertheim B, van Baalen EJ, Dicke M, Vet LE. Pheromone-mediated aggregation in nonsocial arthropods: an evolutionary ecological perspective. Annu Rev Entomol. 2005;50:321–346. doi: 10.1146/annurev.ento.49.061802.123329. [DOI] [PubMed] [Google Scholar]
- 66.Hoffmann AA. A laboratory study of male territoriality in the sibling species Drosophila melanogaster and D. simulans. Animal Behaviour. 1987;35:807–818. http://dx.doi.org/10.1016/S0003-3472(87)80117-3. [Google Scholar]
- 67.Kamyshev NG, et al. Plasticity of social behavior in Drosophila. Neurosci Behav Physiol. 2002;32:401–408. doi: 10.1023/a:1015832328023. [DOI] [PubMed] [Google Scholar]
- 68.Wang L, Dankert H, Perona P, Anderson DJ. A common genetic target for environmental and heritable influences on aggressiveness in Drosophila. Proc Natl Acad Sci U S A. 2008;105:5657–5663. doi: 10.1073/pnas.0801327105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.