Abstract
Characterization of life processes at the molecular level requires structural details of protein–protein interactions (PPIs). The number of experimentally determined protein structures accounts only for a fraction of known proteins. This gap has to be bridged by modeling, typically using experimentally determined structures as templates to model related proteins. The fraction of experimentally determined PPI structures is even smaller than that for the individual proteins, due to a larger number of interactions than the number of individual proteins, and a greater difficulty of crystallizing protein–protein complexes. The approaches to structural modeling of PPI (docking) often have to rely on modeled structures of the interactors, especially in the case of large PPI networks. Structures of modeled proteins are typically less accurate than the ones determined by X-ray crystallography or nuclear magnetic resonance. Thus the utility of approaches to dock these structures should be assessed by thorough benchmarking, specifically designed for protein models. To be credible, such benchmarking has to be based on carefully curated sets of structures with levels of distortion typical for modeled proteins. This article presents such a suite of models built for the benchmark set of the X-ray structures from the Dockground resource (http://dockground.bioinformatics.ku.edu) by a combination of homology modeling and Nudged Elastic Band method. For each monomer, six models were generated with predefined Cα root mean square deviation from the native structure (1, 2, . . ., 6 Å). The sets and the accompanying data provide a comprehensive resource for the development of docking methodology for modeled proteins.
Keywords: protein recognition, protein modeling, benchmark sets, structure prediction, protein interactions
INTRODUCTION
Genome sequencing efforts have determined a massive amount of protein sequences. At the same time, the number of corresponding three-dimensional (3D) structures is far lagging, due to the limitations of the experimental techniques for protein structure determination. This gap is supposed to be bridged by computational approaches, using experimentally determined structures as templates to model related proteins. The rapidly growing PDB provides an opportunity to model a large part of the “protein universe.”1–3 When it comes to protein–protein interactions (PPIs), high-throughput experimental techniques (two-hybrid analysis, mass spectroscopy, etc.) provide data for recreating interaction networks for many organisms and/or biochemical pathways. To understand the mechanisms of these interactions, it is essential to have the structures of the protein–protein complexes. However, the fraction of experimentally determined PPI structures is even smaller than that for the individual proteins, due to a larger number of interactions than the number of individual proteins, and a greater difficulty of crystallizing protein–protein complexes.
Computational methods for structural modeling of PPI (docking) historically started with ab initio methods based on physical potentials (primarily, van der Waals interactions,4 currently increasingly supplemented by knowledge-based approaches (e.g., statistical potentials,5,6 constraints-driven docking,7 etc.). Following a long-standing pattern in individual protein structure prediction, PPI modeling is increasingly employing template-based methods. Efforts of several groups8–11 working on sequence-similarity-based PPI modeling have concluded that this methodology yields accurate PPI models, given suitable templates. The template pool for PPI modeling can be significantly expanded by exploiting structural similarity between protein complexes.12 The structural similarity methodology for PPI modeling is becoming increasingly popular.13
These efforts have paved the way to large-scale structural PPI modeling.13 However, the majority of structures to be docked in such studies will themselves be models of limited accuracy. Thus, to directly address the widespread skepticism about the meaningfulness of such “double modeling,” comprehensive benchmark studies on a carefully selected set of model structures are needed.13 Sets of protein models (“decoys”) are used in structural studies of individual proteins14,15 and small ligand–receptor interactions.16 However, the existing protein–protein benchmark sets,17,18 are restricted to the X-ray structures, which are generally not representative of the potentially limited accuracy of protein models.
In our previous study on the applicability of low-resolution, template-free, protein–protein docking to modeled structures,19 a representative nonredundant set of cocrystallized protein–protein complexes was used to build an array of models of each protein in the set. A procedure was developed to generate the models with root mean square deviation (RMSD) of 1, 2, 3, . . ., 10 Å from the crystal structure, by repacking of the secondary structure elements. Because of the limited availability of the templates for individual proteins, such templates were not utilized in the procedure. Thus, the resulting “simulated models” of the proteins, while reflecting the general structural accuracy of the homology models, were not necessarily structurally similar to those.
A much greater current availability of the templates provides an opportunity to generate a new benchmark set of models, explicitly utilizing the actual homology models of the proteins, and thus providing a more adequate benchmarking resource. This article presents a set of structures with several levels of controlled inaccuracy, which mimic high-throughput homology models. The distortions are 1, 2, . . ., 6 Å Cα RMSD from the X-ray structures of proteins in the Dockground benchmark set.17,20 The models were generated by a combination of homology modeling (HM), simulated annealing (SA), and Nudged Elastic Band (NEB) method.21,22 The sets and the accompanying data provide a comprehensive resource for the development of docking methodology for modeled proteins.
METHODS
The set of complexes is a tool for benchmarking the performance of docking procedures on protein models. Docking programs take the 3D structures of two separate proteins as an input and predict the structure of their complex. To evaluate the prediction, the structure of the correct (X-ray) complex should be available. Thus, the benchmark set consists of models of the individual proteins (not models of complexes) generated from the corresponding structures in cocrystallized complexes. The binary complexes from Dockground were split into two chains, and models were built independently for each of the monomers.
From the initial set of 100 protein complexes (Dockground benchmark 3), we excluded 37 complexes with multichain interactors. Six models were built for each of the remaining 126 single proteins (63 complexes) within the preset accuracy limits (±0.2 Å from 1, 2, . . ., 6 Å), resulting in 126 × 6 = 756 models in the final set. Our previous study indicated that proteins with RMSD > 6 Å, typically, to a significant extent lose structural recognition characteristics at the binding sites. Thus, 6 Å was used as the upper limit in this study.
Each protein sequence in the dataset was first subjected to single-template HM procedure with the corresponding native structure excluded from the template pool. Templates for the homology models were identified by aligning profile of the target sequence against profiles of all nonredundant sequences in PDB using Needleman-Wunsch dynamic programing algorithm23 with affine gap penalty24 as implemented in our in-house program.11 Sequence profiles were extracted from position-specific scoring matrices obtained by five-iteration PSI-BLAST25 search against nonredundant sequence database with the substitution matrix BLOSUM62.26 Alignments of identical sequences from the same organism were excluded from consideration. The model structures were built by the NEST program from JACKAL package27 with default parameters. Assignment of proteins’ secondary structures was by DSSP.28 The HM resulted in ~10,000 full-length models, out of which 290 satisfied our accuracy criteria (38% of the intended 756 structures in the model set).
The remaining 466 models were generated using NEB method,21,22 in which a low-energy pathway between two protein conformations is approximated by a series of images of the molecule, with the endpoint images fixed in space. All atoms of each image are connected to the corresponding atoms of the previous and next images by virtual elastic “springs” that keep the image from sliding down the energy landscape onto adjacent images. The NEB pathway was represented by 16 images including endpoints. The first eight frames were copies of the starting point, whereas the last eight were copies of the end structure. Pathway minimization by a combination of heating and equilibration, as in SA, but applied to the entire multi-image system, generated structures with RMSD between the end point RMSD values. The procedure started with heating of the system from 0 to 300 K within 20 ps with the spring constant between images kNEB = 1 kcal/mol (Stage 1). To increase the linkage between images, three short (10 ps) molecular dynamics runs were performed with kNEB = 5, 10, and 50 kcal/mol (Stage 2), and the last value was used during all subsequent steps. The system was then heated from 300 to 400 K, and from 400 to 500 K and then cooled from 500 to 300 K (Stage 3). Each heating and cooling run was conducted within 50 ps interval and followed by 50 ps molecular dynamics equilibration run. Finally, the system was cooled from 300 to 0 K within 12 ps (Stage 4). Langevin thermostat with collision frequency 1000 ps−1 was used for temperature coupling in all NEB calculations, and a simple leapfrog integrator was exploited to propagate the dynamics. The generalized Born implicit solvent model29 was used in all computations. The nonbonded cutoff distance was set at 12 Å. During initial heating and SA stages in 300–500 K temperature range the time step of 0.5 fs was utilized; otherwise 1.0 fs value was chosen. The NEB calculations were performed by the program sander.MPI from the Amber 10 package30 with Amber ff03 force field.31
The models with RMSD within the set limits were selected for further consideration. Otherwise the NEB procedure was repeated with new end points selected from the intermediate structures of the previous trajectory. As the starting point of the NEB trajectory, we used the homology model with the closest RMSD below the intended accuracy level, or the native structure of the protein. For the final point of the NEB trajectory, we chose the homology model with the closest RMSD above the intended accuracy level. If such model was not available, the structure was generated by SA from the starting-point homology model (this was the case for 55 monomers in our dataset). We did not use just SA for model generation because the absence of the NEB “springs” makes it difficult to control the distortion level of the final structure, and also causes considerable distortions of the secondary structure elements at high annealing temperatures.
RESULTS AND DISCUSSION
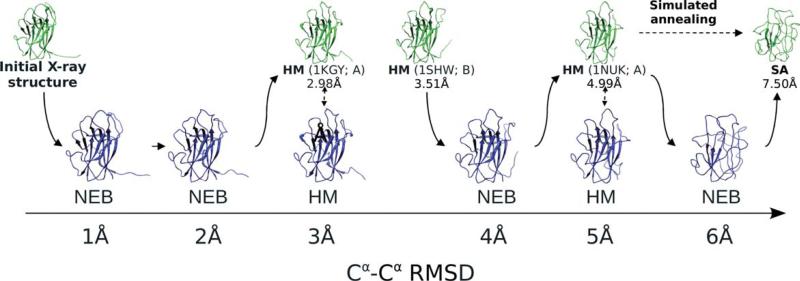
The outline of the procedure is shown in Figure 1 for 2hle, Chain A. The initial HM yielded two models. The first one (with 2.98 Å RMSD) was built using Chain A of 1kgy with sequence identity 43.1%. The template for the second model (with 4.99 Å RMSD) was Chain A of 1nuk with 42.6% sequence identity. The remaining four models were generated by three NEB runs. The start and the end points of the first NEB trajectory were the X-ray structure and the homology model with 2.98 Å RMSD, correspondingly. This NEB run yielded models with 0.90 and 1.95 Å RMSD. The starting point for the second NEB trajectory was the homology model with 3.51 Å RMSD built using Chain B of 1shw (sequence identity 42.5%). The final trajectory point was the 4.99 Å model. This run yielded the model with 4.04 Å RMSD. The third NEB run had 4.99 Å model, as the starting point. The end point (7.50 Å RMSD) was generated from this model by SA with the annealing temperature 500 K and heating, equilibration and cooling times 100, 300, and 100 ps, correspondingly. The run produced the model with 6.02 Å RMSD. As seen in Figure 1, all models have β strands and a globular structure, characteristic to the native structure. The 6 Å model has most of the strands with the dihedral angles twisted out of the exact β strand range and thus displayed as loops. Free terminal fragment, observed in the native structure (Fig. 1), gradually disappears with increasing distortion. Such fragments may introduce a bias for shape-complementarity-based docking procedures, and thus were removed from the structures.
Figure 1.
Model-generating procedure. Models for 2hle, Chain A, are generated by the Nudged Elastic Band technique (NEB), homology modeling (HM), and simulated annealing (SA). The base structures (green) are used for building the final models (blue). For homology models, the templates are shown in parentheses, along with the corresponding Cα RMSD values. Solid arrows show the path obtained by the NEB procedure connecting two fixed end points with the intermediate structures at intended accuracy levels.
Assessment of models
Protein models may have inaccuracies, in principle, anywhere in the structure. Thus, to avoid bias in docking benchmarking, models should have distortions distributed along the polypeptide chain. Thus, we considered distribution of distances between Cα atoms of corresponding residues in the model and the native structure, as shown in Figure 2 for the Chain A of 1r8s. This protein has significant conformational change upon binding, such that the secondary structure patterns in bound and unbound states are different [Fig. 2(C)]. The interface consists of Residues 28–68 (the residue numbers are from the bound structures) and contains a double-stranded β sheet and an α helix [Fig. 2(A)]. Residues 28–35 form a loop that enters the binding cleft of the interacting protein. In the unbound protein, this loop is assembled in a β strand forming a β sheet with two neighboring interface β strands (Residues 36–40 and 45–50). The short interface α helix (Residues 58–64), visible in the bound structure, becomes a loop in the unbound structure. The identified templates resembled mostly the unbound protein and, consequently, the resulting homology models were primarily distorted in the interface area, as can be seen in the 6 Å model in Figure 2(A) [shadowed areas in Fig. 2(B)]. The peak in the Cα–Cα diagram for this model, between Residues 120 and 140, is caused by insertions and deletions in the alignment with the template [Chain C of 1a2k; Fig. 2(C)]. Of the 46 homology models in the 6 ± 0.2 Å RMSD range, only three were considered for the final set. The other 43 models were rejected after visual inspection of superimposed models and the native structure within the complex. The interface loop in these models either had substantial clashes with the partner protein or deviated from the X-ray structure such that it did not enter the binding site. The final selected model has RMSD 6.01 Å, the closest one to 6 Å.
Figure 2.
Assessment of model quality. (A) 1 Å and 6 Å models of 1r8s, Chain A, are shown with the native structure, along with (B) distributions of Cα–Cα distances between the native and the model structures. (C) Secondary structure patterns of bound (1r8s, Chain A) and unbound (1rrf, Chain A) states, along with the sequence alignments of 1 and 6 Å models with their corresponding templates, show α helices in cyan and β strands in orange. Residues in the SEQRES section of the PDB files, which are missing in the ATOM section, are in gray. The interface is shown by the mesh surface in the native 3D structure (A), by the shaded regions (B), and by transparent boxes in the alignments (C).
The binding site distortion (albeit a smaller one) is also observed in the 1 Å model, which was obtained from the NEB trajectory with the native X-ray structure and 3.13 Å homology model as the start and the end points, respectively. Both peaks in the Cα–Cα distance distribution [left-hand panel in Fig. 2(B)] for this model are caused by crystallographically unresolved regions in the template 2h16 [gray regions in Fig. 2(C)], which caused these parts of the model to be built ab initio (and thus with lower accuracy).
The peaks in the Cα distance distributions, corresponding to nonaligned residues, were observed in all models. Such relatively big local distortions are characteristic in homology models and cannot be completely avoided. On the other hand, it was shown previously that due to the stronger conservation of protein–protein binding sites, alignments of the interface sequence fragments tend to contain fewer gaps compared to the rest of alignments.32 Thus, for further consideration, we chose models with the least pronounced peaks and, thus, the lowest level of distortion in the binding region. Finally, all candidate models were visually inspected to exclude those with large distorted parts, corresponding to structural segments built ab initio, due to big alignment gaps, or structural defects in the template PDB files.
In the majority of cases, to build the low-energy path between two protein conformations, homology models of the same protein were used as the endpoints for NEB. The intermediate NEB structures should inevitably reproduce (some) structural properties of the endpoints. However, we realize that such correspondence is not strict and may depend on the similarity between the endpoints. In this sense, NEB models are not exactly homology models but “homology-like” models.
In our analysis, we investigated the effect of potentially mis-charged residues on the structure deformations in our models. Our benchmark set has to contain plausible (typical) homology-like models, but not necessarily high-quality ones. The initial set of models was obtained by single-template HM using NEST to mimic high-throughput real-case scenario. The program uses the default parameters and does not allow user control of the charge state of individual residues. The inaccuracies in conformations of individual residues obtained by NEST should follow those inherent in homology models. However, this may not hold for the NEB models. Comparison of Cα deviations in homology and NEB structures [see 6 Å models in Supporting Information, Fig. S1(A)] showed that the histidines in NEB models are on average more distorted than in homology models. This difference is statistically significant according to two-sample Kolmogorov–Smirnov (K–S) test at 95% confidence level.
However, such analysis cannot unambiguously answer the question whether these differences are caused by improperly set charges or the modeling procedure itself. To better understand the results for histidines, we performed the same analysis for the other 19 amino acids. In most cases (92.5%), the K–S test showed statistically significant differences between homology and NEB models [Supporting Information Fig. S1(B)]. At the same time, the distortions in histidines were similar to the average distortions in all other types of residues in NEB models (6 Å models are shown in Supporting Information Fig. S2), confirmed by the K–S test. Thus, the modeling procedure itself (NEB) is likely the main source of the distortions.
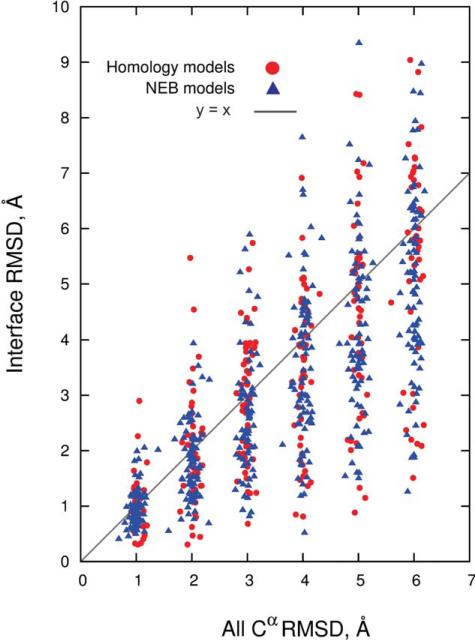
All models were also evaluated based on Cα RMSD values for the interface residues alone (Fig. 3). The interface residues in each of the 126 proteins in the set were extracted at 6 Å cutoff from the corresponding X-ray structures of the complexes, and superimposed with the equivalent residues in the models. The results in Figure 3 show that distortions at the interfaces are generally smaller than in full structures, although variations in RMSD are high. The correlation coefficient between Cα RMSD of the entire structure and the interface is 0.72, which is statistically significant.
Figure 3.
Cα RMSD of the entire structure versus interface.
Current modeling approaches are assessed in the Critical Assessment of Structure Prediction (CASP).33 To show that our final models are similar to those that could be obtained in real-case scenario, we compared our models with the latest CASP results in terms of correlation between overall RMSD and the global distance test GDT_TS score34 (the score is a major criterion in CASP for accessing model quality). The GDT algorithm reflects both local and global structural distortions by several superimpositions with different cutoff values. At each cutoff, the procedure finds superimposition that maximizes the number of Cα–Cα pairs within the cutoff. If some distortions are tolerated at large cutoffs, they should still appear at smaller ones. The similarity of correlation for both datasets (Fig. 4) indicates appropriateness of our procedures for generating structures resembling the real-case scenario protein models. The data in Figure 4 also show that each RMSD range contains models of different quality (wide distribution of GDT_TS scores) pointing to the overall representativeness of the set. More even distributions of Cα–Cα distances usually correspond to the lower values of the GDT_TS score.
Figure 4.
Comparison of the quality of the distorted protein structures with CASP predictions. CASP server predictions are in orange and human predictions are in gray. Models built in this study are in red (homology) and blue (NEB).
Web interface
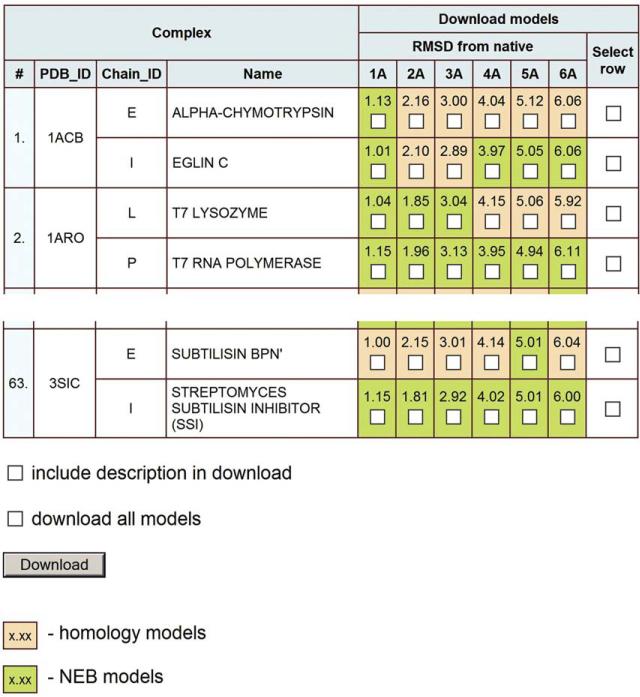
The benchmark set of protein models for 63 binary complexes is available within the Dockground resource at http://dockground.bioinformatics.ku.edu/MODEL/request.php (Fig. 5). The first four columns of the table contain brief information on the complexes, followed by six columns with exact RMSD values for the generated models, along with checkboxes to select the models for customized download. Cells are colored according to the model type: orange for the homology models and green for the NEB models. An option to select all six models for a particular protein chain is provided in the last column. The “download all models” box downloads the entire benchmark set. The selected models are downloaded as a single ZIP file containing PDB files of the models. The ATOM section of the model files contains only residues in the initial X-ray structure, but the entire sequence of the chain is included in the SEQRES section. Brief information on the model (the model type, HM or NEB, RMSD, and GDT_TS values, templates for homology models or end points for the NEB trajectory) is in the REMARK section.
Figure 5.
Web interface for the benchmark set of protein models.
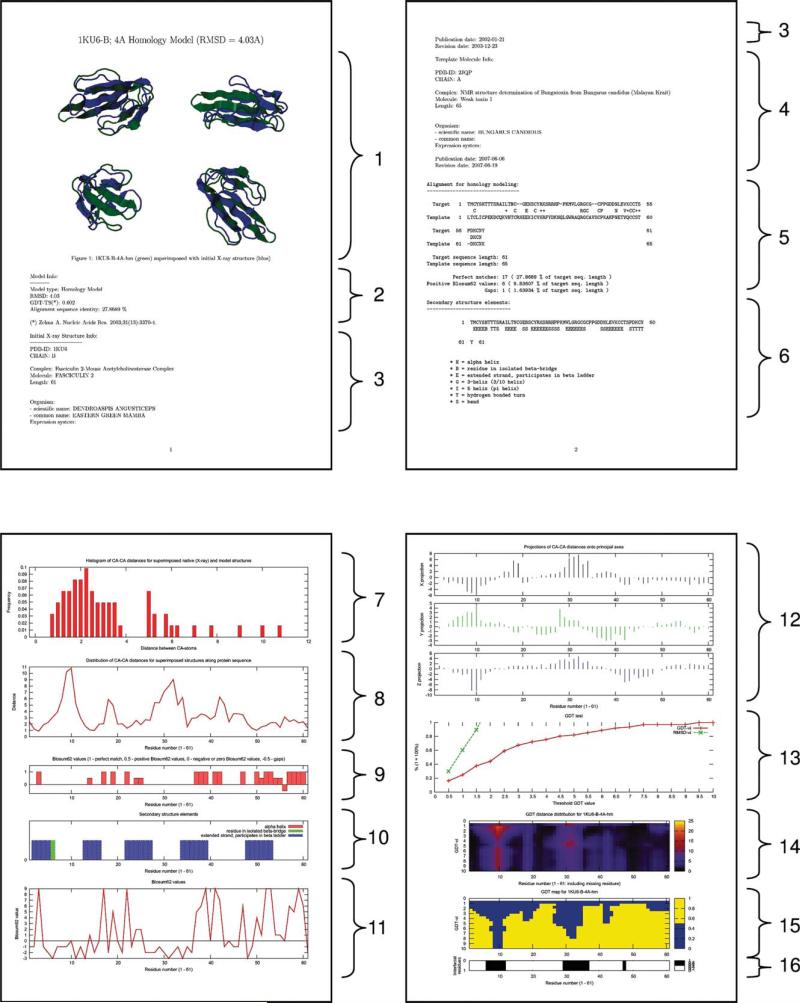
If the box “include description in download” is checked, each PDB file is accompanied by a PDF file with a detailed description of the model. The PDF file includes a description of proteins used as templates for modeling as well as extensive data on the results of the model analysis. The file (example in Fig. 6) contains images of superimposed native X-ray and modeled structures, information on the model type (HM or NEB), RMSD and GDT_TS values, data on the initial X-ray structure and the template used for HM, target/template sequence alignment, secondary structure elements, start and end points for the low-energy path in NEB models, Cα–Cα distances for superimposed native and model structures, distribution of Cα–Cα distances for superimposed structures along the protein sequence, BLOSUM62 values for the amino acid sequence of the model, graphical representation of the secondary structure elements distribution along the protein sequence, distribution of Cα–Cα distances for superimposed native and model structures in projections onto the principal axes of the molecule, visual representation of the GDT_TS test results, and the location of interface residues.
Figure 6.
Information on the complex from the accompanying downloadable file. The 4 Å homology model of 1ku6, Chain B, is characterized by: (1) images of the superimposed native X-ray and modeled structures; (2) information on the model type (HM or NEB), RMSD and GDT_TS values; data on the initial X-ray structure (3) and the template (4) used in homology modeling, both retrieved from PDB; (5) target/template sequence alignment; (6) secondary structure elements in the model structure as defined by DSSP (in Sections 4–6, PDF files for NEB models contain information on both proteins, which were used as start and end points of the low-energy path); (7) histogram of Cα–Cα distances for superimposed native (X-ray) and modeled structures; (8) distribution of Cα–Cα distances for superimposed structures along the protein sequence; (9 and 11) BLOSUM62 values for the amino acid sequence of the model from the alignment (5) (Sections 9 and 11 are provided for HM models only); (10) graphical representation of the secondary structure elements distribution (6) along the protein sequence; (12) distribution of Cα–Cα distances for superimposed native and model structures along the protein sequence (8) in projections onto the principal axes of the molecule; (13–15) visual representation of the GDT_TS test results; and (16) location of the interface residues within the protein sequence.
CONCLUSIONS AND FUTURE DIRECTIONS
The docking approaches often have to rely on modeled rather than experimentally determined structures of the interactors. Structures of modeled proteins are typically less accurate than the ones determined by X-ray crystallography or nuclear magnetic resonance. Thus the utility of approaches to dock these structures should be assessed by thorough benchmarking specifically designed for protein models. To be credible, such benchmarking has to be based on carefully curated sets of structures with levels of distortion typical for the modeled proteins. This article presents such a suite of models based on the benchmark set of the X-ray structures from the Dockground resource (http://dockground.bioinformatics.ku.edu) by a combination of HM and NEB method. For each monomer, six models were generated with predefined Cα RMSD from the native structure (1, 2, . . ., 6 Å). The sets and the accompanying data provide a comprehensive resource for the development of docking methodology for modeled proteins.
Our future research will focus on two major directions. First, a larger, more representative set of protein models, based on the bound Dockground benchmark will consist of several hundreds of protein–protein complexes, with corresponding arrays of models, as opposed to 63 in the current set, which is based on the much smaller Dockground unbound benchmark. We will also explore alternative methods for model generation (e.g., threading combined with refinement trajectories), which may potentially provide a larger percentage of actual models, and decrease or eliminate the fraction of the artificially generated intermediate distorted structures. Second, we will systematically benchmark the template-free and template-based docking methods to determine their applicability to modeled proteins of various accuracies. The results obtained on the smaller set presented in this article will allow comparison of the models docking to the docking of unbound X-ray structures (traditional benchmark of docking methodologies), whereas the results on the larger set will assure greater statistical significance. This will also facilitate the development of the docking approaches adequately accommodating the limited accuracy of the protein models.
Supplementary Material
Acknowledgments
Grant sponsor: NIH; grant number: R01GM074255.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Levitt M. Nature of the protein universe. Proc Natl Acad Sci USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang Y, Hubner IA, Arakaki AK, Shakhnovich E, Skolnick J. On the origin and highly likely completeness of single-domain protein structures. Proc Natl Acad Sci USA. 2006;103:2605–2610. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skolnick J, Arakaki AK, Lee SY, Brylinski M. The continuity of protein structure space is an intrinsic property of proteins. Proc Natl Acad Sci USA. 2009;106:15690–15695. doi: 10.1073/pnas.0907683106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vakser IA, Kundrotas P. Predicting 3D structures of protein–protein complexes. Curr Pharm Biotechnol. 2008;9:57–66. doi: 10.2174/138920108783955209. [DOI] [PubMed] [Google Scholar]
- 5.Kozakov D, Brenke R, Comeau SR, Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 6.Mintseris J, Pierce B, Wiehe K, Anderson R, Chen R, Weng Z. Integrating statistical pair potentials into protein complex prediction. Proteins. 2007;69:511–520. doi: 10.1002/prot.21502. [DOI] [PubMed] [Google Scholar]
- 7.de Vries SJ, van Dijk M. Bonvin AMJJ. The HADDOCK web server for data-driven biomolecular docking. Nat Protoc. 2010;5:883–897. doi: 10.1038/nprot.2010.32. [DOI] [PubMed] [Google Scholar]
- 8.Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Mirkovic N, Rossi A, Marti-Renom MA, Fiser A, Webb B, Greenblatt D, Huang CC, Ferrin TE, Sali A. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2004;32:D217–222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc Natl Acad Sci USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimm V, Zhang Y, Skolnick J. Benchmarking of dimeric threading and structure refinement. Proteins. 2006;63:457–465. doi: 10.1002/prot.20878. [DOI] [PubMed] [Google Scholar]
- 11.Kundrotas PJ, Lensink MF, Alexov E. Homology-based modeling of 3D structures of protein–protein complexes using alignments of modified sequence profiles. Int J Biol Macromol. 2008;43:198–208. doi: 10.1016/j.ijbiomac.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 12.Kundrotas PJ, Zhu Z, Janin J, Vakser IA. Templates are available to model nearly all complexes of structurally characterized proteins. Proc Natl Acad Sci USA. 2012;109:9438–9441. doi: 10.1073/pnas.1200678109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakser IA. Low-resolution structural modeling of protein interactome. Curr Opin Struct Biol. 2013;23:198–205. doi: 10.1016/j.sbi.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Samudrala R, Levitt M. Decoys ‘R’ Us: a database of incorrect conformations to improve protein structure prediction. Protein Sci. 2000;9:1399–1401. doi: 10.1110/ps.9.7.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbajo D, Tramontano A. A resource for benchmarking the usefulness of protein structure models. BMC Bioinformatics. 2012;13:188. doi: 10.1186/1471-2105-13-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brylinski M, Skolnick J. Q-DockLHM: low-resolution refinement for ligand comparative modeling. J Comput Chem. 2010;31:1093–1105. doi: 10.1002/jcc.21395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douguet D, Chen HC, Tovchigrechko A, Vakser IA. DOCK-GROUND resource for studying protein–protein interfaces. Bioinformatics. 2006;22:2612–2618. doi: 10.1093/bioinformatics/btl447. [DOI] [PubMed] [Google Scholar]
- 18.Hwang H, Vreven T, Janin J, Weng Z. Protein–protein docking benchmark version 4.0. Proteins. 2010;78:3111–3114. doi: 10.1002/prot.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tovchigrechko A, Wells CA, Vakser IA. Docking of protein models. Protein Sci. 2002;11:1888–1896. doi: 10.1110/ps.4730102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao Y, Douguet D, Tovchigrechko A, Vakser IA. DOCKGROUND system of databases for protein recognition studies: unbound structures for docking. Proteins. 2007;69:845–851. doi: 10.1002/prot.21714. [DOI] [PubMed] [Google Scholar]
- 21.Elber R, Karplus M. A method for determining reaction paths in large molecules—application to myoglobin. Chem Phys Lett. 1987;139:375–380. [Google Scholar]
- 22.Chu JW, Trout BL, Brooks BR. A super-linear minimization scheme for the Nudged Elastic Band method. J Chem Phys. 2003;119:12708–12717. [Google Scholar]
- 23.Needleman S, Wunsch CD. A general method applicable to search for similarities in amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 24.Gotoh O. An improved algorithm for matching biological sequences. J Mol Biol. 1982;162:705–708. doi: 10.1016/0022-2836(82)90398-9. [DOI] [PubMed] [Google Scholar]
- 25.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of database programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Henikoff S, Henikoff JG. Performance evaluation of amino acid substitution matrices. Proteins. 1993;17:49–61. doi: 10.1002/prot.340170108. [DOI] [PubMed] [Google Scholar]
- 27.Petrey D, Xiang ZX, Tang CL, Xie L, Gimpelev M, Mitros T, Soto CS, Goldsmith-Fischman S, Kernytsky A, Schlessinger A, Koh IYY, Alexov E, Honig B. Using multiple structure alignments, fast model |building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53:430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- 28.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 29.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized Born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 30.Case DA, Darden TA, Cheatham TE, Simmerling C, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossviry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. AMBER 10. University of California; San Francisco: 2008. [Google Scholar]
- 31.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman PA. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 32.Kundrotas PJ, Vakser IA. Accuracy of protein–protein binding sites in high-throughput template-based modeling. PLoS Comp Biol. 2010;6:e1000727. doi: 10.1371/journal.pcbi.1000727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moult J, Fidelis K, Kryshtafovych A, Tramontano A. Critical assessment of methods of protein structure prediction (CASP)—round IX. Proteins. 2011;79(Suppl 10):1–5. doi: 10.1002/prot.23200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zemla A. LGA: a method for finding 3D similarities in protein structures. Nucl Acid Res. 2003;31:3370–3374. doi: 10.1093/nar/gkg571. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.