Abstract
Cryptosporidium causes significant diarrhea worldwide, especially among children and immunocompromised individuals, and no effective drug treatment is currently available for those who need it most. In this report, previous volunteer infectivity studies have been extended to examine the association between fecal indole and indole-producing (IP) gut microbiota on the outcome of a Cryptosporidium infection. Fecal indole concentrations (FICs) of 50 subjects and 19 taxa of common gut microbiota, including six IP taxa (11 subjects) were determined in stool samples collected before and after a challenge with Cryptosporidium oocysts. At the baseline, the mean FIC (± the standard deviation) was 1.66 ± 0.80 mM in those who became infected after a challenge versus 3.20 ± 1.23 mM in those who remained uninfected (P = 0.0001). Only 11.1% of the subjects with a FIC of >2.5 mM became infected after a challenge versus 65.2% of the subjects with a FIC of <2.5 mM. In contrast, the FICs of infected subjects at the baseline or during diarrhea were not correlated with infection intensity or disease severity. The relative abundances (percent) of Escherichia coli, Bacillus spp., and Clostridium spp. were greater ≥2.5-fold in volunteers with a baseline FIC of >2.5 mM, while those of Bacteroides pyogenes, B. fragilis, and Akkermansia muciniphila were greater in those with a baseline FIC of <2.5 mM. These data indicate that some IP bacteria, or perhaps indole alone, can influence the ability of Cryptosporidium to establish an infection. Thus, preexisting indole levels in the gut join the oocyst dose and immune status as important factors that determine the outcome of Cryptosporidium exposure.
INTRODUCTION
Cryptosporidium exposure is a health hazard in almost all populations, particularly among children (1, 2). While the infection is typically self-limited in immunocompetent adults, the very young and the immunocompromised often experience persistent and sometimes life-threatening infections. The only approved drug (nitazoxanide) is largely ineffective, particularly in immunocompromised persons (reviewed in reference 3). Thus, there is renewed interest in new treatment and prevention strategies.
The human gut microbiota exerts a significant influence in a variety of illnesses and conditions, but few studies regarding gut bacteria have been done with Cryptosporidium. Early work noted that young animals are susceptible to Cryptosporidium infection but become refractory after maturation of the gut with attendant changes in the microbiota (4). Cryptosporidium studies using probiotic formulations reported varied results but generally showed a decreased intensity and/or duration of oocyst shedding rather than prevention of infection. For example, a delay in oocyst production was seen in SCID mice colonized with a mixture of anaerobic bacteria (5). Further, immunosuppressed adult mice fed lactobacilli became infected after a Cryptosporidium parvum challenge but had reduced numbers of oocysts and enhanced clearance (6, 45). In contrast, feeding of probiotics containing lactobacilli to neonatal immunocompetent rats had no effect on oocyst shedding (7).
The only positive effect on a human was in a young celiac patient with persistent cryptosporidiosis. A 4-week treatment with Lactobacillus GG and Lactobacillus casei was associated with resolution of diarrhea and oocyst shedding (8), but no positive effect on duration or severity of diarrhea was found when lactobacilli were fed to infants (9) or children with cryptosporidiosis (10). However, other gut microbiota and/or their products may be important. Although limited, evidence suggests an active role for gut microbiota in the host response. In neonatal mice, control of and protection from Cryptosporidium infection depended on innate responses requiring microbiota (11). Further, in Toxoplasma, a TLR9 response that contributed to TH1 development depended on bacterial antigens (12). Similarly, neonatal mice fed TLR9 agonists had an enhanced TH1 response after a Cryptosporidium challenge (13).
The human gut is host to >1,000 taxa, which vary in abundance and composition among individuals. Thus, the identification of particular microbial species that influence Cryptosporidium infections is daunting. One approach is to use a biomarker such as indole, which represents the 85 known Gram-positive and Gram-negative gut bacteria capable of producing it (reviewed in reference 14). In a recent study, the fecal indole concentrations (FICs) in a majority (75%) of 53 healthy persons ranged from 1.0 to 4.0 mM (15), presumably reflecting differences in the number of indole-producing (IP) bacteria or the regulatory microenvironment controlling indole expression (16, 17). Indole's effects include regulation of bacterial motility, epithelial barrier integrity, and biofilm formation (reviewed in references 14 and 18), as well as changes in the virulence and drug resistance of enteric pathogens (19–22). Further, oral administration of indole to germfree mice enhanced intestinal integrity (23) and indole added to cell cultures was anti-inflammatory (24). Lastly, cells exposed to indole showed differential regulation of >4,000 genes involved in a variety of functions, including barrier enhancement (25, 26).
Little is known about any direct or indirect effect of indole on Cryptosporidium, although some aspects of the related tryptophan metabolism have been examined. Interestingly, Cryptosporidium is one of the few protists that can utilize indole to synthesize tryptophan (27). Parasite replication is inhibited by gamma interferon (IFN-γ) treatment of enterocytes, which depletes intracellular tryptophan (28). However, exogenous tryptophan did not enhance Cryptosporidium survival after invasion (29) nor did the parasite apparently synthesize tryptophan to replace depleted levels in IFN-γ-treated enterocytes. Instead, during infection, Cryptosporidium circumvents the host response by inhibiting IFN-γ production and thus avoids tryptophan depletion (30).
We hypothesized that if the parasite is not using available indole as a substrate, then high indole levels might exist in the gut and play a role at some stage of the life cycle. To investigate this hypothesis, we assessed IP gut bacteria by measuring the FIC at the time of a Cryptosporidium challenge and then examined the clinical and microbiological outcomes of the challenge. To this end, we examined stool samples from 50 well-characterized, healthy individuals previously challenged with Cryptosporidium (reviewed in references 31 and 32). Specifically, we (i) examined the association of baseline FICs with exposure outcomes, (ii) measured changes in the FIC at two time points during infection and illness, (iii) explored associations between the FICs at the baseline and during illness by using various infection and disease parameters, and (iv) determined the percent relative abundances (%RAs) of common gut bacterial taxa in infected and uninfected subjects at the baseline and at postchallenge time points.
MATERIALS AND METHODS
Study population and selection of samples.
The stool samples used in the present study were selected from volunteers who were originally enrolled in Cryptosporidium infectivity studies from 1993 to 2004 (reviewed in references 31 and 32). Selected subjects had received one Cryptosporidium hominis isolate or one of five C. parvum isolates. The volunteer study number, challenge isolate, challenge outcome, and day of stool sample collection are shown in Fig. S1 in the supplemental material. A full set of three stool samples was available for 49 (98%) of the volunteers. All stool samples collected from infected subjects during the illness period were obtained at the time of diarrhea, except for that of one subject (number 81), who had diarrhea on the day prior to stool sample collection. The ages, genders, and ethnicities of all of the subjects were known. The Cryptosporidium volunteer studies were approved by the Committee for Protection of Human Subjects at The University of Texas Health Science Center at Houston and the U.S. Food and Drug Administration Research Involving Human Subjects Committee.
The present study was limited to two groups, uninfected and asymptomatic (i.e., no oocysts detected and no gastrointestinal symptoms) versus infected with diarrhea (i.e., oocysts detected in one or more stool samples and a diarrheal illness). Two other groups ([i] oocyst positive and asymptomatic and [ii] oocyst negative with diarrhea) from the original Cryptosporidium volunteer study were not included. Of the 50 subjects included in the present study, 32 were uninfected after a Cryptosporidium challenge, while 24 were infected. Stool samples were selected from time periods corresponding to the following stages: baseline (prechallenge to day 2 postchallenge; stage 1), illness period (days 3 to 10; stage 2), and resolution of the infection and illness (day ≥27; stage 3). Eleven volunteers were further studied for 19 bacteria common in the human gut. Six (volunteers 5, 29, 119, 120, 132, and 178) were infected, and five (volunteers 9, 123, 130, 156, and 185) were uninfected. Full sets of stool samples from nine volunteers were available. Stage 1 and 3 data from volunteers 123 and 178, respectively, were missing because of failed PCR amplification.
Indole measurement.
Stool samples collected from subjects were stored at −80°C. Stage 1 samples were previously tested for FIC (15), while samples from stages 2 and 3 were tested for the present study. All samples were prepared for indole testing immediately after thawing. Briefly, a 250-mg stool sample was diluted in 750 μl of 70% ethanol (1:4, wt/vol) and extracts were assayed in duplicate. Absorbances (530 nm) were compared to a standard curve of known indole concentrations (0 to 100 μM). All standard curves for indole measurement had an r2 value of ≥0.98.
Selection of bacteria, DNA extraction, and 16S rRNA gene amplification.
Common bacterial taxa (n = 19) found in adults (33–35) were selected for this study (Table 1). Genomic DNA was isolated from 250-mg stool samples with the QIAamp DNA Stool minikit (Qiagen, Germantown, MD) according to the manufacturer's instructions. The 16S rRNA gene was amplified with broad-range, bacterium-specific primers 8F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1391R (5′-GACGGGCGGTGTGTRCA-3′) (36), which amplify 90% or more of the full-length bacterial 16S rRNA gene sequence (∼1,400 bases). Extracted DNA (2 μl) was used as the template in 20-μl PCR mixtures containing 1× MyTaq mix (Bioline USA, Inc., Taunton, MA) and 0.5 pmol each of the forward and reverse primers. The PCR conditions used were 5 min at 95°C; 22 cycles of 30 s at 94°C, 30 s at 55°C, and 90 s at 72°C; and 8 min at 72°C (36). Amplification was done with a Thermo Hybaid PCR Express Thermal Cycler (ThermoFisher Scientific, Waltham, MA). Two 50-μl reconditioning PCRs were done under the same conditions with 5 μl of the first PCR product mixture. The products of the two reconditioning PCRs were pooled and purified with 96-well, 0.7-ml filter plates filled with Sephacryl S-300 high-resolution suspension (GE Healthcare, Chicago, IL). Purified products were quantitated with a NanoDrop ND-1000 spectrophotometer (ThermoFisher Scientific, Wilmington, DE) and stored at −20°C.
TABLE 1.
Common human gut microbiota detected in stool samples from healthy adultse
| Organism(s) | Code | rRNA copy no. | Target sequence |
|---|---|---|---|
| Acidaminococcus intestinalis/intestini | R1, 2 | 3 | GAAGGTCTTCGGATTGTAAAACTCTGTTGTTAGGGACGAAAGCACCGT |
| Akkermansia muciniphila | R3 | 3 | TGAAGGTCTTCGGATTGTAAACCCCTGT |
| Anaerostipes butyraticus | R4 | 6 | GAAGTATTTCGGTATGTAAAGC |
| Bacillus spp. | R5 | 9.5a | GAAGGTTTTCGGATCGTAAAACTCTGTTGTGAGGGAAGAACAAGTAC |
| Bacteroides fragilis | R6 | 6 | GAAGGCTCTATGGGTCGTAAACTTCTTTTATATAAGAATAAA |
| Bacteroides pyogenes | R7 | 6 | GACTGCCCTCTGGGTTGTAAACTTCTTTTATACGGGAATAACA |
| Bifidobacterium longum | R8 | 4 | GGAGGCCTTCGGGTTGTAAACCTCTTT |
| Citrobacter gillenii | R9 | 7 | GAAGGCCTTCGGGTTGTAAAGTACTTTCAGCGAGGAGGAAGGGGATG |
| Clostridium spp. | R10 | 7b | AGGCCTTCGGGTCGTAAAGC |
| E. coli CFT073 | R11 | 7 | AAGAAGGCCTTCGGGTTTGTAAAGTACTTTCAGCGGGGAGGAAGGGAGTA |
| Faecalibacterium prausnitzii | R12 | 1 | GAAGGTCTTCGGATTGTAAACT |
| Lactobacillus plantarum | R13 | 5 | GAAGGGTTTCGGCTCGTAAAACTCTGTTGTTAAAGAAGAACATATCT |
| Prevotella bryantii | R14 | 4 | GACGGCCCTATGGGTTGTAAACTGCTTTTTTAGGGGAATAAA |
| Raoultella spp. | R15, 16 | 8 | GAAGGCCTTCGGGTTGTAAAGTACTTTCAGCGAGGAGGAAGGCGTTA |
| Ruminococcus albus | R17 | 4 | GAAGGTTTTAGGATTGTAAACC |
| Subdoligranulum variable strain BI 114 | R18 | 6 | GAAGGTTTTCGGATTGTAAACTCC |
| Uncultured Dorea | R19 | 6.5c | GAAGTATTTCGGTATGTAAACT |
| Uncultured Firmicutes | R20 | 12.5d | GAAGGCTTTCGGGTTGTAAACTTCT |
| Uncultured Mollicutes | R21 | 2 | GACGTATTTCGGTATGTAAAATTCTTTTATTAGGGAAGAAC |
Range of 6 to 14 genes, with the majority having 8 to 11 genes. The median value is shown.
Range of 4 to 15 genes, with the majority having 6 to 11 genes. The value shown is for the predominant species.
Range of 5 to 8 genes. The median value is shown.
Range of 2 to 15 genes, with the majority having 6 to 15 genes. The value shown is for the predominant species.
Identifications of bacterial species/groups were based on the target sequences listed. The copy number of ribosomal genes for each bacterial species/group was determined on the basis of data from the NCBI database.
Preparation of sequencing library.
PCR products were concatenated via ligation before the construction of a resequencing library. Concatenated products (∼1 μg) were digested with the NEBNext dsDNA Fragmentase kit (New England BioLabs, Inc., Ipswich, MA) as instructed by the manufacturer. Fragmented DNA was end repaired with the NEBNext End Repair Module (New England BioLabs, Inc.). End-repaired DNA was ligated with the P1 and unique barcode adaptors (5500 SOLiD Fragment Library Barcode Adaptors 1 to 96; Applied Biosystems, ThermoFisher Scientific); this was followed by purification and size selection as recommended. All reaction products were purified with the Isolate II PCR kit (Bioline USA, Inc.). Agencourt AMPure XP beads (Beckman Coulter, Inc., Brea, CA) were used to selectively capture DNA between 100 and 250 bp. These libraries underwent nick translation and seven cycles of amplification. Experion DNA chips (Bio-Rad Laboratories, Inc., Hercules, CA) were used to confirm fragment lengths and concentrations. Equal molar amounts of all libraries were pooled and subjected to resequencing analysis on a 5500W Series Genetic Analysis System (ThermoFisher Scientific) to generate sequence reads of 55 bp.
Data analysis. (i) Indole measurements.
The cumulative percent infection of each of the various FIC categories was estimated (37). The mean (± standard deviation [SD]) FICs of the outcome groups (infected and uninfected) were compared by using the Mann-Whitney test. The FIC changes during stages 2 and 3 were calculated by subtracting the postchallenge FIC from the stage 1 FIC. The percent change for each individual and time point was calculated by dividing the postchallenge FIC by the stage 1 value. Student's t test was used with or without Welch's correction to compare measurements of change and to analyze various parameters describing the severity of illness and the intensity of infection. A P value of <0.05 was considered significant. All statistical analyses were done with Prism 6 software (GraphPad Instat 3.01; GraphPad Software, Inc.).
(ii) Sequence analysis.
Data analyses were performed with LifeScope software (version 2.5.1; ThermoFisher Scientific). Sequence reads were aligned with a reference sequence of each bacterial species downloaded from the National Center for Biotechnology Information (NCBI) database. The sequencing reads were passed through quality filters to reduce the overall error rate, and reads shorter than 25 nucleotides were discarded from the alignment with the reference genome. The unique target region of each bacterial species was generated by reducing 55 nucleotides from each end of the V3 region to identify the individual read.
(iii) %RA measurements.
To represent the %RA of each taxon, the read depth (RD) was divided by the number of ribosomal genes present. When a range of copy numbers occurred within a genus, the predominant number was used. When there was no predominance, the median value was calculated from the range. The adjusted RD was then used to calculate the %RA of each taxon. The median %RAs of the two outcome groups were calculated and compared by using the Mann-Whitney test since a limited number of individuals were studied and there was a nonnormal distribution of values within each group.
RESULTS
Earlier Cryptosporidium volunteer studies provided a unique bank of stool samples for use with new indole assays and techniques to study microbiota. Subjects whose stool samples were collected before and after a challenge were selected to represent two outcome groups, those with no evidence of infection and those exhibiting both infection and illness. Demographic information for each Cryptosporidium challenge outcome group indicated that the groups were similar in age but had more females than males, especially in the infected group (Table 2). Ethnic representation was similar for Caucasian and Hispanic/Asian subjects, but black subjects were slightly underrepresented in the infected group. Stage 1 FICs did not significantly differ between the outcome groups according to age, gender, or ethnicity or among the C. parvum isolates used (data not shown). Therefore, the data from all C. parvum isolates were collapsed into one group for analyses.
TABLE 2.
Numbers of subjects in the study population in different age, sex, and ethnicity groupsa
| Characteristic | No. (%) of subjects |
Mean baseline indole concn (mM) ± SD | |
|---|---|---|---|
| Uninfectedb | Infectedc | ||
| Total | 32 (64.0) | 18 (36.0) | |
| Age (yr) | |||
| <20 | 0 (0.0) | 1 (5.6) | 1.36 |
| 20–29 | 16 (50.0) | 9 (50.0) | 2.74 ± 1.55 |
| 30–39 | 8 (25.0) | 4 (22.2) | 2.31 ± 1.07 |
| >40–50 | 8 (25.0) | 4 (22.2) | 2.84 ± 0.99 |
| Sex | |||
| Male | 15 (46.9) | 8 (44.4) | 2.65 ± 1.55 |
| Female | 17 (53.1) | 10 (65.6) | 2.64 ± 1.14 |
| Ethnicity | |||
| Caucasian | 15 (46.9) | 9 (50.0) | 2.70 ± 1.49 |
| Black | 10 (31.3) | 4 (22.2) | 2.84 ± 0.91 |
| Hispanic/Asian | 7 (21.9) | 5 (27.8) | 2.30 ± 1.35 |
| Challenge species/isolate | |||
| C. parvum | 26 (81.3) | 15 (83.3) | 2.57 ± 1.41 |
| C. hominis | 6 (18.8) | 3 (16.7) | 2.96 ± 0.73 |
Total n = 50. Volunteers were challenged with C. parvum or C. hominis and monitored for outcomes (no oocysts or diarrhea, oocysts plus diarrhea). One Asian subject was grouped with Hispanic volunteers. Mean FICs and standard deviations were measured in stool samples collected before or within 48 h of the challenge.
No oocysts or diarrhea.
Oocysts plus diarrhea.
Stage 1 FICs and infection outcomes.
New information regarding the various effects microbiota and their products have on many illnesses has expanded dramatically in the last few years. We could now examine if indole, the product of several common gut bacteria, could have an effect on the susceptibility of individuals to infection and/or illness caused by Cryptosporidium. Previously, healthy subjects showed a wide distribution of FICs, with ∼75% in the 1 to 4 mM range (15), and 80% of the baseline indole levels in a subset of 50 subjects used in the present study were in the same range. We then went a step further and examined stage 1 FICs with regard to the outcome of a Cryptosporidium challenge (Fig. 1A and B). The percentage of volunteers who became infected after a challenge decreased as preexisting FICs increased (Fig. 1A). Fifteen (65.2%) subjects with a stage 1 FIC of ≤2.5 mM versus three (11.1%) with a FIC of >2.5 mM became infected after a challenge. Further, none of 11 subjects with a FIC of ≥4.0 mM became infected. The mean (± SD) stage 1 FICs in infected versus uninfected subjects after Cryptosporidium exposure were 1.66 ± 0.80 and 3.20 ± 1.23 mM, respectively (Fig. 1B, P = 0.0001). These data suggest that the stage 1 FIC may be a marker of susceptibility to Cryptosporidium infection.
FIG 1.

FICs in healthy adults at stage 1 (before or within 48 h of a challenge) and outcomes of a challenge with Cryptosporidium oocysts. (A) Cumulative percent infection was calculated as described previously (40) and plotted against the mean indole concentration. The number of subjects at each indole concentration stratum is in parentheses. (B) The indole concentrations and outcome categories of the subjects are shown. The mean indole concentration ± the standard error of the mean are indicated for the infected (oocysts plus diarrhea; n = 18) and uninfected (no oocysts or diarrhea; n = 32) challenge outcome groups. Outcome groups were compared by using the Mann-Whitney test (P = 0.0001).
To check for unintended bias, we further examined the oocyst dose received by the subjects in the FIC categories (i.e., ≤2.5 mM and >2.5 mM) and found no differences in the number of oocysts received in the challenge (data not shown). Also, stage 1 FICs were similar for volunteers who received any of the C. parvum isolates used in the study. Finally, we compared eight pairs of volunteers given the same isolate and oocyst dose and observed that 75% with an FIC of >2.5 mM remained uninfected, while volunteers with an FIC of ≤2.5 mM became infected and ill.
FIC after a challenge.
Changes in the FIC during the course of the study were examined (Table 3). The FICs of 17 infected subjects and 32 uninfected, asymptomatic subjects were determined during stage 2. In addition, stool samples were collected at stage 3. Like the stage 1 FICs, the differences between the two outcome groups at stages 2 and 3 were also significant (P = 0.0001). FICs remained relatively stable in the uninfected group during the study. However, at stage 2, infected individuals had a decrease in the FIC (P = 0.053), a trend that reached statistical significance (P = 0.028) when the percent change in the FIC was calculated. At stage 3, indole levels increased and were not significantly different from those at stage 1.
TABLE 3.
Change in FIC after a Cryptosporidium challengea
| Outcome category | Illness periodb | P value | Resolution periodb | P value |
|---|---|---|---|---|
| Indole concn (mM) | ||||
| No oocysts or diarrhea | 3.43 ± 1.20 | 0.0001 | 2.92 ± 1.30 | 0.0001 |
| Oocysts + diarrhea | 1.41± 0.91 | 1.60 ± 0.68 | ||
| Change in indole concn (mM) | ||||
| No oocysts or diarrhea | 0.023 ± 1.21 | 0.053 | −0.31 ± 1.44 | 0.61 |
| Oocysts + diarrhea | −0.34 ± 0.75 | −0.13 ± 0.92 | ||
| Change in indole concn (%) | ||||
| No oocysts or diarrhea | 7.9 ± 9.3 | 0.028 | −2.10 ± 43.1 | 0.41 |
| Oocysts + diarrhea | −16.3 ± 9.5 | 33.1 ± 39.5 |
Subjects in each outcome category (no oocysts or diarrhea, n = 32; oocysts plus diarrhea, n = 18) were tested during the illness period and/or after resolution of the infection and diarrhea. The median indole concentration, change in indole concentration, and percent change are shown along with 95% confidence intervals for each parameter. For each volunteer, the change and percent change in indole concentration were calculated on the basis of the baseline indole concentration. P values were calculated by using Student's t test with or without Welch's correction.
Values are means ± standard deviations.
FICs in infected subjects were further examined for various outcome parameters. Stage 1 FICs showed no significant association with any illness severity (onset or duration of diarrhea, number of unformed stool samples, total stool weight) or infection intensity (onset or duration of oocyst shedding, total oocysts shed) measurements (see Table S1 in the supplemental material). The same analyses of stage 2 FICs were done, and no significant effects were noted, except for higher FICs, which were associated with a delayed onset of shedding (P = 0.017). Thus, the FIC had an association with susceptibility to infection rather than with the course of the infection once it was established.
Gut microbiota prior to a challenge.
Given the FIC findings, we examined the stool samples of 11 volunteers for the most common bacteria in the human gut, including five IP taxa (Table 1). These included bacteria from five phyla (Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, and Verrucomicrobia). The Firmicutes were dominant (45 to 79%RA) in both outcome groups and at all stages (see Fig. S2 in the supplemental material). Except for Actinobacteria, the other phyla varied between outcome groups, especially at stage 1, with distinct patterns noted. The uninfected group had a greater abundance of Proteobacteria (5.5%) with lower Bacteriodetes (3.4%) and Verrucomicrobia (0.5%) levels than infected subjects, who had greater abundances of Bacteriodetes (16.2%) and Verrucomicrobia (1.6%) and a lower abundance of Proteobacteria (2.2%). The ratio of Firmicutes to Bacteriodetes at stage 1 was approximately 27:1 in uninfected subjects versus 4:1 in infected subjects.
The median %RA (stage 1) per taxon in the two outcome groups was calculated. Seven taxa showed differences of ≥2.5-fold between the two groups. Specifically, uninfected subjects had increased %RAs of two IP bacteria, Escherichia coli CFT073 and Bacillus spp., as well as Clostridium spp. (Fig. 2). In contrast, infected subjects had increased %RAs of Bacteroides fragilis, B. pyogenes, and Prevotella bryantii (all IP bacteria), as well as Akkermansia muciniphila (Fig. 3). Thus, E. coli and Bacillus were the only IP bacteria studied that were associated with high FICs, indicating that one or both may be the major, but not necessarily the only, source of fecal indole.
FIG 2.
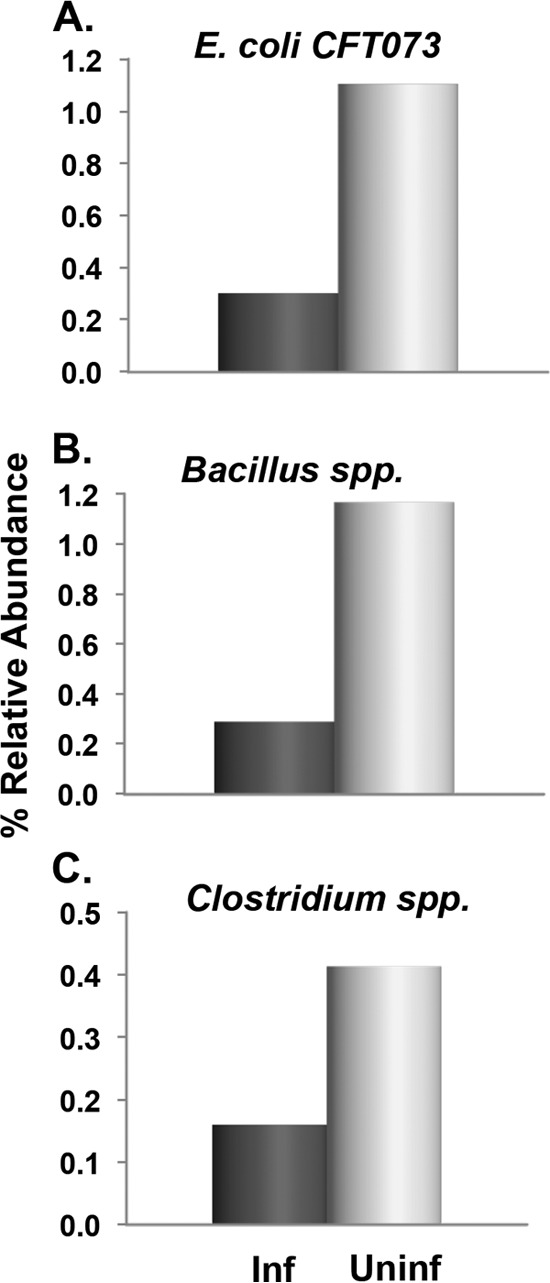
Fecal bacteria associated with protection from Cryptosporidium infection. Species with ≥2.5-fold difference in %RA are shown. Stool samples from healthy adults were collected prior to (or within 48 h of) exposure to Cryptosporidium oocysts. The median %RAs of the infected (Inf; n = 6) and uninfected (Uninf; n = 5) outcome groups are shown. IP bacteria include those in panels A and B.
FIG 3.
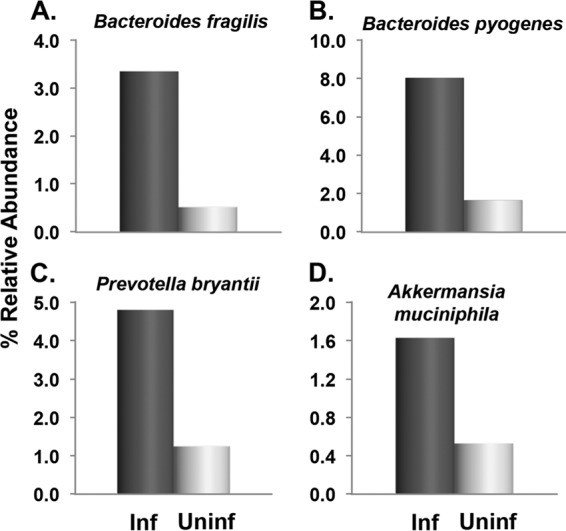
Fecal bacteria associated with susceptibility to Cryptosporidium infection. Species with ≥2.5-fold difference in %RA are shown. Stool samples from healthy adults were collected prior to (or within 48 h of) exposure to Cryptosporidium oocysts. The median %RAs of the infected (Inf; n = 6) and uninfected (Uninf; n = 5) outcome groups are shown. IP bacteria include those in panels A to C.
Gut microbiota during and after diarrhea.
The median %RAs of all 19 taxa were compared between the two outcome groups and at stages 2 and 3 (see Fig. S3 in the supplemental material). Faecalibacterium prausnitzii was dominant (>27%) in both outcome groups and at each stage. Shifts in the microbial population were most prominent during stage 2. Infected subjects had a 6-fold decrease in Clostridium spp., while all other taxa remained relatively unchanged from stage 1. Appreciable (i.e., ≥3-fold) changes were also noted in uninfected subjects, including increases in the %RA of Acidaminococcus spp. and decreases in those of Citrobacter gillenii, E. coli CFT073, and Raoultella spp. This suggests that oocyst exposure may influence the microbiota either directly or perhaps via a successful host response to the challenge. By stage 3, outcome groups were more similar to each other than previously with the levels of only two taxa, Bifidobacterium longum and Lactobacillus plantarum, ≥3-fold higher in the uninfected group. Further, the overall %RA of IP bacteria decreased from stage 1 values, especially in uninfected subjects, and were in contrast to the little change in the FICs over the same time. Lastly, there were too few infected subjects for meaningful comparisons between %RAs and the various outcome parameters.
DISCUSSION
In this study, we compared the FICs in healthy adults with the outcomes of a subsequent challenge with Cryptosporidium oocysts. Our findings demonstrate a highly significant association (P = <0.0001) between the stage 1 FIC and exposure outcome. Specifically, FICs of >2.5 mM were associated with protection from infection even when the oocyst dose exceeded the 50% infective dose (ID50). However, the association was strong but not absolute. That is, there were three infected individuals who had high stage 1 FICs (>2.5 mM) and three uninfected subjects with low FICs (<1.5 mM). The former received an oocyst dose in the ID48 to ID97 range, possibly sufficient in number to overcome the “protective” effect of the FIC noted in these subjects. In comparison, the latter three received relatively small challenge doses (ID8 to ID48), suggesting that the dose may have been insufficient to result in infection despite the low FICs.
We then examined a subset of these same stool samples for common gut bacteria found in humans. While six of these taxa were capable of producing indole, only two, E. coli and Bacillus spp., were associated with increased FICs. Interestingly, both are aerobic and expected to be in greater abundance in the small bowel, the typical site of Cryptosporidium infection. Thus, we presume that one or both could be the major source of fecal indole associated with protection. However, we cannot rule out the FIC contributions of other, less abundant, taxa. In contrast, two IP taxa, Bacteroides spp. and Prevotella bryantii, were associated with a low FIC. Both of these are anaerobic bacteria that mainly populate the colon.
Our earlier dose-response studies concluded that the variability found in the Cryptosporidium ID50 was indicative of differences in the diverse Cryptosporidium isolates used. We now suggest that IP microbiota and perhaps indole itself have important roles in the outcome. Dose-response curves were based on groups of volunteers whose FICs were unknown at the time. The present study consisted of subjects selected from each of these groups and is thus an incomplete representation of any one dose-response curve. Thus, the degree of influence that fecal indole has on the estimation of ID50 cannot be known at this point and will require the evaluation of stool samples from all of the volunteers included in each isolate's dose-response curve.
Our current findings support the notion that IP microbiota and/or indole are important but do not point to a particular mechanism. Several possibilities exist. First, indole may have a direct adverse effect on the parasite and/or invasion, an idea that is testable in cell culture. In this regard, we also recognize that diet plays an important role in the FIC by providing the necessary substrate (tryptophan) for indole production. Unfortunately, no dietary information was collected during the Cryptosporidium volunteer studies, so no further analysis of these individuals is possible. Alternatively, indole may have an indirect effect by acting as a regulatory molecule for other species of gut microbiota. This may have been evident, at least in part, in the positive effects seen previously in some studies testing probiotics (5, 6, 8, 11). Another possibility is that indole acts on host tissues to enhance the innate response by increasing epithelial integrity (23) and/or stimulating anti-inflammatory pathways (24).
Finally, it is possible that a high FIC alone is insufficient to provide protection and other factors or non-IP bacterial species are also required. Interestingly, under some conditions, Clostridium can regulate IP bacteria to increase indole production in vitro (C.D., unpublished data). This notion is supported by the increased abundance of Clostridium bacteria seen in uninfected subjects, which may be needed to reach the high levels of indole associated with protection. We further noted an increased abundance of A. muciniphila in infected subjects. This bacterium has been implicated in mucin degradation (38, 39) and increased susceptibility to infection (40, 41). Since Cryptosporidium sporozoites initiate infection via contact with enterocyte membranes, an increase in the %RA of Akkermansia found may erode the protective mucin covering, thus increasing the opportunity for sporozoite binding. Lastly, we cannot exclude the possibility that indole has no effect but is simply a convenient and easily measured biomarker of other unknown factors that impact host susceptibility to Cryptosporidium infection.
Changes in the microbial population were seen in both outcome groups despite the fact that indole levels remained remarkably stable in each group. The microbiota disturbances seen in infected individuals are consistent with recent observations in mice, where changes in gut microbiota were noted during infection but reverted to a prechallenge profile after the infection was resolved (42). However, the changes seen in mice were independent of symptoms since mice do not develop diarrhea and their infection may be quite different from human infection.
There are several limitations to our study. The stool samples we used had been stored at −80°C for 10 to 20 years, and to our knowledge, there is no published literature regarding indole stability under such conditions. However, our stage 1 indole levels were consistent with values from control individuals in two earlier studies using freshly collected stool samples (43, 44), although direct comparisons could not be made since stool samples were prepared, extracted, and tested by different techniques. Further, all of the stool samples we used were stored under the same conditions and the indole concentrations in samples from 1993 to 1998 were no different from those in samples from 1999 to 2004. Thus, we conclude that indole is relatively stable under these conditions. Second, we selected two groups of subjects based on the outcome of a Cryptosporidium challenge and reasoned that if no differences between these two extreme outcomes were seen, then any differences in those who were infected but asymptomatic were unlikely. Given the findings of the present study, we plan to further examine a group of infected, asymptomatic individuals. A pilot experiment with three individuals from the latter group showed that the stage 1 FICs (mean of 1.66 mM) were consistent with the group of infected subjects with diarrhea. Finally, we were unable to compare substrate (tryptophan) availability to the FIC since no dietary information was collected during the original study. We do know, however, that these subjects were healthy, well-nourished adults. The relevance of these data to undernourished children is not known and would depend on the availability of foodstuffs, such as meats, eggs, fish, lentils, tofu, and other foods that are significant sources of tryptophan.
In summary, the present study indicates that the FIC existing in the gut at the time of Cryptosporidium exposure is an important predictor of the outcome of the encounter between the host and parasite. The FIC can be added to two other factors (oocyst dose and host immune status) previously demonstrated to influence susceptibility to infection. We believe the findings presented here should stimulate further in vitro and in vivo investigations that may eventually lead to new nonantibiotic treatments (e.g., indole) and/or prevention strategies by altering the microbial community and/or its products.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christopher Gonzales for his excellent technical assistance. We also appreciate the contributions of Peter Teunis, who provided insightful comments and helpful discussions.
The Cryptosporidium volunteer studies were supported in part by Environmental Protection Agency cooperative agreements (CR-819814 and CR-824759, C.L.C.), the Environmental Protection Agency National Center for Environmental Research STAR Program (GR-828035-01-0, C.L.C.), the National Institutes of Health General Clinical Research Center (MO1-RR-02558, P.C.O.), the National Institute of Allergy and Infectious Diseases (AI-41735, P.C.O.; AI-52781, G. Widmer; AI-25466, S. Tzipori), and the Food and Drug Administration (FD-U-001621-01, S. Tzipori). The indole studies were supported by the National Institutes of Allergy and Infectious Diseases (R01AI-116914, C.D.). The Environmental Protection Agency was a major funder of the Cryptosporidium volunteer studies; however, because the research described was not subject to EPA peer or administrative review, it may not reflect the views of the agency and no official endorsement should be inferred.
None of us have any conflict of interest or direct financial gain from the study reported here.
Funding Statement
We acknowledge additional grants whose PIs were part of the previous volunteer dose response studies. Some samples collected from these studies were used in the present report. These funding sources are not included above since the PIs are not included as authors on the present report, but they are listed in Acknowledgments.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00336-16.
REFERENCES
- 1.Kotloff KL, Nataro JP, Blackwelder WC. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 2.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, McCormick BJ, McGrath M, Olortegui MP, Samie A, Shakoor S, Mondal D, Lima IF, Hariraju D, Rayamajhi BB, Qureshi S, Kabir F, Yori PP, Mufamadi B, Amour C, Carreon JD, Richard SA, Lang D, Bessong P, Mduma E, Ahmed T, Lima AA, Mason CJ, Zaidi AK, Bhutta ZA, Kosek M, Guerrant RL, Gottlieb M, Miller M, Kang G, Houpt ER, MAL-ED Network Investigators. 2015. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 3:e564–75. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Checkley W, White AC Jr, Jaganath D, Arrowood MJ, Chalmers RM, Chen XM, Fayer R, Griffiths JK, Guerrant RL, Hedstrom L, Huston CD, Kotloff KL, Kang G, Mead JR, Miller M, Petri WA Jr, Priest JW, Roos DS, Striepen B, Thompson RC, Ward HD, Van Voorhis WA, Xiao L, Zhu G, Houpt ER. 2015. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect Dis 15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis CP, McAllister JS, Savage DC. 1973. Microbial colonization of the intestinal epithelium in suckling mice. Infect Immun 7:666–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harp JA, Chen W, Harmsen A. 1992. Resistance of severe combined immunodeficient mice to infection with Cryptosporidium parvum: the importance of intestinal microflora. Infect Immun 60:3509–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alak JIB, Wolf BW, Mdurvwa EG, Pimentel-Smith GE, Adeyemo O. 1997. Effect of Lactobacillus reuteri on intestinal resistance to Cryptosporidium parvum infection in a murine model of acquired immunodeficiency syndrome. J Infect Dis 175:218–221. doi: 10.1093/infdis/175.1.218. [DOI] [PubMed] [Google Scholar]
- 7.Guitard J, Menotti J, Desveaux A, Alimardani P, Porcher R, Derouin F, Kapel N. 2006. Experimental study of the effects of probiotics on Cryptosporidium parvum infection in neonatal rats. Parasitol Res 99:522–527. doi: 10.1007/s00436-006-0181-4. [DOI] [PubMed] [Google Scholar]
- 8.Pickerd N, Tuthill D. 2004. Resolution of cryptosporidiosis with probiotic treatment. Postgrad Med J 80:112–113. doi: 10.1136/pmj.2003.014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salazar-Lindo E, Miranda-Langschwager P, Campos-Sanchez M, Chea-Woo E, Sack RB. 2004. Lactobacillus casei strain GG in the treatment of infants with acute watery diarrhea: a randomized, double-blind, placebo controlled clinical trial [ISRCTN67363048]. BMC Pediatr 4:18–26. doi: 10.1186/1471-2431-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sindhu KN, Sowmyanarayanan TV, Paul A, Babji S, Ajjampur SS, Priyadarshini S, Sarkar R, Balasubramanian KA, Wanke CA, Ward HD, Kang G. 2014. Immune response and intestinal permeability in children with acute gastroenteritis treated with Lactobacillus rhamnosus GG: a randomized, double-blind, placebo-controlled trial. Clin Infect Dis 58:1107–1115. doi: 10.1093/cid/ciu065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lantier L, Drouet F, Guesdon W, Mancassola R, Merton C, Lo-man, Werts RC, Laurent F, Lacroix-Lamande S. 2014. Poly(I:C)-induced protection of neonatal mice against intestinal Cryptosporidium parvum infection requires an additional TLR5 signal provided by the gut flora. J Infect Dis 209:457–467. doi: 10.1093/infdis/jit432. [DOI] [PubMed] [Google Scholar]
- 12.Benson A, Pifer R, Behrendt C, Hooper LV, Yarovinsky F. 2009. Gut commensal bacteria direct a protective immune response against the human pathogen Toxoplasma gondii. Cell Host Microbe 6:187–196. doi: 10.1016/j.chom.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrier M, Lacroix-Lamande S, Mancassola R, Auray GI, Bernardet N, Chausse AM, Uematsu S, Akira S, Laurent F. 2006. Oral and intraperitoneal administration of phosphorothioate oligodeoxynucleotides leads to control of Cryptosporidium parvum infection in neonatal mice. J Infect Dis 193:1400–1407. doi: 10.1086/503748. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Lee J. 2010. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 34:426–444. doi: 10.1111/j.1574-6976.2009.00204.x. [DOI] [PubMed] [Google Scholar]
- 15.Darkoh C, Chappell CL, Gonzales C, Okhuysen P. 2015. A rapid and specific method for the detection of indole in complex biological samples. Appl Environ Microbiol 81:8093–8097. doi: 10.1128/AEM.02787-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han TH, Lee J-H, Cho MH, Wood TK, Lee J. 2011. Environmental factors affecting indole production in Escherichia coli. Res Microbiol 162:108–116. doi: 10.1016/j.resmic.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li G, Young KD. 2013. Indole production by the tryptophanase TnaA in Escherichia coli is determined by the amount of exogenous tryptophan. Microbiology 159:402–410. doi: 10.1099/mic.0.064139-0. [DOI] [PubMed] [Google Scholar]
- 18.Lee JH, Kim YG, Cho MH, Wood TK, Lee J. 2011. Transcriptomic analysis for genetic mechanisms of the factors related to biofilm formation in Escherichia coli O157:H7. Curr Microbiol 62:1321–1330. doi: 10.1007/s00284-010-9862-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J, Attila C, Cirillo SL, Cirillo JD, Wood TK. 2009. Indole and 7-hydroxyindole diminish Pseudomonas aeruginosa virulence. Microb Biotechnol 2:75–90. doi: 10.1111/j.1751-7915.2008.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nikaido E, Giraud E, Baucheron S, Yamasaki S, Wiedemann A, Okamoto K, Takagi T, Yamaguchi A, Cloeckaert A, Nishino K. 2012. Effects of indole on drug resistance and virulence of Salmonella enterica serovar Typhimurium revealed by genome-wide analyses. Gut Pathog 4:5–17. doi: 10.1186/1757-4749-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oh S, Go GW, Mylonakis E, Kim Y. 2012. The bacterial signalling molecule indole attenuates the virulence of the fungal pathogen Candida albicans. J Appl Microbiol 113:622–628. doi: 10.1111/j.1365-2672.2012.05372.x. [DOI] [PubMed] [Google Scholar]
- 22.Raut JS, Shinde RB, Karuppayil MS. 2012. Indole, a bacterial signaling molecule, exhibits inhibitory activity against growth, dimorphism and biofilm formation in Candida albicans. Afr J Microbiol Res 6:6005–6012. [Google Scholar]
- 23.Shimada Y, Kinoshita M, Harada K, Mizutani M, Masahata K, Kayama H, Takeda K. 2013. Commensal bacteria-dependent indole production enhances epithelial barrier function in the colon. PLoS One 8:e80604. doi: 10.1371/journal.pone.0080604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin U-H, Lee S-O, Sridharan G, Lee K, Davidson LA, Jayaraman A, Chapkin RS, Alaniz R, Safe S. 2014. Microbiome-derived tryptophan metabolites and their aryl hydrocarbon receptor-dependent agonist and antagonist activities. Mol Pharmacol 85:777–788. doi: 10.1124/mol.113.091165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bansal T, Alaniz RC, Wood TK, Jayaraman A. 2010. The bacterial signal indole increases epithelial-cell tight-junction resistance and attenuates indicators of inflammation. Proc Natl Acad Sci U S A 107:228–233. doi: 10.1073/pnas.0906112107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bansal T, Alaniz R, Jayaraman A. 2012. Role for the bacterial signal indole in promoting epithelial cell barrier function. J Epithel Biol Pharmacol 5:32–38. doi: 10.2174/1875044301205010032. [DOI] [Google Scholar]
- 27.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, Lancto CA, Deng M, Liu C, Widmer G, Tzipori S, Buck GA, Xu P, Bankier AT, Dear PH, Konfortov BA, Spriggs HF, Iyer L, Anantharaman V, Aravind L, Kapur V. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–445. doi: 10.1126/science.1094786. [DOI] [PubMed] [Google Scholar]
- 28.Gupta SL, Carlin JM, Pyati P, Dai W, Pfefferkorn ER, Murphy MJ Jr. 1994. Antiparasitic and antiproliferative effects of indoleamine 2,3-dioxygenase enzyme expression in human fibroblasts. Infect Immun 62:2277–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pollok RC, Farthing MJ, Bajaj-Elliott M, Sanderson IR, McDonald V. 2001. Interferon gamma induces enterocyte resistance against infection by the intracellular pathogen Cryptosporidium parvum. Gastroenterology 120:99–107. doi: 10.1053/gast.2001.20907. [DOI] [PubMed] [Google Scholar]
- 30.Choudhry N, Korbel DS, Edwards A, Bajaj-Elliott M, McDonald V. 2009. Dysregulation of interferon-γ-mediated signaling pathway in intestinal epithelial cells by Cryptosporidium parvum infection. Cell Microbiol 11:1354–1364. doi: 10.1111/j.1462-5822.2009.01336.x. [DOI] [PubMed] [Google Scholar]
- 31.Chappell CL, Okhuysen PC, and White AC Jr. 2004. Cryptosporidium parvum: infectivity, pathogenesis and the host-parasite relationship, p 19–50. In Thompson RCA, Armson A, Ryan UM (ed), Cryptosporidium: from molecules to disease. Elsevier, Amsterdam, The Netherlands. [Google Scholar]
- 32.Chappell CL, Okhuysen PC, Langer-Curry RC, Widmer G, Akiyoshi DE, Tanriverdi S, Tzipori S. 2006. Cryptosporidium hominis: experimental challenge of healthy adults. Am J Trop Med Hyg 75:851–857. [PubMed] [Google Scholar]
- 33.Zoetendal EG, Akkermans AD, De Vos WM. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64:3854–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dethlefsen L, Huse S, Sogin ML, Relman DA. 2008. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reed LJ, Muench H. 1934. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 38.Hooper LV, Midtvedt T, Gordon JI. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr 22:283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 39.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54(Pt 5):1469–1476. [DOI] [PubMed] [Google Scholar]
- 40.Derrien M, Van Baarlen P, Hooiveld G, Norin E, Müller M, deVos WM. 2011. Modulation of mucosal immune response, tolerance, and proliferation in mice colonized by the mucin-degrader Akkermansia muciniphila. Front Microbiol 2:166. doi: 10.3389/fmicb.2011.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tailford LE, Crost EH, Kavanaugh D, Juge N. 2015. Mucin glycan foraging in the human gut microbiome. Front Genet 6:81. doi: 10.3389/fgene.2015.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ras R, Huynh K, Desoky E, Badawy A, Widmer G. 2015. Perturbation of the intestinal microbiota of mice infected with Cryptosporidium parvum. Int J Parasitol 45:567–573. doi: 10.1016/j.ijpara.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. 1985. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol 109:135–141. doi: 10.1007/BF00391888. [DOI] [PubMed] [Google Scholar]
- 44.Zuccato E, Venturi M, Di Leo G, Colombo L, Bertolo C, Doldi SB, Mussini E. 1993. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci 38:514–519. doi: 10.1007/BF01316508. [DOI] [PubMed] [Google Scholar]
- 45.Alak JI, Wolf BW, Mdurvwa EG, Pimentel-Smith GE, Kolaviaia S, Abdelrahman H, Suppiramaniam V. 1999. Supplementation with Lactobacillus reuteri or L. acidophilus reduced intestinal shedding of Cryptosporidium parvum oocysts in immunodeficient C57BL/6 mice. Cell Mol Biol (Noisy-le-grand) 45:855–863. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


