Abstract
Leucine-rich repeat (LRR) proteins are widely distributed in bacteria, playing important roles in various protein-protein interaction processes. In Yersinia, the well-characterized type III secreted effector YopM also belongs to the LRR protein family and is encoded by virulence plasmids. However, little has been known about other LRR members encoded by Yersinia genomes or their evolution. In this study, the Yersinia LRR proteins were comprehensively screened, categorized, and compared. The LRR proteins encoded by chromosomes (LRR1 proteins) appeared to be more similar to each other and different from those encoded by plasmids (LRR2 proteins) with regard to repeat-unit length, amino acid composition profile, and gene expression regulation circuits. LRR1 proteins were also different from LRR2 proteins in that the LRR1 proteins contained an E3 ligase domain (NEL domain) in the C-terminal region or an NEL domain-encoding nucleotide relic in flanking genomic sequences. The LRR1 protein-encoding genes (LRR1 genes) varied dramatically and were categorized into 4 subgroups (a to d), with the LRR1a to -c genes evolving from the same ancestor and LRR1d genes evolving from another ancestor. The consensus and ancestor repeat-unit sequences were inferred for different LRR1 protein subgroups by use of a maximum parsimony modeling strategy. Structural modeling disclosed very similar repeat-unit structures between LRR1 and LRR2 proteins despite the different unit lengths and amino acid compositions. Structural constraints may serve as the driving force to explain the observed mutations in the LRR regions. This study suggests that there may be functional variation and lays the foundation for future experiments investigating the functions of the chromosomally encoded LRR proteins of Yersinia.
INTRODUCTION
YopM plays a dominant role in the infection processes of different Yersinia strains in different host organs (1–4). The yopM gene sequence was first reported for Yersinia pestis strain KIM5, and subsequently the encoded protein was demonstrated to be a possible virulence factor that mimicked the host homolog human platelet glycoprotein 1b (GPIα) and influenced platelet aggregation (5–7). Boland et al. sequenced a homologous yopM gene from Y. enterocolitica strain W22703 and demonstrated that the N-terminal 100 residues gave YopM the ability to translocate into the host cell cytoplasm through the Ysc type III secretion system (T3SS) (8). YopM also showed the ability to enter the host cell cytoplasm by autonomous translocation independently of T3SSs (9, 10). In another study, YopM-specific antibodies did not slow the progression of experimental plague, indicating that YopM may have a major intracellular function (11). Subcellular localization studies demonstrated that YopM was targeted to the nuclei of host cells, and nuclear targeting signals were found in both the N-terminal and C-terminal regions (12–14). YopM was further shown to interact with protein kinase C-like 2 (PRK2) and ribosomal S6 kinase 1 (RSK1), causing sustained activation of the kinases by shielding them from phosphatase activity toward two serine residues (15, 16). The C terminus and an internal portion of YopM were essential for the interactions with RSK1 and RSK2, respectively, and for the expression of proinflammatory cytokines (17–19). The interaction of YopM with RSKs and the resultant consequences may explain the observation of interference of YopM with host innate immunity (10, 20–23). LaRock and Cookson showed that YopM inhibits caspase-1 activity and alters inflammasome assembly and processing (24). IQGAP1 was further identified as a YopM binding partner important for caspase-1 activity in macrophages infected with Yersinia (25). Caspase-3 was also reported as another target of YopM in Yersinia's interference with host inflammatory processes (2).
The structure of the Y. pestis YopM protein was determined in two different crystal forms: at 2.4- and 2.1-Å resolutions (26). The protein contains a large leucine-rich repeat (LRR) domain comprised of ∼15 LRR units (5, 26). Heterogenic YopM isoforms with varied numbers of LRR units have also been reported (27, 28). Barrick and collaborators performed a lot of work in an endeavor to clarify the possible evolutionary mechanisms of YopM LRR unit amplification and shrinkage with respect to structure, thermostability, and function (29–31). It should be noted that all of the YopM proteins for which function and structure have been investigated experimentally in Yersinia are encoded by the virulence plasmids, that is, pCD1 in Y. pestis and pYV in Y. enterocolitica and Y. pseudotuberculosis (32). There are also other YopM family proteins encoded by Yersinia chromosomal genes (33–36). Both the YopM proteins encoded by virulence plasmids and those encoded by chromosomes contain the LRR domain, and they differ mainly in the presence or absence of the C-terminal E3 ligase domain (NEL domain) (32). Soundararajan et al. used computational methods to model and reason out the possible functional and interaction network of new Yersinia YopM isoforms, which elicited a debate (32–34). Computational modeling acts as a double-edged sword, providing important clues to gene function but also potentially being misleading without experimental support; this is likely a major reason for the debate on the functions of Yersinia YopM isoforms (32). However, there are heterogenic forms of yopM and yopM-like genes in Yersinia. The sequences are quite similar to each other, so the encoded proteins can be considered a superfamily (32). The yopM-like genes should neither be neglected nor considered simply to have the same functions as those of the well-studied Yersinia yopM genes carried on virulence plasmids.
Due to the previous lack of available Yersinia genomes, detailed identification and comparison of Yersinia yopM sequences were not possible (5). The only knowledge about the potential origins and evolution of yopM in Yersinia was provided by the identification of two insertion elements flanking the yopM gene in Y. intermedia and by the analysis of homologs from other bacteria and even eukaryotic cells (5, 37, 38). A large number of Yersinia whole-genome sequences are now publically available, which enabled us to perform a systemic investigation and comparison of the distribution and evolution of the YopM protein family in Yersinia. Furthermore, online expression databases were mined to examine the possible expression of these genes. Finally, the tertiary structures of the proteins were modeled and compared to explore the potential relationships between sequence evolution, structure, and function.
MATERIALS AND METHODS
Screening and sequence analysis of LRR protein-encoding genes (LRR genes) from Yersinia genomes.
Yersinia genomes and annotation files were downloaded from the National Center for Biotechnology Information (NCBI) genome database based on which protein-encoding genes and corresponding protein sequences were retrieved with in-house scripts. The local BLAST suite was downloaded from the NCBI website, installed, and used for LRR protein screening, with Y. pestis CO92 YopM (accession no. NP_395161.1) as the query sequence and with an identity cutoff of 30%. The alignment hits were further analyzed for possible conserved domains annotated in the CDD database (39). The protein sequences were tracked back to their corresponding genomes, and the flanking genes were identified by in-house scripts or checked manually. The interesting genes and their flanking sequences were retrieved from the corresponding genomes, and PipMaker was used for synteny and collinearity analysis (40). The position-specific amino acid composition of LRR units was analyzed with WebLogo (41). The protein sequences were aligned and phylogenetic analysis performed with MEGA6.0, and the neighbor-joining distance was calculated and bootstrapping tests performed with 1,000 replicates (42).
RNA-seq data analysis.
The NCBI SRA database was screened with the key word “Yersinia,” followed by a manual check for each data set. Only two experiments were found for transcriptome sequencing (RNA-seq) analysis of Yersinia, both for Y. pseudotuberculosis YPIII cultured at different temperatures (25, 26, and 37°C), analyzed during different growth phases (exponential and stationary phases), or mutated at an important regulatory gene (crp gene) (43, 44). The raw reads were downloaded and reanalyzed for chromosomally encoded LRR protein expression. Geneious 6.1.8 was used for read mapping, and the protocols for analysis, normalization, and comparison were described previously (45).
Structure modeling and analysis.
The experimentally resolved structure of Y. pestis YopM was used as the main reference, and the structures of other LRR proteins were predicted with PHYRE2, which uses the alignment of hidden Markov models via HHsearch to significantly improve accuracy and incorporates Poing to perform ab initio folding simulations for fragments with no detectable homology (26, 46). Structure visualization and analysis were performed with PyMOL (http://www.pymol.org).
RESULTS
Distribution of LRR proteins in Yersinia.
The Y. pestis KIM5 YopM protein, encoded by the virulence plasmid, was used for alignment against the NCBI protein sequence database by use of the BLASTP program (http://www.ncbi.nlm.nih.gov), generating 317 homologous hits: 180 encoded in chromosomes and 137 encoded in plasmids (see Data Set S1 in the supplemental material). The proteins were together called LRR proteins instead of YopM, since the Yersinia YopM proteins are specifically those encoded by virulence plasmids (32). Only one copy of a plasmid-encoded LRR protein was found for each Y. pestis, Y. pseudotuberculosis, Y. enterocolitica, and Y. intermedia strain. More species showed one or more copies of chromosomally encoded LRR proteins for each strain, including Y. pestis, Y. pseudotuberculosis, Y. intermedia, Y. similis, and Y. wautersii (see Data Set S1). Y. enterocolitica did not have chromosomally encoded LRR proteins (see Data Set S1). The genomes (chromosomes and plasmids) of other species were also examined, including Y. aldovae, Y. ruckeri, Y. bercovieri, Y. rohdei, Y. mollaretii, Y. massiliensis, Y. pekkanenii, Y. nurmii, and Y. aleksiciae, and no LRR genes or associated DNA relics were detected.
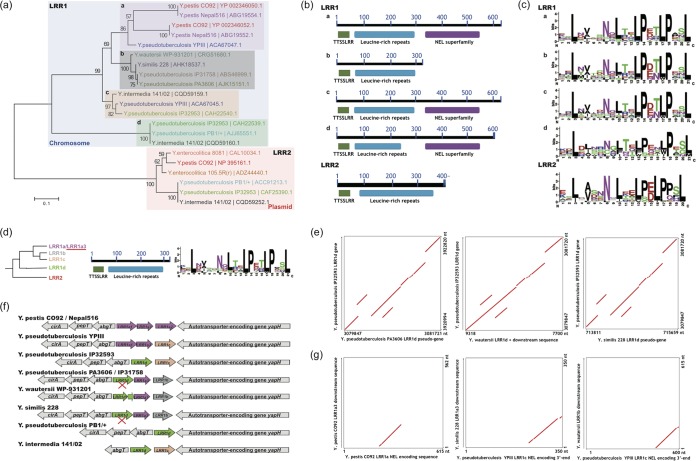
Representative LRR proteins were selected for comparison and evolutionary analysis. The neighbor-joining tree for the protein sequences demonstrated that the LRR proteins fell into two major clusters: LRR1 for chromosomally encoded proteins and LRR2 for plasmid-encoded proteins (Fig. 1a). The LRR1 proteins were further classified into 4 subgroups (LRR1a to -d) (Fig. 1a). Domain analysis indicated that all LRR2 proteins were comprised of an N-terminal signal region (TTSSLRR) and an LRR domain of various lengths, while the LRR1 proteins often had an extra C-terminal domain of the NEL superfamily (LRR1a, -c, and -d but not LRR1b) (Fig. 1b). Amino acid composition profiles of the repeats of the LRR domains disclosed an evolutionary relationship generally concordant with that demonstrated by the phylogenetic tree (Fig. 1c). The LRR2 profile was most different from that for the LRR1 proteins in that the unit length of consensus repeats was one residue shorter (20 versus 21 amino acids [aa]), with a loss of the 16th position of the LRR1 repeat (Fig. 1c). In addition, there were other positions with apparently different amino acid preferences; for example, proline was enriched at position 18 of the LRR2 repeat units (Fig. 1c). Within LRR1 proteins, the LRR1d repeat unit appeared to be more degenerate in the amino acid composition for most positions (Fig. 1c). The LRR1b and -c profiles appeared to be more similar to each other, and both differed strikingly from the LRR1a profile at the 10th position, where a leucine appeared to be more conserved in LRR1a proteins (Fig. 1c). The LRR1b and -c proteins could further be distinguished from each other according to the absence and presence, respectively, of a C-terminal NEL domain (Fig. 1b).
FIG 1.
Screening and sequence analysis of Yersinia LRR proteins. (a) Distribution and phylogenetic analysis of Yersinia LRR proteins. Proteins from the same strain are highlighted in the same color. Sequences with different background colors represent different subgroups. The neighbor-joining distance was calculated and bootstrapping tests performed with MEGA6.0 (see Materials and Methods), and the bootstrapping consistence percentage is indicated for each node. (b) LRR protein domain analysis. Conserved domains and their within-protein locations are shown. (c) Repeat-unit consensus motifs of Yersinia LRR proteins. (d) LRR1a3 sequence features. (Left) Phylogenetic diagram; (middle) domain composition diagram; (right) repeat-unit consensus motif. (e) Comparison of LRR1d loci between Yersinia strains with an LRR1d gene or pseudogene. The collinear relationship is shown with red lines or plots. (f) Synteny analysis of LRR loci of different Yersinia strains. Pseudogenes are marked with “X's.” LRR genes of different subgroups are highlighted in different colors. (g) Nucleotide-level NEL domain relics identified from genome comparisons. The collinear relationship between compared sequences is shown in red.
To elucidate the evolutionary history of LRR proteins in Yersinia, the representative chromosomally encoded LRR proteins were mapped to the respective genomes. Meanwhile, the LRR consensus motifs disclosed in Fig. 1c were used to seek for possible missed LRR proteins in Yersinia strains. The NCBI tblastn program was also applied to find possible LRR pseudogenes. A group of new LRR proteins was consequently identified (see Data Set S1 in the supplemental material). These new LRR proteins (named LRR1a3 proteins) were all encoded in Yersinia chromosomes, with sequences and repeat-unit amino acid composition profiles very similar to those of LRR1a proteins, but without the C-terminal NEL domain (Fig. 1d). Several LRR gene-like pseudogenes were also traced, for which the nucleotide sequences showed high similarity to those of LRR1d protein-encoding genes, but the frames were interrupted (Fig. 1e, left and right panels). Interestingly, Y. wautersii WP-931201 also had a genomic fragment showing high similarity to the LRR1d pseudogenes, but two consecutive gene frames were detected within the region, encoding an N-terminal and a C-terminal LRR1d peptide fragment. The DNA fragment covering the two genes together with an interlinking sequence could be well aligned against the whole LRR1d gene (Fig. 1e, middle panel).
All the LRR protein genes, including pseudogenes, were found within the same locus in each strain (Fig. 1f). The locus-flanking genes showed good collinearity among different Yersinia strains. It should be pointed out that for Y. enterocolitica and other species without chromosomally encoded LRR proteins, the LRR loci and flanking loci could not be traced in the genome either, and therefore the evolutionary analysis of LRR genes in these species could not be well traced. In different species with chromosomally encoded LRR proteins, the locus was often about 63 to 66° away from the conserved chromosome replication initiator dnaA gene, indicating the vertical heredity of the genes after the diversification of these species (see Fig. S1 in the supplemental material). Within each LRR locus, the genes showed polymorphous amplification (Fig. 1f).
The comparative genomic analysis indicated a unique origin of the chromosomal LRR genes. However, it is still an enigma why some LRR genes encode an NEL domain in the C termini of the corresponding proteins, whereas others do not. In other bacteria, such as Salmonella or Shigella, the LRR proteins identified always showed an NEL domain (35, 36, 47, 48). In a recent comparative analysis, many T3SS genes were found to separate into two genes encoding different peptide domains, as occurred for the Y. wautersii WP-931201 LRR1d protein (Y. Hu, H. Huang, A. P. White, W. Koster, G. Zhu, J. Stavrinides, Z. Zhao, and Y. Wang, unpublished data) (Fig. 1f). It is possible that the NEL-free LRR protein-encoding genes also followed this phenomenon. Therefore, the nucleotide sequences adjacent to the LRR1b and LRR1a3 genes were collected and observed for NEL domain-encoding capacity. As shown in Fig. 1g, some NEL domain-encoding relics and/or downstream peptide-encoding sequences could be detected in the downstream genome sequences for all the investigated LRR1b and LRR1a3 genes. Therefore, we drew the conclusion that the ancient Yersinia chromosomal LRR gene potentially encoded the NEL domain before gene amplifications. For LRR2 proteins, i.e., the YopMs encoded in plasmids, however, no NEL domain-encoding sequence trace could be detected in the context of genome regions (data not shown).
In summary, we identified a hot spot in most Yersinia (except Y. enterocolitica) chromosomes where a diverse group of LRR protein-encoding genes are located. The locus is unstable, and the LRR genes are often amplified, tandemly arrayed, split, or found without protein-encoding frames. Proteins encoded by the genes are significantly different from YopMs expressed from virulence plasmids in two major respects, i.e., different LRR unit lengths and the presence of the NEL domain. Based on the comparative genomic and phylogenetic analysis, a hypothesis on the diversifying history of representative Yersinia chromosomal LRR protein-encoding genes was tentatively proposed (see Fig. S2 in the supplemental material).
Expression of LRR1 genes with transcription regulation circuits different from those for yopM genes.
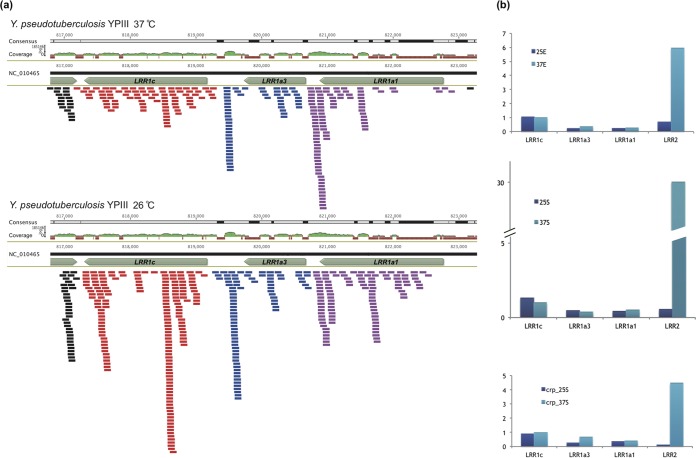
The mRNAs corresponding to LRR1 protein-encoding genes were detected in Y. pseudotuberculosis YPIII cultured at both 37°C and 26°C, but only at low to moderate expression levels (Fig. 2a). No apparent expression level difference was detected for each of the LRR1 genes between 37°C and 26°C. The expression levels of LRR genes were also compared during different growth phases, with or without crp regulation; this time, LRR1c gene expression was higher than that of LRR1a3 and LRR1a1 genes (Fig. 2a and b). Unlike LRR1 genes, the LRR2 gene showed a dramatic response to temperature, with more expression at 37°C for exponential- or stationary-phase growth, with or without CRP regulation (Fig. 2b). The results indicated different expression regulation circuits and maybe different functions of the LRR1 and LRR2 genes in Yersinia.
FIG 2.
Expression of Yersinia chromosomally carried LRR genes. (a) Diagrams for LRR gene expression at 26°C and 37°C as disclosed by RNA-seq read mapping. The genome coordinates, LRR genes, read coverage, and mapped reads are shown. Reads mapped for different gene models are shown in different colors. (b) LRR gene expression comparison between different temperatures and different growth stages. The normalized read count for LRR1c genes of wild-type Y. pseudotuberculosis YPIII cultured at 37°C and at exponential phase was set at 1.0, while the normalized read counts for other genes and different conditions were scaled appropriately. 25E and 37E represent the combination of 25 and 37°C and the exponential phase; similarly, 25S and 37S represent the combination of 25 and 37°C and the stationary phase, while crp_25S and crp_37S represent the combination of crp gene mutation, 25 and 37°C, and the stationary phase. The virulence plasmid-carried yopM (LRR2) gene was also included for comparison.
Evolution of LRR1 repeats in Yersinia.
At the sequence level, the LRR proteins are characterized by the presence of multiple leucine-rich repeats. The number of repeats was found to vary among plasmid-encoded YopMs from different strains (27, 28). The repeats within Yersinia chromosomally encoded LRR proteins were analyzed and compared in the present study.
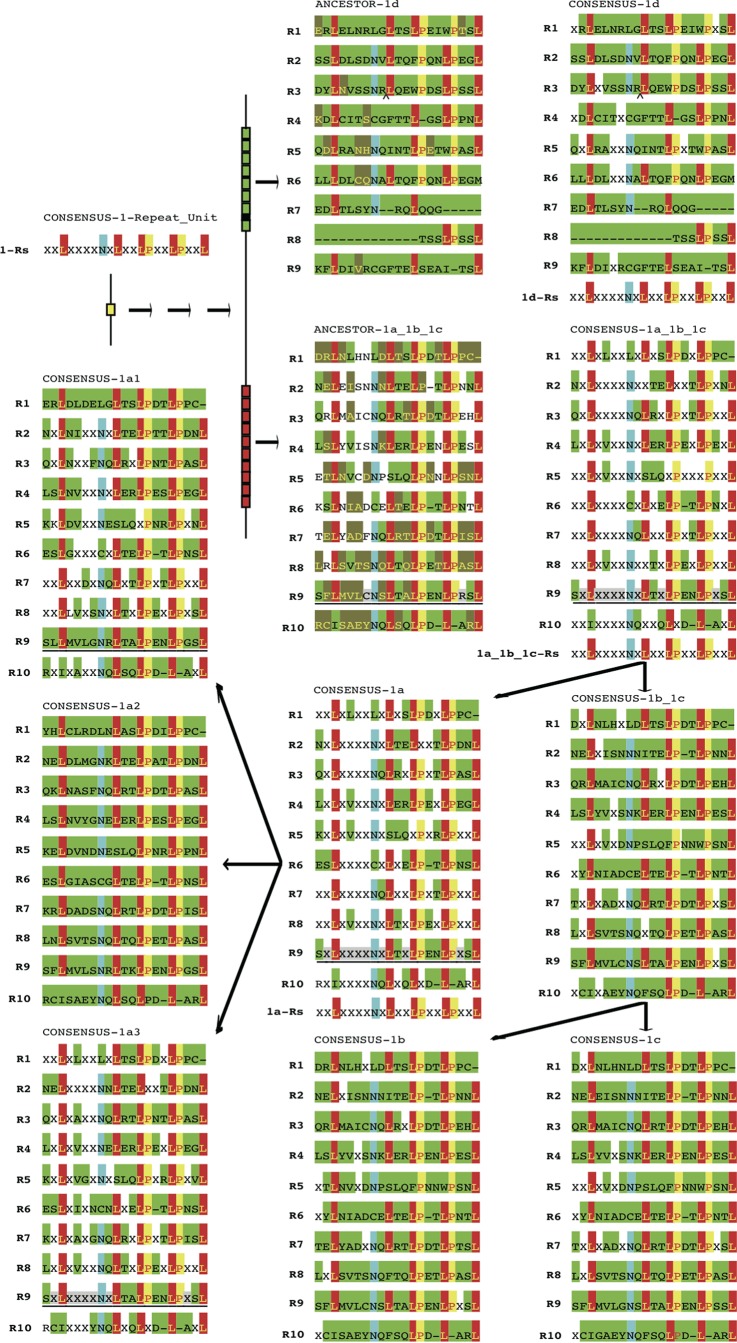
For the 3 subgroups of LRR1a proteins, LRR1a1 proteins were detected in Y. pseudotuberculosis YPIII and all Y. pestis strains, LRR1a2 proteins were found only in Y. pestis strains, and LRR1a3 proteins were widely distributed in strains from different species, e.g., Y. pestis, Y. pseudotuberculosis, Y. wautersii, and Y. similis (Fig. 1f). In most strains, there were 10 repeat motifs in LRR1a proteins, but there were only 9 in the LRR1a1 and LRR1a3 proteins of Y. pestis due to a loss of the R9 motif (Fig. 3, CONSENSUS-1a, -1a1, -1a2, and -1a3). The consensus sequence for each repeat unit between the LRR1a proteins had higher similarity than the consensus sequences for repeats within an individual protein. This suggested a more ancient intragene amplification of repeat units rather than whole-LRR-gene amplification (Fig. 3, CONSENSUS-1a). The 10 most frequent repeats in LRR1a1 to -3 proteins of different species further indicated that the ancestor of LRR1a proteins should have contained 10 repeats (Fig. 3, CONSENSUS-1a, -1a1, -1a2, and -1a3). The consensus sequences for the whole repeat domain for LRR1a2, LRR1a1, and LRR1a3 proteins showed the highest, moderate, and lowest levels of conservation, respectively, in concordance with the widened distribution of corresponding proteins in Yersinia species/strains, probably reflecting the ancient evolutionary history of the proteins (Fig. 3, CONSENSUS-1a, -1a1, -1a2, and -1a3; see Fig. S2 in the supplemental material).
FIG 3.
Consensus and ancestor sequences for each repeat unit of each LRR1 protein subgroup/supersubgroup. Peptide sequences in the same repeat unit of the same LRR1 protein subgroup or supersubgroup (e.g., 1b_1c and 1a_1b_1c) were compared. For consensus sequences, identical residues in the orthologous positions for all compared sequences are shown with red, yellow, blue, and green backgrounds. Residues with a white background indicate the consensus but not a unique amino acid composition, and “X” indicates an unconserved amino acid. The gaps are shown with dashes. Conserved positions among repeat units are shown with red, yellow, and blue backgrounds for L, P, and N, respectively. The repeat unit R9 is underscored frequently, indicating that some of the compared sequences did not contain this repeat. For ancestor sequences, the representation is similar to that for consensus sequences, with the exception that residues with a dark green background indicate the ancestor composition inferred by the maximum parsimony method, with a high confidence.
LRR1b and LRR1c amino acid sequences were similar to each other, though LRR1b protein genes experienced gene splitting events and lost NEL domain-encoding potential (Fig. 1b). The presence of LRR1c proteins in Y. intermedia (Fig. 1f), the collinearity of the LRR1 locus among different Yersinia species (see Fig. S1 in the supplemental material), and the relatively ancient divergence of Y. intermedia from other Yersinia species (49) indicated that the LRR1c (and LRR1b) proteins were present more anciently than the LRR1a proteins. Like the LRR1a consensus sequence, both the LRR1b and LRR1c proteins also contained 10 leucine-rich repeats (Fig. 3, CONSENSUS-1b and -1c). The LRR1b and -c repeat consensus sequences showed a high level of conservation (Fig. 3, CONSENSUS-1b_1c), which also showed high consistency with the consensus for LRR1a proteins (Fig. 3, CONSENSUS-1a_1b_1c versus CONSENSUS-1a).
LRR1d proteins contained 7 complete leucine-rich repeats (R1 to R6 and R9) and 2 degenerate repeats (R7 and R8) according to sequence alignment results (Fig. 3, CONSENSUS-1d). The LRR1d proteins from distantly diverged species showed high similarity, but the repeat consensus sequences were quite different from those for LRR1a to -c proteins, indicating the more ancient event of divergence of LRR1d proteins from the other LRR1 groups (Fig. 3, CONSENSUS-1a, -b, -c, and -d). The consensus between repeats within an individual LRR1d protein was identical to that for LRR1a to -c proteins; therefore, the repeat could have been amplified within the protein before the divergence of LRR1d and the other LRR1 groups. According to the consensus repeat sequences and the phylogenetic relationships, the ancestor protein sequences of LRR1a to -c and LRR1d proteins were inferred, and a possible hypothesis was proposed to model the evolutionary process for LRR1 proteins (Fig. 3). The oldest LRR1 protein may have had only one leucine-rich repeat copy, with the consensus X2LX4NXLX2LPX2LPX2L, which further amplified and evolved into two proteins: the oldest ancestor of LRR1 proteins detected in the Yersinia genome, i.e., the tandemly arrayed LRR1d ancestor, and the LRR1a to -c ancestor (Fig. 3). The evolution from the two different ancestor proteins subsequently resulted in a burst that created various forms of LRR1 proteins.
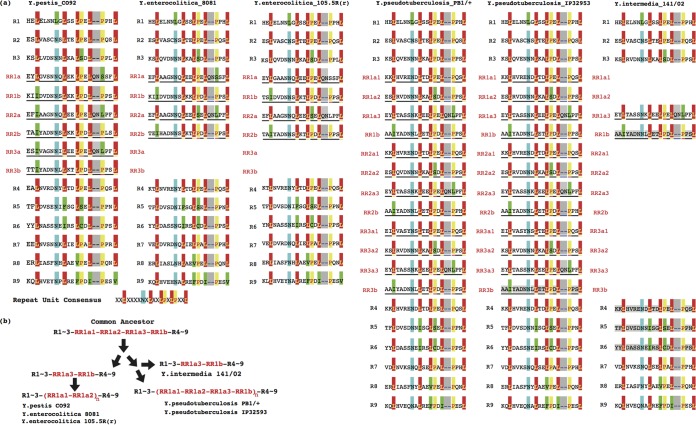
Evolution of LRR2 repeats in Yersinia.
Despite the different sizes of repeat units (20 aa for LRR2 proteins versus 21 aa for LRR1 proteins), the consensus repeat sequence for LRR2 proteins was similar to that for LRR1 proteins (Fig. 4a). Although strains had large variations in the number of repeats, from 11 for Y. intermedia 141/02 to 21 for Y. pseudotuberculosis PB1/+ and IP32593, the N-terminal 3 repeats (R1 to R3) and C-terminal 5 repeats (R4 to R9) were conserved (Fig. 4a). For the variable, internal repeats between R1 to R3 and R4 to R9, there were often conserved amino acid insertions (successive QN insertions within RR1a/2a/3a). Based on the insertion pattern and sequence similarity between repeat units, it was noted that the gain/loss unit was a combination of 2 or 4 contiguous repeats rather than a single repeat (Fig. 4a). The two LRR2 protein groups (Fig. 1a) were divided according to the repeat number of the gain/loss units: Y. enterocolitica 105.5R(r), Y. enterocolitica 8081, and Y. pestis CO92 showed various amplifications or deletions of 2 contiguous LRR units, while Y. pseudotuberculosis PB1/+, Y. pseudotuberculosis IP32593, and Y. intermedia 141/02 had an amplification/deletion unit of 4 contiguous LRR units (Fig. 4a). Several models were proposed to explain the Yersinia LRR2 repeat amplification or deletion in different strains, among which the most plausible one is shown in Fig. 4b. The common ancestor of LRR2 proteins in Yersinia was likely to have the N-terminal R1 to R3 repeats and the C-terminal R4 to R9 repeats as well as a single and hierarchical 4-repeat amplification/deletion unit. The ancestor evolved into two phylogenetic groups. In one group, two N-terminal LRR units in the hierarchical amplification/deletion unit were deleted, while the rest of the LRRs were amplified as a unit, generating the LRR2 proteins of Y. enterocolitica 105.5R(r), Y. enterocolitica 8081, and Y. pestis CO92. In the other group, the branch ancestor further evolved into two subgroups, one of which lost the two N-terminal LRRs in the hierarchical amplification/deletion unit and formed the LRR2 protein of Y. intermedia 141/02 and the other of which amplified the hierarchical 4-repeat unit and generated the LRR2 proteins of Y. pseudotuberculosis PB1/+ and Y. pseudotuberculosis IP32593 (Fig. 4b).
FIG 4.
Consensus sequences and evolution of LRR2 protein repeat units. (a) Consensus sequences of LRR2 repeat units. Insertions and deletions are indicated with a gray background. The other background colors indicate conserved residues among repeat units, except for the green background, which represents a composition different from the expected consensus. The hierarchically replicated repeats are underscored. (b) The amplification/deletion history hypothesis of LRR2 repeat units.
Maximum parsimony models of amino acid variations in LRR1 repeats.
A maximum parsimony model was used for reference to the ancient peptide sequences of LRR1 repeats. From the position-specific amino acid composition profiles shown in Fig. 1c, we noticed that the conserved positions that were dominated by leucine also contained mutations. However, the mutations consisted of only six possible amino acids: phenylalanine, isoleucine, tryptophan, methionine, arginine, and serine. Each of these amino acids was encoded by nucleotide codons that were closely related to those for leucine. Therefore, a maximum parsimony model was proposed for the mutations. The comparison of nucleotide sequences encoding the LRR validated this model; for most mutations from leucine, only one base was changed, and there were only two examples of mutations that required two base substitution steps (Fig. 5). The maximum parsimony models also explained other amino acid variations in LRR1 (Fig. 5) and LRR2 (data not shown) proteins.
FIG 5.

Maximum parsimony models of LRR protein amino acid substitutions among different strains or repeat units. Amino acid substitutions, codon changes, and examples are shown in different columns.
Tertiary structure of repeat domains in Yersinia LRR proteins.
The tertiary structure has been resolved for several LRR proteins, whose LRR units are frequently composed of 20 amino acids rather than 21 as in Yersinia LRR1 proteins (26, 35, 36). Using the structures of Y. pestis YopM (i.e., an LRR2 protein), Salmonella SlrP/SspH1/SspH2, and the Shigella IpaH family as templates, the Yersinia LRR1 proteins and other LRR2 proteins were examined for determination of the structure of the LRR units.
The LRR2 repeat domains showed very similar overall folding topologies, with moderate differences in the strand copy numbers and consequent interface sizes (see Fig. S3 in the supplemental material). Consistent with the sequence conservation we observed for N-terminal and C-terminal repeats (Fig. 4), the LRR2 repeat domain generally showed two conserved subdomains, in the N terminus and the C terminus, with repeat copy and interface size variation in the middle (see Fig. S3).
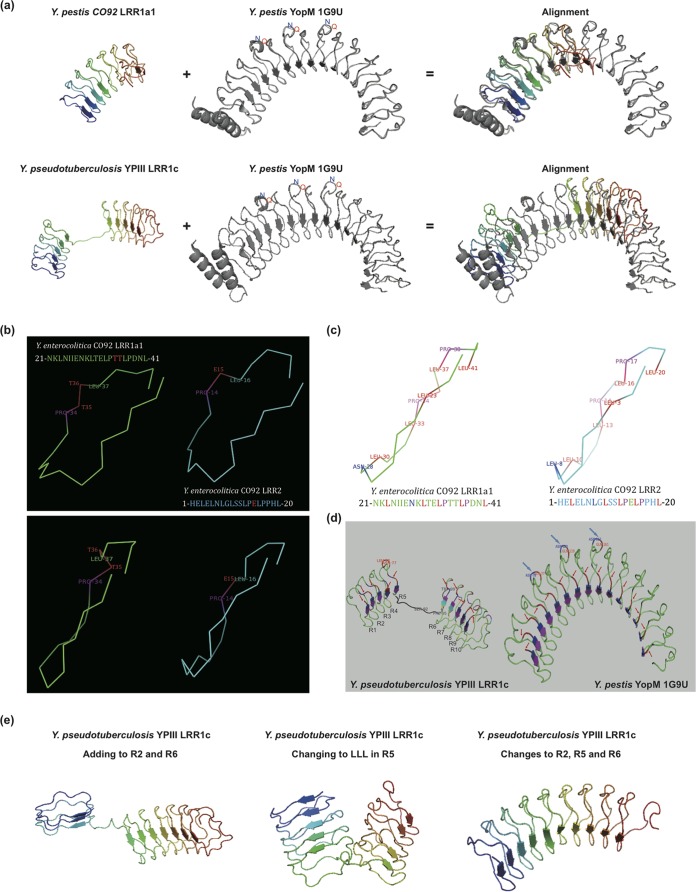
The LRR1 repeat domains showed more diverse tertiary structures than the LRR2 domains (see Fig. S3 and S4 in the supplemental material). Although the whole domain was often organized into an N-terminal and a C-terminal subdomain as in LRR2 proteins, the twist angle between each pair of subdomains varied a lot, leading to general structural differences (see Fig. S4). Structural diversity was also observed frequently among the LRR1 proteins, with orthologous relationships for proteins from different species (e.g., Y. pestis LRR1a1 and Y. pseudotuberculosis LRR1a1 proteins or Y. pestis LRR1a3 and Y. pseudotuberculosis LRR1a3 proteins) (see Fig. S4) or among the LRR1 protein paralogs of the same phylogenetic group and in the same strain (e.g., Y. pestis LRR1a1, LRR1a2, and LRR1a3 proteins or Y. pseudotuberculosis LRR1a1 and LRR1a3 proteins) (see Fig. S4). LRR1c and Y. pestis LRR1a1 proteins showed an LRR domain structure most similar to that of Y. pestis YopM, but with a smaller overall surface area (Fig. 6a).
FIG 6.
LRR protein structure comparison. (a) Structural comparison of Y. pestis CO92 LRR1a1 protein or Y. pseudotuberculosis YPIII LRR1c protein and the reference structure 1G9U for Y. pestis YopM. (b) Structural comparison of the repeat units of LRR1 and LRR2 proteins. The top and side faces of the units are shown in the upper and lower panels, respectively. The key position difference is shown in red. The Y. enterocolitica CO92 LRR1a1 and LRR2 repeat units are used as representatives. (c) Shape-maintaining residues of repeat units (indicated in red, purple, and blue). (d) Repeat sequence starting difference in Y. pseudotuberculosis YPIII LLR1c protein. Each repeat sequence start is indicated with a red arrow; QN loops are indicated with blue arrows. Repeat sequence starts of the reference YopM structure (1G9U) are also shown. (e) Structure of Y. pseudotuberculosis YPIII LRR1c with the gaps in repeats R2 and R6 filled with D residues (left), with the three residues (S, F, and W) in R5 consensus positions replaced by L residues (middle), and with both the gaps in repeats R2 and R6 filled with D residues and the three residues (S, F, and W) in R5 consensus positions replaced by L residues (right).
As observed before, each LRR1 repeat unit contained one more residue than those of LRR2 proteins. However, each repeat unit in LRR1 proteins also formed a strand or loop and a cylinder interface unit with a shape and size very similar to those for LRR2 proteins (Fig. 6a to c). The single-residue difference between the two repeat consensus sequences changed the local structure only slightly and had no influence on the complete repeat structure (Fig. 6b). The proline and leucine residues flanking TT in LRR1 proteins and E in LRR2 proteins appeared to be very important for maintaining the local conformation (Fig. 6b). Similarly, the local QN insertions in partial LRR2 repeat units did not change the overall structure (Fig. 6a). For each repeat unit of LRR1 or LRR2 proteins, the two conserved prolines near the C terminus (Fig. 6c, purple residues) appeared to be important for forming a forward-extension angle, while the conserved asparagine (or leucine in every first repeat unit of LRR proteins; blue) and leucines (red) were important for constraining the complete repeat-unit conformation.
The Y. pseudotuberculosis YPIII LRR1c structure was further compared with the YopM structure (PDB accession no. 1G9U). In the 1G9U structure, all the repeat units started and ended uniformly and conformed to corresponding repeat sequence units (Fig. 6d [starting positions of sequence units are indicated by red arrows]). The repeats with additional residues (QN) formed local larger loops, and the whole repeat structure was not influenced (Fig. 6d, blue arrows). However, in Y. pseudotuberculosis YPIII LRR1c proteins, the repeat structure appeared to not be uniform, and the starting and ending positions of repeat units were not always consistent with the corresponding repeat sequence units (Fig. 6d [starting positions of sequence units are indicated by red arrows]). The structure of repeats R1 to R3 and R8 to R10 was similar to that for the repeats in the 1G9U structure, while R4, R6, and R7 started from residues different from those of sequence repeats (Fig. 5d). Although the repeat-unit structures were similar overall, the conformation may have been unstable, causing the striking R5 conformation change due to torsion and tension. R5 was predicted to become a loop, interlinking the N- and C-terminal subdomains while maintaining the complete domain conformation (Fig. 6d).
LRR1c sequence analysis indicated that there were two deletions, in the R2 and R6 repeats (Fig. 3), and three mutations involving conserved leucines in R5 (L10S, L13F, and L17W). It was speculated that these changes could influence the thermodynamics of folding of the LRR domain. A structure modeling experiment was performed to estimate the influence of the R5 mutations and R2/R6 deletions. In one model with the 10th, 13th, and 17th residues of R5 restored back to the conserved leucine residues and another model with consensus residues added back to the R2 and R6 repeats, the overall structure appeared to be unstable and different from the 1G9U structure or the wild-type LRR1a3 structure (Fig. 6e; compare to Fig. 6d for the wild-type LRR1c structure and the 1G9U structure). However, if the R2, R5, and R6 repeats were restored together, the combination generated a very stable structure similar to the 1G9U structure (Fig. 6e, right panel). We hypothesized that the deletions within LRR1 repeat units (R2 and/or R6) imposed a structural selection pressure which prompted mutations in R5 residues, making the structure more stable and maintaining overall protein function.
DISCUSSION
In this study, we systematically investigated the distribution and evolution of LRR proteins in Yersinia. In contrast to the multiple copies of LRR family effector proteins that have been analyzed in other bacteria, such as Shigella and Salmonella, until now, only one such protein, YopM, was well studied for Yersinia (5, 35, 36, 47, 48). YopM is encoded by virulence plasmids and is sequentially similar to but still different from other LRR proteins in that most of the latter effectors contain an NEL domain in the C terminus and therefore can mimic E3 ligase activity in host cells (35, 36). In this study, through a comprehensive genome survey, we identified a large number of varied LRR proteins encoded in Yersinia chromosomes. Interestingly, nearly all of the proteins contain an NEL domain or have NEL domain-encoding nucleotide relics in the gene-flanking regions of the genome. The LRR proteins with a complete NEL domain may also have E3 ligase activity, like their homologs in other bacteria. Unlike those in Salmonella or Shigella, the chromosomally carried LRR protein genes in Yersinia were located in the same locus. Therefore, we hypothesize that the multiple copies of Yersinia LRR genes originated through gene amplifications rather than independent horizontal gene transfer events. Although many LRR proteins identified in bacteria have been validated to be T3SS effectors, the different copies of Yersinia LRR proteins may have varied functions, as do SlrP, SspH1, and SspH2 in Salmonella (47, 48). The expression of chromosomally encoded LRR proteins was partly detected in Y. pseudotuberculosis and was different from expression of the type III secreted effector YopM. It should be noted that there are two T3SSs: the chromosomally encoded Ysa system and the plasmid-encoded Ysc system (50). Therefore, the lack of coexpression of the genes with yopM does not exclude possible translocation of the LRR proteins through the Ysa T3SS. Alternatively, the LRR proteins may also enter host cells and act as effectors by other secretion mechanisms, such as autotranslocation (10). The LRR genes were not detected in the chromosomes of all Yersinia species, possibly due to acquisition of the ancestral genes after species diversification had occurred. The orthologous pairs in different species also seemed not to have identical copies of repeat units. Therefore, it would be interesting to investigate whether the genes function in the virulence specificity of different Yersinia species or serotypes.
The Yersinia LRR proteins are also attractive in evolutionary and structural biology due to their wide distribution among bacterial and eukaryotic cells, the highly conserved repeat consensus, and the varied number of tandem repeat copies (51, 52). It is clear that Yersinia chromosomally and plasmid-encoded LRR proteins originated from two different ancestors. The ancestor of the LRR1 genes encoded an NEL domain and 21-residue LRR units. However, the ancestor of the LRR2 genes acquired by Yersinia may have lost the NEL-encoding potential and encoded LRR units of only 20 residues. Despite many subgroups and members of LRR1 proteins in Yersinia, these proteins originated from two discernible common ancestors with amplified repeat units: one similar to LRR1d proteins and the other similar to LRR1a to -c proteins. However, it is not yet clear whether the form of the LRR1 gene(s) in the oldest Yersinia chromosome is that of the two tandemly arrayed LRR1 ancestor genes or their common older ancestor. Based on current data, the Yersinia chromosomally carried LRR1 and plasmid-carried LRR2 ancestors appear to have diverged for a much longer time, before amplification of the repeat units. The LRR proteins identified in other genera, such as Salmonella, Shigella, Escherichia, Providencia, and others, all contain NEL domains similar to those in Yersinia LRR1 proteins, but the repeat units have 20 residues, similar to those in Yersinia LRR2 proteins (data not shown). It is likely that the ancient LRR genes in bacteria possessed an NEL domain but lost the potential in the Yersinia plasmid ancestor.
Bacteria can adapt to specific environments by utilizing various gene-based mechanisms, such as whole-protein modification and/or domain modification. This was potentially observed within the Yersinia LRR proteins. We detected frequent LRR1 gene amplification and variation but a relatively stable repeat-unit copy number, in contrast to infrequent gene amplification and variation of LRR2 genes but frequent changes in the number of repeat units. The LRR2 proteins, despite the various numbers of repeat units, were predicted to have conserved protein conformation and topology. The LRR1 proteins were predicted to have a variable overall tertiary structure; however, in strains with multiple LRR1 proteins, there appeared to be at least one member displaying an overall topology similar to that of LRR2 proteins (e.g., Y. pestis CO92 LRR1a1 and Y. pseudotuberculosis YPIII LRR1c proteins). We hypothesize that these proteins have conserved and important functions. In general terms, there was a wide variety of mutations in the LRR1/2 repeat units, but the backbone of the repeat units appeared to be quite stable. Even for the mutations, maximum parsimony mutation steps were detected. This further implied the functional conservation of the repeats. We also detected potential synergistic mutations to maintain structural conservation in Yersinia LRR proteins. Future experiments could be designed to test whether the synergistic mutations are important for maintenance of the conserved LRR protein function.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by a National Natural Science Foundation of China grant to Y.W. (grant NSFC 81301390). X.H. was supported by Chinese National Undergraduate Training Programs for Innovation and Entrepreneurship (grant 201510590049).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.00324-16.
REFERENCES
- 1.Ye Z, Uittenbogaard AM, Cohen DA, Kaplan AM, Ambati J, Straley SC. 2011. Distinct CCR2(+) Gr1(+) cells control growth of the Yersinia pestis ΔyopM mutant in liver and spleen during systemic plague. Infect Immun 79:674–687. doi: 10.1128/IAI.00808-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ye Z, Gorman AA, Uittenbogaard AM, Myers-Morales T, Kaplan AM, Cohen DA, Straley SC. 2014. Caspase-3 mediates the pathogenic effect of Yersinia pestis YopM in liver of C57BL/6 mice and contributes to YopM's function in spleen. PLoS One 9:e110956. doi: 10.1371/journal.pone.0110956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McPhee JB, Mena P, Zhang Y, Bliska JB. 2012. Interleukin-10 induction is an important virulence function of the Yersinia pseudotuberculosis type III effector YopM. Infect Immun 80:2519–2527. doi: 10.1128/IAI.06364-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stasulli NM, Eichelberger KR, Price PA, Pechous RD, Montgomery SA, Parker JS, Goldman WE. 2015. Spatially distinct neutrophil responses within the inflammatory lesions of pneumonic plague. mBio 6:e01530-15. doi: 10.1128/mBio.01530-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leung KY, Straley SC. 1989. The yopM gene of Yersinia pestis encodes a released protein having homology with the human platelet surface protein GPIb alpha. J Bacteriol 171:4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leung KY, Reisner BS, Straley SC. 1990. YopM inhibits platelet aggregation and is necessary for virulence of Yersinia pestis in mice. Infect Immun 58:3262–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Skrzypek E, Straley SC. 1996. Interaction between Yersinia pestis YopM protein and human alpha-thrombin. Thromb Res 84:33–43. doi: 10.1016/0049-3848(96)00159-4. [DOI] [PubMed] [Google Scholar]
- 8.Boland A, Sory MP, Iriarte M, Kerbourch C, Wattiau P, Cornelis GR. 1996. Status of YopM and YopN in the Yersinia Yop virulon: YopM of Y. enterocolitica is internalized inside the cytosol of PU5-1.8 macrophages by the YopB, D, N delivery apparatus. EMBO J 15:5191–5201. [PMC free article] [PubMed] [Google Scholar]
- 9.Rüter C, Buss C, Scharnert J, Heusipp G, Schmidt MA. 2010. A newly identified bacterial cell-penetrating peptide that reduces the transcription of pro-inflammatory cytokines. J Cell Sci 123:2190–2198. doi: 10.1242/jcs.063016. [DOI] [PubMed] [Google Scholar]
- 10.Scharnert J, Greune L, Zeuschner D, Lubos ML, Alexander Schmidt M, Rüter C. 2013. Autonomous translocation and intracellular trafficking of the cell-penetrating and immune-suppressive effector protein YopM. Cell Mol Life Sci 70:4809–4823. doi: 10.1007/s00018-013-1413-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemeth J, Straley SC. 1997. Effect of Yersinia pestis YopM on experimental plague. Infect Immun 65:924–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skrzypek E, Cowan C, Straley SC. 1998. Targeting of the Yersinia pestis YopM protein into HeLa cells and intracellular trafficking to the nucleus. Mol Microbiol 30:1051–1065. doi: 10.1046/j.1365-2958.1998.01135.x. [DOI] [PubMed] [Google Scholar]
- 13.Skrzypek E, Myers-Morales T, Whiteheart SW, Straley SC. 2003. Application of a Saccharomyces cerevisiae model to study requirements for trafficking of Yersinia pestis YopM in eucaryotic cells. Infect Immun 71:937–947. doi: 10.1128/IAI.71.2.937-947.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benabdillah R, Mota LJ, Lützelschwab S, Demoinet E, Cornelis GR. 2004. Identification of a nuclear targeting signal in YopM from Yersinia spp. Microb Pathog 36:247–261. doi: 10.1016/j.micpath.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 15.McDonald C, Vacratsis PO, Bliska JB, Dixon JE. 2003. The yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J Biol Chem 278:18514–18523. doi: 10.1074/jbc.M301226200. [DOI] [PubMed] [Google Scholar]
- 16.Hentschke M, Berneking L, Belmar Campos C, Buck F, Ruckdeschel K, Aepfelbacher M. 2010. Yersinia virulence factor YopM induces sustained RSK activation by interfering with dephosphorylation. PLoS One 5:e13165. doi: 10.1371/journal.pone.0013165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCoy MW, Marré ML, Lesser CF, Mecsas J. 2010. The C-terminal tail of Yersinia pseudotuberculosis YopM is critical for interacting with RSK1 and for virulence. Infect Immun 78:2584–2598. doi: 10.1128/IAI.00141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McPhee JB, Mena P, Bliska JB. 2010. Delineation of regions of the Yersinia YopM protein required for interaction with the RSK1 and PRK2 host kinases and their requirement for interleukin-10 production and virulence. Infect Immun 78:3529–3539. doi: 10.1128/IAI.00269-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Höfling S, Scharnert J, Cromme C, Bertrand J, Pap T, Schmidt MA, Rüter C. 2014. Manipulation of pro-inflammatory cytokine production by the bacterial cell-penetrating effector protein YopM is independent of its interaction with host cell kinases RSK1 and PRK2. Virulence 5:761–771. doi: 10.4161/viru.29062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kerschen EJ, Cohen DA, Kaplan AM, Straley SC. 2004. The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells. Infect Immun 72:4589–4602. doi: 10.1128/IAI.72.8.4589-4602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marketon MM, DePaolo RW, DeBord KL, Jabri B, Schneewind O. 2005. Plague bacteria target immune cells during infection. Science 309:1739–1741. doi: 10.1126/science.1114580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye Z, Kerschen EJ, Cohen DA, Kaplan AM, van Rooijen N, Straley SC. 2009. Gr1+ cells control growth of YopM-negative Yersinia pestis during systemic plague. Infect Immun 77:3791–3806. doi: 10.1128/IAI.00284-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uittenbogaard AM, Chelvarajan RL, Myers-Morales T, Gorman AA, Brickey WJ, Ye Z, Kaplan AM, Cohen DA, Ting JP, Straley SC. 2012. Toward a molecular pathogenic pathway for Yersinia pestis YopM. Front Cell Infect Microbiol 2:155. doi: 10.3389/fcimb.2012.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.LaRock CN, Cookson BT. 2012. The Yersinia virulence effector YopM binds caspase-1 to arrest inflammasome assembly and processing. Cell Host Microbe 12:799–805. doi: 10.1016/j.chom.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung LK, Philip NH, Schmidt VA, Koller A, Strowig T, Flavell RA, Brodsky IE, Bliska JB. 2014. IQGAP1 is important for activation of caspase-1 in macrophages and is targeted by Yersinia pestis type III effector YopM. mBio 5:e01402–14. doi: 10.1128/mBio.01402-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evdokimov AG, Anderson DE, Routzahn KM, Waugh DS. 2001. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J Mol Biol 312:807–821. doi: 10.1006/jmbi.2001.4973. [DOI] [PubMed] [Google Scholar]
- 27.Boland A, Havaux S, Cornelis GR. 1998. Heterogeneity of the Yersinia YopM protein. Microb Pathog 25:343–348. doi: 10.1006/mpat.1998.0247. [DOI] [PubMed] [Google Scholar]
- 28.Foultier B, Cornelis GR. 2003. DNA sequence and analysis of the pYVa127/90 virulence plasmid of Yersinia enterocolitica strain A127/90. Res Microbiol 154:553–557. doi: 10.1016/S0923-2508(03)00147-5. [DOI] [PubMed] [Google Scholar]
- 29.Kloss E, Barrick D. 2008. Thermodynamics, kinetics, and salt dependence of folding of YopM, a large leucine-rich repeat protein. J Mol Biol 383:1195–1209. doi: 10.1016/j.jmb.2008.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kloss E, Barrick D. 2009. C-terminal deletion of leucine-rich repeats from YopM reveals a heterogeneous distribution of stability in a cooperatively folded protein. Protein Sci 18:1948–1960. doi: 10.1002/pro.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vieux EF, Barrick D. 2011. Deletion of internal structured repeats increases the stability of a leucine-rich repeat protein, YopM. Biophys Chem 159:152–161. doi: 10.1016/j.bpc.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McPhee JB, Bliska JB. 2011. Letter to the editor and response. Innate Immun 17:558–559. doi: 10.1177/1753425911426251. [DOI] [PubMed] [Google Scholar]
- 33.Soundararajan V, Raman R, Raguram S, Sasisekharan V, Sasisekharan R. 2010. Atomic interaction networks in the core of protein domains and their native folds. PLoS One 5:e9391. doi: 10.1371/journal.pone.0009391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Soundararajan V, Patel N, Subramanian V, Sasisekharan V, Sasisekharan R. 2011. The many faces of the YopM effector from plague causative bacterium Yersinia pestis and its implications for host immune modulation. Innate Immun 17:548–557. doi: 10.1177/1753425910377099. [DOI] [PubMed] [Google Scholar]
- 35.Zhu Y, Li H, Hu L, Wang J, Zhou Y, Pang Z, Liu L, Shao F. 2008. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15:1302–1308. doi: 10.1038/nsmb.1517. [DOI] [PubMed] [Google Scholar]
- 36.Singer AU, Rohde JR, Lam R, Skarina T, Kagan O, Dileo R, Chirgadze NY, Cuff ME, Joachimiak A, Tyers M, Sansonetti PJ, Parsot C, Savchenko A. 2008. Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol 15:1293–1301. doi: 10.1038/nsmb.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strauch E, Hoffmann B, Heins G, Appel B. 2000. Isolation of a new insertion element of Yersinia intermedia closely related to remnants of mobile genetic elements present on Yersinia plasmids harboring the Yop virulon. FEMS Microbiol Lett 193:37–44. doi: 10.1111/j.1574-6968.2000.tb09399.x. [DOI] [PubMed] [Google Scholar]
- 38.Reisner BS, Straley SC. 1992. Yersinia pestis YopM: thrombin binding and overexpression. Infect Immun 60:5242–5252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. 2002. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res 30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W. 2000. PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586. doi: 10.1101/gr.10.4.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nuss AM, Heroven AK, Waldmann B, Reinkensmeier J, Jarek M, Beckstette M, Dersch P. 2015. Transcriptomic profiling of Yersinia pseudotuberculosis reveals reprogramming of the Crp regulon by temperature and uncovers Crp as a master regulator of small RNAs. PLoS Genet 11:e1005087. doi: 10.1371/journal.pgen.1005087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Avican K, Fahlgren A, Huss M, Heroven AK, Beckstette M, Dersch P, Fällman M. 2015. Reprogramming of Yersinia from virulent to persistent mode revealed by complex in vivo RNA-seq analysis. PLoS Pathog 11:e1004600. doi: 10.1371/journal.ppat.1004600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, MacKenzie KD, White AP. 2015. An empirical strategy to detect bacterial transcript structure from directional RNA-seq transcriptome data. BMC Genomics 16:359. doi: 10.1186/s12864-015-1555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsolis RM, Townsend SM, Miao EA, Miller SI, Ficht TA, Adams LG, Bäumler AJ. 1999. Identification of a putative Salmonella enterica serotype Typhimurium host range factor with homology to IpaH and YopM by signature-tagged mutagenesis. Infect Immun 67:6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miao EA, Scherer CA, Tsolis RM, Kingsley RA, Adams LG, Bäumler AJ, Miller SI. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol Microbiol 34:850–864. doi: 10.1046/j.1365-2958.1999.01651.x. [DOI] [PubMed] [Google Scholar]
- 49.Reuter S, Connor TR, Barquist L, Walker D, Feltwell T, Harris SR, Fookes M, Hall ME, Petty NK, Fuchs TM, Corander J, Dufour M, Ringwood T, Savin C, Bouchier C, Martin L, Miettinen M, Shubin M, Riehm JM, Laukkanen-Ninios R, Sihvonen LM, Siitonen A, Skurnik M, Falcão JP, Fukushima H, Scholz HC, Prentice MB, Wren BW, Parkhill J, Carniel E, Achtman M, McNally A, Thomson NR. 2014. Parallel independent evolution of pathogenicity within the genus Yersinia. Proc Natl Acad Sci U S A 111:6768–6773. doi: 10.1073/pnas.1317161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Huang H, Sun M, Zhang Q, Guo D. 2012. T3DB: an integrated database for bacterial type III secretion system. BMC Bioinformatics 13:66. doi: 10.1186/1471-2105-13-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buchanan SG, Gay NJ. 1996. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol 65:1–44. [DOI] [PubMed] [Google Scholar]
- 52.Rämisch S, Weininger U, Martinsson J, Akke M, André I. 2014. Computational design of a leucine-rich repeat protein with a predefined geometry. Proc Natl Acad Sci U S A 111:17875–17880. doi: 10.1073/pnas.1413638111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.