Abstract
Traumatic brain injuries (TBIs) present a chief public health threat affecting nations worldwide. As numbers of patients afflicted by TBI are expected to rise, the necessity to increase our understanding of the pathophysiological mechanism(s) as a result of TBI mounts. TBI is known to augment the risk of developing a number of neurodegenerative diseases (NDs) such as Alzheimer’s disease (AD) and Parkinson’s disease (PD). Hence, it is rational to assume that a common mechanistic ground links the pathophysiology of NDs to that of TBIs. Through this review, we aim to identify the protein–protein interactions, differential proteins expression, and PTMs, mainly glycosylation, that are involved in the pathogenesis of both ND and TBI. OVID and PubMed have been rigorously searched to identify studies that utilized advanced proteomic platforms (MS based) and systems biology tools to unfold the mechanism(s) behind ND in an attempt to unveil the mysterious biological processes that occur postinjury. Various PTMs have been found to be common between TBI and AD, whereas no similarities have been found between TBI and PD. Phosphorylated tau protein, glycosylated amyloid precursor protein, and many other modifications appear to be common in both TBI and AD. PTMs, differential protein profiles, and altered biological pathways appear to have critical roles in ND processes by interfering with their pathological condition in a manner similar to TBI. Advancement in glycoproteomic studies pertaining to ND and TBI is urgently needed in order to develop better diagnostic tools, therapies, and more favorable prognoses.
Keywords: Glycans, Glycomics, Neurodegenerative diseases, PTMs, TBI
1 Introduction
After the recent advancement of the now indispensable tool of mass spectrometry (MS), the field of large-scale analysis of proteins, termed proteomics [1], has allowed the scientific community to further our understanding of molecular and cellular biology and has thoroughly impacted the emerging field of systems biology [2]. Proteomics has greatly increased our knowledge of human physiology and disease. Clinical implications of proteomics have led to a better understanding of disease processes, the development of novel biomarkers discovery for diagnosis and early detection of diseases, as well as the development of new drugs [3]. Of particular interest are the protein–protein interactions, differential protein expression profiles, protein quantification, and post translational modifications (PTMs) related to neurodegenerative diseases (NDs) and traumatic brain injury (TBI).
In this paper, we aimed to review the different types of PTMs in two major NDs, Alzheimer’s disease (AD) and Parkinson’s disease (PD), along with TBI, with a main focus on glycosylation changes. For this purpose, articles published between 1946 and November 2015 were retrieved via the online databases PubMed and MEDLINE. Keywords and combinations related to NDs (AD and PD), TBI (brain damage, brain injury, concussion, etc.), proteomics, and PTMs (glycans, glycosylation, glycomics, etc.) were used to perform the search. Two independent reviewers evaluated the abstracts of the articles, and data extraction was then performed on the relevant articles that met our objectives for the review.
2 PTMs
Proteins are large macromolecules that comprise a specific sequence of amino acids. After a protein is synthesized, its function is modulated by PTMs and structural changes such as folding and refolding [4]. PTMs comprise the addition of various functional groups to amino acids such as acetate and phosphate [5]. Currently, more than 300 different types of PTMs are known, ranging from single atom modifications (e.g., oxide) to small protein modifiers (e.g., ubiquitin) [6]. The activity and function of most proteins in molecular pathways are modulated by various PTMs [7], which are known to influence protein turnover and localization, protein–protein interactions, signaling cascades, enzymatic activity, and cell division [4, 8]. Nevertheless, the function of most PTMs is still unknown since the rate of their discovery surpasses the rate at which any one modification can be studied empirically [6]. Approximately 80% of mammalian proteins are modified posttranslationally. For instance, phosphorylation is implicated in cell signaling [9, 10], acetylation in epigenetics [11] and NDs [12], ubiquitination in regulation of cellular protein homeostasis [13], hydroxylation in collagen formation [14], and glycosylation in the pathophysiology of neurotrauma [15–17]. The basic findings from studies that demonstrated the role of PTMs in the relevant diseases are summarized in Table 1.
Table 1.
List of glycoproteins, corresponding pathologies, and suggested therapeutic interventions
| Glycoprotein | Significance | PTM | Disease(s) and associated pathologies |
Underlying mechanism |
Potential therapeutic modalities |
Research model(s) |
Reference (s) |
|---|---|---|---|---|---|---|---|
| Lectin targets (galectin, siglec) |
Development, axonal tracing and neurite fasciculation |
N-acetyllactosamine | Impaired repair and regeneration.Liver cirrhosis.Heart failure.Neoplastic transformation |
Upregulation of lectins | Galectin-3 inhibitor; TD139 | RatMouseSTEM cells |
53,58 |
| Neural cell adhesion molecules (neuroserpin) |
Neurogenesis | Polysialic acid and N-glycosylation |
Alzheimer’s diseaseCancer (myeloma, myeloid leukemia, lymphoma, and others) |
Signaling through axon guidance receptors (EphA2, neuropilin-1, Robo, and DCC) |
Radioimmunolocalization of metastases using NCAM-binding radioim- munoconjugates |
RatCells | 142,143 |
| β-Amyloid | Pro-inflammatory and cholesterol regulation |
Bisecting N-GlcNAc | Alzheimer’s dis- ease.Dementia.Amyloid angiopathy |
Increase in N-acetylglucosaminyltransferase III (GnT-III) producingβ-amyloid that causes oxidative stress, lipid peroxidation and mitochondrial impairment |
Upregulation of the expression of bisecting N-glycans |
Mouse | 93 |
| Tau | Cytoskeletal stabilization |
N-glycosylation and hyperphosphoryla- tion |
Alzheimer’s disease.Parkinson’s dis- ease.Dementia.Traumatic brain injury |
Defective tau weakly stabilizes cytoskeletal microtubules.Tau activity decreases with age |
New treatment with curcumin is under investigation.Early detection by associated biomarkers |
Saliva samples | [144] |
| Transferrin (ceruloplasmin) |
Iron homeostasis | N-glycosylation | Alzheimer’s disease.Parkinson’s disease.Iron overload anemia.Protein malnutrition |
Abnormal binding to available iron | Targeting transferrin (Trf) and its receptor |
CSF samples | 145 |
| Neuronal pentraxin | Pattern recognition receptor; acute immune response and neuronal plasticity |
N-glycosylation | Parkinson’s disease | Immune response and acute inflammation, acute phase proteins |
Goeckerman’s therapy (GT) |
Human SN and frontal cortex |
104 |
| Nicastrin | Part of the complex that cleaves APP |
O-GlcNAcylated | Alzheimer’s disease | Reduction of γ-secretase activity and Aβ production |
Antinicastrin antibodies | CSF samples | 90 |
| α-Synuclein | Synaptic maintenance | O-glycosylation | Parkinson’s dis- ease.Dememtia.System atro- phy.Synucleinopathies |
Interacts with tubulin as a potential microtubule-associated protein |
Inhibit aggregation of α-synuclein-like cuminaldehyde |
MouseCells | 140 |
| Biglycan | Antoi-apototic | Glycosaminoglycan- like glycosylation |
Alzheimer’s disease.Parkinson’s disease.Amyotrophic lateral sclerosis.Multiple sclerosis.Huntington’s disease |
NF-κB mediated NO-induced apoptotic cell death |
Biglycan gene therapy | Cells | 148 |
| Acetylcholinesterase (also butyryl- cholinesterase) |
Cleaves Ach | Amphiphilic dimeric and monomeric AChE isoforms |
Alzheimer’s disease (AChE is almost specific).Muscular paralysis and convulsions |
Increased Ach in the frontal cortex.Increase in amyloidStress and inflammation |
Pharmacological intervention |
CSF samples | 89 |
| PP2A | Major brain tau phosphatase |
Methylation and glycosylation |
Parkinson’s disease.Alzheimer’s disease.Traumatic brain injury.Cancer |
Reduce tau hyperphosphorylation | PP2 as a biological target but recent evidence suggest its role as a tumor suppressor protein |
Rat | 149 |
2.1 Introduction to glycosylation
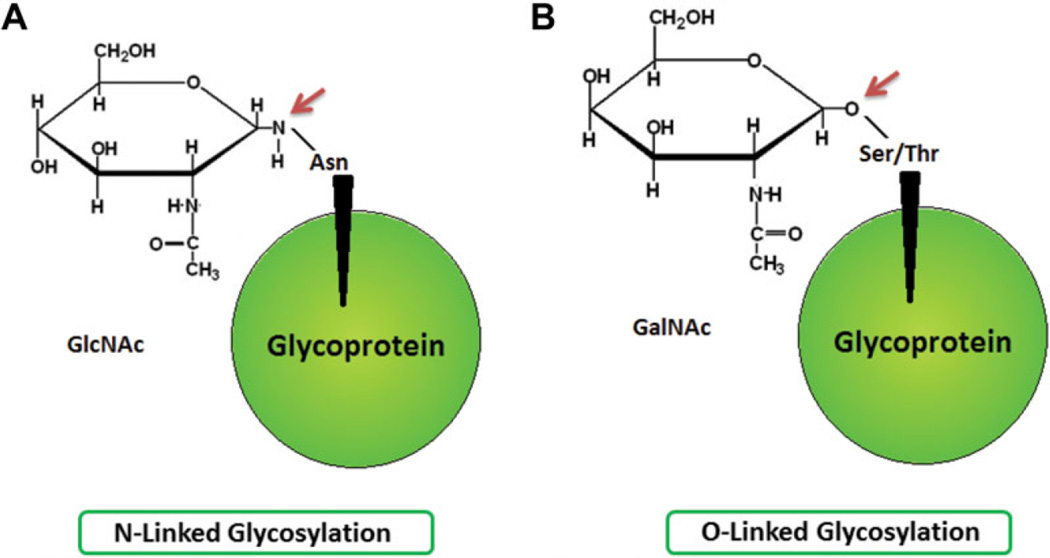
The most common PTM of proteins is glycosylation, as protein sequencing data suggest that it represents more than half of all mammalian cell protein modifications [18, 19]. Glycosylation is a cotranslational and/or posttranslational mechanism by which a carbohydrate moiety is added to a lipid, protein, or other organic molecule within or outside of the cell. The glycosylation process is tightly regulated as it is an enzymatic modification that is site and substrate specific [4]. Indeed, glycans of secreted glycoproteins are proven to affect various protein properties such as solubility, whereas cell surface glycosylated proteins are shown to be implicated in various cellular processes such as cell–cell communication. [20]. Two major classification groups of the major glycans of glycoproteins exist and are categorized according to their glycan–peptide bonds, namely the N- and O-glycans. N-glycans represent the linkage of the amino sugar derivative of glucose N-acetylglucosamine (GlcNAc) to the amide group of asparagine, whereas O-glycans represent the linkage of N-acetylgalactosamine, an amino sugar derivative of galactose, to the hydroxyl groups of serine or threonine amino acid residues of polypeptides (Fig. 1) [19].
Figure 1.
Biochemical structures of different N- and O-glycans. (A) Linkage of N-acetylglucosamine to asparagine amino acid via an N-linked bond. (B) Linkage of N-acetylgalactosamine to serine or threonine amino acids via an O-linked bond.
2.2 Characterization of glycoproteins
The initial step for characterization of glycosylated proteins is by isolating them from their complex biological samples that contains both nonglycosylated and diversely glycosylated proteins. After isolation, glycoproteins/glycopeptides are enriched, subjected to proteolytic digestion, and finally detected/identified using MS-based techniques via glycoproteomics platforms, which are a subset of the proteomics discipline [19]. In general, various forms of HPLC such as ion exchange methods, hydrophobic interactions, size exclusions, and affinity chromatography can be used to purify and separate glycoproteins. The required analytical methods should be fast and robust in order to study the altered glycosylation profiles induced by the specific diseases. [21]. Several techniques have been previously used for the characterization of glycoproteins, namely, methods based on lectin affinity, hydrazide chemistry, and enrichment at the glycopeptide or protein level; the latter method comprises deglycosylation and other chemical methods [21,22].
2.3 Characterization of glycoproteins by MS
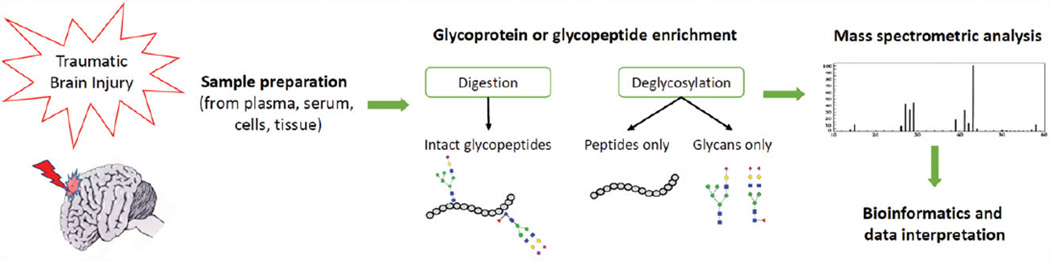
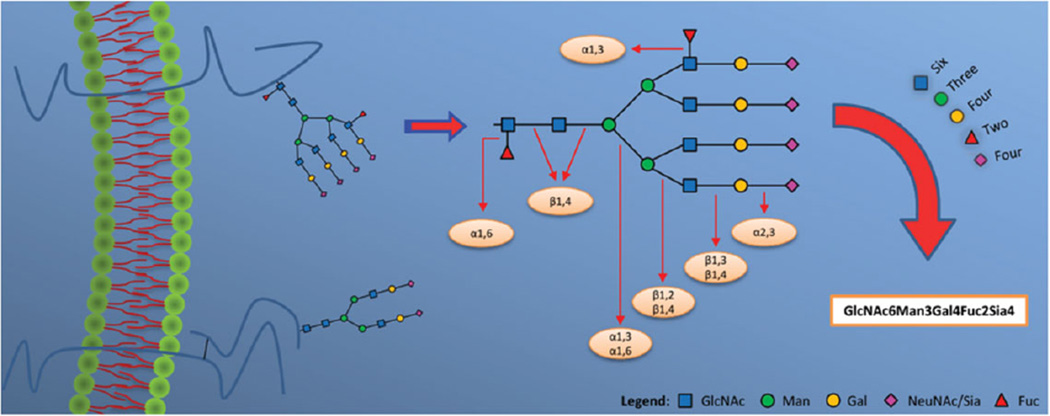
MS provides significant pieces of information on proteins with PTMs. This is especially important when it comes to quantitatively comparing two or more samples. To examine disease-associated changes in glycosylation patterns, sensitive, quick, reliable, and robust analytical techniques are desirable. Despite the many methods that can be used to identify glycoproteins, the field of glycoproteomics remains challenging, yet very promising. MS techniques have significantly reduced the limitations of glycoprotein profiling, especially in complex types of samples such as plasma, serum, and body fluids [23, 24]. Recent advances in MS technology have also allowed for the use of more accurate approaches in the characterization of glycoproteins. By analyzing the glycopeptide-based results, site-specific information reflecting the site of attachment of the glycan on the protein can be determined, which subsequently indicates the putative functional roles and properties of those modifications. As shown in Fig. 2, glycoproteins are initially enzymatically cleaved by an endoprotease followed by performing separation techniques coupled with mass analysis. Figure 3 shows an example on the nomenclature and topology of glycans.
Figure 2.
An overview on the workflow of MS-based glycoproteomics. After taking a brain sample following TBI, several steps are performed to enrich and digest the proteins in order to administer the sample into the MS.
Figure 3.
An example on the nomenclature, topology, and glycosylation patterns of glycans. The glycoprotein depicted is an example of a transmembrane protein. The possible bond linkages between glycan residues are shown. GlcNAc: N-acetylglucosamine; Man: mannose; Gal: galactose; NeuNAc/Sia: N-acetylneuraminic acid/sialic acid; Fuc: fucose.
2.4 Quantitative glycoproteomics
Quantitative glycoproteomics is beneficial in assessing the level of glycoproteins reflecting certain cellular states, whereby isotope-labeling techniques are commonly used for this purpose. For instance, specific functional groups can be tagged isotopically via certain chemical reactions, particularly ICATs, isobaric tags for relative and absolute quantitation , or enzymatic 18O incorporation [25–27]. Cysteinyl residues are primarily labeled by ICAT at their side chains to generate the specific mass values, thus facilitating identification and quantification of specific proteins [28]. However, ICAT selectively searches for cysteine residues and therefore an enormous percentage of proteins that are deficient of this amino acid might be missed. In addition, some residues are inaccessible to the reagent [28].
3 Glycoproteomics in NDs
Long-term effects of TBI have been demonstrated to increase the risk of ND [29], suggesting that a common ground unites TBI and ND. PTMs of AD and PD will be sought to address this idea. PTM alterations have been reported to contribute to the variations in health and disease homeostasis, as it has been assumed that they regulate protein–protein interactions and cellular signaling pathways. However, once the physiology of PTM has been impaired, deleterious consequences could occur [30]. AD is the most common type of dementia in aging adults [31] and is followed by PD [32]. Rates of both diseases are projected to increase significantly over the next few years as the global population ages. Both diseases impose a substantial burden to patients, caregivers, and healthcare systems worldwide [31,32]. AD is characterized by the presence of two histopathologically brain lesions, namely senile plaques, where β-amyloid plaque is the major constituent, and neurofibrillary tangles, where tau protein is the major component [33]. As for PD, it results from degeneration of dopaminergic neurons in the substantia nigra of the midbrain and the deposition of neuronal Lewy bodies [32]. Clinically, AD manifests by cognitive deterioration and memory loss [34], whereas in PD both motor and nonmotor symptoms may be observed, such as resting tremors, rigidity, bradykinesia, stooping posture, and cognitive impairment [35]. The pathophysiological and clinical findings of AD, PD, Huntington’s disease (HD), amyotrophic lateral sclerosis (ALS), and multiple sclerosis are summarized in Table 2.
Table 2.
Neurodegenerative diseases: Description of pathophysiology and clinical picture
| Disease | Pathophysiology | Clinical picture |
|---|---|---|
| Alzheimer disease [31, 33, 34] |
|
|
| Parkinson’s disease [32, 35] |
Degeneration of dopaminergic neurons in
|
|
| Amyotrophic lateral sclerosis [124, 125] |
Mutation in Cu/Zn superoxide dismutase (SOD1)
|
|
| Huntington’s disease [126] |
Elongated CAG repeat (36 repeats or more) on the short arm of chromosome 4p16.3 in the Huntingtin gene |
|
| Multiple sclerosis [127] |
Autoimmune diseases that causes inflammatory demyelination, axon degeneration, and neuronal loss |
|
The relevance of glycoproteins to ND has been widely demonstrated in the literature. In the following section, we discuss the implication of different glycoprotein modifications and their implications on neural activity. Gene knockout studies in mice have shown that some glycosyltransferase enzymes that are known to be responsible for the biosynthesis of glycans are indispensible for neural development and that their dysfunction may lead to neurological manifestations [36]. Endo reported that aberrant O-mannosylation has altered the normal neural migration, leading to several congenital disorders such as muscular dystrophy [37].
Moreover, the role of lectins, which are glycan-binding proteins, in neurological maintenance has been tackled formerly. In fact, lectins have been classified into galectin, a β-galactose-binding lectin proposed to regulate cellular apoptosis [38], mannose-binding lectin with implications for immunity [39], and the sialic acid binding immunoglobulin superfamily lectin, termed “Siglec,” that serves a role on cell surface of immune cells [40]. Research studies on galectins have identified their key role during brain development, involving axonal tracing and neurite fasciculation [41–43] along with developmental regulation in terms of expression and localization [44]. For example, galectin-1 knockout mice have been shown to prevent neurite processes outgrowth in olfactory neurons [45].
In addition, several reports have pointed out the contribution of O-GlcNAcylation to neurogenesis and neuronal morphology. Indeed, O-GlcNAcylation, also referred to as O-linked β-N-acetylglucosamine (O-GlcNAc) modification, involves the transfer of a single sugar (GlcNAc) to the hydroxyl group of Ser and Thr residues of proteins. O-GlcNAcylation can be intracellularly mediated by O-GlcNAc transferase or extracellularly mediated by epidermal growth factor domain-specific O-GlcNAc transferase [46–48]. O-GlcNAc transferase expression, enriched at neuronal synapses [49, 50], has been found to dynamically modify various neuronal proteins related to synaptic function, learning, memory, and neurodegeneration [51–53]. A recent neuroproteomic study has identified 249 O-GlcNAcylated proteins in mouse cerebral cortex [54]. Several pre- and postsynaptic proteins including synapsin, piccolo, bassoon, and shank2 proteins have been found to be extensively O-GlcNAcylated [55].
3.1 Glycosylation in AD
As previously mentioned, alterations in protein glycosylation have been implicated in AD pathogenesis [56,57]. A follow-up study by Sato et al. that has investigated the primary structure of N-glycans obtained from paired helical filaments tau and AD phosphorylated-tau has showed that paired helical filaments tau was more rich in truncated glycans than is AD phosphorylated-tau, which has been suspected to be involved in stabilizing the pathological fibrils and promoting their assembly in AD [58].
Several studies have also showed that tau, known to function in microtubule stabilization [59,60], can be glycosylated by O-GlcNAc, which has been shown to be inversely proportional to the amount of phosphorylation [61, 62]. Yuzwa et al. have demonstrated that increasing O-GlcNAc slows neurodegeneration and stabilizes tau against aggregation [63]. In the same context, Borghgraef et al. have validated that increasing O-GlcNAcylation in mice brains by pharmacological inhibition of O-GlcNAc-ase has improved clinical conditions and mitigated mortality of Tau.P301L mice [63, 64]. Therefore, O-GlcNAcase inhibition might be a potential therapeutic strategy for attenuating the propagation of tau pathology in AD and other tauopathies.
Moreover, different studies have assessed glycosylation of the amyloid precursor protein (APP) that can alter its physiological role in AD. Cleavage of APP, catalyzed by α-secretase, β-secretase, and γ-secretase complex [65–67], is known to occur either via a nonamyloidogenic pathway producing nontoxic peptide fragments, or via the amyloidogenic pathway generating Aβ40 or Aβ42, which are major components of the senile plaque amyloids [68]. It has been reported that in SH-SY5Y neuronal cells PUGNAc, an inhibitor of β-N-acetylglucosaminidase, which catalyzes the removal of O-GlcNAc, increases O-GlcNAcylation and nonamyloidogenic processing of APP, resulting in a decrease in secreted Aβ40 [69]. An important impact of glycosylation in AD disease is emphasized by a study on acetylcholinesterase (AChE) by Saez-Valero et al., where it was shown that AChE possesses an altered glycosylation pattern in postmortem brain tissue and cerebrospinal fluid (CSF) of AD patients [70,71]. Notably, glycosylation of AChE has not been seen in other NDs yet, so it is considered specific to AD. In fact, AChE is one of the critical enzymes targeted in the current clinical management of AD, as it is widely distributed in brain regions since it hydrolyzes the neurotransmitter acetylcholine at cholinergic synapses. More recently, the glycosylation of a related enzyme, butyrylcholinesterase, appears to be altered in CSF of patients with AD [72].
Several PTMs besides glycosylation have been identified in AD. Sumoylation, a PTM catalyzed by small ubiquitin like modifier, has been shown to regulate APP and tau proteins and may modulate other proteins in AD [73]. Morris et al. have shown that endogenous tau protein from transgenic mice expressing human APP versus that of its wild-type littermate can be subject to different PTMs by the use of MS such as acetylation [74–76], ubiquitination [77], or methylation of lysine residues [78–80].
3.2 Glycosylation in PD
As in AD, PTMs are implicated in a similar manner in PD. PD is a progressive neurodegenerative disorder that occurs sporadically in most cases. It is recognized as the second most prevalent neurodegenerative disorder, currently affecting more than 1% of the worldwide population over the age of 65 [81], and the most prevalent degenerative movement disorder.
A study by Hwang et al. characterized overlapping glycoproteins from CSF and human brain using MALDI-TOF-TOF analysis [82]. The prospective study revealed a total of 243 nonredundant glycoproteins in human CSF and 34 nonredundant glycoproteins in brain tissue with known or probable glycosylation sites. Remarkably, the classification of glycoproteins by gene ontology analysis showed that several overlapping glycoproteins between human CSF and brain tissue are linked to PD pathogenesis such as pentraxin and neuroserpin [82]. Similarly, Moran et al. previously showed that an overlapping glycoprotein, neuronal pentraxin II, is vastly up-regulated in PD [83]. Another study by Pisani et al. has showed that another overlapping glycoprotein, neuroserpin, has been predominantly expressed in neuronal tissues during the late stages of neurogenesis [84].
Knowing that sodium channel β4 contains four potential N-linked glycosylation sites in the extracellular region sub-unit and its expression increases with PD progression, Zhou et al. have characterized the impact of glycosylation of β4 on PD pathogenesis in Neuro2a cells through comparing the expression of the β4-WT and β4-MUT plasmids [85]. They have reported that overexpression of β4-MUT was correlated with extension of neurites and an increase in the number of filopodia-like protrusions when compared with cells expressing β4-WT, and this has indicated that the alteration of glycosylation of β4 may be involved in the pathogenesis of PD [85].
3.3 Glycosylation in multiple sclerosis
Vital scientific cues link multiple sclerosis, an autoimmune disease, and a neurodegenerative disorder of the CNS, to amendments in glycoproteins [86]. Lee et al. have demonstrated that experimental autoimmune encephalomyelitis susceptible mice have reduced GlcNAc branching, which had been subsequently justified by hypomorphisms in multiple N-glycan GlcNAc-branching enzymes [87]. Nutritional and environmental factors, such as vitamin D metabolism, has also shown to converge with multiple genetic factors to dysregulate a common pathway, Golgi N-glycosylation, and consequently alter cytotoxic T-lymphocyte antigen 4 surface retention, which may lead to pathological manifestations such as inflammatory demyelination and neurodegeneration characteristic to multiple sclerosis [88]. Thus, such findings have warranted the possibility of therapeutic supplementation to N-glycan biosynthesis as a possible therapy to suppress phenotypic expression of multiple sclerosis.
3.4 Glycosylation in HD
HD is an inherited ND caused by a mutation of the huntingtin (HTT) gene [89]. At the glycoproteomics level, an imbalance in ganglioside metabolism has been found in HD caused by an altered expression of the genes encoding glycosyltransferases that are involved in the synthesis of gangliosides [90]. Gizaw et al. have conducted the first study on the total glycomics in HD brain tissue and sera of HD transgenic and control mice by using the glycoblotting method and MALDI-TOF/MS where significant alterations in the total glycome expression levels between HD transgenic and control group mice have been found. Interestingly, they have reported gender-specific differences in the expression level of the brain glycome, with an increase in brain tissue levels of core-fucosylated and bisecting-GlcNAc types of N-glycans and an increase in the sera levels of core-fucosylated and sialic acid for biantennary-type glycans [91].
3.5 Glycosylation in ALS
ALS is an ND that is characterized by a progressive loss of motor neurons and subsequent muscular atrophy. Ludemann et al. have shown that O-glycosylation of the tail domain of mammalian neurofilament protein (NF-M) is decreased in an ALS mouse model as compared to the wild type [92]. Interestingly, evidence exists on Golgi apparatus disruption in motor and this has suggested alterations in the glycosylation patterns of secretory proteins in ALS [93]. Furthermore, Edri-Brami et al. have investigated the glycome of ALS using patient sera and found high levels of sialylated glycans versus low levels of core fucosylated glycans in sera of ALS patients as compared to sera of healthy volunteers [94].
4 Traumatic brain injury
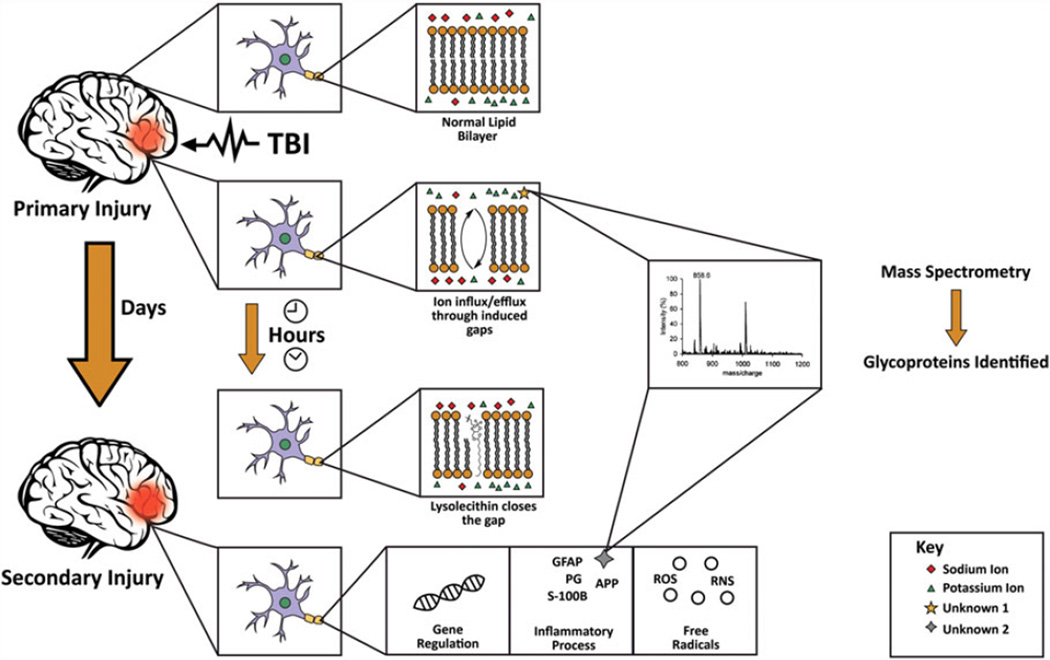
TBI represents a major public health problem causing significant long-term disability worldwide. TBI has been projected to surpass many diseases and become the third leading cause of death and disability by the year 2020 [95]. TBI, as described by the Centers for Disease Control and Prevention (CDC), “is caused by a bump, blow or jolt to the head or a penetrating head injury that disrupts the normal function of the brain” [96]. Immediately following TBI, primary insult causes a distortion in the lipid bilayer in the brain parenchyma [97] resulting in an efflux of potassium and influx of sodium, calcium, and chloride across the disrupted cellular membrane [98]. Delayed cellular dysfunction following this initial mechanical event occurs by four main mechanisms. The mechanisms are carried out primarily through inflammation, reactive oxygen and nitrogen free radical species and oxidative damage, calcium or other ion-mediated damage, and receptor-mediated dysfunction [99]. These processes modulate gene expression in turn and ultimately lead to the aggravation of neural injury. Secondary injury is initiated several hours to days after the initial injury and induces several intracerebral sequelae including intracranial hypertension and ischemia due to vascular failure [100] through commencing a cluster of biochemical reactions and cascades that are attributed to neuroproteomic alterations and PTMs (Fig. 4). Table 3 presents different biomarkers isolated from different bodily sources following either primary or secondary brain injuries.
Figure 4.
Schematic diagram showing the pathophysiology of TBI and the use of MS to identify different altered glycoproteins. Immediately after TBI, primary injury causes a distortion in the phospholipid bilayer of neural cells resulting in ion influx/efflux through the cell membrane. This is followed within minutes to hours by closure of the defect by lysolecithin to prevent further ion exchange. Subsequently, secondary injury occurs igniting a set of biochemical reactions and cascades that may take several hours to days to manifest. Mass spectrometric analysis is later used to identify and characterize the different altered glycoproteins.
Table 3.
List of TBI biomarkers isolated from different bodily sources following either primary or secondary injury
| Cerebrospinal fluid |
Blood | Saliva and tears |
|---|---|---|
| Tau [27, 128] | S100-B [138] | NGAL [143] |
| NFL [129] | GFAP [138] | S100-B [144] |
| NSE [130] | NSE [139] | TNF-α [145] |
| S100-B [131] | MBP [140,141] | |
| GFAP [131–133] | UCHL 1 [142] | |
| APP [134] | ||
| Aβ42 isoform [135] | ||
| α-II spectrin [136] | ||
| UCHL 1 [137] |
NFL : neurofilament light polypeptide; NSE : neuron-specific enolase; GFAP : glial fibrillary acidic protein; APP : amyloid precursor protein; UCHL 1 : ubiquitin carboxyl-terminal hydrolase isoenzyme L1; MBP : myelin basic protein; NGAL : neutrophil gelatinase-associated lipocalin.
4.1 PTMs and TBI
Similar to what was previously mentioned about the crucial role of PTMs in determining the functions of proteins in NDs as AD, a limited number of studies applied glycoproteomic analyses to screen whether those altered neuroproteins show changes also in protein glycosylation following TBI [15–17]. Yi et al. conducted a study in 2012 on mice to investigate the secondary cascade of events and PTMs that are believed to cause gradual neuronal death following experimental TBI, specifically elucidating the role of chondroitin sulfate proteoglycans (CSPGs) in this context [16]. CSPGs, which constitute most of the glial scars that form post-TBI, are thought to serve as inhibitors of axon growth and regeneration [101]. Results showed that the addition of glycosaminoglycan residues to CSPGs boost this function, while removal of these residues enhanced neurite sprouting and development [16]. This was also established in vivo, where removal of glycosaminoglycan chains with chondroitinase ABC following injury had shown to permit axonal budding [102]. Moreover, Yi et al. found that sulfation of CSPGs was elevated following injury, promoting amelioration in the function of these proteins and hence worsening neural injury [16]. Accordingly, further research into the role of these PTMs in the tight band around the injury core will help us understand its significance in TBI, and thus lead to a potential therapeutic strategy in brain injury.
Additionally, phosphorylation of proteins appears to increase following TBI. Similar to AD, hyperphosphorylation of tau is observed in TBI and leads to plaque formation [103]. Shultz et al. conducted a TBI study on rats using the fluid percussion injury method and interestingly found a decrease in the expression of PP2A, one of the main enzymes that de-phosphorylate tau protein. Consequently, this points toward not only an increase in kinase activity following TBI but also a parallel decrease in the phosphatase activity as a result of such injurious insult. Henceforward, targeting tau pathology may be an essential part of TBI therapy because it has significant implications on neurotransmission and cognitive function. For this reason, Shultz et al. also tested sodium selenate, a potential molecule that could prevent these alterations. Results showed that this molecule successfully activated PP2A and more importantly, improved behavioral/cognitive outcome in the rats [104].
In addition, Lazarus et al. studied the role of carbonylation in a rat-controlled cortical impact injury model [105]. They reported that carbonylation post-TBI affected specific proteins, namely dihydropyrimidase-related protein 2, glial fibrillary acidic protein, fructose-bisphosphate aldolase A, and fructose bisphosphate aldolase C at specific brain sites distant from the induced injury location with males more susceptible to these modifications than females suggesting a hormonal role [105].
No single study was found to tackle glycosylation post-TBI. The few studies that were found exploring the role of glycosylation in neurotrauma were illustrated in a part of the central nervous system, the spinal cord. Spinal cord injuries are no less important than TBI. Studies targeting PTMs, especially glycosylation, in spinal cord injuries validate our assumption about the role of these modifications in neurotraumas [17,106]. One such example is that of sialic acid, a carbohydrate molecule that is commonly added to membrane proteins, especially ion channels. This addition can affect the membrane potential as it can alter ion conductance across the neuronal membrane. In a study involving chronic constrictive injury in rats, Li et al. found that the dorsal root ganglion neurons had a higher concentration of membrane proteins and a parallel increase in glycosylation [17]. More importantly, this alteration was found to be associated with excessive neuronal excitability and more spontaneous firing due to the effect of sialic acid on voltage-gated channels specifically. This is extremely undesirable, as it can exacerbate the excitotoxic environment that spreads following TBI [106]. It is interesting to note that sialic acid has also been investigated for its potential to be added as a cap to medications for it may increase their efficiency in vivo [107].
5 AD and TBI: A common ground
It is evident that many PTMs identified in AD, but not PD, are similar to the ones identified in TBI. Glycosylation and sulfation of the CSPG [16,108], phosphorylation of tau protein [104,109], N-glycosylation, O-glycosylation [110,111] and sialylation of APP [112], carbonylation due to oxidative stress imposed by reactive oxygen species and reactive nitrogen species on glial fibrillary acid protein and MAPK-1 protein [105,113], and mannose-binding lectins [114,115] are some of the PTMs identified that are shared between TBI and AD.
The link between TBI and AD is of high complexity, involving common pathologies of cytoskeletal changes, neuronal loss, and inflammation. Nevertheless, it is not farfetched that TBI predisposes to earlier clinical and molecular neurodegenerative manifestations of AD. Gerson et al. recently reported AD-like neurodegenerative tauopathies, such as tau aggregation, in animal models of TBI with a significant increase in the hippocampus and cerebellum [116]. Since tau oligomers may be partially responsible for the pathophysiology seen in both TBI and AD, targeting tau in therapy holds a promising approach in the treatment of both of neurotrauma-induced damage and NDs. In the same scope, amyloid-β (Aβ) plaques, a hallmark of AD, were shown to be found in TBI patients a few hours postinjury [117]. In an interesting and reciprocal approach, inducing TBI in a mouse model with an AD-like genetic background led to a worsening of post-TBI outcomes at both molecular and behavioral levels [118]. At the genetic level, APOE polymorphisms are associated with TBI and NDs. A strong association between the apolipoprotein E allele, APOE ε4, and AD was found, but not with PD [119]. The same allele was not reported as a risk factor in PD patients [120]. However, patients with a history of TBI and are APOE ε4 carriers were found to have a tenfold risk increase for the development of AD [121]. TBI patients who are APOE ε4 carriers were also found to have an earlier onset of neurodegenerative pathologies [122]. Moreover, earlier onset of HD was observed in patients carrying the APOE ε2/ε3 genotype but not the APOE ε4 genotype [123].
6 Conclusion and future perspectives
In conclusion, glycoproteomic alterations appear to play critical roles in the disease processes of AD and PD through interfering with their pathogenesis. Advances should thus be made to identify more modified proteins involved in these processes in order to better understand their role in this context. Similarly, to obtain a thorough understanding of the molecular pathologies of neurotrauma, it is important to study each of these PTMs alterations, mainly glycosylation following TBI, and not merely evaluate the changes at the levels of protein expression. This may help in obtaining novel future therapies through blocking these intracellular pathways in order to prevent additional damage at the cellular level in neurotrauma and hence prevent further exacerbation.
Acknowledgments
This work was supported by the National Council for Scientific Research (CNRS), Lebanon, for the grant entitled “The Effect of Aspirin and Clopidogrel on Bleeding, platelet aggregation and Neuronal Damage after Moderate Brain Injury”; PI: Professor Samir Atweh, MD.
Abbreviations
- AChE
acetylcholinesterase
- AD
Alzheimer’s disease
- ALS
amyotrophic lateral sclerosis
- APP
amyloid precursor protein
- CSF
cerebrospinal fluid
- CSPGs
chondroitin sulfate proteoglycans
- GlcNAc
N-acetylglucosamine
- HD
Huntington’s disease
- MS
mass spectrometry
- ND
neurodegenerative disease
- PD
Parkinson’s disease
- TBI
traumatic brain injury
Footnotes
The authors have declared no conflict of interest.
References
- 1.Zhu H, Bilgin M, Snyder M. Annu. Rev. Biochem. 2003;72:783–812. doi: 10.1146/annurev.biochem.72.121801.161511. [DOI] [PubMed] [Google Scholar]
- 2.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 3.Hanash S. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 4.Karve TM, Cheema AK. J. Amino Acids. 2011;2011:2076–2091. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalivaeva NN, Turner AJ. Proteomics. 2001;1:735–747. doi: 10.1002/1615-9861(200106)1:6<735::AID-PROT735>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 6.Dewhurst HM, Choudhury S, Torres MP. Mol. Cell. Proteomics. 2015;14:2285–2297. doi: 10.1074/mcp.M115.051177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hwang H, Zhang J, Chung KA, Leverenz JB, Zabetian CP, Peskind ER, Jankovic J, Su Z, Hancock AM, Pan C, Montine TJ, Pan S, Nutt J, Albin R, Gearing M, Beyer RP, Shi M, Zhang J. Mass Spectrom. Rev. 2010;29:79–125. doi: 10.1002/mas.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Jensen ON. Proteomics. 2009;9:4632–4641. doi: 10.1002/pmic.200900398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rigatti M, Le AV, Gerber C, Moraru II, Dodge-Kafka KL. Cell. Signal. 2015;27:1807–1815. doi: 10.1016/j.cellsig.2015.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han Y, Yuan ZF, Molden RC, Garcia BA. Mol. Cell. Proteomics. 2016;15:834–853. doi: 10.1074/mcp.M115.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sartor GC, Powell SK, Brothers SP, Wahlestedt C. J. Neurosci. 2015;35:15062–15072. doi: 10.1523/JNEUROSCI.0826-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Min SW, Chen X, Tracy TE, Li Y, Zhou Y, Wang C, Shirakawa K, Minami SS, Defensor E, Mok SA, Sohn PD, Schilling B, Cong X, Ellerby L, Gibson BW, Johnson J, Krogan N, Shamloo M, Gestwicki J, Masliah E, Verdin E, Gan L. Nat. Med. 2015;21:1154–1162. doi: 10.1038/nm.3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Udeshi ND, Mertins P, Svinkina T, Carr SA. Nat. Protoc. 2013;8:1950–1960. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arsenault PR, Heaton-Johnson KJ, Li LS, Song D, Ferreira VS, Patel N, Master SR, Lee FS. Proteomics. 2015;15:1259–1267. doi: 10.1002/pmic.201400398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barreda-Manso MA, Yanguas-Casas N, Nieto-Sampedro M, Romero-Ramirez L. Exp. Cell Res. 2015;335:82–90. doi: 10.1016/j.yexcr.2015.04.002. [DOI] [PubMed] [Google Scholar]
- 16.Yi JH, Katagiri Y, Susarla B, Figge D, Symes AJ, Geller HM. J. Comp. Neurol. 2012;520:3295–3313. doi: 10.1002/cne.23156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li CX, Jing YL, Xie YK. Brain Res. 2007;1139:201–209. doi: 10.1016/j.brainres.2006.12.039. [DOI] [PubMed] [Google Scholar]
- 18.Apweiler R, Hermjakob H, Sharon N. Biochim. Biophys. Acta. 1999;1473:4–8. doi: 10.1016/s0304-4165(99)00165-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim EH, Misek DE. Int. J. Proteomics. 2011;2011:1–10. doi: 10.1155/2011/343582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobata A. Biochimie. 2003;85:13–24. doi: 10.1016/s0300-9084(03)00003-8. [DOI] [PubMed] [Google Scholar]
- 21.Wei X, Li L. Brief. Funct. GenomicProteomic. 2009;8:104–113. doi: 10.1093/bfgp/eln053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ongay S, Boichenko A, Govorukhina N, Bischoff R. J. Sep. Sci. 2012;35:2341–2372. doi: 10.1002/jssc.201200434. [DOI] [PubMed] [Google Scholar]
- 23.Hood BL, Malehorn DE, Conrads TP, Bigbee WL. Methods Mol. Biol. 2009;520:107–128. doi: 10.1007/978-1-60327-811-9_8. [DOI] [PubMed] [Google Scholar]
- 24.Yang H, Zhou B, Deng H, Prinz M, Siegel D. Int. J. Legal Med. 2013;127:1065–1077. doi: 10.1007/s00414-013-0848-1. [DOI] [PubMed] [Google Scholar]
- 25.Beck F, Burkhart JM, Geiger J, Zahedi RP, Sick-mann A. Methods Mol. Biol. 2012;893:101–113. doi: 10.1007/978-1-61779-885-6_8. [DOI] [PubMed] [Google Scholar]
- 26.Haqqani AS, Kelly JF, Stanimirovic DB. Methods Mol. Biol. 2008;439:225–240. doi: 10.1007/978-1-59745-188-8_16. [DOI] [PubMed] [Google Scholar]
- 27.Kaji H, Yamauchi Y, Takahashi N, Isobe T. Nat. Protoc. 2006;1:3019–3027. doi: 10.1038/nprot.2006.444. [DOI] [PubMed] [Google Scholar]
- 28.D’Ascenzo M, Choe L, Lee KH. Brief. Funct. Genomic Proteomic. 2008;7:127–135. doi: 10.1093/bfgp/eln007. [DOI] [PubMed] [Google Scholar]
- 29.Chauhan NB. Restor. Neurol. Neurosci. 2014;32:337–365. doi: 10.3233/RNN-130354. [DOI] [PubMed] [Google Scholar]
- 30.Reimand J, Wagih O, Bader GD. PLoS Genet. 2015;11:e1004919. doi: 10.1371/journal.pgen.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salawu FK, Umar JT, Olokoba AB. Annals of African Medicine. 2011;10:73–79. doi: 10.4103/1596-3519.82057. [DOI] [PubMed] [Google Scholar]
- 32.Beitz JM. Front. Biosci. (Scholar edition) 2014;6:65–74. doi: 10.2741/s415. [DOI] [PubMed] [Google Scholar]
- 33.Sato Y, Akimoto Y, Kawakami H, Hirano H, Endo T. J. Histochem. Cytochem. 2001;49:1311–1319. doi: 10.1177/002215540104901014. [DOI] [PubMed] [Google Scholar]
- 34.Angelucci F, Spalletta G, di Iulio F, Ciaramella A, Salani F, Colantoni L, Varsi AE, Gianni W, Sancesario G, Caltagirone C, Bossu P. Curr. Alzheimer Res. 2010;7:15–20. doi: 10.2174/156720510790274473. [DOI] [PubMed] [Google Scholar]
- 35.Massano J, Bhatia KP. Cold Spring Harb. Perspect. Med. 2012;2:a008870. doi: 10.1101/cshperspect.a008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe JB, Marth JD. Annu. Rev. Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 37.Endo T. Glycoconj. J. 2004;21:3–7. doi: 10.1023/B:GLYC.0000043740.26062.2c. [DOI] [PubMed] [Google Scholar]
- 38.Liu FT, Patterson RJ, Wang JL. Biochim. Biophys. Acta. 2002;1572:263–273. doi: 10.1016/s0304-4165(02)00313-6. [DOI] [PubMed] [Google Scholar]
- 39.Ip WK, Takahashi K, Ezekowitz RA, Stuart LM. Immunol. Rev. 2009;230:9–21. doi: 10.1111/j.1600-065X.2009.00789.x. [DOI] [PubMed] [Google Scholar]
- 40.Bochner BS. Clin. Exp. Allergy. 2009;39:317–324. doi: 10.1111/j.1365-2222.2008.03173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Joubert R, Kuchler S, Zanetta JP, Bladier D, Avellana-Adalid V, Caron M, Doinel C, Vincendon G. Dev. Neurosci. 1989;11:397–413. doi: 10.1159/000111916. [DOI] [PubMed] [Google Scholar]
- 42.Kuchler S, Joubert R, Avellana-Adalid V, Caron M, Bladier D, Vincendon G, Zanetta JP. Dev. Neurosci. 1989;11:414–427. doi: 10.1159/000111917. [DOI] [PubMed] [Google Scholar]
- 43.Mahanthappa NK, Cooper DN, Barondes SH, Schwarting GA. Development. 1994;120:1373–1384. doi: 10.1242/dev.120.6.1373. [DOI] [PubMed] [Google Scholar]
- 44.Zanetta JP. Acta Anat. (Basel) 1998;161:180–195. doi: 10.1159/000046457. [DOI] [PubMed] [Google Scholar]
- 45.Puche AC, Poirier F, Hair M, Bartlett PF, Key B. Dev. Biol. 1996;179:274–287. doi: 10.1006/dbio.1996.0257. [DOI] [PubMed] [Google Scholar]
- 46.Bond MR, Hanover JA. J. Cell Biol. 2015;208:869–880. doi: 10.1083/jcb.201501101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ogawa M, Sawaguchi S, Furukawa K, Okajima T. Biochim. Biophys. Acta. 2015;1850:1319–1324. doi: 10.1016/j.bbagen.2015.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Ogawa M, Sawaguchi S, Kawai T, Nadano D, Matsuda T, Yagi H, Kato K, Furukawa K, Okajima T. J. Biol. Chem. 2014;290:2137–2149. doi: 10.1074/jbc.M114.598821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akimoto Y, Comer FI, Cole RN, Kudo A, Kawakami H, Hirano H, Hart GW. Brain Res. 2003;966:194–205. doi: 10.1016/s0006-8993(02)04158-6. [DOI] [PubMed] [Google Scholar]
- 50.Cole RN, Hart GW. J. Neurochem. 2001;79:1080–1089. doi: 10.1046/j.1471-4159.2001.00655.x. [DOI] [PubMed] [Google Scholar]
- 51.Dias WB, Hart GW. Mol. Biosyst. 2007;3:766–772. doi: 10.1039/b704905f. [DOI] [PubMed] [Google Scholar]
- 52.Yanagisawa M, Yu RK. J. Neurosci. Res. 2009;87:3535–3545. doi: 10.1002/jnr.22170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu Y, Shan X, Yuzwa SA, Vocadlo DJ. J. Biol. Chem. 2014;289:34472–34481. doi: 10.1074/jbc.R114.601351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alfaro JF, Gong CX, Monroe ME, Aldrich JT, Clauss TR, Purvine SO, Wang Z, Camp DG, 2nd, Shabanowitz J, Stanley P, Hart GW, Hunt DF, Yang F, Smith RD. Proc. Natl. Acad. Sci. USA. 2012;109:7280–7285. doi: 10.1073/pnas.1200425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vosseller K, Trinidad JC, Chalkley RJ, Specht CG, Thalhammer A, Lynn AJ, Snedecor JO, Guan S, Medzihradszky KF, Maltby DA, Schoepfer R, Burlingame AL. Mol. Cell. Proteomics. 2006;5:923–934. doi: 10.1074/mcp.T500040-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.Saez-Valero J, Fodero LR, Sjogren M, Andreasen N, Amici S, Gallai V, Vanderstichele H, Vanmechelen E, Parnetti L, Blennow K, Small DH. J. Neurosci. Res. 2003;72:520–526. doi: 10.1002/jnr.10599. [DOI] [PubMed] [Google Scholar]
- 57.Silveyra MX, Cuadrado-Corrales N, Marcos A, Barquero MS, Rabano A, Calero M, Saez-Valero J. J. Neurochem. 2006;96:97–104. doi: 10.1111/j.1471-4159.2005.03514.x. [DOI] [PubMed] [Google Scholar]
- 58.Sato Y, Naito Y, Grundke-Iqbal I, Iqbal K, Endo T. FEBS Lett. 2001;496:152–160. doi: 10.1016/s0014-5793(01)02421-8. [DOI] [PubMed] [Google Scholar]
- 59.Iqbal K, Alonso Adel C, Chen S, Chohan MO, El-Akkad E, Gong CX, Khatoon S, Li B, Liu F, Rahman A, Tanimukai H, Grundke-Iqbal I. Biochim. Biophys. Acta. 2005;1739:198–210. doi: 10.1016/j.bbadis.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 60.Lazarus BD, Love DC, Hanover JA. Int. J. Biochem. Cell Biol. 2009;41:2134–2146. doi: 10.1016/j.biocel.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Donnell N, Zachara NE, Hart GW, Marth JD. Mol. Cell. Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ, Vocadlo DJ. Nat. Chem. Biol. 2008;4:483–490. doi: 10.1038/nchembio.96. [DOI] [PubMed] [Google Scholar]
- 63.Yuzwa SA, Shan X, Macauley MS, Clark T, Skorobogatko Y, Vosseller K, Vocadlo DJ. Nat. Chem. Biol. 2012;8:393–399. doi: 10.1038/nchembio.797. [DOI] [PubMed] [Google Scholar]
- 64.Borghgraef P, Menuet C, Theunis C, Louis JV, Devijver H, Maurin H, Smet-Nocca C, Lippens G, Hilaire G, Gijsen H, Moechars D, Van Leuven F. PLoS One. 2014;8:e84442. doi: 10.1371/journal.pone.0084442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vassar R, Bennett BD, Babu-Khan S, Kahn S, Mendiaz EA, Denis P, Teplow DB, Ross S, Amarante P, Loeloff R, Luo Y, Fisher S, Fuller J, Edenson S, Lile J, Jarosinski MA, Biere AL, Curran E, Burgess T, Louis JC, Collins F, Treanor J, Rogers G, Citron M. Science. 1999;286:735–741. doi: 10.1126/science.286.5440.735. [DOI] [PubMed] [Google Scholar]
- 66.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, Von Figura K, Van Leuven F. Nature. 1998;391:387–390. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 67.Lammich S, Kojro E, Postina R, Gilbert S, Pfeiffer R, Jasionowski M, Haass C, Fahrenholz F. Proc. Natl. Acad. Sci. USA. 1999;96:3922–3927. doi: 10.1073/pnas.96.7.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Estus S, Golde TE, Kunishita T, Blades D, Lowery D, Eisen M, Usiak M, Qu XM, Tabira T, Greenberg BD. Science. 1992;255:726–728. doi: 10.1126/science.1738846. [DOI] [PubMed] [Google Scholar]
- 69.Jacobsen KT, Iverfeldt K. Biochem. Biophys. Res. Commun. 2010;404:882–886. doi: 10.1016/j.bbrc.2010.12.080. [DOI] [PubMed] [Google Scholar]
- 70.Saez-Valero J, Barquero MS, Marcos A, McLean CA, Small DH. J. Neurol. Neurosurg. Psychiatry. 2000;69:664–667. doi: 10.1136/jnnp.69.5.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saez-Valero J, Sberna G, McLean CA, Small DH. J. Neurochem. 1999;72:1600–1608. doi: 10.1046/j.1471-4159.1999.721600.x. [DOI] [PubMed] [Google Scholar]
- 72.Saez-Valero J, Small DH. J. Alzheimers Dis. 2001;3:323–328. doi: 10.3233/jad-2001-3307. [DOI] [PubMed] [Google Scholar]
- 73.Hoppe JB, Salbego CG, Cimarosti H. Aging Dis. 2015;6:322–330. doi: 10.14336/AD.2014.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohen TJ, Guo JL, Hurtado DE, Kwong LK, Mills IP, Trojanowski JQ, Lee VM. Nat. Commun. 2011;2:252. doi: 10.1038/ncomms1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cook C, Carlomagno Y, Gendron TF, Dunmore J, Scheffel K, Stetler C, Davis M, Dickson D, Jarpe M, DeTure M, Petrucelli L. Hum. Mol. Genet. 2013;23:104–116. doi: 10.1093/hmg/ddt402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Min SW, Cho SH, Zhou Y, Schroeder S, Haroutunian V, Seeley WW, Huang EJ, Shen Y, Masliah E, Mukherjee C, Meyers D, Cole PA, Ott M, Gan L. Neuron. 2010;67:953–966. doi: 10.1016/j.neuron.2010.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cripps D, Thomas SN, Jeng Y, Yang F, Davies P, Yang AJ. J. Biol. Chem. 2006;281:10825–10838. doi: 10.1074/jbc.M512786200. [DOI] [PubMed] [Google Scholar]
- 78.Funk KE, Thomas SN, Schafer KN, Cooper GL, Liao Z, Clark DJ, Yang AJ, Kuret J. Biochem. J. 2014;462:77–88. doi: 10.1042/BJ20140372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, Ren J, Beausoleil SA, Silva JC, Vemulapalli V, Bedford MT, Comb MJ. Mol. Cell. Proteomics. 2013;13:372–387. doi: 10.1074/mcp.O113.027870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morris M, Knudsen GM, Maeda S, Trinidad JC, Ioanoviciu A, Burlingame AL, Mucke L. Nat. Neurosci. 2015;18:1183–1189. doi: 10.1038/nn.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hatcher JM, Pennell KD, Miller GW. Trends Pharmacol. Sci. 2008;29:322–329. doi: 10.1016/j.tips.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hwang H, Zhang J, Chung KA, Leverenz JB, Zabetian CP, Peskind ER, Jankovic J, Su Z, Hancock AM, Pan C, Montine TJ, Pan S, Nutt J, Albin R, Gearing M, Beyer RP, Shi M. Mass Spectrom. Rev. 2009;29:79–125. doi: 10.1002/mas.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moran LB, Hickey L, Michael GJ, Derkacs M, Christian LM, Kalaitzakis ME, Pearce RK, Graeber MB. Acta Neuropathol. 2008;115:471–478. doi: 10.1007/s00401-007-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pisani A, Centonze D, Bernardi G, Calabresi P. Mov. Disord. 2005;20:395–402. doi: 10.1002/mds.20394. [DOI] [PubMed] [Google Scholar]
- 85.Zhou TT, Zhang ZW, Liu J, Zhang JP, Jiao BH. Int. J. Biol. Sci. 2012;8:630–639. doi: 10.7150/ijbs.3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pirko I, Lucchinetti CF, Sriram S, Bakshi R. Neurology. 2007;68:634–642. doi: 10.1212/01.wnl.0000250267.85698.7a. [DOI] [PubMed] [Google Scholar]
- 87.Lee SU, Grigorian A, Pawling J, Chen IJ, Gao G, Mozaffar T, McKerlie C, Demetriou M. J. Biol. Chem. 2007;282:33725–33734. doi: 10.1074/jbc.M704839200. [DOI] [PubMed] [Google Scholar]
- 88.Mkhikian H, Grigorian A, Li CF, Chen HL, Newton B, Zhou RW, Beeton C, Torossian S, Tatarian GG, Lee SU, Lau K, Walker E, Siminovitch KA, Chandy KG, Yu Z, Dennis JW, Demetriou M. Nat. Commun. 2011;2:334. doi: 10.1038/ncomms1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li SH, Li XJ. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 90.Desplats PA, Denny CA, Kass KE, Gilmartin T, Head SR, Sutcliffe JG, Seyfried TN, Thomas EA. Neurobiol. Dis. 2007;27:265–277. doi: 10.1016/j.nbd.2007.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gizaw ST, Koda T, Amano M, Kamimura K, Ohashi T, Hinou H, Nishimura S. Biochim. Biophys. Acta. 2015;1850:1704–1718. doi: 10.1016/j.bbagen.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 92.Ludemann N, Clement A, Hans VH, Leschik J, Behl C, Brandt R. J. Biol. Chem. 2005;280:31648–31658. doi: 10.1074/jbc.M504395200. [DOI] [PubMed] [Google Scholar]
- 93.Gonatas NK, Stieber A, Gonatas JO. J. Neurol. Sci. 2006;246:21–30. doi: 10.1016/j.jns.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 94.Edri-Brami M, Rosental B, Hayoun D, Welt M, Rosen H, Wirguin I, Nefussy B, Drory VE, Porgador A, Lichtenstein RG. PLoS One. 2012;7:e35772. doi: 10.1371/journal.pone.0035772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gean AD, Fischbein NJ. Neuroimag. Clin. North Am. 2010;20:527–556. doi: 10.1016/j.nic.2010.08.001. [DOI] [PubMed] [Google Scholar]
- 96.CDC. Report to Congress on traumatic brain injury in the United States: epidemiology and rehabilitation. Atlanta, GA: Centers for Disease Control and Prevention; 2014. [Google Scholar]
- 97.Gennarelli T, Thibault L, Graham D. Neuroscientist. 1998;4:202–215. [Google Scholar]
- 98.Yawo H, Kuno M. Science. 1983;222:1351–1353. doi: 10.1126/science.6658457. [DOI] [PubMed] [Google Scholar]
- 99.Graham DI, Gennarelli TA, McIntosh TK. In: Greenfield’s Neuropathology. Greenfield JG, Graham DI, Lantos PL, editors. London: Arnold; 2002. pp. 823–898. [Google Scholar]
- 100.DeKosky S, Kochanek P, Clark R, Ciallella J, Dixon C. Semin. Clin. Neuropsychiatry. 1998;3:176–185. [PubMed] [Google Scholar]
- 101.Myer DJ, Gurkoff GG, Lee SM, Hovda DA, Sofroniew MV. Brain. 2006;129:2761–2772. doi: 10.1093/brain/awl165. [DOI] [PubMed] [Google Scholar]
- 102.Bradbury EJ, Moon LDF, Popat RJ, King VR, Bennett GS, Patel PN, Fawcett JW, McMahon SB. Nature. 2002;416:636–640. doi: 10.1038/416636a. [DOI] [PubMed] [Google Scholar]
- 103.Johnson VE, Stewart W, Smith DH. Brain Pathol. (Zurich, Switzerland) 2012;22:142–149. doi: 10.1111/j.1750-3639.2011.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shultz SR, Wright DK, Zheng P, Stuchbery R, Liu S-J, Sashindranath M, Medcalf RL, Johnston LA, Hovens CM, Jones NC, O’Brien TJ. Brain. 2015;138:1297–1313. doi: 10.1093/brain/awv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lazarus RC, Buonora JE, Jacobowitz DM, Mueller GP. Free Radical Biol. Med. 2015;78:89–100. doi: 10.1016/j.freeradbiomed.2014.10.507. [DOI] [PubMed] [Google Scholar]
- 106.Papastefanaki F, Matsas R. Glia. 2015;63:1101–1125. doi: 10.1002/glia.22809. [DOI] [PubMed] [Google Scholar]
- 107.Shriver Z, Raguram S, Sasisekharan R. Nat. Rev. Drug Discov. 2004;3:863–873. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- 108.DeWitt DA, Silver J, Canning DR, Perry G. Exp. Neurol. 1993;121:149–152. doi: 10.1006/exnr.1993.1081. [DOI] [PubMed] [Google Scholar]
- 109.Hampel H, Blennow K, Shaw LM, Hoessler YC, Zetterberg H, Trojanowski JQ. Exp. Gerontol. 2010;45:30–40. doi: 10.1016/j.exger.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pahlsson P, Shakin-Eshleman SH, Spitalnik SL. Biochem. Biophys. Res. Commun. 1992;189:1667–1673. doi: 10.1016/0006-291x(92)90269-q. [DOI] [PubMed] [Google Scholar]
- 111.Schedin-Weiss S, Winblad B, Tjernberg LO. FEBS J. 2014;281:46–62. doi: 10.1111/febs.12590. [DOI] [PubMed] [Google Scholar]
- 112.Nakagawa K, Kitazume S, Oka R, Maruyama K, Saido TC, Sato Y, Endo T, Hashimoto Y. J. Neurochem. 2006;96:924–933. doi: 10.1111/j.1471-4159.2005.03595.x. [DOI] [PubMed] [Google Scholar]
- 113.Sultana R, Perluigi M, Newman SF, Pierce WM, Cini C, Coccia R, Butterfield DA. Antioxid. Redox Sign. 2010;12:327–336. doi: 10.1089/ars.2009.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Longhi L, Orsini F, De Blasio D, Fumagalli S, Ortolano F, Locatelli M, Stocchetti N, De Simoni MG. Crit. Care Med. 2014;42:1910–1918. doi: 10.1097/CCM.0000000000000399. [DOI] [PubMed] [Google Scholar]
- 115.Sjolander A, Minthon L, Nuytinck L, Vanmechelen E, Blennow K, Nilsson S. J. Alzheimers Dis. 2013;35:121–127. doi: 10.3233/JAD-122044. [DOI] [PubMed] [Google Scholar]
- 116.Gerson JE, Castillo-Carranza DL, Sengupta U, Bodani R, Prough DS, DeWitt D, Hawkins BE, Kayed R. J. Neurotrauma. 2016 doi: 10.1089/neu.2015.4262. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Johnson VE, Stewart W, Smith DH. Nat. Rev. Neurosci. 2010;11:361–370. doi: 10.1038/nrn2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Miszczuk D, Debski KJ, Tanila H, Lukasiuk K, Pitkanen A. Mol. Neurobiol. 2015 doi: 10.1007/s12035-015-9578-3. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 119.Hardy J, Crook R, Prihar G, Roberts G, Raghavan R, Perry R. Neurosci. Lett. 1994;182:1–2. doi: 10.1016/0304-3940(94)90190-2. [DOI] [PubMed] [Google Scholar]
- 120.Egensperger R, Bancher C, Kösel S, Jellinger K, Mehraein P, Graeber MB. Biochem. Biophys. Res. Commun. 1996;224:484–486. doi: 10.1006/bbrc.1996.1053. [DOI] [PubMed] [Google Scholar]
- 121.Tang MX, Maestre G, Tsai WY, Liu XH, Feng L, Chung WY, Chun M, Schofield P, Stern Y, Tycko B, Mayeux R. Ann. N Y Acad. Sci. 1996;802:6–15. doi: 10.1111/j.1749-6632.1996.tb32593.x. [DOI] [PubMed] [Google Scholar]
- 122.Luukinen H, Viramo P, Herala M, Kervinen K, Kesaniemi YA, Savola O, Winqvist S, Jokelainen J, Hillbom M. Eur. J. Neurol. 2005;12:86–92. doi: 10.1111/j.1468-1331.2004.00953.x. [DOI] [PubMed] [Google Scholar]
- 123.Verpillat P, Camuzat A, Hannequin D, Thomas-Anterion C, Puel M, Belliard S, Dubois B, Didic M, Lacomblez L, Moreaud O, Golfier V, Campion D, Brice A, Clerget-Darpoux F. Eur. J. Hum. Genet. 2002;10:399–405. doi: 10.1038/sj.ejhg.5200820. [DOI] [PubMed] [Google Scholar]
- 124.Boillee S, Vande Velde C, Cleveland DW. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 125.Gordon PH. Aging Dis. 2013;4:295–310. doi: 10.14336/AD.2013.0400295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Roos RAC. Orphanet J. Rare Dis. 2010;5:1–8. doi: 10.1186/1750-1172-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gelfand JM. Handbook Clin. Neurol. 2014;122:269–290. doi: 10.1016/B978-0-444-52001-2.00011-X. [DOI] [PubMed] [Google Scholar]
- 128.Blennow K, Nellgard B. Neurology. 2004;62:159. doi: 10.1212/wnl.62.1.159. author reply 159–160. [DOI] [PubMed] [Google Scholar]
- 129.Friede RL, Samorajski T. Anat. Rec. 1970;167:379–387. doi: 10.1002/ar.1091670402. [DOI] [PubMed] [Google Scholar]
- 130.Olsson B, Zetterberg H, Hampel H, Blennow K. Prog. Neurobiol. 2011;95:520–534. doi: 10.1016/j.pneurobio.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 131.Neselius S, Brisby H, Theodorsson A, Blennow K, Zetterberg H, Marcusson J. PLoS One. 2012;7:e33606. doi: 10.1371/journal.pone.0033606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Czeiter E, Mondello S, Kovacs N, Sandor J, Gabrielli A, Schmid K, Tortella F, Wang KK, Hayes RL, Barzo P, Ezer E, Doczi T, Buki A. J. Neurotrauma. 2012;29:1770–1778. doi: 10.1089/neu.2011.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zetterberg H, Hietala MA, Jonsson M, Andreasen N, Styrud E, Karlsson I, Edman A, Popa C, Rasulzada A, Wahlund LO, Mehta PD, Rosengren L, Blennow K, Wallin A. Arch. Neurol. 2006;63:1277–1280. doi: 10.1001/archneur.63.9.1277. [DOI] [PubMed] [Google Scholar]
- 134.McKenzie KJ, McLellan DR, Gentleman SM, Maxwell WL, Gennarelli TA, Graham DI. Acta Neuropathol. 1996;92:608–613. doi: 10.1007/s004010050568. [DOI] [PubMed] [Google Scholar]
- 135.Olsson A, Csajbok L, Ost M, Hoglund K, Nylen K, Rosengren L, Nellgard B, Blennow K. J. Neurol. 2004;251:870–876. doi: 10.1007/s00415-004-0451-y. [DOI] [PubMed] [Google Scholar]
- 136.Mondello S, Robicsek SA, Gabrielli A, Brophy GM, Papa L, Tepas J, Robertson C, Buki A, Scharf D, Jixiang M, Akinyi L, Muller U, Wang KK, Hayes RL. J Neurotrauma. 2010;27:1203–1213. doi: 10.1089/neu.2010.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Carr W, Yarnell AM, Ong R, Walilko T, Kamimori GH, da Silva U, McCarron RM, LoPresti ML. Front. Neurol. 2015;6:49. doi: 10.3389/fneur.2015.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mondello S, Muller U, Jeromin A, Streeter J, Hayes RL, Wang KK. Expert Rev. Mol. Diagn. 2010;11:65–78. doi: 10.1586/erm.10.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Berger RP, Adelson PD, Pierce MC, Dulani T, Cassidy LD, Kochanek PM. J. Neurosurg. 2005;103:61–68. doi: 10.3171/ped.2005.103.1.0061. [DOI] [PubMed] [Google Scholar]
- 140.Zurek J, Bartlova L, Fedora M. Brain Inj. 2011;25:221–226. doi: 10.3109/02699052.2010.541895. [DOI] [PubMed] [Google Scholar]
- 141.Zurek J, Fedora M. Acta Neurochir. (Wien) 2011;154:93–103. doi: 10.1007/s00701-011-1175-2. [DOI] [PubMed] [Google Scholar]
- 142.Papa L, Lewis LM, Silvestri S, Falk JL, Giordano P, Brophy GM, Demery JA, Liu MC, Mo J, Akinyi L, Mondello S, Schmid K, Robertson CS, Tortella FC, Hayes RL, Wang KK. J. Trauma Acute Care Surg. 2012;72:1335–1344. doi: 10.1097/TA.0b013e3182491e3d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Li N, Zhao WG, Xu FL, Zhang WF, Gu WT. J. Nephrol. 2013;26:1083–1088. doi: 10.5301/jn.5000282. [DOI] [PubMed] [Google Scholar]
- 144.Rodriguez-Rodriguez A, Egea-Guerrero JJ, Leon-Justel A, Gordillo-Escobar E, Revuelto-Rey J, Vilches-Arenas A, Carrillo-Vico A, Dominguez-Roldan JM, Murillo-Cabezas F, Guerrero JM. Clin. Chim. Acta. 2012;414:228–233. doi: 10.1016/j.cca.2012.09.025. [DOI] [PubMed] [Google Scholar]
- 145.Comoglu SS, Guven H, Acar M, Ozturk G, Kocer B. Neurosci. Lett. 2013;553:63–67. doi: 10.1016/j.neulet.2013.08.019. [DOI] [PubMed] [Google Scholar]