Abstract
Super-enhancers comprise of dense transcription factor platforms highly enriched for active chromatin marks. A paucity of functional data led us to investigate their role in the mammary gland, an organ characterized by exceptional gene regulatory dynamics during pregnancy. ChIP-Seq for the master regulator STAT5, the glucocorticoid receptor, H3K27ac and MED1, identified 440 mammary-specific super-enhancers, half of which were associated with genes activated during pregnancy. We interrogated the Wap super-enhancer, generating mice carrying mutations in STAT5 binding sites within its three constituent enhancers. Individually, only the most distal site displayed significant enhancer activity. However, combinatorial mutations showed that the 1,000-fold gene induction relied on all enhancers. Disabling the binding sites of STAT5, NFIB and ELF5 in the proximal enhancer incapacitated the entire super-enhancer, suggesting an enhancer hierarchy. The identification of mammary-specific super-enhancers and the mechanistic exploration of the Wap locus provide insight into the complexity of cell-specific and hormone-regulated genes.
The sole purpose of the mammary gland is to produce large quantities of milk to support newborns. The milk secreting alveolar epithelium, absent in the virgin state, is established during pregnancy and completely remodeled upon cessation of lactation1. Proliferation and differentiation of mammary alveoli during pregnancy is controlled by progesterone2 and the cytokine prolactin3,4 through the transcription factors STAT55,6 and ELF57. STAT58,9 has emerged as the critical transcription factor activating genes encoding milk proteins, and other differentiation associated proteins linked to secretion, up to 10,000 fold during pregnancy10. The ten most abundant mRNAs encoding milk proteins, account for more than 90% of the mRNA11,12 and expression of the respective genes are synergistically induced by prolactin and glucocorticoids13, making them an ideal system to investigate underlying transcriptional regulation.
Traditionally, transgenic mice had been used to identify sequences conveying mammary-specificity and hormonal responsiveness13-19. Although informative, these transgenes contained only limited promoter and upstream sequences, which conveyed mammary-restricted expression but normal hormonal regulation during pregnancy and expression levels were not obtained. This suggests that enhancers and insulators were located outside the sequences used in these transgenes. Putative regulatory elements within the genome can now be identified using ChIP-Seq analyses for transcription factors and specific histone modifications, such H3K27ac for enhancers20-23. This permits a more surgical approach in the genetic analysis of these sequences.
The emerging concept of super-enhancers proposes that lineage-specific genes are under control of enhancer clusters, which are characterized by several densely occupied transcription factor platforms and extended H3K27ac marks24,25. Although super-enhancers have been reported in ES cells and other systems26-46 there is scant genetic support of their biological relevance. Several hundred genes are expressed specifically in mammary tissue under the control of prolactin10 making them an ideal test system for the concept of super-enhancers. Here we have integrated ChIP-Seq for the mammary master regulator STAT5, GR, MED1 and H3K27ac to identify putative mammary enhancers. To validate the biological significance of super-enhancers we focused on one associated with Wap47, a gene highly expressed in mammary tissue and induced more than 1,000-fold during pregnancy48. We mutated constituent enhancers, individually and in combination, within the Wap super-enhancer, determined their respective importance and identified a hierarchy among them.
RESULTS
Identification of mammary-specific super-enhancers
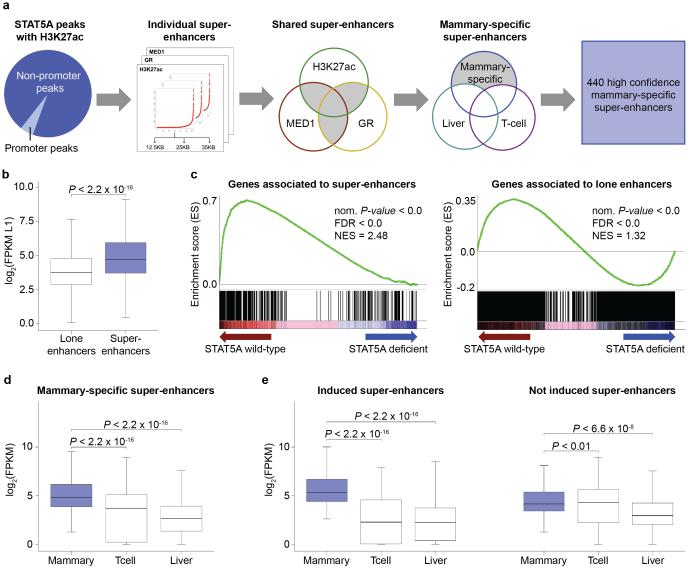
In search of mammary-specific super-enhancers, ChIP-Seq experiments were conducted for STAT5A, a master regulator in mammary epithelium6,49, the active chromatin mark H3K27ac, the glucocorticoid receptor (GR) and MED1. Mammary tissue was initially analyzed at lactation when mammary-specific genes are highly expressed. Based on the co-localization of TF binding and H3K27ac marks and defined stitch sizes46 (Fig. 1a and Supplementary Figs. 1-3) we identified approximately 580 super-enhancers. After subtracting STAT5 super-enhancers common to mammary tissue and either liver or T cells, we obtained 440 mammary-specific super-enhancers, encompassing a total of 2,712 individual enhancers (Fig. 1a and Supplementary Table 1). These 440 super-enhancers were exclusively identified based on ChIP-Seq experiments, and their biological function has not been experimentally validated.
Figure 1.
Identification of mammary-specific super-enhancers. (a) Out of 10,953 peaks coinciding with H3K27ac marks, 549 are located in promoter regions and 10,404 within non-promoter regions. The 10,404 STAT5 peaks, together with H3K27ac, GR and MED1, served as basis for the super-enhancer analysis37,46 using three stitched sizes as parameter for the calculations. The final steps comprised overlapping them per size and subtracting super-enhancers shared with T cells and liver and a total of 440 mammary-specific bona fide super-enhancers were identified. (b) The boxplot depicts the significant higher expression level of 384 genes associated with super-enhancers (mean ~5,931 FPKM) compared to 4,384 genes linked to lone enhancers (mean ~31 FPKM) at day one of lactation (L1) (cutoff of 5 FPKM). Median, middle bar inside the box; IQR, 50% of the data; whiskers, 1.5 times the IQR. (c) Gene Set Enrichment Analysis shows that super-enhancers are less enriched in STAT5A deficient samples (70% reduction) suggesting that those genes are more sensitive to changes of STAT5 levels and further implying that the super-enhancers are mammary-specific (Nom P-value, nominal P-value; FDR, false discovery rate; NES, normalized enrichment score). (d) Boxplot shows that the expression of the 384 genes associated with mammary-specific super-enhancer is significantly elevated at L1, compared to T cells and liver tissue. (e) Super-enhancer associated genes were categorized into genes induced at least 2-fold between pregnancy day 6 (p6) and L1 (198), and not induced genes (186). Notably, induced genes exhibit lineage specificity and their expression in T cell and liver was greatly lower.
To establish the functional significance in lactating mammary tissue, we used RNA-Seq data10 and compared expression of genes associated with super-enhancers to genes associated with lone STAT5 enhancers (Fig. 1b). While the median expression of genes linked to lone enhancers was ~14 FPKM (mean ~31 FPKM), it was ~27 FPKM (mean ~5,931 FPKM) for genes associated with super-enhancers. Gene Set Enrichment Analysis (GSEA) demonstrated that super-enhancers were preferentially associated with STAT5-dependent genes (Fig. 1c). Expression of approximately 35% of super-enhancers associated genes was induced more than 2-fold by STAT5 (Supplementary Table 1) and approximately 50% of the genes associated with super-enhancers were also induced during pregnancy. Notably while most highly induced genes are associated with super-enhancers, some, such as Aldoc, are linked to lone enhancers (Supplementary Fig. 3).
Next we examined whether expression of super-enhancer associated genes was specifically elevated in mammary tissue. We analyzed expression in mammary tissue at day one of lactation, in Th1 cells and liver tissue, all of which are targets of cytokines that activate STAT5. While expression of the super-enhancer associated genes in mammary tissue averaged 27 FPKM (mean ~ 5,931 FPKM), it was ~12 FPKM (mean ~31 FPKM) in T cells and ~6 FPKM (mean ~28 FPKM) in liver (Fig. 1d). Although this demonstrates a mammary preference of genes associated with mammary super-enhancers, it also reveals that some were expressed in non-mammary cells under cytokine control. Approximately 50% of super-enhancer associated genes are induced during pregnancy through STAT5 and their expression is highly enriched in mammary tissue (Supplementary Table 1) at a median of ~43 FPKM (mean ~11,388 FPKM) (Fig. 1e, left panel). In contrast expression of genes not induced during pregnancy was only slightly enriched in mammary tissue (Fig. 1e, right panel).
Assembly of mammary super-enhancers during pregnancy
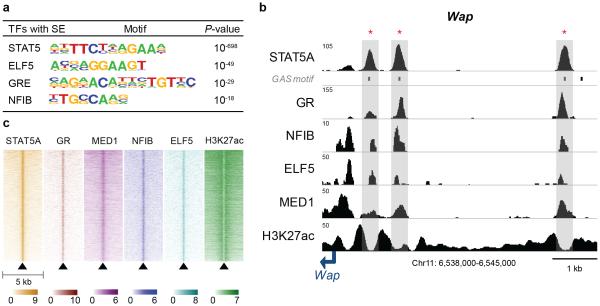
Enhancers are characterized by the binding of several transcription factors, so called hot spots43,50. A motif search (see M&M for details) established an enrichment of predicted binding sites for ELF5 and NFIB (Fig. 2a), transcription factors critical to mammary development7,13,51. ChIP-Seq experiments validated that NFIB and ELF5 bound to enhancers also occupied by STAT5, GR and MED1 (Fig. 2b and 2c), as exemplified for the Wap and Olah genes (Fig. 2b and Supplementary Fig. 4).
Figure 2.
Transcription factor binding signatures in mammary-specific super-enhancers. (a) Motif analysis within mammary-specific super-enhancers. Predicted motifs for transcription factors critical for mammary development were highly enriched in mammary-specific super-enhancers (±200 bp). (b) Transcription factor binding profiles of constituent enhancers within a mammary-specific super-enhancer. The Wap mammary-specific super-enhancer was shown as a representative example and the constituent enhancers were indicated as red asterisk. The data represent the biological duplicates. (c) Heatmap of transcription factor binding, STAT5A, GR, MED1, NFIB, ELF5, and H3K27ac within a 5 kb region around the center of hotspots in mammary-specific super-enhancers. The y-scale is sorted by the row sum of the STAT5A values for all transcription factors.
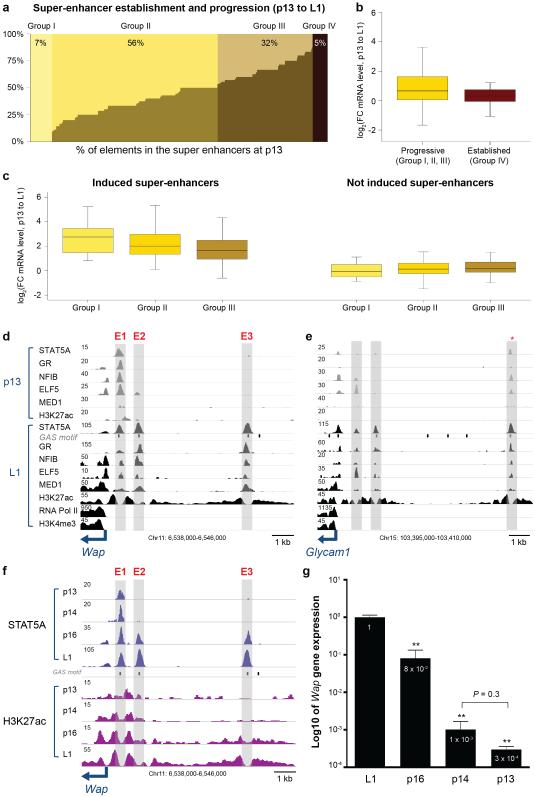
It can be hypothesized that mammary super-enhancers are established during pregnancy, a time frame during which expression of mammary-specific genes greatly increases. To address this we performed ChIP-Seq for STAT5, GR, NFIB, ELF5, MED1 and H3K27ac on mammary tissue at day 13 of pregnancy (p13), prior to the activation of key genes, including Wap. While 7% (32) of all super-enhancers were not occupied at p13 and were established only at day 1 of lactation (L1), some constituent enhancers within the remaining 93% (408) super-enhancers showed already limited TF occupancy at p13 (Fig. 3a). In 56% of the super-enhancers less than one half of the ChIP-Seq peaks are established at p13, 32% had coverage of more than one half and in 5% of the super-enhancers all peaks were already fully established. The sensitivity of ChIP-Seq experiments depends on the affinity of antibodies and it is possible that the data in Figure 3a underestimate TF binding. Next we assessed to what extent the temporal gain of TF binding to these enhancers translated into the activation of associated genes (Fig. 3b). Genes associated with super-enhancers void of TF binding (group I) or only partially occupied (group II and III) at p13 are characterized by higher induction levels than those already fully occupied (groups IV) at p13. Furthermore, genes associated to fully established super-enhancers at p13 show a higher gene expression level (Supplementary Fig. 5). Super-enhancers without apparent STAT5 occupancy at p13 show the highest induction followed by those with less than 50% occupancy and those with more than 50% pre-occupied enhancers (Fig. 3c, left panel). In contrast, super-enhancers associated with non-induced genes show equivalent expression patterns (Fig. 3c, right panel).
Figure 3.
Assembly of constituent enhancers in mammary-specific super-enhancers during pregnancy. (a) Seven percent of super-enhancers have no established peaks at day 13 of pregnancy (p13), 56% are occupied less than half, 32% are more than half, and 5% are already established at p13. (b) Genes associated with progressive super-enhancers (418) show higher induction levels than those fully occupied at p13 (22). Median, middle bar inside the box; IQR, 50% of the data; whiskers, 1.5 times the IQR. (c) Progressive enhancers associated with genes induced during pregnancy show a higher induction than those associated to not induced genes. Super-enhancers having no established enhancers at p13 show the highest induction followed by those with less than 50% and more than 50% pre-occupied enhancers (left). (d) Establishment of enhancers within the mammary-specific Wap super-enhancer. Only enhancer 1 (E1) is occupied by mammary-enriched transcription factors at p13, whereas E2 and E3 are exclusively occupied at day 1 of lactation (L1). Data represent biological duplicates. (e) Progressive establishment of the mammary-specific Glycam1 super-enhancer. One (asterisk) out of three individual enhancers is already occupied by mammary transcription factors at p13 and all three enhancers are fully occupied at lactation. (f) Establishment of individual enhancers within the mammary-specific Wap super-enhancer across different pregnancy stages. Only E1 is occupied by STAT5A and H3K27ac at p13 and p14, whereas all three enhancers are fully occupied at p16 and L1. (g) Wap mRNA levels in mammary tissue at different pregnancy stages. L1, n = 6; p16, p14, p13, n = 3. The casein locus served as a control (Supplementary Fig 6).
Individual enhancers within a given super-enhancer were frequently not established in concert during pregnancy but in a defined temporal order, suggesting that they differentially sense prolactin and possibly transcription factor concentrations. Notably, some individual enhancers within super-enhancers were already occupied during pregnancy prior to the activation of the associated genes. To attain a better understanding of the temporal progression of mammary-specific super-enhancers, we focused on the tripartite enhancer of Wap47, a gene induced more than 1,000-fold during pregnancy13,52. The Wap super-enhancer consists of three constituent enhancers, with one (E1) already being occupied by STAT5A, GR, NFIB, ELF5 and MED1 at mid pregnancy (Fig. 3d, Supplementary Fig. 6a). In contrast, E2 and E3 were fully occupied only at the onset of lactation. Similarly, one of the three individual enhancers within the Glycam1 super-enhancer was prominent already during pregnancy prior to its transcriptional activation (Fig. 3e). These findings suggest that individual enhancers within mammary-specific super-enhancers have unique capacities and differentially sense hormonal cues as pregnancy progresses. Moreover, enhancers already occupied in early pregnancy, such as E1 in the Wap super-enhancer, are generally unable to activate mammary-specific genes by themselves. The window between p13 and L1 is characterized by epithelial differentiation and devoid of significant proliferation suggesting that the establishment of super-enhancers is the result of cellular differentiation. Expression of Krt8 and Krt18, markers of mammary secretory epithelium, is equivalent between p13 and L1 (Supplementary Table 1). To further strengthen this, we conducted STAT5 and H3K27ac ChIP-Seq experiments at p14 and p16, stages that are distinguished by their differentiation status as evidenced by the 100-fold activation of the Wap gene (Fig. 3f, 3g, Supplementary Fig. 6b). While E1, but not E2 and E3, was occupied by STAT5 at p14, binding to all sites was secured by p16, demonstrating that the Wap super-enhancer is fully activated within this narrow time window. Although the expansion of mammary epithelium between p14 and p16 is negligible, the respective tissue has an equivalent appearance (Supplementary Fig. 7) and we analyzed the gain of signals specific to mammary epithelium, we still decided to compare specific chromatin marks in intact tissue and enriched mammary epithelial cells (Supplementary Fig. 8).
Hierarchy within the mammary-specific Wap super-enhancer
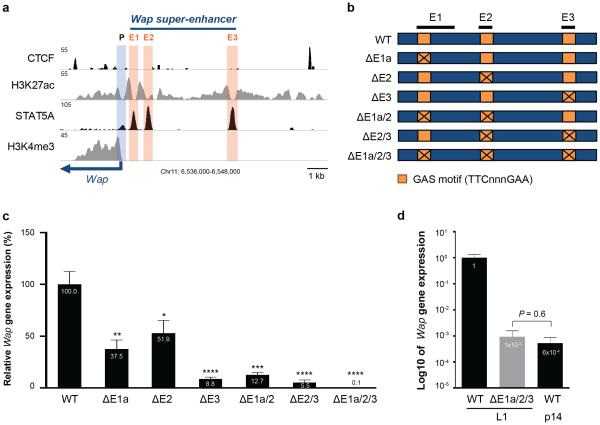
The progressive establishment of mammary-specific super-enhancers during pregnancy parallels the activation of some, but not all, associated genes suggesting a possible causal relationship. Since the creation of mammary alveoli is strictly dependent upon STAT56,53, it has not been possible to investigate it’s in vivo role in the establishment of individual mammary-specific enhancers10 in tissue devoid of STAT5. To unequivocally recognize the contribution of individual STAT5 sites within a mammary-specific super-enhancer we focused on Wap, which is activated more than 1,000-fold during pregnancy13,52. First, we individually deleted GAS motifs in each of the three putative STAT5 enhancers (E1 – E3) (Fig. 4a and Supplementary Fig. 9). The proximal E1 site was mutated using CRISPR/Cas9 genome editing and the E2 and E3 sites were deleted using TALEN technology (Fig. 4b). Mammary tissue from homozygous mutant mice was analyzed at the onset of lactation. Deletion of the proximal STAT5 enhancer site (ΔE1) resulted in 62% reduction of Wap mRNA levels, loss of E2 (ΔE2) in 48% and inactivation of the distal site (ΔE3) led to a reduction of 91% (Fig. 4c). This demonstrates that the three STAT5 enhancer units, despite their equivalent transcription factor occupancy and H3K27ac profiles, possess different in vivo strength, suggesting unique contributions of STAT5 at these sites.
Figure 4.
In vivo functions of individual enhancers within the mammary-specific super-enhancer of the Wap gene. (a) Genomic features of E1, E2, and E3 in the Wap locus. (b) Schematics of single enhancer mutations (ΔE1a, ΔE2 and ΔE3) and combined mutations (ΔE1a/2, ΔE2/3, ΔE1a/2/3) in the mouse genome. Exact deletions are shown in Supplementary Fig 9. (c) Wap mRNA levels in mammary tissues from mice carrying individual enhancer mutants (ΔE1a, ΔE2 and ΔE3) and combined mutants (ΔE1a/2, ΔE2/3, ΔE1a/2/3) at day 1 of lactation (L1). Wap mRNA levels were measured by qRT-PCR and normalized to Gapdh. Results are means of independent biological replicates with s.e.m. (WT, n = 9; ΔE1a, n = 7; ΔE2, n = 10; ΔE3, n = 7; ΔE1a/2, n = 3; ΔE2/3, n = 3; ΔE1a/2/3, n = 3). A two-tailed unpaired t-test was used to evaluate the statistical significance of differences between WT and each mutant group (*P < 0.05, **P < 0.001, ***P < 0.0001, ****P < 0.00001). Wap expression was reduced by approximately 91% in ΔE3 mutant mice and over 99.9% in ΔE1a/2/3 mutant mice. (d) Comparison of Wap mRNA levels in mammary tissues from ΔE1a/2/3 mutant mice at L1 and from WT controls at different stages (L1 and p14). Results are shown in log10 of means (error bars, s.e.m; n = 3). Wap expression was reduced 1,000-fold in ΔE1a/2/3 mutant mice compared to WT at L1 and was equivalent to WT p14.
Although highly informative, this study did not address potential additive or synergistic interactions between the three constituent enhancers that would account for the 1,000-fold induction of the Wap gene. To investigate this, we conducted successive gene targeting using CRISPR/Cas9 and generated mice carrying different combinations of mutants. Since E1 is established prior to Wap gene activation and precedes the formation of E2 we explored functional synergy between them. E1 mutant embryos were targeted at site E2 and ΔE1a/2 mice were generated. The combined loss of these two enhancers had a modest impact with an 87% reduction of Wap expression (Fig. 4c), less than what had been observed in the sole absence of E3. This provides further evidence of the prominent status of the distal E3 enhancer. Indication that sites E2 and E3 might hold the key to the extraordinary activation of Wap came from the observation that STAT5 occupation of these sites coincided with the 100-fold activation of Wap between days 14 and 16 of pregnancy (Fig. 3f and 3g). To test this supposition we targeted E2 within E3 mutant embryos and generated mice (ΔE2/3) lacking both sites. Wap expression in these mice was reduced by ~95% (Fig. 4c) demonstrating additive, yet no synergistic, interaction between these two enhancers. Although this induction is extraordinary compared to other enhancer-dependent genes it still did not account for the 1,000-fold induction of the endogenous Wap gene. We surmised that the combined entity of all three STAT5 enhancers was necessary and sufficient to impose the full induction, a hypothesis we tested in mice devoid of all three STAT5 enhancer sites. E1 mutant embryos were targeted for E2 and E3 and homozygous mice (ΔE1a/2/3) were generated and analyzed. In the absence of the three STAT5 enhancers Wap expression at day one of lactation was reduced by a remarkable 1,000-fold compared to wild type controls (Fig. 4c). To determine whether this unprecedented enhancer activity mirrored the induction of Wap at mid pregnancy, the stage when E2 and E3 recruit STAT5 and other transcription factors and acquire extensive H3K27ac, we compared Wap expression in ΔE1a/2/3 mutants and control tissues. In the absence of its tripartite enhancer Wap levels at day one of lactation were equivalent to those found for the wild type gene at day 14 of pregnancy (Fig. 4d), providing evidence that the tripartite STAT5 enhancer is responsible for the activation of Wap at mid pregnancy.
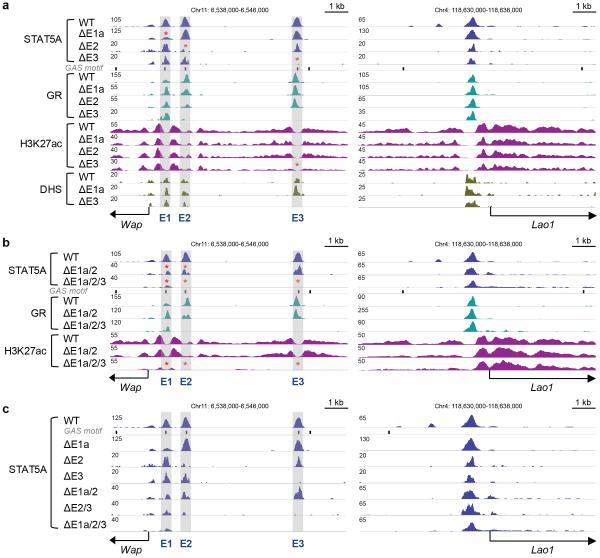
Although mutational analyses have validated the exceptional role of the tripartite STAT5 super-enhancer, the consequences of individual and combined mutations on the binding of other transcription factors and the establishment of H3K27ac islands remained unknown. We addressed this and investigated chromatin configuration and TF binding of the Wap super-enhancer in mammary tissue devoid of individual or combined constituent enhancers. The ΔE3 mutation not only led to loss of STAT5A binding but unexpectedly also to the complete absence of GR binding, H3K27ac and DNaseI hypersensitivity at E3 (Fig. 5a, left panel). The mammary-specific Lao1 gene served as a control and was not affected (Fig. 5a, right panel). In contrast, some STAT5 binding was retained upon deletion of the GAS motif in E1 (ΔE1a) and GR and MED1 binding were largely intact as was the H3K27ac profile (Fig. 5a) suggesting that this site had retained limited integrity. Similarly, the structural integrity of E2, including H3K27ac, remained largely intact upon deletion of the underlying STAT5 binding (Fig. 5a). The retention of structural integrity of E1 and E2 in the respective mutants is in agreement with their modest contribution to Wap expression. In contrast, E3 is exceptional in that loss of STAT5 binding prevented the establishment of visible enhancer features at this site, in agreement with a more than 90% reduced activity. We propose that STAT5 is the pioneer factor in the establishment of E3 but E1 and E2 are established, at least partially, in its absence. The rather modest functional contributions of E1a and E2, alone or combined, are mirrored by the retention of GR binding and H3K27ac marks in mutant tissue (Fig. 5b). The combined mutations E1a/2 and E2/3 were additive, but not synergistic, between two constituent enhancers. STAT5 ChIP-Seq experiments emphasized that loss of individual STAT5 binding sites had limited consequences on the remaining intact constituent enhancers (Fig. 5c). In contrast combined loss of the dominant E3 and E2 profoundly affected occupancy of the proximal E1. Most importantly, loss of all three STAT5 sites was incompatible with the formation of TF complexes over E2 and E3 and the establishment of active chromatin (Fig. 5b), in agreement with the complete loss of Wap activation during pregnancy (Fig. 4c). The residual binding of GR in the E1 region was insufficient to convey any meaningful activation.
Figure 5.
Structural consequences resulting from the loss of STAT5 binding at individual and combined enhancers in the mammary-specific Wap super-enhancer. (a) ChIP-Seq profiles and DNaseI hypersensitive sites (DHS) at the Wap locus in mammary tissue from single enhancer mutants at day one of lactation (L1). The Lao1 locus served as a ChIP-Seq control. The data for STAT5A and H3K27ac ChIP-Seq represent biological duplicates. STAT5A binding was reduced at site E1 in ΔE1a mutants and at E2 in ΔE2 mutants. GR binding and H3K27ac marks were retained in ΔE1a and ΔE2 mutants. STAT5A binding, GR binding and H3K27ac marks were lost at site E3 in ΔE3 mutants. (b) ChIP-Seq profiles in combined enhancer mutant mice. The data for STAT5A, GR and H3K27ac ChIP-Seq represent biological duplicates. STAT5A binding was reduced at site E1 and E2 and GR binding and H3K27ac marks were retained in mammary tissue from ΔE1a/2 mutants. Complete absence of STAT5A and GR binding, H3K27ac marks at E2 and E3 in mammary tissue from ΔE1a/2/3 mutants. Residual marks were retained at E1. (c) STAT5A ChIP-Seq profiles in single and combined enhancer mutant mice. STAT5A binding at the E3 site plays the most prominent role in Wap super-enhancer.
A seed enhancer in mammary-specific Wap super enhancer
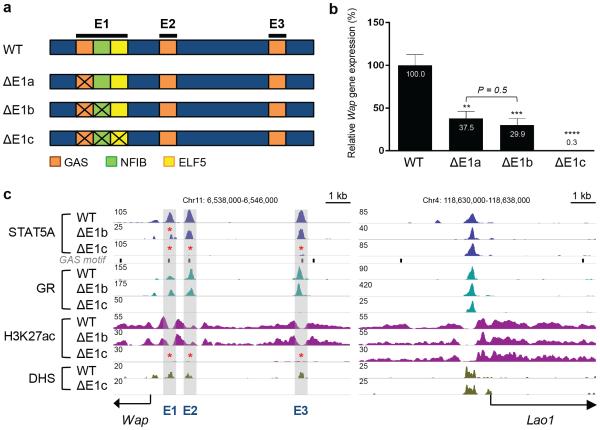
The importance of STAT5 within E1 is limited and its loss did not affect the overall enhancer structure suggesting that establishment of E1 might require the presence of additional transcription factors, possibly, NFIB and ELF5, which bind close to STAT514,15,54,55 (Fig. 6a, Supplementary Fig. 10). CRISPR/Cas9 genome editing was used to generate mice lacking the GAS and the juxtaposed NFIB motifs (ΔE1b) and homologous recombination in ES cells was used to introduce point mutations into the GAS, NFIB and ELF5 motifs (ΔE1c) (Fig. 6a). Combined inactivation of the GAS and NFIB motifs resulted in the reduction of Wap mRNA by approximately 70% (Fig. 6b) suggesting that NFIB is not essential for the activity of E1. Although studies using a Wap transgene with only 800 bp of 5’ flanking sequences, and thus lacking E2 and E3, had concluded that NFIB was essential for gene activity14,15,48,56 this was not the case for the endogenous gene. ChIP-Seq confirmed loss of NFIB binding to the mutant E1 and also demonstrated that ELF5 binding was unaffected (Supplementary Fig. 11).
Figure 6.
In vivo function of the epicenter within E1 of the Wap super-enhancer. (a) Diagram of mutations within E1, inactivating binding of STAT5 alone, STAT5 and NFIB, and STAT5, NFIB and ELF5. (b) Wap mRNA levels in mammary tissues from ΔE1a, ΔE1b and ΔE1c mutant mice at day 1 of lactation (L1). Wap mRNA levels were measured by qRT-PCR and normalized to Gapdh. Results are means of independent biological replicates with s.e.m. (WT, n = 9; ΔE1a, n = 7; ΔE1b, n = 7; ΔE1c, n = 5). A two-tailed unpaired t-test was used to evaluate the statistical significance of differences between WT and each mutant group (**P < 0.001, ***P < 0.0001, ****P < 0.00001). Wap expression levels between ΔE1a and ΔE1b were not significantly different (P = 0.5). Wap expression was reduced by 99.7% in ΔE1c mutant mice. (c) Genomic features of ΔE1b and ΔE1c mutant mice. The data for STAT5A and H3K27ac ChIP-Seq represent biological duplicates. STAT5A binding and H3K27ac marks were reduced at E1 in mammary tissue from ΔE1b mutants. STAT5A and GR binding, H3K27ac marks and DHS were absent at the three individual enhancers (E1, E2 and E3) in mammary tissue from ΔE1c mutants.
The combined roles of STAT5, NFIB and ELF5 were investigated in mice carrying point mutations in all three sites (ΔE1c) (Fig. 6a, Supplementary Fig. 11). Unexpectedly, mutations in these three sites incapacitated the entire Wap locus (Fig. 6b). Moreover, E2 and E3 failed to be established during pregnancy as indicated by the absence of transcription factor binding and H3K27ac marks (Fig. 6c). The complete absence of DNaseI hypersensitivity suggested that it had not undergone any priming and was unable to respond to pregnancy hormones. Collectively, these data indicate that the joint binding of three TFs within E1 provides an epicenter required for the activation of this mammary-specific locus and the recruitment of additional enhancers in response to hormonal stimuli during pregnancy. The super-enhancer that conduces the 1,000-fold induction during pregnancy is built on STAT5 that binds to three individual enhancers with distinct properties. Notably, the Nfib and Elf5 genes appear also to be under the control of STAT5-driven enhancers (Supplementary Fig. 12).
DISCUSSION
Although super-enhancers have been identified in diverse cell systems4,26-35,37-46,50,57-60 there has been scant genetic proof of their biological significance in genuine in vivo settings in mice (Supplementary Table 5). Similarly, individual and combined contributions of constituent enhancers within super-enhancers has not been investigated using mouse genetics24,25,57. Our study now provides compelling genetic evidence that a unique mammary-specific super-enhancer controls hormone-regulated expression of the Wap gene during pregnancy and that embedded constituent enhancers deliver distinct contributions. In the case studied, the most distal enhancer does the heavy lifting, while the proximal ones have modest activity by themselves. Importantly, additive effects but no synergism was observed upon mutating two out of the three constituents, and all three constituents were required to achieve the 1,000-fold induction. Additional genetic studies are needed to determine whether this is a general strategy used by other cell-specific super-enhancers in vivo. Mutational studies of enhancers within an erythroid super-enhancer in a cell line also demonstrated an enhancer with the most distal one being most potent34. Based on this cell line study and our mouse study a hierarchy within super-enhancers can be predicted and only the loss of all constituent enhancers might abolish induction of the respective target genes. Establishment of the seed enhancer within the Wap super-enhancer during pregnancy depends on three mammary-enriched TFs and its presence is required to launch the two accessory enhancers leading to the full induction of gene expression.
Our study strongly suggests that super-enhancers, as defined by ChIP-Seq experiments and specific algorithms to identify peak patterns, do not necessarily provide a meaningful biological concept. Despite equivalent patterns of TF binding, only one half of the super-enhancers were associated with highly expressed genes induced during pregnancy by STAT5, the principal TF used to define mammary-specific super-enhancers. Moreover, the range of gene induction covered four orders of magnitude and there was no obvious correlation between TF occupancy and gene induction. Although increased and progressive occupancy of super-enhancers was observed during pregnancy, this did not necessarily parallel induction of the associated genes. This questions the concept that the degree of TF loading is a predictor of gene expression.
Although STAT family members are key in controlling cell lineages, likely through cell-restricted enhancers10,57,61-64, studies with knock-out mice have only provided limited insight into the contributions of individual STATs to specific super-enhancers or its embedded constituents. Loss of either STAT3 or STAT5 from the mouse genome results in the absence of specific T cell populations65-68 and mammary epithelium6 making it impossible to assess their contributions to lineage-specific super-enhancers. Using genome editing to target individual enhancers avoids the systemic pitfalls encountered in mice lacking transcription factors, either in the germline or specific cell types. Such an approach provides an unbiased functional appraisal of predicted enhancer structures in the context of an otherwise uncompromised organism69. This is particularly relevant for cytokine-responsive systems, including T cells and mammary epithelium, whose lineages fail to develop in the absence of specific cytokine-sensing transcription factors. In summary, our study provides evidence that an unbiased genome-wide survey of putative enhancers, coupled with enhancer editing, can provide mechanistic insight into complex organ-specific and cytokine-regulated genetic circuits.
URLs
MIT CRISPR Design tool, http://crispr.mit.edu/; R Project for Statistical Computing, https://www.R-project.org/; dplyr, https://CRAN.R-project.org/package=dplyr; Power Analysis, https://CRAN.R-project.org/package=pwr.
ONLINE METHODS
Mice
Six to eight-week-old C57BL/6 female mice were purchased from Charles River and used as Wild Type controls. CRISPR/Cas9 targeted mice were generated by the transgenic core of the National Heart Lung and Blood Institute (NHLBI) and TALEN targeted mice were generated by Cyagen Biosciences (2255 Martin Avenue, CA 95050). Mice carrying point mutations in the E1 region were generated by Ingenious Targeting Laboratory (2200 Smithtown Ave, NY 11779). All animal procedures were in accordance with NIH, NIDDK guidelines for the care and use of laboratory animals.
ChIP-Seq
Frozen-stored mammary tissues harvested at day 13 of pregnancy (p13), p14, p16 and day 1 of lactation (L1) were ground into powder with a mortar and pestle. Chromatin was fixed with 1% formaldehyde at room temperature for 10 min and the fixation was quenched with Glycine at a final concentration of 0.125M. Samples were processed as previously described70. The following antibodies were used for ChIP-Seq: anti-STAT5A (Santa Cruz, sc-1081), anti-GR (Thermo Scientific, PA1-511A), anti-NFIB (Santa Cruz, sc-5567), anti-ELF5 (Santa Cruz, sc-9645), anti-MED1 (Bethyl Laboratory, A300-793A), anti-H3K27ac (Abcam, ab4729), anti-H3K4me3 (Millipore, 17-614), and anti-RNA Polymerase II (Abcam, ab5408). Libraries for the next generation sequencing (NGS) were prepared as previously described70 and sequenced with HiSeq 2000 (Illumina).
ChIP-Seq data analysis
ChIP-Seq signals were trimmed using trimmomatic71 (version 0.33) for filtering low quality reads (using following parameters: LEADING:20, TRAILING:20, SLIDINGWINDOW:4:20, MINLEN:20, HEADCROP:15). The subsequent ChIP-Seq reads were aligned to the mouse reference genome (mm9) using Bowtie72 aligner (version 1.1.2) with the −m 1 parameter to get only uniquely mapped reads. The average alignment rate was 93% and around 24% were discarded due to the −m parameter (Supplementary Table 4). The correlation of all replicates was calculated using deepTools73 with default parameters and spearman correlation, as it is more stable if outliers occur, with the caveat that the correlation value is less sensitive. ChIP-Seq data that have shown in this study were highly reproducible (spearman’s correlation coefficient > 0.7). The HOMER software74 (default settings) was used for the visualization. To identify the regions of ChIP-Seq enrichment over background, MACS275 peak-finding algorithms (version 2.1.0) was used. As data were from different resources the q-value/p-value parameter was adjusted individually for each file to optimize STAT5 peak calling (STAT5A L1 replicate one q-value cutoff = 1×10−5, STAT5A L1 replicate two p-value cutoff = 1×10−2, STAT5A p13 replicate one q-value cutoff = 1×10−2, STAT5A p13 replicate two q-value cutoff = 1×10−3, STAT5A liver q-value cutoff = 1×10−3, STAT5B T cell q-value cutoff = 5×10−2). For all STAT5A L1 and p13 samples also adequate input files were used. To receive high confident peaks, replicates were used for STAT5A L1 and p13, which were identified by overlapping the files using bedOps76 genome analysis toolkit (version 2.4.14) using one base pair as overlapping criteria. For H3K27ac of wild type L1 tissue the broad peak calling option with a q-value cutoff of 5×10−2 was selected. Only STAT5A L1 peaks, validated using both replicates, which coincide with H3K27ac marks within ±500 bp were taken into account for further analysis. The final step comprised the extraction of promoter peaks using ±500 bp to the TSS. Those verified and filtered peaks were used for all subsequent analyses.
Identification of super-enhancers
The non-promoter STAT5 peaks served as the basis for the super-enhancer analysis calculation applying the ROSE algorithm 37,46 using default stitching size of 12.5 kb, as well as 25 kb and 35 kb since mammary-specific super-enhancers might be different from super-enhancers identified in embryonic stem cells. To obtain only mammary-specific super-enhancers each stitched size was used as a parameter for the calculations based on STAT5A peaks, H3K27ac, GR and MED1. The next step comprised the overlap of H3K27ac, GR and MED1 super-enhancers per size. The super-enhancers identified and reproduced for at least two factors were maintained for further analysis. The subsequent subtraction of liver and T-cell super-enhancers (12.5 kb) was applied using bedOps with the criteria that minimum 30% of the mammary super-enhancer needed to overlap with the liver or T cell super-enhancers. In the final step nested super-enhancers, which had been annotated to the same gene, were removed.
RNA-Seq data analysis and gene annotation
RNA-Seq data were trimmed in the same manner as the ChIP-Seq data. The mapping was carried out using STAR RNA-Seq aligner77 with default settings and Mus_musculus.NCBIM37.67 as GTF file. In order to assign the super-enhancers and lone enhancers only to high confident genes, the GTF file was filtered to get only protein-coding genes, and predicted genes (LOC, Rik and BC) were excluded. R (version 3.2.3), Bioconductor78 and the packages Rsubread79 (default settings) and DESeq280 (default settings) were used for RNA-Seq analysis. Data from mammary tissue at days 6 (p6) and 13 (p13) of pregnancy, lactation day one and STAT5A deficient tissue at lactation day one as well as liver and T cell data were analyzed. Mammary-specific super-enhancers were annotated taking into account the two nearest genes and choosing the one with the higher FPKM value based on expression data obtained at L1.
Additional bioinformatics analysis
HOMER74 was used for motif analysis using the default settings with an individually generated background, based on concatenated DNase-Seq data from B cell, cerebellum, kidney, liver, lung, spleen and thymus. The heatmap was created using a width of 5 kb. The analysis of L1 specific enhancers and enhancers already established at p13 was done using bedOps76 genome analysis toolkit, with the criteria that the peaks need to be inside the super-enhancer. Graph plotting was performed using R with the packages dplyr and ggplot281.
Data
STAT5 ChIP-Seq data for liver and T-cells were obtained from the GEO accession No. GSE31578 and GSE27158, respectively. H3K27ac ChIP-Seq data for liver and T-cells were obtained from the GEO accession No GSE31039. The RNA-Seq data for wild type at p6 and L1 and deficient STAT5a at L1 are deposited under GSE37646. RNA-Seq data for wild type at p13 are obtained from GSE70440. T cell and liver RNA-Seq data are available under GSE48138 and GSE66140.
Generation of CRISPR/Cas9 targeted mice
The sgRNA constructs were designed to target specific GAS motif at E1 (−0.7 kb from Wap TSS), E2 (−1.4 kb from Wap TSS), and E3 (−5.6 kb from Wap TSS). The off-target scores were evaluated by the online tool. The specific sgRNA sequences are described in the Supplementary Table 2. Target-specific sgRNA and Cas9 mRNA were in vitro transcribed and microinjected into the cytoplasm of fertilized eggs for the founder mice production.
Generation of TALEN targeted mice
The specific GAS motif (−1.4 kb from Wap TSS or −5.6 kb from Wap TSS) was selected as a TALEN target site. TALEN mRNA was generated in vitro transcription, which was then injected into fertilized eggs for the founder mice production.
Generation of knockin mice
Point mutations were introduced in the GAS motif, NFIB motif, and ELF5 motif in E1 (−0.7 kb from Wap TSS) using homologous recombination in embryonic stem cells.
Generation of homozygous mice and genotyping
Founder (F0) of CRISPR/Cas9 targeted mice and TALEN targeted mice were bred to C57BL/6 wild type mice to segregate the mosaicism and generate F1 heterozygous mice, which were inter-bred to generate F2 homozygous mice. Founders of knockin mice were interbred for the mouse colony expansion. All mice were genotyped by PCR amplification of genomic DNA isolated from mouse tail snips followed by Sanger sequencing. Details of PCR primers and sequencing primers are described in the Supplementary Table 3. The two lines carrying deletions within the GAS site in E1 were named ΔE1a (11 bp deletion) and ΔE1b (27 kb). The knockin mice were named ΔE1c. TALEN targeted mice were named ΔE2 and ΔE3 according to the location of specific GAS deletion sites. The mice that have combined mutations at GAS site in E1, E2, and E3 by CRISPR/Cas9 genome editing named ΔE1a/2, ΔE2/3, and ΔE1a/2/3 according to the location of specific GAS deletion sites. The specific deletion sequences in each mouse are shown in the Supplementary Figure 9.
RNA isolation and quantitative RT-PCR
RNA was isolated using the PureLink RNA Mini kit (Ambion) according to the manufacturer’s protocol. Complementary DNA was synthesized from total RNA using SuperScript II (Invitrogen) and the quantitative PCR was performed using the Taqman probe-based system (Wap, Mm00839913_m1; mouse Gapdh endogenous control, 4352339E, Applied Biosystem) on the CFX384 Real-Time PCR Detection System (Bio-Rad). Wap mRNA levels were measured by qRT-PCR and normalized to Gapdh.
DNase-Seq
DNase-Seq in mammary glands of WT and mutant mice was performed as previously described70 .
Statistical analyses
All samples that were used for qRT-PCR, ChIP-Seq and DNase-Seq were randomly selected and the blind test was not applied. The significance of the boxplots was calculated using a Student’s t-test with a one-sided alternative hypothesis in case of a two sample comparison. For more samples ANOVA was applied to see whether the comparison is significant or not. In case of significance an additional pairwise t-test was used to compare the groups. Prior to any statistical analysis a Shapiro-Wilk Normality test was applied to validate the normal distribution of the data. Statistical power was calculated using R and the package pwr. The effect size was calculated for each comparison based on the estimated means and standard deviations of each group and the sample size for each group was therefore determined via power analysis of Wap expression change with significance level 0.05 and power level 0.9. Gene expression data were presented with the mean of independent biological replicates. To evaluate the gene expressions that are statistically different between WT and each mutant group, a two-tailed unpaired t-test was used.
Mammary epithelial cell (MECs) isolation
Mammary tissue was obtained at p13 and L1 from wild type mice. Tissue was digested for 2 hours at 37°C in complete EpiCult-B medium (EpiCult-B medium with 5% fetal bovine serum) supplemented with 300 U/mL collagenase and 100 U/mL hyaluronidase. After lysis of red blood cells in NH4Cl, single cell suspensions were obtained by sequential dissociation with pre-warmed 0.25% Trypsin-EDTA, followed by pre-warmed 5 mg/mL Dispase and 0.1 mg/mL DNase I, and filtration through a 70 μm cell strainer. All reagents were from Stemcell Technologies unless otherwise specified. Obtained single cell suspensions were isolated to mammary epithelial cells by the removal of non-epithelial cell compartments with the biotinylated antibodies against the cell surface antigens of murine hematopoietic, endothelial, and fibroblast cells (CD45, CD31, TER119, and BP-1) using EasySep Mouse Epithelial Cell Enrichment Kit (Stemcell Technologies).
Histological analysis
Mammary tissues from wild type mice were harvested at p13, p14, p16, and L1. Harvested mammary tissues were fixed in 10% formalin and dehydrated in ethanol. Paraffin sections were stained with hematoxylin and eosin by standard methods (Histoserve).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Harold Smith from the NIDDK genomics core for never-ending help with NGS and Dr. Chengyu Liu from the NHLBI transgenic core for generating the CRISPR/Cas9-based mouse mutants. M.W. is a graduate student of the Individual Graduate Partnership Program (GPP) between NIH/NIDDK and the Medical University of Innsbruck (Innsbruck, Austria). This work was performed in partial fulfillment of the graduate requirements for M.W. We thank Dr. Keunsoo Kang and Sumin Oh (Dankook University, Cheonan, South Korea) for discussions in the early stage of this project and Dr. Zlatko Tajanoski (Medical University of Innsbruck) for advising M.W. during her graduate studies. This research was funded by the IPR of the NIDDK/NIH.
Footnotes
Accession codes. All ChIP-Seq and DNase-seq data sets have been deposited in the Gene Expression Omnibus (GEO) under accession GSE74826. Liver and T cell data: GSE31578, GSE27158 and GSE31039. RNA-Seq from mammary tissue: GSE70440. The T cell and liver RNA-Seq data are available under GSE48138 and GSE66140.
AUTHOR CONTRIBUTIONS
H.Y.S. designed, executed and supervised genotyping, identified and validated all CRISPR/Cas9-based mutant founders, ChIP-Seq, gene expression experiments, histological analysis, analyzed data and wrote the manuscript. M.W. designed experiments, analyzed ChIP-Seq and RNA-Seq data, performed computational and statistical analyses and wrote the manuscript. K.H.Y. designed and conducted ChIP-Seq, gene expression experiments, histological analysis, and analyzed data. X.Z. conducted genotyping and performed and analyzed gene expression experiments on mutant mice. C.W. performed DHS and ChIP-Seq experiments and analyzed data. G.M. genotyped mutant mice and performed and analyzed gene expression experiments and identified founder mice carrying combined mutations. L.H. conceived and supervised the study, analyzed data and wrote the manuscript. H.Y.S., M.W. and L.H. wrote and finalized the manuscript and all authors reviewed and approved the submitted version.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–25. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- 2.Lydon JP, et al. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–78. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- 3.Ormandy CJ, et al. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 1997;11:167–78. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- 4.Horseman ND, et al. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. EMBO J. 1997;16:6926–35. doi: 10.1093/emboj/16.23.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waller A, Adunski A, Hershkowitz M. Terminal dehydration and intravenous fluids. Lancet. 1991;337:745. doi: 10.1016/0140-6736(91)90342-m. [DOI] [PubMed] [Google Scholar]
- 6.Cui Y, et al. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol Cell Biol. 2004;24:8037–47. doi: 10.1128/MCB.24.18.8037-8047.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, et al. Elf5 is essential for early embryogenesis and mammary gland development during pregnancy and lactation. EMBO J. 2005;24:635–44. doi: 10.1038/sj.emboj.7600538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wakao H, Gouilleux F, Groner B. Mammary gland factor (MGF) is a novel member of the cytokine regulated transcription factor gene family and confers the prolactin response. EMBO J. 1994;13:2182–91. doi: 10.1002/j.1460-2075.1994.tb06495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu X, Robinson GW, Gouilleux F, Groner B, Hennighausen L. Cloning and expression of Stat5 and an additional homologue (Stat5b) involved in prolactin signal transduction in mouse mammary tissue. Proc Natl Acad Sci U S A. 1995;92:8831–5. doi: 10.1073/pnas.92.19.8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamaji D, Kang K, Robinson GW, Hennighausen L. Sequential activation of genetic programs in mouse mammary epithelium during pregnancy depends on STAT5A/B concentration. Nucleic Acids Res. 2013;41:1622–36. doi: 10.1093/nar/gks1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hennighausen LG, Sippel AE. Characterization and cloning of the mRNAs specific for the lactating mouse mammary gland. Eur J Biochem. 1982;125:131–41. doi: 10.1111/j.1432-1033.1982.tb06660.x. [DOI] [PubMed] [Google Scholar]
- 12.Richards DA, Rodgers JR, Supowit SC, Rosen JM. Construction and preliminary characterization of the rat casein and alpha-lactalbumin cDNA clones. J Biol Chem. 1981;256:526–32. [PubMed] [Google Scholar]
- 13.Pittius CW, Sankaran L, Topper YJ, Hennighausen L. Comparison of the regulation of the whey acidic protein gene with that of a hybrid gene containing the whey acidic protein gene promoter in transgenic mice. Mol Endocrinol. 1988;2:1027–32. doi: 10.1210/mend-2-11-1027. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Rosen JM. Distal regulatory elements required for rat whey acidic protein gene expression in transgenic mice. J Biol Chem. 1994;269:14235–43. [PubMed] [Google Scholar]
- 15.Li S, Rosen JM. Nuclear factor I and mammary gland factor (STAT5) play a critical role in regulating rat whey acidic protein gene expression in transgenic mice. Mol Cell Biol. 1995;15:2063–70. doi: 10.1128/mcb.15.4.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKnight RA, Wall RJ, Hennighausen L. Expression of genomic and cDNA transgenes after co-integration in transgenic mice. Transgenic Res. 1995;4:39–43. doi: 10.1007/BF01976500. [DOI] [PubMed] [Google Scholar]
- 17.Burdon TG, Maitland KA, Clark AJ, Wallace R, Watson CJ. Regulation of the sheep beta-lactoglobulin gene by lactogenic hormones is mediated by a transcription factor that binds an interferon-gamma activation site-related element. Mol Endocrinol. 1994;8:1528–36. doi: 10.1210/mend.8.11.7877621. [DOI] [PubMed] [Google Scholar]
- 18.Greenberg NM, Reding TV, Duffy T, Rosen JM. A heterologous hormone response element enhances expression of rat beta-casein promoter-driven chloramphenicol acetyltransferase fusion genes in the mammary gland of transgenic mice. Mol Endocrinol. 1991;5:1504–12. doi: 10.1210/mend-5-10-1504. [DOI] [PubMed] [Google Scholar]
- 19.Gordon K, et al. Production of human tissue plasminogen activator in transgenic mouse milk. Biotechnology. 1992;24:425–8. [PubMed] [Google Scholar]
- 20.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–86. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 21.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–93. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ong CT, Corces VG. Enhancers: emerging roles in cell fate specification. EMBO Rep. 2012;13:423–30. doi: 10.1038/embor.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet. 2012;46:1–19. doi: 10.1146/annurev-genet-110711-155459. [DOI] [PubMed] [Google Scholar]
- 24.Heinz S, Romanoski CE, Benner C, Glass CK. The selection and function of cell type-specific enhancers. Nat Rev Mol Cell Biol. 2015;16:144–54. doi: 10.1038/nrm3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pott S, Lieb JD. What are super-enhancers? Nat Genet. 2015;47:8–12. doi: 10.1038/ng.3167. [DOI] [PubMed] [Google Scholar]
- 26.Adam RC, et al. Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature. 2015;521:366–70. doi: 10.1038/nature14289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown JD, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:219–31. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapuy B, et al. Discovery and characterization of super-enhancer-associated dependencies in diffuse large B cell lymphoma. Cancer Cell. 2013;24:777–90. doi: 10.1016/j.ccr.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chipumuro E, et al. CDK7 inhibition suppresses super-enhancer-linked oncogenic transcription in MYCN-driven cancer. Cell. 2014;159:1126–39. doi: 10.1016/j.cell.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Z, et al. Transcription factor co-occupied regions in the murine genome constitute T-helper-cell subtype-specific enhancers. Eur J Immunol. 2015;45:3150–7. doi: 10.1002/eji.201545713. [DOI] [PubMed] [Google Scholar]
- 31.Gosselin D, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–40. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hnisz D, et al. Convergence of developmental and oncogenic signaling pathways at transcriptional super-enhancers. Mol Cell. 2015;58:362–70. doi: 10.1016/j.molcel.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J, et al. Dynamic Control of Enhancer Repertoires Drives Lineage and Stage-Specific Transcription during Hematopoiesis. Dev Cell. 2016;36:9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, et al. CRISPR reveals a distal super-enhancer required for Sox2 expression in mouse embryonic stem cells. PLoS One. 2014;9:e114485. doi: 10.1371/journal.pone.0114485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu CF, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015;43:8183–203. doi: 10.1093/nar/gkv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–34. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansour MR, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–7. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohba S, He X, Hojo H, McMahon AP. Distinct Transcriptional Programs Underlie Sox9 Regulation of the Mammalian Chondrocyte. Cell Rep. 2015;12:229–43. doi: 10.1016/j.celrep.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parker SC, et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc Natl Acad Sci U S A. 2013;110:17921–6. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pelish HE, et al. Mediator kinase inhibition further activates super-enhancer-associated genes in AML. Nature. 2015;526:273–6. doi: 10.1038/nature14904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinz S, Unser S, Rascle A. Signal transducer and activator of transcription STAT5 is recruited to c-Myc super-enhancer. BMC Mol Biol. 2016;17:10. doi: 10.1186/s12867-016-0063-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siersbaek R, et al. Transcription factor cooperativity in early adipogenic hotspots and super-enhancers. Cell Rep. 2014;7:1443–55. doi: 10.1016/j.celrep.2014.04.042. [DOI] [PubMed] [Google Scholar]
- 44.Thakurela S, Sahu SK, Garding A, Tiwari VK. Dynamics and function of distal regulatory elements during neurogenesis and neuroplasticity. Genome Res. 2015;25:1309–24. doi: 10.1101/gr.190926.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vahedi G, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–62. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–19. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hennighausen LG, Sippel AE. Mouse whey acidic protein is a novel member of the family of 'four-disulfide core' proteins. Nucleic Acids Res. 1982;10:2677–84. doi: 10.1093/nar/10.8.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burdon T, Sankaran L, Wall RJ, Spencer M, Hennighausen L. Expression of a whey acidic protein transgene during mammary development. Evidence for different mechanisms of regulation during pregnancy and lactation. J Biol Chem. 1991;266:6909–14. [PubMed] [Google Scholar]
- 49.Liu X, et al. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–86. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 50.Siersbaek R, et al. Extensive chromatin remodelling and establishment of transcription factor 'hotspots' during early adipogenesis. EMBO J. 2011;30:1459–72. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson GW, et al. Coregulation of genetic programs by the transcription factors NFIB and STAT5. Mol Endocrinol. 2014;28:758–67. doi: 10.1210/me.2012-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bayna EM, Rosen JM. Tissue-specific, high level expression of the rat whey acidic protein gene in transgenic mice. Nucleic Acids Res. 1990;18:2977–85. doi: 10.1093/nar/18.10.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miyoshi K, et al. Signal transducer and activator of transcription (Stat) 5 controls the proliferation and differentiation of mammary alveolar epithelium. J Cell Biol. 2001;155:531–42. doi: 10.1083/jcb.200107065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li S, Rosen JM. Glucocorticoid regulation of rat whey acidic protein gene expression involves hormone-induced alterations of chromatin structure in the distal promoter region. Mol Endocrinol. 1994;8:1328–35. doi: 10.1210/mend.8.10.7854350. [DOI] [PubMed] [Google Scholar]
- 55.McKnight RA, et al. An Ets site in the whey acidic protein gene promoter mediates transcriptional activation in the mammary gland of pregnant mice but is dispensable during lactation. Mol Endocrinol. 1995;9:717–24. doi: 10.1210/mend.9.6.8592517. [DOI] [PubMed] [Google Scholar]
- 56.McKnight RA, Spencer M, Wall RJ, Hennighausen L. Severe position effects imposed on a 1 kb mouse whey acidic protein gene promoter are overcome by heterologous matrix attachment regions. Mol Reprod Dev. 1996;44:179–84. doi: 10.1002/(SICI)1098-2795(199606)44:2<179::AID-MRD6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 57.Witte S, O'Shea JJ, Vahedi G. Super-enhancers: Asset management in immune cell genomes. Trends Immunol. 2015;36:519–26. doi: 10.1016/j.it.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gonzalez AJ, Setty M, Leslie CS. Early enhancer establishment and regulatory locus complexity shape transcriptional programs in hematopoietic differentiation. Nat Genet. 2015;47:1249–59. doi: 10.1038/ng.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu CF, Lefebvre V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou H, et al. Epstein-Barr virus oncoprotein super-enhancers control B cell growth. Cell Host Microbe. 2015;17:205–16. doi: 10.1016/j.chom.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vahedi G, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–93. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang XP, et al. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat Immunol. 2011;12:247–54. doi: 10.1038/ni.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang K, Yamaji D, Yoo KH, Robinson GW, Hennighausen L. Mammary-specific gene activation is defined by progressive recruitment of STAT5 during pregnancy and the establishment of H3K4me3 marks. Mol Cell Biol. 2014;34:464–73. doi: 10.1128/MCB.00988-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li P, Spolski R, Liao W, Leonard WJ. Complex interactions of transcription factors in mediating cytokine biology in T cells. Immunol Rev. 2014;261:141–56. doi: 10.1111/imr.12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yao Z, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–5. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao Z, et al. Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood. 2007;109:4368–75. doi: 10.1182/blood-2006-11-055756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–81. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 68.Wei L, Laurence A, Elias KM, O'Shea JJ. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J Biol Chem. 2007;282:34605–10. doi: 10.1074/jbc.M705100200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Metser G, et al. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res. 2016;44:1052–63. doi: 10.1093/nar/gkv999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Metser G, et al. An autoregulatory enhancer controls mammary-specific STAT5 functions. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–91. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neph S, et al. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012;28:1919–20. doi: 10.1093/bioinformatics/bts277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huber W, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat Methods. 2015;12:115–21. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Research. 2013;41 doi: 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Ggplot2: Elegant Graphics for Data Analysis. 2009:1–212. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.