Abstract
Objective
Although several genome-wide association studies (GWAS) of non-cardia gastric cancer have been published, more novel association signals could be exploited by combining individual studies together, which will further elucidate the genetic susceptibility of non-cardia gastric cancer.
Design
We conducted a meta-analysis of two published Chinese GWAS studies (2031 non-cardia gastric cancer cases and 4970 cancer-free controls) and followed by genotyping of additional 3564 cases and 4637 controls in two stages.
Results
The overall meta-analysis revealed two new association signals. The first was a novel locus at 5q14.3 and marked by rs7712641 (per-allele OR=0.84, 95% CI 0.80 to 0.88; p=1.21×10−11). This single-nucleotide polymorphism (SNP) marker maps to the intron of the long non-coding RNA, lnc-POLR3G-4 (XLOC_004464), which we observed has lower expression in non-cardia gastric tumour compared with matched normal tissue (Pwilcoxon signed-rank=7.20×10−4). We also identified a new signal at the 1q22 locus, rs80142782 (per-allele OR=0.62; 95% CI 0.56 to 0.69; p=1.71×10−19), which was independent of the previously reported SNP at the same locus, rs4072037 (per-allele OR=0.74; 95% CI 0.69 to 0.79; p=6.28×10−17). Analysis of the new SNP conditioned on the known SNP showed that the new SNP remained genome-wide significant (Pconditional=3.47×10−8). Interestingly, rs80142782 has a minor allele frequency of 0.05 in East Asians but is monomorphic in both European and African populations.
Conclusion
These findings add new evidence for inherited genetic susceptibility to non-cardia gastric cancer and provide further clues to its aetiology in the Han Chinese population.
Keywords: GASTRIC CANCER, GENETIC POLYMORPHISMS, EPIDEMIOLOGY
Significance of this study.
What is already known on this subject?
Approximately 40% of all cases of gastric cancer worldwide occur in China, and this form of cancer remains one of the key public health issues in cancer prevention and control.
Several previous genome-wide association studies (GWAS) of gastric cancer have reported several associations for common single-nucleotide polymorphisms (SNPs).
Combining together studies of moderate sample sizes will increase statistical power, so more novel signals can be exploited.
What are the new findings?
Based on a GWAS meta-analysis approach to pool two published Chinese GWAS, and followed by two-stage replications (more than 10 000 samples), we identified two novel signals associated with the risk of non-cardia gastric cancer. The first one rs7712641 maps to the intron of the long non-coding RNA, lnc-POLR3G-4 (XLOC_004464). Further analysis showed that rs7712641 had significantly lower expression in non-cardia gastric tumour compared with matched normal tissue. In addition, we observed a new signal marked by rs80142782 at the 1q22 locus. It was independent of the previously reported SNP rs4072037 and is a common SNP in East Asians but is monomorphic in both European and African populations.
Introduction
Globally, gastric cancer remains the third-leading cause of cancer death in both sexes,1 with more than half of gastric cancer cases worldwide occurring in East Asia, predominantly in China. Most cases of gastric cancer are sporadic,2 and its aetiology is related to both genetic susceptibility and epidemiological risk factors3 such as age, sex, Helicobacter pylori infection,4 5 family history, excessive salt intake and tobacco smoking. Anatomically, gastric cancer is classified into cardia gastric cancer and non-cardia gastric cancer, which are characterised by distinct risk factors and clinical features.4 6–8
Recently, several genome-wide association studies (GWAS) of gastric adenocarcinoma were conducted in East Asians.9–12 Notable findings include single-nucleotide polymorphism (SNP) markers mapping to 8q24.3, for both an intronic SNP (rs2976392) and an exonic SNP (rs2294008) in the Prostate Stem Cell Antigen gene (PSCA); and two markers rs2075570 and rs2070803 near the Mucin 1 gene (MUC1) on 1q22.9 The findings on 1q22 locus and gastric cancer risk were further replicated in several follow-up studies and additional evidence pointed to the non-synonymous SNP, rs4072037, as the functional variant underlying the observed association.13–18 In addition, 3q13.31 marked by rs9841504, 5p13.1 marked by rs13361707 or rs10074991 and 6p21.1 marked by rs2294693 were reported to be associated with non-cardia gastric adenocarcinoma in China,11 12 whereas 10q23 marked by rs2274223, a non-synonymous SNP located in PLCE1, was associated with cardia but not non-cardia gastric cancer.10 Together, these data indicate that five chromosomal regions, 1q22, 3q13.31, 5p13.1, 6p21.1 and 8q24.3, have strong evidence of harbouring one or more susceptibility alleles of non-cardia gastric cancer. Based on the experience of other cancer sites, additional loci will likely be found by interrogation of increasingly larger studies.
Materials and methods
Primary GWAS scan data
For the National Cancer Institute (NCI) GWAS, subjects were drawn from four prospective cohort studies and one large case–control study as reported in Abnet et al.10 In addition, all subjects used in replication in the original paper were subsequently genotyped using the Illumina 660W-Quad microarray; this included scanning of 725 additional gastric cancer cases and 608 additional controls. The current analysis included all 1025 non-cardia gastric cancer cases and 2697 controls from the NCI Upper Gastrointestinal Cancer GWAS.
For the Nanjing and Beijing GWAS, individuals were derived from separate case–control studies conducted in Nanjing (565 cases and 1162 controls) and Beijing (468 cases and 1123 controls), as previously reported in Shi et al,11 where individuals were genotyped using the Affymetrix Genome-Wide Human SNP Array (V.6.0).
Replication samples
The first-stage replication including 1145 cases and 2253 controls were derived from Jiangsu Province. The second-stage replication included 2419 cases and 2384 controls, which were derived from Beijing, Hubei and Shandong province.
Gastric cancer cases tested for expression of the lncRNA associated with the GWAS SNPs came from our UGI Cancer Genetic Studies (URL: http://dceg.cancer.gov/about/staff-directory/biographies/O-Z/taylor-philip). Genotypes for all these cases are known because they were also all participants in our previous GWAS.10
All study individuals provided informed consent and both the institutional review boards of NCI and Nanjing Medical University approved all procedures and all experiments, which were conducted in accordance with the approved guidelines.
Genotype imputation
In addition to the quality control procedures performed in the previous primary publications for previous GWAS, SNPs with call rate of <95%, p value for Hardy–Weinberg Equilibrium (HWE) in controls ≤1.0×10−6 or minor allele frequency (MAF) of <1% in controls were further removed before imputation. Imputation was conducted separately for the NCI (Illumina 660W) and the Nanjing+Beijing (Affymetrix 6.0) scan data taking all populations in the 1000 Genomes Project Phase I (V.3) as the reference set and using IMPUTE2 software (V.2.2.2), which automatically finds haplotypes from the best matching population from the entire reference set to do the imputation. First, genomic coordinates for National Center for Biotechnology Information (NCBI) human genome Build 36 were converted to those of NCBI human genome Build 37 using the UCSC liftOver tool. The few loci for which coordinates could not be converted were also excluded from imputation. Second, the strand of the inference data was aligned with the 1000 Genomes Project data by simple allele state comparison or allele frequency matching for adenine/thymine (A/T) and guanine/cytosine (G/C) SNPs. We implemented a 4 Mb sliding window to impute across the genome, resulting in 744 windows. A pre-phasing strategy with SHAPEIT software (V.2) was adopted to improve the imputation performance. The phased haplotypes from SHAPEIT were fed directly into IMPUTE2.
SNP selection and replication genotyping
The meta-analysis included 6 223 896 SNPs based on the intersection of the three imputed datasets. Individual SNPs for the stage 2A replication were selected based on the following criteria: (1) SNPs with INFO score of ≥0.5; (2) MAF in control set ≥0.01; (3) p value for HWE in control set >1.0×10−4 in each set; (4) Phet >1.0×10−4 and I2 <75% in meta-analysis; (5) linkage disequilibrium (LD) pruning: included only one SNP with the lowest p value when the pair-wise r2 ≥0.3 within a distance of 200 kb; (6) exclusion of previously identified loci associated with risk for non-cardia gastric cancer. After applying the above criteria, we then picked the top 48 SNPs (Pmeta≤2.58×10−5). For 1q22 and 8q24, there were two SNPs (rs80142782; rs76845414) retained in our LD filtered list, so we included back two more SNPs (rs4072037; rs2294008) previously reported for each of these regions in order to search for potential secondary signals for these two known loci to derive an initial list of 50 SNPs. Subsequently, 13 SNPs failed either Sequenom assay design or genotyping studies. As a result, a total of 37 SNPs (see online supplementary table S2) were successfully genotyped using iPLEX Sequenom MassARRAY platform (Sequenom, California, USA) in stage 2A replication (1145 cases and 2253 controls).
Five SNPs with p <0.05 in stage 2A without significant heterogeneity (Phet>1.0×10−4 and I2<75%) were advanced for TaqMan assays (Applied Biosystems) in stage 2B replication (2419 cases and 2384 controls) (see online supplementary table S3). Further information on primers and probes are available on request. For quality control purposes: (1) case and control samples were mixed on each plate; (2) genotyping was performed blind to case/control status and (3) two water controls were used in each plate as blank controls.
Quantitative reverse transcription-PCR
Total RNA was extracted from each patient's matched frozen tumour and normal surgical resection tissues using All Prep DNA/RNA/Protein kit (QIAGEN) in accordance with the manufacturer's instructions. RNA quality and quantity were determined using the RNA Nano Chip/Agilent 2100 Bioanalyzer (Agilent Technologies). Reverse transcription of RNA was done by adding 0.2–2 µg total RNA, 1 µL of oligo(dT)12–18 (500 µg/mL), 1 µL (200 units) of SuperScript II reverse transcriptase, 1 µL (2 units) of E-coli RNase and 1 µL of 10 mmol/L deoxynucleotide triphosphate (Invitrogen) in total volume of 20 µl. All real-time PCRs (RT-PCRs) were done using an ABI 7300 Sequence Detection System. Primer and probe for the target gene and the internal control gene (GAPDH, glyceraldehyde-phosphate dehydrogenase) were designed and ordered from ABI (Assay ID: AJBJXUX; Part Number: 4441114). A singleplex reaction mix was prepared according to the manufacturer's protocol of ‘Assays-on-Demand Gene Expression Products’, including 10 µL Taqman Universal PCR Master Mix, No AmpErase UNG (2X), 1 µL of 20X Assays on-Demand Gene Expression Assay Mix (all Gene Expression assays have a 6-carboxyfluorescein (FAM) reporter dye at the 5′ end of the TaqMan minor groove binder (MGB) probe and a non-fluorescent quencher at the 3′ end of the probe) and 9 µL of cDNA (1000 ng) diluted in RNase-free water to a total volume of 20 µL. Each sample for the gene was run in triplicate and the expression level was averaged over all runs. The thermal cycling conditions included an initial denaturation step at 95°C for 10 min, 40 cycles at 95°C for 15 s and 60°C for 1 min.
Statistical analysis
Association testing was performed using SNPTEST software (V.2.2), with adjustment for age, sex and study variables for NCI. Two eigenvectors (ev4 and ev8) were significantly associated with case status (p<0.05) in the baseline model (not including SNP effects) which was adjusted for age, sex, study and all top 10 eigenvectors, and therefore these two significant eigenvectors were also included to adjust for population stratification in final association models. For Nanjing and Beijing, age, sex, smoking and alcohol consumption were adjusted in baseline models. Three eigenvectors (ev1, ev4 and ev9) for Nanjing and five eigenvectors (ev1, ev3, ev7, ev9 and ev10) for Beijing were adjusted for population stratification separately, which were also significantly associated with case status (p<0.05). In each replication study, we adjusted for gender, age, smoking and alcohol consumption only.
For the meta-analysis we used the meta-module implemented in genotype library utilities (GLU) (see URLs). Strand flipping was handled by comparing alleles either with direct matching or with reverse complement matching. For A/T or G/C SNPs, strand matching was based on allele frequency checking. The fixed-effects inverse variance method was used to combine the β estimates and SEs from each GWAS scan as well as the replication stages. The p value for heterogeneity was calculated using Cochran’s Q, which is distributed as a χ2 statistic with (n−1) degrees of freedom, where n is the number of sets included in the meta-analysis. I2 was calculated as 100%×(Q–(n–1))/Q. Data analysis and management was performed with GLU or PLINK (see URLs).
PLINK was also used for the conditional haplotype analysis. Wilcoxon signed-rank test (R package) was applied to the tumour/normal paired quantitative RT-PCR data to assess the RNA expression level.
In silico bioinformatics analysis
We used GTEx (see URLs) for expression quantitative trait loci (eQTL) information for associated SNPs (see online supplementary table S5). We also searched HaploReg (V.3)19 to explore potential functional annotations within Encyclopedia of DNA Elements (ENCODE) data for the genomic regions surrounding our lead SNPs (see online supplementary table S6).
Results
To discover additional susceptibility alleles for non-cardia gastric cancer in the Han Chinese population, we conducted a combined analysis of two previously published GWAS10 11 after imputing the genetic data with the 1000 Genome Project data Phase I release (V.3).20 The combined dataset included a total of 6 223 896 SNPs for a fixed-effects meta-analysis of 2031 cases and 4970 cancer-free controls (see online supplementary table S1). Quantile-quantile and Manhattan plots based on stage 1 meta-analysis p values are shown in online supplementary figures S1 and S2, respectively. We further followed up 37 promising loci (see Methods) and genotyped them in an independent set of 1145 cases and 2253 controls in stage 2A (see online supplementary table S2). Finally, for stage 2B, we advanced five loci that were nominally significant in stage2A to a second independent set of 2419 cases and 2384 controls (see online supplementary table S3).
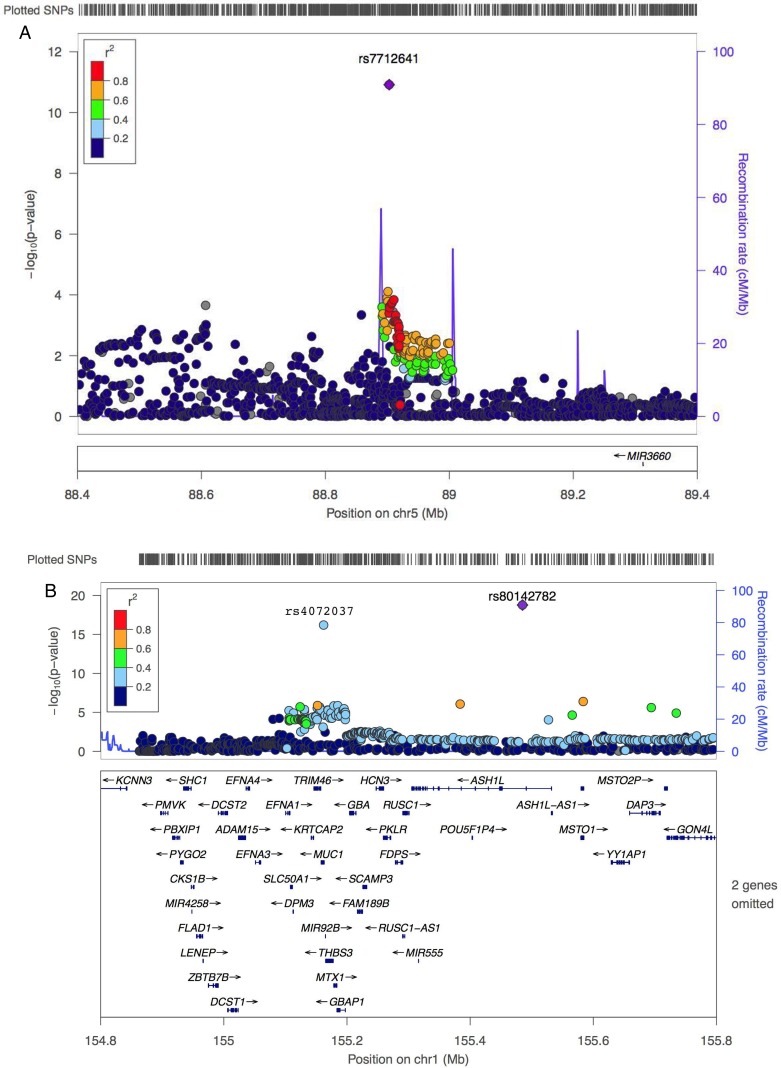
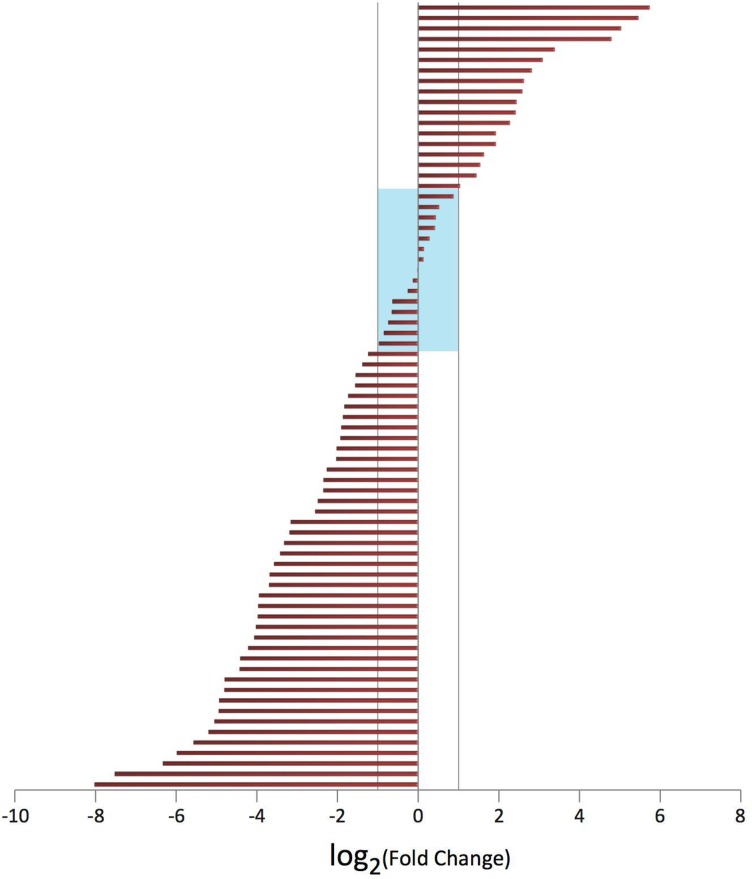
Based on the overall meta-analysis including two discovery GWAS scans and two replication studies, we identified two novel risk loci for non-cardia gastric cancer, the first one is rs7712641 at 5q14.3 (per-allele OR=0.84, 95% CI 0.80 to 0.88; p=1.21×10−11). No heterogeneity was observed across the two GWAS scans and two replication studies (Phet=0.56) (table 1). There are no protein-coding genes within the 1 Mb of the associated SNP (chr5: 88 346 298–89 459 630; hg19) (figure 1A). However, rs7712641 is located in the intron of lnc-POLR3G-4 (XLOC_004464), a long non-coding RNA (lncRNA) which is poorly characterised. To explore the possible effect of the SNP marker on the lncRNA, lnc-POLR3G-4, we extracted total RNA from 75 matched gastric non-cardia adenocarcinoma and adjacent normal tissue pairs and performed a quantitative RT-PCR analysis to measure its expression abundance. We found that expression differed between tumour and normal tissues (Pwilcoxon signed-rank=7.2×10−4); with the majority of pairs (50 of 75) showing lower expression in tumour compared with normal tissue (figure 2). These data provided preliminary evidence that this lncRNA could function in a manner resembling a tumour suppressor gene. However, the association between rs7712641and expression of lnc-POLR3G-4 in normal tissue was negative (p=0.99), which does not support the notion of a functional role for this SNP. More functional studies are warranted to clarify the complicated phenomena.
Table 1.
Four single-nucleotide polymorphisms (SNPs) associated with non-cardia gastric cancer risk above genome-wide significance level
| Cytoband (BP) | NCBI dbSNP identifier, build 138 (referent, effect allele) | Nearest gene | Group | Effect allele frequency (case, control) | OR (95% CI) | p Value | Phet | I2 |
|---|---|---|---|---|---|---|---|---|
| 1q22 | rs80142782 | ASH1L | NCI | 0.06, 0.07 | 0.68 (0.55 to 0.86) | 7.44E-04 | ||
| 155485027 | (T,C) | Beijing | 0.03, 0.07 | 0.47 (0.27 to 0.80) | 5.39E-03 | |||
| Nanjing | 0.04, 0.08 | 0.56 (0.38 to 0.82) | 2.55E-03 | |||||
| Stage 2a | 0.05, 0.09 | 0.61 (0.50 to 0.73) | 1.46E-07 | |||||
| Stage 2b | 0.05, 0.07 | 0.64 (0.54 to 0.75) | 1.16E-07 | |||||
| Combined | 0.62 (0.56 to 0.69) | 1.71E-19 | 0.66 | 0.00 | ||||
| 1q22 | rs4072037 | MUC1 | NCI | 0.14, 0.16 | 0.76 (0.65 to 0.87) | 1.31E-04 | ||
| 155162067 | (T,C) | Beijing | 0.10, 0.14 | 0.87 (0.61 to 1.22) | 4.17E-01 | |||
| Nanjing | 0.13, 0.16 | 0.84 (0.66 to 1.08) | 1.70E-01 | |||||
| Stage 2a | 0.13, 0.19 | 0.68 (0.59 to 0.77) | 9.63E-09 | |||||
| Stage 2b | 0.12, 0.16 | 0.75 (0.67 to 0.84) | 1.22E-06 | |||||
| Combined | 0.74 (0.69 to 0.79) | 6.28E-17 | 0.44 | 0.00 | ||||
| 5q14.3 | rs7712641 | NA | NCI | 0.44, 0.46 | 0.88 (0.79 to 0.97) | 1.14E-02 | ||
| 88902964 | (C,T) | Beijing | 0.41, 0.47 | 0.75 (0.60 to 0.93) | 9.41E-03 | |||
| Nanjing | 0.45, 0.48 | 0.79 (0.67 to 0.94) | 7.54E-03 | |||||
| Stage 2a | 0.43, 0.48 | 0.81 (0.73 to 0.90) | 4.91E-05 | |||||
| Stage 2b | 0.42, 0.46 | 0.86 (0.79 to 0.94) | 4.94E-04 | |||||
| Combined | 0.84 (0.80 to 0.88) | 1.21E-11 | 0.56 | 0.00 | ||||
| 8q24.3 | rs2294008 | PSCA | NCI | 0.31, 0.29 | 1.16 (1.03 to 1.31) | 1.18E-02 | ||
| 143761931 | (C,T) | Beijing | 0.39, 0.34 | 1.29 (1.01 to 1.64) | 4.30E-02 | |||
| Nanjing | 0.36, 0.32 | 1.22 (1.00 to 1.48) | 4.78E-02 | |||||
| Stage 2a | 0.29, 0.25 | 1.16 (1.04 to 1.30) | 9.04E-03 | |||||
| Stage 2b | 0.31, 0.26 | 1.26 (1.15 to 1.39) | 1.24E-06 | |||||
| Combined | 1.20 (1.15 to 1.28) | 5.95E-11 | 0.76 | 0.00 |
Bold indicates combined results using meta-analysis based method.
BP, base pair; C, cytosine; dbSNP, the Single Nucleotide Polymorphism database; NA, not available; NCI, National Cancer Institute; PSCA, prostate stem cell antigen; T, thymine.
Figure 1.
Regional plots of association results (A) 5q14.3:88 400 000–89 400 000; (B) 1q22:152 940 379–153 897 376. Association results are based on a trend test in which –log10 p values (y axis, left) were plotted against the chromosomal positions based on hg19 (x axis). All p values were based on the discovery meta-analysis stage 1 data except for three index single-nucleotide polymorphisms (SNPs) (rs7712641 in 1a; rs80142782 and rs4072037 in (B) which were based on all data (stages 1, 2A and 2B). The line graph shows recombination rate (y axis, right). The linkage disequilibrium (LD) (r2) is colour-coded (figure legend) based on estimates from the 1000 Genomes Mar 2012 release Asian (ASN) population. The plots were generated using LocusZoom online version (URLs).
Figure 2.
Differential expression of lnc-POLR3G-4 between non-cardia gastric tumour and normal tissue based on a quantitative real-time PCR analysis. X axis is the log2 of tumour:normal fold change. All 75 pairs were sorted in increasing order of fold change and plotted along the Y axis. Two-thirds of pairs show lower expression in tumour compared with normal, while expression is higher in normal compared with tumour in one-third of pairs. Highlighted in the middle are 15 pairs with fold changes between 0.5 and 2 (or −1 to 1 in log2 scale). Based on all 75 pairs, the Wilcoxon signed-rank test was P=7.2×10−4 and the median tumour:normal fold change is 0.3.
As anticipated, the current study, which included samples from these previous GWAS reports,10 11 also replicated the association with rs4072037 (table 1; per-allele OR=0.74; 95% CI 0.69 to 0.79; p=6.28×10−17) at 1q22. However, we also identified a second strong signal in this region and by doing so established an independent, new genome-wide significant SNP rs80142782 (table 1 and figure 1B; per-allele OR=0.62; 95% CI 0.56 to 0.69; p=1.71×10−19). Based on the evidence at hand, it seems that rs80142782 is likely an independent primary signal (rs4072037 as a secondary signal) at this locus. This evidence includes the following: (1) both SNPs are about 323 kb apart and have moderately low (r2=0.3) pair-wise LD in 1000 Genome s Project data for Asians; (2) rs80142782 conditioned on rs4072037 remained genome-wide significant (Pconditional=3.47×10−8, see online supplementary table S5), although rs4072037 conditioned on rs80142782 did not (Pconditional=2.95×10−6, see online supplementary table S4); (3) we used the haplotype inference method implemented in the plink haplotype test ‘—chap’ option. For rs4072037 and rs80142782, there are three inferred haplotypes with frequencies greater than 1% from all four possible ones. The two models compared in the conditional haplotype likelihood ratio test are the null model: {CC} {CT, TT} and the alternative model: {CC} {CT} {TT}, where each {set} allows a unique effect. The conditional haplotype analysis demonstrated that the effect size of haplotype CT differed from that of TT among the possible haplotypes formed by these two SNPs (Plikelihood ratio=9.11×10−9) and(4) finally, it is notable that rs80142782 is Asian-specific with a MAF of 0.05 in Asians but monomorphic in both European and African populations. Thus, rs80142782 appears to be a better association signal at 1q22 in Asian populations. Further validation in additional studies with even larger sample sizes will be required to determine if these two SNPs are truly independent signals tagging two different causal variants.
The previously reported SNP marker, rs4072037 at 1q22, is a synonymous SNP in MUC1 which is a member of the mucin family that collectively forms the protective mucous barrier on epithelial surfaces. Its expression was highest in stomach among all normal tissues examined by the Genotype-Tissue Expression (GTEx) project (see URLs). Evidence suggests that rs4072037 is the functional variant for this locus because it alters transcriptional regulation and determines splice variants in MUC1.15 Although MUC1 is a putative candidate gene for gastric cancer risk, it is also interesting to note that GTEx data show that rs4072037 is an eQTL for several neighbouring genes (including THBS3, GBAP1, GBA and RP11-263K19.4) in other tissues (see online supplementary table S5). The rs80142782 may act on the ASH1L gene based on its close proximity. ASH1L encodes a member of the trithorax group of transcriptional activators and functions as an epigenetic regulator by histone methylation (H3K4 methyltransferase) and is frequently altered in lung cancer tumours and cell lines,21 22 oesophageal squamous cell carcinoma tumour tissue23 and colorectal cancer cell lines.24 It was also implicated in inflammatory autoimmune disease.25
Discussion
Our study identified a new risk locus at 5q14.3 marked by rs7712641 which lies in the intron of a lncRNA with little known prior functional characterisation. Other lncRNAs implicated in cancers include PCA3 and PCGEM1 in prostate tumour,26 and MALAT1 in tumours of the colorectum, liver, pancreas, lung, breast and prostate.27 28 It is remarkable that a recent comprehensive transcriptome analysis nominated a total of 7942 lineage-associated or cancer-associated lncRNA genes,29 for which further functional investigations are warranted. Overexpression or knockdown of this lncRNA may be informative in identifying target genes through analysis of differential gene expression profiles in non-cardia gastric tumour cell lines.
Our analysis also revealed an apparently stronger associated SNP (rs80142782) at 1q22 than the previously identified rs4072037. Our data indicate that the association with rs80142782 is independent of rs4072037. Both the new locus at 5q14.3 marked by rs7712641 and the new independent signal at 1q22 marked by rs80142782 could contribute to epigenome regulation. Haploreg data show that both of these SNPs (or SNPs in high LD with them) locate to sites of multiple regulatory elements, including promoter histone marks, enhancer histone marks and DNAse hypersensitivity (see online supplementary table S6). Further functional validation studies are warranted to understand the contribution of these susceptibility alleles to gastric carcinogenesis.
Online supplementary table S7 shows the results from our meta-analysis of two GWAS scans for previously reported variants from the literature. Notably we confirmed a prior independent GWAS report9 of an association between multiple SNPs in PSCA at 8q24.3 and risk of non-cardia gastric cancer. We also confirmed the association for rs13361707 in PRKAA1,11 but there was no additional evidence to support an association for rs9841504 in ZBTB20 with non-cardia gastric cancer in NCI data (p=0.27). Recently, Mocellin et al30 collected published data and nominated a list of 11 SNPs at eight loci with a high level of cumulative evidence for susceptibility to gastric cancer. Among these 11 SNPs, 4 (at 2q33.1, 3p24.1, 6p21.33 and 11q13.2, respectively) were identified beyond those previously established in GWAS findings, but none of these SNPs was associated (p<0.05) with gastric non-cardia cancer risk in the current meta-analysis.
In summary, by combining two pre-existing GWAS scans of non-cardia gastric cancer to increase the sample size for the discovery stage and adding over 8000 individuals for further replication, we identified two novel loci. In the future, additional studies are warranted with larger sample size and/or with a design that considers heterogeneity of the gastric cancer where, for example, the Cancer Genome Atlas (TCGA) recently reported four molecular subtypes for gastric cancer based on multi-omics profiling analyses.31
URLs
GTEx, http://www.gtexportal.org/;
GTEx MUC1 expression, http://www.gtexportal.org/home/gene/MUC1
IMPUTE2 software (V.2.2.2), http://mathgen.stats.ox.ac.uk/impute/impute_v2.html;
LocusZoom, http://csg.sph.umich.edu/locuszoom/;
SHAPEIT software (V.2), https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html;
SNPTEST software (V.2.2), https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html
UCSC liftOver tool, http://hgdownload.cse.ucsc.edu/downloads.html
Footnotes
Contributors: HBS, PRT, GJ, AMG, YS, SJC, ZW, JCD, NH, XM, CCA, MY, NDF and JFC organised and designed the study; ZW, JCD, HBS, PRT, AMG and SJC wrote the first draft of the manuscript; ZW and JCD contributed to the design and execution of statistical analysis; HBS, PRT, GJ, AMG, YS, SJC, ZW, JCD, NH, XM, CCA, MY, NDF and JFC contributed to the writing of the manuscript; ZW, JCD, HBS, GJ, ZH and LB conducted and supervised the genotyping of samples; HS, LW and NH conducted the quantitative real-time PCR experiments; MY, XZ, CCC, CR, SMD, MW, TD, JBD, YTG, RZ, CG, WP, WPK, ND, LML, CY, YLQ, YJ, XOS, JPC, CW, HM, ZZ, CW, YBX, ZH, JMY, LX, WZ and DL contributed to the conduct of the epidemiological studies or contributed samples to the GWAS or follow-up genotyping.
Funding: This work was supported by the Intramural Research Program of the National Cancer Institute (NCI), the National Institutes of Health, the Division of Cancer Epidemiology and Genetics and the Center for Cancer Research. This work was also supported in part by the National Basic Research Program (973) (2013CB910304), Program of National Natural Science Foundation of China (81230067), National Natural Science Foundation of China (81573228, 81521004. 81422042, 81373090); Science Foundation for Distinguished Young Scholars in Jiangsu (BK20130042); Jiangsu Natural Science Foundation (BK2012443, BK2012841); Jiangsu Province Clinical Science and Technology Projects (Clinical Research Center, BL2012008); National Program for Support of Top-notch Young Professionals, National Natural Science Foundation of China (81222038); Key Grant of Natural Science Foundation of Jiangsu Higher Education Institutions (15KJA330002); National High-Tech Research and Development Program of China (2015AA020950) and Priority Academic Program for the Development of Jiangsu Higher Education Institutions (Public Health and Preventive Medicine).
Competing interests: None declared.
Patient consent: Obtained.
Ethics approval: Both the institutional review boards of NCI and Nanjing Medical University approved all procedures and all experiments, which were conducted in accordance with the approved guidelines.
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Ferlay Jea. GLOBOCAN 2012 v1.0, Cancer incidence and Mortality Worldwide: IARC CancerBase No. 11. International Agency for Research on Cancer, 2013.
- 2.McLean MH, El-Omar EM. Genetics of gastric cancer. Nat Rev Gastroenterol Hepatol 2014;11:664–74. 10.1038/nrgastro.2014.143 [DOI] [PubMed] [Google Scholar]
- 3.Karimi P, Islami F, Anandasabapathy S, et al. . Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev 2014;23:700–13. 10.1158/1055-9965.EPI-13-1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kamangar F, Qiao YL, Blaser MJ, et al. . Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. Br J Cancer 2007;96:172–6. 10.1038/sj.bjc.6603517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawsey SM, Mark SD, Taylor PR, et al. . Gastric cancer and H pylori. Gut 2002;51:457–8. 10.1136/gut.51.3.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Helicobacter and Cancer Collaborative Group. Gastric cancer and Helicobacter pylori: a combined analysis of 12 case control studies nested within prospective cohorts. Gut 2001;49:347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mark SD, Qiao YL, Dawsey SM, et al. . Prospective study of serum selenium levels and incident esophageal and gastric cancers. J Natl Cancer Inst 2000;92:1753–63. 10.1093/jnci/92.21.1753 [DOI] [PubMed] [Google Scholar]
- 8.Taylor PR, Qiao YL, Abnet CC, et al. . Prospective study of serum vitamin E levels and esophageal and gastric cancers. J Natl Cancer Inst 2003;95:1414–16. 10.1093/jnci/djg044 [DOI] [PubMed] [Google Scholar]
- 9.Sakamoto H, Yoshimura K, Saeki N, et al. . Genetic variation in PSCA is associated with susceptibility to diffuse-type gastric cancer. Nat Genet 2008;40:730–40. 10.1038/ng.152 [DOI] [PubMed] [Google Scholar]
- 10.Abnet CC, Freedman ND, Hu N, et al. . A shared susceptibility locus in PLCE1 at 10q23 for gastric adenocarcinoma and esophageal squamous cell carcinoma. Nat Genet 2010;42:764–7. 10.1038/ng.649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Hu Z, Wu C, et al. . A genome-wide association study identifies new susceptibility loci for non-cardia gastric cancer at 3q13.31 and 5p13.1. Nat Genet 2011;43:1215–18. 10.1038/ng.978 [DOI] [PubMed] [Google Scholar]
- 12.Hu N, Wang Z, Song X, et al. . Genome-wide association study of gastric adenocarcinoma in Asia: a comparison of associations between cardia and non-cardia tumours. Gut Published Online First: 30 June 2015. 10.1136/gutjnl-2015-309340 10.1136/gutjnl-2015-309340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Yuan Y, Sun LP, et al. . Risk of gastric cancer is associated with the MUC1 568 A/G polymorphism. Int J Oncol 2009;35:1313–20. [DOI] [PubMed] [Google Scholar]
- 14.Jia Y, Persson C, Hou L, et al. . A comprehensive analysis of common genetic variation in MUC1, MUC5AC, MUC6 genes and risk of stomach cancer. Cancer Causes Control 2010;21:313–21. 10.1007/s10552-009-9463-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saeki N, Saito A, Choi IJ, et al. . A functional single nucleotide polymorphism in mucin 1, at chromosome 1q22, determines susceptibility to diffuse-type gastric cancer. Gastroenterology 2011;140:892–902. 10.1053/j.gastro.2010.10.058 [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Jin G, Li H, et al. . Genetic variants at 1q22 and 10q23 reproducibly associated with gastric cancer susceptibility in a Chinese population. Carcinogenesis 2011;32:848–52. 10.1093/carcin/bgr051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palmer AJ, Lochhead P, Hold GL, et al. . Genetic variation in C20orf54, PLCE1 and MUC1 and the risk of upper gastrointestinal cancers in Caucasian populations. Eur J Cancer Prev 2012;21:541–4. 10.1097/CEJ.0b013e3283529b79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song HR, Kim HN, Kweon SS, et al. . Common genetic variants at 1q22 and 10q23 and gastric cancer susceptibility in a Korean population. Tumour Biol 2014;35:3133–7. 10.1007/s13277-013-1409-4 [DOI] [PubMed] [Google Scholar]
- 19.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 2012;40:D930–4. 10.1093/nar/gkr917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genomes Project C, Abecasis GR, Auton A, et al. . An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osada H, Tomida S, Yatabe Y, et al. . Roles of achaete-scute homologue 1 in DKK1 and E-cadherin repression and neuroendocrine differentiation in lung cancer. Cancer Res 2008;68:1647–55. 10.1158/0008-5472.CAN-07-5039 [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Lee W, Jiang Z, et al. . Genome and transcriptome sequencing of lung cancers reveal diverse mutational and splicing events. Genome Res 2012;22:2315–27. 10.1101/gr.140988.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Song Y, Li L, Ou Y, et al. . Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014;509:91–5. 10.1038/nature13176 [DOI] [PubMed] [Google Scholar]
- 24.Mouradov D, Sloggett C, Jorissen RN, et al. . Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 2014;74:3238–47. 10.1158/0008-5472.CAN-14-0013 [DOI] [PubMed] [Google Scholar]
- 25.Xia M, Liu J, Wu X, et al. . Histone methyltransferase Ash1l suppresses interleukin-6 production and inflammatory autoimmune diseases by inducing the ubiquitin-editing enzyme A20. Immunity 2013;39:470–81. 10.1016/j.immuni.2013.08.016 [DOI] [PubMed] [Google Scholar]
- 26.Fu X, Ravindranath L, Tran N, et al. . Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol 2006;25:135–41. 10.1089/dna.2006.25.135 [DOI] [PubMed] [Google Scholar]
- 27.Lin R, Maeda S, Liu C, et al. . A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 2007;26:851–8. 10.1038/sj.onc.1209846 [DOI] [PubMed] [Google Scholar]
- 28.Xu C, Yang M, Tian J, et al. . MALAT-1: a long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int J Oncol 2011;39:169–75. 10.3892/ijo.2011.1007 [DOI] [PubMed] [Google Scholar]
- 29.Iyer MK, Niknafs YS, Malik R, et al. . The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 2015;47:199–208. 10.1038/ng.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mocellin S, Verdi D, Pooley KA, et al. . Genetic variation and gastric cancer risk: a field synopsis and meta-analysis. Gut 2015;64:1209–19. 10.1136/gutjnl-2015-309168 [DOI] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]