Abstract
Enteroaggregative Escherichia coli (EAEC) causes acute or persistent diarrhea. The aggR gene is widely used as a marker for typical EAEC. The heterogeneity of EAEC is well known; however, there are few reports on the phylogenetic relationships of EAEC. Recently, CTX-M extended-spectrum β-lactamase (ESBL)-producing EAEC strains have been reported worldwide. To characterize EAEC strains in Japan, we investigated the population structure of EAEC. A total of 167 aggR-positive strains isolated from stool specimens from diarrheal patients in Kagoshima (139 strains) and Osaka (28 strains), Japan, between 1992 and 2010 were examined for the prevalence of EAEC virulence markers, the blaCTX-M gene, and the capacity to form biofilms. Multilocus sequence typing was also conducted. EAEC strains were widely distributed across four major E. coli phylogroups. Strains of O111:H21/clonal group 40 (CG40) (30 strains), O126:H27/CG200 (13 strains), and O86a:H27/CG3570 (11 strains) in phylogroup B1 are the historical EAEC clones in Japan, and they exhibited strong biofilm formation. Twenty-nine strains of EAEC O25:H4/CG131 were identified in phylogroup B2, 79% of which produced CTX-M-14. This clone has emerged since 2003. The clone harbored plasmid-encoded EAEC virulence genes but not chromosomal virulence genes and had lower biofilm-forming capacity than historical EAEC strains. This clone most likely emerged from a pandemic uropathogenic O25:H4/sequence type 131 clone by acquiring an EAEC virulence plasmid from canonical EAEC. Surveillance of the horizontal transfer of both virulence and ESBL genes among E. coli strains is important for preventing a worldwide increase in antimicrobial drug resistance.
INTRODUCTION
Enteroaggregative Escherichia coli (EAEC) causes acute or persistent diarrhea in children and adults, in both developing (1) and industrialized (2, 3) countries. Several outbreaks of gastroenteritis linked to EAEC have been reported (4, 5). EAEC is also the second most common cause of travelers' diarrhea (6). EAEC is defined by its characteristic aggregative adherence (AA) to HEp-2 cells in culture (7). Although detection of the AA phenotype in the HEp-2 cell adherence assay is still the gold standard for EAEC identification, the transcriptional regulator AggR, which controls the expression of multiple EAEC virulence genes, is also used as a typical EAEC marker for PCR detection (8).
The pathogenesis of EAEC infection involves the adherence of the bacterium to the intestinal mucosa, followed by the secretion of one or more enterotoxins and/or cytotoxins that induce mucosal inflammation (9). EAEC adherence to the mucosa is characterized by the formation of a thick biofilm (10), which may promote persistent infection. The majority of EAEC strains carry a large (∼100-kb) plasmid, which encodes most putative EAEC virulence factors, such as AggR, aggregative adherence fimbriae (AAF), the dispersin protein Aap (11), its transporter Aat (12), the plasmid-encoded toxin Pet (13), enteroaggregative heat-stable toxin 1 (EAST-1) (14), and the Shigella extracellular protease Sep (15). AAF is a major EAEC adhesin, and at least five variants have been identified, as follows: AAF/I, AAF/II, AAF/III, AAF/IV, and AAF/V (16). In addition, chromosomally encoded factors such as the mucinase Pic (17), Shigella enterotoxin 1 (ShET1) (17), proteins secreted through the type VI secretion system (18), and the secreted autotransporter toxin Sat (19) are also considered potential virulence factors. Furthermore, EAEC plasmid-borne virulence genes have been identified in certain uropathogenic E. coli (UPEC) strains isolated from patients with urinary tract infections (UTIs) (20).
Worldwide dissemination of plasmid-borne extended-spectrum β-lactamases (ESBLs) among E. coli isolates is a public concern (21). E. coli strains that produce the CTX-M type of ESBLs are increasingly being isolated from adults and children with UTIs and bacteremia; however, CTX-M ESBLs have rarely been found among diarrheagenic E. coli (DEC) strains. We previously reported that 36.8% of the aggR gene-positive EAEC strains isolated from diarrheal children in Kagoshima, Japan, were cefotaxime resistant (22), suggesting ESBL production. Subsequently, some CTX-M-producing EAEC strains were reported worldwide (23–26), including an EAEC-Shiga toxin-producing E. coli (STEC) hybrid clone of O104:H4 that caused a large outbreak in Germany in 2011 (27).
Although the genetic heterogeneity of EAEC is well known, the entire genomic sequences are available for only two strains (28, 29). In addition, although there have been two reports containing phylogenetic analyses of clinical EAEC isolates from the United States and Nigeria (30, 31), a phylogenetic analysis of EAEC strains in Japan has not yet been conducted. The genetic background of CTX-M-producing EAEC also has not been elucidated. Therefore, we conducted a large-scale phylogenetic analysis of EAEC clinical isolates from two regions (Kagoshima and Osaka) in Japan, coupled with an analysis of the prevalence of EAEC virulence markers and the CTX-M gene among the isolates.
MATERIALS AND METHODS
Subjects and strains.
A total of 167 typical (aggR-positive) EAEC strains were used; they were isolated from stool specimens from diarrheal patients in Kagoshima (138 strains) or Osaka (29 strains), Japan. Detailed information about the strains examined is presented in Table S1 in the supplemental material. The Kagoshima strains were identified by aggR-directed PCR from a collection of E. coli strains isolated from diarrheal children who visited clinics of the Kagoshima Prefecture between 1998 and 2010. The Osaka strains were selected from a collection of aggR-positive EAEC strains isolated in the Osaka Prefectural Institute of Public Health between 1992 and 2010. The selected Osaka strains included five EAEC strains isolated from different overseas travelers. The EAEC prototype strain 042 was kindly provided by J. P. Nataro (University of Virginia) and was used as a control strain.
Culture conditions and DNA extraction.
Bacteria were routinely grown either in Luria-Bertani (LB) broth (MP Biomedicals), with shaking, or on LB agar plates at 37°C. DNA templates for PCR amplification were prepared by resuspending fresh bacterial colonies in 100 μl of Tris-EDTA, heating the resuspended cells for 10 min at 100°C, and then centrifuging the mixture for 5 min at 10,000 × g.
Detection of virulence genes and the blaCTX-M gene.
The EAEC strains were evaluated by PCR for the possession of 14 EAEC-associated virulence genes, i.e., aggR (AggR), aap (Aap), aatA (AatA), aggA (AAF/I), aafA (AAF/II), agg3A (AAF/III), hdaA/agg4A (AAF/IV), agg5A (AAF/V), pet (PET), astA (EAST-1), sepA (SepA), sat (Sat), pic (Pic), and aaiC (AaiC) (15, 32, 33). The strains were also examined by PCR for the prevalence of five UPEC virulence factors (34), i.e., S fimbriae (sfa), P fimbriae (pap), afimbrial adhesin (afa), aerobactin (aer), and cytotoxic necrotizing factor 1 (cnf1). Primer sequences and PCR conditions are presented in Table S2 in the supplemental material. Amplification reactions were performed using a TProfessional Basic thermocycler (Biometra). The products were separated on 1.5% agarose gels, stained with ethidium bromide, and visualized with UV transillumination.
CTX-M-producing strains were screened using the blaCTX-M-directed PCR, as described elsewhere (35) (see Table S2 in the supplemental material). CTX-M subtypes were determined by direct sequencing of blaCTX-M PCR amplicons.
Confirmation of ESBL production.
ESBL production of blaCTX-M-positive strains was evaluated in an antibiotic susceptibility test using the ESBL MicroScan Neg MIC 3.31E confirmation panel (Siemens Healthcare Diagnostics, Tokyo, Japan), according to Clinical and Laboratory Standards Institute criteria (36).
Multilocus sequence typing and phylogenetic analysis.
Multilocus sequence typing (MLST) was performed using the nucleotide sequences of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA). Target genes were amplified and sequenced according to the protocol provided at the University of Warwick website (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/documents/primersColi_html), with a slight modification of the primer sequences (37). Using the concatenated nucleotide sequences of the seven genes and the maximum composite likelihood model, a neighbor-joining tree was constructed using MEGA5 software (38). Six EcoR collection strains (39) and 16 whole-genome-sequenced E. coli and Escherichia albertii strains (40) were included in a phylogenetic representation. Sequence types (STs) were determined from the MLST databases at the University of Warwick website.
Phylogenetic analysis of the aggR gene.
Using the nucleotide sequences obtained by direct sequencing of aggR PCR amplicons, a phylogenetic analysis was performed using MEGA5 software (38).
Serotyping.
Strains were O serogrouped in a slide agglutination test using 50 commercially available antisera (Denka Seiken, Tokyo, Japan). Strains that did not demonstrate agglutination with any antiserum were defined as untypeable (UT). For the ESBL-producing UT strains, we determined the O serogroups by O-genotyping PCR (41, 42). The H serogroup was determined by sequencing of the fliC gene (43) (see Table S2 in the supplemental material).
Quantitative biofilm assay.
The capacity to form biofilms was assessed using a quantitative biofilm assay in a polystyrene microtiter plate, as described previously (44). Briefly, the bacterial cells were grown overnight in LB broth at 37°C. Overnight cultures (5 μl for each strain) were inoculated into a 96-well flat-bottom polystyrene microtiter plate (Becton Dickinson, Franklin Lakes, NJ) containing 200 μl of Dulbecco's modified Eagle's medium (DMEM) with 0.45% glucose. The plate was then incubated at 37°C. After 18 h of incubation, each well was washed with distilled water, and biofilm formation was visualized by staining with 0.5% crystal violet for 5 min. Two hundred microliters of 95% ethanol was added, and the biofilm was quantified in an enzyme-linked immunosorbent assay plate reader (iMark microplate reader; Bio-Rad) at 570 nm. The biofilm index was defined as the average of the values for the optical density at 570 nm (OD570). EAEC 042 and E. coli HB101 were used as positive and negative control strains, respectively.
HEp-2 cell adherence assay.
The HEp-2 cell adherence assay was performed using the previously described method, with slight modifications (7). Briefly, after 16 h of incubation at 37°C, 20-μl aliquots of static E. coli cultures (2 × 106 bacteria) in LB broth were incubated for 3 h with nonconfluent HEp-2 cell monolayers (estimated coverage, 25 to 50%) in DMEM, on round coverslips in a 24-well microtiter plate. The medium was supplemented with 0.5% methyl-α-d-mannoside to prevent attachment by type 1 somatic fimbriae. The patterns of bacterial adherence to HEp-2 cells on glass coverslips were observed by light microscopy after Giemsa staining.
Statistical analysis.
Statistical analyses were performed using the Fisher exact probability test and the Mann-Whitney U test, in SPSS v. 17. Two-tailed P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
All nucleotide sequences obtained in this study have been deposited into the DDBJ/EMBL/GenBank database. The accession numbers are LC120396 to LC120563 (for the aggR genes) and LC119139 to LC120307 (for the 7 housekeeping genes [adk, fumC, gyrB, icd, mdh, purA, and recA] used for MLST).
RESULTS
Phylogenetic analysis of EAEC strains.
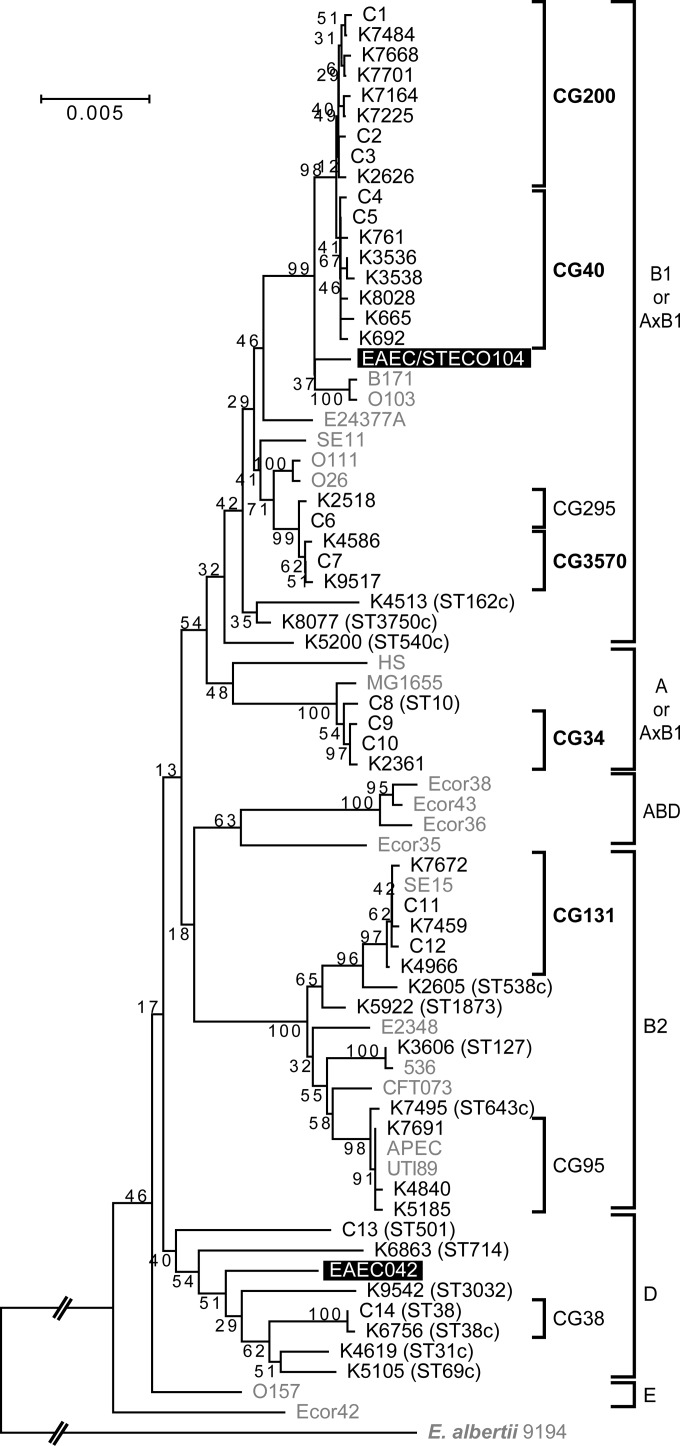
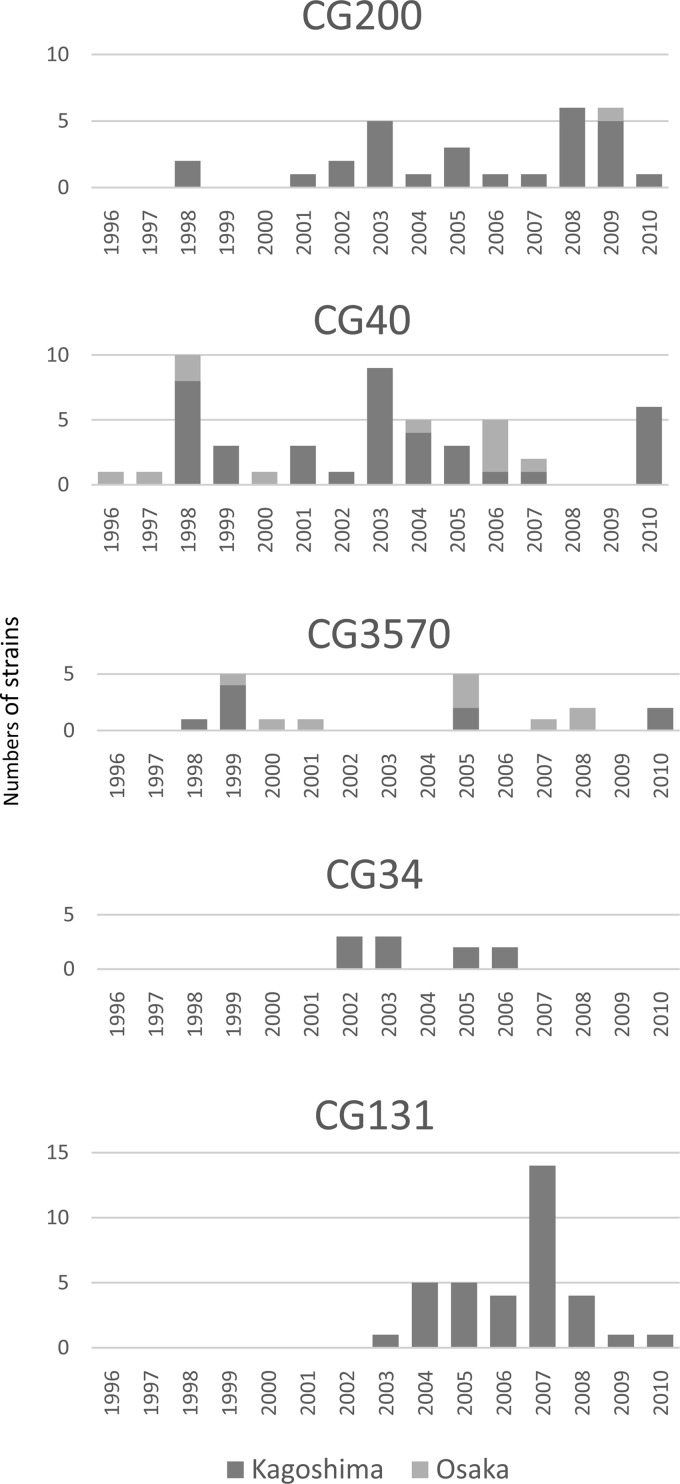
To determine the phylogeny of the 167 aggR-positive EAEC strains, we performed an MLST-based phylogenetic analysis. EAEC strains were distributed across all four classical phylogroups (A, B1, B2, and D) (Fig. 1; also see Table S3 in the supplemental material). By defining a group of strains showing the same sequence type (ST) as a cluster, we identified 14 clusters (C1 to C14) and 34 singletons, i.e., 48 STs in total. A clonal group (CG) was defined as a group of closely related STs, and groups were named in accordance with the dominant ST. The predominant CGs, which contained more than 10 strains, were CG40 (29.9%), CG131 (21.0%), CG200 (17.4%), CG3570 (10.8%), and CG34 (6.0%). CG200, CG40, and CG3570 belonged to phylogroup B1 and each group included both Kagoshima strains and Osaka strains, whereas CG34 and CG131 strains, belonging to phylogroups A and B2, respectively, were isolated only in Kagoshima. The annual changes in the numbers of the major CG strains are shown in Fig. 2. CG200, CG40, and CG3570 strains were found during the whole study period, whereas CG34 strains were isolated between 2002 and 2006 and CG131 strains were isolated in 2003 and thereafter. The major serotypes of each group were as follows: CG200, O126:H27 (13 strains); CG40, O111:H21 (30 strains); CG3570, O86a:H27 (11 strains); CG34, OUT:H33 (9 strains); CG131, O25:H4 (29 strains).
FIG 1.
Phylogenetic tree of aggR-positive EAEC strains. A clonal group (CG) was defined as a group of closely related STs, and groups were named in accordance with the dominant ST. A cluster (C) was defined as a group of strains showing the same ST. Major CGs are indicated by bold type. The scale bar shows 0.005 nucleic acid substitutions per site. EAEC reference strains are highlighted, and reference strains other than EAEC are in gray.
FIG 2.
Yearly changes in the numbers of major CG strains.
Prevalence of virulence genes.
The PCR screening results are shown in Tables S1 and S3 in the supplemental material. Among the EAEC plasmid-derived virulence genes, the aatA gene (147 strains [88.0%]) and the aap gene (146 strains [87.4%]) were highly conserved in the 167 strains tested. Regarding AAFs (the major EAEC adhesins), 139 strains (83.2%) possessed one of the five known variants, and the distribution of variants clearly showed a lineage dependency. CG200 strains mainly possessed AAF/II. CG295 and CG3570 strains possessed AAF/IV, whereas CG40 and CG131 strains predominantly possessed AAF/V. Several strains did not harbor any AAF/I to AAF/V genes. Additionally, different AAF/I to AAF/V gene types were observed within the same group. The pet gene was mostly found in CG200 strains.
Among the chromosomally encoded EAEC virulence genes, the pic and aaiC genes were detected in all STs or CGs except for CG131. Intriguingly, whereas the five UPEC virulence genes examined were sporadically present in the EAEC strains with the exception of CG131, most of the CG131 strains possessed the afa and aer genes (91.4% [32/35 strains] and 88.6% [31/35 strains], respectively).
CTX-M ESBL gene prevalence.
Among the 167 strains examined, 32 strains (19.2%) harbored the blaCTX-M gene and were confirmed to produce ESBLs (see Tables S1 and S3 in the supplemental material). The 32 strains belonged to multiple lineages, and three subtypes of CTX-M were identified. The CTX-M types were as follows: CTX-M-14 in 30 strains (94%), CTX-M-15 in 1 strain (3%), and CTX-M-55 in 1 strain (3%). Notably, 77.1% of the CG131 strains (27/35 strains) produced CTX-M-14-type ESBLs. The blaCTX-M-15 and blaCTX-M-55 genes were detected in each strain of C14 and C8, respectively, both of which were isolated from patients in Osaka.
Biofilm formation and the HEp-2 cell adherence assay.
The average biofilm indexes of each CG (or of individual singleton strains) are shown in Table S3 in the supplemental material. The capacity to form biofilms varied between the groups. A comparison of the biofilm index values for five groups is shown in Fig. 3. The biofilm indexes of CG40 and CG34 were significantly higher than those of the other three groups (P ≤ 0.001 and P ≤ 0.009, respectively). The biofilm index of CG131 was significantly lower than those of the other groups (P ≤ 0.001). Although the CG40 and CG131 strains possessed the same AAF/V, there was a significant difference in the biofilm indexes between the two groups (P < 0.0001). Most singleton strains in phylogroups B2 and D exhibited lower biofilm index values than did the strains belonging to phylogroups B1 and A (average ± standard deviation [SD], 0.25 ± 0.32 versus 0.99 ± 0.74; P < 0.0001). In the HEp-2 cell adherence assay, most of the strains examined showed an AA pattern (87.6% [141/161 strains]). Diffuse adherence patterns were observed for five strains (four ST131 strains and one ST3570 strain).
FIG 3.
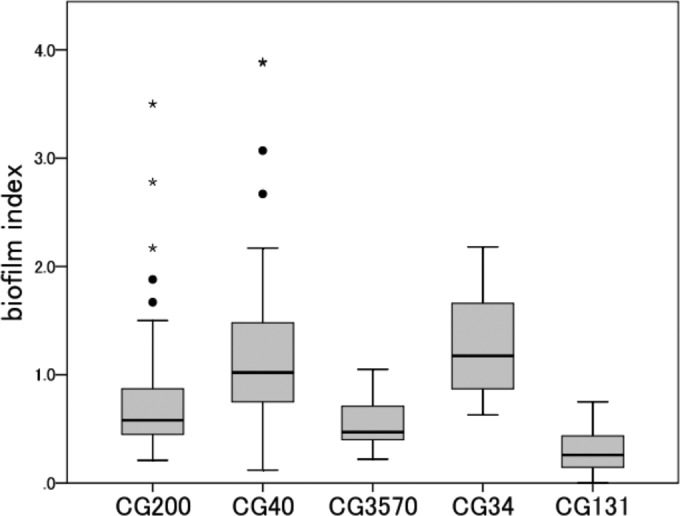
Comparison of biofilm index values for five groups. The biofilm index was defined as the average optical density (OD570). Boxplots show the medians (horizontal lines) and the 10th, 25th, 75th, and 90th percentiles (boxes and whiskers). ●, outliers; ⭑, extreme values. The numbers in the five groups were as follows: CG200, 29 strains; CG40, 49 strains; CG3570, 16 strains; CG34, 10 strains; CG131, 35 strains. The average biofilm index for EAEC 042, which was used as the positive control, was 3.61.
Phylogenetic analysis of the aggR gene.
To clarify the origin of EAEC plasmids in each strain, nucleotide sequences of the aggR gene (which is known to be on the EAEC plasmids) were compared among the strains isolated in Kagoshima. The aggR gene phylogenetic tree demonstrated that CG131 strains isolated in 2003 through 2005 clustered with CG40 and CG34 strains and not with CG200 and CG3570 strains (see Fig. S1 in the supplemental material). CG131 strains isolated in 2006 through 2010 showed one single-nucleotide polymorphism (SNP) with a nonsynonymous substitution, compared with those isolated in 2003 through 2005. Thus, the EAEC virulence plasmid of CG131 is likely to have originated from CG40 or CG34.
DISCUSSION
The phylogenetic analysis showed that the EAEC strains in this study were phylogenetically diverse and originated from multiple lineages (phylogroups A, B1, B2, and D), as described in a previous report (31). O126:H27/CG200, O111:H21/CG40, and O86a:H27/CG3570 strains were prevalent in both Kagoshima and Osaka. These three clones were isolated continuously during the study period and possessed both EAEC chromosomal and EAEC plasmid virulence genes. Although the repertoire of virulence genes, particularly those encoded by plasmids, varied between these groups, they had the capacity to form strong biofilms and showed the typical AA pattern in the HEp-2 adherence assay. Thus, we considered them canonical EAEC strains that were major historical EAEC clones in Japan. In fact, EAEC O126:H27 caused an outbreak in Shizuoka, Japan, in 2005 (5). Ito et al. also recently reported that these three serotypes are frequently found in Japan (45). However, these three serotypes have not been reported to be prevalent in other countries (15, 24, 46, 47), suggesting that the dominant EAEC serotypes differ geographically.
All aggR-positive O25 strains in phylogroup B2 were ST131 or related to ST131, and 83% (29/35 strains) contained the H4 flagellin. These strains have emerged since 2003 (Fig. 2) and harbor EAEC plasmid virulence genes. However, they do not possess the EAEC chromosomal virulence genes pic and aaiC, which are highly conserved in the historical EAEC clones in Japan. This observation suggests that the genetic backgrounds of these CG131 strains are different from those of the historical EAEC strains in Japan. In fact, this group belongs to phylogroup B2, in which the pandemic clone UPEC O25:H4/ST131 is distributed, whereas the historical EAEC clones in Japan belong to phylogroup A or B1. The CG131 strains possessed the sat gene, which is also chromosomally encoded; however, Sat was originally reported as the autotransporter toxin of UPEC. In fact, most ST131 strains have been reported to possess the sat gene intrinsically (48). In addition, most of the CG131 strains in our study harbored the afa gene, which is also encoded within some UPEC O25:H4/ST131 strains (49).
The UPEC O25:H4/ST131 clone has become increasingly prevalent worldwide (48). Thus, we assumed that the pandemic UPEC O25:H4/ST131 clone acquired an EAEC virulence plasmid from historical EAEC strains. The aggR genes of CG131 strains isolated in 2003 through 2005 showed exactly the same nucleotide sequences as the CG34 and CG40 strains. Thus, it is suggested that the EAEC virulence plasmid was horizontally transferred from either of these two groups to the UPEC O25:H4/ST131 clone. CG131 strains isolated later contained one SNP, which can be considered a form of microdiversity. However, further investigations of EAEC plasmids in CG131, CG34, and CG40 are needed to clarify the hypothesis. The number of CG131 strains peaked around 2007 and then declined (Fig. 2). The prevalence of ESBL-producing E. coli O25 among E. coli strains isolated from diarrheal children in Kagoshima peaked around 2007 and then declined from 2008 to 2010 (data not shown). We speculated that the prevalence of CG131 reflected a change in ESBL-producing E. coli O25 prevalence.
An interesting finding is that, although the EAEC O25:H4/CG131 and O111:H21/CG40 strains both possessed the AAF/V gene (aaf5A), the biofilm index of the former was significantly lower than that of the latter. This result suggests the importance of EAEC chromosomal factors in biofilm formation. However, the genetic difference between the EAEC virulence plasmids of the two groups (if it exists) may also be attributable to the difference in the capacity to form biofilms.
EAEC O25 strains in Japan have not been reported thus far. Three EAEC O25:H4 strains in Vietnam have been reported, although the ST and virulence gene patterns have not been described (46). Nuesch-Inderbinen et al. reported that three EAEC O25:H4/ST131 strains in Germany possessed agg3C and sat but not aggR or aap (24). EAEC O25:H4/CG131 strains cannot form strong biofilms, compared to canonical EAEC strains, and rarely harbor the toxin genes tested in this study. Therefore, they may not frequently cause severe enterocolitis or food-borne illness. These features may partly explain why EAEC O25 was rarely isolated from stool specimens as a diarrheal pathogen in either clinical laboratories or public health institutes. Although EAEC O25:H4/CG131 strains were isolated from diarrheal children in this study, it has not been determined that these strains are the causative agents of enterocolitis. Rather, the emergence of this clone may be important for UTIs, because increased biofilm-forming capacity or unknown specific adhesins may enhance their pathogenicity as UPEC. Further study is needed to clarify the pathogenicity of EAEC O25:H4/CG131.
UPEC O25:H4/CG131 is likely to have acquired a blaCTX-M-encoding plasmid (48). In this study, most of the CG131 strains produced CTX-M-14. CTX-M-14 is a prevalent CTX-M type in Japan, and the prevalence of CTX-M-14-producing E. coli ST131 increased from 2002 through 2012 (50). It is likely that the O25:H4/CG131 clone acquired both a plasmid harboring blaCTX-M-14 and an EAEC virulence plasmid in the intestines of people in our district.
In conclusion, this study shows the population structure of EAEC clones in Japan and the emergence of a hybrid clone, EAEC-UPEC O25:H4/CG131, producing CTX-M-14. Although further study is needed to clarify the pathogenicity of this hybrid clone as EAEC and/or UPEC, the aggregative characteristics of this clone can enhance conjugational transfer of both virulence and antibiotic resistance genes to other clones. The surveillance of horizontal transfer of both virulence and antibiotic resistance genes between diarrheagenic E. coli and UPEC strains is important for preventing increasing antimicrobial drug resistance worldwide.
Supplementary Material
ACKNOWLEDGMENTS
All authors declare that they have no conflicts of interest to disclose.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.00711-16.
REFERENCES
- 1.Sarantuya J, Nishi J, Wakimoto N, Erdene S, Nataro JP, Sheikh J, Iwashita M, Manago K, Tokuda K, Yoshinaga M, Miyata K, Kawano Y. 2004. Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol 42:133–139. doi: 10.1128/JCM.42.1.133-139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nataro JP, Mai V, Johnson J, Blackwelder WC, Heimer R, Tirrell S, Edberg SC, Braden CR, Glenn Morris J Jr, Hirshon JM. 2006. Diarrheagenic Escherichia coli infection in Baltimore, Maryland, and New Haven, Connecticut. Clin Infect Dis 43:402–407. doi: 10.1086/505867. [DOI] [PubMed] [Google Scholar]
- 3.Cohen MB, Nataro JP, Bernstein DI, Hawkins J, Roberts N, Staat MA. 2005. Prevalence of diarrheagenic Escherichia coli in acute childhood enteritis: a prospective controlled study. J Pediatr 146:54–61. doi: 10.1016/j.jpeds.2004.08.059. [DOI] [PubMed] [Google Scholar]
- 4.Itoh Y, Nagano I, Kunishima M, Ezaki T. 1997. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J Clin Microbiol 35:2546–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada T, Hiroi M, Kawamori F, Furusawa A, Ohata K, Sugiyama K, Masuda T. 2007. A food poisoning diarrhea outbreak caused by enteroaggregative Escherichia coli serogroup O126:H27 in Shizuoka, Japan. Jpn J Infect Dis 60:154–155. [PubMed] [Google Scholar]
- 6.Paschke C, Apelt N, Fleischmann E, Perona P, Walentiny C, Loscher T, Herbinger KH. 2011. Controlled study on enteropathogens in travellers returning from the tropics with and without diarrhoea. Clin Microbiol Infect 17:1194–1200. doi: 10.1111/j.1469-0691.2010.03414.x. [DOI] [PubMed] [Google Scholar]
- 7.Nataro JP, Kaper JB, Robins-Browne R, Prado V, Vial P, Levine MM. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr Infect Dis J 6:829–831. doi: 10.1097/00006454-198709000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Morin N, Santiago AE, Ernst RK, Guillot SJ, Nataro JP. 2013. Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect Immun 81:122–132. doi: 10.1128/IAI.00676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebbelstrup Jensen B, Olsen KE, Struve C, Krogfelt KA, Petersen AM. 2014. Epidemiology and clinical manifestations of enteroaggregative Escherichia coli. Clin Microbiol Rev 27:614–630. doi: 10.1128/CMR.00112-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tzipori S, Montanaro J, Robins-Browne RM, Vial P, Gibson R, Levine MM. 1992. Studies with enteroaggregative Escherichia coli in the gnotobiotic piglet gastroenteritis model. Infect Immun 60:5302–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheikh J, Czeczulin JR, Harrington S, Hicks S, Henderson IR, Le Bouguenec C, Gounon P, Phillips A, Nataro JP. 2002. A novel dispersin protein in enteroaggregative Escherichia coli. J Clin Invest 110:1329–1337. doi: 10.1172/JCI16172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishi J, Sheikh J, Mizuguchi K, Luisi B, Burland V, Boutin A, Rose DJ, Blattner FR, Nataro JP. 2003. The export of coat protein from enteroaggregative Escherichia coli by a specific ATP-binding cassette transporter system. J Biol Chem 278:45680–45689. doi: 10.1074/jbc.M306413200. [DOI] [PubMed] [Google Scholar]
- 13.Eslava C, Navarro-Garcia F, Czeczulin JR, Henderson IR, Cravioto A, Nataro JP. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun 66:3155–3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Savarino SJ, Fasano A, Robertson DC, Levine MM. 1991. Enteroaggregative Escherichia coli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J Clin Invest 87:1450–1455. doi: 10.1172/JCI115151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boisen N, Scheutz F, Rasko DA, Redman JC, Persson S, Simon J, Kotloff KL, Levine MM, Sow S, Tamboura B, Toure A, Malle D, Panchalingam S, Krogfelt KA, Nataro JP. 2012. Genomic characterization of enteroaggregative Escherichia coli from children in Mali. J Infect Dis 205:431–444. doi: 10.1093/infdis/jir757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jonsson R, Struve C, Boisen N, Mateiu RV, Santiago AE, Jenssen H, Nataro JP, Krogfelt KA. 2015. Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect Immun 83:1396–1405. doi: 10.1128/IAI.02820-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson IR, Czeczulin J, Eslava C, Noriega F, Nataro JP. 1999. Characterization of Pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect Immun 67:5587–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dudley EG, Thomson NR, Parkhill J, Morin NP, Nataro JP. 2006. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol Microbiol 61:1267–1282. doi: 10.1111/j.1365-2958.2006.05281.x. [DOI] [PubMed] [Google Scholar]
- 19.Guyer DM, Radulovic S, Jones FE, Mobley HL. 2002. Sat, the secreted autotransporter toxin of uropathogenic Escherichia coli, is a vacuolating cytotoxin for bladder and kidney epithelial cells. Infect Immun 70:4539–4546. doi: 10.1128/IAI.70.8.4539-4546.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe CM, Salvador FA, Falsetti IN, Vieira MA, Blanco J, Blanco JE, Blanco M, Machado AM, Elias WP, Hernandes RT, Gomes TA. 2008. Uropathogenic Escherichia coli (UPEC) strains may carry virulence properties of diarrhoeagenic E. coli. FEMS Immunol Med Microbiol 52:397–406. doi: 10.1111/j.1574-695X.2008.00388.x. [DOI] [PubMed] [Google Scholar]
- 21.Pitout JD, Laupland KB. 2008. Extended-spectrum β-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis 8:159–166. doi: 10.1016/S1473-3099(08)70041-0. [DOI] [PubMed] [Google Scholar]
- 22.Tokuda K, Nishi J, Imuta N, Fujiyama R, Kamenosono A, Manago K, Kawano Y. 2010. Characterization of typical and atypical enteroaggregative Escherichia coli in Kagoshima, Japan: biofilm formation and acid resistance. Microbiol Immunol 54:320–329. doi: 10.1111/j.1348-0421.2010.00210.x. [DOI] [PubMed] [Google Scholar]
- 23.Amaya E, Reyes D, Vilchez S, Paniagua M, Mollby R, Nord CE, Weintraub A. 2011. Antibiotic resistance patterns of intestinal Escherichia coli isolates from Nicaraguan children. J Med Microbiol 60:216–222. doi: 10.1099/jmm.0.020842-0. [DOI] [PubMed] [Google Scholar]
- 24.Nuesch-Inderbinen MT, Hofer E, Hachler H, Beutin L, Stephan R. 2013. Characteristics of enteroaggregative Escherichia coli isolated from healthy carriers and from patients with diarrhoea. J Med Microbiol 62:1828–1834. doi: 10.1099/jmm.0.065177-0. [DOI] [PubMed] [Google Scholar]
- 25.Guiral E, Mendez-Arancibia E, Soto SM, Salvador P, Fabrega A, Gascon J, Vila J. 2011. CTX-M-15-producing enteroaggregative Escherichia coli as cause of travelers' diarrhea. Emerg Infect Dis 17:1950–1953. doi: 10.3201/eid1710.110022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khoshvaght H, Haghi F, Zeighami H. 2014. Extended spectrum β-lactamase producing Enteroaggregative Escherichia coli from young children in Iran. Gastroenterol Hepatol Bed Bench 7:131–136. [PMC free article] [PubMed] [Google Scholar]
- 27.Frank C, Werber D, Cramer JP, Askar M, Faber M, an der Heiden M, Bernard H, Fruth A, Prager R, Spode A, Wadl M, Zoufaly A, Jordan S, Kemper MJ, Follin P, Muller L, King LA, Rosner B, Buchholz U, Stark K, Krause G. 2011. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany. N Engl J Med 365:1771–1780. doi: 10.1056/NEJMoa1106483. [DOI] [PubMed] [Google Scholar]
- 28.Chaudhuri RR, Sebaihia M, Hobman JL, Webber MA, Leyton DL, Goldberg MD, Cunningham AF, Scott-Tucker A, Ferguson PR, Thomas CM, Frankel G, Tang CM, Dudley EG, Roberts IS, Rasko DA, Pallen MJ, Parkhill J, Nataro JP, Thomson NR, Henderson IR. 2010. Complete genome sequence and comparative metabolic profiling of the prototypical enteroaggregative Escherichia coli strain 042. PLoS One 5:e8801. doi: 10.1371/journal.pone.0008801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buchholz U, Bernard H, Werber D, Bohmer MM, Remschmidt C, Wilking H, Delere Y, an der Heiden M, Adlhoch C, Dreesman J, Ehlers J, Ethelberg S, Faber M, Frank C, Fricke G, Greiner M, Hohle M, Ivarsson S, Jark U, Kirchner M, Koch J, Krause G, Luber P, Rosner B, Stark K, Kuhne M. 2011. German outbreak of Escherichia coli O104:H4 associated with sprouts. N Engl J Med 365:1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 30.Czeczulin JR, Whittam TS, Henderson IR, Navarro-Garcia F, Nataro JP. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect Immun 67:2692–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okeke IN, Wallace-Gadsden F, Simons HR, Matthews N, Labar AS, Hwang J, Wain J. 2010. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages. PLoS One 5:e14093. doi: 10.1371/journal.pone.0014093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerna JF, Nataro JP, Estrada-Garcia T. 2003. Multiplex PCR for detection of three plasmid-borne genes of enteroaggregative Escherichia coli strains. J Clin Microbiol 41:2138–2140. doi: 10.1128/JCM.41.5.2138-2140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prager R, Lang C, Aurass P, Fruth A, Tietze E, Flieger A. 2014. Two novel EHEC/EAEC hybrid strains isolated from human infections. PLoS One 9:e95379. doi: 10.1371/journal.pone.0095379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamamoto S, Terai A, Yuri K, Kurazono H, Takeda Y, Yoshida O. 1995. Detection of urovirulence factors in Escherichia coli by multiplex polymerase chain reaction. FEMS Immunol Med Microbiol 12:85–90. doi: 10.1111/j.1574-695X.1995.tb00179.x. [DOI] [PubMed] [Google Scholar]
- 35.Boyd DA, Tyler S, Christianson S, McGeer A, Muller MP, Willey BM, Bryce E, Gardam M, Nordmann P, Mulvey MR. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob Agents Chemother 48:3758–3764. doi: 10.1128/AAC.48.10.3758-3764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; M100-S24. Table 3A. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.Garcia BG, Ooka T, Gotoh Y, Vieira MA, Yamamoto D, Ogura Y, Girão DM, Sampaio SC, Melo AB, Irino K, Hayashi T, Gomes TA. 2016. Genetic relatedness and virulence properties of enteropathogenic Escherichia coli strains of serotype O119:H6 expressing localized adherence or localized and aggregative adherence-like patterns on HeLa cells. Int J Med Microbiol 306:152–164. doi: 10.1016/j.ijmm.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ochman H, Selander RK. 1984. Standard reference strains of Escherichia coli from natural populations. J Bacteriol 157:690–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ooka T, Seto K, Kawano K, Kobayashi H, Etoh Y, Ichihara S, Kaneko A, Isobe J, Yamaguchi K, Horikawa K, Gomes TA, Linden A, Bardiau M, Mainil JG, Beutin L, Ogura Y, Hayashi T. 2012. Clinical significance of Escherichia albertii. Emerg Infect Dis 18:488–492. doi: 10.3201/eid1803.111401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iguchi A, Iyoda S, Seto K, Morita-Ishihara T, Scheutz F, Ohnishi M. 2015. Escherichia coli O-genotyping PCR: a comprehensive and practical platform for molecular O serogrouping. J Clin Microbiol 53:2427–2432. doi: 10.1128/JCM.00321-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li D, Liu B, Chen M, Guo D, Guo X, Liu F, Feng L, Wang L. 2010. A multiplex PCR method to detect 14 Escherichia coli serogroups associated with urinary tract infections. J Microbiol Methods 82:71–77. doi: 10.1016/j.mimet.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 43.Fields PI, Blom K, Hughes HJ, Helsel LO, Feng P, Swaminathan B. 1997. Molecular characterization of the gene encoding H antigen in Escherichia coli and development of a PCR-restriction fragment length polymorphism test for identification of E. coli O157:H7 and O157:NM. J Clin Microbiol 35:1066–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wakimoto N, Nishi J, Sheikh J, Nataro JP, Sarantuya J, Iwashita M, Manago K, Tokuda K, Yoshinaga M, Kawano Y. 2004. Quantitative biofilm assay using a microtiter plate to screen for enteroaggregative Escherichia coli. Am J Trop Med Hyg 71:687–690. [PubMed] [Google Scholar]
- 45.Ito K, Matsushita S, Yamazaki M, Moriya K, Kurazono T, Hiruta N, Narimatsu H, Ueno N, Isobe J, Yatsuyanagi J, Kumagai N, Hashimoto M, Ratchtrachenchai OA. 2014. Association between aggregative adherence fimbriae types including putative new variants and virulence-related genes and clump formation among aggR-positive Escherichia coli strains isolated in Thailand and Japan. Microbiol Immunol 58:467–473. doi: 10.1111/1348-0421.12173. [DOI] [PubMed] [Google Scholar]
- 46.Hien BT, Scheutz F, Cam PD, Serichantalergs O, Huong TT, Thu TM, Dalsgaard A. 2008. Diarrheagenic Escherichia coli and Shigella strains isolated from children in a hospital case-control study in Hanoi, Vietnam. J Clin Microbiol 46:996–1004. doi: 10.1128/JCM.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Regua-Mangia AH, Gomes TA, Vieira MA, Irino K, Teixeira LM. 2009. Molecular typing and virulence of enteroaggregative Escherichia coli strains isolated from children with and without diarrhoea in Rio de Janeiro city, Brazil. J Med Microbiol 58:414–422. doi: 10.1099/jmm.0.006502-0. [DOI] [PubMed] [Google Scholar]
- 48.Nicolas-Chanoine MH, Bertrand X, Madec JY. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blanco J, Mora A, Mamani R, Lopez C, Blanco M, Dahbi G, Herrera A, Marzoa J, Fernandez V, de la Cruz F, Martinez-Martinez L, Alonso MP, Nicolas-Chanoine MH, Johnson JR, Johnston B, Lopez-Cerero L, Pascual A, Rodriguez-Bano J. 2013. Four main virotypes among extended-spectrum-β-lactamase-producing isolates of Escherichia coli O25b:H4-B2-ST131: bacterial, epidemiological, and clinical characteristics. J Clin Microbiol 51:3358–3367. doi: 10.1128/JCM.01555-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsumura Y, Johnson JR, Yamamoto M, Nagao M, Tanaka M, Takakura S, Ichiyama S. 2015. CTX-M-27- and CTX-M-14-producing, ciprofloxacin-resistant Escherichia coli of the H30 subclonal group within ST131 drive a Japanese regional ESBL epidemic. J Antimicrob Chemother 70:1639–1649. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.