Abstract
Purpose of Review
Red cell receptors provide unique entry points for Plasmodium parasites to initiate blood-stage malaria infection. Parasites encode distinct ligands that bind specifically to both highly abundant and low copy receptors. Recent advances in the understanding of molecular and structural mechanisms of these interactions provide fundamental insights into receptor-ligand biology and molecular targets for intervention.
Recent Findings
This review focuses on the requirements for known interactions, insight derived from complex structures, and mechanisms of receptor/ligand engagement. Further, novel roles for established red cell membrane proteins, parasite ligands and associated interacting partners have recently been established in red cell invasion.
Summary
This new knowledge underlines the intricacies involved in invasion by a eukaryotic parasite into a eukaryotic host cell demonstrated by expanded parasite ligand families, redundancy in red cell receptor engagement, multi-tiered temporal binding, and the breadth of receptors engaged.
Keywords: Receptor, Malaria, Ligand, Interaction, Host-pathogen
Introduction
Forty percent of the world’s population is at risk for malaria leading to 800,000 deaths each year, predominantly in children. Malaria is caused by Plasmodium parasites which have a complicated lifecycle involving two different hosts, the Anopheles mosquito and the human, and three distinct stages of development: exo-erythrocytic, erythrocytic, and sporogonic stage. All stages are required to establish an infection and continue the cycle of transmission.
Within the human host, the erythrocytic stage consists of multiple cycles of red blood cell (RBC) invasion, replication, and lysis that lead to the anemia and fatigue associated with this disease. RBC invasion occurs in four distinct steps: low affinity interactions between the merozoite and RBC; apical reorientation and tight junction formation; active invasion and surface shedding; and formation of the parasitophorous vacuole for parasite replication. The first two steps rely on distinct receptor-ligand interactions between the host RBC and Plasmodium merozoite that are essential for parasite survival. Recently, great strides in understanding these interactions have been made and those efforts are described here.
Band 3
Band 3 (anion exchanger-1) is the most abundant integral membrane protein on the surface of RBCs with approximately 1.2 million copies per cell (Table 1) [2]. It is a 100 kDa multipass transmembrane channel for anions that forms dimers, tetramers, and higher-order oligomers as a result of interactions with cytoskeletal proteins [2].
Table 1.
Molecular and Functional Description of RBC Receptors for Plasmodium Parasites
| RBC receptor | Alternate Names | Malaria Ligand(s) Engaged | Copies per RBC | Blood Groups | Domains | Endogenous Function | Refs |
|---|---|---|---|---|---|---|---|
| Band 3 | AE-1 | PfMSP119, PfRhopH3, PfMSP9 | 1.2 million | Di, Wr | N-terminal cytoplasmic C-terminal transmembrane spanning |
Binding to cytoskeletal proteins, Oligomerization Anion transport channel for bicarbonate and chloride anions |
[1–3] |
| Glycophorin A | CD235A | PfEBA-175, PfMSP183 | 1 million | M, N, S, s | Extracellular Transmembrane |
Unknown Oligomerization |
[1–4] |
| Glycophorin B | CD235B | PfEBL-1 | 150,000 | N, S, s | Extracellular Transmembrane |
Unknown Oligomerization |
[1–4] |
| Glycophorin C | CD236 | PfEBA-140 | 143,000 | Ge | Extracellular Cytoplasmic |
Unknown Binding to cytoskeletal proteins |
[1–3,5] |
| CD55 | DAF | Unknown | 20,000 | CROM | Control protein repeats (CCP) 2–4 | Complement decay-accelerating activity | [1,3] |
| Duffy antigen/Receptor for chemokines | CD234 | PvDBP | 6,000–13,000 | Fy | Seven-span transmembrane GPCR | Receptor for CC and CXC cheomkines | [1,3] |
| Intracellular adhesion molecule 4 | CD242 | Unknown | 2,800–5,100 | LW | Immunoglobulin-like | Intercellular adhesion | [1,3] |
| Basigin | CD147, EMMPRIN, TCSF, Neurothelin, 5A11, HT7, OX47, CE9, gp42 | PfRh5 | 3,000 | Ok | Extracellular | Transport and function of monocarboxylate transporters, binding to integrins, cyclophilins, and caveolin-1 | [1,3,6] |
| Complement Receptor 1 | CD35, C3b/C4b adherence receptor, immune adherence receptor | PfRh4 | 50–1200 | Kn | CCP 1–3, CCP 7–9, CCP 15–17 CCP 1–3 CCP 7–9 and CCP 15–17 |
Binding of C4b Complement decay-accelerating activity Binding of C3b |
[1,3,7] |
| Semaphorin 7A | CDw108 | PfMTRAP | unknown | JMH |
Ectodomain Sema Immunoglobulin-like |
T-cell maturation, T-cell/macrophage interactions, regulation of TGF-β1 Binding to PlexinC1 and β1-Integrin and oligomerization Oligomerization |
[1,3,8,9] |
Plasmodium parasites exploit the high abundance of Band 3 and employ a variety of ligands to engage this receptor (Figure 1, Table 2) including the multimeric Plasmodium falciparum (Pf) Merozoite surface protein 1 (MSP1) complex. The PfMSP1 complex is composed of PfMSP1, PfMSP6, PfMSP7, PfMSP9, PfMSPDBL1, PfMSPDBL2, PfRhopH3, PfRhopH1/Clag, PfRAP-1, and PfRAP-2 proteins [10,12,15,49]. PfMSP1, a 200 kDa protein, facilitates the majority of the interactions of the complex with Band 3 and undergoes proteolytic processing to produce the truncated proteins identified by their molecular weight as PfMSP183, PfMSP130, PfMSP142, PfMSP138, and PfMSP119. Of these products, PfMSP119 is proposed to mediate the principal interaction with Band 3 and binds to amino acids (aa) 720–761 with an apparent nanomolar affinity [10]. Additional proteins including PfMSP9, aa 77–183 and 364–528, and PfRhopH3 also interact with aa 720–761 of Band 3 albeit with lesser affinities [12,15]. PfMSP119 also contacts PfRhopH3 by immuno-precipitation and is able to compete for binding of PfRhopH3 to Band 3 [12].
Figure 1.
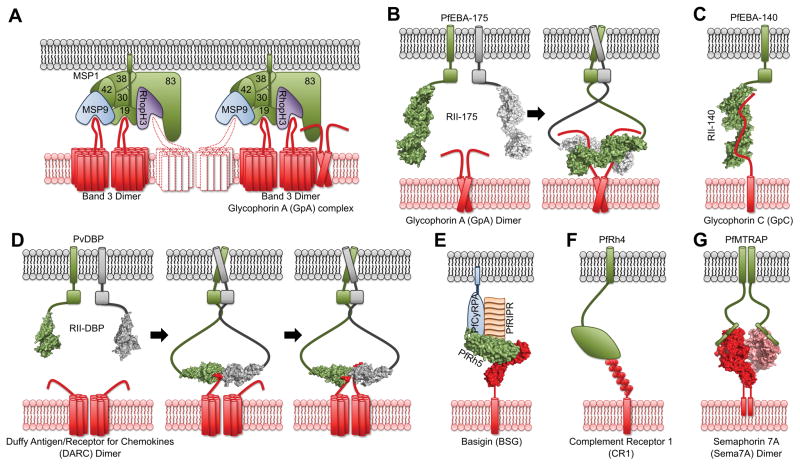
Molecular interactions between RBC receptors and malaria parasite ligands. A) Band 3 (left) and Band 3/GpA (right) bind the PfMSP1 complex. PfMSP119, PfMSP9 and PfRhopH3 can independently bind Band 3 leading to multiple binding configurations raising the possibility for engagement of multiple Band 3 dimers (dotted lines), B) Glycophorin A (GpA) drives dimerization of PfEBA-175 upon engagement, C) Glycophorin C binding to PfEBA-140, D) Duffy antigen receptor for chemokines (DARC) engages PvDBP via a model of receptor-induced ligand dimerization, E) Basigin (BSG) interacts with the multiprotein complex of PfRh5, PfRIPR and PfCyRPA, F) Complement receptor 1 (CR1) in complex with PfRh4, G) Each monomer of the Semaphorin 7A (Sema7A) dimer binds a monomer of PfMTRAP. Available structures are shown in surface representation. Domains, transmembrane segments (long cylinders) and GPI anchors (short cylinders) are shown as cartoons wherever structural information is lacking.
Table 2.
Malaria Parasite Ligand/RBC Receptor Pairs and Their Interactions
| RBC receptor | Rceptor binding region (aa) | Receptor MW (kDa) | Malaria Ligand | Ligand MW (kDa) | Ligand binding region (aa) | PDB ID of receptor/ligand complex structure | Refs |
|---|---|---|---|---|---|---|---|
| Band 3 | 720–761 | 100 | PfMSP119 | 19 | 1517–1640 | [10,11] | |
| 720–761 | PfRhopH3 | 110 | 620–897 | [12] | |||
| 720–761 | PfMSP138 | 38 | 931–1348 | [10,13] | |||
| 720–761 | PfMSP142 | 42 | 1255–1640 | [10,11,14] | |||
| 720–761 | PfMSP9 | 88.6 | 77–183, 364–528 | [15] | |||
|
| |||||||
| Glycophorin A | 50–84 | 43 | PfEBA-175 | 175 | 145–760 | 1ZRO [16] | [16–22] |
| 53–84 | PfMSP183 | 83 | 194–296 | [23] | |||
|
| |||||||
| Glycophorin B | Unknown | 25 | PfEBL-1 | 300 | 601–669 | [24,25] | |
|
| |||||||
| Glycophorin C | Unknown | 40 | PfEBA-140 | 140 | 143–740 | 4JNO [26] | [26–30] |
|
| |||||||
| Duffy antigen/Receptor for chemokines | 14–43 | 35–45 | PvDBP | 120 | 211–525 | 4NUU, 4NUV [31] | [31–35] |
|
| |||||||
| Basigin | 22–205 | 35–65 | PfRH5 | 45–63 | 140–526 | 4U0Q [36] | [36–40] |
|
| |||||||
| Complement receptor 1 | 7–9, 18–20 | 190–280 | PfRH4 | 205 | 104–465 | [41–43] | |
|
| |||||||
| Semaphorin 7A | 54–491 | 80 | PfMTRAP | 58 | 23–105 | [44,45] | |
|
| |||||||
| CD55 | Unknown | 60–70 | Unknown | [46] | |||
|
| |||||||
| ICAM 4 | Unknown | 40 | Unknown | [47] | |||
|
| |||||||
| Unknown | PfEBA-181 | 181 | Unknown | [48] | |||
|
| |||||||
| Unknown | PfMSPDBL1 | 80 | 143–443, 542–696 | [49] | |||
|
| |||||||
| Unknown | PfMSPDBL2 | 88 | 161–457, 571–672 | [49] | |||
|
| |||||||
| Unknown | PfRh2 | 370 | 495–860 | [50] | |||
|
| |||||||
| Unknown | PfTRAMP | 37 | 26–307 | [51] | |||
An unanswered question remains how do these competing proteins interact with Band 3 as a complex? The first model relies on the multimeric state of Band 3 (Figure 1A). In the example of binding to a dimer, two independent binding sites on Band 3 exist in the right configuration to support independent binding. PfMSP119, PfMSP9, and PfRhopH3 could each bind an independent Band 3 monomer resulting in increased avidity that drives attachment. An alternate model is that each component binds the same Band 3 monomer. While this is possible, the competitive binding of PfMSP119 and PfRhopH3 suggests this is unlikely [12].
Glycophorin A and B
GpA is the most abundant glycoprotein on the surface of the RBC with 1 million copies per cell (Table 1) [2]. GpA is a single span transmembrane protein and part of a gene family that includes Glycophorin B (GpB) and Glycophorin E (GpE). GpB and GpE arose from gene duplication and share 95% sequence similarity and exon structure with GpA [4]. GpA is comprised of seven exons and exons 2 through 6 form the mature receptor. In contrast, the mature form of GpB consists of exons 2, 4, and 5. Exon 5 forms a constitutive transmembrane dimer that allows for both homodimers and heterodimers of GpA and GpB on the RBC surface [4].
Mature GpA is heavily glycosylated with 16 O-linked glycans and 1 N-linked glycan on exons 2 and 3 that make up 50% of the molecular weight of the receptor [2,4,52]. Five O-linked glycans are located in exon 3 and are unique to GpA [2,4,52]. O-linked glycosylation on GpA is essential for binding to P. falciparum EBA-175, a member of the Erythrocyte Binding Like (EBL) family of proteins (Figure 1B, Table 2) [17]. Region II of PfEBA-175 (RII-175) contains two Duffy Binding Like (DBL) domains that bind to GpA [18]. The crystal structure of RII-175 in complex with α-2,3-sialyllactose, a trisaccharide similar to O-linked glycans from GpA, revealed that the glycan binding sites are located at the dimer interface of RII-175 [16] implicating multimeric assembly of PfEBA-175 around a dimeric GpA during invasion [19,21].
Recently, production of fully glycosylated recombinant GpA (rGpA) in a mammalian tissue culture system enabled careful interrogation of the binding requirements for malaria infection [20,22]. This system overcomes the poor yield, poor solubility, and contamination with GpB observed when GpA is extracted from RBCs. Multiple glycans in exon 3 of GpA are critical for P. falciparum invasion as mutation of three glycosylation sites within, or elimination of, exon 3 from rGpA abrogated binding to RII-175 [20]. These glycans are also critical during in vitro infection as loss of these glycosylation sites prevented inhibition of P. falciparum growth observed with rGpA [20]. Therefore, PfEBA-175 differentiates between GpA and GpB by engaging glycans unique to GpA [20]. It is possible that GpB arose due to evolutionary pressure driven by malaria infection to lose the PfEBA-175 binding site in GpA [20]. Finally, GpB is engaged by PfEBL-1, a related protein to PfEBA-175, [24,25] although little is currently known about the mechanisms of this interaction (Table 2).
PfMSP183, a member of the PfMSP1 complex, also binds to GpA purified from RBCs (Figure 1A, Table 2) [23]. Binding is not affected by trypsin or neuraminidase treatment and contrasts that of PfEBA-175 binding to GpA [18]. Trypsin treatment cuts GpA at two arginine residues in exon 3 and removes all but three glycans while neuraminidase removes sialic acid from glycans of GpA. Further analysis using protease treatment of GpA indicated that the PfMSP183 binding site is in amino acids 53 to 84 of GpA [49] and overlaps with, but is not identical to, the PfEBA-175 binding site [20]. GpA can form a complex with Band 3 [2] and the PfMSP183:GpA interaction likely augments the Band 3 contacts mediated by other PfMSP1 complex components (Figure 1A). It is not yet clear how these two Plasmodium proteins can bind the same receptor region during invasion. However, it is plausible that PfMSP-1 binding occurs first during the initial cell-cell contact and is supplanted by PfEBA-175 during tight junction formation and invasion.
Glycophorin C and D
Glycophorin C (GpC) and Glycophorin D (GpD) are ~40kDa single span transmembrane glycoproteins unrelated to GpA/B/E and are present on the surface of RBCs in levels of 143,000 and 82,000 copies/cell respectively (Table 1) [2,5]. GpD results from an alternate start codon in exon 2 generating a protein that is 22 amino acids shorter, however, both proteins retain the cytoplasmic domain that is responsible for binding to the cytoskeleton [2]. GpC is heavily glycosylated with 1 N-linked glycan and 12 O-linked glycans. The alternate start codon of GpD eliminates the N-linked glycan and 6 O-linked glycans [2].
GpC, but not GpD, is the receptor for PfEBA-140, another member of the EBL family (Figure 1C, Table 2) [27]. PfEBA-140 is unable to bind RBCs lacking GpC or lacking exon 2 of GpC, and antibodies targeting exon 2 of GpC block binding of PfEBA-140 to normal RBCs [27]. Together, this data argues that exon 2 is critical for binding. However, PfEBA-140 cannot bind GpC lacking the single N-linked glycan, indicating that this glycan in exon 1 also contributes to the GpC:PfEBA-140 interaction [28].
Crystal structures of the minimal RBC binding domain of PfEBA-140 (region II, RII-140) were solved with and without sialyllactose [26,29]. RII-140 consists of two DBL domains named F1 and F2 that share an overall fold with PfEBA-175, although RII-140 is monomeric instead of dimeric [26,29]. F1 and F2 use the same region of the DBL domain to bind receptor glycans and these receptor binding pockets are structurally similar to each other, but distinct from the pockets in PfEBA-175 [26]. However, mutational data revealed that the binding pocket in F1, but not F2, is essential for receptor binding [26].
Polymorphisms in the F1 domain of PfEBA-140 alter receptor specificity and affinity [30,53]. The polymorphism I185V is located in the F1 binding pocket [26], and parasite strains with V185 have reduced binding to RBCs compared to strains with I185 [30,53]. The larger Ile creates a tight binding pocket for glycans in F1, and substitution by the smaller Val creates a cavity in the glycan binding pocket increasing solvent accessibility and reducing affinity for the receptor [26]. The structure of EBA-140 has answered a number of questions relevant for engagement of GpC although the exact mechanism remains to be determined.
Duffy antigen/Receptor for chemokines
The Duffy antigen/Receptor for chemokines (DARC) is a 35–50 kDa glycoprotein with 7 transmembrane spanning regions and three N-linked glycans (Table 1) [1]. It is expressed on the surface of RBCs and endothelial tissue and belongs to the G protein-coupled receptor family [1,3]. DARC exists with either a glycine or an aspartate at amino acid position 42 resulting in the Duffy blood groups Fya and Fyb, respectively. A Fy-null phenotype is also observed and is caused by a mutation of the GATA consensus translation sequence that eliminates DARC expression in RBCs but not in endothelial tissues. In DARC expressing erythrocytes, DARC is present at levels of 6,000–13,000 copies per cell. Antibody staining using the DARC specific antibody Fy6 that recognizes aa 19–26 of DARC shows that levels of DARC are 50% greater in reticulocytes than erythrocytes [1].
DARC is the receptor for Duffy Binding Protein (DBP) ligand from P. vivax (Pv) and P. knowlesi (Pk), and PvDBP is the sole member of the EBL family found in P. vivax (Figure 1D, Table 2) [32]. Loss of DARC on RBCs is protective against P. vivax malaria making DBP a high priority vaccine candidate [54]. DBP contains a single DBL domain in Region II (RII-DBP) that is sufficient to bind DARC on RBCs.
Residues 1–60 form the N-terminal ectodomain of DARC (DARC1–60) and are sufficient for DBP binding [32,33]. In the absence of DARC1–60, PvDBP is monomeric in solution [34] consistent with the monomeric structure of PkDBPα, which represents the unbound monomeric form of DBP [35]. Strikingly, addition of DARC1–60 induces dimerization of PvDBP to form an oligomeric complex [34]. Two distinct crystal structures of the PvDBP:DARC complex have been determined: a heterotrimer (2 RII-DBP: 1 DARC) and a heterotetramer (2 RII-DBP: 2 DARC) [31]. In both structures, only residues 19–30 of DARC are engaged by PvDBP, and this segment includes the Fy6 antigen [1,3]. Isothermal titration calorimetry demonstrated a stepwise assembly mechanism in solution: the heterotrimer forms first with micromolar affinity followed by a second binding event to form the heterotetramer with nanomolar affinity. This stepwise binding mechanism results in a stable high affinity complex to initiate invasion [31].
Although these structural and biophysical studies provide deep mechanistic insight into DARC engagement by DBP, several unanswered questions remain. The glycine to aspartate change at residue 42 (FyB) increased DBP binding and P. vivax infection [55] and sulfation at residues 30 and 41 of DARC also increase binding to DBP [33]. However, residues 41 and 42 of DARC do not make direct contacts with DBP in the crystal structures [31]. It is therefore plausible that these modifications to DARC may alter the surface exposure or presentation of the DARC N-terminus rather than facilitating a direct contact with DBP. Further studies are required to address these models.
Basigin
Basigin (BSG) is a 35–65 kDa glycoprotein composed of two to three extracellular Immunoglobin-like domains, a single span transmembrane domain, and a short cytoplasmic domain (Table 1) [1,6]. BSG is expressed on number of cell types including erythrocytes with 3,000 copies per RBC [1,3,6] and has a broad range of functions. In RBCs, BSG is necessary for the correct surface targeting and activity of monocarboxylate trasporters [1,6].
An avidity-based screen optimized to detect low affinity interactions identified BSG as the receptor for P. falciparum reticulocyte-binding homology 5 (PfRh5), a member of the reticulocyte binding like homolog proteins (RBPs or Rhs) (Figure 1E, Table 2) [37]. PfRh5 binds to the short isoform of BSG that contains two Ig-like domains with a Kd of 0.43–1.3 μM [37,38]. Crystal structures of PfRh5 [36,38] and PfRh5 in complex with BSG [36] were also recently reported. PfRh5 is a 60 kDa protein that is processed upon secretion to remove 139 residues from the N-terminus [36,38]. The mature form of PfRh5 is composed of two alpha-helix bundles that form a flat kite like shape [36,38]. BSG binds the end of the alpha-helical bundles closest to the C-terminus of PfRh5 and engages both Ig-like domains of BSG forming a 1:1 complex [36]. Surprisingly, interactions in the complex are primarily between the backbone of BSG and the loops of PfRh5 [36].
PfRh5 does not contain a transmembrane domain or a GPI-anchor but still localizes to the surface of merozoites. PfRh5 immunoprecipitated from parasite cultures revealed a multiprotein complex of 200 kDa [39,40] that includes PfRipr, a large 123 kDa protein containing multiple EGF-like domains [40], and PfCyRPA, a 35 kDa GPI-anchored protein [39]. These weights suggest the 200 kDa complex is a 1:1:1 association of PfRH5, PfRIPR and PfCyRPA. However, the molecular interactions within this complex are still unknown. BSG is an essential receptor for invasion by all strains of P. falciparum investigated [37] making this interaction central for malaria infection despite its relatively low abundance.
Complement Receptor 1
Complement Receptor 1 (CR1) is a relatively low abundance, 50–1200 copies/cell, type 1 transmembrane protein on the surface of RBCs (Table 1) [7]. CR1 consists of up to 44 control protein repeats (CCPs) organized into long homologous repeats (LHRs) composed of seven CCPs resulting in a protein of 190–280 kDa. CR1 binds C3b and C4b immune complexes resulting in transport of the complexes to the spleen and liver for clearance [7].
CR1 is the receptor for PfRh4, a member of the RBP/Rh family, [41–43] as anti-CR1 antibodies block native PfRh4 binding to RBCs and soluble CR1 binds soluble PfRh4 with a Kd of 2.9 uM (Figure 1F, Table 2) [41]. PfRh4 binds to CCP1–3, which are also the repeats responsible for binding C4b and convertase decay-accelerating activity (DAA) [42]. Surprisingly, C4b and PfRh4 are able to bind to CR1 at the same time. However, PfRh4 engagement of CR1 disrupts DAA activity in a dose dependent manner [42].
More recently, the PfRh4 binding site was narrowed to CCP1 [43]. Amino acid residues 18 and 20 are essential for binding and mutation of amino acids 7–9 and 19 greatly reduced binding to PfRH4. These residues form a continuous patch on one face of CCP1 [43]. While the regions of CR1 that interact with PfRh4 have been identified, the molecular architecture of the interaction remains unknown.
Semaphorin 7A
Semaphorin 7A (Sema7A) is an 80 kDa GPI-anchored member of the semaphorin family [8,9] composed of three domains: a sema domain, PSI domain, and Ig domain (Table 1) [8]. Sema7A is the receptor for the P. falciparum MTRAP (Figure 1G, Table 2) [44]. PfMTRAP, a member of the thrombospondin-related anonymous protein (TRAP) family, is composed of receptor-binding extracellular domains, a transmembrane domain, and a cytoplasmic domain that facilitates active invasion utilizing an actin-myosin motor complex.
PfMTRAP’s extracellular domain contains two thrombospondin repeat (TSR)-like domains that are required to bind RBCs [45]. Direct binding studies showed the two TSR domains alone were necessary and sufficient to bind Sema7A with similar kinetics as the full length PfMTRAP ectodomain (Kd = 1.96 uM and 1.18 uM respectively) [44]. The interaction is dependent on the sema domain, which promotes homodimerization of Sema7A [8], and each PfMTRAP molecule binds a Sema7A monomer [44].
Novel receptors and ligands with roles in invasion
There are currently RBC receptors for which no parasite ligand has been defined and vice versa. CD55 and ICAM-4 are RBC receptors without known parasite ligands identified through forward genetics [46] and parasite inhibition studies [47], respectively (Table 2). Conversely, parasite ligands without known receptors include PfMSPDBL 1 & 2 [49], PfRh2 [50], PfEBA-181 [48], and PfTRAMP (Table 2) [51]. These proteins remain an area of active study to fully define the range of RBC interactions available to the malaria parasite.
Consequences for therapeutic design
A mechanistic understanding of host-parasite receptor-ligand interactions and a mechanistic framework for antibody neutralization are crucial for the design of effective vaccines that will prevent malaria. Antibodies are a critical component of naturally acquired immunity to malaria. While antibodies that target blood-stage parasites can prevent interactions, not all antibodies are inhibitory [36,44,56]. It is clear that antibodies that target protein function by engaging to receptor-binding pockets or dimerization interfaces are potently neutralizing while antibodies that engage non-functional or decoy epitopes have no effect on parasite viability [56]. In addition, multiple interactions for RBC invasion indicate that targeting multiple parasite ligands will be necessary to prevent blood-stage infection. Lastly, polymorphic variation in parasite ligands results in immune evasion, and this phenomenon must also be addressed during vaccine design. This is best exemplified in PvDBP where recent efforts have proven successful in eliminating highly immunogenic polymorphic segments that result in strain-specific responses while retaining broadly neutralizing segments [57,58].
Conclusion
Recent advances have defined some of the interactions involved in malaria parasite invasion into RBCs. Receptors have been identified for four major families of Plasmodium invasion ligands (EBL, Rh, MSP, and TRAP) and the mechanisms of the interactions have been determined for a handful of the family members. Crystal structures of Plasmodium ligands alone and in complex with receptors have contributed greatly to the mechanisms of invasion. For those receptor/ligand pairs that have not been crystallized, mutagenesis studies have further elucidated how these pairs interact. In conclusion, red cell receptor engagement by malaria parasites is far more intricate than initially anticipated. Molecular mechanistic studies of host-parasite interactions remain an active area of research, and are desperately needed not only for the fundamental understanding of receptor biology but also for the rational design of vaccines and therapeutics for malaria.
Keypoints.
Malaria parasites must engage distinct red cell receptors for disease pathogenesis
Multimeric assembly is critical for receptor-ligand engagement
Parasites engage a wide range of red cell receptors to ensure invasion
Identification of novel receptors and ligands inform receptor biology and host-pathogen interactions.
Recent advances have defined the molecular mechanisms that underpin receptor-ligand interactions
Acknowledgments
We thank M. Paing, J. Park, J. Jimah, D. Urusova, and R.D Etheridge for constructive comments on the manuscript.
Financial support and sponsorship
This work was supported by the Burroughs Wellcome Fund (to NHT), NIH/NIAID R56AI080792 (to NHT), and NIH/NIAID R01 AI064478 (to NHT).
Footnotes
Conflicts of Interest
None Declared.
References
- 1.Daniels G. Human blood groups. 3. John Wiley and Sons; 2013. [Google Scholar]
- 2.Yawata Y. Cell membrane : the red blood cell as a model. Weinheim: Wiley-VCH; 2003. [Google Scholar]
- 3.Reid ME, Lomas-Francis C, Olsson ML. The blood group antigen factsbook. 3. Amsterdam: Elsevier/AP; 2012. [Google Scholar]
- 4.Blumenfeld OO, Huang CH. Molecular genetics of glycophorin MNS variants. Transfus Clin Biol. 1997;4:357–365. doi: 10.1016/s1246-7820(97)80041-9. [DOI] [PubMed] [Google Scholar]
- 5.Colin Y. Gerbich blood groups and minor glycophorins of human erythrocytes. Transfus Clin Biol. 1995;2:259–268. doi: 10.1016/s1246-7820(05)80092-8. [DOI] [PubMed] [Google Scholar]
- 6.Iacono KT, Brown AL, Greene MI, Saouaf SJ. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol. 2007;83:283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krych-Goldberg M, Atkinson JP. Structure-function relationships of complement receptor type 1. Immunol Rev. 2001;180:112–122. doi: 10.1034/j.1600-065x.2001.1800110.x. [DOI] [PubMed] [Google Scholar]
- 8.Jongbloets BC, Ramakers GM, Pasterkamp RJ. Semaphorin7A and its receptors: pleiotropic regulators of immune cell function, bone homeostasis, and neural development. Semin Cell Dev Biol. 2013;24:129–138. doi: 10.1016/j.semcdb.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 9.Angelisová P, Drbal K, Cerný J, Hilgert I, Horejsí V. Characterization of the human leukocyte GPI-anchored glycoprotein CDw108 and its relation to other similar molecules. Immunobiology. 1999;200:234–245. doi: 10.1016/s0171-2985(99)80073-4. [DOI] [PubMed] [Google Scholar]
- 10.Goel VK, Li X, Chen H, Liu SC, Chishti AH, Oh SS. Band 3 is a host receptor binding merozoite surface protein 1 during the Plasmodium falciparum invasion of erythrocytes. Proc Natl Acad Sci U S A. 2003;100:5164–5169. doi: 10.1073/pnas.0834959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blackman MJ, Ling IT, Nicholls SC, Holder AA. Proteolytic processing of the Plasmodium falciparum merozoite surface protein-1 produces a membrane-bound fragment containing two epidermal growth factor-like domains. Mol Biochem Parasitol. 1991;49:29–33. doi: 10.1016/0166-6851(91)90127-r. [DOI] [PubMed] [Google Scholar]
- 12*.Baldwin M, Yamodo I, Ranjan R, Li X, Mines G, Marinkovic M, Hanada T, Oh SS, Chishti AH. Human erythrocyte band 3 functions as a receptor for the sialic acid-independent invasion of Plasmodium falciparum. Role of the RhopH3-MSP1 complex. Biochim Biophys Acta. 2014;1843:2855–2870. doi: 10.1016/j.bbamcr.2014.08.008. This is the first study to identify PfRhopH3 binding to Band 3 and PfMSP119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stafford WH, Blackman MJ, Harris A, Shai S, Grainger M, Holder AA. N-terminal amino acid sequence of the Plasmodium falciparum merozoite surface protein-1 polypeptides. Mol Biochem Parasitol. 1994;66:157–160. doi: 10.1016/0166-6851(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 14.Heidrich HG, Miettinen-Baumann A, Eckerskorn C, Lottspeich F. The N-terminal amino acid sequences of the Plasmodium falciparum (FCB1) merozoite surface antigens of 42 and 36 kilodalton, both derived from the 185–195-kilodalton precursor. Mol Biochem Parasitol. 1989;34:147–154. doi: 10.1016/0166-6851(89)90006-6. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Chen H, Oo TH, Daly TM, Bergman LW, Liu SC, Chishti AH, Oh SS. A Co-ligand Complex Anchors Plasmodium falciparum Merozoites to the Erythrocyte Invasion Receptor Band 3. J Biol Chem. 2004;279:5765–5771. doi: 10.1074/jbc.M308716200. [DOI] [PubMed] [Google Scholar]
- 16.Tolia NH, Enemark EJ, Sim BK, Joshua-Tor L. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell. 2005;122:183–193. doi: 10.1016/j.cell.2005.05.033. [DOI] [PubMed] [Google Scholar]
- 17.Orlandi PA, Klotz FW, Haynes JD. A malaria invasion receptor, the 175-kilodalton erythrocyte binding antigen of Plasmodium falciparum recognizes the terminal Neu5Ac(alpha 2–3)Gal- sequences of glycophorin A. J Cell Biol. 1992;116:901–909. doi: 10.1083/jcb.116.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 19*.Paing MM, Tolia NH. Multimeric assembly of host-pathogen adhesion complexes involved in apicomplexan invasion. PLoS Pathog. 2014;10:e1004120. doi: 10.1371/journal.ppat.1004120. This is a review on host-pathogen invasion complexes in P. falciparum, P. vivax, and T. gondii and how multimeric assembly effects invasion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20**.Salinas ND, Paing MM, Tolia NH. Critical Glycosylated Residues in Exon Three of Erythrocyte Glycophorin A Engage Plasmodium falciparum EBA-175 and Define Receptor Specificity. MBio. 2014:5. doi: 10.1128/mBio.01606-14. This study identifies the ligand binding determinants on GpA for PfEBA-175. This study also reports a new method for the production of recombinant GpA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21*.Malpede BM, Tolia NH. Malaria adhesins: structure and function. Cell Microbiol. 2014;16:621–631. doi: 10.1111/cmi.12276. This is a review on how structure of malaria adhesion proteins effects the function and binding to host receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salinas ND, Tolia NH. A quantitative assay for binding and inhibition of Plasmodium falciparum Erythrocyte Binding Antigen 175 reveals high affinity binding depends on both DBL domains. Protein Expr Purif. 2014;95:188–194. doi: 10.1016/j.pep.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23*.Baldwin MR, Li X, Hanada T, Liu SC, Chishti AH. Merozoite surface protein 1 recognition of host glycophorin A mediates malaria parasite invasion of red blood cells. Blood. 2015;125:2704–2711. doi: 10.1182/blood-2014-11-611707. This is the first study to identify a Plasmodium ligand other than PfEBA-175 that binds to GpA and increases the number of receptors that the PfMSP1 complex can bind. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Marinkovic M, Russo C, McKnight CJ, Coetzer TL, Chishti AH. Identification of a specific region of Plasmodium falciparum EBL-1 that binds to host receptor glycophorin B and inhibits merozoite invasion in human red blood cells. Mol Biochem Parasitol. 2012;183:23–31. doi: 10.1016/j.molbiopara.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mayer DC, Cofie J, Jiang L, Hartl DL, Tracy E, Kabat J, Mendoza LH, Miller LH. Glycophorin B is the erythrocyte receptor of Plasmodium falciparum erythrocyte-binding ligand, EBL-1. Proc Natl Acad Sci U S A. 2009;106:5348–5352. doi: 10.1073/pnas.0900878106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26**.Malpede BM, Lin DH, Tolia NH. Molecular basis for sialic acid-dependent receptor recognition by the Plasmodium falciparum invasion protein erythrocyte-binding antigen-140/BAEBL. J Biol Chem. 2013;288:12406–12415. doi: 10.1074/jbc.M113.450643. This study reports the structure of PfEBA-140 in complex with GpC glycans and identifies new regions for receptor binding distinct from other family members. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobo CA, Rodriguez M, Reid M, Lustigman S. Glycophorin C is the receptor for the Plasmodium falciparum erythrocyte binding ligand PfEBP-2 (baebl) Blood. 2003;101:4628–4631. doi: 10.1182/blood-2002-10-3076. [DOI] [PubMed] [Google Scholar]
- 28.Mayer DC, Jiang L, Achur RN, Kakizaki I, Gowda DC, Miller LH. The glycophorin C N-linked glycan is a critical component of the ligand for the Plasmodium falciparum erythrocyte receptor BAEBL. Proc Natl Acad Sci U S A. 2006;103:2358–2362. doi: 10.1073/pnas.0510648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin DH, Malpede BM, Batchelor JD, Tolia NH. Crystal and solution structures of Plasmodium falciparum erythrocyte-binding antigen 140 reveal determinants of receptor specificity during erythrocyte invasion. J Biol Chem. 2012;287:36830–36836. doi: 10.1074/jbc.M112.409276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayer DC, Mu JB, Feng X, Su XZ, Miller LH. Polymorphism in a Plasmodium falciparum erythrocyte-binding ligand changes its receptor specificity. J Exp Med. 2002;196:1523–1528. doi: 10.1084/jem.20020750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Batchelor JD, Malpede BM, Omattage NS, DeKoster GT, Henzler-Wildman KA, Tolia NH. Red blood cell invasion by Plasmodium vivax: structural basis for DBP engagement of DARC. PLoS Pathog. 2014;10:e1003869. doi: 10.1371/journal.ppat.1003869. This study reports the structure of PvDBP bound to the receptor DARC, and elucidates a multistep mechanism for receptor binding. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chitnis CE, Chaudhuri A, Horuk R, Pogo AO, Miller LH. The domain on the Duffy blood group antigen for binding Plasmodium vivax and P. knowlesi malarial parasites to erythrocytes. J Exp Med. 1996;184:1531–1536. doi: 10.1084/jem.184.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choe H, Moore MJ, Owens CM, Wright PL, Vasilieva N, Li W, Singh AP, Shakri R, Chitnis CE, Farzan M. Sulphated tyrosines mediate association of chemokines and Plasmodium vivax Duffy binding protein with the Duffy antigen/receptor for chemokines (DARC) Mol Microbiol. 2005;55:1413–1422. doi: 10.1111/j.1365-2958.2004.04478.x. [DOI] [PubMed] [Google Scholar]
- 34.Batchelor JD, Zahm JA, Tolia NH. Dimerization of Plasmodium vivax DBP is induced upon receptor binding and drives recognition of DARC. Nat Struct Mol Biol. 2011;18:908–914. doi: 10.1038/nsmb.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh SK, Hora R, Belrhali H, Chitnis CE, Sharma A. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature. 2006;439:741–744. doi: 10.1038/nature04443. [DOI] [PubMed] [Google Scholar]
- 36**.Wright KE, Hjerrild KA, Bartlett J, Douglas AD, Jin J, Brown RE, Illingworth JJ, Ashfield R, Clemmensen SB, de Jongh WA, et al. Structure of malaria invasion protein RH5 with erythrocyte basigin and blocking antibodies. Nature. 2014;515:427–430. doi: 10.1038/nature13715. This is one of two studies that reports the crystal structure of PfRh5. In addition, this study also provides the crystal structure of PfRh5 in complex with its receptor BSG and two inhibitory antibodies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crosnier C, Bustamante LY, Bartholdson SJ, Bei AK, Theron M, Uchikawa M, Mboup S, Ndir O, Kwiatkowski DP, Duraisingh MT, et al. Basigin is a receptor essential for erythrocyte invasion by Plasmodium falciparum. Nature. 2011;480:534–537. doi: 10.1038/nature10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Chen L, Xu Y, Healer J, Thompson JK, Smith BJ, Lawrence MC, Cowman AF. Crystal structure of PfRh5, an essential P. falciparum ligand for invasion of human erythrocytes. Elife. 2014:3. doi: 10.7554/eLife.04187. This is one of two studies to report the crystal structure of PfRh5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Reddy KS, Amlabu E, Pandey AK, Mitra P, Chauhan VS, Gaur D. Multiprotein complex between the GPI-anchored CyRPA with PfRH5 and PfRipr is crucial for Plasmodium falciparum erythrocyte invasion. Proc Natl Acad Sci U S A. 2015;112:1179–1184. doi: 10.1073/pnas.1415466112. This study identifies the membrane anchoring protein, PfCyRPA, for the PfRh5 complex. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen L, Lopaticki S, Riglar DT, Dekiwadia C, Uboldi AD, Tham WH, O’Neill MT, Richard D, Baum J, Ralph SA, et al. An EGF-like protein forms a complex with PfRh5 and is required for invasion of human erythrocytes by Plasmodium falciparum. PLoS Pathog. 2011;7:e1002199. doi: 10.1371/journal.ppat.1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tham WH, Wilson DW, Lopaticki S, Schmidt CQ, Tetteh-Quarcoo PB, Barlow PN, Richard D, Corbin JE, Beeson JG, Cowman AF. Complement receptor 1 is the host erythrocyte receptor for Plasmodium falciparum PfRh4 invasion ligand. Proc Natl Acad Sci U S A. 2010;107:17327–17332. doi: 10.1073/pnas.1008151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tham WH, Schmidt CQ, Hauhart RE, Guariento M, Tetteh-Quarcoo PB, Lopaticki S, Atkinson JP, Barlow PN, Cowman AF. Plasmodium falciparum uses a key functional site in complement receptor type-1 for invasion of human erythrocytes. Blood. 2011;118:1923–1933. doi: 10.1182/blood-2011-03-341305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Park HJ, Guariento M, Maciejewski M, Hauhart R, Tham WH, Cowman AF, Schmidt CQ, Mertens HD, Liszewski MK, Hourcade DE, et al. Using mutagenesis and structural biology to map the binding site for the Plasmodium falciparum merozoite protein PfRh4 on the human immune adherence receptor. J Biol Chem. 2014;289:450–463. doi: 10.1074/jbc.M113.520346. This study identifies the minimal binding domains on CR1 for PfRh4 and how it differs from other highly related domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartholdson SJ, Bustamante LY, Crosnier C, Johnson S, Lea S, Rayner JC, Wright GJ. Semaphorin-7A is an erythrocyte receptor for P. falciparum merozoite-specific TRAP homolog, MTRAP. PLoS Pathog. 2012;8:e1003031. doi: 10.1371/journal.ppat.1003031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uchime O, Herrera R, Reiter K, Kotova S, Shimp RL, Miura K, Jones D, Lebowitz J, Ambroggio X, Hurt DE, et al. Analysis of the conformation and function of the Plasmodium falciparum merozoite proteins MTRAP and PTRAMP. Eukaryot Cell. 2012;11:615–625. doi: 10.1128/EC.00039-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46**.Egan ES, Jiang RH, Moechtar MA, Barteneva NS, Weekes MP, Nobre LV, Gygi SP, Paulo JA, Frantzreb C, Tani Y, et al. Malaria. A forward genetic screen identifies erythrocyte CD55 as essential for Plasmodium falciparum invasion. Science. 2015;348:711–714. doi: 10.1126/science.aaa3526. This is the first study to utilize a forward genetic screen in erythrocytes to identify erythrocyte receptors for Plasmodium falciparum. The screen conducted identified CD55 as a novel receptor for P. falciparum. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47*.Bhalla K, Chugh M, Mehrotra S, Rathore S, Tousif S, Prakash Dwivedi V, Prakash P, Kumar Samuchiwal S, Kumar S, Kumar Singh D, et al. Host ICAMs play a role in cell invasion by Mycobacterium tuberculosis and Plasmodium falciparum. Nat Commun. 2015;6:6049. doi: 10.1038/ncomms7049. This is the first study to identify a role for an ICAM family member in invasion by P. falicparum and not cytoadherance. [DOI] [PubMed] [Google Scholar]
- 48.Gilberger TW, Thompson JK, Triglia T, Good RT, Duraisingh MT, Cowman AF. A novel erythrocyte binding antigen-175 paralogue from Plasmodium falciparum defines a new trypsin-resistant receptor on human erythrocytes. J Biol Chem. 2003;278:14480–14486. doi: 10.1074/jbc.M211446200. [DOI] [PubMed] [Google Scholar]
- 49*.Lin CS, Uboldi AD, Marapana D, Czabotar PE, Epp C, Bujard H, Taylor NL, Perugini MA, Hodder AN, Cowman AF. The merozoite surface protein 1 complex is a platform for binding to human erythrocytes by Plasmodium falciparum. J Biol Chem. 2014;289:25655–25669. doi: 10.1074/jbc.M114.586495. This study identifies PfMSPDBL-1 and PfMSPDBL-2 as additional Plasmodium falciparum invasion ligands. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sahar T, Reddy KS, Bharadwaj M, Pandey AK, Singh S, Chitnis CE, Gaur D. Plasmodium falciparum reticulocyte binding-like homologue protein 2 (PfRH2) is a key adhesive molecule involved in erythrocyte invasion. PLoS One. 2011;6:e17102. doi: 10.1371/journal.pone.0017102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siddiqui FA, Dhawan S, Singh S, Singh B, Gupta P, Pandey A, Mohmmed A, Gaur D, Chitnis CE. A thrombospondin structural repeat containing rhoptry protein from Plasmodium falciparum mediates erythrocyte invasion. Cell Microbiol. 2013;15:1341–1356. doi: 10.1111/cmi.12118. [DOI] [PubMed] [Google Scholar]
- 52.Pisano A, Redmond JW, Williams KL, Gooley AA. Glycosylation sites identified by solid-phase Edman degradation: O-linked glycosylation motifs on human glycophorin A. Glycobiology. 1993;3:429–435. doi: 10.1093/glycob/3.5.429. [DOI] [PubMed] [Google Scholar]
- 53.Maier AG, Baum J, Smith B, Conway DJ, Cowman AF. Polymorphisms in erythrocyte binding antigens 140 and 181 affect function and binding but not receptor specificity in Plasmodium falciparum. Infect Immun. 2009;77:1689–1699. doi: 10.1128/IAI.01331-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 55.King CL, Adams JH, Xianli J, Grimberg BT, McHenry AM, Greenberg LJ, Siddiqui A, Howes RE, da Silva-Nunes M, Ferreira MU, et al. Fy(a)/Fy(b) antigen polymorphism in human erythrocyte Duffy antigen affects susceptibility to Plasmodium vivax malaria. Proc Natl Acad Sci U S A. 2011;108:20113–20118. doi: 10.1073/pnas.1109621108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen E, Paing MM, Salinas N, Sim BK, Tolia NH. Structural and functional basis for inhibition of erythrocyte invasion by antibodies that target Plasmodium falciparum EBA-175. PLoS Pathog. 2013;9:e1003390. doi: 10.1371/journal.ppat.1003390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Chen E, Salinas ND, Ntumngia FB, Adams JH, Tolia NH. Structural analysis of the synthetic Duffy Binding Protein (DBP) antigen DEKnull relevant for Plasmodium vivax malaria vaccine design. PLoS Negl Trop Dis. 2015;9:e0003644. doi: 10.1371/journal.pntd.0003644. This study structurally anlayzes an parasite antigen engineered for vaccine development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58*.Ntumngia FB, Barnes SJ, McHenry AM, George MT, Schloegel J, Adams JH. Immunogenicity of a synthetic vaccine based on Plasmodium vivax Duffy binding protein region II. Clin Vaccine Immunol. 2014;21:1215–1223. doi: 10.1128/CVI.00205-14. This study described the engineering of parasite antigens for vaccine development. [DOI] [PMC free article] [PubMed] [Google Scholar]