Abstract
Myosin VI functions in endocytosis and cell motility. Alternative splicing of myosin VI mRNA generates two distinct isoform types, myosin VIshort and myosin VIlong, which differ in the C-terminal region. Their physiological and pathological role remains unknown. Here we identified an isoform-specific regulatory helix, named α2-linker that defines specific conformations and hence determines the target selectivity of human myosin VI. The presence of the α2-linker structurally defines a novel clathrin-binding domain that is unique to myosin VIlong and masks the known RRL interaction motif. This finding is relevant to ovarian cancer, where alternative myosin VI splicing is aberrantly regulated, and exon skipping dictates cell addiction to myosin VIshort for tumor cell migration. The RRL interactor optineurin contributes to this process by selectively binding myosin VIshort. Thus the α2-linker acts like a molecular switch that assigns myosin VI to distinct endocytic (myosin VIlong) or migratory (myosin VIshort) functional roles.
Molecular motors exert critical roles in virtually all cell processes. Myosin VI belongs to a superfamily of more than 20 different classes of actin-based motor proteins1, 2 that hydrolyze ATP for mechanical work and movement of cargos along actin tracks. The ability to walk towards the minus end of actin filaments makes myosin VI unique among the myosin family3. This unusual property appears to bestow on myosin VI a number of specific cellular functions, such as clathrin-mediated endocytosis and vesicular transport4, 5. Myosin VI was originally discovered in Drosophila, where it participates in cell migration during ovary development6, spermatogenesis7, 8 and neuroblast asymmetric division9, 10. In mammalian cells, myosin VI localizes to the Golgi complex, membrane ruffles at the leading edges of migrating cells, clathrin coated pits at plasma membranes, APPL1-positive early endosomes and autophagosomes (reviewed in11). Detailed mechanisms underlying the function of myosin VI in these diverse processes are currently unclear.
At a molecular scale, myosin VI consists of two functional domains (Fig. 1a), a motor–IQ domain sufficient for pointed-end (minus-end) directed movement, and a tail domain, which can be further divided into a long helical region, reported as a coiled-coil domain, and a C-terminal globular region, known as a cargo-binding domain (CBD). A MIU (Motif Interacting with Ubiquitin)12 is located at the boundaries. The CBD contains two myosin VI ligand interaction surfaces, the “RRL” and “WWY” motifs11. The RRL triplet is responsible for myosin VI binding to endocytic adaptors, including GAIP-interacting protein C-terminus (GIPC)13, 14, and autophagy receptors, such as optineurin15, Nuclear Dot Protein 52 (NDP52)16, and Traf6-binding protein (T6BP)16.
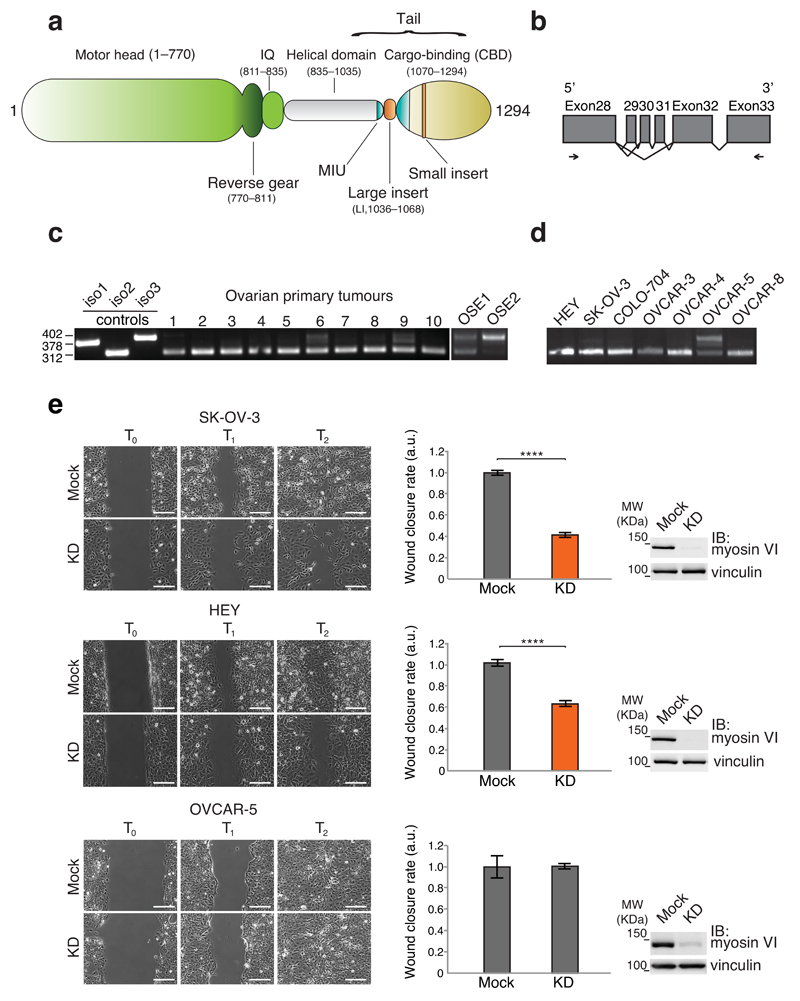
Figure 1. Myosin VIshort is selectively expressed in ovarian cancer cells and is critical for cell migration.
(a) Myosin VI domain structure. (b) Schematic representation of the region amplified by RT-PCR. Coding exons are represented by grey boxes. Alternative splicing events are depicted. Oligos used for the PCR mapped in Exon 28 and Exon 33 and are indicated by arrows. (c,d) RT-PCR from cDNA prepared from the indicated primary cells (c) and cell lines (d). Controls are from plasmids carrying the tails of the different isoforms. (e) Wound healing assay. The indicated cell lines were knocked down for myosin VI (KD) or mock treated. Left panel, sample images: T0 first frame, T1 and T2 arbitrary points identical for control and KD of the same cell line. Scale bars, 200μm. Central panel: quantification of the wound closure speed relative to control. Error bars, s.d. (n=6 movies for SK-OV-3 and HEY, n=8 movies for OVCAR-5. Data are from three independent experiments). **** P<0.0001; ns, no significant difference by two-tailed T-test. Right panel: anti-myosin VI immunoblotting (IB) performed at T0.
An intrinsic level of complexity in deciphering the functions of myosin VI comes from the presence of different isoforms. Two regions are alternatively spliced within the tail (Fig. 1a), leading to an insertion between the helical domain and the CBD (large insert, LI)17, 18, as well as additional residues at the C-terminal (small insert, SI)19. Although myosin VI is widely expressed in most tissues, isoforms containing the LI are specifically found in polarized epithelial cells with well-developed apical microvilli17, 20. How the presence of LI in the tail influences the functions and intracellular targeting of myosin VI is not known. Up to now, isoforms lacking the LI present no specific expression and localization but are required for polarized transport of tyrosine motif containing basolateral membrane proteins20 and for maintaining an active pool of secretory granules near the plasma membrane of neurosecretory cells19.
The role of myosin VI in cell migration has been linked to ovarian21 and prostate cancers22–24, where myosin VI overexpression correlates with clinically aggressive behavior. Notably, silencing of myosin VI expression decreases the migratory potential and the malignant properties of ovarian21 and prostate cancer cell lines21, 22.
In this study, we set out to investigate the major splicing event occurring in myosin VI from a molecular and a functional perspective. By analyzing the interactomes and the conformational structure of myosin VI isoforms, we provided mechanistic insights into how myosin VI function becomes pathological in human cancers and demonstrated the importance of an isoform-specific helix in assigning myosin VI to distinct functional roles.
Results
Selective expression of myosin VIshort in ovarian cancer
Several studies report overexpression of myosin VI in prostate22–24 and ovarian cancers21, which positively correlates with their grade and metastatic potential. No information is available concerning specificity of isoform expression. To shed light on this issue, we analyzed human myosin VI transcripts in detail. The gene consists of 36 coding exons, three of which (exons 29, 30 and 31) generate the LI (Supplementary Fig. 1a). We focused on the main myosin VI isoforms expressed in human cell lines17, 18, 20, namely isoforms 1 and 3, which have different LI lengths (23- or 32-residues long, respectively, due to the presence or absence of exon 29, Supplementary Fig. 1a), and isoform 2, which lacks the LI entirely (exons 29, 30 and 31 are absent, Supplementary Fig. 1a). Interestingly, these alternative exons are conserved throughout evolution (Supplementary Fig. 1b), suggesting a relevant function for the regulated splicing in myosin VI.
To gain insight into possible differential expression of myosin VI isoforms in tumors, we analyzed their expression by RT-PCR using primers positioned in exon 28 and 33 (Fig. 1b). The PCR reaction was able to discriminate between the different isoforms, as demonstrated in the control lanes (Fig. 1c). RT-PCR was performed using RNA extracted from high-grade primary ovarian cancer cells cultured in vitro for two passages. In contrast to their normal counterpart (ovarian surface epithelium, OSE), ten out of ten tumors expressed almost exclusively isoform 2, suggesting a positive selection for this isoform in tumor progression and metastasis (Fig. 1c). We then examined a number of ovarian cancer cell lines for myosin VI isoform expression in order to identify possible cellular models for in vitro studies. OVCAR-5 is unique among them as it expresses all myosin VI isoforms, while the remaining 6 cell lines tested selectively expressed isoform 2 (Fig. 1d).
Knocking-down myosin VI substantially inhibits migration in vitro and reduces dissemination of ovarian tumor cells propagated in nude mice21. To gain insight into the possible advantages derived from overexpression of myosin VI isoform 2 in cancer cells, we looked at cell migration as an important feature of cancer metastasis. We analyzed the effect of myosin VI depletion in SK-OV-3, HEY and OVCAR-5 cell lines in wound-healing assay. Depletion of myosin VI was complete and comparable in all cell lines (Fig. 1e). In the absence of myosin VI, SK-OV-3 and HEY cells were severely impaired in their ability to migrate and to close wounds, whereas the migration efficiency of OVCAR-5 cells was unaffected (Fig. 1e and Supplementary movies 1-3). Comparable results were obtained with different oligos (Supplementary Fig. 2a,b). Hence, only ovarian cancer cells that selectively express myosin VI isoform 2 were deficient in cell migration upon myosin VI depletion.
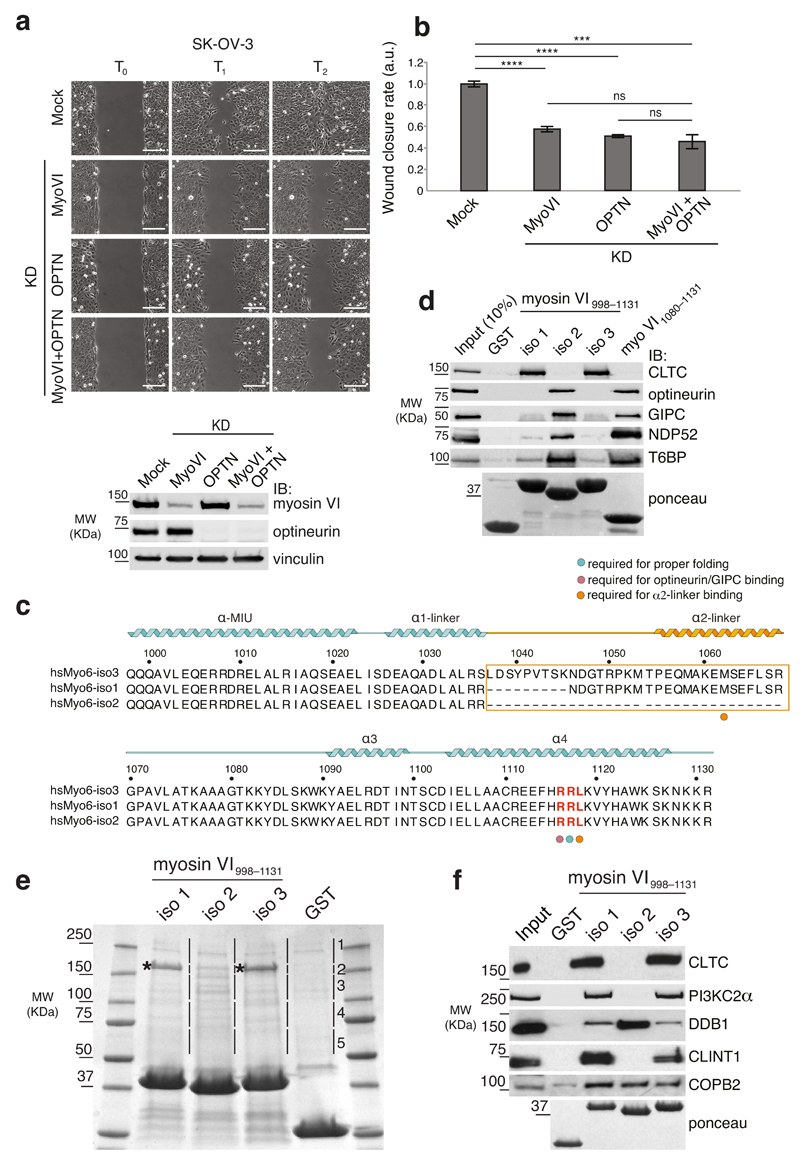
Next, we looked at possible partners of myosin VI in this process. In A549 cells, ablation of myosin VI or its interactor optineurin15 inhibits polarized delivery of the epidermal growth factor receptor into the leading edge and leads to profound defects in lamellipodia formation25. This prompted us to test the effect of optineurin depletion in our model system. Optineurin knockdown reduced the migration ability of SK-OV-3 but did not further decrease cell speed in myosin VI-depleted cells (Fig. 2a,b and Supplementary movie 4). The lack of additive effects suggests that the two proteins are acting in the same axis.
Figure 2. The RRL binder optineurin affects cell migration and selectively binds myosin VIshort.
(a) Wound healing assay. SK-OV-3 cells were mock treated or KD for myosin VI, optineurin, or for both. Left panel, sample images: T0 first frame, T1 and T2 arbitrary points identical for control and KD cells. Scale bars, 200μm. Bottom panel: anti-myosin VI and anti-optineurin IBs performed at T0. (b) Quantification of the wound closure speed relative to control. Error bars, s.d. (n=36 movies for mock and myosin VI KD, n=39 movies for optineurin and n=25 for double KD. Data are from three independent experiments). **** P<0.0001; *** P<0.001; ns, no significant difference by two-tailed T-test. (c) Amino acid sequence alignment covering the region amplified by RT-PCR. The RRL motif is highlighted in red. Secondary structure elements, predicted using http://www.predictprotein.org and confirmed by structural data are depicted above the sequence. The large insert (LI) corresponds to the linker region that varies between the short (iso 2) and the long (iso 1,3) isoforms, and is boxed in orange. Numbering on top of the sequence alignment refers to isoform 3. (d) GST pull-down assay using the indicated myosin VI998–1131 constructs and lysates from HEK293T cells transfected with GFP-optineurin, Flag-NDP52, His-GIPC, Flag-T6BP1 constructs. IB was performed with the respective anti-TAG antibodies. Ponceau as indicated. (e) Colloidal-blue stained SDS-PAGE of the pull-down performed with GST-myosin VI998–1131 of isoform variants. Numbering and lines refer to the slices cut for the mass spectrometry identification. Asterisks indicate clathrin heavy chain bands. (f) Validation analysis performed as in d. using lysate from HEK293T (clathrin and PI3KC2α), or from HEK293T transfected with tagged constructs.
Our functional data suggested that optineurin may specifically interact with myosin VI isoform 2. Optineurin binding requires the RRL motif15 of myosin VI which lies outside of the splicing region (Fig. 2c). Nonetheless, we tested its binding to the different isoforms. A sequence alignment of the utilized region of myosin VI (myosin VI998–1131, amino acid numbering from isoform 3) together with its features is shown in Figure 2c. As shown in Figure 2d, optineurin binds specifically to isoform 2 and to a shorter construct lacking the alternatively spliced region (1080–1131). This prompted us to test for a possible preference of the other known RRL interactors; GIPC13, NDP52 (ref.16) and T6BP16. Surprisingly, all three interactors showed a clear preference for isoforms 2, which lacks the large insert (Fig. 2c).
Myosin VI isoforms display different interaction partners
Distinct functions for myosin VI isoforms are described but no molecular rationale for such differences is provided17, 20. The results obtained with the RRL interactors prompted us to hypothesize that the various myosin VI isoforms may have different interactomes. Thus, we performed a mass spectrometry-based proteomic screen, aimed at comparing the interaction profiles of the various isoforms. GST-tagged myosin VI998–1131 constructs were used in pull-down assays with HEK293T cellular lysates (Fig. 2e). A label-free quantitative experiment was performed, and resulting data were analysed by MaxQuant software. Inspection of the gel (Fig. 2e), confirmed by data presented in Table 1, indicated that isoforms 1 and 3 have almost identical binding partners. Most of these binding partners are implicated in endocytosis and vesicular trafficking and are not shared by isoform 2. Isoform 2-specific interactors were more heterogeneous and comprised nuclear factors, an E3 ligase (DDB1–DCAF1) and few GEFs, among which the previously identified myosin VI binding partner DOCK7 (ref.26). Few interactors common to all three isoforms (e.g. COPB2) were also identified. Table 1 reported examples of the top hits for each class with their total peptide counts. None of the previously characterized RRL interactors were present. Since we cut the gel higher than GST fusion proteins, GIPC was automatically excluded from the analysis whereas NDP52 and T6BP were identified with a single peptide and thus filtered out. Supplementary Table 1 reported the complete list of proteins that differentially interact with isoform 1, isoform 2 and 3 vs GST control confirmed by additional proteomics experiments.
Table 1. List of the top hits of interactors identified by label-free quantitative mass spectrometry.
Gene name, description and total peptide count (TPC) is reported for each protein. *The same total peptide count was reported for the control (GST alone), see also Supplementary Table 1.
| Gene name | Iso1 (TPC) | Iso2 (TPC) | Iso3 (TPC) | |
|---|---|---|---|---|
| CLTC | clathrin, heavy chain (CHC) | 1050 | *153 | 1068 |
| SEC16A | SEC16 homolog A (S. cerevisiae) | 175 | 0 | 145 |
| AP2B1 | adaptor-related protein complex 2, beta 1 subunit | 91 | 0 | 107 |
| AP2A2 | adaptor-related protein complex 2, alpha 2 subunit | 87 | 0 | 89 |
| AP2A1 | adaptor-related protein complex 2, alpha 1 subunit | 85 | 0 | 90 |
| AP1G1 | adaptor-related protein complex 1, gamma 1 subunit | 28 | 0 | 32 |
| AP2M1 | AP-2 complex subunit mu | 35 | 0 | 51 |
| CLINT1 | clathrin interactor 1 (EPSIN4) | 138 | 0 | 146 |
| PIK3C2A | Class 2 phosphatidylinositol-4-phosphate 3-kinase | 73 | 0 | 90 |
| PICALM | Phosphatidylinositol-binding clathrin assembly protein | 15 | 0 | 19 |
| EPS15 | Epidermal growth factor receptor substrate 15 | 6 | 0 | 5 |
| REPS | RalBP1-associated Eps domain-containing protein 1 | 10 | 0 | 5 |
| COPB2 | Coatomer subunit beta' | 45 | 50 | 57 |
| DDB1 | DNA damage-binding protein 1 | 137 | 251 | 123 |
| EPRS | Bifunctional glutamate, proline, tRNA ligase | 206 | 686 | 183 |
| VPRBP | Vpr (HIV-1) binding protein (DCAF) | 0 | 140 | 0 |
| DCTN1 | dynactin 1 | 0 | 105 | 0 |
| GTPBP4 | GTP binding protein 4 | 5 | 44 | 0 |
| ARHGEF12 | Rho guanine nucleotide exchange factor 12 | 6 | 64 | 0 |
| YTHDC2 | Probable ATP-dependent RNA helicase | 4 | 37 | 2 |
| DCTN2 | Dynactin subunit 2 | 2 | 37 | 0 |
| DOCK7 | Dedicator of cytokinesis protein 7 | 2 | 32 | 4 |
Experimental validation confirmed the most abundant identified interactors (5 out of 5 tested) and their specificity (Fig. 2f). Since isoforms 1 and 3 showed no difference in terms of interaction networks, we referred to them collectively as myosin VIlong, while using myosin VIshort to indicate isoform 2.
Structure of Myosin VIlong clathrin-binding domain
The interactome analysis identified clathrin heavy chain (CLTC) as a major and specific binding partner of myosin VIlong (Table 1 and Fig. 2f). It is well-known that myosin VI and clathrin form a ternary complex with the Dab-2 adaptor27, 28. However, neither the WWY motif of myosin VI (ref.27), or the larger surfaces implicated in binding to Dab-2 (ref.28) were present in the constructs used for the proteomic screening. Thus, we inferred that myosin VI could also interact with clathrin directly.
To map this novel interaction surface, we generated a panel of GST-tagged deletion constructs and tested their interaction with endogenous clathrin starting from isoform 3 myosin VI998–1131, which was used for the proteomic screen (Supplementary Fig. 3a-c). Regions deleted from the C-terminal end did not show any clathrin binding (Supplementary Fig. 3a), whereas fragments deleted at the N-terminal end retained clathrin interaction (Supplementary Fig. 3b). The minimal binding region appeared to be 1055–1131, which includes the isoform-specific LI region (Fig. 2c) and the region directly following it. Further N-terminal shortening, or even small internal deletion disrupted binding (Supplementary Fig. 3b,c).
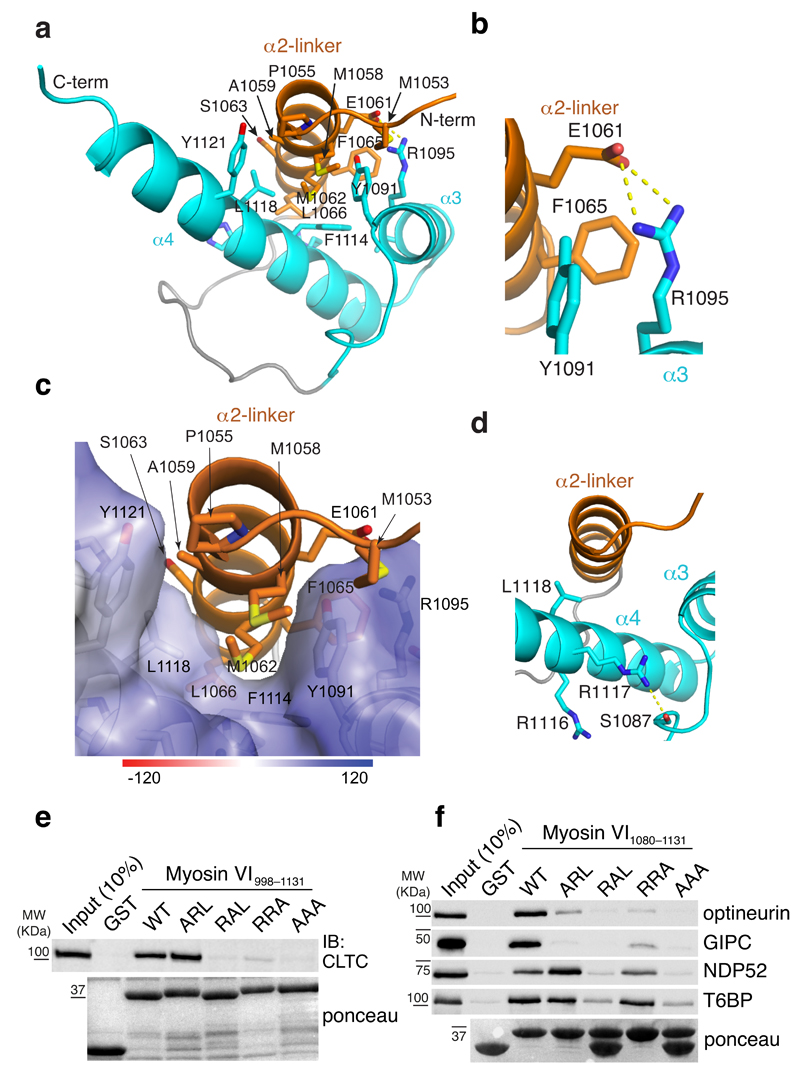
We used NMR techniques to solve the structure of the clathrin binding surface (Fig. 3a). The twenty calculated NMR structures with lowest energy from 100 starting structures converged well (stereoview showed in Supplementary Fig. 3d) with a backbone root mean square deviation (RMSD) of 0.18 Å, following the exclusion of an 11-amino acid flexible loop that directly follows the LI (Supplementary Fig. 3d and Table 2). In the isoform-specific LI region, residues 1055–1068 form an amphipathic α-helix (α2-linker, Fig. 2c and Supplementary Fig. 3e) that packs against two other helices present in all isoforms (Fig. 3a), named α3 and α4. Interaction between the isoform-specific α2-linker and the common helices α3 and α4 (α3/α4) is defined by 106 NOE interactions, as exemplified for L1118 (Supplementary Fig. 3f). A hydrophobic surface of the α2-linker formed by M1058, M1062, and L1066 inserts into a cleft formed by the α3/α4 interface and interacts with hydrophobic residues Y1091, F1114, L1118, and Y1121 (Fig. 3a and 3c). In addition, a salt bridge is formed between α2-linker E1061 and α3 R1095 (Fig. 3b).
Figure 3. Clathrin interacts specifically with myosin VIlong.
(a) Ribbon representation of the myosin VIlong (R1050–R1131) structure. The isoform-specific α2-linker is in orange while the α3 and α4 helices are displayed in blue. Hydrophobic interactions between the three helices are highlighted. (b) Expanded view of a that illustrates the salt bridge formed between α2-linker E1061 and α4 R1095. (c) Expanded view of a with α2-linker displayed on an electrostatic surface representation of the α3 and α4 helices. (d) Expanded view of a that illustrates the position of the RRL motif residues. L1118 is directed towards α2-linker while R1117 forms a hydrogen bond with S1087. (e,f) GST pull-down assay with myosin VI998–1131 (e) and myosin VI1080–1131 (f) iso3 constructs carrying the indicated mutations in the RRL motif. IB and ponceau as indicated. Except for clathrin (e), tagged constructs were transiently transfected in HEK293T cells and IB were performed with anti-tag antibody.
Table 2. Structural statistics for myosin VI1050–1131.
| Protein | |
|---|---|
| NMR distance and dihedral constraints | |
| Distance constraints | |
| Total NOE | 1846 |
| Intraresidue | 483 |
| Inter-residue | |
| Sequential (|i – j| = 1) | 537 |
| Medium range (2 ≤ |i – j| ≤ 4) | 566 |
| Long range (|i – j| ≥ 5) | 260 |
| Intermolecular | |
| Hydrogen bonds | 42 |
| Total dihedral-angle restraints | |
| ϕ | 51 |
| ψ | 51 |
| Structure statistics | |
| Violations (mean ± s.d.) | |
| Distance constraints (Å) | 0.0077±0.0010 |
| Dihedral-angle constraints (°) | 0.12±0.041 |
| Max. dihedral-angle violation (°) | 1.13 |
| Max. distance-constraint violation (Å) | 0.22 |
| Deviations from idealized geometry | |
| Bond lengths (Å) | 0.0016±0.000073 |
| Bond angles (°) | 0.39±0.0042 |
| Impropers (°) | 0.23±0.0067 |
| Average pairwise r.m.s. deviation (Å)a | |
| Heavy | 0.68±0.06 |
| Backbone | 0.18±0.05 |
Values were calculated for residues T1054-R1068 and Y1084-S1126 from the twenty lowest energy structures.
Mutually exclusive association of clathrin and RRL binders
The RRL motif is embedded in helix α4 of the clathrin-binding domain (Fig. 2c). As visible in the structure, two residues of the RRL motif were not accessible for binding with interactors (Fig. 3d), thus explaining their specificity for binding only myosin VIshort (Fig. 2d). L1118 is buried in the interface between α2-linker and α3/α4 (Fig. 3c and 3d) while R1117 formed a salt bridge with S1087 of the α3 (Fig. 3d). We generated single point mutations of the RRL motif as well as the triple AAA mutation previously used to validate interactions with known myosin VI binders13, 16 in the context of a myosin VIlong fragment. As shown in Figure 3e, R1117A (RAL), L1118A (RRA) and the triple AAA mutation abrogated the ability of myosin VI998–1131 to pull-down clathrin from cell lysates, whereas the R1116A (ARL) mutant behaved as wild type. Circular dichroism data of the R1117A-containing fragment (Supplementary Fig. 3g) revealed that R1117 is required for structural integrity of this myosin VI region.
The structural data provided an explanation for the myosin VIshort selective interaction of RRL binders (Fig. 2d) but raised an issue related the triple AAA mutation. Thus, we probed the ability of all RRL interactors to interact with a myosin VI fragment carrying mutations in the RRL motif. We used a minimal fragment (G1080–R1131) that includes α3/α4 but lacks the α2-linker and retains binding (Fig. 2d). As for clathrin, and consistent with the structural data, the destabilizing R1117A mutation either alone or in the context of the AAA mutant impaired binding to partners (Fig. 3f). Analyses performed with the other single point mutants demonstrated that R1116 and L1118 are critical for GIPC and optineurin binding, whereas T6BP and NDP52 associate with myosin VI1080–1131 in an RRL-independent fashion (Fig. 3f).
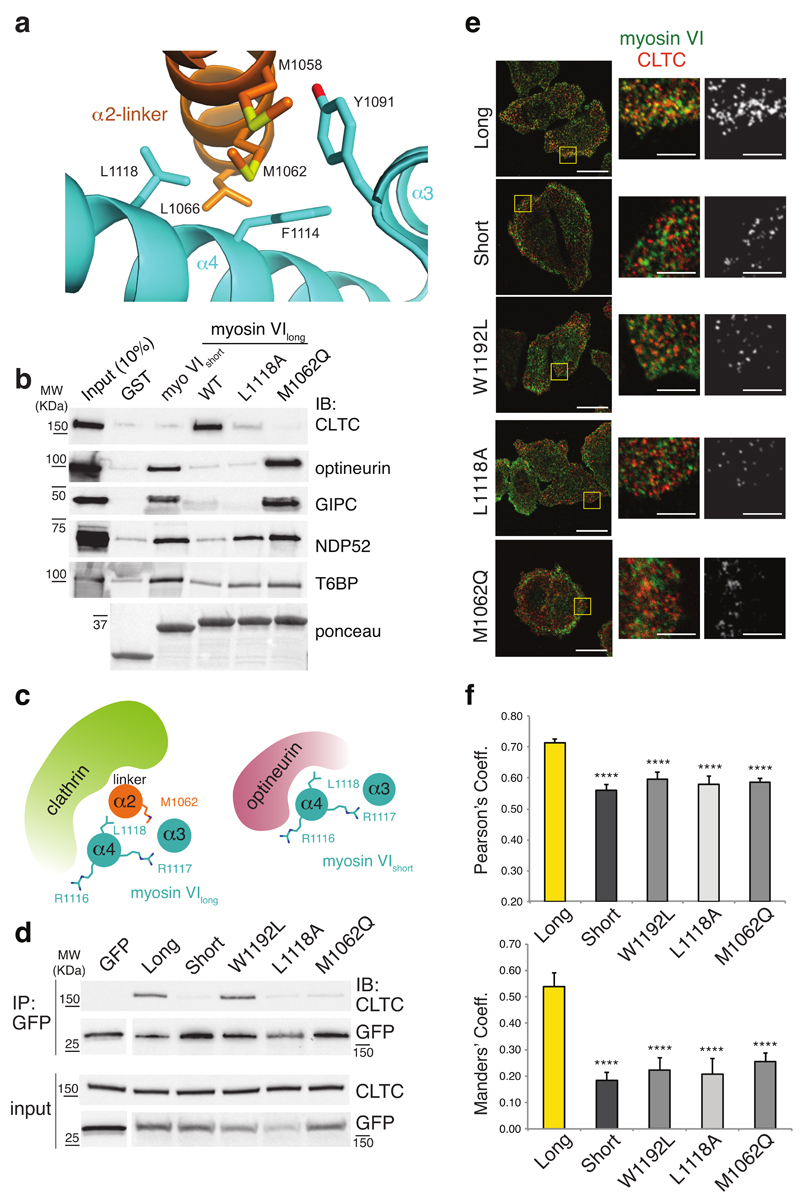
Taken together, these data implied that packing of the α2-linker against the α3/α4 helices in myosin VIlong determined: i) its exquisite clathrin-binding ability and ii) its impairment in RRL-mediated binding (i.e. optineurin). We endeavored to validate the functional impact of the myosin VIlong conformation by testing point mutation(s) capable of disrupting interaction between α3/α4 and the α2-linker. We replaced M1062, which is sandwiched between α3 and α4 (Fig. 4a), with glutamine, and evaluated this variant together with the previously characterized L1118A mutant (Fig. 3e), for binding to RRL-specific interactors by pull-down assays. As shown in Fig. 4b, M1062Q was impaired in clathrin binding, similarly to L1118A. Strikingly, GIPC and optineurin gained binding to M1062Q but not to L1118A (Fig. 4b), consistent with the evidence that L1118 belongs to their interaction surface (Fig. 3f). Finally, NDP52 and T6BP interacted with both mutants at levels similar to myosin VIshort (Fig. 4b). Thus, these point mutations were able to induce a conformational change that converts a myosin VIlong into a myosin VIshort-like protein. NMR data of the L1118A mutant further validated this conclusion (Supplementary Fig. 3h).
Figure 4. Myosin VI isoforms have mutually exclusive interactors.
(a) Expanded view of the myosin VI R1050–R1131 structure that illustrates the interaction surface between α2-linker and α3/α4 and highlights M1062 and L1118. (b) GST pull-down assay with the myosin VI1080–1131 and lysates from HEK293T cells transfected with GFP-optineurin, Flag-NDP52, His-GIPC, Flag-T6BP1 constructs. Ponceau as indicated. (c) Model representation of myosin VIlong–clathrin (left panel) and myosin VIshort–optineurin (right panel) interaction. (d) Immunoprecipitation (IP) analysis of HeLa cells stably knocked down for myosin VI transfected with the indicated GFP-myosin VI full-length constructs. IP and IB as indicated. (e) Immunofluorescence analysis of HeLa cells stably knocked down for myosin VI transfected as in d. Endogenous CLTC is in red. Scale bars, 20µm, 5 µm for magnifications. Right panels show co-localization generated by ImageJ plugin colocalization highlighter (pseudocolored in grey). (f) Quantification of the co-localization using Pearson’s and Manders’ coefficients. Error bars, s.d. (n= 15 cells from two independent experiments) **** P<0.0001 by two-tailed T-test.
These results confirmed the conformational differences between the myosin VI isoforms induced by the presence of the α2-linker and established their mutually exclusive association with clathrin and RRL interactors (i.e. optineurin) (Fig. 4c).
Functional analysis of Myosin VIlong–clathrin interaction
To functionally evaluate these findings, we generated GFP-tagged versions of full-length myosin VIlong wild type, L1118A and M1062Q mutants. As controls, we used myosin VIshort and a myosin VIlong mutated in the WWY motif (W1192L) and impaired for Dab-2 binding13. We transfected the constructs in HeLa cells where we knocked-down endogenous myosin VI to avoid confounding effects due to the presence of the endogenous protein (e.g., dimerization). Results showed that endogenous clathrin was immunoprecipitated by myosin VIlong but not by myosin VIshort and that L1118A and M0162Q mutations were sufficient to abrogate this interaction (Fig. 4d). In this context, the W1192L mutant showed binding comparable to that of wild type myosin VIlong (Fig. 4d), suggesting that Dab-2 is dispensable for a stable interaction with clathrin. We also analyzed the behavior of the isoforms by immunofluorescence analysis. As previously reported17, myosin VIlong strongly co-localized with clathrin, whereas myosin VIshort exhibited no significant co-localization (Fig. 4e,f). Notably, W1192L, L1118A and M1062Q mutants failed to co-localize with clathrin, similarly to myosin VIshort (Fig. 4e,f). Similar results were obtained using myosin VI tails (Supplementary Fig. 4), which recapitulate myosin VI interaction and localization17. Taken together, these findings led us to conclude that, in addition to the known Dab-2-binding motif, a second isoform-specific binding surface drives myosin VIlong interaction with clathrin.
Exon skipping is a common event in cancer
Our findings revealed that the isoform-specific α2-linker defines specific conformation (Fig. 3a), interaction (Fig. 4d) and localization (Fig. 4e,f) of myosin VI isoforms. During in vitro epithelial cell polarization a myosin VI isoform switch occurs towards longer isoforms17, 18, suggesting a need of myosin VIlong for clathrin-mediated endocytosis in fully polarized cells.
As loss of cell polarity is a hallmark of cancer, we decided to extend our analyses of myosin VI isoform expression to other cancer types using an in silico approach. We evaluated the relative expression of the isoforms by exploiting the presence of exon 31, which encodes for the α2-linker (outlined in Supplementary Figure 1a) as a discriminating marker of myosin VIlong. As a source of data, we used The Cancer Genome Atlas (TCGA) database and included all experiments performed using RNAseq technology. The resulting dataset consisted of 293 control and 1646 tumor samples, belonging to 14 different tumor types. The relative abundance of exon 31 (E31RA) was obtained as the ratio of its RPKM (Reads Per Kilobase of Exon Model per Million Mapped Reads, see Materials and Methods for details) and the average RPKM values of the four flanking constitutive exons (namely E27, E28, E32 and E33, Supplementary Fig. 1b).
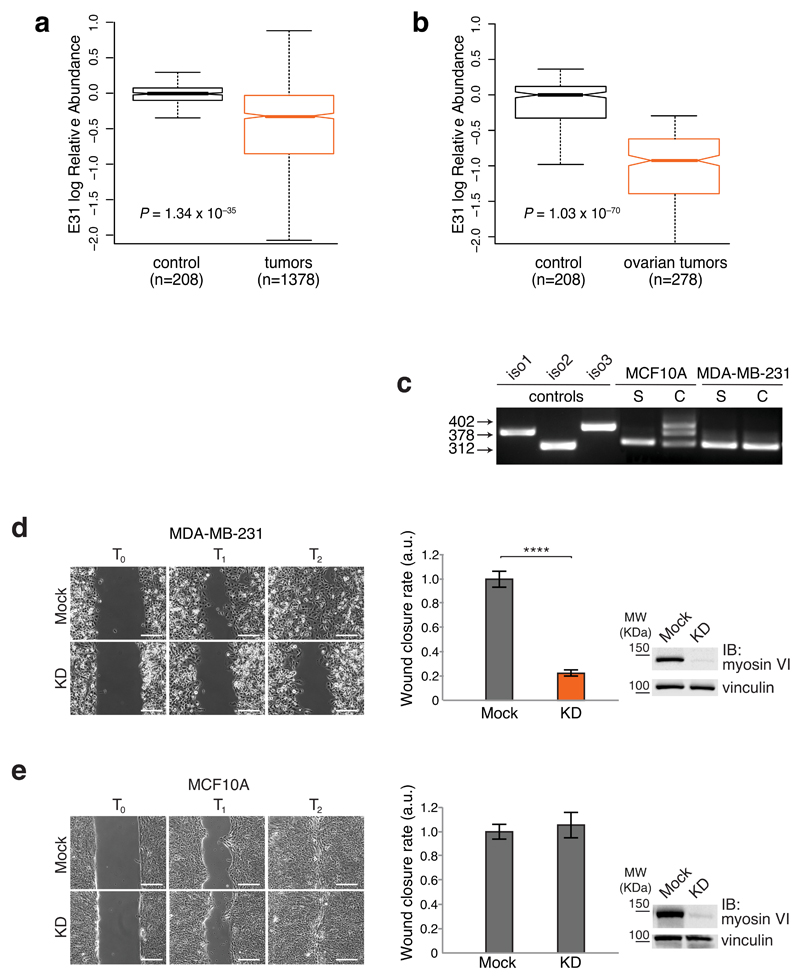
The box plot relative to the overall analysis (Fig. 5a) showed that exon 31 skipping is a common event occurring with significantly increased frequency in tumor compared to normal samples. Thus, myosin VIshort is selectively expressed in tumors. The analysis conducted at the single cancer type level showed that exon skipping is highly frequent among certain tumors, but not others (Supplementary Fig. 5), and suggested that the function of myosin VIshort may be critical and positively selected only in specific types of cancers. Of note, in the case of ovarian cancer, no ‘normal’ controls were present in the database, but compared to the average of normal samples, exon E31RA is barely measurable (Fig. 5b), further confirming the data obtained in primary tumors (Fig. 1c). As proof of principle, we selected two breast cell lines, MCF10A and MDA-MB-231. This latter cancer cell line showed high levels of myosin VIshort-only in both sparse and confluent conditions (Fig. 5c). Upon myosin VI depletion, MDA-MB-231 cells displayed a clear impairment in wound closure (Fig. 5d and Supplementary movie 5), while MCF10A cells were unaffected (Fig. 5e and Supplementary movie 6). Thus, cancer cells that selectively express myosin VIshort-only are addicted to myosin VI for cell migration.
Figure 5. Cancer cells that selectively express myosin VIshort-only are addicted to myosin VI for cell migration.
(a,b) Box plots represent Exon 31 log Relative Abundance (E31RA). Box limits: 25th and 75th percentile; center line: median; whiskers: 1.5 interquartile range IQR; notches: ±1.58 IQR/sqrt(n). P value from Wilcoxon rank-sum test, lower tail. (a) values have been normalized by their median in the tumor matched control samples. (b) E31RA of ovarian cancer is reported together with the average E31RA of normal samples from all available cancer types. (c) RT-PCR analysis performed on the indicated cell lines in growing (S, sparse) or confluent (C, confluent) condition. (d,e) Wound healing assay. The indicated cell lines were KD for myosin VI or mock treated. Left panel, sample images: T0 first frame, T1 and T2 arbitrary points identical for control and KD of the same cell line. Scale bars, 200μm. Central panel: quantification of the wound closure speed relative to control. Error bars, s.d. (n=6 movies for MDA-MB-231, n=13 movies for MCF10A from three independent experiments). **** P<0.0001; ns, no significant difference by two-tailed T-test. Right panel: anti-myosin VI IB performed at T0.
Discussion
The presence of myosin VI splice variants lead to the suggestion that they are directly responsible for disparate myosin VI functions, but no data confirm this hypothesis11, 29. Dab-2 is considered as the adaptor that links myosin VI to clathrin-mediated function13, 27, 30. The Dab-2-myosin VI interaction, however, fails to explain the myosin VIlong-specific interaction with clathrin17, as the Dab-2 binding region lies outside of the alternatively spliced regions13, 27. We provided for the first time a rationale for clathrin selectivity, as the presence of the isoform-specific α2-linker, together with the following region defines a novel clathrin-binding domain and dictates isoform-specific interactions. This finding has important functional implications. During epithelial cell polarization, an isoform switch of myosin VI occurs17, 20. In this context, cells acquire myosin VIlong that is recruited, possibly via clathrin interaction, to the apical part of polarized cells17. Here, myosin VIlong could provide the coated vesicles with the required mechanical force to pass through the intricate meshwork of cortical actin29.
Our study also offers a new perspective on the molecular mechanism that regulates the monomer-dimer conversion of this anchor-motor protein31, 32. It is tempting to speculate that the existence of distinct interaction surfaces for clathrin and Dab-2, both of which are required for co-localization with clathrin-coated pits (Fig. 4e,f) may provide critical synergy that regulates the myosin VI motor ability. Clathrin binding may recruit and concentrate myosin VI at clathrin-coated pits where Dab2 could tether it into a stable and active dimer, crucial to convert myosin VI into a cellular transporter28.
Unexpectedly, our data showed a clear preference of RRL interactors towards myosin VIshort. In the case of optineurin and GIPC, structural data elucidated this specificity as R1116 and L1118, which belong to their interaction surface, are masked in the myosin VIlong conformation. A similar explanation could also apply for NDP52 and T6BP, although their precise interaction surface remains to be established. Whatever the case, our data suggested that myosin VIshort is the isoform specifically involved in the functional interaction with these adaptor proteins that have an established role in autophagy33.
Myosin VI is implicated in cell migration both in Drosophila and mammals (reviewed in 34). However published data lead to an intriguing conundrum. On the one hand, myosin VI depletion from epithelial cells leads to loss of vinculin from adhesion sites and weakens E-cadherin complexes35. Such rearrangement results in loss of cohesive forces in epithelial layers, a well-known prerequisite for invasive cancer cell migration. On the other hand, myosin VI is remarkably upregulated in prostate and ovarian cancer cells, where its overexpression correlates with aggressive clinical behavior21, 22. In this context, silencing of myosin VI expression in ovarian and prostate cancer cell lines decreases the migratory potential of these cells in vitro and ovarian tumor dissemination in vivo21, 22. Our results provided a molecular explanation for this conundrum, ascribing the cohesive and migratory defects to the two different myosin VI isoforms. We found that myosin VIshort is overexpressed in ovarian and other invasive cancers, whereas myosin VIlong is expressed in polarized epithelial cells, in which it likely promotes E-cadherin stabilization at cell-cell junctions.
In Drosophila, the border cell migration occurring during oogenesis is reminiscent of tumor cell invasion21. Initially, the border cells are polarized epithelial cells with strong cell-cell contacts. For migration to begin, cell polarity changes from apico-basal to front-rear, and proteins found at cell-cell junctions reorganize at the invasive edge of the cell to facilitate motility and invasion36. During these events, myosin VI changes localization, from the baso-lateral membrane to membrane ruffles at the front edge of moving cells36. An interesting possibility is that at the onset of border cell migration, an isoform switch towards the shorter myosin VI isoform occurs. Nothing is known about the differential expression and functions of myosin VI isoforms in Drosophila. However, we noticed that different splicing variants also exist in the fly gene. Secondary structure prediction analysis suggested that, even if the primary sequence is not conserved, the exon that is skipped in the shortest isoforms encodes an alpha helix with length and position similar to the α2-linker in mammals (Supplementary Fig. 6).
Our data suggest that cancer cells positively select myosin VIshort because it confers them with a migratory advantage. The exact molecular mechanism through which myosin VIshort modulates cell migration remains to be established. Here, we substantiated previous findings25, providing genetic evidence that the autophagic RRL binder optineurin acts in the same pathway (Fig. 2a,b). In addition, we have demonstrated that myosin VI has a novel ubiquitin-binding domain37 that most likely contributes to optinerurin interaction and to myosin VI short migratory function.
The contribution of alternative splicing in human diseases is widely recognized38. Unbalanced expression of splicing variants or failure to properly express the correct isoform is clearly part of cancer cell biology39. Here, we found that exon skipping determines the expression of myosin VIshort-only in in several types of tumors. Cancer-associated alternative splicing variants are considered new tools for the diagnosis and classification of cancers and could become targets for innovative therapeutic interventions based on highly specific splicing correction approaches. The selective expression of myosin VIshort in cancer may very well fit in this context. This unexpected finding paves the way for defining the splicing mechanism of myosin VI and provides an exciting starting point to understand and therapeutically exploit novel key events in pathological cell migration.
Online Methods
Cell lines, Media and Constructs
| Cell line | Source | Medium |
|---|---|---|
| HEK293T | ICLC | DMEM (Lonza), 10% FBS S.A. (Biowest), 2 mM L-glutamine |
| HEY | CELLution biosystems | DMEM, 10% FBS S.A., 2 mM L-glutamine |
| MCF10A | ATCC | DMEM/Ham’s F12 1:1 (Gibco), 5% horse serum (Thermo Fisher Scientific), hydrocortisone (0.5 μg/ml), insulin (10 μg/ml), cholera toxin (50 ng/ml), EGF (20 ng/ml) |
| OVCAR-3 | NCI-60 | RPMI-1640 (Lonza), 10% FBS N.A. (SIGMA), 2 mM L-glutamine |
| OVCAR-4 | NCI-60 | RPMI-1640, 10% FBS N.A., 2 mM L-glutamine |
| OVCAR-5 | NCI-60 | RPMI-1640, 10% FBS N.A., 2 mM L-glutamine |
| OVCAR-8 | NCI-60 | RPMI-1640, 10% FBS N.A., 2 mM L-glutamine |
| MDA-MB-231 | ATCC | RPMI-1640, 10% FBS N.A., 2 mM L-glutamine |
| COLO-704 | DSMZ | RPMI-1640, 10% FBS N.A., 2 mM L-glutamine |
| SK-OV-3 | NCI-60 | McCoy’s 5A (Gibco), 10% FBS S.A. |
| HeLa | ATCC | MEM (Gibco), 10% FBS S.A., 0.1 mM non-essential amino acids, 2 mM L-glutamine, 1 mM Na-Pyruvate |
At each batch freezing all cell lines were authenticated by STR profiling (StemElite ID System, Promega) and tested for mycoplasma using PCR and biochemical test (MycoAlert, Lonza).
Human myosin VI cDNA KIAA0389 (isoform 1) was obtained from Kazusa DNA Research Institute (Kisarazu, Japan); PCR amplified and cloned into pGEX 6P1 (Amersham Pharmacia Biotech), FLAG-pcDNA3.1 (Invitrogen) and pEGFP-C2 (Clontech). Myosin VI isoform 2 and isoform 3 were generated by mutagenesis (deletion and insertion, respectively) using isoform 1 as template and the primers shown in Supplementary Table 2. All other constructs described were engineered by site-directed mutagenesis or recombinant PCR and sequence verified. Details are available upon request.
GFP-PI3KC2α was kindly provided by Emilio Hirsch, SFB-DDB1 by Maddika Subba Reddy, GFP-optineurin by Alain Israel, FLAG-T6BP by Folma Buss, His-GIPC by Guido Serini, FLAG-NDP52 by Felix Randow.
Antibodies
| Antibody | Species | Supplier | Code | WB | IF |
|---|---|---|---|---|---|
| anti-FLAG M2 | Mouse | Sigma | F3165 | 1:10000 | |
| anti-HA | Mouse | Babco | 16B12 | 1:1000 | |
| anti-His | Mouse | GE Healthcare | 27-4710-01 | 1:3000 | |
| anti-GFP | Rabbit | Sigma | G1544 | 1:5000 | |
| anti-GFP | Rabbit | generated by EUROGENTEC S.A., purified by Cogentech | SI470 | 1:400 | |
| anti-myosin VI | Mouse | Sigma | MUD-19 | 1:1000 | |
| anti-myosin VI | Rabbit | generated by EUROGENTEC S.A., purified by Cogentech | 1295 | 1:2000 | |
| anti-CLCT, clone X22 | Mouse | Pierce | MA1-065 | 1:2000 | |
| anti-CLTC, clone 23 | Mouse | BD bioscience | 610499 | 1:1000 | |
| anti-CLINT1 | Rabbit | Novus biological | 05991 | 1:2000 | |
| anti-COPB2 | Goat | Santa Cruz Biotechnologies | sc13332 | 1:1000 | |
| anti-hVinculin | Mouse | Sigma | V9131 | 1:5000 | |
| anti-optineurin | Rabbit | AbCam | ab-23666 | 1:1000 | |
| anti-Dab-2 | Mouse | BD bioscience | 610464 | 1:1000 | |
| anti-Mouse IgG HRP | Goat | Bio-Rad | 1721011 | 1:5000 | |
| anti-Rabbit IgG HRP | Goat | Bio-Rad | 1706515 | 1:5000 | |
| anti-Goat IgG HRP | Rabbit | Dako | P044902 | 1:10000 | |
| anti-mouse A647 | Donkey | LifeTechnologies | A31571 | 1:400 | |
| anti-rabbit A488 | Donkey | LifeTechnologies | A21206 | 1:400 |
Commercial antibodies were validated by the manufacturer. The home-made anti-myosinVI and anti-GFP rabbit polyclonal antibodies were produced by Eurogentech S.A., affinity purified by Cogentech and validated by immunoblot (Fig. 1e, right panels) or by immunofluorescence (Supplementary Fig. 4a), respectively.
Myosin VI knockdown
Transient myosin VI and optineurin knock-down were performed using Stealth siRNA oligonucleotides (ThermoFisher Scientific). Cells were transfected using RNAiMax (Invitrogen) at 8nM final concentration according to manufacturer’s instruction. For double knock-down single oligos were used at a final 4nM each. Two oligonucleotides (shown in Supplementary Table 2) were used with comparable results. HeLa stably knocked down for Myosin VI were generated using a pSUPER vector carrying the shRNA shown in Supplementary Table 2, targeting a sequence in the motor domain.
Immunofluorescence and Co-localization experiments
For immunofluorescence analysis, cells were fixed with 4% paraformaldehyde for 10 minutes and permeabilized at room temperature with 0.1% Triton-X100. Cells were incubated with primary antibodies for 1 hour followed by secondary antibodies for 30 minutes, at room temperature. Coverslips were mounted in a glycerol solution (20% glycerol, 50 mM Tris pH=8.4) to avoid mechanical deformation of the sample. Images were captured using a Leica inverted SP2 microscope with a laser scanning confocal system. Analysis was performed with ImageJ (http://imagej.nih.gov/ij/).
For co-localization experiments in HeLa cells, to remove soluble cellular proteins, cells were extracted with 0.03% saponin in cytosolic buffer (25 mM Hepes-KOH, pH 7.4, 25 mM KCl, 2.5 mM magnesium acetate, 5 mM EGTA, 150 mM K-glutamate) for 1 minute prior to fixation. For co-localization analysis ROIs were drawn around individual cells (n=15 from 2 independent experiments) were analyzed. The Pearson’s and Manders’ coefficients obtained using JACoP plugin and processed for statistical analysis with Prism (GraphPad software). Statistical significance was determined by non-parametric two-tailed t-tests. Sample size was chosen arbitrary with no inclusion and exclusion criteria. The investigators were not blinded to the group allocation during the experiments and data analysis.
Isoform detection by PCR
Expression of myosinVI isoforms in various cell lines was assessed by PCR. Messenger RNA was isolated from cells grown on plastic dishes using TRIzol reagent (Invitrogen) and RNeasy Mini Kit (Qiagen) according to the manufacturer’s protocols. Genomic DNA and RNA retro-transcription was performed with QuantiTect Reverse Transcription Kit (Qiagen). cDNA obtained was used in PCR reactions with primers flanking the spliced region (Supplementary Table 2).
Original images of gels showed in this study can be found in Supplementary Data Set 1.
Wound-healing assay
3x104 cells were plated into each chamber of a Culture–Insert for wound healing (Ibidi) 48-72 hours before the beginning of the experiment. After insert removal, cells pictures were acquired every 5 minutes for 24 hours. Live-imaging was performed using an ORCA-ER camera (Hamamatsu) on an Olympus IX81 automatic microscope equipped with closed heating and CO2 perfusion devices. Data analysis was performed automatically using ImageJ (http://rsb.info.nih.gov/ij/), and a script designed in order to distinguish and measure the wound area from the area covered by cells. Wound closure rate values correspond to normalized slopes of linear equations derived from regression analyses of the area over time plots. Statistical significance was determined by non-parametric two-tailed t-tests. Sample size was chosen arbitrary with no inclusion and exclusion criteria. The investigators were not blinded to the group allocation during the experiments and data analysis.
Protein expression and purification
GST fusion proteins were expressed in E.coli Bl21 (DE3) Rosetta at 18°C for 16 hours after induction with 1 mM IPTG at an OD600 of 0.5. Cell pellets were resuspended in lysis buffer (50 mM Na-HEPES pH 7.5, 150 mM NaCl, 1 mM EDTA, 5% Glycerol, 0.1% NP40, Protease Inhibitor Cocktail set III, Calbiochem). Sonicated lysates were cleared by centrifugation at 20,000 rpm for 30 minutes. Supernatants were incubated with 1 ml of glutathione-sepharose-beads (GE Healthcare) per liter of bacterial culture. After 2 hours at 4°C, the beads were washed with PBS/0.1% Triton and equilibrated in storage buffer (50 mM Tris pH 7.4, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 10% glycerol).
Biochemical Experiments
For pull-down experiments, 2 μM of GST-fusion proteins immobilized onto GSH beads were incubated for 1 hour at 4°C in JS buffer (50 mM Hepes pH 7.5, 50 mM NaCl, 1.5mM MgCl2, 5mM EGTA, 5% glycerol and 1% Triton X-100) with either 1 mg of HEK293T cellular lysate or 200 µg of transfected HEK293T cellular lysate. Detection was performed by immunoblotting using specific antibody. Ponceau-stained membrane was used to show loading of GST-fusion proteins.
For co-immunoprecipitation (co-IP) experiments, HeLa cells stably knocked-down for myosin VI were transfected using Lipofectamine 2000 (Life Technologies) according to manufacturer’s instruction and IP was performed using 500 μg of lysate and GFP-Trap (Chromotek) in JS buffer.
Original images of blots showed in this study can be found in Supplementary Data Set 1.
Liquid chromatography–tandem MS (LC–MS/MS) analysis
Proteins were resolved by SDS-PAGE on a gradient gel (4-12 % TGX Precast Gel, Biorad) and stained with Colloidal Blue (Colloidal Blue Staining Kit, Invitrogen).
Each lane was divided into 5 slices, higher than GST fusion proteins (Fig. 2e) and digested with trypsin. Proteins with a molecular mass lower than 50 kDa were automatically excluded from the analysis (i.e. the RRL interactor GIPC, 36 kDa). Briefly, samples were subjected to reduction in 10 mM DTT for 1 hour at 56ºC. Digestion was carried out saturating the gel with 12.5 ng/μL sequencing grade modified trypsin (Promega) in 50 mM ammonium bicarbonate overnight. Peptide mixtures were acidified with tri-fluoro acetic acid (TFA, final concentration 3%), extracted from gel slices with 30% acetonitrile (ACN)/ 3% TFA, dried in a Speed-Vac and resuspended in 20 µL of 0.1% FA. Three technical replicates of 5 µL injected for each sample were analyzed on a Fourier transformed-LTQ mass spectrometer (Thermo Electron, San Jose, CA). Peptides separation was achieved by a linear LC gradient from 100% solvent A (5 % ACN, 0.1% formic acid) to 20% solvent B (ACN, 0.1% formic acid) over 33 minutes and from 20% to 80% solvent B in 4 minutes at a constant flow rate of 0.3µL/minutes on Agilent chromatographic separation system 1100 (Agilent Technologies, Waldbronn, Germany) equipped with a 15 cm fused-silica emitter of 75 µm inner diameter (New Objective, Inc. Woburn, MA USA), packed in-house with ReproSil-Pur C18-AQ 3 µm beads (Dr. Maisch GmbH, Ammerbuch, Germany) using a high-pressure bomb loader (Proxeon, Odense, Denmark). Survey MS scans were acquired in the FT from m/z 350-1650 with 100,000 resolution. The five most intense doubly and triply charged ions were automatically selected for fragmentation. Target ions already selected for the MS/MS were dynamically excluded for 60s.
Data processing and protein quantitation analysis
Raw MS files were converted into peaklist (.msm files); all MS/MS samples were analyzed using Mascot (Matrix Science, London, UK; version 2.3.02) set up with the following parameters: UniProt_CP_Human_20140416 database (88708 entries), Taxonomy Homo sapiens, enzyme Trypsin, maximum missed cleavages 2, fixed modification carbamidomethyl cysteine, variable modifications methionine oxidation and acetyl (protein N-terminus), peptide tolerance 10 ppm, MS/MS tolerance 0.5 Da, instrument ESI-TRAP.
Interactomics results were generated with Scaffold_4.3.4 (Proteome Software Inc., Portland, OR) and protein quantitation was displayed as Total Spectral Count (best candidates are reported in Table 1). Peptide identifications were accepted if they could be established at greater than 95.0% probability by the Peptide Prophet algorithm with Scaffold delta-mass correction40. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least 2 identified peptides. Two RRL interactors, T6BP and NDP52, were excluded at this step due to single peptide identification. Protein probabilities were assigned by the Protein Prophet algorithm41. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing marked peptide evidence were grouped into clusters.
Label-free quantification was obtained using MaxQuant software (1.4.0.5). MS/MS peak lists were searched against the Uniprot_cp_human_2013_07 in which trypsin specificity was used with up to two missed cleavages allowed. Searches were performed selecting alkylation of cysteine by carbamidomethylation as fixed modification, and oxidation of methionine and N-terminal acetylation as variable modifications. Mass tolerance was set to 20 ppm and 0.5 Da for parent and fragment ions, respectively. The false discovery rate for both peptides and proteins was set at 0.01. Additionally, we required at least two peptide identifications per protein, of which at least one unique. “LFQ intensities”, which are the intensity values normalized across the entire dataset, were used for quantification based on unique plus razor peptides. Statistical analysis was performed using Perseus. Proteins were filtered applying Benjamini Hochberg and t-test. Proteins that differentially interact with isoform 1, isoform 2 and 3 vs GST control are reported in Supplementary Table 1. The proteomic data as raw files, total proteins and peptides identified with relative intensities and search parameters have been deposited on Peptide Atlas repository (accession number PASS00591).
NMR spectroscopy
The NMR experiments were acquired at 10 °C on Bruker 700, 800, and 850 MHz spectrometers equipped with gradient cryoprobes. 15N- or 13C,15N-labeled samples used for titration experiments ranged from 0.1 to 0.2 mM, whereas experiments acquired for structure determination were performed with 0.5 to 0.8 mM samples; all experiments were conducted in 20 mM sodium phosphate buffer (pH 6.5), 50 mM sodium chloride, 2 mM DTT, 0.01 % NaN3, and 10 % 2H2O/90 % 1H2O. 2D [1H, 15N]-HSQC and 3D HNCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB and CBCA(CO)NH spectra were used for backbone 1H, 15N, and 13C assignments. Side chain 1H and 13C assignments were obtained by 2D [1H, 13C]-HSQC and 3D HBHA(CO)NH, H(CCCO)NH, (H)CC(CO)NH, HCCH-COSY, HCCH-TOCSY and (H)CCH-TOCSY spectra. Assignments were checked for consistency with 3D 15N/13C –NOESY-HSQC spectra recorded with mixing times of 120 or 100 ms.
Structure determination
Statistics for the structure calculations for myosin VI1050-1131 are displayed in Table 2. The regions for Ramachandran plot statistics and RMSD calculations include T1054-R1068 and Y1084-S1126. NMR spectra were processed with NMRPipe and analyzed with KUJIRA42 and SPARKY. CYANA2.1 (ref.43) was used for automated NOE assignment and to calculate the structures by torsion angle dynamics; each NOE assignment was manually inspected and confirmed or corrected. Dihedral angle restraints were derived by TALOS44. A total of 100 structures were independently calculated and the twenty conformers with the lowest target-function values were selected for refinement with Xplor-NIH. Structures were evaluated with PROCHECK-NMR45, visualized with MOLMOL46, and figures generated by PYMOL (The PyMOL Molecular Graphics System, http://www.pymol.org/). The quality of the structures is also reflected by the distribution of 94.6% and 5.4% of the backbone torsion angles in the most favored and additionally allowed regions of the Ramachandran plot, respectively, as calculated by PROCHECK_NMR.
Isolation, culture and processing of primary epithelial ovarian cancer cells
Patient-derived tissue samples were collected with the approval of the Ethical Committee of the European Institute of Oncology. Fresh biopsies of ovarian cancer were obtained from patients with high-grade epithelial ovarian serous carcinoma who underwent surgical tumor debulking. Fresh biopsies of normal ovaries were obtained upon informed consent from patients undergoing adnexectomy for non-ovarian gynecological pathologies.
All tumors were digested in DMEM/F12 medium (Gibco) containing 2 mM glutamine, 1% Penicillin/Streptomycin, 200 U/ml collagenase IA and 100 U/ml hyaluronidase. Normal ovaries were digested in 5U/ml Dispase for 30 minutes at 37°C and then the organ surfaces were scraped to isolate the epithelial cells. The derived primary epithelial cells were maintained in monolayer adherent cultures in collagen I Cellware coated flask (Corning) in DMEM/F12 medium (Gibco) containing 1% fetal bovine serum, 2 mM glutamine, 1% Penicillin/Streptomycin, 0.2% gentamicin, 0.2% amphotericin, 10 mg/ml transferrin, 1 mg/ml insulin, 1 mg/ml hydrocortisone, 10 mM HEPES pH 7.5, 50 mM ascorbic acid, 15 nM sodium selenite, 50 ng/ml cholera toxin, 10 nM EGF, 35 mg/ml bovine pituary extract, 10 nM T3 and 10 nM β-estradiol. For RNA extraction, 5x106 primary cells were washed in PBS and the cell pellets were snap frozen in dry ice.
RNA-Seq Analysis
RNA-Seq exons quantification data were downloaded from 'The Cancer Genome Atlas' Data Portal (https://tcga-data.nci.nih.gov/tcga/dataAccessMatrix.htm). Downloaded files included experiments which were analyzed with the pipeline named RNAseq and conducted on neoplastic cells belonging to ten different tissues: bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), esophageal carcinoma (ESCA), head and neck squamous cell carcinoma (HNSC), kidney renal clear cell carcinoma (KIRC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), stomach adenocarcinoma (STAD) and uterine corpus endometrial carcinoma (UCEC). Control experiments were also downloaded for each neoplastic counterpart. Additionally, experiment files relative to four additional neoplasic tissues were considered: colon adenocarcinoma (COAD), kidney renal papillary cell carcinoma (KIRP), serous cystadenocarcinoma (OV) and rectum adenocarcinoma (READ). For these last four sets, control counterparts were not available. Despite the availability of RNA-Seq data from acute myeloid leukemia (LAML), such files were discarded, due to peculiar non-comparable exon notation. The resulting dataset consisted of 293 control and 1646 tumor samples, belonging to 14 different tumor types. For each experiment, the exon quantification file was considered. The 'raw counts' (RC) and 'Reads Per Kilobase of Exon Model per Million Mapped Reads’ (RPKM) were extracted 47 for the genomic positions corresponding to exons 27 to 33 of the MYO VI gene, which follow
E27: chr6:76600945-76601023:+
E28: chr6:76602247-76602407:+
E31: chr6:76608090-76608128:+
E32: chr6:76617322-76617425:+
E33: chr6:76618213-76618344:+
All positions are relative to the hg19 genome assembly. Exons E29 and E30 were not individually annotated due to the presence of a confounding overlapping first exon (chr6:76603648-76604977:+). A detailed representation of the isoforms and the considered exons is reported in Supplementary Figure 2a. Exon expression changes between cancer and normal samples were estimated using RPKM, a measure of expression that reflects the molar concentration of a transcript in the sample by normalizing read counts for mRNA length and for the total read number in the sample. Exon E31 relative abundances (E31RA) were then obtained for each sample as the ratio of their RPKM and the average RPKM values of the four flanking constitutive exons (namely E27, E28, E32 and E33). E31 Normalized Relative Abundances (E31NRA) were obtained for each cancer type as the ratio between each sample E31RA and the median E31RA value of the corresponding control samples. These normalized values can be compared among different cancer types. For cancer types with no matched control set, we obtained E31NRA as the ratio between each sample E31RA and the median E31RA value of all the control samples. Wilcoxon test48 has been performed by using the R-package (http://www.R-project.org/.) to verify the hypothesis that the logRatio between tumors and control samples E31NRA values is negative (i.e. the median E31NRA is less in tumors than in controls). This analysis has been performed for cancer types for which a control set was available as well as overall among all cancer types.
Supplementary Material
Acknowledgments
We thank Folma Buss for critically reading the manuscript and for helpful discussions and advice, Markus Ladwein for performing initial wound–healing experiments. We also thank Emilio Hirsch (Università di Torino), Maddika Subba Reddy (Centre for DNA Fingerprinting and Diagnostics), Alain Israel (Institute Pasteur), Folma Buss (Cambridge Institute for Medical Research), Guido Serini (Università di Torino) and Felix Randow (Medical Research Council) for DNA constructs. This work was supported by the Association for International Cancer Research (AICR), grant 11-0051 (S.P.); from Italian Ministry of Education, Universities and Research, grant PRIN 20108MXN2J (S.P.) and by the Intramural Research Program of the US National Cancer Institute (K.J.W.). H-P.W. was supported by fellowship from the Associazione Italiana per la Ricerca sul Cancro (AIRC) co-funded by Marie Curie Actions. M.B was supported by fellowship from Fondazione Umberto Veronesi (FUV).
Footnotes
Accession numbers
The structural coordinates and chemical shift data have been deposited in the Protein Data Bank (PDB) and Biological Magnetic Resonance Data Bank (BMRB) with respective accession codes 2N12 and 25544. The proteomic data as raw files, total proteins and peptides identified with relative intensities and search parameters have been deposited on Peptide Atlas repository (accession number PASS00591).
Author contributions
H-P.W., M.B and El.M. designed and performed the experiments and analyzed the data; F.H. and K.J.W. designed and interpreted CD and NMR experiments, which F.H. performed and analyzed; E.V. carried out FP analysis; P.S. carried out MS analysis; M.L. and U.C. generated primary cells from high-grade ovarian cancer; Er.M and U.P. conducted exons analysis; K.R. participated in setting up and interpreting the migration assays; M.M. participated in the experimental design and data analysis; S.P. conceived the project, interpreted the results and wrote the paper with the contribution of all authors.
References
- 1.Foth BJ, Goedecke MC, Soldati D. New insights into myosin evolution and classification. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:3681–3686. doi: 10.1073/pnas.0506307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 3.Wells AL, et al. Myosin VI is an actin-based motor that moves backwards. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 4.Buss F, Spudich G, Kendrick-Jones J. Myosin VI: cellular functions and motor properties. Annual review of cell and developmental biology. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 5.Hasson T. Myosin VI: two distinct roles in endocytosis. Journal of cell science. 2003;116:3453–3461. doi: 10.1242/jcs.00669. [DOI] [PubMed] [Google Scholar]
- 6.Geisbrecht ER, Montell DJ. Myosin VI is required for E-cadherin-mediated border cell migration. Nature cell biology. 2002;4:616–620. doi: 10.1038/ncb830. [DOI] [PubMed] [Google Scholar]
- 7.Hicks JL, Deng WM, Rogat AD, Miller KG, Bownes M. Class VI unconventional myosin is required for spermatogenesis in Drosophila. Molecular biology of the cell. 1999;10:4341–4353. doi: 10.1091/mbc.10.12.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rogat AD, Miller KG. A role for myosin VI in actin dynamics at sites of membrane remodeling during Drosophila spermatogenesis. Journal of cell science. 2002;115:4855–4865. doi: 10.1242/jcs.00149. [DOI] [PubMed] [Google Scholar]
- 9.Petritsch C, Tavosanis G, Turck CW, Jan LY, Jan YN. The Drosophila myosin VI Jaguar is required for basal protein targeting and correct spindle orientation in mitotic neuroblasts. Developmental cell. 2003;4:273–281. doi: 10.1016/s1534-5807(03)00020-0. [DOI] [PubMed] [Google Scholar]
- 10.Erben V, et al. Asymmetric localization of the adaptor protein Miranda in neuroblasts is achieved by diffusion and sequential interaction of Myosin II and VI. Journal of cell science. 2008;121:1403–1414. doi: 10.1242/jcs.020024. [DOI] [PubMed] [Google Scholar]
- 11.Tumbarello DA, Kendrick-Jones J, Buss F. Myosin VI and its cargo adaptors - linking endocytosis and autophagy. Journal of cell science. 2013;126:2561–2570. doi: 10.1242/jcs.095554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Penengo L, et al. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell. 2006;124:1183–1195. doi: 10.1016/j.cell.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 13.Spudich G, et al. Myosin VI targeting to clathrin-coated structures and dimerization is mediated by binding to Disabled-2 and PtdIns(4,5)P2. Nature cell biology. 2007;9:176–183. doi: 10.1038/ncb1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bunn RC, Jensen MA, Reed BC. Protein interactions with the glucose transporter binding protein GLUT1CBP that provide a link between GLUT1 and the cytoskeleton. Molecular biology of the cell. 1999;10:819–832. doi: 10.1091/mbc.10.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sahlender DA, et al. Optineurin links myosin VI to the Golgi complex and is involved in Golgi organization and exocytosis. The Journal of cell biology. 2005;169:285–295. doi: 10.1083/jcb.200501162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morriswood B, et al. T6BP and NDP52 are myosin VI binding partners with potential roles in cytokine signalling and cell adhesion. Journal of cell science. 2007;120:2574–2585. doi: 10.1242/jcs.007005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buss F, Arden SD, Lindsay M, Luzio JP, Kendrick-Jones J. Myosin VI isoform localized to clathrin-coated vesicles with a role in clathrin-mediated endocytosis. The EMBO journal. 2001;20:3676–3684. doi: 10.1093/emboj/20.14.3676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dance AL, et al. Regulation of myosin-VI targeting to endocytic compartments. Traffic. 2004;5:798–813. doi: 10.1111/j.1600-0854.2004.00224.x. [DOI] [PubMed] [Google Scholar]
- 19.Tomatis VM, et al. Myosin VI small insert isoform maintains exocytosis by tethering secretory granules to the cortical actin. The Journal of cell biology. 2013;200:301–320. doi: 10.1083/jcb.201204092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Au JS, Puri C, Ihrke G, Kendrick-Jones J, Buss F. Myosin VI is required for sorting of AP-1B-dependent cargo to the basolateral domain in polarized MDCK cells. The Journal of cell biology. 2007;177:103–114. doi: 10.1083/jcb.200608126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida H, et al. Lessons from border cell migration in the Drosophila ovary: A role for myosin VI in dissemination of human ovarian cancer. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8144–8149. doi: 10.1073/pnas.0400400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn TA, et al. A novel role of myosin VI in human prostate cancer. The American journal of pathology. 2006;169:1843–1854. doi: 10.2353/ajpath.2006.060316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puri C, et al. Overexpression of myosin VI in prostate cancer cells enhances PSA and VEGF secretion, but has no effect on endocytosis. Oncogene. 2010;29:188–200. doi: 10.1038/onc.2009.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szczyrba J, et al. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–538. doi: 10.1158/1541-7786.MCR-09-0443. [DOI] [PubMed] [Google Scholar]
- 25.Chibalina MV, Poliakov A, Kendrick-Jones J, Buss F. Myosin VI and optineurin are required for polarized EGFR delivery and directed migration. Traffic. 2010;11:1290–1303. doi: 10.1111/j.1600-0854.2010.01101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majewski L, Sobczak M, Havrylov S, Jozwiak J, Redowicz MJ. Dock7: a GEF for Rho-family GTPases and a novel myosin VI-binding partner in neuronal PC12 cells. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2012;90:565–574. doi: 10.1139/o2012-009. [DOI] [PubMed] [Google Scholar]
- 27.Morris SM, et al. Myosin VI binds to and localises with Dab2, potentially linking receptor-mediated endocytosis and the actin cytoskeleton. Traffic. 2002;3:331–341. doi: 10.1034/j.1600-0854.2002.30503.x. [DOI] [PubMed] [Google Scholar]
- 28.Yu C, et al. Myosin VI undergoes cargo-mediated dimerization. Cell. 2009;138:537–548. doi: 10.1016/j.cell.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 29.Buss F, Kendrick-Jones J. How are the cellular functions of myosin VI regulated within the cell? Biochemical and biophysical research communications. 2008;369:165–175. doi: 10.1016/j.bbrc.2007.11.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Inoue A, Sato O, Homma K, Ikebe M. DOC-2/DAB2 is the binding partner of myosin VI. Biochemical and biophysical research communications. 2002;292:300–307. doi: 10.1006/bbrc.2002.6636. [DOI] [PubMed] [Google Scholar]
- 31.Lu Q, Li J, Zhang M. Cargo recognition and cargo-mediated regulation of unconventional myosins. Accounts of chemical research. 2014;47:3061–3070. doi: 10.1021/ar500216z. [DOI] [PubMed] [Google Scholar]
- 32.Sweeney HL, Houdusse A. Myosin VI rewrites the rules for myosin motors. Cell. 2010;141:573–582. doi: 10.1016/j.cell.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 33.Tumbarello DA, et al. Autophagy receptors link myosin VI to autophagosomes to mediate Tom1-dependent autophagosome maturation and fusion with the lysosome. Nature cell biology. 2012;14:1024–1035. doi: 10.1038/ncb2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chibalina MV, Puri C, Kendrick-Jones J, Buss F. Potential roles of myosin VI in cell motility. Biochemical Society transactions. 2009;37:966–970. doi: 10.1042/BST0370966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maddugoda MP, Crampton MS, Shewan AM, Yap AS. Myosin VI and vinculin cooperate during the morphogenesis of cadherin cell cell contacts in mammalian epithelial cells. The Journal of cell biology. 2007;178:529–540. doi: 10.1083/jcb.200612042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinheiro EM, Montell DJ. Requirement for Par-6 and Bazooka in Drosophila border cell migration. Development. 2004;131:5243–5251. doi: 10.1242/dev.01412. [DOI] [PubMed] [Google Scholar]
- 37.He F, et al. Myosin VI contains a structural motif that binds to ubiquitin chains. Cell reports. 2016 doi: 10.1016/j.celrep.2016.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh RK, Cooper TA. Pre-mRNA splicing in disease and therapeutics. Trends in molecular medicine. 2012;18:472–482. doi: 10.1016/j.molmed.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biamonti G, Catillo M, Pignataro D, Montecucco A, Ghigna C. The alternative splicing side of cancer. Seminars in cell & developmental biology. 2014;32:30–36. doi: 10.1016/j.semcdb.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 40.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 41.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical chemistry. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 42.Kobayashi N, et al. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. Journal of biomolecular NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 43.Guntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 44.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. Journal of biomolecular NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 45.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. Journal of biomolecular NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 46.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 47.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 48.Bauer DF. Constructing confidence sets using rank statistics. Journal of the American Statistical Association. 1972;67:687–690. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.