Abstract
In pre-clinical studies, combination therapy with gemcitabine and targeted radioimmunotherapy (RIT) using 212Pb-trastuzumab showed tremendous therapeutic potential in the LS-174T tumor xenograft model of disseminated intraperitoneal disease. To better understand the underlying molecular basis for the observed cell killing efficacy, gene expression profiling was performed after a 24 h exposure to 212Pb-trastuzumab upon gemcitabine (Gem) pre-treatment in this model. DNA damage response genes in tumors were quantified using a real time quantitative PCR array (qRT-PCR array) covering 84 genes. The combination of Gem with α-radiation resulted in the differential expression of apoptotic genes (BRCA1, CIDEA, GADD45α, GADD45γ, IP6K3, PCBP4, RAD21, and p73), cell cycle regulatory genes (BRCA1, CHK1, CHK2, FANCG, GADD45α, GTSE1, PCBP4, MAP2K6, NBN, PCBP4, and SESN1), and damaged DNA binding and repair genes (BRCA1, BTG2, DMC1, ERCC1, EXO1, FANCG, FEN1, MSH2, MSH3, NBN, NTHL1, OGG1, PRKDC, RAD18, RAD21, RAD51B, SEMA4G, p73, UNG, XPC, and XRCC2). Of these genes, the expression of CHK1, GTSE1, EXO1, FANCG, RAD18, UNG and XRCC2 were specific to Gem/212Pb-trastuzumab administration. In addition, the present study demonstrates that increased stressful growth arrest conditions induced by Gem/212Pb-trastuzumab could suppress cell proliferation possibly by up-regulating genes involved in apoptosis such as p73, by down-regulating genes involved in cell cycle check point such as CHK1, and in damaged DNA repair such as RAD51 paralogs. These events may be mediated by genes such as BRCA1/MSH2, a member of BARC (BRCA-associated genome surveillance complex). The data suggest that up-regulation of genes involved in apoptosis, perturbation of checkpoint genes, and a failure to correctly perform HR-mediated DSB repair and mismatch-mediated SSB repair may correlate with the previously observed inability to maintain the G2/M arrest, leading to cell death.
Introduction
Combination therapy with radiation and chemotherapeutics, a commonly used regimen for the treatment of cancer, highly improves therapeutic response. Due to a high linear transfer (LET) and a short range in tissue, alpha (α)-particles induce clusters of DNA strand breaks, leading to cell death [1–5]. Thus, high-LET radiation with less damage to surrounding normal tissue is more specific and effective in cell killing than low-LET radiation such as β−-particles [6–8]. Several α-emitting radionuclides have been successfully used in radioimmunotherapy (RIT) for targeted therapy of cancer [9–12]. When applied as a monotherapy or in combination with chemotherapeutics, radioimmunotherapies with 212Pb have shown the high therapeutic efficacy of this isotope in targeted α-particle therapy for disseminated peritoneal diseases [9, 13–16].
Gemcitabine (Gem), a well-defined FDA approved chemotherapeutic, is a nucleoside analogue widely used as the first-line chemotherapy against cancer. It has demonstrated the therapeutic feasibility as a single modality against tumors [17–20]. As such, Gem in conjunction with 212Pb-trastuzumab was evaluated as one of chemotherapeutics, the combination of which was reported to significantly enhance therapeutic response [15, 16].
In response to DNA breaks, catastrophic cellular injury that causes failure in maintaining the genetic integrity, leading to cell death results via a variety of mechanisms such as apoptosis, autophagy, necrosis, and mitotic catastrophe. Radiation-induced complex signaling pathways and alterations in gene expression may provide valuable information to identify potential biomarkers of human response to radiation [21]. Tissue response and associated gene modulations have, however, not been clearly defined following exposure of tumors to α-particle RIT unlike the many possibilities that are described for chemotherapy. Recently, gene expression profiles in different biological systems have been identified following exposure to high-LET radiation such as α-particles. In comparison with 60Co in human fibroblasts, biological processes such as mitosis, spindle assembly checkpoint, and apoptotic chromosome condensation were uniquely modified after exposure to α-particle radiation (211At-labeled trastuzumab), suggesting α-particle radiation clearly influenced tumor protein p53-activated and repressed genes [22]. Pathway analysis associated with differentially modulated genes in human lung epithelial cells exposed to α-particle radiation (222Rn) suggested that α-particle radiation inhibits DNA synthesis and subsequent mitosis, and caused cell cycle arrest via p53 signaling. Seidl and colleagues demonstrated that cell killing by α-particle radiation (213Bi-d9MAb) in human gastric cancer cells (HSC45-M2) was evident in the formation of micronuclei and severe chromosomal aberrations. In gene expression profiling for the whole genome, up-regulated genes (COL4A2, NEDD9, and C3) and down-regulated genes (WWP2, RFX3, HIST4H4, and JADE1) were unique, which were not related to any biological processes [23–25].
In response to α-particle RIT combined with an established chemotherapeutic agent such as Gem, application of gene expression profiling may reveal potential clinical targets by providing novel information for further biomedical and clinical research. For this purpose, the gene modulation in tumors that received Gem combined with specifically targeted α-particle RIT (212Pb-trastuzumab) in the LS-174T i.p. xenograft model is described using a real time quantitative PCR (qRT-PCR) array to investigate key biological processes such as apoptosis, cell cycle arrest, and DNA repair with regard to gene expression.
Materials and Methods
Cell line
All of the in vivo studies were conducted using the human colon carcinoma cell line (LS-174T; provided by Dr. J. Greiner, NCI, Bethesda, MD) grown in supplemented Dulbecco’s Modified Eagle’s Medium (DMEM) as previously described by Tom BH et al [26] with all media and supplements being purchased from Lonza (Walkersville, MD) unless otherwise indicated. The cell line was screened for mycoplasma and other pathogens before in vivo use according to National Cancer Institute (NCI) Laboratory Animal Sciences Program policy without any further cell line authentication.
Chelate synthesis, mAb conjugation, and radiolabeling
The synthesis, characterization, and purification of the bifunctional ligand TCMC have been previously described [27]. Conjugation of trastuzumab (Herceptin®; Genentech, South San Francisco, CA) was conducted with TCMC by established methods using a 10-fold molar excess of ligand to mAb. A 10 mCi 224Ra/212Pb generator (AlphaMed, Lakewood, NJ) was washed with 2 M HCl to remove any impurities and any unbound 224Ra. 212Pb was eluted from the generator with 1 M HCl and dried. The residue dissolved in 0.1 M HCl was used for radiolabeling of mAb. The radiolabeled mAb was purified using a desalting column (GE Healthcare, Piscataway, NJ) with PBS. Purified polyclonal IgG (HuIgG) fraction was similarly conjugated with TCMC and radiolabeled with 212Pb as described above, providing a non-specific control antibody for the experiments.
Tumor model, treatment and tumor harvesting
All animal protocols were approved by the National Cancer Institute (NCI) Animal Care and Use Committee for all experiments. To provide ample space to mice, five female mice were housed per autoclaved cage at the NCI vivarium with bedding and nesting materials provided in each cage. The mice were also provided with sterile mouse chow and drinking water. The mouse chow and water were stored in clean, dedicated areas of the vivarium. All equipment and supplies entering the facilities were sterilized for animal health and well-being. Monitoring animals for health problems were performed on a daily basis. Any animal experiencing rapid weight loss, debilitating diarrhea, rough hair coat, hunched posture, labored breathing, lethargy, persistent recumbence, jaundice, anemia, significantly abnormal neurological signs, bleeding from any orifice, self-induced trauma, impaired mobility, or difficulty eating or drinking were immediately euthanized. Mice bearing i.p. xenografts may manifest additional clinical signs of disease progression such as sizeable abdominal distention, ascites or generalized subcutaneous edema and were euthanized. Mice experiencing significant weight loss or gain (10%, determined by weekly weighings) were also determined to reach the experimental/humane endpoints and were euthanized. Euthanasia was performed by removing the animal(s) from the home cage, and placing it in a chamber with a specialized euthanasia lid attached to a CO2 line. CO2 was allowed to flood the chamber at a rate of 2 L/min. When breathing ceased for all mice, the mice were removed from the chamber.
In vivo studies were performed with 19–21 g female athymic mice (NCI-Frederick). Athymic mice were injected i.p. with 1 x 108 LS-174T cells in 1 mL of DMEM as previously reported [27]. The 212Pb-TCMC-trastuzumab (10 μCi) was administrated to the mice (n = 10–15) 3 days post-implantation of tumor in 0.5 mL PBS. HuIgG labeled with 212Pb served as the non-specific control. The α-radiation was administrated 3 d after tumor implantation. Gemcitabine (Eli Lilly, Indianapolis, IN), obtained through the NIH Division of Veterinary Resources Pharmacy, was prepared for injection at 1 mg/ 0.5 mL phosphate-buffered saline (PBS) and given by i.p. injection to the mice 2 d after injection of the LS-174T cells. This treatment group was compared with sets of tumor bearing mice that received gemcitabine alone, Gem/212Pb-HuIgG, or no treatment. Mice were euthanized 24 h after receiving the Gem/212Pb-RIT, the tumors harvested and stored at -80°C until use.
RNA purification
To produce high quality RNA from tumor tissues (212Pb-trastuzumab treated or non-specific controls), total RNA isolation from tissue was performed using the RNeasy mini kit (Qiagen, Santa Clarita, CA) in accordance with the manufacturer’s instructions. Quantity and quality of isolated total RNA were assessed using Nano-drop spectrophotometer (Thermo Scientific, Wilmington, DE) using OD260 for calculation of concentration. Only that total RNA with an A260/A280 ratio > 1.9 and without detectable contamination of DNA (PCR) was employed in the gene expression array (qRT-PCR array).
Human DNA damage PCR array
The human DNA damage PCR array (SABiosciences, Frederick, MD) profiles expression of 84 genes involved in apoptosis, cell cycle and damaged DNA binding and repair (S1 Table). cDNA was prepared from RNA using the First strand cDNA synthesis Kit (SABiosciences, Fredrick, MD). Comparison of the relative expression of 84 genes was characterized (RT2 real-time SYBR Green/Rox PCR master mix, SABiosciences) in 96 well microtiter plates on a 7500 real time PCR system (Applied Biosystems, Rockville, MD). Data was analyzed using the RT2 profiler PCR Array Data Analysis v3.5 software (Qiagen). The fold change in gene expression was calculated using the equation 2(-ΔΔCT). If the fold change was greater than 1, the result was considered as an up-regulation. For down-regulated (less than 1-fold change) genes the value was reported as the negative inverse.
Chromatin immunoprecipitation
The chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology, Billerica, MA) was performed in accordance with the manufacturer’s instructions with minor adjustments. In brief, lysates from tumor tissues were prepared and aliquoted. Chromatin was immunoprecipitated with 10 μL (1:100) of antibody for E2F1 (Upstate Biotechnology). Antibody was incubated overnight with chromatin on a rotator at 4°C; the resulting DNA-protein complexes were isolated using protein G agarose magnetic beads. The samples were subjected to 65°C for 5 h, the DNA extracted, and dissolved in the elution reagent. The PCR-amplified DNAs using CHK1, MSH2 and p73 promoter specific primers (Applied Biosystems) were analyzed by electrophoresis using 2% agarose gels.
Immunoblot analysis
Total protein isolates using tissue protein extraction reagent (T-PER) (Thermo Scientific, Asheville, NC) containing protease inhibitors (Roche, Indianapolis, IN) were prepared for immunoblot analysis. Equivalent amounts of protein extracts were resolved on a 4–20% tris-glycine gel electrophoresis system and transferred to a nitrocellulose membrane. For immmunodetection, antibodies against RAD51B and XRCC2 (Abcam, Cambridge, MA) were used at a dilution of 1:1000 in PBS containing 5% BSA and 0.05% Tween-20 for 1 h. Horseradish peroxidase conjugated rabbit secondary antibodies were used at a 1:5000 dilution prepared in PBS with 3% non-fat dry milk. The immunoblots were developed using the enhanced chemoluminescent detection kit (GE Healthcare, Pascataway, NJ).
Statistics
A minimum of at least three independent experiments were conducted for each treatment described. Statistical differences between the groups were determined using Student t test. For multiple comparisons, the ANOVA was performed. Statistically significant difference between datasets was determined at p-value < 0.05.
Results
Gemcitabine may potentiate α-radiation-induced cell killing by regulation of genes involved in apoptosis
Significantly up- or down-regulated genes 24 h after exposure of tumors to 212Pb-RIT in combination with Gem (n = 3) were identified through application of a 2-fold change threshold using qRT-PCR array as compared to the untreated group as a control. Thirteen of the 84 genes of DNA damage signaling pathway investigated in this study are associated with the regulation of the apoptotic process. Of these affected genes, six genes (CIDEA, GADD45α, GADD45γ, IP6K3, PCBP4, and p73) were up-regulated and two genes (BRCA1 and Rad21) were down-regulated to varying degrees among the various treatment groups (Table 1). The expression of p73 of the up-regulated genes appeared to exhibit the greatest impact from Gem/212Pb-trastuzumab (8.6-fold increase, p < 0.0021) and Gem/212Pb-HuIgG (10.1-fold increase, p < 0.0005) treatment. Clear differences were observed between these groups and the group that received only Gem (2.7-fold increase, p < 0.1584). The increase in the expression of GADD45α for the radiation treatment groups was also greater than the group that received Gem alone. The expression of BRCA1 was significantly down-regulated after treatment with Gem/212Pb-trastuzumab (-3.2-fold decrease, p < 0.002) and Gem/212Pb-HuIgG (-3.1 fold decrease, p < 0.0028) compared to the group that received Gem alone (-1.1-fold decrease, p < 0.6248) (Table 1).
Table 1. Differential expression of genes involved in apoptosis in LS-174T i.p. xenografts following treatment with Gemcitabine and α-treatment.
| Symbol | Gene name | GeneBank ID | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|
| Gemcitabine-212Pb-trastuzumab | p | Gemcitabine-212Pb-HuIgG | p | Gemcitabine | p | |||
| BRCA1 | Breast Cancer 1, early onset | NM007294 | -3.2 | 0.0020 | -3.1 | 0.0028 | -1.1 | 0.6268 |
| CIDEA | Cell death-inducing DEFA-like effector a | NM001279 | 2.7 | 0.1447 | 3.9 | 0.0145 | 3.0 | 0.0001 |
| GADD45α | Growth arrest and DNA-damage-inducible, alpha | NM001924 | 4.5 | 0.0047 | 5.8 | 0.0004 | 3.0 | 0.0005 |
| GADD45γ | Growth arrest and DNA-damage-inducible, gamma | NM006705 | 5.0 | 0.0001 | 5.9 | 0.0003 | 6.2 | 0.0143 |
| IP6K3 | Inositol hexakisphosphate kinase 3 | NM054111 | 2.1 | 0.0241 | 1.2 | 0.4338 | 2.8 | 0.0005 |
| PCBP4 | Poly(rC)binding protein 2 | NM020418 | 2.7 | 0.0048 | 3.2 | 0.0001 | 3.0 | 0.0011 |
| RAD21 | RAD21 homolog | NM006265 | -2.4 | 0.0003 | -2.2 | 0.0002 | -2.2 | 0.0012 |
| p73 | Tumor protein p73 | NM005427 | 8.6 | 0.0021 | 10.1 | 0.0005 | 2.7 | 0.1584 |
Mice bearing i.p. LS-174T xenografts were treated by Gem/212Pb-trastuzumab for 24h. qRT-PCR array was used for gene expression analysis in three independent experiments. The numbers indicate fold change compared to untreated control (2-fold change cut-off). Additional groups included gemcitabine alone and Gem/212Pb-HuIgG as a nonspecific control antibody. Results represent the average of a minimum of three replicates. A p-value < 0.05 was considered significantly significant.
Gem/α-radiation treatment-induced tumor cytotoxicity may be associated with differentially expression of genes in the regulation of cell cycle arrest and cell cycle check point
The panel of genes in this study contained 15 cell cycle arrest and 8 cell cycle checkpoint regulatory genes. Of the 23 genes in these two categories, 6 genes (CHK1, CHK2, GTSE1, BRCA1, FANCG, and NBN) showed a >2 fold decrease and 4 genes (GADD45α, MAP2K6, PCBP4, and SESN) showed a >2 fold increase in expression from Gem/212Pb-trastuzumab treatment (Table 2). For tumors treated with Gem/212Pb-HuIgG, four genes (CHK1, GTSE1, BRCA1, and FANCG) decreased >2 fold while another 4 genes (GADD45α, MAP2K6, PCBP4, and SESN) showed a > 2 fold increase in expression. For those that decreased in expression, the level of fold change tended to be greater following the Gem/212Pb-trastuzumab treatment than from Gem/212Pb-HuIgG treatment. The inverse effect was exhibited for the genes whose expression increased whereby Gem/212Pb-HuIgG treatment tended to result in an enhanced level of effect versus that from Gem/212Pb-trastuzumab treatment. The greatest difference in the expression of CHK1 and GTSE1 was associated with Gem/212Pb-trastuzumab treatment versus tumors that received Gem/212Pb-HuIgG treatment. Additionally, five genes showed a change in expression that was > 2 fold due to treatment with Gem alone. With the exception of NBN, the level of expression tended to be lower than both the Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treatments.
Table 2. Differential expression of genes involved in cell cycle in LS-174T i.p. xenografts by Gemcitabine and α-treatment.
| Symbol | Gene name | GeneBank ID | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|
| Gemcitabine-212Pb-trastuzumab | p | Gemcitabine-212Pb-HuIgG | p | Gemcitabine | p | |||
| BRCA1 | Breast Cancer 1, early onset | NM007294 | -3.2 | 0.0020 | -3.1 | 0.0028 | -1.1 | 0.6268 |
| CHK1 | CHK1 checkpoint homolog | NM001274 | -4.5 | 0.0001 | -3.4 | 0.0002 | -1.6 | 0.0029 |
| CHK2 | CHK2 checkpoint homolog | NM007194 | -2.6 | 0.0015 | -1.9 | 0.0051 | -1.0 | 0.8729 |
| FANCG | Francomianemia, complementation group G | NM004629 | -2.8 | 0.0007 | -2.1 | 0.0019 | 1.5 | 0.0788 |
| GADD45α | Growth arrest and DNA-damage-inducible, alpha | NM001924 | 4.5 | 0.0047 | 5.8 | 0.0004 | 3.0 | 0.0005 |
| GTSE1 | G-2 and S-phase expressed 1 | NM016426 | -5.2 | 0.0020 | -4.3 | 0.0025 | -2.0 | 0.0152 |
| MAP2K6 | Mitogen activated protein kinase kinase 6 | NM002758 | 2.1 | 0.0032 | -2.4 | 0.0047 | 1.7 | 0.0078 |
| NBN | Nibrin | NM002485 | -2.1 | 0.0068 | -1.8 | 0.0093 | -3.0 | 0.0032 |
| PCBP4 | Poly(rC)binding protein 2 | NM020418 | 2.7 | 0.0048 | 3.2 | 0.0001 | 3.0 | 0.0011 |
| SESN1 | Sestrin1 | NM014454 | 3.7 | 0.0011 | 3.9 | 0.0023 | 2.9 | 0.0181 |
Four genes associated with cell cycle arrest (GADD45α, MAP2K6, PCBP4, and SESN1) demonstrated increased expression while three genes (CHK1, CHK2, and GTSE1) decreased in expression (Table 2). Three genes (BRCA1, FANCG, and NBN) associated with cell cycle checkpoint elicited a decrease in gene expression. Of those genes, an alteration in BRCA1 (-3.2-fold decrease, p < 0.0020) and FANCG (-2.8-fold decrease, p < 0.0007) gene expression was noted when compared to those tumors that were treated with Gem (Gem/212Pb-trastuzumab vs Gem, p < 0.05). In contrast, no significant differences in gene expression were observed for those same genes for Gem/212Pb-trastuzumab versus Gem/212Pb-HuIgG treated tumors.
α-Radiation plus gemcitabine-induced cell killing is associated with a decrease in expression of damaged DNA repair genes
The profiling study using the PCR array also demonstrated that several genes associated with DNA repair pathways were significantly affected after exposure to GEM/212Pb-trastuzumab (Table 3). Genes pivotal in major DNA repair pathways including nucleotide excision repair (NER), base-excision (BER), mismatch repair (MMR), and double-strand break repair (DSB) are categorized in S1 Table. A total of twelve genes (BRCA1, DMC1, EXO1, FANCG, FEN1, MSH2, PRKDC, RAD18, RAD51B, p73, UNG, and XRCC2) were found to be clearly impacted in those tumors treated with Gem/212Pb-trastuzumab versus those treated with Gem alone. Interestingly, only three genes (RAD18, XRCC2, and p73) among these twelve demonstrated a significant difference between the tumors treated with Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG. RAD18 and XRCC2 fall into a category of genes related to damaged DNA binding (DDB) while p73 is involved in MMR. As noted previously for p73, tumors from the Gem/212Pb-HuIgG group demonstrated a somewhat higher increase (Gem/212Pb-trastuzumab vs. Gem/212Pb-HuIgG, p < 0.05) than those tumors treated with Gem/212Pb-trastuzumab (a 8.6-fold increase, p < 0.00219 vs. a 10.7-fold increase, p < 0.0005). RAD18 and XRCC2 demonstrated a decrease (-3.2-fold decrease, p < 0.0001; -3.2-fold decrease, p < 0.0006) in expression as well as a significant difference between Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG, (p < 0.05) treated tumors. The rest of the genes among these twelve were all down-regulated. However, the differences in gene expression for those genes between the Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treated tumor tissue were negligible. Seven genes (BRCA1, DMC1, FANCG, FEN1, MSH2, RAD18, and RAD51B) are involved in DDB while EXO1 and MSH2 are associated with MMR. FEN1 and PRKDC are involved in DSB repair while UNG is the only gene related to BER.
Table 3. Differential expression of gene expression involved in DNA repair in LS-174T i.p. xenografts by Gemcitabine and α-treatment.
| Symbol | Gene name | GeneBank ID | Fold Change | |||||
|---|---|---|---|---|---|---|---|---|
| Gemcitabine-212Pb-trastuzumab | p | Gemcitabine-212Pb-HuIgG | p | Gemcitabine | p | |||
| BRCA1 | Breast Cancer 1, early onset | NM007294 | -3.2 | 0.0020 | -3.1 | 0.0028 | -1.1 | 0.6268 |
| BTG2 | BTG family, member 2 | NM006763 | 4.6 | 0.0001 | 4.9 | 0.0004 | 4.8 | 0.0001 |
| DMC1 | DNC1 dose suppressor of mck1 homolog | NM007068 | -3.0 | 0.0009 | -2.9 | 0.0046 | -1.2 | 0.0519 |
| ERCC1 | Excision repair cross-complementing rodent repair efficiency, complementation group1 | NM001983 | 2.6 | 0.0289 | 3.0 | 0.0001 | 1.8 | 0.0436 |
| EXO1 | Exonuclease 1 | NM130398 | -3.9 | 0.0003 | -3.2 | 0.0003 | -2.1 | 0.0030 |
| FANCG | Fanconi anemia, complementation group G | NM004629 | -2.8 | 0.0007 | -2.1 | 0.0019 | 1.5 | 0.0788 |
| FEN1 | Flap structure-specific endonuclease 1 | NM004111 | -3.1 | 0.0035 | -2.9 | 0.0039 | -1.3 | 0.1582 |
| MSH2 | MutS homolog 2 | NM000251 | -3.3 | 0.0059 | -3.0 | 0.0068 | -2.1 | 0.0151 |
| MSH3 | MutS homolog 3 | NM002439 | -2.0 | 0.0752 | -1.8 | 0.0898 | -1.8 | 0.0113 |
| NBN | Nibrin | NM002485 | -2.1 | 0.0068 | -1.8 | 0.0093 | -3.0 | 0.0032 |
| NTHL1 | Nth endonuclease III-like 1 | NM002528 | -2.1 | 0.0006 | -1.8 | 0.0001 | -1.9 | 0.0002 |
| OGG1 | 8-oxoguanine DNA glycosylase | NM002542 | -2.0 | 0.1458 | -1.9 | 0.1657 | -1.5 | 0.2915 |
| PRKDC | Protein kinase, DNA-activated, catalytic polypeptide | NM006904 | -3.0 | 0.0100 | -2.6 | 0.0112 | -2.0 | 0.0204 |
| RAD18 | RAD18 homolog | NM020165 | -3.2 | 0.0001 | -2.2 | 0.0001 | -1.0 | 0.9928 |
| RAD21 | RAD21 homolog | NM006265 | -2.4 | 0.0003 | -2.2 | 0.0002 | -2.2 | 0.0012 |
| RAD51B | RAD51 homolog B | NM133509 | -2.4 | 0.0072 | -1.9 | 0.0098 | -1.1 | 0.4491 |
| SEMA4A | Semadomain, immunoglobulin domain, cycloplastic domain 4A | NM022367 | 2.2 | 0.0795 | 2.9 | 0.0030 | 3.7 | 0.0002 |
| p73 | Tumor protein p73 | NM005427 | 8.6 | 0.0021 | 10.7 | 0.0005 | 2.7 | 0.1584 |
| UNG | Uracil-DNA glycosylase | NM003362 | -2.9 | 0.0010 | -2.2 | 0.0017 | -1.2 | 0.0450 |
| XPC | Xeroderma pigmentosum, complementation group C | NM004628 | 4.9 | 0.0004 | 5.1 | 0.0003 | 5.7 | 0.0211 |
| XRCC2 | X-ray repair complementing defective repair in Chinese hamster cells 2 | NM005431 | -3.2 | 0.0006 | -2.2 | 0.0007 | -1.2 | 0.2123 |
212Pb-trastuzumab with gemcitabine pre-treatment may interfere with DNA damage repair
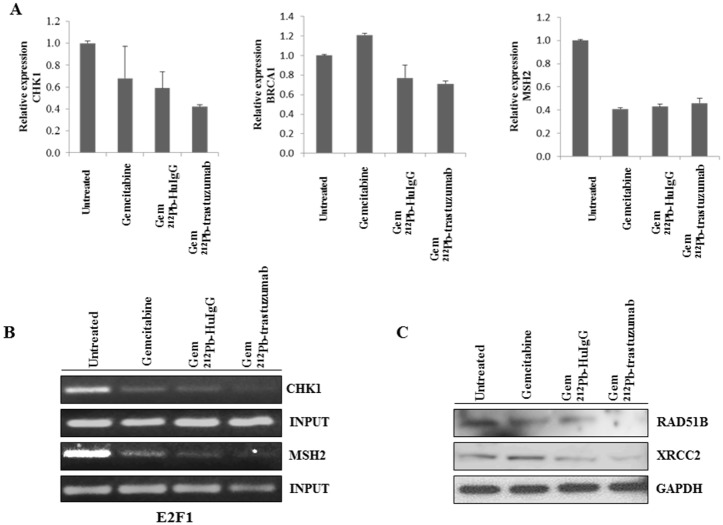
Based on the differentially expressed genes, further inquiry into possible pathways involved in the cell killing effect of Gem/212Pb-trastuzumab was initiated. Among those genes identified in the gene expression profile, BRCA1, MSH2, MSH3, and NBN were found down-regulated after exposure to α-radiation with Gem pretreatment. These genes are involved in BRCA1-associated genome surveillance complex (BASC) complex composed of MSH2, MSH3, MSH6 and MLH1, as well as ATM, NBN, MRE11, and BLM [28]. The expression of BRCA1 and MSH2 was determined at the transcriptional level to investigate the effect of targeted α-radiation on BASC. In response to Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treatment, expression of BRCA1 at the transcriptional level was attenuated to a greater degree than the treatment of Gem only suggesting that defects in transcription-coupled repair systems including mismatch repair (MSH2) and DNA double stand repair (BRCA1) might occur (Fig 1A).
Fig 1. Expression of genes related to BASC (BRCA1-associated genome surveillance complex) and HRR in response to sequential treatment with Gem and 212Pb-trastuzumab.
Mice bearing i.p. LS-174T xenografts were pre-treated with Gem followed 24 h thereafter with 212Pb-RIT. (A) Expression of BRCA1, CHK1, MSH2 and was determined by qRT-PCR. Results represent the average of a minimum of three replications. (B) Binding abundance to E2F1 was determined by ChIP using specific primers for CHK1 and MSH2. (C) Immunoblot analysis for RAD51B and XRCC2 was performed with tumor tissue collected as described. RAD51B and XRCC2 were detected at 32 kDa and 42 kDa, respectively. Equal protein loading control was GAPDH.
A greater reduction in the expression of CHK1 is also evident in the LS-174T tumors that had been treated with Gem/212Pb-trastuzumab (p < 0.05) and Gem/212Pb-HuIgG (p < 0.05). CHK1 and MSH2 have binding sites for E2F, a transcription factor which is involved in DNA replication and DNA damage repairs [29, 30]. To investigate whether E2F may mediate an expression of those genes by recruitment of E2Fs to their promoter regions following the combined treatment of GEM/212Pb-trastuzumab, the binding of E2F1 to the CHK1 and MSH2 promoters were evaluated using a ChIP assay. As shown in Fig 1B, the association of E2F1 on CHK1 and MSH2 promoters appeared to be attenuated by Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treatment, suggesting that modulation of these genes may occur via a decrease in binding of the active transcription factor, E2F1, to the promoter region.
Among genes identified in the gene expression profile, XRCC2 and RAD51B, which are RAD51 paralogs [31], appeared to be down-regulated after exposure to Gem/212Pb-trastuzumab. To examine the effect of Gem/212Pb-trastuzumab on damaged DNA repair, the expression of RAD51B and XRCC at the protein level were determined using immunoblot analysis. The results indicated that Gem/212Pb-trastuzumab attenuated expression in both proteins, suggesting the inefficient HR repair by Gem/212Pb-trastuzumab may be involved (Fig 1C).
Cell killing induced by Gem/α-radiation treatment may be associated with p73 signaling
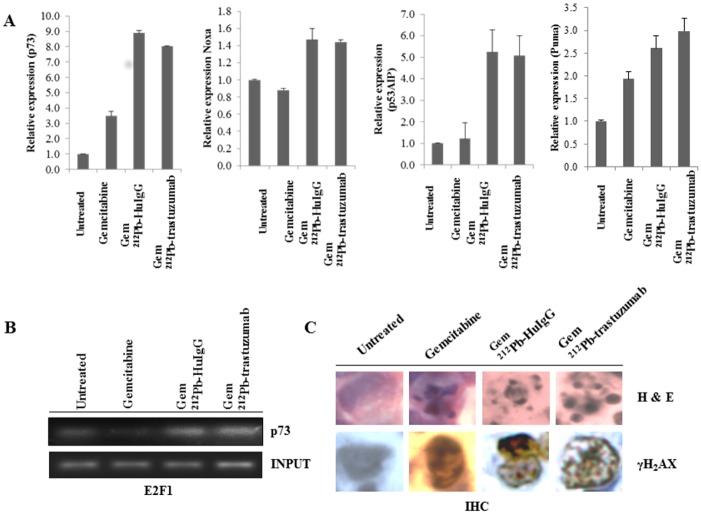
Gem/212Pb-trastuzumab treatment significantly altered the expression of p73 (8.6-fold increase, p < 0.0021) as demonstrated in the gene profiling study. To investigate the role of p73 induced apoptosis in LS-174T i.p. xenografts harvested from mice treated with Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG, the expression of p73 at the transcriptional level was first determined using PCR. Expression of p73 was significantly increased in the tumors treated with Gem/212Pb-trastuzumab as compared the ones treated with Gem only (Gem/212Pb-trastuzumab vs. Gem, p < 0.05). Expression of down-stream effectors of p73 including NOXA, PUMA, and P53AIP1 was also examined at the transcription level (Fig 2A). Gem/212Pb-trastuzumab increased the expression of NOXA, PUMA, and P53AIP1, compared to Gem only treated tumor (Gem/212Pb-trastuzumab vs. Gem, NOXA and PUMA, p < 0.05; P53AIP1, p < 0.01). There were only modest to negligible differences between Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treated tumors amongst these genes. p73 is also an E2F target gene [32]. ChIP analysis revealed abundant E2F1 on the p73 promoter to effect increased expression in both the Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treatment groups, suggesting the E2F1/p73 signaling may be activated after exposure to α-radiation with Gem pretreatment (Fig 2B). Next, to determine whether Gem/212Pb-trastuzumab induces DNA damage and apoptosis, immunohistochemistry (IHC) was performed using γH2AX and Haemotoxylin and Eosin (H&E) staining. DNA double strand damage and multi-micronuclei was evident from α-radiation with Gem pretreatment at 24 h as compared to the control groups (Fig 2C), indicating that DNA damage by α-radiation with 212Pb potentiates cell death to a greater extent than a Gem mono-therapy.
Fig 2. Gem/212Pb-trastuzumab may induce expression of p73, resulting in apoptosis.
Mice bearing i.p. LS-174T xenografts were pre-treated with Gem followed 24 h thereafter with 212Pb-RIT. (A) Expression of p73, NOXA, p53AIP1, and PUMA was determined by qRT-PCR. Results represent the average of a minimum of three replications. (B) Binding abundance to E2F1 was determined by ChIP using specific primer for p73. (C) Immunohistochemical analysis using γH2AX and H&E staining was performed with tumor tissue collected as described.
Discussion
There is no guarantee that conventional radiation therapy procedures will consistently result in an efficient therapeutic response for the treatment of undetected metastatic or disseminated cancers. Targeted α-radiation therapy using biological vectors such as monoclonal antibodies (mAbs) against tumor associated antigens, may serve as magic bullets in a coordinated strategy to cure these diseases [9]. Targeted α-particle therapy with 212Pb-trastuzumab was successfully applied for the treatment of disseminated i.p. disease in murine xenograft models [14–16]. Based on these preclinical results, clinical translation to a Phase I trial has been successfully performed without toxicity at the University of Alabama [33, 34]. Gemcitabine is a clinically proven radiation sensitizer and improves therapeutic response in the treatment of locally advanced, metastatic and non-metastatic diseases [17]. Therapeutic efficacy of 212Pb-trastuzumab was even greater when employed with addition of Gem to the treatment protocol in the LS-174T i.p. tumor xenograft model [15]. Yong et al recently demonstrated that application of the combined modality of Gem/212Pb-trastuzumab not only abrogated G2 arrest but also impaired DNA damage repair in the same model [35]. To further understand in vivo mechanisms on a molecular basis, gene expression profiling was performed in LS-174T i.p. tumor xenografts after exposure to 212Pb-trastuzumab and gemcitabine.
Herein, a total of 84 genes associated with DNA damage response were analyzed using a real-time quantitative PCR (qRT-PCR) array 24 h after Gem/212Pb-RIT treatment of LS-174T tumor xenografts. In each of the functionally classified categories such as apoptosis, cell cycle regulation, and damaged DNA repair (S1 Table), differentially expressed genes by Gem/212Pb-trastuzumab were compared to Gem mono-therapy. In many of these instances the level of gene expression was similar to Gem/212Pb-HuIgG, an indication that a strong α-radiation effect occurs in the presence of Gem.
Six genes (CIDEA, GADD45α, GADD45γ, IP6K3, PCBP4, and p73) involved in apoptosis were affected in the α-particle radiation treatment groups. Increased expression following α-particle radiation treatment was greater for p73 and GADD45α than for the group that received just Gem. In response to DNA damage, p73/GADD45 has been known to induce cell cycle arrest and cell death. Indeed, the induction of G2/M arrest and apoptosis through the p73/GADD45 signaling pathway by 212Pb-trastuzumab treatment has been recently reported from Yong et al [36].
Ten genes involved in the cell cycle were differentially regulated by Gem/212Pb-trastuzumab compared to Gem mono-therapy. The effect of Gem alone was not pronounced in those genes. BRCA1, CHK1, CHK2, GTSE1, and FANCG were down-regulated by Gem/212Pb-trastuzumab treatment. While alteration in gene expression between Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG was negligible for some of the genes, expression of CHK1 (-4.5-fold, p < 0.0001) after Gem/212Pb-trastuzumab treatment was substantially lower compared to either the Gem/212Pb-HuIgG or Gem treatments. Sensitization of tumor cells to cell death through inhibition of the DNA damage response is a promising strategy for enhancing therapeutic efficacy in the treatment of cancers. As a mediator of DNA damage response, checkpoint kinase 1 (CHK1) generally coordinates cell cycle arrest and DNA damage repair. CHK1 and CHK2 have been found to play pivotal roles in checkpoint functions of ATR and ATM. In fact, CHK1 deficiency has been found to inhibit the activation of G2/M resulting in suppression of proliferation in response to radiation [37–39]. Thus, decreased CHK1 and CHK2 expression by a combined Gem/212Pb-trastuzumab treatment may be significant to the response of the cancer cells.
Among those genes associated with DNA repair, twelve genes (BRCA1, DMC1, EXO1, FANCG, FEN1, MSH2, PRKDC, RAD18, RAD51B, p73, UNG, and XRCC2) were differentially expressed in the LS-174T tumor xenografts following Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG treatments. Gem mono-therapy resulted in negligible effects on these twelve genes in this study. Compared to results in a previous study that related treatment with 212Pb-trastuzumab alone [36], more genes were down-regulated in their expression by the Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG, suggesting compromised efforts to overcome the stressful conditions invoked by a combined modality of targeted α-radiation and Gem. Comparison of the differential expression of the DNA damage repair genes shows a greater negative expression for XRCC2 and RAD18 for the Gem/212Pb-trastuzumab group than the Gem/212Pb-HuIgG group. However, for most of the other genes, the difference in the gene expression response between the two groups was negligible.
Among those genes identified in the profile, the four involved in the BRCA1-associated genome surveillance complex (BASC), BRCA1, MSH2, MSH3, and NBN, were down-regulated by Gem/212Pb-trastuzumab and Gem/212Pb-HuIgG. In response to DNA damage, BASC may play an important role as sensors of abnormal DNA structure or as effectors of DNA damage repair [28]. Loss of BRCA1 function results in abnormal G2/M checkpoint, causing genetic instability [40, 41]. The defect in MSH2 function is associated with inhibition of CHK1 and CHK2 and abrogated RAD51, leading to suppression of cell proliferation in response to radiation [42, 43]. As indicated in the results, aberrant regulation of the BRCA1-associated target genes such as CHK1 and MSH2 was observed by ChIP analysis. The observed results here suggest that a defective ATR/CHK1 signaling pathway mediated by BRCA1/MSH2 may be involved in suppression of cell proliferation by Gem/212Pb-trastuzumab. Interaction of CHK1 and RAD51, which are required for HR, may be disrupted in BRCA1 deficient cells [44]. Defects in the RAD51 paralog genes result in abnormal recombinational repair, causing genomic instability [31]. Treatment with Gem/212Pb-trastuzumab also down-regulated expression of RAD51B, and XRCC2 as evidenced by the gene expression profiling and immunoblot analysis. These observations suggest that maintenance of genomic integrity through recombinational repair may be impaired by Gem/212Pb-trastuzumab. Previously, sensitization of tumor treated with Gem/212Pb-trastuzumab was shown to result in inhibition of checkpoint and impaired DNA damage repair [35]. As observed here, the lower expression of CHK1, MSH2, BRCA1, and RAD51 palalog genes together bolsters the earlier findings. The failure to correctly perform checkpoint response and DNA repair could also correlate with the observed inability to maintain the G2/M arrest by Gem/212Pb-trastuzumab. Therefore, targeting genes associated with the checkpoint signaling pathway and also DNA damage repair may be an attractive therapeutic strategy to take advantage of these two interlinked processes.
p73 is functionally and structurally related to p53 [45]. The α-particle radiation and Gem combined modality effects a greater cell killing more than likely through activation of the p73 signaling pathway as previously observed when tumors were treated with just 212Pb-trastuzumab [36]. Indeed, up-regulation of p73 also induced expression of its downstream effectors (NOXA, PUMA, and p53AIP1) by Gem/212Pb-trastuzumab, suggesting that the GADD45/p73 signaling pathway is activated after exposure to the combination of Gem and 212Pb-trastuzumab. ChIP analysis elicited an increased binding capacity of E2F1 on the p73 promoter, suggesting that the enhancement of apoptosis may be associated with active E2F1/p73 signaling. In p53 inactivated cells, up-regulation of p73 expression is mediated through E2F-1, suggesting an intrinsic rescuing mechanism may occur to compromise the loss of p53 function [46]. In vivo cell death mechanisms by the α-particle radiation are tremendously complex in the interlinked biological processes. Among those genes modulated after exposure to α-particle radiation, GADD45, IP6K3 (inositol hexakisphosphate kinase 3), and PCBP4 (Poly(rC)-binding protein 4) have been previously known to be mediated by p53-regulated signaling pathway, leading to apoptosis. However, gene expression of p53 has not been observed in gene profiling after either exposure to 212Pb-TCMC-trastuzumab [36] or Gem/212Pb-TCMC-trastuzumab. It has been known that RIT may induce lethal impact on radio-resistant tumors regardless of p53 gene status. Therefore, activation of p73 may play a pivotal role in the interlinked biological processes, leading to cell death in tumors that lack a p53-regulated signaling pathway.
The possibly predicted pathways that control cell cycle arrest and DNA damage repair, resulting in cell death have been demonstrated as depicted in Fig 3. DNA repair and checkpoint response are two interlinked processes. In response to the combined treatment of Gem and 212Pb-trastuzumab, one must note that there is an extensive interplay between the signaling pathways of checkpoint and DNA damage repair leading to severe growth arrest. While a need to improve the therapeutic efficacy of α-particle RIT combined with chemotherapy exists, the successful development and application of new tools such as a gene expression profiling and the elucidation of the fundamental molecular mechanisms in action during these combination therapies could aid in optimization of the combinations of chemotherapy reagents with radiation therapy as well as the sequence of their administration leading to augmented and enhanced radiotherapy choices for future clinical trials.
Fig 3. Interplay between DNA damage repair and check point signaling in the stressful growth arrest conditions by Gem/ 212Pb-trastuzumab.
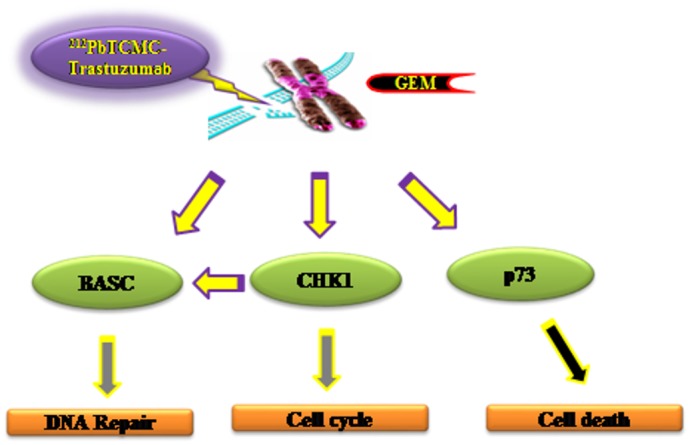
See text for details.
Supporting Information
Comparison of the relative expression of 84 DNA damage related genes involved in apoptosis (Table 1), cell cycle (Table 2), and DNA damage repair (Table 3) was characterized with the human DNA damage signaling pathway PCR array.
(PPT)
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research and AREVA Med LLC.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, and AREVA Med LLC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Goodhead DT. (1994) Initial events in the cellular effects of ionizing: clustered damage in DNA. Int J Radiat Biol 65:7–17. [DOI] [PubMed] [Google Scholar]
- 2.Soyland C, Hassfjell SP. (2000) Survival of human lung epithelial cells following in vitro alpha particle irradiation with absolute determination of the number of alpha-particle traversals of individual cells. Int J Radiat Biol 76:1315–1322. [DOI] [PubMed] [Google Scholar]
- 3.Hei TK, Wu LJ, Liu SX, Vannais D, Randers-Pehrson G. (1997) Mutagenic effects of a single and an exact number of alpha particles in mammalian cells. Proc Natl Acad Sci USA 94:3765–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakely EA, Kronenberg A. (1998) Heavy-ion radiobiology: new approaches to delineate mechanisms underlying enhanced biological effectiveness. Radiat Res 150:S126–145. [PubMed] [Google Scholar]
- 5.Azure MT, Archer RD, Sastry KS, Rao DV, Howell RW. (1994) Biological effect of lead-212 localized in the nucleus of mammalian cells: role of recoil energy in the radiotoxicity of internal alpha-particle emitters. Radiat Res 140:276–283. [PMC free article] [PubMed] [Google Scholar]
- 6.Barbet J, Bardies M, Bourgeois M, Chatal JF, Cherel M, Davodeau F, et al. (2012) Radiolabeled antibodies for cancer imaging and therapy. Methods Mol Biol 907:681–697. 10.1007/978-1-61779-974-7_38 [DOI] [PubMed] [Google Scholar]
- 7.Navarro-Teulon I, Lozza C, Pelegrin A, Vives E, Pouget JP. (2013) General overview of radioimmunotherapy of solid tumors. Immunotherapy 5:467–487. 10.2217/imt.13.34 [DOI] [PubMed] [Google Scholar]
- 8.Jurcic JG. (2013) Radioimmunotherapy for hematopoietic cell transplantation. Immunotherapy 5:383–394. 10.2217/imt.13.11 [DOI] [PubMed] [Google Scholar]
- 9.Yong K, Brechbiel MW. (2011) Towards translation of 212Pb as a clinical therapeutic; getting the lead in! Dalton Trans 40:6068–6076. 10.1039/c0dt01387k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dahle J, Abbas N, Bruland OS, Larsen RH. (2011) Toxicity and relative biological effectiveness of alpha emitting radioimmunoconjugates. Curr Radiopharm 4:321–328. [DOI] [PubMed] [Google Scholar]
- 11.Kim YS, Brechbiel MW. (2012) An overview of targeted alpha therapy. Tumour Biol 33:573–590. 10.1007/s13277-011-0286-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seidl C. (2014) Radioimmunotherapy with α-particle-emitting radionuclides. Immunotherapy 6:431–458. 10.2217/imt.14.16 [DOI] [PubMed] [Google Scholar]
- 13.Yong KJ, Milenic DE, Baidoo KE, Brechbiel MW. (2012) 212Pb-Radioimmunotherapy induces G2 cell cycle arrest and delays DNA damage repair in tumor xenografts in a model for disseminated intraperitoneal disease. Mol Cancer Ther 11:639–648. 10.1158/1535-7163.MCT-11-0671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Milenic DE, Garmestani K, Brady ED, Albert PS, MA D, Abdulla A, et al. (2004) Targeting of HER2 antigen for the treatment disseminated peritoneal disease. Clin Cancer Res 10:7834–7841. [DOI] [PubMed] [Google Scholar]
- 15.Milenic DE, Garmestani K, Brady ED, Albert PS, Abdulla A, Flynn J, et al. (2007) Potentiation of high-LET radiation by gemcitabine: targeting HER2 with trastuzumab to treat disseminated peritoneal disease. Clin Cancer Res 13:1926–1935. [DOI] [PubMed] [Google Scholar]
- 16.Milenic DE, Garmestani K, Brady ED, Baidoo KE, Albert PS, Wong KJ, et al. (2008) Multimodality therapy: potentiation of high linear energy transfer radiation with paclitaxel for the treatment of disseminated peritoneal disease. Clin Cancer Res, 14:5108–5115. 10.1158/1078-0432.CCR-08-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toschi L, Finocchoaro G, Bartolini S, Gioia V, Cappuzzo F. (2005) Role of gemcitabine in cancer therapy. Future Oncol 1:7–17. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence TS, Eisbruch A, Shewach DS. (1997) Gemcitabine-mediated radiosensitization. Semin Oncol 24:S7-24–S7-28. [PubMed] [Google Scholar]
- 19.Milas L, Fujii T, Hunter N, Elshaikh M, Mason K, Plunkett W, et al. (1999) Enhancement of tumor radioresponse in vivo by gemcitabine. Cancer Res 59:107–114. [PubMed] [Google Scholar]
- 20.Schwash DS, Hahn TM, Chang E, Hertel LW, Lawence TS. (1994) Metabolism of 2’,2’-difluoro-2’-deoxycytidine and radiation sensitization of human colon carcinoma cells. Cancer Res 54:3218–3223. [PubMed] [Google Scholar]
- 21.Chaudhry MA. (2008) Biomarkers for human radiation exposure. J Biomed Sci 15:557–563. 10.1007/s11373-008-9253-z [DOI] [PubMed] [Google Scholar]
- 22.Danielsson A, Claesson K, Parris TZ, Helou K, Nemes S, Elmroth K, et al. (2013) Differential gene expression in human fibroblasts after alpha-particle emitter 211At compared with 60Co irradiation. Int J Radiat Biol 89: 250–258. 10.3109/09553002.2013.746751 [DOI] [PubMed] [Google Scholar]
- 23.Chauhan V, Howland M, Mendenhall A, O’Hara S, Stocki TJ, McNamee JP, et al. (2012) Effect of alpha particle radiation on gene expression in human pulmonary epithelial cells. Int J Hyg Environ Health 215:522–535. 10.1016/j.ijheh.2012.04.004 [DOI] [PubMed] [Google Scholar]
- 24.Seidl C, Port M, Apostolidis C, Bruchertseifer F, Schwaiger M, Senekowitsch-Schmidtke R, et al. (2010) Differential gene expression triggered by highly cytotoxic alpha-emitter-immunoconjugates in gastric cancer cells. Invest New Drugs 28:49–60. 10.1007/s10637-008-9214-4 [DOI] [PubMed] [Google Scholar]
- 25.Seidl C, Port M, Gilbertz KP, Morgenstern A, Bruchertseifer F, Schwaiger M, et al. (2007) 213Bi-induced death of HSC-M2 gastric cancer cells is characterized by G2 arrest and up-regulation of genes known to prevent apoptosis but induce necrosis and mitotic catastrophe. Mol Cancer Ther 6:2346–2359. [DOI] [PubMed] [Google Scholar]
- 26.Tom BH, Rutzky LP, Jakstys MM, Oyasu R, Kaye CI, Kahan BD. (1976) Human colonic adenocarcinoma cells. I. Establishment and description of a new cell line. In Vitro 12:180–191. [DOI] [PubMed] [Google Scholar]
- 27.Milenic DE, Garmestani K, Brady ED, Albert PS, Ma D, Abdulla A, et al. (2005) Alpha-particle radioimmunotherapy of disseminated peritoneal disease using a 212Pb-labeled radioimmunoconjugate targeting HER2. Cancer Biother Radiopharm 20:557–568. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J. (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 927–939. [PMC free article] [PubMed] [Google Scholar]
- 29.Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, et al. (2007) The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res 67:6574–6581. [DOI] [PubMed] [Google Scholar]
- 30.Chang IY, Jin M, Yoon SP, Youn CK, Yoon Y, Moon SP, et al. (2008) Senescence-dependent MutS alpha dysfunction attenuates mismatch repair. Mol Cancer Res 6:978–989. 10.1158/1541-7786.MCR-07-0380 [DOI] [PubMed] [Google Scholar]
- 31.Yokoyama H, Sarai N, Kagawa W, Enomoto R, Shibata T, Kurumizaka H, et al. (2004) Preferential binding to branched DNA strands and strand-annealing activity of the human Rad51B, Rad51C, Rad51D and Xrcc2 protein complex. Nucleic Acids Res 32:2556–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seelan RS, Irwin M, van der Stoop P, Qian C, Kaelin WG Jr, Liu W. (2002) The human p73 promoter: characterization and identification of functional E2F binding sites. Neoplasia 4:195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meredith RF, Torgue J, Azure MT, Shen S, Saddekni S, Banaga E, et al. (2014) Pharmacokinetics and imaging of 212Pb-TCMC-trastuzumab after intraperitoneal administration in ovarian cancer patients. Cancer Biother Radiopharm 29:12–17. 10.1089/cbr.2013.1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meredith RF, Torgue J, Shen S, Fisher DR, Banaga E, Bunch P, et al. (2014) Dose escalation and dosimetry of first-in-human α-radioimmunotherapy with 212Pb-TCMC-trastuzumab. J Nucl Med 55:1636–1642. 10.2967/jnumed.114.143842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yong KJ, Milenic DE, Baidoo KE, Brechbiel MW. (2012) Sensitization of tumor to 212Pb-radioimmunotherapy by gemcitabine involves initial abrogation of G2 arrest and blocked DNA damage repair by interference with Rad51. Int J Radiat Onc 85:1119–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yong KJ, Milenic DE, Baidoo KE, Kim YS, Brechbiel MW. (2013) Gene expression profiling upon 212Pb-TCMC-trastuzumab treatment in the LS-174T i.p. xenograft model. Cancer Med 2:646–653. 10.1002/cam4.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, et al. (2005) The-cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7:195–201. [DOI] [PubMed] [Google Scholar]
- 38.Bucher N, Britten C. (2008) G2 checkpoint abrogation and checkpoint kinase I targeting in the treatment of cancer. Br J Cancer 98:523–528. 10.1038/sj.bjc.6604208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith J, Tho LM, Xu N, Gillespie DA. (2010) The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res 108:73–82. 10.1016/B978-0-12-380888-2.00003-0 [DOI] [PubMed] [Google Scholar]
- 40.Deng CX. (2006) BRCA1: cell cycle checkpoint, genetic instability, DNA damage response and cancer evolution. Nucleic Acids Res 34:1416–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamane K, Schupp JE, Kinsella TJ. (2007) BRCA1 activates a G2-M cell cycle checkpoint following 6-thioguanine-induced DNA mismatch damage. Cancer Res 67:6286–6292. [DOI] [PubMed] [Google Scholar]
- 42.Franchitto A, Pichierri P, Piergentili R, Crescenzi M, Bignami M, Palitti F. (2003) The mammalian mismatch repair protein MSH2 is required for correct MRE11 and Rad51 relocalization and for efficient cell cycle arrest induced by ionizing radiation in G2 phase. Oncogene 22:2110–2120. [DOI] [PubMed] [Google Scholar]
- 43.Hong Z, Jiang J, Hashiguchi K, Hoshi M, Lan L, Yasui A. (2008) Recruitment of mismatch repair proteins to the site of DNA damage in human cells. J Cell Sci 121:3146–3154. 10.1242/jcs.026393 [DOI] [PubMed] [Google Scholar]
- 44.Jia Y, Song W, Zhang F, Yan J, Yang Q. (2013) Akt1 inhibits homologous recombination in Brca1-deficient cells by blocking the Chk1-Rad51 pathway. Oncogene 32:1943–1949. 10.1038/onc.2012.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossi M, Sayan AE, Terrinoni A, Melino G, Knight RA. (2004) Mechanism of induction of apoptosis by p73 and its relevance to neuroblastoma biology. Ann NY Acad Sci 1028:143–149. [DOI] [PubMed] [Google Scholar]
- 46.Tophkane C, Yang SH, Jiang Y, Ma Z, Subramaniam D, Anant S, et al. (2012) p53 inactivation up-regulates p73 expression through E2F1 mediated transcription. Plos One 7:e43564 10.1371/journal.pone.0043564 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of the relative expression of 84 DNA damage related genes involved in apoptosis (Table 1), cell cycle (Table 2), and DNA damage repair (Table 3) was characterized with the human DNA damage signaling pathway PCR array.
(PPT)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.