Abstract
Recent work in sequential fluorescent in-situ hybridization (FISH) has demonstrated the ability to uniquely encode a large number of molecular species in single cells. However, the multiplexing capacity is practically limited by the density of the barcoded objects in the cell. Here, we present a general method using image correlation to resolve the temporal barcodes in sequential hybridization experiments, allowing high density objects to be decoded. Using this correlation FISH (corrFISH) approach, we profiled the gene expression of ribosomal proteins in single cells in cell cultures and in mouse thymus tissue sections. In tissues, corrFISH revealed cell type specific gene expression of ribosomal proteins. The combination of sequential barcoding FISH and correlation analyses provides a general strategy for multiplexing a large number of RNA molecules and potentially other high copy number molecules in single cells.
Profiling molecules such as RNAs or proteins in cells is the key to explore cell identities and can reveal patterns in gene regulatory networks. The ability to barcode a large number of different mRNA species in single cells has been solved by the Sequential Coding anALYSis of Fluorescent In Situ Hybridizations (FISH SCALYS) approach1. A recent implementation using binary sequential barcodes demonstrated the multiplex of 100–1,000 transcripts in single cells2. Exciting in-situ sequencing approaches can also decode the RNA species with nucleotide resolution3,4. However, barcoding a large number of transcripts can result in a high-density of spots within the cell, which makes resolving individual spots and barcode readout across the hybridizations difficult. While we had previously demonstrated that super-resolution microscopy improved the detection of abundant RNA molecules6, and others have demonstrated the use of advanced algorithms including compressed sensing to assist super-resolution microscopy7,8, a robust method to decode high copy number RNAs in highly multiplexed, sequentially barcoded FISH experiments with conventional fluorescence microscopy is needed.
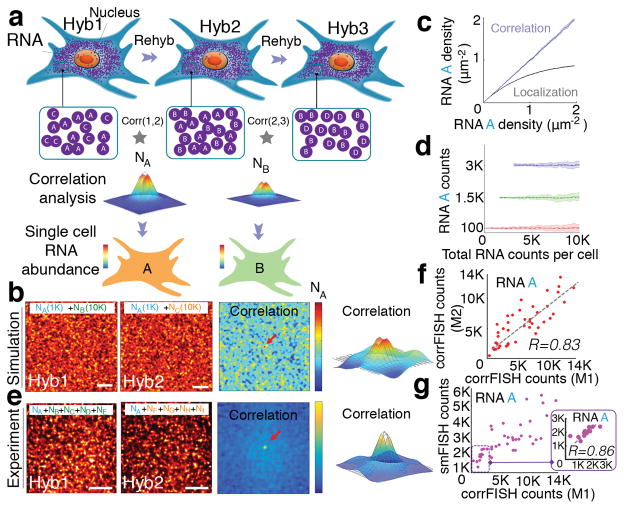
Here, we present an image correlation method to decode the temporal barcode in FISH SCALYS experiments. The principle can be illustrated by a simple example: we encode RNA species such that each RNA species appears in only two of out of a total of 3 rounds of hybridization (Fig 1a). For example, RNA B appears in rounds 2 and 3. While there are other RNAs also labeled in round 2 or round 3, no other RNAs are labeled in both round 2 and 3. Thus, only FISH spots that correspond to RNA B are in the same positions between hyb 2 and hyb 3. By cross-correlating the images from hyb 2 and hyb 3, only RNA B will generate a positive correlation (Fig. 1b) with an amplitude that is proportional to the number of RNA B molecules (Fig. 1c) while the other RNA species will not be correlated and will not contribute to the cross-correlation. Thus, the copy number of RNA B can be extracted from the cross-correlation of the images corresponding to the RNA B barcode assignment even when the images are dense. Similarly, RNA A appears only in rounds 1 and 2, the abundance of which can be decoded from the cross-correlation of hyb1 and hyb2 images. Since each gene is assigned a unique temporal barcode, the correlation of the images in the channels corresponding to the barcode provides the copy number of that gene.
Figure 1.
Correlation FISH. (a) Schematic of sequential hybridizations and barcoding of dense mRNA molecules in a cell. Correlation analysis of any two subsets of hybridization images reveals the amount of common RNA molecules. Hyb 1 and Hyb 2 images share the RNA A in the presence of other uncorrelated RNA B and C in each channel. Pair-wise correlations across Hybs (1,2) and (2,3) peak values are estimators of the copy number of RNA A (NA) and B (NB), respectively in that cell (b, c). (d) Simulated images of a 30 μm cell with 1000 molecule A in the presence of 10,000 molecule B or molecule C in hybridizations 1 and 2. Correlation of these images provided a peak value (denoted by the red arrow and 3D plot) that corresponds to the copy number of A. (e) Simulated RNA molecules are measured using both correlation and spatial localization. Correlation accurately estimates RNA levels even at high density of RNAs. (f) Detection accuracy analysis with 100, 1500, 3000 copies of A in the presence of increasing concentrations of B and C in simulated data. RNA A levels were kept constant while increasing RNA B and C density (n=20 simulation runs). Dashed line shows the expected value of A transcripts. Even 100 molecules can be accurately detected out of a total of 10,000 molecules at a density of 10 molecule μm−2. (g) Experimental images of a 20 μm cellular regions with A gene (NA) in the presence of other 4 uncorrelated genes (NB, NC, ND, and NE) and (NF, NG, NH, and NI) in hybridizations 1 and 2, respectively. Corresponding correlation functions yield transcript abundance of gene A. Scale bars are 5 μm. (h) Transcript detection of A gene in five gene experiments using two different correlation FISH barcodes (M1: H1*H2 and M2: H2*Rps2 from H3) with a linear fit of R = 0.83 and P<0.002 (Student’s t-test) for n=43 cells. (i) Comparison of transcript counting of A molecules based on single molecular FISH counting and correlation FISH quantification. Inset shows a correlation with R= 0.86 and P<0.0006 (Student’s t-test) in the linear regime, between 0–3,000 transcripts, where smFISH quantification is not saturated.
To simulate the conditions of a FISH SCALYS experiment, we generated images corresponding to two rounds of hybridization with only RNA A in common between the two images (mimicking 30 μm×30 μm cell size) with 10 fold excess of uncorrelated RNAs (Fig. 1d and Supplementary Fig. 1). We then calculated cross-correlation in between hyb1 and hyb2 images9–11. The cross-correlation shows a distinct peak at the center (Fig. 1d) and provides the RNA A copy number (NA) even when the spots overlap and no obvious colocalization occurs between the images. In our previous measurements, we observed that mRNA distributions in cells follow a Poisson distribution5 spatially. As the variance of the Poisson distribution is equal to its mean, the peak value at the cross-correlation is a good estimator for the copy number of NA. In addition, given that the higher order cumulants of the Poisson distribution are all equal to the mean, the higher order cross-correlation peaks are also good estimators for the corresponding barcode copy numbers. For simplicity, we will only implement pair-wise correlation for the proof-of-principle demonstrations.
The corrFISH counts scale linearly with the increasing density, while localization based method of counting FISH spots yield significant errors in estimating the copy number of RNAs at a density of more than 1 molecule μm−2 (Fig. 1e and Supplementary Fig. 2) In simulated data, target transcripts were measured at concentrations as low as 1% of the total molecule density: 100 A molecules in 30 μm cells were accurately measured in the presence of 10,000 B or C molecules with a coefficient of variation (CV) of 1.76±1.54 (S.E., n=20) (Fig 1f and Supplementary Fig. 3). Thus, the copy number of A molecules which comprise of only 1% of the total labeled RNAs (at a density of more than 10 molecules μm−2) can be estimated within a factor of 2 of the mean value.
For experimental demonstrations, we applied corrFISH to the quantification of highly abundant ribosomal protein transcripts in single cells. It has been shown that mice embryos have mosaic distribution of ribosomal protein gene expression in different tissue types12. Thus, it would be interesting to profile the abundances of these genes at the single cell level. Initially, we compared the quantification of corrFISH with smFISH measurement in the same cell. We performed a proof of concept demonstration to decode a transcript, Rps2, in mammalian cell cultures in the presence of other dense transcripts using sequential hybridizations. In the first hybridization, Rps2, denoted by A, is hybridized along with four other genes (B, C, D, and E). In hyb 2, A is hybridized with another four different genes (F, G, H, and I) (Fig. 1g and Supplementary Fig. 4). The cross-correlation of these two hybridization images provides an estimate for the Rps2 transcript abundance. As a control, in the third hybridization, we hybridized only Rps2 gene. This allows us to perform two controls. First, we compared the detection of Rps2 using corrFISH barcoding of H1*H2 (Method 1) to the H2*Rps2 from H3 (Method 2), providing a linear regression with R=0.83 and P<0.002 (Fig. 1h). Next, we compared corrFISH barcoding of H1*H2 to the smFISH counting in H3 that yielded an R= 0.86 value at the lower copy range (Fig. 1i). At high copy numbers, the overlapping FISH dots underestimate the transcript copy number in smFISH quantification. These experiments show that accurate quantitation of high copy number genes by corrFISH in the presence of other high copy number transcripts. In fact, scaling this approach to all 79 ribosomal protein mRNAs requires similar densities of 6 genes per hybridization image (Supplementary Fig. 5).
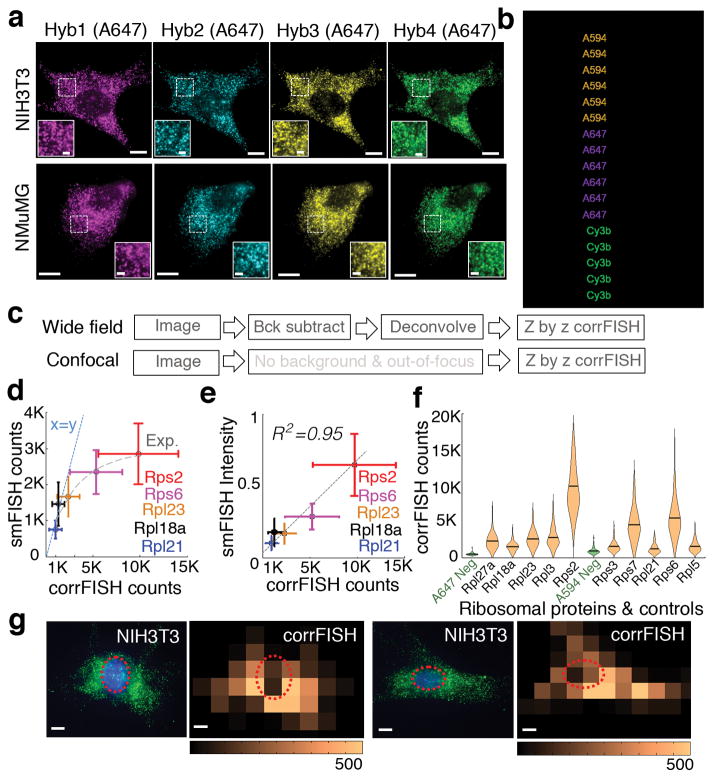
To test the performance of corrFISH, we targeted transcripts for ten ribosomal proteins in two phenotypically different cell lines: mouse embryonic fibroblast cells (NIH3T3) and mammary gland epithelial cells (NMuMG) (Fig. 2a). We used wide field and confocal microscopy for our binary barcoded transcript measurements (Fig. 2b, 2c, and Supplementary Figs. 6–8). To assess the dynamic range of corrFISH, we compared single molecule FISH counting to corrFISH quantification for five different ribosomal proteins of Rps2, Rps6, Rpl23, Rpl18a, and Rpl21 (Supplementary Fig. 9). While smFISH counting tended to underestimate the transcripts due to the overlapping FISH spots (Fig. 2d), the integrated intensity correlated well with corrFISH with an R2 value of 0.95 (Fig. 2e). Both of these smFISH comparisons validated that corrFISH achieves reliable detection over high dynamic range of up to 15,000 counts per cell. In our experiments, a target RNA molecule (1 out of 3 species) with 0.5–9 μm−2 density can be accurately quantified within a total density of 5–15 molecule μm−2 per hybridization by corrFISH within less than 10% error (Supplementary Fig. 10). To determine the sensitivity of corrFISH, we measured the amount of empty barcodes detected (Fig. 2f). The negative controls (the correlation of hybs 1 and 2) showed negligible correlation values and provided limit of detection (LOD) of 0.08 molecule μm−2 density (222±18S.E. counts/average 2,674 μm2 cellular area).
Figure 2.
corrFISH works accurately in cell cultures. (a) Images of ribosomal protein transcripts in NIH3T3 cells across four hybridizations in NIH3T3 and NMuMG cell types. Scale bars 10 μm and 2 μm (insets). (b) Barcode scheme for ten genes using two fluorophores and four rounds of hybridization. 1 corresponds to the probes with Alexa 594 dyes, 2 corresponds to probes labeled with Alexa 647. Additional color 3 corresponds to cy3b labeled probes that were used to perform one gene at a time as a control experiment for single molecule localization based counting. (c) Experimental images generated using a workflow of corrFISH processing for either wide field or confocal microscope. (d) Experimental characterization of dynamic range based on the comparison of single molecule FISH counting and corrFISH quantification. Five ribosomal protein genes, covering Rpl21, Rpl18a, Rpl23, Rps6, and Rps2, were plotted for smFISH counting versus corrFISH quantitation. smFISH significantly underestimates the transcripts at higher densities due to the overlapping FISH spots. At the highest copy numbers, 5000 smFISH counts corresponded to the 15000 corrFISH counts. A linear x=y line (dashed blue) and an exponential fit (grey dash) are shown for comparison. (e) Comparison of the summed intensity of single molecule FISH dots and correlation FISH quantification, agreeing well with each other (R2 = 0.95) for n=85 cells. (f) Single cell RNA distributions of 10 ribosomal protein genes with negative controls. The negative control (green), which is empty barcode, is detected at 0.08 molecule μm−2. This corresponds to the sensitivity limit, of 222 ±18 s.e. mRNAs per cell for 2,674 μm2 cellular area. (g) Spatial analysis via corrFISH using subregion analysis. NIH3T3 cell with ribosomal protein transcripts (Rpl3 and Rpl27a in hybridization round 2) in green and nucleus stained by DAPI shown in blue. Correlation based transcript abundance mapping over 25 sub-regions within the cell. Repeat hybridizations, both contain Rpl3 and Rpl27a, are correlated to generate the map. Another fibroblast with original image and the corresponding correlation map. Dashed circle shows the nuclear region in both of the microscopic images and correlation based transcript quantification result. Scale bar 10 μm.
corrFISH can also provide spatial maps of gene expression profiles. Subcellular details can be resolved by performing correlation analysis on subregions within cells (Supplementary Fig. 11). Transcript distribution within a cell was profiled in 25 subcellular regions for Rpl3 and Rpl27a mRNAs (Fig. 2g). In most of the fibroblast cells, ribosomal protein transcripts are detected outside of the nuclear region (circled), which is detected in the sub-region correlation analysis. We varied subregion window size and showed that it does not significantly affect the results (Supplementary Fig. 12).
When the distribution of RNAs is not Poisson distributed inside a cell or across the entire sample, corrFISH utilizes subregion analysis. The heterogeneous image from a sample is subdivided into regions where the mRNA distribution is relatively homogeneous and Poisson distributed. We then compile the final analysis from these sub-regions (Supplementary Fig. 13). We demonstrated the successful implementation of spatial analysis in the tissue sections and quantified ribosomal protein transcripts at single cell resolution (Fig. 3). The case when corrFISH quantitation breaks down is if there are RNA granules that contain multiple RNAs within a diffraction-limited region. These problematic regions can always be cropped out of the image for correlation analysis if they occur.
Figure 3.
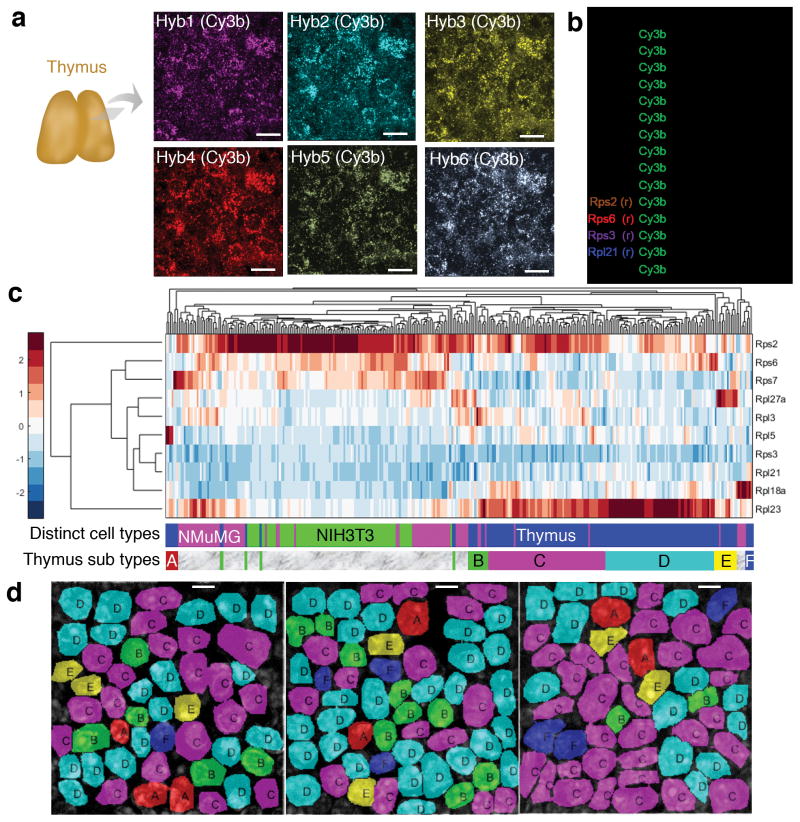
corrFISH reveals cell specific ribosomal protein gene expression in tissues and cell cultures. (a) 10 μm thick thymus sections were used for corrFISH experiments over six hybridizations with only Cy3B labeled probes. Scale bar 10 μm. (b) Barcodes were assigned to ten ribosomal protein genes, four repeats as a positive control (Rps2, Rps6, Rps3, and Rpl21), and empty as a negative control (Ctl). (c) Single cell gene expression profiles of ten ribosomal proteins in the combined data from cell cultures and thymus tissue sections. Distinct clusters were obtained for each cell type (NMuMG: 80 cells, NIH3T3: 85 cells, and Thymus: 162 cells). This indicates that there are distinct patterns of usage of ribosomal proteins in different cells. Six distinct groups (A–F) of cells in the thymus were determined based on clustering of ribosomal protein transcription profiles. (d) Spatial mapping of the cell clusters in the thymus section at 3 different locations showed heterogeneous distribution and layering of cell types. Scale bar 5 μm.
We quantified gene expression of ribosomal proteins with corrFISH in 10 μm thick thymus tissue sections (Fig. 3). It has been recently shown that ribosome function plays an important role in hematopoiesis14 and immune system15. Thus, it will be rather informative to explore ribosomal proteins in thymus. To barcode ten ribosomal proteins, we performed six hybridizations with a single color (Fig. 3b). The transcript distributions for these ribosomal protein genes are heterogeneous (Figs. 3d and Supplementary Figs. 14). The repeat barcodes were plotted against their counterparts, providing a linear correlation with R=0.9 value for 162 thymus cells (Supplementary Fig. 15), indicating that the quantitation in tissues are accurate.
To evaluate the molecular differences in phenotypically distinct cells, we clustered the gene expression of all the cellular populations covering 80 NMuMG, 85 NIH3T3, and 162 Thymus cells using a hierarchical heat map. These results exhibited unique clusters for NMuMG (Magenta), NIH3T3 (Green), and Thymus Cells (Blue) (Fig. 3c). The ability to define cell types based on their ribosomal protein composition supports the specialized ribosome theory13. Besides, NIH3T3 and NMuMG gene expression profiles for ten genes in each cell line showed significant expression variability in single cells and most exhibited a unimodal distribution (Supplementary Figs. 16 and 17). Rps2 had the highest expression level (Mean: 9800 copy/cell in NIH3T3 and 5000 copy/cell in NMuMG) while Rpl21 and Rpl18a had the lowest (Mean: 975 and 1284 copy/cell in NIH3T3; 981 and 1400 copy/cell in NMuMG for Rpl21 and Rpl18a, respectively). We observed combinatorial gene expression patterns for each cell type (Fig. 3c). Rps2 (high copy regulator) and Rpl21 (low copy regulator) were shared across clusters for both cell types, while other genes exhibit mosaic expression patterns that are unique for each cell type.
To ask whether thymus has spatial patterns of cell types with distinct gene expression profiles, we mapped the transcriptional profile back to the position of single cells on the microscopic image. Unique layering and grouping of six different subtypes (A, B, C, D, E, and F) of thymus cells were obtained on three distinct regions of the same thymus section (Fig. 3d and Supplementary Figs. 18). Cellular interactions in thymus play an important role to create proper immune cell repertoire, and thus, our direct observation of cellular subtypes will pave the way for studying spatial organization of developmental processes in immunology.
The presented corrFISH technique shows that resolving single molecule FISH spots is not required to quantify RNA abundances, removing the constraints on the FISH SCALYS approach to allow targeting RNAs with much higher expression levels. Conceptually, this is similar to the ability of fluorescence correlation spectroscopy (FCS)16 to quantify molecular concentration down to the single molecule level even in high concentration solutions. In our case, the time is generated by sequential rounds of hybridization rather than real time. In principle, this image correlation approach can be applied to decode high density images to multiplex molecular species other than RNA, including proteins17–20 and metabolites21.
Correlation analysis shows that high density transcripts to be multiplexed without super-resolution microscopy in culture and tissue samples. To cover a large dynamic range of transcript abundances, high copy number can be first barcoded by FISH SCALYS and analyzed by correlation method. Then, the probes can be stripped, and a second barcoding experiment in the same cell can be performed on the low abundance transcripts. Thus, this hybrid approach allows both low abundance genes to be accurately detected with single molecule resolution, and high copy number genes to be profiled in the same cell. Future experiments extending corrFISH to all ribosomal proteins as well as additional marker genes would allow the study of ribosome code in single cells within native tissue architectures.
ONLINE METHODS
Theory
For quantification of transcripts, we used correlation analyses on a series of FISH SCALYS images. We perform a pair-wise correlation from the subset of hybridization images for quantitation of each gene. For instance, we used equation 1 to compute cross-correlation in between hyb1 and hyb2 images9–11 (see Supplementary note for derivation).
| (1) |
where G12 is the cross-correlation of the images from hyb1 and hyb2. ℱ−1 is the inverse Fourier Transform and ℱ is for the Fourier Transform operations. H1 and H2 are the images. 〈H1〉 and 〈H2〉 are the mean value of the corresponding images (Brackets are expected values).
These correlation results were then converted to the transcript abundance by using equations 2 and 3. The amplitude of cross-correlation was normalized by the auto-correlation amplitudes as an estimator of RNA copy number in a cell per point spread function (PSF) area.
| (2) |
where 〈d12〉 is the abundance of transcripts in an PSF area that are common across hybridizations 1 and 2 (Brackets are expected values). G12 is the amplitude of the cross-correlations peak between the hyb 1 and 2 images. G11 and G12 are the auto-correlation peak amplitudes of hyb1 and hyb2, respectively. Transcript count per PSF area was then converted to the total number of transcripts for a single cell:
| (3) |
where 〈N12〉 is the number of transcripts that are common across hybridizations 1 and 2. Here, 〈d12〉 is the copy number of transcripts per PSF area from Equation 2. APSF is the PSF area (π × PSFwidth2), where PSFwidth is about 0.3 μm. ACell is the area of the single cell image (N × p2), where N is the total number of pixels in a cell and p is the pixel size of 0.13 μm in our corrFISH analysis.
Simulations
We used digital data to imitate the realistic FISH experiments at various densities and conditions. Using a custom written MATLAB algorithm, point emitters were randomly distributed in a digital image of size 230×230 pixels (corresponding to 30 μm × 30 μm cell area). Emitters were convolved with 0.3 μm width point-spread function (PSF) corresponding to a 100x wide field fluorescence microscope (0.13 μm pixel size), matching the cell sizes and pixel resolution in the FISH experiments. The obtained images mimic the molecules labeled with fluorescent dyes. Adjusting the total number of emitters per simulation area changed concentration of the molecules. Sequences of these images were correlated in spatial or Fourier domains to compute the abundances of molecules that are common across these image arrays (Supplementary note). In addition, we performed super resolution microscopy simulations with 0.05 μm pixel size to show that correlation decoding can even higher density multiplexing (Supplementary Fig. 19).
Cultures
NIH3T3 cells (ATCC CRL-1658) were cultured in Dulbecco’s Modified Eagle’s medium (DMEM) with GlutaMAX, high glucose, and pyruvate (Thermo Fisher Scientific, Gibco, 10569), Bovine Calf Serum (%10), and Penicillin Streptomycin (PenStrep) media and passaged every few days. Cells were plated on 24×50 mm fibronectin coated slides overnight within culture media in a petri dish. After 15 hours, glass slides were washed with phosphate-buffered saline (PBS) 1X and then fixed with 4% formaldehyde for 4 minutes. Cell were then placed in 70% ethanol for permeabilization and stored at −20°C. NMuMg cells (ATCC CRL-1636) were cultured in DMEM with Glutamine and 4.5 g/L glucose (Thermo Fisher Scientific, Gibco, 11965), Fetal Bovine Serum (10%), insulin (10 mcg/ml), and PenStrep media. Rest of the fixation and permeabilization protocols was the same. For experiments, glass slides with either cell types were removed from −20°C storage and then dried with air blow. A custom flow-cell chamber (Grace Bio-Labs, SecureSeal, 3 mm × 11 mm size × 0.5 mm depth, RD478682) was then bound to this glass slide for sequential FISH measurements.
Tissue
Intact thymus was extracted from a four weeks old female mouse. Animal handling was done in Rothenberg’s laboratory with the approval by Caltech’s Institutional Animal Care and Use Committee. Immediately after extraction, thymus was embedded into 4% paraformaldehyde for three hours at room temperature for fixation of cells within the organ. Thymus was then rinsed with PBS1X for 10 minutes. For slicing, thymus was then embedded in 10% sucrose and kept at 4°C for more than 12 hours. Saline coated glass slides (24 mm × 50 mm) was then used to host 10 μm tissue sections after cutting at the City of Hope Pathology Core. Thymus tissue sections were kept at −80°C deep freezer for storage. The day before the FISH experiment, tissue section was removed from the freezer and thawed at the room temperature for one hour. To clear the section and reduce non-specific binding, tissue was first treated with 8% sodium dodecyl sulfate (SDS) for 10 minutes. Tissue section was then treated with 70% ethanol overnight at 4°C for permeabilization. Next day, the glass slide with tissue section was then bound to the hybridization flow chamber (Grace Bio-Labs, SecureSeal, 8–9 mm diameter × 0.6 mm depth, GBL621101) for sequential labeling and imaging experiments.
FISH SCALYS
Twenty-four FISH probes were used for each gene. Cy3B, Alexa 594, and Alexa 647 were coupled to the probes according to the barcode scheme. Cells or tissue sections within flow chambers were hybridized at a concentration of 1nM per probe overnight in a hybridization buffer of 10% Dextran Sulfate (Sigma D8906), 30% Formamide, 2X Saline-Sodium Citrate (SSC) buffer at room temperature. Labeled cells were then washed with 30% formamide for 30 minutes. For tissues, cells were washed second time with 50% formamide for 10 minutes. Samples were rinsed several times with 2X SSC to remove leftover probes. Samples were then imaged within an antibleaching buffer consisting of Tris-HCL (20mM), NaCl (50mM), Glucose (0.8%), Saturated Trolox (Sigma, 53188-07-1), Pyranose oxidase (Sigma: P4234, OD405nm: 0.05), and catalase (Sigma, 9001-05-2, 1:1000 dilution). A piece of glass slide or parafilm was used to cover the top facet of the flow cell, preventing the imaging or hybridization buffer from evaporation. After imaging the samples, FISH probes were removed by 100 Units of DNase I enzymatic treatment (Sigma-Aldrich, 04716728001 ROCHE) for 4 hours followed by a post wash with 30% formamide and 2X SSC wash for 10 minutes. The cells were subsequently hybridized by another probe set at 1 nM concentration for more than 12 hours at room temperate in the hybridization buffer. These presented imaging, probe stripping, and hybridization protocols are repeated based on the barcoding scheme1.
Barcoding
In cultures, we targeted transcripts for ten ribosomal proteins with corrFISH with two color binary codes (Fig. 2b). We barcoded five of the ribosomal protein genes (Rpl5, Rps6, Rpl21, Rps3, and Rps7) by sequential hybridizations of probes labeled by Alexa 594 fluorophores, and then we coded the second set of genes (Rps2, Rpl3, Rpl27a, Rpl23, and Rpl18a) by Alexa 647 probes. This implementation of corrFISH keeps the barcoding within each fluorophore channel to avoid errors introduced by chromatic aberration and spectral cross-talk. For validation of corrFISH, we used an additional color (in this case Cy3b) for single molecule FISH experiments to validate the corrFISH results (Fig. 2b). In tissues, we barcoded transcripts for ten ribosomal proteins with a single binary code (Cy3b), in which additional four repeats for Rps2, Rps6, Rpl21, and Rps3 as a positive control, and 1 empty barcode as a negative control are included in the barcoding scheme (Fig. 3b). Adding more colors and correlation dimensions to the barcoding scheme linearly increase the number of target RNA species for corrFISH (Supplementary Fig. 5). Incorporating chromatic corrections can further scale up the multiplexing capacity to the exponential rate seen in the FISH SCALYS method1.
Imaging
Cell cultures were imaged by a custom wide field fluorescence microscope with single molecule imaging capability (Supplementary Fig. 20). A simple fiber combiner (CNI Laser, 7 pieces to 1 piece connect with ferrule connector (FC) end, or alternatively Oz Optics, 1×6 pigtail wavelength division multiplexer) was used to merge laser beams creating a single output beam for illumination. Each illumination laser was fiber coupled and had about 1-Watt power with 405, 473, 533, 589, 640, and 730 nm center wavelengths (CNI Lasers). This fiber based design removed the free space optics alignment requirement for multi-color illumination of cells. Fiber output (>100mW) was then collimated with a simple convex lens and got vibrated by a custom fiber shaker all the times to avoid speckle issues. To enlarge the beam, a 1.5X telescope (f1:200 mm and f2:300mm) was inserted after collimation lens. Expanded illumination was then focused at the back aperture of the objective lens using another 200 mm convex lens. All of these lenses (Thorlabs) were 2″ diameter to reduce aberrations. 60X objective (1.42 numerical aperture) lens was used to collect fluorescence from the cells. We obtained 96X magnification with the use of 1.6X tube lens magnification providing 0.135 μm pixel size. High quantum efficiency Andor Ikon M camera was used to record the fluorescence from single molecules. We used up to 500 milliseconds exposure time per color channel to capture FISH images. All the devices were controlled by Micromanager.
Tissue sections were imaged by a spinning disk confocal microscope (Andor, WD model). In our experiments, 561 nm laser illumination was utilized to image Cy3b labeled probes and 405 nm laser was used to capture DAPI signal. 100X objective lens was used to collect the fluorescence. We tuned into relatively higher exposure time of up to 20 seconds per FISH image to obtain high signal to noise ratio FISH dots. A Metamorph software was used to control the devices and capture images. Compared to the point confocal imaging, spinning disk allowed us to reduce the photobleaching of our single molecules and increased imaging speed.
Analysis
Sequential FISH images were processed in MATLAB using a digital image-processing pipeline. Raw single molecule images were segmented to create a mask to cover the entire cell. Cell autofluorescence background was subtracted by using the image of the cell without FISH probes. Cell background image was registered to the original cell image with FISH probes to improve the subtraction quality. To remove the out-of-focus light, background subtracted images were deconvolved using a Lucy-Richardson iterative algorithm. The binary mask was then multiplied by deconvolved cell images from sequential hybridizations to define the cell areas. corrFISH was then processed z by z plane on the cell images. These transcript counts were then combined by summing the results from each section (Supplementary Fig. 8). Here, two subsequent planes were summed to increase the robustness of corrFISH processing. The final single-cell transcript counts were then plotted as a heat map, violin plot, and hierarchical cluster of gene expression. Additional control experiments with various experimental conditions were analyzed by box plots (Supplementary Figs. 21 and 22).
Single molecule FISH counting was performed based on a localization and counting algorithm5. Specifically, a Laplacian of Gaussian (LoG) filter was applied to the background subtracted and deconvolved images (Supplementary note). Individual transcripts were then identified and counted z-by-z plane. These counts were then summed to compute the gene expression in single cells. As the density of molecules increased, the FISH dots overlapped creating underestimation for most of the ribosomal protein transcripts.
Tissue analysis was performed by segmenting the cells based on their DAPI image and propagating to subsequent hybridizations. We processed 16 pixels by 16 pixel subregions and the subregions within a cell mask were correlated individually. Then the correlation values were summed together to provide the gene expression value for that cell (Supplementary Fig. 13).
For statistical analysis, we used Origin and Excel programs with regression and Student’s t-test modules.
Code availability
Custom MATLAB source codes (Supplementary Software) with test images are available and updated at https://github.com/singlecelllab/correlationFISH.
Supplementary Material
Acknowledgments
We thank J. Linton from Elowitz Laboratory (Caltech) for providing cell lines and M. Yui from Rothenberg Laboratory (Caltech) for the intact thymus organ. We appreciate the help of City of Hope Pathology Core to slice thymus into sections. This work is funded by US National Institute of Health single-cell analysis program award R01HD075605.
Footnotes
AUTHOR CONTRIBUTIONS
A.F.C. and L.C. designed the project and wrote the manuscript. L.C. supervised the project.
COMPETING FINANCIAL INTERESTS
L.C. and A.F.C. declare conflict of interests and have filed a patent application.
References
- 1.Lubeck E, Coskun AF, Zhiyentayev T, Ahmad M, Cai L. Nat Methods. 2014;11:360–361. doi: 10.1038/nmeth.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Science. 2015;348:aaa6090. doi: 10.1126/science.aaa6090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JH, et al. Science. 2014;343:1360–1363. doi: 10.1126/science.1250212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ke R, et al. Nat Methods. 2013;10:857–860. doi: 10.1038/nmeth.2563. [DOI] [PubMed] [Google Scholar]
- 5.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lubeck E, Cai L. Nat Methods. 2012;9:743–748. doi: 10.1038/nmeth.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holden SJ, Uphoff S, Kapanidis AN. Nat Methods. 2011;8:279–280. doi: 10.1038/nmeth0411-279. [DOI] [PubMed] [Google Scholar]
- 8.Zhu L, Zhang W, Elnatan D, Huang B. 2012;9:721–723. doi: 10.1038/nmeth.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwille P. Cell biochemistry and biophysics. 2001;34:383–408. doi: 10.1385/CBB:34:3:383. [DOI] [PubMed] [Google Scholar]
- 10.Kettling U, Koltermann A, Schwille P, Eigen M. Proceedings of the National Academy of Sciences. 1998;95:1416–1420. doi: 10.1073/pnas.95.4.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costantino S, Comeau JWD, Kolin DL, Wiseman PW. Accuracy and Dynamic Range of Spatial Image Correlation and Cross-Correlation Spectroscopy. Biophys J. 2005;89:1251–1260. doi: 10.1529/biophysj.104.057364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondrashov N, et al. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xue S, Barna M. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Signer RA, Magee JA, Salic A, Morrison SJ. Nature. 2014;509:49–54. doi: 10.1038/nature13035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito Y, Hashimoto M, Hirota K, Ohkura N, Morikawa H, Nishikawa H, Tanaka A, Furu M, Ito H, Fujii T, Nomura T. Science. 2014;346:363–368. doi: 10.1126/science.1259077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Magde D, Elson E, Webb WW. Phys Rev Lett. 1972;29:705–708. [Google Scholar]
- 17.Gerdes MJ, Sevinsky CJ, Sood A, Adak S, Bello MO, Bordwell A, Can A, Corwin A, Dinn S, Filkins RJ, Hollman D. Proceedings of the National Academy of Sciences. 2013;29:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray E, Cho JH, Goodwin D, Ku T, Swaney J, Kim SY, Choi H, Park YG, Park JY, Hubbert A, McCue M. Cell. 2015;6:1500–1514. doi: 10.1016/j.cell.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gong H, Holcomb I, Ooi A, Wang X, Majonis D, Unger MA, Ramakrishnan R. Bioconjugate chemistry. 2016;1:217–225. doi: 10.1021/acs.bioconjchem.5b00613. [DOI] [PubMed] [Google Scholar]
- 20.Fan R, Vermesh O, Srivastava A, Yen BK, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nature biotechnology. 2008;12:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xue M, Wei W, Su Y, Kim J, Shin YS, Mai WX, Nathanson DA, Heath JR. Journal of the American Chemical Society. 2015;12:4066–4069. doi: 10.1021/jacs.5b00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.