Abstract
Purpose of review
There has been increasing interest in the contents and function of the microbiota, as it relates to pediatric inflammatory diseases. Here we discuss the factors underlying the development of the microbiota, its role in juvenile idiopathic arthritis (JIA), and prospects for therapeutic interventions in the microbiota.
Recent findings
The human microbiota undergoes a succession of changes, until it reaches a mature form. A variety of early-life exposures, including mode of delivery and form of feeding, can affect the contents of the microbiota and possibly impact upon long-term risk of developing autoimmune diseases. The microbiota is altered in children with JIA, including elevated Bacteroides genus in JIA as a whole and decreased Faecalibacterium prausnitzii in pediatric spondyloarthritis. Although there is limited data so far indicating that microbiota-based therapies can result in therapeutic improvement of arthritis, most of the data is in adults and thus may not be applicable to children.
Summary
Perturbations of the microbiota during childhood may result in the development of a microbiota associated with increased risk of pediatric rheumatic illness. Whether the microbiota can be targeted is a focus of ongoing research.
Keywords: Juvenile idiopathic arthritis, microbiota, spondyloarthritis
Introduction
The human microbiota consists of 100 trillion bacteria, the vast majority of which are located in the intestines. Among them, they contain 3 million genes, 100 times their human host (1). Arguably, the first observation that microbes may promote arthritis was several millennia ago, when Hippocrates observed, “A youth does not get gout before sexual intercourse”, a possible though not definite reference to reactive arthritis (2). In addition, it is has been more than a century since Elie Metchkinoff suggested that that alterations in the intestinal may promote improved overall health (3). However, the tools to explore in depth the contents and function of the gut microbiota have only existed for about 10-15 years. This review will focus on the nature of the microbiota in pediatric rheumatic diseases, their potential role in disease pathogenesis, factors that influence its development, and the potential for treatment through manipulation of the microbiome. To the extent that the microbiota may play a pathogenic role in the etiology of juvenile idiopathic arthritis (JIA), it is important to recognize factors influencing microbial colonization.
Development of the microbiome
Early events
A developing fetus lacks a microbiome. Initial colonizers of the gut consist of aerobic organisms, such as Enterococcus and Streptococcus, which are then replaced with anaerobes such as Bifidobacterium and Lactobacillus (4). Subsequently, the infant begins to develop a more mature (adult-like) microbiota dominated by the Bacteroidetes and Firmicutes phyla. The age at which this transition begins to take place is variable, and appears to be affected by when the child transitions from a formula / breast-milk diet to table foods (4, 5). Data from the Human Microbiome Project indicated the microbiota has largely reached an adult steady-state by age 3 (6), although a more recent study indicated older children still have a distinct microbiota as compared to adults (7).
The type of delivery (vaginal vs Cesarean section) affects the initial colonization and may have long-term implications on the microbiota. The initial microbiota of infants delivered vaginally bears strong resemblance to their maternal microbiota, while that of infants delivered via Cesarean section is more consistent with an adult skin microbiota (8). These alterations can last for at least one year (9). Of note, there are varying indications for C-Sections, and differences were also seen between children born via elective versus urgent C-Section, speaking to other perinatal factors that may influence the microbiota, including prematurity, perinatal illness, and antibiotic use (9). Several studies have evaluated whether mode of delivery is associated with autoimmune disease risk, with conflicting results. A Canadian study did not show any such effect (10), while a Danish registry study showed C-section was associated with small yet significant increases in the risk of a variety of conditions, including IBD and JIA (11). Thus, mode of delivery may result in altered programming of the developing microbiota, with these changes potentially manifesting themselves for many years.
Just as delivery mode can affect the neonatal microbiota, so can the infant's early diet, with differences in breast vs formula-fed infants observed (12). Although such studies were by necessity observational, large differences were also observed in macaques assigned to formula- vs breast-fed diets (13). While there are no long term follow-up studies evaluating whether these early-life differences are sustained, a recent study showed adult patients with ankylosing spondylitis were less likely to have been breast-fed as compared to healthy controls, even among their own siblings (14). Thus, early life exposures have the potential to alter the microbiota, and these alterations may impact disease in the long-term.
Beyond infancy, it is well-established that diet and weight can affect the microbiota. Obese individuals tend to have less diversity in their microbiota (15, 16), and the obese state appears to potentiate itself. The obese microbiota is enriched for genes producing proteins with enhanced capacity to extract nutrients from the diet (17). That the microbiota can potentiate obesity was demonstrated by fecal transfer studies of microbiota from genetically obese leptin-deficient mice to naïve germ-free mice, who gained more weight as compared to those that received microbiota from non-obese mice (17). Thus, environment clearly impacts the gut microbiota in health and disease.
Genetic influences
Undoubtedly, the microbiota results from a complex interplay of both genetic and environmental factors. Genetic influences are in most studies difficult to evaluate fully, as closely related individuals typically share the same environment. One of the most ambitious efforts to study environmental versus genetic effects on the microbiota was published by Gordon and colleagues, who recruited 54 twin pairs aged 21 - 32 years of age [31 monozygotic (MZ) and 21 dizygotic (DZ)], plus 46 mothers (16). They demonstrated the microbiotas of the MZ twin pairs were no more similar to each other as were the DZ twin pairs were to each other, implying a strong environmental effect. In contrast, Si et al. (2015) compared 8 MZ and 4 DZ twin pairs, concluding that the former microbiotas were indeed more similar (18); however, the numbers were smaller and the manuscript did not indicate whether the difference was statistically significant. Thus, the evidence from these twin studies points to strong environmental influences on the microbiota.
On the other hand, genome wide association studies in humans and mice (19, 20) show associations between the microbiota and host genetics. Additional studies have linked abundance of certain taxa to specific polymorphisms associated with inflammatory diseases (21-24) (Table 1), potentially accounting for the effects of these polymorphisms on disease risk. Nevertheless, while some genes may affect certain taxa, the overall importance of genetic versus environmental effects remains to be determined.
Table 1.
Genetic polymorphisms associated with specific bacterial taxa. Original Table.
| Study | Polymorphism | Affected taxa |
|---|---|---|
| Khachatryan (2008)21 | MEFV | Increased Enterobacteriaceae, Acidaminococcaceae, Ruminococcus and Megasphaera; Decreased Roseburia |
| De Palma (2010)22 | HLA | HLA alleles associated with increased risk of CD resulted in increased E. coli, Bacteroides-Prevotella, Streptococcus-Lactococcus, E. rectale-C. coccoides, sulphate-reducing bacteria, C. lituseburense, and C. histolyticum among infants up to age four months |
| Knights (2014)23 | NOD2 risk alleles for CD | Increased Enterobacteriaceae |
| Lin (2014)24 | HLA-B27 (rats) | Increased Bacteroides vulgatus, Paraprevotella |
Demographic influences
To explore the impact of ethnicity and sex on microbiota diversity, Chen et al. (2016) studied 118 healthy adults from the Midwestern United States (15). They reported increased diversity of the microbiota in Caucasians as compared to non-Whites, as well as different community structures (as evidenced by principal coordinates plots) in males as compared to females, along with specific bacteria taxa whose abundance differed by race and sex. Likewise, a study of 82 healthy adults living in the Washington, D.C. area showed altered microbial community structure based upon sex, as well as different abundances of organisms even at the phyla level (25). A study of the oral microbiota also identified differences in diversity based upon ethnicity (26). There are clearly multiple potential reasons for these differences, and their overall significance to health is yet unknown. However, animal studies indicate that sex-based differences in the microbiota may help contribute to disease. Specifically, in the non-obese diabetic model of disease, males are relatively protected as compared to females. Transfer of the microbiota from adult males to immature females resulted in alterations of the recipient's microbiota, increased production of androgens, and decreased islet inflammation (27). Thus, ethnic and sex differences affect the microbiota, and these differences may account for some of the sexual differences in autoimmune disease risk.
Alterations in the microbiome in pediatric rheumatic diseases
Prior to next generation sequencing, there was evidence that the intestinal milieu was altered in children with JIA. Specifically, Picco et al. (2000) used the urinary lactulose/mannitol test to demonstrate altered intestinal permeability in children with JIA (28); they found it was higher in children with juvenile spondyloarthritis (SpA) compared to those with other forms of JIA, who themselves had higher levels than healthy control subjects. Several additional studies have identified evidence of sub-clinical intestinal inflammation in children with spondyloarthritis using colonoscopy (29), fecal calprotectin (30), or MRI (31); all of these studies suggest children with at least the SpA variant of JIA may have qualitatively similar, while quantitatively less, intestinal abnormalities as patients with inflammatory bowel disease (IBD). Conversely, it is well-established that up to one quarter of patients with IBD has arthritis, typically SpA (32).
Two recent studies used 16S sequencing to evaluate the contents of the fecal microbiota in children with JIA. One was an unselected group of children with newly diagnosed JIA, comprised of 13 children with oligoarticular JIA, 16 with polyarticular (15 rheumatoid factor negative), and one child with enthesitis-related arthritis (ERA), which constitutes juvenile SpA (33). This study revealed a modest but statistically significant (44% vs 34%, p = 0.04) increase in the Bacteroides genus in patients compared to controls. Stoll et al. (2014) studied a group of children with ERA (34). The primary difference identified in this study was decreased abundance of the Firmicutes, Faecalibacterium prausnitzii, in patients compared to controls (3.8% vs 10%, p = 0.008). Additionally two distinct subsets of patients were identified, one with markedly elevated abundance of Bacteroides genus and the other with high abundance of Akkermansia muciniphlia. This finding of decreased F. prausnitzii was consistent with previous findings in both pediatric and adult IBD (35), and is consistent with observations this organism may have anti-inflammatory properties through direct effects on cytokine production (36), as well as increased production of short-chain fatty acids such as butyrate (37). Butyrate and other short-chain fatty acids (SCFAs) appear to promote regulatory T cell function (38). Therefore, decreased abundance of F. prausnitzii may result in decreased regulation of inflammation, thus contributing to both IBD and JIA/ERA.
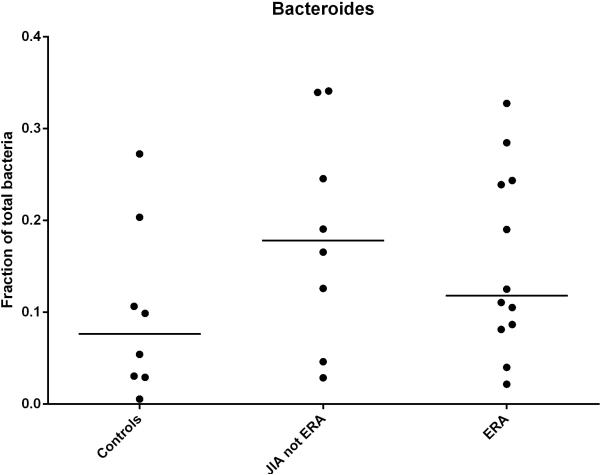
Among children with JIA, alterations in F. praunitzii levels appear to be at least partially specific to the ERA subgroup, as evidenced by a subsequent study showing intermediate levels of this organism in JIA children without ERA (39), as well as the finding by Tejesvi et al. (33), in which there were no reported alterations in this organism. In contrast, increased abundance of the Bacteroides genus may be a more universal finding in JIA, as demonstrated both by the Tejesvi study and by a comparison with non-ERA JIA subjects (Figure 1). Although the differences shown in Figure 1 were not statistically significant (likely due to insufficient numbers of patients evaluated), they do appear to support the Tejesvi findings.
Figure 1. Abundance of Bacteroides genus in the feces of children with JIA as assessed by 16S sequencing.
The JIA category included 5, 2, and 1 children/child with oligoarticular, rheumatoid factor-negative polyarticular, and early-onset psoriatic JIA, respectively. This is an original figure (previously unpublished data).
This association of the Bacteroides genus with JIA is intriguing. Although Bacteroides is not generally considered to be pro-inflammatory, and indeed the polysaccharide A tail of B. fragilis appears to preferentially activate regulatory T cells (40), studies in animal models of SpA have suggested the Bacteroides genus may be contributory to the disease. Specifically, mouse (41) and rat (42) models of SpA were both shown to have protection against the disease in the germ-free state, with reemergence of disease when the Bacteroides genus was added back. Neither of the studies in JIA was able to speciate the Bacterodes; this is a limitation of 16S sequencing in general.
Finally, the significance of increased abundance of A. muciniphila, a member of the Verrucomicrobia phylum, in a subset of JIA/ERA patients, is unclear; in a study of adult psoriatic arthritis patients, controls had elevated levels as compared to patients (43). Stoll et al. (2014) postulated that as this species is known for its capacity to degrade intestinal mucins (44), decreased production of mucin could potentially be damaging to the gut wall, resulting in increased permeability; indeed, increased intestinal permeability has been observed in adults with ankylosing spondylitis (45), as well as in children with JIA (28).
To study the functional consequences of an altered microbiota, Stoll et al. (2015) performed whole genome sequencing and fecal water metabolomics on children with ERA and healthy control subjects, the results of which were presented at the 2015 American College of Rheumatology conference (46). They found patients had decreased taxonomic diversity, decreased genetic coverage of the metabolic map, and decreased content of fecal water metabolites, including those involved in the butanoate pathway, which makes butyrate. Although decreased F. prausnitzii was not observed, it appears plausible similar alterations in the functional potential of the microbiota may have occurred, and ultimately may be more important than the abundance of any single organism.
Therapeutic potential of alterations in the microbiota
A recent review (47) concluded most interventions targeting the microbiota, including dietary alterations and probiotics, have not yielded dramatic results. It bears emphasis, however, that most of these studies were conducted in adults. This may be quite significant, as it appears two interventions in adult patients with IBD that attempt to target the microbiota, exclusive enteral nutrition (EEN) and fecal microbial transplantation (FMT), both work better in younger populations (48, 49). Similarly, there is a case report of EEN as effective therapy for a child with polyarticular JIA refractory to methotrexate and multiple tumor necrosis factor inhibitors (50). Therefore, dietary interventions may have promise in targeting the microbiota in pediatric rheumatic diseases.
Nevertheless, it is unclear why interventions targeting the microbiota might be more effective in the pediatric population. It may be for mundane reasons, such as better adherence to the dietary intervention or shorter disease duration. However, it might also reflect the developing nature of the pediatric microbiota. That is, interventions such as dietary changes that result in reversible changes in the microbiota in adults (51) may result in long-lasting changes in young children; as an illustration, the effects of breast-milk vs formula in young macaques was observed at 12 months, despite that both groups ate the same diet beginning at six months of age (13).
It may be the case, then, that childhood presents a unique opportunity to alter the course of the disease through interventions targeting the microbiota. In short, the development of the microbiota may be analogous to “original antigenic sin,” in that once the mature steady-state is reached, long-term alterations may be difficult to sustain; while alterations in the immature microbiota might have greater likelihood of success. This possibility is consistent with observations discussed above showing even events in the first few days of life can result in changes in the microbiota that may affect future disease risk, while dietary interventions in adults result in only transient changes. Likewise, early use of antibiotics may be a risk factor for development of both JIA and pediatric IBD (52-55), albeit not psoriasis (56); Table 2. These observations are consistent with the possibility interventions that alter the development of the microbiota can have long-lasting effects.
Table 2.
Risk of pediatric IBD, JIA, or psoriasis associated with exposure to antibiotics.
| Study | Disease | Risk of abx exposure | Dose dependence | Category-specific |
|---|---|---|---|---|
| Kronman (2012)52 | IBD | HR of 5 | Yes | Strong association seen with anti-anaerobics and penicillins; week association with cephalosporins; not with macrolides or sulfonamides |
| Virta (2012)53 | CD | OR 1.42 for ≥ exposures | Yes | Association largely limited to cephalosporins |
| Virta (2012)53 | UC | Non-significant association | No | Not assessed |
| Arvonen (2015)54 | JIA | OR of 1.6 | Yes | Strong association seen with anti-anaerobics; weak association with all other classes |
| Horton (2015a)55 | JIA | OR of 2.1 | Yes | Association with all classes except cephalosporins |
| Horton (2015b)56 | Psoriasis | OR of 1.2 | N/A | No |
Abbreviations: CD = Crohn Disease, IBD = inflammatory bowel disease, JIA = juvenile idiopathic arthritis, N/A = not assessed, OR = odds ratio, UC = ulcerative colitis. Original Table.
However, all interventions will not likely be equally effective. Simply setting out to alter the microbiota, without solid rationale for the change, could just as easily change it for the worse as for the better, or have no effect. Along those lines, a randomized controlled trial of probiotics in children with ERA (57) was no more effective than similar studies conducted in adults with SpA (58, 59). Future studies will benefit from knowledge gained on the role of specific alterations in the microbiota of diseased humans and experimental animal models.
Conclusion
We are just beginning to explore the potential of microbiota-based therapy in the management of pediatric rheumatic diseases. To the extent animal models of disease reflect the human counterparts, there is reason for optimism such approaches may have a potential to alter if not ameliorate disease. However, there is much to be learned about the contents of the normal pediatric microbiota, the microbiota in a variety of disease states, and safe and effective means of effecting long-term microbiota changes to benefit pediatric rheumatic disorders.
Key points.
Early-life environmental exposures, such as delivery mode, diet, and antibiotics, can have long-lasting effects that may influence the future risk of autoimmune diseases
Children with juvenile idiopathic arthritis have altered fecal microbiota which may contribute to disease
Therapeutic alterations of the microbiota are a promising but currently largely unfulfilled avenue of treatment for chronic inflammatory arthritis
Acknowledgements
None
Financial support and sponsorship
Dr. Stoll was supported by the NIH and the American College of Rheumatology.
Footnotes
Conflicts of Interest
The authors declare no conflicts of interest pertaining to this work.
References
- 1.Qin J, Li R, Raes J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464(7285):59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nuki G, Simkin PA. A concise history of gout and hyperuricemia and their treatment. Arthritis Res Ther. 2006;8(Suppl 1):S1. doi: 10.1186/ar1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Podolsky SH. Metchnikoff and the microbiome. Lancet. 2012;380(9856):1810–1. doi: 10.1016/s0140-6736(12)62018-2. [DOI] [PubMed] [Google Scholar]
- 4**.Backhed F, Roswall J, Peng Y, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17(5):690–703. doi: 10.1016/j.chom.2015.04.004. [The authors performed serial shotgun metagenomics sequencing of fecal DNA of 98 infants over one year. Mode of delivery, feeding practice, and age all contribute significantly to the contents of the fecal microbiome.] [DOI] [PubMed] [Google Scholar]
- 5.Koenig JE, Spor A, Scalfone N, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4578–85. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hollister EB, Riehle K, Luna RA, et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome. 2015;3:36. doi: 10.1186/s40168-015-0101-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–5. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Azad MB, Konya T, Persaud RR, et al. Impact of maternal intrapartum antibiotics, method of birth and breastfeeding on gut microbiota during the first year of life: a prospective cohort study. BJOG. 2015 doi: 10.1111/1471-0528.13601. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein CN, Banerjee A, Targownik LE, et al. Cesarean Section Delivery Is Not a Risk Factor for Development of Inflammatory Bowel Disease: A Population-based Analysis. Clin Gastroenterol Hepatol. 2016;14(1):50–7. doi: 10.1016/j.cgh.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 11*.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015;135(1):e92–8. doi: 10.1542/peds.2014-0596. [This study showed that Ceserean section delivery is associated with an increased risk of a variety of autoimmune diseases, including JIA.] [DOI] [PubMed] [Google Scholar]
- 12.Fan W, Huo G, Li X, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in infants during the six months of life. Journal of microbiology and biotechnology. 2014;24(2):133–43. doi: 10.4014/jmb.1309.09029. [DOI] [PubMed] [Google Scholar]
- 13.Ardeshir A, Narayan NR, Mendez-Lagares G, et al. Breast-fed and bottle-fed infant rhesus macaques develop distinct gut microbiotas and immune systems. Sci Transl Med. 2014;6(252):252ra120. doi: 10.1126/scitranslmed.3008791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14*.Montoya J, Matta NB, Suchon P, et al. Patients with ankylosing spondylitis have been breast fed less often than healthy controls: a case-control retrospective study. Ann Rheum Dis. 2015 doi: 10.1136/annrheumdis-2015-208187. [This study showed that compared to both healthy controls as well as siblings, patients with ankylosing spondylitis were less likely to have been breast-fed.] [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Ryu E, Hathcock M, et al. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ. 2016;4:e1514. doi: 10.7717/peerj.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–31. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 18.Si J, Lee S, Park JM, et al. Genetic associations and shared environmental effects on the skin microbiome of Korean twins. BMC Genomics. 2015;16(1):992. doi: 10.1186/s12864-015-2131-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–99. doi: 10.1016/j.cell.2014.09.053. [This study of 416 twin pairs showed the heredibility of multiple bacterial taxa.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benson AK, Kelly SA, Legge R, et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci U S A. 2010;107(44):18933–8. doi: 10.1073/pnas.1007028107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khachatryan ZA, Ktsoyan ZA, Manukyan GP, et al. Predominant role of host genetics in controlling the composition of gut microbiota. PLoS One. 2008;3(8):e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knights D, Silverberg MS, Weersma RK, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome medicine. 2014;6(12):107. doi: 10.1186/s13073-014-0107-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Palma G, Capilla A, Nadal I, et al. Interplay between human leukocyte antigen genes and the microbial colonization process of the newborn intestine. Current issues in molecular biology. 2010;12(1):1–10. [PubMed] [Google Scholar]
- 24.Lin P, Bach M, Asquith M, et al. HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One. 2014;9(8):e105684. doi: 10.1371/journal.pone.0105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominianni C, Sinha R, Goedert JJ, et al. Sex, body mass index, and dietary fiber intake influence the human gut microbiome. PLoS One. 2015;10(4):e0124599. doi: 10.1371/journal.pone.0124599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mason MR, Nagaraja HN, Camerlengo T, et al. Deep sequencing identifies ethnicity-specific bacterial signatures in the oral microbiome. PLoS One. 2013;8(10):e77287. doi: 10.1371/journal.pone.0077287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–8. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 28.Picco P, Gattorno M, Marchese N, et al. Increased gut permeability in juvenile chronic arthritides. A multivariate analysis of the diagnostic parameters. Clin Exp Rheumatol. 2000;18(6):773–8. [PubMed] [Google Scholar]
- 29.Mielants H, Veys EM, Cuvelier C, et al. Gut inflammation in children with late onset pauciarticular juvenile chronic arthritis and evolution to adult spondyloarthropathy--a prospective study. J Rheumatol. 1993;20(9):1567–72. [PubMed] [Google Scholar]
- 30.Stoll ML, Punaro M, Patel AS. Fecal calprotectin in children with the enthesitis-related arthritis subtype of juvenile idiopathic arthritis. J Rheumatol. 2011;38(10):2274–5. doi: 10.3899/jrheum.110508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stoll ML, Patel AS, Punaro M, Dempsey-Robertson M. MR enterography to evaluate sub-clinical intestinal inflammation in children with spondyloarthritis. Pediatr Rheumatol Online J. 2012;10:6. doi: 10.1186/1546-0096-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salvarani C, Vlachonikolis IG, van der Heijde DM, et al. Musculoskeletal manifestations in a population-based cohort of inflammatory bowel disease patients. Scand J Gastroenterol. 2001;36(12):1307–13. doi: 10.1080/003655201317097173. [DOI] [PubMed] [Google Scholar]
- 33**.Tejesvi MV, Arvonen M, Kangas SM, et al. Faecal microbiome in new-onset juvenile idiopathic arthritis. Eur J Clin Microbiol Infect Dis. 2015 doi: 10.1007/s10096-015-2548-x. [This was the first study evaluating the microbiota of newly-diagnosed JIA subjects, showing increased abundance of Bacteroides genus in patients as compared to controls.] [DOI] [PubMed] [Google Scholar]
- 34*.Stoll ML, Kumar R, Morrow CD, et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther. 2014;16(6):486. doi: 10.1186/s13075-014-0486-0. [This study showed decreased Faecalibacterium prausnitzii in children with ERA compared to healthy control subjects, analogous to findings in pediatric and adult IBD. Elevated Bacteroides genus and Akkermansia muciniphila were also seen in subsets of ERA subjects.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cao Y, Shen J, Ran ZH. Association between Faecalibacterium prausnitzii Reduction and Inflammatory Bowel Disease: A Meta-Analysis and Systematic Review of the Literature. Gastroenterol Res Pract. 2014;2014:872725. doi: 10.1155/2014/872725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sokol H, Pigneur B, Watterlot L, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105(43):16731–6. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hold GL, Schwiertz A, Aminov RI, et al. Oligonucleotide probes that detect quantitatively significant groups of butyrate-producing bacteria in human feces. Appl Environ Microbiol. 2003;69(7):4320–4. doi: 10.1128/AEM.69.7.4320-4324.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–73. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stoll ML. Gut microbes, immunity, and spondyloarthritis. Clinical immunology. 2015;159(2):134–42. doi: 10.1016/j.clim.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 40.Round JL, Lee SM, Li J, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332(6032):974–7. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinkorova Z, Capkova J, Niederlova J, et al. Commensal intestinal bacterial strains trigger ankylosing enthesopathy of the ankle in inbred B10.BR (H-2(k)) male mice. Hum Immunol. 2008;69(12):845–50. doi: 10.1016/j.humimm.2008.08.296. [DOI] [PubMed] [Google Scholar]
- 42.Rath HC, Herfarth HH, Ikeda JS, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98(4):945–53. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43*.Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis & rheumatology. 2015;67(1):128–39. doi: 10.1002/art.38892. [Decreased bacterial diversity and increased abundance of rare taxa were observed in subjects with newly diagnosed psoriatic arthritis, compared to adult healthy controls.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004;54(Pt 5):1469–76. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 45.Martinez-Gonzalez O, Cantero-Hinojosa J, Paule-Sastre P, et al. Intestinal permeability in patients with ankylosing spondylitis and their healthy relatives. Br J Rheumatol. 1994;33(7):644–7. doi: 10.1093/rheumatology/33.7.644. [DOI] [PubMed] [Google Scholar]
- 46.Stoll ML, Wilson L, Barnes S, et al. Multi-omics study of gut microbiota in enthesitis-related arthritis identify diminished microbial diversity and altered typtophan metabolism as potential factors in disease pathogenesis [abstract]. Arthritis Rheum. 2015;67:S10. [Google Scholar]
- 47.Bravo-Blas A, Wessel H, Milling S. Microbiota and arthritis: correlations or cause? Curr Opin Rheumatol. 2016;28(2):161–7. doi: 10.1097/BOR.0000000000000261. [DOI] [PubMed] [Google Scholar]
- 48.Wall CL, Day AS, Gearry RB. Use of exclusive enteral nutrition in adults with Crohn's disease: a review. World J Gastroenterol. 2013;19(43):7652–60. doi: 10.3748/wjg.v19.i43.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Colman RJ, Rubin DT. Fecal microbiota transplantation as therapy for inflammatory bowel disease: a systematic review and meta-analysis. J Crohns Colitis. 2014;8(12):1569–81. doi: 10.1016/j.crohns.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Berntson L. Anti-inflammatory effect by exclusive enteral nutrition (EEN) in a patient with juvenile idiopathic arthritis (JIA): brief report. Clin Rheumatol. 2014;33(8):1173–5. doi: 10.1007/s10067-014-2672-5. [DOI] [PubMed] [Google Scholar]
- 51.Martinez I, Muller CE, Walter J. Long-term temporal analysis of the human fecal microbiota revealed a stable core of dominant bacterial species. PLoS One. 2013;8(7):e69621. doi: 10.1371/journal.pone.0069621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kronman MP, Zaoutis TE, Haynes K, et al. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. 2012;130(4):e794–803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virta L, Auvinen A, Helenius H, et al. Association of repeated exposure to antibiotics with the development of pediatric Crohn's disease--a nationwide, register-based finnish case-control study. American journal of epidemiology. 2012;175(8):775–84. doi: 10.1093/aje/kwr400. [DOI] [PubMed] [Google Scholar]
- 54*.Arvonen M, Virta LJ, Pokka T, et al. Repeated exposure to antibiotics in infancy: a predisposing factor for juvenile idiopathic arthritis or a sign of this group's greater susceptibility to infections? J Rheumatol. 2015;42(3):521–6. doi: 10.3899/jrheum.140348. [Prior exposure to antibiotics was identified as a risk factor for subsequent development of JIA.] [DOI] [PubMed] [Google Scholar]
- 55*.Horton DB, Scott FI, Haynes K, et al. Antibiotic Exposure and Juvenile Idiopathic Arthritis: A Case-Control Study. Pediatrics. 2015;136(2):e333–43. doi: 10.1542/peds.2015-0036. [This study also demonstrated that prior exposure to antibiotics was a risk factor for JIA, even after adjustments for infections themselves.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton DB, Scott FI, Haynes K, et al. Antibiotic Exposure, Infection, and the Development of Pediatric Psoriasis: A Nested Case-Control Study. JAMA dermatology. 2015:1–9. doi: 10.1001/jamadermatol.2015.3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57*.Shukla A, Gaur P, Aggarwal A. Double Blind Placebo Controlled Randomized Trial of Probiotics in Enthesitis-Related-Arthritis Category of JIA: Effect on Clinical and Immunological Parameters. Arthritis Rheum. 2015;67:S10. [Unsuccessful trial of probiotics in children with ERA.] [Google Scholar]
- 58.Jenks K, Stebbings S, Burton J, et al. Probiotic therapy for the treatment of spondyloarthritis: a randomized controlled trial. J Rheumatol. 2010;37(10):2118–25. doi: 10.3899/jrheum.100193. [DOI] [PubMed] [Google Scholar]
- 59.Brophy S, Burrows CL, Brooks C, et al. Internet-based randomised controlled trials for the evaluation of complementary and alternative medicines: probiotics in spondyloarthropathy. BMC Musculoskelet Disord. 2008;9:4. doi: 10.1186/1471-2474-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]