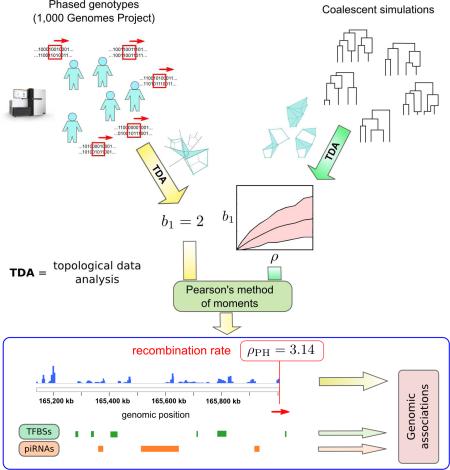
Summary
Meiotic recombination is a fundamental evolutionary process driving diversity in eukaryotes. In mammals, recombination is known to occur preferentially at specific genomic regions. Using topological data analysis (TDA), a branch of applied topology that extracts global features from large datasets, we developed an efficient method for mapping recombination at fine scales. When compared to standard linkage-based methods, TDA can deal with a larger number of SNPs and genomes without incurring prohibitive computational costs. We applied TDA to 1,000 Genomes Project data and constructed high-resolution whole-genome recombination maps of seven human populations. Our analysis shows that recombination is generally under-represented within transcription start sites. However, the binding sites of specific transcription factors are enriched for sites of recombination. These include transcription factors that regulate the expression of meiosis- and gametogenesis-specific genes, cell cycle progression and differentiation blockage. Additionally, our analysis identifies an enrichment for sites of recombination at repeat-derived loci matched by piwi-interacting RNAs.
Graphical Abstract
Introduction
The maintenance of genetic diversity in a species can promote survival during times of unpredictable environmental change. Germline mutations, inherited by the offspring, are the raw material of genetic diversity in sexually reproducing organisms. Meiotic recombination enables a population to explore and maintain this genetic diversity by allowing for the rapid generation of new allele combinations. An excessive amount of genetic linkage due to insufficient meiotic recombination can preclude removal of deleterious variants from the genome over successive generations, leading to a substantial fitness reduction.
Meiotic recombination is initiated by the induction of programmed DNA double-strand breaks (DSBs) during meiosis. These initiating lesions can be repaired through various pathways involving the formation of heteroduplex DNA. Consequently, meiotic recombination is usually accompanied by GC-biased gene conversion tracts (Duret and Galtier, 2009). Additionally, some of the repair pathways lead to the formation of chromosomal crossovers, required for proper chromosomal disjunction (Koehler et al., 1996). The aggregate effect of all these biochemical processes over evolutionary time defines the recombination landscape of the genome.
Studies of the recombination landscape in eukaryotes have revealed that recombination is highly regulated, with ~80% of recombination events in humans occurring at narrow (~2 kb) regions known as recombination hotspots (Crawford et al., 2004; Kauppi et al., 2004; McVean et al., 2004; Myers et al., 2005). The enrichment of non-allelic homologous recombination (NAHR) variants observed at recombination hotspots (Mills et al., 2011) suggests that the programmed DSBs required for recombination initiation can also serve as a source for NAHR. Regulation of the location and frequency of recombination can therefore potentially reduce the impact of DSB repair problems and target recombination to genomic regions where genetic diversity is more advantageous.
The specific biological mechanisms that regulate meiotic recombination are largely unknown. In mammals, the meiosis-specific Histone 3 Lysine 4 (H3K4) tri-methyltransferase PRDM9 binds chromosomes at recombination hotspots through a tandem-array of C2H2 zinc-fingers that recognizes a specific DNA binding motif (Baudat et al., 2010; Myers et al., 2010; Parvanov et al., 2010). In Prdm9 knockout mice, meiotic DSBs occur at different locations than in wild type, suggesting that this gene determines the location of meiotic DSBs (Brick et al., 2012). Accordingly, variants of the Prdm9 gene coding different numbers of zinc-finger domains are associated with variation in hotspot location, both between human populations and between mammalian species (Baudat et al., 2010; Myers et al., 2010), as well as between human individuals (Pratto et al., 2014). It is an open question whether factors other than PRDM9 modulate recombination in mammals.
Several methods have been proposed to study the landscape of recombination (Kirkness et al., 2013; Lu et al., 2012; McVean et al., 2004; Pan et al., 2011; Pratto et al., 2014; Smagulova et al., 2011; Wang et al., 2012). Population-based recombination maps capture the recombination history of populations using genome-wide genomic data and have become a valuable tool in the study of human recombination during the last decade (Hinch et al., 2011; International HapMap et al., 2007; Kong et al., 2010; Myers et al., 2005). Sub-kilobase scale mapping and annotation of human recombination is now possible due to the large number of genomes published by consortia such as the 1,000 Genomes Project (1000 Genomes Project Consortium et al., 2012) and ENCODE (Encode Project Consortium, 2012). Nucleotide-resolution datasets, such as those obtained by chromatin immunoprecipitation (ChIP-seq), bisulfite, or RNA sequencing methods, reveal a gamut of biological features associated to small genomic regions, often spanning mere handfuls of bases. How these fine scale, nucleotide-level features influence the structure and position of recombination hotspots is not understood.
A key step towards this understanding is the development of methods that can accurately estimate fine-scale meiotic recombination rates genome-wide, so that relationships with narrow (and often clustered) biological features of the genome can be assessed statistically. Such high-resolution recombination maps are only attainable through the analysis of large numbers of sequences and segregating sites, becoming an important challenge for current methods of recombination rate estimation. Widely used methods (Crawford et al., 2004; Hudson, 2001; Li and Stephens, 2003; McVean et al., 2002) are based on the non-random association of alleles at different loci, that is, linkage disequilibrium. Analysis based on these methods, however, becomes computationally expensive when the number of sequences is on the order of one hundred. New mathematical and computational approaches are needed to meet this challenge.
Topological data analysis (TDA) is a new branch of applied topology that extracts global features from large datasets. TDA has been successfully utilized in cross-sectional studies of complex genetic diseases (Li et al., 2015) and cancer (Nicolau et al., 2011). Persistent homology, a framework within TDA for deriving and classifying topological features associated to data (discussed in detail below), has been shown to capture instances of recombination and re-assortment in viral populations (Chan et al., 2013). These results suggest that it may be also used for quantifying recombination in human populations.
Here, we introduce an estimator of recombination rates at fine scales (0.5 – 1 kb) that uses persistent homology and is tailored to the analysis of very large genomic samples. We make use of this estimator to build fine-scale recombination maps of seven human populations sequenced by the 1,000 Genomes Project. Comparison of these recombination maps with recent fine-scale annotations of the human genome (Gerstein et al., 2012; Sai Lakshmi and Agrawal, 2008) reveals that although transcription start sites are generally depleted for recombination (Coop et al., 2008; Lu et al., 2012), specific transcription factor (TF) binding sites are frequently associated with PRDM9 binding motifs and recombination. These include TFs that regulate expression of meiosis- and gametogenesis-specific genes, cell cycle progression, and differentiation blockage. We also observe that repeat-derived loci targeted by piwi-interacting RNAs (piRNAs), coding some recent families of transposable elements known to be expressed during gametogenesis (Guo et al., 2015) and early embryogenesis (Smith et al., 2014), are also enriched for recombination.
Results
Topology and evolution
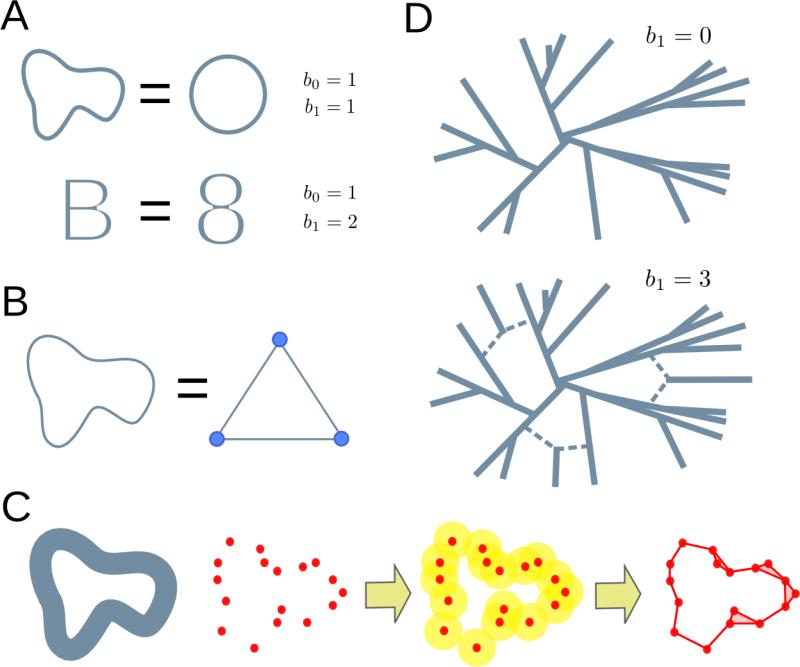
Topology is the branch of mathematics concerned with properties of spaces that are preserved under continuous deformations (deformations that do not involve cutting or pasting), such as the number of loops or connected components of a space. For example, a “B”-shaped space can be continuously deformed into an “8”-shaped space without changing its topology (Figure 1A). A frequently used approach in topology is the replacement of the original space by a simpler one, known as a simplicial complex, which has the same topological features as the original space but consists of a finite set of elements (Figure 1B). A simplicial complex is a generalization of a network that, in addition to nodes and edges, includes higher dimensional elements like triangles and tetrahedra. Simplicial complexes are powerful because they allow the implementation of algebraic operations to extract the topological features of the space. These topological features of a space can be arranged in homology groups, that is, algebraic structures which encompass and classify all gaps or holes in the space. Note that throughout this work the term homology refers to topological homology, which is unrelated to the notion of sequence homology. Elements of the 0th homology group correspond to disconnected parts of the space; elements of the 1st homology group correspond to loops; elements of the 2nd homology group correspond to hollow spheres, and, in general, elements of the nth homology group correspond to (n+1)-dimensional voids of the space. The number of independent elements of the nth homology group is called the nth Betti number. For instance, the first Betti number of an “8”-shaped space is 2 (Figure 1A). We refer to (Ghrist, 2014; Hatcher, 2002) for an extended introduction to the basic concepts of algebraic topology.
Figure 1. Topology and evolution.
(A) Topology is concerned with properties of objects that are invariant under continuous deformations. For instance, a “B”-shaped space can be continuously deformed into an “8”-shaped space. Both have one connected piece and two inequivalent loops. These topological invariants are counted by Betti numbers, bn. Similarly, a circumference always has one connected component and a loop, no matter how it is deformed, as long as nothing is cut or pasted.
(B) A prominent tool in algebraic topology are simplicial complexes. These are finite set representations of the original space that share the same topology. Here, we present a simplicial complex that describes the topology of a circumference. The simplicial complex is given in terms of a finite set of elements (3 points and 3 segments). Algebraic operations on the simplicial complex can extract the topological features of the original circumference.
(C) Topological data analysis infers the topological features of a space from a finite set of sampled points by assigning simplicial complexes to the data. One such construction consists of taking balls of fixed radius ε centered on the points. Points at the center of intersecting balls are connected in the simplicial complex. From the resulting complex, it is possible to extract topological features associated to the data at scale ε.
(D) In the context of evolution, genomic sequences can be represented as points in a high dimensional space, where the distance between points is given by the Hamming distance between the corresponding sequences. In the absence of back-mutation and recombination, subsequent mutations can only increase the distance between genomes, and the evolutionary space of the system does not have loops. When recombination events are present, the evolutionary space contains loops, whose presence can be inferred from the finite genomic sample using TDA methods.
Motivated by the fact that actual data is rarely given in the form of topological spaces, recent mathematical developments have expanded the realm of algebraic topology to point cloud data, that is, any set of data points with a notion of distance between them (Carlsson, 2009). Starting from a set of points sampled from an unknown space, topological data analysis (TDA) aims to infer the topological features of the underlying space (Figure 1C). TDA provides the necessary tools to build simplicial complexes starting from point cloud data. One such construction builds a simplicial complex by taking balls of radius ε, centered on the data points. If two balls intersect, the points at the center of the balls are connected in the simplicial complex. In this way, there is a simplicial complex (and hence a set of topological features) associated to the data at each value of ε. Tracking how homology groups change with ε permits their generalization to point cloud data. The resulting mathematical structures are known as persistent homology groups (Edelsbrunner et al., 2002; Zomorodian and Carlsson, 2005).
TDA can be used to infer evolutionary relations from a sample of genomic sequences (Chan et al., 2013). We consider high-dimensional spaces where each point corresponds to a genomic sequence and distances between points are given by the genetic distance (e.g. Hamming distance) between sequences. The evolutionary history of a sample of genomic sequences can be represented as such a space, consisting of all the genomic sequences that occur from the most common recent ancestor of the sample to the present. Assuming that each genomic site mutates at most once across the entire sample history, the genetic distance between two sequences can only increase with the acquisition of new mutations (Figure 1D). Hence, the only way of “closing” a loop in this space is by means of a recombination event. In populations evolving clonally without recombination or back-mutation, the first Betti number of the evolutionary space of the sample is zero (Chan et al., 2013), as a phylogenetic tree suffices to describe ancestry. More generally, the number of loops of the evolutionary space is related to the number of recombination events in the sample history. Since we only have access to a sample of points, we can make use of the persistent first Betti number (b1) of the sample to infer the amount of recombination in the sample history. In what follows, we make use of this approach to build an estimator of recombination. As noted, our estimator does not rely on genetic linkage, but rather on the topology of spaces formed by genomic sequences.
Persistent homology estimator of recombination (ρPH)
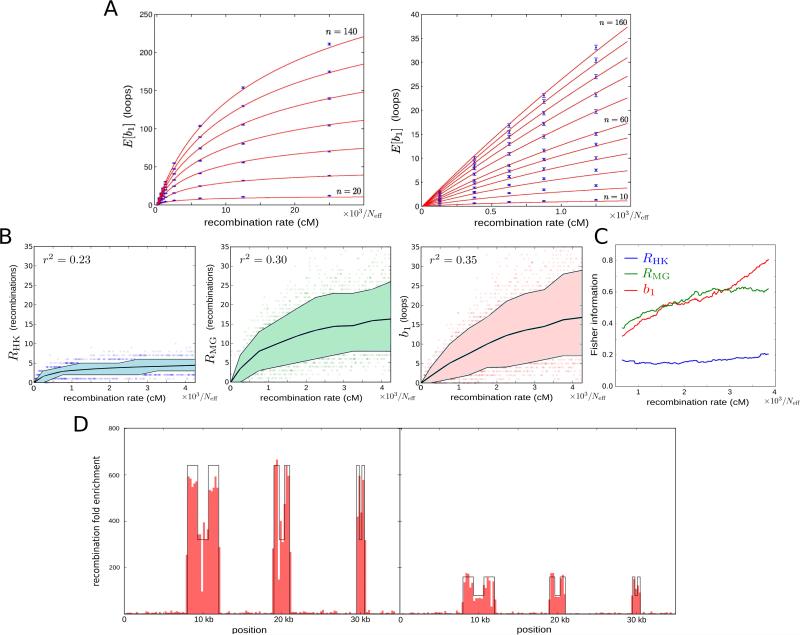
A suitable approach to recombination rate estimation in very large datasets is the use of estimators based on summary statistics of the data (Wall, 2000). As we have argued, the persistent first Betti number of a sample of genomic sequences is expected to be a concise mathematical summary of recombination. To check this hypothesis, we performed extensive coalescent simulations with recombination (Table S1) and observed that the expected value of b1 increases monotonically with the population recombination rate parameter ρ (Figure 2A), which is defined as four times the product of the effective population size (Neff, the number of diploid individuals in a coalescent simulation that produces a level of genetic diversity similar to that of the population of interest) and the per-meiosis recombination rate. At low values of this parameter, b1 is proportional to ρ. Intuitively this behavior is expected, as the number of topology-changing events in coalescent models of evolution scales as ρ log(n) for large n, where n is the number of sequences in the sample (Hein et al., 2004). At large values, b1 saturates due to the limit in the number of loops that a finite set of sequences can generate. We found that this behavior of b1 is well described by a logarithmic function (equation (1) in Experimental Procedures and Figure 2A).
Figure 2. Persistent homology estimator of recombination.
(A) Estimated expected first Betti number E[b1] as a function of the recombination rate (expressed in terms of the effective population size Neff) and sample size n. Each point is based on 500 simulations. Error bars represent 95% confidence level intervals. Red curves correspond to the best fit according to expression (1) in Experimental Procedures.
(B) Dependence of Hudson-Kaplan (left), Myers-Griffiths (center), and b1 (right) summaries of recombination on the recombination rate at fixed number of segregating sites (s = 14). Each plot is based on 4,000 coalescent simulations of a sample of 160 sequences. Colored bands represent the interdecile range and central lines correspond to the mean. Squared Pearson's correlation coefficient is shown in each case.
(C) Fisher information for each of the 3 summaries in (B) as a function of the recombination rate. Information was computed in increments of 12.5/Neff cM. A smoothed trend is plotted by averaging windows of 101 computed values, weighted by the number of simulations.
(D) Distribution of 500 bp segments with ρPH > 0 in simulated samples of 160 sequences, 35 kbp long. The background recombination rate is 500/Neff cM/Mb. Six recombination hotspots of widths 4 kbp, 2 kbp, and 1 kbp are simulated. The local recombination rate is enhanced at hotspots by a factor 640 (left) and 160 (right). Intra-hotpot recombination rate variation is also simulated, with a ½ decay of the local recombination rate at the central region of hotspots.
To evaluate the utility of b1 as a summary of recombination, we compared it to other summaries of recombination available in the literature. Specifically, we considered the lower bounds on the minimum number of recombination events of a sample history developed in (Hudson and Kaplan, 1985) and (Myers and Griffiths, 2003). We refer to those as RHK and RMG, respectively. These summaries combine simple local bounds across different genomic regions to generate a more stringent global bound. Based on simulated samples of sequences with a small number of segregating sites (s = 14), we observed that among the three summaries, b1 has the largest fraction of variance explained by the recombination rate (Figure 2B). In particular, b1 has better sensitivity to high recombination rates, as it does not saturate as early as other summaries of recombination. This conclusion was also confirmed by comparing Fisher information of the three summaries as a function of the recombination rate (Figure 2C). For samples with a large number of segregating sites (s = 40), the performance of RMG and b1 was comparable, with RMG having a larger fraction of explained variance than b1 (Figure S1A) but similar Fisher information (Figure S1B).
To investigate whether saturation of b1 occurs in practical applications, we assumed a mutation rate of 10−8 mutations per base per generation and an effective population size of Neff = 25,000 individuals. With these assumptions, saturation effects for a sample of 200 sequences became important at genetic map distances above ~0.2 centimorgans (cM) over 2.6 kb (for s = 14), or ~0.4 cM over 7.5 kb (for s = 40). These recombination rates are rarely found in the human genome (Kong et al., 2010).
Taking these results together, we concluded that b1 is a robust summary of recombination at fine-scales, where the number of segregating sites is small, and used Pearson's method of moments to build an estimator of recombination rate, ρPH, based on b1 (Supplemental Experimental Procedures).
Comparison to Linkage Methods
Currently, the most commonly used methods to estimate recombination rates are based on linkage disequilibrium, as defined above. Practical methods implement Markov Chain Monte Carlo algorithms to approximate a likelihood function, built from the observed genetic linkage between pairs of sites (Hudson, 2001; McVean et al., 2002), or from the reconstructed segments of exchanged genetic material (Crawford et al., 2004; Li and Stephens, 2003). These methods have great accuracy, producing low-variance unbiased recombination rate estimates, but their applicability to very large datasets can be hindered by their computational cost.
We compared our estimator, ρPH, to the ones generated by widely used software packages LDhat (McVean et al., 2002; McVean et al., 2004) and PHASE (Crawford et al., 2004; Li and Stephens, 2003). The three estimators produced comparable results on simulated data at constant recombination rate and fixed number (s = 14) of segregating sites (Figure S1C), observing some advantage of PHASE over the other two estimators in terms of accuracy. Our estimator, ρPH, however, was on average 12 times faster than LDhat and 30 times faster than PHASE (Figure S1D). At a large number (s = 40) of segregating sites, both LDhat and PHASE offered some advantage over ρPH in terms of precision (Figure S1C), although ρPH was still 2 – 6 times faster than these estimators.
We evaluated the power of ρPH to resolve variation in the recombination rate across narrow genomic loci, using simulations at non-constant recombination rate. Our estimator ρPH produced lower variance estimates than LDhat and PHASE on a sliding window of variable length and fixed number (s = 14) of segregating sites (Figure S1E). To enhance the sensitivity of our estimator to variation in the recombination rate, we implemented a second sliding window with fixed length (L = 500 bp). Counting the number of times that each 500 bp segment had ρPH > 0, over multiple simulations, was sensitive to relative fine-scale variation in the recombination rate, allowing for detection of sub-kilobase scale variations (Figure 2D). In these simulations, our persistent homology estimator was 150 – 1,500 times faster than LDhat and PHASE, making it uniquely suited to very large genomic samples. We exploited this property of our approach and built recombination maps across human populations from the 1,000 Genomes Project.
Recombination maps of seven human populations
We built recombination maps of seven human populations (one African, two Asian, and four of European ancestry, Table S2), using phased genotype data of ~38 million single nucleotide polymorphisms (SNPs) from the 1,000 Genomes Project (1000 Genomes Project Consortium et al., 2012). Our dataset included a total of 647 individuals. We scanned the entire genome three times for each population, using two sliding windows with fixed number of segregating sites (s = 14 and 40) and a sliding window with fixed length (L = 500 bp). For each window and genomic position, a Hamming distance matrix was obtained, from which b1 and ρPH were computed. Windows with a large number of segregating sites give accurate estimates of the recombination rate over relatively large (5-10 kbp) genomic intervals; whereas windows with a small number of segregating sites provide information about the precise genomic location of recombination events. As a first consistency check of our method, we detected no signature of recombination along the Y and mitochondrial chromosomes with either window, as expected from the predominantly uniparental inheritance of these chromosomes. Nevertheless, the relative dearth of SNPs in these chromosomes may also reduce detection sensitivity.
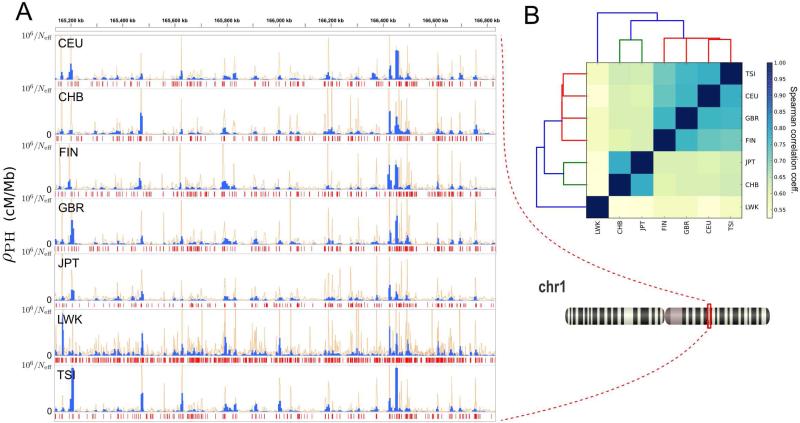
A snapshot of the output produced by this method is shown in Figure 3A. Median detected recombination rates for non-African populations are ~15,000/Neff cM/Mb, and the highest 10% of recombination rates are ≥ 100,000/Neff cM/Mb (Table S2). Population recombination rates ρ were approximately doubled in the African population. This is consistent with a larger effective population size and an out-of-Africa human expansion model (Templeton, 2002). Our method was therefore able to capture the expected differences in population recombination rates due to known population structure effects.
Figure 3. Recombination rate estimates across distant human populations.
(A) Position-wise recombination rates for each of the 7 populations, for the cytogenic band 1q24.1. Blue and orange line plots represent recombination rates estimated with sliding windows with fixed number of segregating sites (s = 40 and s = 14, respectively). Below each track, red segments represent genomic regions where a 500bp sliding window detects recombination (b1 > 0).
(B) Spearman correlation matrix for the position-wise recombination rate of different populations across the entire genome. Maps were binned at 10 kbp and correlation was computed for bins with an average recombination rate of at least 25,000/Neff cM/Mb in each of the maps. Tiles are colored according to the degree of correlation. Hierarchical clustering of the matrix components is also shown, with colored leaves corresponding to African (blue), Asian (green) and European (red) populations.
See also Figure S2 and Table S2.
To check the consistency of the TDA approach across different datasets, we performed a pairwise comparison between the seven maps. We found a high degree of consistency between the location and intensities of recombination peaks identified in distant populations (Figure 3). Position-dependent recombination rates were correlated between pairs of populations, with Spearman's r ranging from 0.53 to 0.78. Hierarchical clustering of populations based on these correlations followed known ancestral relationships, with African, Asian, and European populations grouped in different clusters (Figure 3B).
We also compared our recombination maps to other maps in the literature. Specifically, we considered the deCODE map (Kong et al., 2010), based on a half million crossovers identified in 15,000 Icelandic meioses; the African-American (AA) map (Hinch et al., 2011), based on more than 2 million crossovers in 30,000 unrelated African-Americans; and the HapMap recombination map (International HapMap et al., 2007), based on linkage disequilibrium breakdown using 3 million SNPs genotype data. Although the nature, content, and underlying assumptions of each of these maps differ from each other, comparison with our recombination maps across ~300 kbp regions within the major histocompatibility complex and the MS32 mini-satellite loci revealed a large degree of consistency between different maps (Figures S2A and S2B). To perform a more quantitative comparison, we binned all maps at 10 kbp. Whole-genome Spearman's correlation with our recombination maps was in the range 0.54 – 0.63, 0.53 – 0.61, and 0.43 – 0.48, respectively for HapMap, AA, and deCODE maps. These correlations were comparable to those observed between HapMap and deCODE (r = 0.60), and between deCODE and AA (r = 0.62) maps. Furthermore, recombination rates at exons, introns, and intergenic regions matched those observed in pedigree-based studies (Table 1), providing additional consistency checks of our maps.
Table 1.
Recombination rate estimates across different genomic loci and comparison to pedigree-based estimates (Kong et al., 2010).
| Type | Recombination rate, TDA (FE) | Recombination rate, deCODE (FE) |
|---|---|---|
| Exon | 0.84 ± 0.02 | 0.85 |
| Intron | 0.99 ± 0.01 | 1.02 |
| Intergenic | 1.02 ± 0.01 | 1.03 |
From these results we inferred the merit of the high-resolution recombination maps produced by TDA.
Recombination enrichment at transcription factor binding sites
Because we hypothesized that the human recombination landscape is largely affected by the epigenome and transcriptome of meiotic germ cells, we focused our analysis on the loci of transcription factor binding sites and piRNAs. We considered the DNA binding sites of 118 TFs detected by ChIP-seq in at least one of 91 cell lines studied by the ENCODE Analysis Working Group (Gerstein et al., 2012). For each cell line and transcription factor we computed the recombination enrichment across binding sites, observing little variation across cell lines. We found TF binding sites to be, on average, depleted of recombination with respect to the whole-genome average (fold enrichment (FE) = 0.96, p < 10−37). This observation is consistent with previous work showing a depletion of recombination at transcription start sites (Coop et al., 2008; Lu et al., 2012).
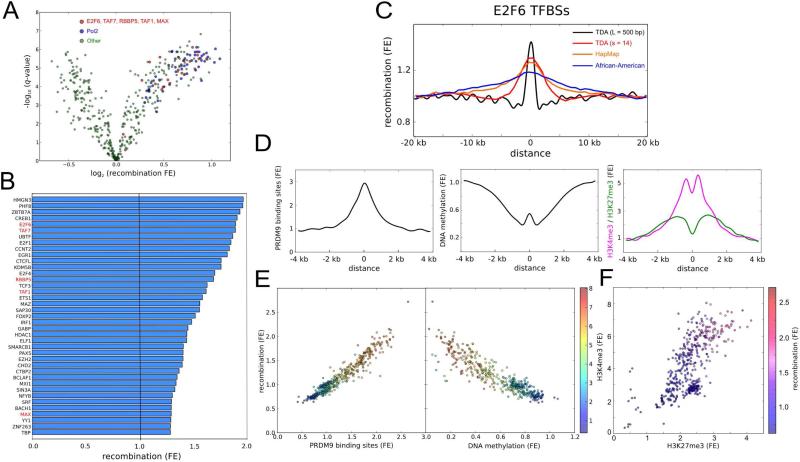
Disaggregating TF binding sites by type of transcription factor, we identified systematic differences across transcription factor families (Figures 4A, 4B, S3A and S3B). Several types of TF binding sites show significant recombination enrichments with respect to the whole-genome average (log-likelihood ratio test as described in Supplemental Experimental Procedures, Benjamini-Hochberg adjusted p < 10−5). These binding sites are found to associate mostly with TFs that have a bias towards proximal promoters according to the ENCODE categorization (Gerstein et al., 2012). Some of those TFs are members of the E2F family, with key roles in the regulation of cell cycle progression and differentiation blockage (DeGregori and Johnson, 2006). In particular, promoters containing binding sites of the transcriptional repressor E2F6, regulating the expression of meiosis-specific genes (Kehoe et al., 2008; Velasco et al., 2010), were found to be enriched for recombination (Figure 4B and 4C). In addition, binding sites of RNA polymerase II and regulatory subunits of MLL1/2 protein complexes were also found to be enriched for recombination (FE 1.28 – 1.68, p ~ 10−2 – 10−7) (Figure 4B). These results were independently confirmed using HapMap and AA recombination maps, despite the lower resolution of these maps (Figure 4C).
Figure 4. Relation of recombination to TF binding sites.
(A) Recombination enrichments at TF binding sites. Each point corresponds to a different combination of TF and cell line. Binding sites are based on ChIP-seq data from ENCODE (Gerstein et al., 2012). In total 118 TFs and 91 cell lines were considered. Recombination enrichments were computed with respect to neighboring regions using the 500bp recombination map of the British (GBR) population. Statistical significances are adjusted for multiple testing using Benjamini-Hochberg procedure. Pol2 and TFs that may for part of MLL1/2 complexes are indicated in blue and red, respectively.
(B) Recombination enrichment with respect to the whole-genome average for the TF binding sites considered in (A). Only TFs with the highest enrichments are shown. ChIP-seq peaks of each TF were merged across all cell lines. TFs that may for part of MLL1/2 complexes (indicated in red) are generally enriched for recombination.
(C) Recombination enrichment at E2F6 binding sites as a function of the distance to the binding site, according to TDA (L = 500bp and s = 14) recombination maps of the GBR population, as well as Hapmap (International HapMap et al., 2007) and African-American (Hinch et al., 2011) recombination maps. E2F6 binding sites were obtained from ENCODE, merging ChIP-seq peaks across K562 and Hela cell lines.
(D) Enrichment for predicted PRDM9 binding sites (defined by the motif CCNCCNTNNCCNC), and sperm CpG methylation, H3K4me3 and H3K27me3 marks as functions of the distance to E2F6 binding sites, for the binding sites considered in (C).
(E) Recombination enrichment at TF binding sites against enrichment for predicted PRDM9 binding sites (left) and sperm CpG methylation (right), for the TFs and cell lines considered in (A). Color scale represents enrichments for sperm H3K4me3 marks at the loci of these TF binding sites.
(F) Enrichments for sperm H3K4me3 and H3K27me3 marks at the loci of TF binding sites, for the TFs and cell lines considered in (A). Color scale represents recombination enrichment at the loci of these TF binding sites. Higher recombination enrichments occur for bivalent TF binding sites.
See also Figure S3 and Table S3.
The above analysis demonstrates that instances of recombination are enriched within the binding sites of specific TF families. This observation is in accord with the local nucleotide sequence. We observed a strong association between recombination and CpG content at these sites (Pearson's r = 0.95, p < 10−50); the observed CpG abundance within regions of recombination enrichment is partially explained by local enrichments for G and C nucleotides (Figure S3C). CpG enrichment at highly recombinant regions is thought to occur through biased gene conversion in the repair of meiotic DSBs (Duret and Galtier, 2009). In addition, we observed a statistically significant association of recombination at TF binding sites with predicted PRDM9 binding sites at these loci (Pearson's r = 0.96, p < 10−50) (Figure 4D and S3B), suggesting that PRDM9 drives recombination towards these loci during meiosis.
Motivated by our findings on the type of TF binding sites that are enriched for recombination, we decided to interrogate whether these binding sites are part of active promoters in the germline. To that end, we assessed the DNA methylation state of these sites in sperm (Molaro et al., 2011) and human primordial germ cells (PGCs) (Gkountela et al., 2015). Our analysis revealed a statistically significant association (Pearson's r = 0.92, p < 10−50) between recombination at TF binding sites and CpG hypo-methylation of these loci in sperm (Figures 4D, 4E and S3B). Hypo-methylation was also detected at earlier stages of gametogenesis, with similar associations in PGCs of male 19.5-week and female 16.1-week embryos (Figure S3D).
DNA hypo-methylated TF binding sites in germ cells and embryonic stem cells have been related to bivalent developmental gene promoters (Bernstein et al., 2006; Hammoud et al., 2009), characterized by simultaneous H3 Lys-4 and Lys-27 tri-methylation (H3K4me3 and H3K27me3) marks on their nucleosomes. The simultaneous presence of these marks defines a poised transcriptional state for the promoters, which remain off until the appropriate developmental stage. To find whether recombination-enriched TF binding sites are part of bivalent promoters in the germline, we considered the profile of these epigenetic marks in sperm (Hammoud et al., 2009). Whereas male germ cells are mostly depleted of nucleosomes after meiosis, we found that loci of recombination-enriched TF binding sites retain nucleosomes (Figure S3E). In addition, our analysis revealed a statically significant association between recombination at TF binding sites and simultaneous H3K4me3 (Pearson's r = 0.82, p < 10−50) and H3K27me3 (Pearson's r = 0.80, p < 10−50) marks in sperm (Figure 4F). Taken together, these results suggest that PRDM9 drives meiotic recombination towards certain active or poised promoters in germ cells.
Recombination enrichment at loci targeted by piRNA
Inspired by the association between developmental promoters and PRDM9-mediated recombination at these loci, we decided to explore recombination rates at the loci of other important transcriptional regulators in the germline. Piwi-interacting RNAs attain their broadest expression in germ cells, and have a central role in post-transcriptional regulation (Watanabe et al., 2015) and transposon control (Aravin et al., 2007). We studied recombination across genomic loci with 100% sequence identity to known human piRNA sequences (Sai Lakshmi and Agrawal, 2008). We observed an enrichment for recombination (FE = 2.67, p < 10−49) at these loci with respect to the whole-genome average. The enrichment was also present, although reduced (FE = 1.31, p ~ 10−9), when restricting to piRNA-producing clusters as defined and annotated by (Ha et al., 2014).
A large fraction of piRNAs are repeat-derived. We estimated the enrichment for recombination at piRNA-matched loci derived from repetitive elements (Figure 5A). All main families (LTR, LINE and SINE) are enriched for recombination compared to neighboring genomic regions. In terms of specific transposable elements, some of the most recent L1 elements (L1Hs, L1PA1-4), human endogenous retroviruses (HERVK, HERVH, HERVL), LTRs (LTR12C, MER11C), and most Alu and SVA elements present the highest recombination rates. These elements have been found to be expressed during germ cell development (Guo et al., 2015) and early embryogenesis (Smith et al., 2014). We observed a systematic pattern for the recombination rate at these loci, peaking in most cases at the 5’- and 3’-ends of the transposon, with the 5’-end presenting the highest recombination rate (Figure 5B). Our analysis also reproduced known recombination enrichments at THE1A/B repeat elements (Myers et al., 2005) (Figure S4A). These findings were also confirmed using linkage-based methods on 1,000 Genomes Project data (Figure S4B).
Figure 5. Relation of recombination to piRNA loci.
(A) Recombination enrichment at repeat-derived piRNA-matched loci with respect to the whole-genome average. Repeat-derived loci with 100% identity to some sequence deposited in piRNA-Bank (Sai Lakshmi and Agrawal, 2008) were classified as belonging to SINE, LINE, SVA or other family of repetitive elements. Recombination enrichments were computed using the British (GBR) recombination map.
(B) Enrichment for recombination, CpG sites and sperm methylation for loci matched by four specific repeat-derived piRNA (piRNA-Bank accession numbers DQ577359, DQ579099, DQ577145 and DQ571358). The location of transposable elements and their 5’-and 3’-ends are shown in black. Predicted PRDM9 binding motifs conserved across different loci are indicated in red. The origin of coordinates corresponds to the location of the piRNA-matched sequence. In the four cases the piRNA sequence is antisense to the transposable element.
(C) Enrichment for recombination (top left), predicted PRDM9 binding sites (top right) according to the motif CCNCCNTNNCCNC, sperm CpG methylation (bottom left), and sperm H3K4me3 and H3K27me3 marks (bottom right) for the loci considered in (A).
See also Figure S4.
Being aware that repetitive elements are particularly prone to misalignment, we looked for additional evidence supporting the enrichment for recombination at these loci. We found that the enrichment is also accompanied by conserved PRDM9 binding motifs (Figures 5B and 5C). Furthermore, recombination at these loci is strongly correlated with CpG abundance (Figures 5B and S4C). As occurred with TF binding sites, a large fraction of the CpG abundance is explained by a local enrichment for G and C nucleotides, being suggestive of extensive biased gene conversion from meiotic DSB repair. All these features added strong support to the observed recombination enrichment at repeat-derived loci targeted by piRNA, and were different in the case of uniquely mapped piRNAs (single locus piRNA genes) (Figure 5C), suggesting that only repeat-derived loci are enriched for recombination.
Discussion
In the last few years, technological advances have allowed the identification of highly localized molecular features such as TF binding sites, methylation sites, loci of noncoding RNAs, and regions of open chromatin, at unprecedented resolution. Population-based methods tailored for the analysis of large datasets at the relevant fine genomic scales are needed to explore the relationship between recombination and these highly localized molecular features. We have presented an efficient method, based on TDA, to estimate recombination rates from very large samples of genomic sequences. Our method detects a topological signature of recombination distinct from the linkage-based information upon which existing methods depend. The performance and accuracy of this approach have allowed us to build high-resolution recombination maps of seven human populations (Figure 3). These maps can be used to establish new associations between recombination and genomic features, as we have demonstrated for TF binding sites (Figure 4) and piRNA loci (Figure 5). These associations are supported by enrichments for predicted PRDM9 binding motifs, CpG content, and epigenetic marks in germ cells. Overall, these findings provide confidence in the application of TDA to human genomic data and demonstrate its utility for overcoming some of the limitations of conventional methods and for generating hypotheses about fine-scale human recombination.
Our results raise broad questions about the control and evolution of recombination in humans. Prdm9 knockout experiments in mice suggest that the protein encoded by this gene plays a role in sequestering recombination away from gene promoters and other functional genomic elements (Brick et al., 2012). We find that hypothesis to be consistent with the overall depletion of recombination at TF binding sites observed in our study. However, by disaggregating different TF binding site types, we have shown that binding sites of specific TFs are systematically enriched for recombination. This constitutes a clear exception to the general rule described above. These TF binding sites include binding sites of the E2F family and regulatory subunits of MLL1/2 complexes, which play prominent regulatory roles in germ cell development and early embryogenesis. This circumstance in humans is analogous to the case of homologous recombination at Saccharomyces cerevisiae promoters, where an H3K4 methyltransferase forms part of the COMPASS protein complex and links H3K4me3 marks to the formation of meiotic DSBs at promoters (Acquaviva et al., 2013). Accordingly, MLL complexes are human homologues of the COMPASS complex in yeast (Miller et al., 2001).
From an evolutionary perspective, an increased recombination rate at gene promoters regulating germ cell development and embryogenesis would render selection at these loci more effective by unlinking selective forces that act on different positions (Hill and Robertson, 2007; Iles et al., 2003). A similar argument can be made regarding the enrichments for recombination observed at repeat-derived loci matched by piRNA sequences, particularly in light of recent work uncovering the role of transposons and piRNAs in post-transcriptional regulation in the germline (Watanabe et al., 2015). The rapid genetic divergence of genes involved in meiosis suggests that these regions undergo extraordinary positive selection (Keeney, 2008; Richard et al., 2005; Schwartz et al., 2014); it stands to reason that elevated recombination at these loci would be selected for. On the other hand, DSBs targeted to regions active during meiosis may deprive the cell of necessary transcripts. Our findings highlight a potential evolutionary trade-off in regulation of recombination that merits further study.
Experimental Procedures
Expected b1
We performed 3.5×105 neutral population simulations using the program ms (Hudson, 2002) and seq-gen (Rambaut and Grassly, 1997). Simulated haplotypes were produced for the population recombination rate, ρ, the population mutation rate, θ, and the number of sampled sequences, n, taking values in the ranges 0 – 25,000/Neff cM, 0 – 62.5/Neff mutations per generation and 0 – 160 sampled sequences, respectively. Pairwise distances were defined using Hamming distance. For each distance matrix we built a filtration of Vietoris-Rips complexes and computed b1 using the software Dionysus (http://www.mrzv.org/software/dionysus/index.html). For each configuration of the parameters, we estimated E[b1] based on 500 simulations (Table S1). Simulated data was empirically described by the equation
| (1) |
where the parameters f, g, and h depend on the number of sequences and segregating sites in the sample. We performed 6.4×104 neutral population simulations of samples with fixed number (s = 14 and 40) of segregating sites, for ρ and n taking values in the ranges 0 – 4,500/Neff cM and 0 – 160 sampled sequences, respectively. Based on these simulations, we determined by least squares fitting the following structure for the parameters in equation (1),
| (2) |
| (3) |
where f1 = 0.04404, f2 = 1.5734, g1 = 0.50663, g2 = −2.3129, for s = 14, and f1 = −0.08225, f2 = 3697124, g1 = 0.11483, g2 = 8.4455, for s = 40. For convenience, we fixed h = 0 in equation (1).
Persistent homology estimator of recombination
From population genetics simulations we observed that the first Betti number b1 of a set of sequences sampled from a Wright-Fisher population with recombination is Poisson distributed. From this fact and relation (1) with h = 0, we proved that
| (4) |
is an estimator of the population recombination rate ρ (see Supplemental Experimental Procedures). Comparison of this estimator to linkage methods was performed as described in the Supplemental Experimental Procedures.
Genome scan implementation
We implemented ρPH in three sliding windows, acting on the phased SNP genotype data of 1,000 Genomes Project. Two of the sliding windows had a fixed number of segregating sites (s = 14 and s = 40), and were moved in steps of 7 and 20 segregating sites, respectively. The other sliding window had fixed length (L = 500 bp), and was moved in steps of 250 bp. We included both chromosomes in the case of autosomal chromosomes, and only one of the two X chromosomes for females. Sources used for recombination map annotation, and methods for estimating relative recombination rates and similarity across human populations are described in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Topological data analysis captures recombination from large genomic samples.
High-resolution recombination maps of seven human populations are presented.
Binding sites of specific transcription factors are enriched for recombination.
Repeat-derived loci matched by piwi-interacting RNAs are enriched for recombination.
Acknowledgments
We thank Debra J. Wolgemuth, Marcia Manterola, Molly Przeworski, Nicholas Parrish, and Suzanne Christen for their useful comments, and Oliver Elliott for technical support. R.R. acknowledges funding from NIH (U54 CA193313-01). P.G.C. acknowledges Universidad de Barcelona for financial support and hospitality during the initial stages of the project. D.I.S.R. acknowledges funding from NIH (R01 GM117591, T15 LM007079).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
P.G.C. conceived of the methodology, formulated the persistent homology estimator of recombination and carried out the genomic analysis. D.I.S.R., K.E. and P.G.C developed and carried out comparisons to linkage methods. R.R. conceived of the methodology. A.L. contributed to the discussion. All authors discussed results and implications and collaborated on the writing of the manuscript.
eTOC Blurb
Camara et al. introduce a new method to estimate recombination rates from large genomic samples and present high-resolution recombination maps of seven human populations. Using these maps, they show evidence of previously unreported associations of recombination with binding sites of specific transcription factors and with repeat-derived loci matched by piRNAs.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acquaviva L, Szekvolgyi L, Dichtl B, Dichtl BS, de La Roche Saint Andre C, Nicolas A, Geli V. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science. 2013;339:215–218. doi: 10.1126/science.1225739. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science. 2007;316:744–747. doi: 10.1126/science.1142612. [DOI] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327:836–840. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485:642–645. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson G. Topology and data. Bulletin of the American Mathematical Society. 2009;46:255–308. [Google Scholar]
- Chan JM, Carlsson G, Rabadan R. Topology of viral evolution. Proc Natl Acad Sci U S A. 2013;110:18566–18571. doi: 10.1073/pnas.1313480110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coop G, Wen X, Ober C, Pritchard JK, Przeworski M. High-resolution mapping of crossovers reveals extensive variation in fine-scale recombination patterns among humans. Science. 2008;319:1395–1398. doi: 10.1126/science.1151851. [DOI] [PubMed] [Google Scholar]
- Crawford DC, Bhangale T, Li N, Hellenthal G, Rieder MJ, Nickerson DA, Stephens M. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nat Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- Edelsbrunner H, Letscher D, Zomorodian A. Topological persistence and simplification. Discrete and Computational Geometry. 2002;28:511–533. [Google Scholar]
- Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. Architecture of the human regulatory network derived from ENCODE data. Nature. 2012;489:91–100. doi: 10.1038/nature11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghrist R. Elementary applied topology (Createspace) 2014 [Google Scholar]
- Gkountela S, Zhang KX, Shafiq TA, Liao WW, Hargan-Calvopina J, Chen PY, Clark AT. DNA Demethylation Dynamics in the Human Prenatal Germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow EJ, Flynn RA, Chavez SL, Bayless NL, Wossidlo M, Wesche DJ, Martin L, Ware CB, Blish CA, Chang HY, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo F, Yan L, Guo H, Li L, Hu B, Zhao Y, Yong J, Hu Y, Wang X, Wei Y, et al. The Transcriptome and DNA Methylome Landscapes of Human Primordial Germ Cells. Cell. 2015;161:1437–1452. doi: 10.1016/j.cell.2015.05.015. [DOI] [PubMed] [Google Scholar]
- Ha H, Song J, Wang S, Kapusta A, Feschotte C, Chen KC, Xing J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15:545. doi: 10.1186/1471-2164-15-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher A. Algebraic topology. Vol. 606. Cambridge University Press; Cambridge: 2002. [Google Scholar]
- Hein J, Schierup M, Wiuf C. Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press; USA: 2004. [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 2007;89:311–336. doi: 10.1017/S001667230800949X. [DOI] [PubMed] [Google Scholar]
- Hinch AG, Tandon A, Patterson N, Song Y, Rohland N, Palmer CD, Chen GK, Wang K, Buxbaum SG, Akylbekova EL, et al. The landscape of recombination in African Americans. Nature. 2011;476:170–175. doi: 10.1038/nature10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Two-locus sampling distributions and their application. Genetics. 2001;159:1805–1817. doi: 10.1093/genetics/159.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kaplan NL. Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics. 1985;111:147–164. doi: 10.1093/genetics/111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iles MM, Walters K, Cannings C. Recombination can evolve in large finite populations given selection on sufficient loci. Genetics. 2003;165:2249–2258. doi: 10.1093/genetics/165.4.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap, C. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Jeffreys AJ, Keeney S. Where the crossovers are: recombination distributions in mammals. Nat Rev Genet. 2004;5:413–424. doi: 10.1038/nrg1346. [DOI] [PubMed] [Google Scholar]
- Keeney S. Spo11 and the Formation of DNA Double-Strand Breaks in Meiosis. Genome Dyn Stab. 2008;2:81–123. doi: 10.1007/7050_2007_026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe SM, Oka M, Hankowski KE, Reichert N, Garcia S, McCarrey JR, Gaubatz S, Terada N. A conserved E2F6-binding element in murine meiosis-specific gene promoters. Biol Reprod. 2008;79:921–930. doi: 10.1095/biolreprod.108.067645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkness EF, Grindberg RV, Yee-Greenbaum J, Marshall CR, Scherer SW, Lasken RS, Venter JC. Sequencing of isolated sperm cells for direct haplotyping of a human genome. Genome Res. 2013;23:826–832. doi: 10.1101/gr.144600.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehler KE, Hawley RS, Sherman S, Hassold T. Recombination and nondisjunction in humans and flies. Hum Mol Genet. 1996;5 Spec No:1495–1504. doi: 10.1093/hmg/5.supplement_1.1495. [DOI] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- Li L, Cheng WY, Glicksberg BS, Gottesman O, Tamler R, Chen R, Bottinger EP, Dudley JT. Identification of type 2 diabetes subgroups through topological analysis of patient similarity. Sci Transl Med. 2015;7:311ra174. doi: 10.1126/scitranslmed.aaa9364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics. 2003;165:2213–2233. doi: 10.1093/genetics/165.4.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zong C, Fan W, Yang M, Li J, Chapman AR, Zhu P, Hu X, Xu L, Yan L, et al. Probing meiotic recombination and aneuploidy of single sperm cells by whole-genome sequencing. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean GA, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci U S A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–1041. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers S, Bottolo L, Freeman C, McVean G, Donnelly P. A fine-scale map of recombination rates and hotspots across the human genome. Science. 2005;310:321–324. doi: 10.1126/science.1117196. [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science. 2010;327:876–879. doi: 10.1126/science.1182363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SR, Griffiths RC. Bounds on the minimum number of recombination events in a sample history. Genetics. 2003;163:375–394. doi: 10.1093/genetics/163.1.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolau M, Levine AJ, Carlsson G. Topology based data analysis identifies a subgroup of breast cancers with a unique mutational profile and excellent survival. Proc Natl Acad Sci U S A. 2011;108:7265–7270. doi: 10.1073/pnas.1102826108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144:719–731. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327:835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346:1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- Richard GF, Kerrest A, Lafontaine I, Dujon B. Comparative genomics of hemiascomycete yeasts: genes involved in DNA replication, repair, and recombination. Mol Biol Evol. 2005;22:1011–1023. doi: 10.1093/molbev/msi083. [DOI] [PubMed] [Google Scholar]
- Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JJ, Roach DJ, Thomas JH, Shendure J. Primate evolution of the recombination regulator PRDM9. Nat Commun. 2014;5:4370. doi: 10.1038/ncomms5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472:375–378. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Chan MM, Humm KC, Karnik R, Mekhoubad S, Regev A, Eggan K, Meissner A. DNA methylation dynamics of the human preimplantation embryo. Nature. 2014;511:611–615. doi: 10.1038/nature13581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton A. Out of Africa again and again. Nature. 2002;416:45–51. doi: 10.1038/416045a. [DOI] [PubMed] [Google Scholar]
- Velasco G, Hube F, Rollin J, Neuillet D, Philippe C, Bouzinba-Segard H, Galvani A, Viegas-Pequignot E, Francastel C. Dnmt3b recruitment through E2F6 transcriptional repressor mediates germ-line gene silencing in murine somatic tissues. Proc Natl Acad Sci U S A. 2010;107:9281–9286. doi: 10.1073/pnas.1000473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall JD. A comparison of estimators of the population recombination rate. Mol Biol Evol. 2000;17:156–163. doi: 10.1093/oxfordjournals.molbev.a026228. [DOI] [PubMed] [Google Scholar]
- Wang J, Fan HC, Behr B, Quake SR. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Cheng EC, Zhong M, Lin H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res. 2015;25:368–380. doi: 10.1101/gr.180802.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomorodian A, Carlsson G. Computing persistent homology. Discrete & Computational Geometry. 2005;33:249–274. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.