Abstract
Loss of 14-3-3σ has been observed in multiple tumor types; however, the mechanisms by which 14-3-3σ loss leads to tumor progression are not understood. The experiments in this report demonstrate that loss of 14-3-3σ leads to a decrease in the expression of epithelial markers and an increase in the expression of mesenchymal markers, which is indicative of an induction of the epithelial to mesenchymal transition (EMT). The EMT was accompanied by an increase in migration and invasion in the 14-3-3σ−/− cells. 14-3-3σ−/− cells show increased stabilization of c-Jun, resulting in an increase in the expression of the EMT transcription factor slug. 14-3-3σ induces the ubiquitination and degradation of c-Jun in an FBW7-dependent manner. c-Jun ubiquitination is dependent on the presence of an intact nuclear export pathway as c-Jun is stabilized and localized to the nucleus in the presence of a nuclear export inhibitor. Furthermore, the absence of 14-3-3σ leads to the nuclear accumulation and stabilization of c-Jun, suggesting that 14-3-3σ regulates the subcellular localization of c-Jun. Our results have identified a novel mechanism by which 14-3-3σ maintains the epithelial phenotype by inhibiting EMT and suggest that this property of 14-3-3σ might contribute to its function as a tumor suppressor gene.
Keywords: 14–3-3 protein, c-Jun transcription factor, E3 ubiquitin ligase, epithelial-mesenchymal transition (EMT), tumor suppressor gene, EMT transcription factors, nuclear export, tumor progression and metastasis, ubiquitin-mediated proteolysis
Introduction
14-3-3 proteins are evolutionarily conserved and ubiquitously expressed in all eukaryotes (1, 2). Seven 14-3-3 isoforms have been identified in mammals: β, γ, ϵ, σ, η, ζ, and τ. They form homodimers and heterodimers that recognize phosphoserine/phosphothreonine-containing consensus motifs (mode 1 (RSXpSXP where pS is phosphoserine) and mode 2 (RXXXpSXP) (3, 4)) in their ligands. Some 14-3-3 ligands do not contain mode 1 or mode 2 consensus sequences but still form a complex with 14-3-3 proteins (5, 6).
14-3-3σ also known as stratifin and HME1 (human mammary epithelium-specific marker) was originally identified as a protein expressed only in epithelial cells (7). Unlike other 14-3-3 isoforms, 14-3-3σ exclusively forms homodimers (8, 9), leading to the hypothesis that 14-3-3σ performs a unique set of functions that are not performed by the other 14-3-3 isoforms. 14-3-3σ expression can be activated by p53 and p63, resulting in a cell cycle arrest in G2 in response to DNA damage in part by sequestering the cdk1-cyclinB complex to the cytoplasm, thus preventing mitotic progression (10–13). In addition, 14-3-3σ also regulates mitotic translation, which is required for accurate progression through mitosis (14). 14-3-3σ can positively regulate p53 transcription and stability, suggesting the presence of a positive feedback loop between p53 and 14-3-3σ (15). 14-3-3σ is a tumor suppressor, and its expression is decreased in multiple tumor types such as breast cancer (16, 17), ovarian cancer (18), hepatocellular carcinoma (19), prostate cancer (20, 21), basal cell carcinoma (22), gastric cancer (23), and lung cancer (24). The decrease in 14-3-3σ levels is due to the inhibition of gene expression due to methylation of the 14-3-3σ promoter or increased degradation of 14-3-3σ by the proteasome (16, 17).
The progression of a primary epithelial tumor to an invasive and metastatic tumor is often accompanied by activation of the epithelial to mesenchymal transition (EMT)3 (25–28). The induction of EMT is also associated with the acquisition of chemoresistance (29, 30), and EMT induction is mediated by a group of transcription factors such as snail, slug, ZEB, and twist (25, 27, 31, 32). These transcription factors have exclusive and redundant functions in terms of regulating expression of epithelial and mesenchymal genes (33–35); both snail and slug can bind to the E box of E-cadherin promoter and repress transcription of E-cadherin (33, 36). In addition to activating/repressing the expression of other gene products, the EMT transcription factors can induce the formation of a positive feedback loop by activating either their own expression (37) or the expression of other EMT transcription factors (38), thus resulting in activation of the EMT cascade.
Expression of the EMT transcription factors can be activated by multiple cellular signaling pathways; e.g. TGFβ-mediated induction of EMT is often observed in epithelial cancers such as breast cancer (39, 40), hepatocellular carcinoma (41), cervical carcinoma (42), and lung cancer (43). Activation of the MAPK and ERK pathways is required for TGFβ-mediated EMT and directly stimulates the expression of snail and slug (44–46). The process of EMT can also be induced by growth factors such as EGF and hepatocyte growth factor, leading to the activation of signaling pathways that stimulate the expression of EMT transcription factors (31). AKT-mediated activation of NF-κB leads to an increase in snail expression (47), whereas the activation of WNT signaling leads to a LEF1- and TCF1-mediated increase in expression of snail, slug, and twist (48–50).
Multiple conserved regions for AP1 and AP4 transcription factors have been identified in the promoter regions of the snail family of transcription factors (51). The AP1 transcription factor c-Jun has been shown to bind to the slug promoter, which can result in an increase in expression of slug and induction of EMT (52). c-Jun expression is found to be elevated in multiple cancer types and shows a significant association with invasion and metastasis (53–55). In epithelial cells, c-Jun is targeted for degradation by the proteasome, and multiple E3 ligases that mediate c-Jun ubiquitination, including COP1, ITCH, and FBW7, have been identified (56–58). Interestingly, it has been observed that loss of FBW7 leads to EMT (59), although it is not clear whether this is due to an increase in c-Jun levels.
Despite a strong correlation between a decrease in 14-3-3σ protein levels and progression of multiple human cancers of epithelial origin, the mechanisms by which 14-3-3σ loss leads to tumor progression are unclear. 14-3-3σ deficiency has been shown to cause deregulation of epithelial cell polarity, which is a hallmark of the activation of the EMT program (60). The results in this report indicate that loss of 14-3-3σ can lead to activation of the EMT program in HCT116 cells. 14-3-3σ binds to c-Jun, resulting in the proteasome-dependent degradation of c-Jun by FBW7. The increased nuclear localization and stability of c-Jun in 14-3-3σ−/− cells leads to an increase in slug expression, leading to the induction of an EMT with an increase in invasion and migration. These data suggest that one mechanism by which loss of 14-3-3σ leads to tumor progression is by the induction of an EMT.
Experimental Procedures
Cell Culture and Transfections
HCT116 (ATCC), HCT116-derived 14-3-3σ−/− cells (10), HEK293, and HCT116 14-3-3σ−/− cell-derived stable cell lines were cultured in complete Dulbecco's modified Eagle's medium (DMEM) as described (61, 62). Transfections were performed by either of the following methods: calcium phosphate precipitation as described (63) or Lipofectamine LTX (Invitrogen), PEI (Polysciences Inc.), or FuGENE Xtremegene HP (Roche Applied Science) according to the manufacturer's instructions.
Plasmids and Generation of Stable Cell Lines
HA-14-3-3σ has been described previously (61). HA-14-3-3σ was cloned in pcDNA3 puro vector. Wild type (WT) c-Jun was cloned in HA-pcDNA3 vector using BamHI and XhoI sites to generate HA-c-Jun WT. c-Jun S58A and c-Jun S267A were generated by site-directed mutagenesis (see supplemental Table 1 for primers) and cloned as described above. Stable clones expressing HA-tagged 14-3-3σ, namely HA-14-3-3σ-1 and HA-14-3-3σ-2, and the respective vector control (vector) clones were generated in HCT116 14-3-3σ−/− cells, and clones were selected in medium containing 1 μg/ml puromycin. Published shRNA sequences for slug (sh-1Slug and sh-2Slug) and c-Jun (sh1-c-Jun and sh2-c-Jun) (64, 65) cloned in pLKO.1 vector were used to generate viral particles in HEK293FT as described previously (66). The viral particles were used to transduce 14-3-3σ−/− cells to generate stable knockdown clones. These clones were selected in medium containing 1 μg/ml puromycin. FBW7, c-Jun WT, c-Jun S263A, and 14-3-3σ were cloned into a mammalian expression vector expressing an S-protein/FLAG/streptavidin-binding protein triple epitope tag (SFB) and a Myc-tagged destination vector described previously (67) using a Gateway cloning system (Invitrogen). Bacterial expression vectors for GST-c-Jun WT, GST-c-Jun S263A, maltose-binding protein (MBP)-FBW7, and MBP-14-3-3σ vectors were generated by transferring the appropriate cDNA into destination vectors as described previously (67). FBW7 constructs (68) were a kind gift from Dr. Sagar Sengupta (National Institute of Immunology, India) and Dr. Markus Welcker (Fred Hutchinson Cancer Research Center).
Immunofluorescence and Confocal Microscopy
Immunofluorescence with antibodies against plakoglobin, desmocollin2/3, desmoglein2, HA, plakophilin3, desmoplakin, ZO1, E-cadherin, β-catenin, α-tubulin, and keratin8 were performed as described (66, 69, 70). Vimentin (Sigma; dilution, 1:500) or N-cadherin (BD Transduction Laboratories; dilution, 1:10) were immunostained using methanol fixation as described (66). Images were obtained by using an LSM 510 Meta Carl Zeiss confocal system.
Immunoprecipitation and Western Blotting
14-3-3σ+/+ cells were treated with MG132 (10 μm) for 6 h, and cell lysates were prepared with EBC lysis buffer. 120 μl of 14-3-3σ antibody (hybridoma supernatant CS112) was used to immunoprecipitate 14-3-3σ as described (71). Myc antibody (9E10; mouse monoclonal) was used as an isotype control. This was followed by Western blotting with c-Jun and plakophilin 3 antibodies as described (62). Suppliers and dilutions of antibodies for Western blotting are described in supplemental Table 2.
Reverse Transcription-PCR and Quantitative Real Time PCR
Reverse transcription assays and quantitative real time PCR were performed as described (70, 72). Primers used for both RT-PCR and quantitative real time PCR for PKP3, CK8, CK18, vimentin, c-Jun, and different 14-3-3 isoforms were described previously (65, 72, 73). Primer sequences are listed in supplemental Table 3.
Wound Healing, Dispase Assays, and Hanging Drop Assays
Wound healing and hanging drop assays were performed as described earlier (62). For Dispase assays, cells were cultured as a confluent monolayer in complete medium. The spent medium was removed, and the cells were washed twice with 1× PBS followed by incubation with 0.6–1.2 units/ml Dispase II (Roche Applied Science) in 1× Dulbecco's PBS for 30 min at 37 °C. The cells were subjected to mechanical orbital shaking at 150 rpm for 10–20 min and imaged under a dissecting microscope. The number of monolayer fragments was counted using ImageJ software.
Matrigel Invasion Assay
Boyden chamber (BD Biosciences) Matrigel invasion assays were performed in 24 -well plates. The inner side of the chamber was washed two to three times with 1× PBS and then once with DMEM. The inner surface of the insert was coated with 100 μl of a 300 μg/ml working solution of Matrigel (BD Biosciences). After incubation at 37 °C for 1 h, unpolymerized Matrigel was removed from the upper chamber. 50,000 cells were resuspended in 200 μl of DMEM without FBS and seeded in the insert. 700 μl of DMEM with FBS was added in the lower chamber to act as a chemoattractant. 36 h postincubation in a 37 °C incubator with 5% CO2, the inserts were cleaned with a cotton swab and fixed in 100% chilled methanol. The inserts were stained with 1% crystal violet and mounted on slides with DPX. Images of 20 random fields were taken with a 10× objective on an Upright Axio Imager Z1 microscope.
Determination of 14-3-3 Binding Motif in c-Jun
The c-Jun sequence was examined for the presence of the 14-3-3 binding consensus motifs ((mode 1 (RSXpSXP) and mode 2 (RXXXpSXP)) (4, 74). Two sequences, which have similarity to the mode 2 site, RAKNSDL and RIAASKC, were identified. These sequences had serine residues at positions 58 and 267, respectively.
In Vitro GST Pulldown Assays
To determine in vitro binding, GST-14-3-3σ and c-Jun WT or point mutant (S58A and S267A) HEK293 cells were transfected with HA-c-Jun WT, HA-c-Jun S58A, and HA-c-Jun S267A. GST pulldown assays were performed as described (70, 71).
In Vivo Ubiquitination Assays
Cells were transfected with various combinations of plasmids. At 24 h post-transfection, cells were treated with MG132 (10 μm) for 6 h, and whole-cell extracts were prepared by NETN lysis or denaturing lysis and subjected to immunoprecipitation of the substrate protein. Ubiquitination was determined by immunoblotting with substrate antibody or anti-ubiquitin antibody.
In Vitro Ubiquitination Assays
The reactions were carried out at 30 °C for 15 min in 25 μl of ubiquitylation reaction buffer (40 mm Tris-HCl, pH7.6, 2 mm dithiothreitol (DTT), 5 mm MgCl2, 0.1 m NaCl, 2 mm ATP) containing the following components: 100 m ubiquitin, 20 nm E1 (UBE1), and 100 nm UbcH5b (all from Boston Biochem). Bacterially purified MBP-FBW7 and MBP-14-3-3σ were added to the reaction mixture. Bacterially purified GST, GST-c-Jun, and GST-c-Jun S267A bound to glutathione-Sepharose beads (Amersham Biosciences) were used as substrates in the reaction mixture. After the reaction, beads were washed three times with NETN buffer and boiled with SDS-PAGE loading buffer, and ubiquitination of substrates was detected by Western blotting with anti-GST antibody.
Nuclear-Cytoplasmic Fractionation Assays
14-3-3σ+/+ and 14-3-3σ−/− cells or 14-3-3σ+/+ cells transfected with plasmids expressing either HA-c-Jun WT, HA-c-Jun S58A, HA-c-Jun S267A, or the vector control were harvested by trypsinization, and nuclear and cytoplasmic fractions were prepared according to the manufacturer's instructions using the NE-PER kit from Promega. Protein lysates were separated by SDS-PAGE. α-Tubulin and lamin A antibodies were used as controls for cytoplasmic and nuclear fractions, respectively.
Results
Loss of 14-3-3σ Leads to Induction of the EMT Program
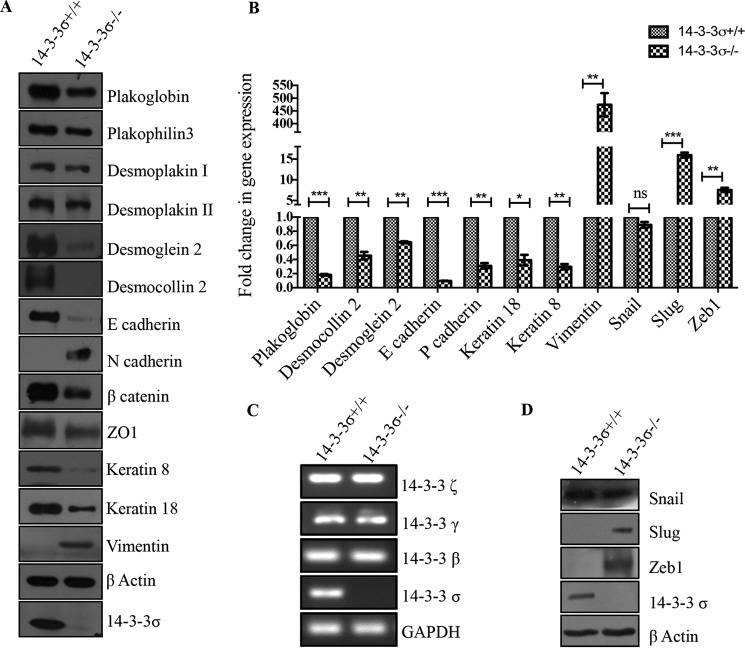
A decrease in the levels of the desmosomal plaque protein plakoglobin and a consequent decrease in cell-cell adhesion have been observed upon loss of 14-3-3σ (70). To determine whether these observations could be extended to other epithelial markers required for cell-cell adhesion, the levels of several epithelial markers were determined in the 14-3-3σ−/− cells. A decrease in the levels of both the protein and mRNA of epithelial markers such as plakoglobin, plakophilin 3, desmoplakin, desmoglein 2, desmocollin 2, E-cadherin, β-catenin, ZO1, keratin 8, and keratin 18 was observed in 14-3-3σ−/− cells as compared with the 14-3-3σ+/+ cells, and this was accompanied by an increase in the protein and mRNA levels of mesenchymal markers such as N-cadherin and vimentin (Fig. 1, A and B). The levels of the other 14-3-3 isoforms were not altered in the 14-3-3σ−/− cells (Fig. 1C), and immunofluorescence assays demonstrated that although the protein levels of plakoglobin, plakophilin 3, desmoplakin, desmocollin 2, E-cadherin, P-cadherin, β-catenin, and ZO1 decreased upon 14-3-3σ loss, no significant change in the localization of these proteins was observed in 14-3-3σ−/− cells (supplemental Fig. S1, A and B). The decreased levels of keratin 8 resulted in the loss of a well defined filamentous structure in 14-3-3σ−/− cells, whereas the mesenchymal intermediate filament protein vimentin was completely absent in 14-3-3σ+/+ cells (69) and formed a filament network in 14-3-3σ−/− cells (supplemental Fig. S1C). These results suggest that the loss of 14-3-3σ leads to an EMT.
FIGURE 1.
Loss of 14-3-3σ leads to an increase in epithelial markers and a decrease in mesenchymal markers. A, protein extracts from 14-3-3σ−/− cells and 14-3-3σ+/+ cells were resolved by SDS-PAGE followed by Western blotting with the indicated antibodies. Western blots for β-actin served as a loading control. B, quantitative PCR was performed using cDNA synthesized from total mRNA isolated from 14-3-3σ+/+ cells and 14-3-3σ−/− cells. ΔΔct values were calculated and normalized against GAPDH. The -fold change in gene expression is plotted on the y axis for the indicated genes. p values were calculated using a non-parametric t test (*** indicates a p value ≤0.001, ** indicates a p value ≤0.01, * indicates a p value ≤0.05, and ns indicates not significant). C, total mRNA was isolated from 14-3-3σ+/+ cells and 14-3-3σ−/− cells. cDNA was synthesized, and reverse transcription-coupled PCR was performed with equal amounts of cDNA using primers for different 14-3-3 isoforms. GAPDH was used as a control. D, protein extracts from 14-3-3σ−/− cells and 14-3-3σ+/+ cells were resolved by SDS-PAGE, and Western blotting was performed with the indicated antibodies. β-Actin served as a loading control. The error bars represent S.D.
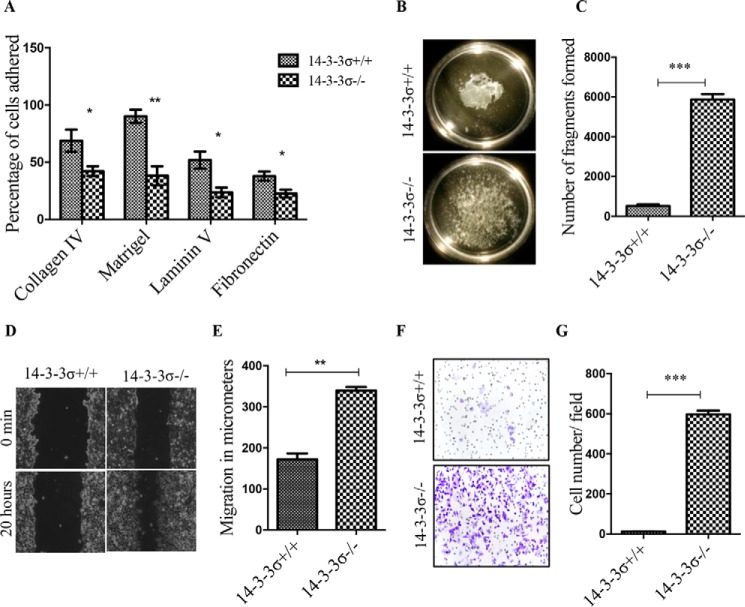
As the changes identified above are indicative of the acquisition of the EMT program, the levels of the EMT transcription factors were determined in the 14-3-3σ+/+ and 14-3-3σ−/− cells. Western blotting (Fig. 1D) and quantitative real time PCR (Fig. 1B) demonstrated that although snail was expressed at relatively low levels in both 14-3-3σ+/+ and 14-3-3σ−/− cells, the expression of slug and ZEB1 was induced only in the 14-3-3σ−/− cells. In addition to the molecular changes observed above, loss of 14-3-3σ led to a decrease in cell-matrix adhesion (Fig. 2A). This was accompanied by a decrease in cell-cell adhesion (Fig. 2, B and C), an increase in cell migration (Fig. 2, D and E), and an increase in cell invasion through Matrigel (Fig. 2, F and G). All these properties are consistent with the induction of the EMT program, tumor progression, and metastasis. These results suggested that loss of 14-3-3σ leads to the induction of an EMT program.
FIGURE 2.
Loss of 14-3-3σ leads to EMT. A, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-based cell-ECM adhesion assays were performed with 14-3-3σ+/+ and 14-3-3σ−/− cells. The percentage of cells adhered to the individual matrix components was calculated as described under “Experimental Procedures,” and the mean and S.D. from three independent experiments were plotted. B and C, to determine whether loss of 14-3-3σ led to changes in cell-cell adhesion, Dispase assays were performed for both 14-3-3σ+/+ and 14-3-3σ−/− cells. Representative images of the different cell types are shown (B). The number of cell aggregates was measured using the particle counting module in ImageJ Particle, and the mean and S.D. are plotted (C). D and E, scratch wound healing assays were performed with 14-3-3σ+/+ and 14-3-3σ−/− cells to determine whether loss of 14-3-3σ affects migration rates. Phase-contrast images of wound closure taken at 10× are shown for 0 and 20 h (D). Cell migration was measured using Axiovision software, and the mean and S.D. were plotted (E). F and G, Boyden chamber invasion assays were performed with 14-3-3σ+/+ and 14-3-3σ−/− cells after coating the inserts with Matrigel. Images of 0.1% crystal violet stained inserts were taken with a Zeiss inverted microscopes using a 10× objective (F). The number of cells invading was determined in 20 random fields per insert, and the mean and S.D. were plotted (G). Where indicated, the p values were calculated using a non-parametric t test (*** indicates a p value ≤0.001, ** indicates a p value ≤0.01, and * indicates a p value ≤0.05). The error bars represent S.D.
Ectopic Expression of 14-3-3σ in 14-3-3σ−/− Cells Leads to Reversal of EMT
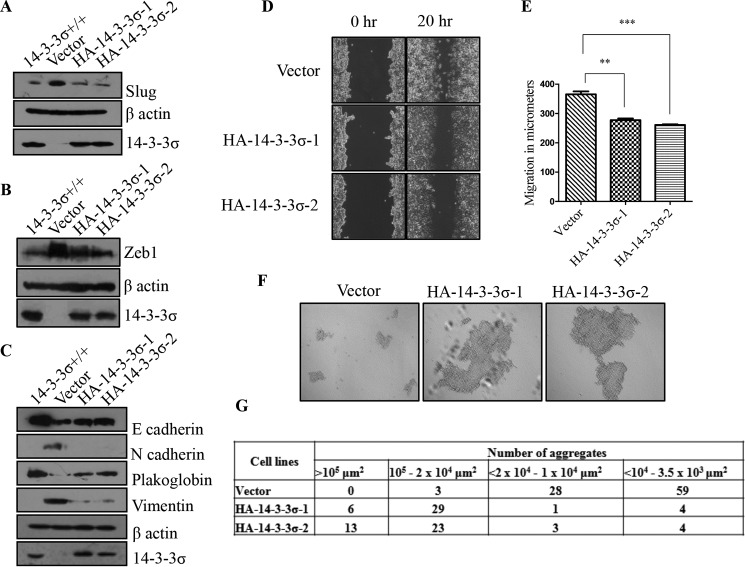
To determine whether the EMT observed in the 14-3-3σ−/− cells was due to 14-3-3σ loss, HA epitope-tagged 14-3-3σ was transfected into these cells, and two stable clones (HA-14-3-3σ-1 and HA-14-3-3σ-2) were generated. The HA-14-3-3σ-expressing clones showed decreased expression of the EMT transcription factors slug and ZEB1 (Fig. 3, A and B) as well as increased expression of the epithelial markers E-cadherin and plakoglobin and decreased expression of the mesenchymal markers N-cadherin and vimentin when compared with the vector control (Fig. 3C). Expression of HA-14-3-3σ in the 14-3-3σ−/− cells led to a decrease in cell migration in wound healing assays (Fig. 3, D and E) and an increase in cell-cell adhesion in hanging drop assays (Fig. 3, F and G). These results suggest that 14-3-3σ inhibits the EMT program.
FIGURE 3.
Ectopic expression of 14-3-3σ in 14-3-3σ−/− cells leads to reversal of EMT. A–C, equivalent amounts of whole cell lysates from 14-3-3σ+/+ cells, 14-3-3σ−/−-derived vector control (Vector), or HA-14-3-3σ-expressing (HA-14-3-3σ-1 and HA-14-3-3σ-2) cells were resolved by SDS-PAGE, and Western blotting was performed with the indicated antibodies for EMT-specific transcription factors (A and B) and epithelial and mesenchymal markers (C). Western blots for β-actin served as a loading control. D and E, scratch wound assays were performed for vector, HA-14-3-3σ-1, and HA-14-3-3σ-2 cells. Phase-contrast images of wound closure taken at 10× are shown for 0 and 20 h (D). Cell migration was measured using Axiovision software, and the mean and S.D. from three independent experiments were plotted (E). p values were calculated using a non-parametric t test (*** indicates a p value ≤0.001, and ** indicates a p value ≤0.01). F and G, to determine whether cell-cell adhesion was increased upon reintroduction of 14-3-3σ into the 14-3-3σ−/− cells, hanging drop assays were performed with vector, HA-14-3-3σ-1, and HA-14-3-3σ-2 cells. Images (F) of cell aggregates were acquired with a Zeiss invested microscope, and the size of the cell aggregates were measured using ImageJ software and are shown in the table (G). Note the increase in size and number of aggregates observed upon restoration of 14-3-3σ expression. The error bars represent S.D.
Loss of 14-3-3σ Leads to EMT and Increase in Slug Expression Due to the Stabilization of c-Jun
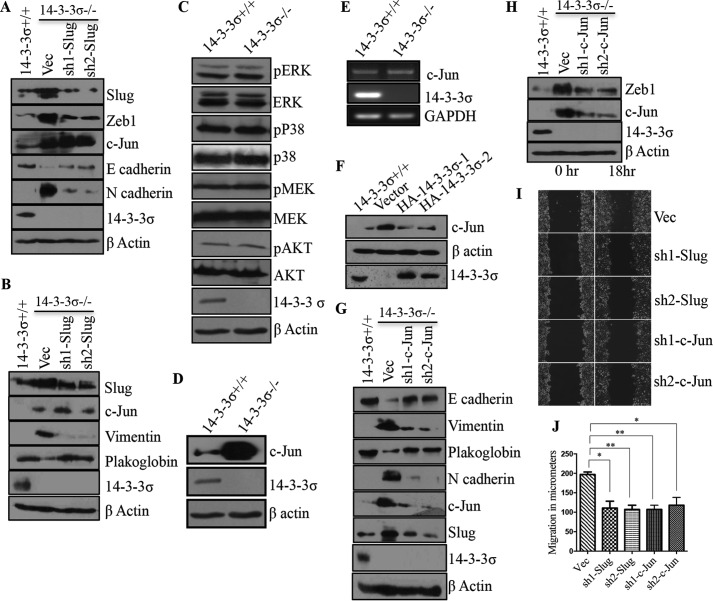
The EMT program in 14-3-3σ−/− cells is associated with an increase in the levels of slug and ZEB1. Previous reports have demonstrated that slug can induce the expression of ZEB1 (38), suggesting that the increase in slug levels might be responsible for the EMT observed in the 14-3-3σ−/− cells. To test this hypothesis, slug expression was inhibited in the 14-3-3σ−/− cells, and two clones, sh-1Slug and sh-2Slug, that had diminished slug levels were generated (Fig. 4, A and B). Western blotting analysis demonstrated that a decrease in slug expression led to an increase in the levels of epithelial markers like E-cadherin and plakoglobin, a decrease in the levels of mesenchymal markers like N-cadherin and vimentin, and a decrease in levels of the EMT transcription factor ZEB1 (Fig. 4, A and B).
FIGURE 4.
Loss of 14-3-3σ leads to an increase in the levels of slug and c-Jun. A and B, 50 μg of protein extracts prepared from 14-3-3σ+/+ or the 14-3-3σ−/−-derived vector control (Vec) and slug knockdown (sh-1Slug and sh-2Slug) cells were resolved by SDS-PAGE followed by Western blotting with the indicated antibodies. β-Actin served as a loading control. C, protein extracts from 14-3-3σ−/− cells and 14-3-3σ+/+ cells were resolved by SDS-PAGE gels followed by Western blotting with the indicated antibodies for various kinases involved in the MAPK signaling pathway. β-Actin served as a loading control. D, protein extracts from 14-3-3σ−/− cells and 14-3-3σ+/+ cells were resolved by SDS-PAGE followed by Western blotting with c-Jun antibody. β-Actin served as a loading control. E, mRNA prepared from the 14-3-3σ−/− cells and 14-3-3σ+/+ knockdown cells was used as a template in reverse transcription-coupled PCRs. GAPDH was used as an internal control. Note that the levels of c-Jun mRNA are not different in the two cell types. F, 50 μg of protein extracts prepared from 14-3-3σ+/+, vector, HA-14-3-3σ-1, and HA-14-3-3σ-2 cells were resolved by SDS-PAGE, and Western blotting was performed with the indicated antibodies. β-Actin served as a loading control G and H, 50 μg of protein extracts prepared from 14-3-3σ+/+, 14-3-3σ−/−-derived vector control (Vec), or c-Jun knockdown (sh1-c-Jun and sh2-c-Jun) cells were resolved by SDS-PAGE followed by Western blotting with the indicated antibodies. β-Actin served as a loading control. Western blots for β-actin served as a loading control in all experiments. I and J, scratch wound assays were performed for the 14-3-3σ−/−-derived vector, sh-1Slug, sh-2Slug, sh1-c-Jun, and sh2-c-Jun cells. Phase-contrast images of wound closure taken at 10× are shown for 0 and 18 h (I). Cell migration was measured using Axiovision software, and the mean and S.D. from three independent experiments were plotted (J). p values were calculated using a non-parametric t test (** indicates a p value ≤0.01, and * indicates a p value ≤0.05). The error bars represent S.D.
Signaling through the MAPK pathway has been reported to result in an increase in slug expression, leading to EMT (44–46). However, 14-3-3σ loss did not lead to any changes in the activation of the MAPK signaling pathway (Fig. 4C). Previous reports suggest that c-Jun can induce slug transcription (51, 52). Western blotting analysis demonstrated that 14-3-3σ-null cells showed increased c-Jun protein levels (Fig. 4D), whereas reverse transcription-PCRs demonstrated that c-Jun mRNA levels were not elevated in 14-3-3σ−/− cells when compared with the 14-3-3σ+/+ cells (Fig. 4E). Expression of HA-14-3-3σ in the 14-3-3σ−/− cells led to a reduction in c-Jun protein levels (Fig. 4F). Furthermore, a stable knockdown of c-Jun generated in 14-3-3σ−/− cells (sh1-c-Jun and sh2-c-Jun) resulted in an increase in the levels of epithelial markers such as E-cadherin and plakoglobin, a decrease in the levels of mesenchymal markers like N-cadherin and vimentin, and a decrease in the levels of the EMT transcription factors slug and ZEB1 (Fig. 4, G and H). Loss of slug in the 14-3-3σ−/− cells did not lead to an alteration in c-Jun levels, suggesting that the increase in c-Jun protein levels was driving the increase in slug expression as reported previously (52) (Fig. 4B). Finally, loss of either slug or c-Jun in the 14-3-3σ−/− cells led to a decrease in cell migration in wound healing assays (Fig. 4, I and J), which is consistent with a reversal of the EMT phenotype. These results suggest that the stabilization of c-Jun in the 14-3-3σ−/− cells leads to the expression of slug, thereby inducing the EMT program.
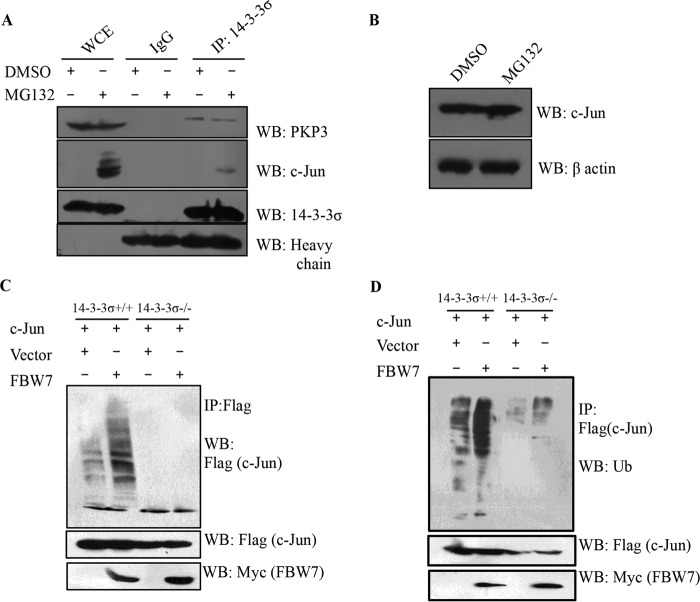
14-3-3σ Induces the FBW7-dependent Ubiquitin-mediated Degradation of c-Jun
The ubiquitin-mediated proteasomal degradation of c-Jun is mediated by multiple E3 ligases such as COP1, ITCH, and FBW7 (56–58). To determine whether 14-3-3σ induced the ubiquitin-mediated degradation of c-Jun, 14-3-3σ+/+ cells were treated with the proteasome inhibitor MG132 (Fig. 5A). c-Jun was stabilized, and higher molecular weight forms of c-Jun were observed in the presence of MG132 in the 14-3-3σ+/+ cells but not in the 14-3-3σ−/− cells (Fig. 5B). In addition, 14-3-3σ co-immunoprecipitated with c-Jun only in the presence of MG132 but not in cells treated with the vehicle (DMSO) control. This suggests that 14-3-3σ forms a complex with c-Jun in 14-3-3σ+/+ cells under conditions where c-Jun is stabilized. In contrast, another 14-3-3σ ligand, plakophilin 3 (75), forms a complex with 14-3-3σ in the presence and absence of MG132 (Fig. 5A), indicating that MG132 does not lead to alterations in the interaction of 14-3-3σ with other ligands. When 14-3-3σ−/− cells were treated with MG132, no increase in c-Jun protein levels was observed in comparison with the vehicle control (Fig. 5B), suggesting that loss of 14-3-3σ led to a decrease in the proteasome-mediated degradation of c-Jun. The loss of the E3 ligase FBW7, which induces the ubiquitination and degradation of c-Jun, has been reported to lead to EMT (59). Ubiquitination assays performed by exogenous expression of FLAG-tagged c-Jun with or without Myc-FBW7 in 14-3-3σ+/+ as well as 14-3-3σ−/− cells demonstrated that c-Jun was ubiquitinated only in the presence of 14-3-3σ, and the ubiquitination of c-Jun in 14-3-3σ+/+ cells was enhanced in the presence of Myc-FBW7 (Fig. 5, C and D). The ubiquitinated species were not observed in 14-3-3σ−/− cells, indicating that 14-3-3σ is essential for the ubiquitination and subsequent degradation of c-Jun in these cells and that 14-3-3σ induces the ubiquitin-mediated proteolysis of c-Jun in an FBW7-dependent manner.
FIGURE 5.
14-3-3σ induces the ubiquitin-mediated degradation of c-Jun. A, 14-3-3σ+/+ cells were treated with either MG132 or DMSO for 6 h, protein extracts were prepared, and immunoprecipitations (IP) were performed with antibody to either 14-3-3σ or an isotype control (IgG). Immune complexes were resolved by SDS-PAGE along with 5% input for whole cell extract (WCE) followed by Western blotting (WB) with the indicated antibodies. Note that 14-3-3σ is observed in a complex with c-Jun only in the presence of MG132. B, 14-3-3σ−/− cells were treated with either MG132 or DMSO for 6 h, protein extracts were prepared, and protein extracts were resolved by SDS-PAGE followed by Western blotting with the indicated antibodies. β-Actin served as a loading control. C and D, 14-3-3σ+/+ and 14-3-3σ−/− cells were co-transfected with SFB-c-Jun in the presence or absence of Myc-FBW7α. At 24 h post-transfection, cells were treated with MG132 for 6 h, and protein extracts were prepared and immunoprecipitated with antibody to FLAG. The reactions were resolved by SDS-PAGE followed by Western blotting with antibody to FLAG (C) or ubiquitin (D). Note that the higher molecular weight forms of c-Jun are only present in 14-3-3σ+/+ cells, and the levels of these bands are elevated in the presence of FBW7.
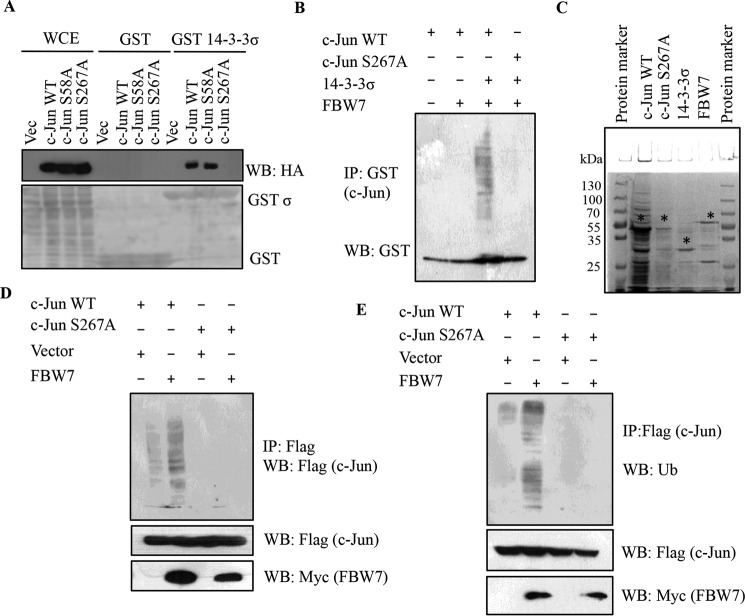
14-3-3 proteins bind to substrates containing phosphoserine or phosphothreonine residues that may or may not match previously identified consensus motifs or may bind to ligands that are not phosphorylated on either serine or threonine (3–6). Two putative 14-3-3 binding sites at serine residues 58 and 267 were identified in c-Jun. The serine residues were altered to alanine (S58A and S267A, respectively) by site-directed mutagenesis, and WT and mutant cDNAs were cloned downstream of the HA epitope tag and transfected into HEK293 cells. Biochemical assays using bacterially purified GST-14-3-3σ or GST alone demonstrated that although WT c-Jun and the S58A mutant formed a complex with 14-3-3σ, the S267A mutant did not form a complex with 14-3-3σ, suggesting that Ser-267 was required for the association between 14-3-3σ and c-Jun (Fig. 6A). In vitro ubiquitination assays with bacterially purified c-Jun WT and c-Jun S267A proteins in the presence of FBW7 and 14-3-3σ confirmed that the presence of 14-3-3σ as well as the Ser-267 residue in c-Jun was necessary for ubiquitination of c-Jun by FBW7 (Fig. 6, B and C). Consistent with the in vitro data, exogenous expression of FLAG epitope-tagged c-Jun WT or FLAG epitope-tagged c-Jun S267A and Myc-FBW7 in 14-3-3σ+/+ cells showed that the Ser-267 residue was necessary for c-Jun ubiquitination (Fig. 6, D and E). Cumulatively, these results suggest that the Ser-267 residue in c-Jun is required for the interaction between c-Jun and 14-3-3σ, and this interaction is sufficient and necessary for c-Jun proteasomal degradation mediated by the E3 ligase FBW7.
FIGURE 6.
14-3-3σ targets c-Jun to proteasomal degradation in an FBW7-dependent manner. A, HEK293 cells were transfected with HA-c-Jun WT, HA-c-Jun S58A, and HA-c-Jun S267A. 24 h post-transfection, cell lysates were prepared and incubated with bacterially purified GST or GST-14-3-3σ, and the reactions were resolved by SDS-PAGE. 5% input for whole cell extract (WCE) served as the input. Western blotting was performed with the indicated antibodies (upper panel), and the levels of GST and GST-14-3-3σ are shown in the Ponceau-stained membrane (lower panel). B, in vitro ubiquitination experiments were performed using bacterially purified GST-c-Jun WT and GST-c-Jun S267A as substrates in the indicated combinations with MBP-tagged 14-3-3σ and MBP-tagged FBW7α along with E1 (UBE1) and E2 (UbcH5B). Full-length and ubiquitinated species of GST-c-Jun WT and GST-c-Jun S267A were detected by immunoblotting with anti-GST antibody. C, the protein levels of GST-c-Jun WT, GST-c-Jun S267A, MBP-14-3-3σ, and MBP-FBW7α shown by Coomassie staining. The * indicates the full-length protein; the lower molecular bands are probably degradation products. D, 14-3-3σ+/+ cells were transfected with FLAG-tagged c-Jun WT and c-Jun S267A with or without Myc-FBW7. 24 h post-transfection, cells were treated with MG132 for 6 h, and immunoprecipitated with anti-FLAG antibody. c-Jun ubiquitination was detected by immunoblotting with FLAG antibody. E, 14-3-3σ+/+ cells were transfected with FLAG-tagged c-Jun WT and c-Jun S267A with or without Myc-FBW7. At 24 h post-transfection, cells were treated with MG132 for 6 h, and immunoprecipitated with anti-FLAG antibody. c-Jun ubiquitination was detected by immunoblotting with ubiquitin (Ub) antibody. IP, immunoprecipitation; WB, Western blotting.
c-Jun Nuclear Export Is Required for Ubiquitin-mediated Proteolysis
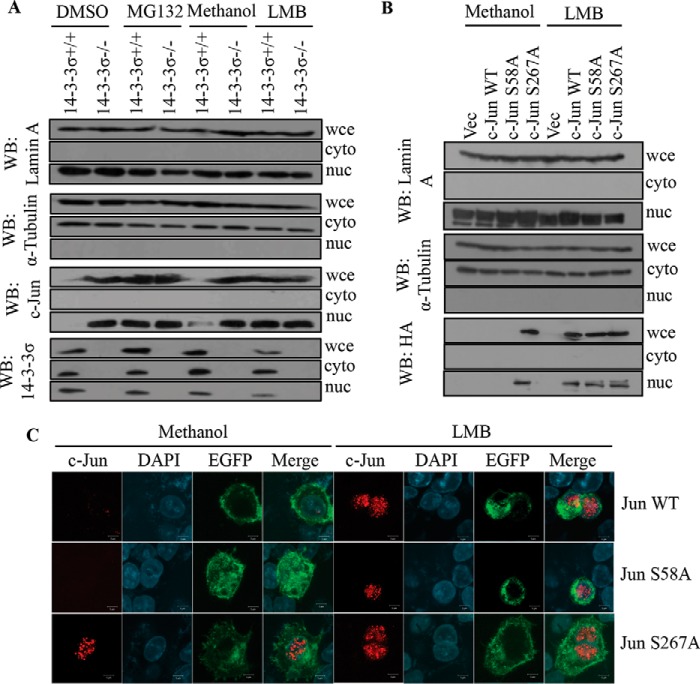
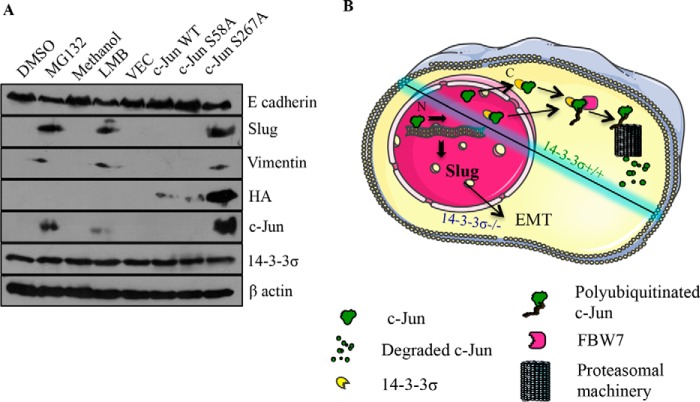
14-3-3 proteins often regulate the subcellular localization of their target proteins, thereby regulating cellular signaling pathways (76, 77). To determine whether 14-3-3σ regulated the subcellular localization of c-Jun, 14-3-3σ+/+ and 14-3-3σ−/− cells were treated with either MG132, the nuclear export inhibitor leptomycin B (LMB), or the corresponding vehicle controls. Western blotting performed on nuclear and cytoplasmic fractions demonstrated that in the 14-3-3σ−/− cells treated with the vehicle controls c-Jun was present in the nuclear fraction, whereas it was not detectable in the 14-3-3σ+/+ cells (Fig. 7A). However, upon treatment with MG132, c-Jun levels in the 14-3-3σ+/+ cells were elevated to levels comparable with those in the 14-3-3σ−/− cells, and c-Jun was present only in the nuclear fraction (Fig. 7A). Similar results were observed when the 14-3-3σ+/+ cells were treated with LMB (Fig. 7A). The integrity of the nuclear and cytoplasmic fractions was tested by Western blotting with antibodies to lamin A and α-tubulin, respectively (Fig. 7A). These results suggested that the enforced nuclear localization of c-Jun resulted in stabilization of c-Jun. To determine whether disruption of complex formation between c-Jun and 14-3-3σ resulted in an increase in the nuclear localization of c-Jun, 14-3-3σ+/+ cells were transfected with constructs expressing HA epitope-tagged versions of WT c-Jun, S58A, or S267A, and the transfected cells were treated with the nuclear export inhibitor LMB or vehicle control. Western blotting performed on nuclear and cytoplasmic fractions prepared from the treated cells demonstrated that LMB treatment resulted in stabilization of WT c-Jun and the S57A mutant in nuclear fractions (Fig. 7B). The similarity of these results with those observed for endogenous 14-3-3σ suggests that the HA epitope tag does not alter cellular localization and that 14-3-3σ might regulate the subcellular localization of c-Jun. However, the S267A mutant was stable and localized to the nucleus both in the presence and absence of LMB (Fig. 7B). Similar results were obtained when immunofluorescence assays using antibody to the HA epitope were performed on the transfected cells (Fig. 7C). The presence of enhanced GFP was used to identify transfected cells (Fig. 7C). Inhibition of nuclear export or proteasome-mediated degradation in 14-3-3σ+/+ cells led to the stabilization of c-Jun and the induction of EMT, leading to an increase in the levels of slug and vimentin and a decrease in the levels of E-cadherin (Fig. 8A). Similarly, the expression of S267A, which localizes to the nucleus and is not targeted for degradation, leads to an increase in slug and vimentin levels and a decrease in E-cadherin levels (Fig. 8A). These results suggest that 14-3-3σ stimulates both the nuclear export of c-Jun and its degradation via FBW7-dependent ubiquitin-mediated proteolysis, thus preventing the activation of the EMT program.
FIGURE 7.
14-3-3σ stimulates the nuclear export of c-Jun, and this is required for c-Jun degradation. A, 14-3-3σ+/+ and 14-3-3σ−/− cells were treated with 10 μm MG132, 20 ng/ml LMB, or the respective vehicle controls (DMSO and methanol, respectively) for 8 h. The cells were harvested by trypsinization, and the whole cell extracts (wce) and nuclear (nuc) and cytoplasmic (cyto) fractions were resolved by SDS-PAGE followed by Western blotting (WB) with the indicated antibodies. Note that c-Jun is present in the nucleus in vehicle-treated 14-3-3σ−/− cells but is not detectable in the vehicle-treated 14-3-3σ+/+ cells. In contrast, c-Jun is stabilized and localized to the nucleus in 14-3-3σ+/+ cells treated with either MG132 or LMB. B, 14-3-3σ+/+ cells were transfected with HA-c-Jun WT, HA-c-Jun S58A, and HA-c-Jun S267A. 24 h post-transfection, cells were either treated or not treated with LMB for 8 h. Post-treatment, whole cell extracts (wce) and nuclear (nuc) and cytoplasmic (cyto) fractions were resolved by SDS-PAGE followed by Western blotting with the indicated antibodies. Note that LMB treatment leads to enrichment of c-Jun and S58A in the nuclear fraction, whereas S267A is always enriched in the nuclear fraction. C, 14-3-3σ+/+ cells were transfected with HA-c-Jun WT, HA-c-Jun S58A, and HA-c-Jun S267A along with a vector expressing GFP to identify transfected cells. 24 h post-transfection, cells were either treated or not treated with LMB for 8 h. Post-treatment, cells were fixed and stained with antibody to the HA epitope tag (red) and counterstained with DAPI (blue). GFP expression (green) was used to identify transfected cells. Merged images are shown in the fourth panel. Magnification, ×630 with 4× optical zoom. Scale bars, 5 μm. Note that WT c-Jun and S58A are only present in the nucleus in the presence of LMB in contrast to the S267A mutant. EGFP, enhanced GFP.
FIGURE 8.
The nuclear localization of c-Jun in 14-3-3σ+/+ cells induces an EMT. A, 14-3-3σ+/+ cells were either treated with vehicle control, MG132, or LMB or transfected with the indicated constructs. 12 h postincubation with vehicle control, MG132, or LMB or 24 h post-transfection, cells were lysed in SDS buffer. 50 μg of protein extracts were resolved by SDS-PAGE followed by Western blotting with the indicated antibodies. β-Actin served as a loading control. B, model depicting the fate and function of c-Jun protein in 14-3-3σ+/+ and 14-3-3σ−/− cells. 14-3-3σ either stimulates the nuclear export of c-Jun or retains c-Jun in the cytoplasm, resulting in the FBW7-mediated degradation of c-Jun by the proteasome. In the absence of 14-3-3σ, c-Jun accumulates in the nucleus and promotes slug transcription. N, nucleus; C, cytoplasm.
Discussion
Loss of 14-3-3σ has been observed in multiple human tumors, suggesting that it might function as a tumor suppressor (16, 18–21, 23, 78). However, the mechanisms by which it might inhibit tumor progression remain unclear. The results in this report suggest that loss of 14-3-3σ leads to the stabilization of c-Jun, leading to increased expression of the EMT transcription factor slug and the activation of EMT, a pathway that is often associated with tumor progression and metastasis (79). Therefore, the inhibition of EMT by 14-3-3σ loss represents a novel pathway by which 14-3-3σ might inhibit tumor progression.
Our data suggest that one of the mechanisms by which 14-3-3σ might execute its tumor suppressive function is by preventing expression of EMT-inducing genes by restricting c-Jun to the cytoplasm and inducing c-Jun degradation via the ubiquitin pathway. This is consistent with previous reports that have shown that loss of 14-3-3σ leads to loss of polarity in MCF10A cells (60). Loss of polarity is a hallmark of EMT (31), but it was unclear whether 14-3-3σ induced an EMT. The results in this report indicate that loss of 14-3-3σ results in the acquisition of mesenchymal markers and the loss of epithelial markers due to the stabilization of c-Jun, which leads to the increased expression of the EMT transcription factor slug. These properties are accompanied by a decrease in cell-cell adhesion and an increase in invasion and migration, all properties of invasive tumor cells. In contrast, two reports have suggested that other 14-3-3 isoforms, particularly 14-3-3ϵ, can form a complex with the EMT transcription factor snail (80) and that 14-3-3ϵ is over-expressed in hepatocellular carcinoma and it contributes to the acquisition of the EMT phenotype (81). Importantly, 14-3-3σ did not form a complex with snail (80), which is consistent with our observations that it preserves the epithelial phenotype. Therefore, these observations suggest that distinct 14-3-3 isoforms have different effects on the acquisition of the EMT phenotype with 14-3-3σ inhibiting EMT progression and 14-3-3ϵ promoting EMT. The two 14-3-3 isoforms affect EMT progression by targeting different components in the EMT pathway, 14-3-3σ by inducing c-Jun degradation and therefore preventing expression of slug (this report) and 14-3-3ϵ by forming a complex with the EMT transcription factor snail and stimulating snail-dependent transcription (80). These results are also consistent with the observation that 14-3-3σ expression is low in multiple tumor types (16–24), whereas 14-3-3ϵ expression is associated with the acquisition of EMT in hepatocellular carcinoma (81). Therefore, these are two distinct molecular mechanisms, and they reflect the diversity of mechanisms by which the 14-3-3 protein family regulates cellular pathways. These results are also consistent with data from our laboratory and from other laboratories suggesting that different 14-3-3 isoforms form complexes with and alter the function of different ligands, leading to differences in phenotype both in cells in culture and in mouse models (70, 82–85).
Previous work has demonstrated that 14-3-3 proteins regulate the nuclear to cytoplasmic transport of their ligands such as the cdc25 phosphatase, COP1, forkhead transcription factors, and many others (76, 77, 86–88). Some of these reports suggest that the 14-3-3 proteins contain a nuclear export signal (NES) in their N termini that seems to be required for nuclear export (87, 88); however, other reports suggest that the NES identified above is required for binding to the phosphopeptide and that 14-3-3 binding exposes a cryptic NES in the ligand, thus promoting nuclear export (76). The results in this report suggest that 14-3-3σ is required for the retention of c-Jun in the cytoplasm either by promoting the nuclear export of c-Jun or the complex between 14-3-3σ and c-Jun is recognized by export factors, leading to the export of the complex to the cytoplasm where 14-3-3σ mediates the FBW7-dependent degradation of c-Jun via the ubiquitin-dependent pathway (Fig. 8B). A nuclear import signal has been identified near the leucine zipper region of c-Jun, but no NES has been reported so far for the c-Jun protein (89). It is possible that 14-3-3σ binding to c-Jun results in exposure of a cryptic NES in c-Jun. An alternative explanation for our data is that c-Jun is exported to the cytoplasm in a 14-3-3σ-independent manner, and it is retained in the cytoplasm by 14-3-3σ followed by c-Jun ubiquitination and degradation (Fig. 8B). Our data demonstrate that 14-3-3σ is present in both the cytoplasm and nucleus (Fig. 7A), which is consistent with either of the alternatives described above. Furthermore, treatment of 14-3-3σ+/+ cells with MG132 leads to the accumulation of c-Jun in the nucleus (Fig. 7A). Although this could suggest that 14-3-3σ is not required to retain c-Jun in the cytoplasm and is consistent with the possibility that 14-3-3σ is required for the nuclear export of c-Jun, it does not completely rule out the possibility that the inhibition of degradation by MG132 could lead to a disruption of complex formation between 14-3-3σ and c-Jun, allowing for c-Jun nuclear import and accumulation of c-Jun in the nucleus.
The results here also suggest that complex formation between 14-3-3σ and c-Jun is required for the ubiquitination of c-Jun by FBW7. The in vivo and in vitro ubiquitination data in our studies also reveal possible functions of 14-3-3σ that are required for proper presentation of substrates to the E3 ligase. This is a novel finding as there have been no previous reports suggesting that a 14-3-3 isoform is required for the ubiquitination of c-Jun. These findings are also consistent with previous reports that suggest that 14-3-3σ is required for the nuclear export of another E3 ligase required for c-Jun degradation, COP1, and that this interaction might contribute to the tumor suppressive function of 14-3-3σ (88, 90). Another study suggests that 14-3-3σ regulates the stability of c-Myc by inducing the ubiquitin-mediated degradation of c-Myc, thus negatively regulating the activation of c-myc genes that are required for tumor progression (91). Taken together with our data, these results suggest that loss of 14-3-3σ might lead to the stabilization of oncogenes such as c-Myc and c-Jun, resulting in tumor progression. Previous studies have identified a role of FBW7 in inducing the proteasome-mediated degradation of c-Jun. The GSK3-mediated phosphorylation of residue Thr-239 in c-Jun is necessary for the recognition of c-Jun by FBW7, which results in subsequent ubiquitination of c-Jun (92). Phosphorylation by GSK3 at the Thr-239 position in v-Jun may require a priming phosphorylation at the Ser-243 position in c-Jun; both are sites for ERK1 and DYRK1 (93, 94). However, the phosphorylation status of c-Jun at Thr-239 and Ser-243was not investigated in this study.
14-3-3 proteins preferably bind to ligands containing a phosphorylated serine residue in one of two consensus motifs (4, 74). The experiments in this report suggest that the sequences surrounding a serine residue at position 267 resembles a 14-3-3 protein binding consensus motif and might be required for either the nuclear export or cytoplasmic retention of c-Jun and subsequently facilitate proper presentation of c-Jun to FBW7 and help in degradation of c-Jun. There is no evidence in the literature or in multiple databases (PhosphoSitePlus, Scansite, Human Protein Reference Database, and UniProt), which either document known sites of phosphorylation or identify putative kinases for a given sequence, to suggest that Ser-267 in c-Jun is phosphorylated or that this sequence can be recognized and phosphorylated by a known kinase. In addition, there are several 14-3-3 ligands that bind to 14-3-3 proteins in a phosphorylation-independent manner, and the Ser-267 site in c-Jun might belong to this category of 14-3-3 binding sites (5, 6). The overexpression of the S267A mutant leads to an increase in expression of mesenchymal markers and a decrease in expression of epithelial markers in 14-3-3σ+/+ cells, a phenotype also observed upon treatment with the export inhibitor LMB and the proteasome inhibitor MG132. Interestingly, an earlier report shows that loss of FBW7 leads to EMT (59). These results are consistent with our data and suggest that in HCT116 cells 14-3-3σ stimulates the nuclear export of c-Jun and targets it for ubiquitination by FBW7 and proteasomal degradation, thus preventing activation of the EMT program. ITCH and COP1 can function as E3 ligases for c-Jun (56, 58), and 14-3-3σ can form a complex with and stimulate the nuclear export of COP1 (88); however, it is not clear whether 14-3-3σ is required for the ubiquitination of c-Jun by either ITCH or COP1.
Our results suggest that loss of 14-3-3σ leads to the induction of a slug-dependent EMT program. Slug and snail operate in a positive feedback loop with each factor stimulating the expression of the other (37). However, in this report, an increase in snail expression was not observed despite the increase in slug expression. Slug has been shown to stimulate the expression of ZEB1 (38), a finding that is consistent with our data, and the increase in ZEB1 was indeed due to an increase in slug expression as a knockdown of slug in 14-3-3σ−/− cells results in a decrease in ZEB1 protein levels. Slug expression has been positively associated with cancer progression, invasion, and metastasis without a parallel increase in snail, which suggests that snail and slug have different functions in tumor progression (95–97). Slug expression can be regulated by multiple transcription factors such as Myb, c-Jun (52, 98), growth factors such as EGF (99) and TGF-β (100), and signaling pathways like the MAPK pathway (46, 101). The results in this report demonstrate that, in 14-3-3σ+/+ cells, the AP1 transcription factor c-Jun regulates slug expression as a decrease in c-Jun levels in 14-3-3σ−/− cells results in a decrease in slug protein levels (Fig. 4). c-Jun expression is found to be elevated in multiple cancer types and is strongly associated with invasion and metastasis (53–55). c-Jun function can be regulated by the ERK pathway (52). No change in activation and expression of ERK or MAPK pathway-related kinases was observed in this study (Fig. 4C), suggesting that the increase in c-Jun activity in 14-3-3σ−/− cells is not dependent on ERK or MAPK but is due to the inhibition of c-Jun proteolysis.
In summary, the results in this report demonstrate that 14-3-3σ is required for maintenance of the epithelial phenotype, and loss of 14-3-3σ leads to EMT in HCT116 cells. 14-3-3σ is required for the ubiquitination and degradation of c-Jun by FBW7. The induction of EMT upon loss of 14-3-3σ is driven by the stabilization and nuclear localization of c-Jun, leading to increased transcription of the EMT transcription factor slug (Fig. 8B). These results identify a novel pathway through which 14-3-3σ maintains the epithelial phenotype and suppresses expression of the mesenchymal phenotype and might contribute to the tumor suppressor function of 14-3-3σ.
Author Contributions
K. R., M. G., N. C., S. M., and S. N. D. designed experiments. K. R., M. G., and S. N. D. wrote the manuscript. K. R. performed the majority of the experiments with contributions from N. C., M. G., R. D., and J. L. N. C. preformed in vivo and in vitro ubiquitination assays. M. G. designed primers for reverse transcription- and real time PCR analysis. M. G. analyzed the processed the real time PCR data. M. G. standardized Dispase assays, and J. L. performed the Dispase assays with the help of K. R. R. D. generated the HA-pcDNA3 puro 14-3-3σ, HA-pcDNA3 c-Jun WT, HA-pcDNA3 c-JunS58A, and HA-pcDNA3 c-JunS267A vectors. J. L. and K. R. performed cell-ECM adhesion assays.
Supplementary Material
Acknowledgments
We thank Dr. S. Sengupta (National Institute of Immunology) and Dr. M. Welcker (Fred Hutchinson Cancer Research Center) for the gift of the FBW7 constructs. We are also thankful to Dr. Sharmila Bapat from National Centre for Cell Science, Pune, India for generously sharing antibodies against snail and slug.
This work was supported in part by an intramural grant from the Advanced Center for Treatment, Research and Education in Cancer, Tata Memorial Centre (to S. N. D.). The authors declare that they have no conflicts of interest with the contents of this article.

This article contains supplemental Fig. S1 and Tables 1–3.
- EMT
- epithelial to mesenchymal transition
- SFB
- S-protein/FLAG/streptavidin-binding protein triple epitope tag
- MBP
- maltose-binding protein
- LMB
- leptomycin B
- NES
- nuclear export signal.
References
- 1. Aitken A. (1996) 14-3-3 and its possible role in co-ordinating multiple signalling pathways. Trends Cell Biol. 6, 341–347 [DOI] [PubMed] [Google Scholar]
- 2. van Heusden G. P., Griffiths D. J., Ford J. C., Chin-A-Woeng T. F., Schrader P. A., Carr A. M., and Steensma H. Y. (1995) The 14-3-3 proteins encoded by the BMH1 and BMH2 genes are essential in the yeast Saccharomyces cerevisiae and can be replaced by a plant homologue. Eur. J. Biochem. 229, 45–53 [PubMed] [Google Scholar]
- 3. Jones D. H., Ley S., and Aitken A. (1995) Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 368, 55–58 [DOI] [PubMed] [Google Scholar]
- 4. Yaffe M. B., Rittinger K., Volinia S., Caron P. R., Aitken A., Leffers H., Gamblin S. J., Smerdon S. J., and Cantley L. C. (1997) The structural basis for 14-3-3:phosphopeptide binding specificity. Cell 91, 961–971 [DOI] [PubMed] [Google Scholar]
- 5. Henriksson M. L., Francis M. S., Peden A., Aili M., Stefansson K., Palmer R., Aitken A., and Hallberg B. (2002) A nonphosphorylated 14-3-3 binding motif on exoenzyme S that is functional in vivo. Eur. J. Biochem. 269, 4921–4929 [DOI] [PubMed] [Google Scholar]
- 6. Zhai J., Lin H., Shamim M., Schlaepfer W. W., and Cañete-Soler R. (2001) Identification of a novel interaction of 14-3-3 with p190RhoGEF. J. Biol. Chem. 276, 41318–41324 [DOI] [PubMed] [Google Scholar]
- 7. Prasad G. L., Valverius E. M., McDuffie E., and Cooper H. L. (1992) Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ. 3, 507–513 [PubMed] [Google Scholar]
- 8. Benzinger A., Popowicz G. M., Joy J. K., Majumdar S., Holak T. A., and Hermeking H. (2005) The crystal structure of the non-liganded 14-3-3σ protein: insights into determinants of isoform specific ligand binding and dimerization. Cell Res. 15, 219–227 [DOI] [PubMed] [Google Scholar]
- 9. Wilker E. W., Grant R. A., Artim S. C., and Yaffe M. B. (2005) A structural basis for 14-3-3σ functional specificity. J. Biol. Chem. 280, 18891–18898 [DOI] [PubMed] [Google Scholar]
- 10. Chan T. A., Hermeking H., Lengauer C., Kinzler K. W., and Vogelstein B. (1999) 14-3-3σ is required to prevent mitotic catastrophe after DNA damage. Nature 401, 616–620 [DOI] [PubMed] [Google Scholar]
- 11. Dohn M., Zhang S., and Chen X. (2001) p63α and ΔNp63alpha can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20, 3193–3205 [DOI] [PubMed] [Google Scholar]
- 12. Hermeking H., Lengauer C., Polyak K., He T.-C., Zhang L., Thiagalingam S., Kinzler K. W., and Vogelstein B. (1997) 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3–11 [DOI] [PubMed] [Google Scholar]
- 13. Laronga C., Yang H. Y., Neal C., and Lee M. H. (2000) Association of the cyclin-dependent kinases and 14-3-3σ negatively regulates cell cycle progression. J. Biol. Chem. 275, 23106–23112 [DOI] [PubMed] [Google Scholar]
- 14. Wilker E. W., van Vugt M. A., Artim S. A., Huang P. H., Petersen C. P., Reinhardt H. C., Feng Y., Sharp P. A., Sonenberg N., White F. M., and Yaffe M. B. (2007) 14-3-3σ controls mitotic translation to facilitate cytokinesis. Nature 446, 329–332 [DOI] [PubMed] [Google Scholar]
- 15. Yang H. Y., Wen Y. Y., Chen C. H., Lozano G., and Lee M. H. (2003) 14-3-3σ positively regulates p53 and suppresses tumor growth. Mol. Cell. Biol. 23, 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ferguson A. T., Evron E., Umbricht C. B., Pandita T. K., Chan T. A., Hermeking H., Marks J. R., Lambers A. R., Futreal P. A., Stampfer M. R., and Sukumar S. (2000) High frequency of hypermethylation at the 14-3-3σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Urano T., Saito T., Tsukui T., Fujita M., Hosoi T., Muramatsu M., Ouchi Y., and Inoue S. (2002) Efp targets 14-3-3σ for proteolysis and promotes breast tumour growth. Nature 417, 871–875 [DOI] [PubMed] [Google Scholar]
- 18. Akahira J., Sugihashi Y., Suzuki T., Ito K., Niikura H., Moriya T., Nitta M., Okamura H., Inoue S., Sasano H., Okamura K., and Yaegashi N. (2004) Decreased expression of 14-3-3σ is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin. Cancer Res. 10, 2687–2693 [DOI] [PubMed] [Google Scholar]
- 19. Iwata N., Yamamoto H., Sasaki S., Itoh F., Suzuki H., Kikuchi T., Kaneto H., Iku S., Ozeki I., Karino Y., Satoh T., Toyota J., Satoh M., Endo T., and Imai K. (2000) Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3σ gene in human hepatocellular carcinoma. Oncogene 19, 5298–5302 [DOI] [PubMed] [Google Scholar]
- 20. Lodygin D., Diebold J., and Hermeking H. (2004) Prostate cancer is characterized by epigenetic silencing of 14-3-3σ expression. Oncogene 23, 9034–9041 [DOI] [PubMed] [Google Scholar]
- 21. Urano T., Takahashi S., Suzuki T., Fujimura T., Fujita M., Kumagai J., Horie-Inoue K., Sasano H., Kitamura T., Ouchi Y., and Inoue S. (2004) 14-3-3σ is down-regulated in human prostate cancer. Biochem. Biophys. Res. Commun. 319, 795–800 [DOI] [PubMed] [Google Scholar]
- 22. Lodygin D., Yazdi A. S., Sander C. A., Herzinger T., and Hermeking H. (2003) Analysis of 14-3-3σ expression in hyperproliferative skin diseases reveals selective loss associated with CpG-methylation in basal cell carcinoma. Oncogene 22, 5519–5524 [DOI] [PubMed] [Google Scholar]
- 23. Suzuki H., Itoh F., Toyota M., Kikuchi T., Kakiuchi H., and Imai K. (2000) Inactivation of the 14-3-3σ gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res. 60, 4353–4357 [PubMed] [Google Scholar]
- 24. Osada H., Tatematsu Y., Yatabe Y., Nakagawa T., Konishi H., Harano T., Tezel E., Takada M., and Takahashi T. (2002) Frequent and histological type-specific inactivation of 14-3-3σ in human lung cancers. Oncogene 21, 2418–2424 [DOI] [PubMed] [Google Scholar]
- 25. Kalluri R., and Weinberg R. A. (2009) The basics of epithelial-mesenchymal transition. J. Clin. Investig. 119, 1420–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. López-Novoa J. M., and Nieto M. A. (2009) Inflammation and EMT: an alliance towards organ fibrosis and cancer progression. EMBO Mol. Med. 1, 303–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nisticò P., Bissell M. J., and Radisky D. C. (2012) Epithelial-mesenchymal transition: general principles and pathological relevance with special emphasis on the role of matrix metalloproteinases. Cold Spring Harb. Perspect. Biol. 4, a011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yang J., Mani S. A., Donaher J. L., Ramaswamy S., Itzykson R. A., Come C., Savagner P., Gitelman I., Richardson A., and Weinberg R. A. (2004) Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117, 927–939 [DOI] [PubMed] [Google Scholar]
- 29. Fischer K. R., Durrans A., Lee S., Sheng J., Li F., Wong S. T., Choi H., El Rayes T., Ryu S., Troeger J., Schwabe R. F., Vahdat L. T., Altorki N. K., Mittal V., and Gao D. (2015) Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527, 472–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zheng X., Carstens J. L., Kim J., Scheible M., Kaye J., Sugimoto H., Wu C. C., LeBleu V. S., and Kalluri R. (2015) Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527, 525–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thiery J. P., Acloque H., Huang R. Y., and Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 32. Tsai J. H., and Yang J. (2013) Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev. 27, 2192–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hajra K. M., Chen D. Y., and Fearon E. R. (2002) The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 62, 1613–1618 [PubMed] [Google Scholar]
- 34. Cano A., Pérez-Moreno M. A., Rodrigo I., Locascio A., Blanco M. J., del Barrio M. G., Portillo F., and Nieto M. A. (2000) The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2, 76–83 [DOI] [PubMed] [Google Scholar]
- 35. Medici D., Hay E. D., and Olsen B. R. (2008) Snail and Slug promote epithelial-mesenchymal transition through β-catenin-T-cell factor-4-dependent expression of transforming growth factor-β3. Mol. Biol. Cell 19, 4875–4887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Batlle E., Sancho E., Francí C., Domínguez D., Monfar M., Baulida J., and García De Herreros A. (2000) The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat. Cell Biol. 2, 84–89 [DOI] [PubMed] [Google Scholar]
- 37. Chen Y., and Gridley T. (2013) The SNAI1 and SNAI2 proteins occupy their own and each other's promoter during chondrogenesis. Biochem. Biophys. Res. Commun. 435, 356–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wels C., Joshi S., Koefinger P., Bergler H., and Schaider H. (2011) Transcriptional activation of ZEB1 by Slug leads to cooperative regulation of the epithelial-mesenchymal transition-like phenotype in melanoma. J. Invest. Dermatol. 131, 1877–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pang M. F., Georgoudaki A. M., Lambut L., Johansson J., Tabor V., Hagikura K., Jin Y., Jansson M., Alexander J. S., Nelson C. M., Jakobsson L., Betsholtz C., Sund M., Karlsson M. C., and Fuxe J. (2016) TGF-β1-induced EMT promotes targeted migration of breast cancer cells through the lymphatic system by the activation of CCR7/CCL21-mediated chemotaxis. Oncogene 35, 748–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wendt M. K., Smith J. A., and Schiemann W. P. (2010) Transforming growth factor-β-induced epithelial-mesenchymal transition facilitates epidermal growth factor-dependent breast cancer progression. Oncogene 29, 6485–6498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fan L. C., Shiau C. W., Tai W. T., Hung M. H., Chu P. Y., Hsieh F. S., Lin H., Yu H. C., and Chen K. F. (2015) SHP-1 is a negative regulator of epithelial-mesenchymal transition in hepatocellular carcinoma. Oncogene 34, 5252–5263 [DOI] [PubMed] [Google Scholar]
- 42. Lee M. Y., and Shen M. R. (2012) Epithelial-mesenchymal transition in cervical carcinoma. Am. J. Transl. Res. 4, 1–13 [PMC free article] [PubMed] [Google Scholar]
- 43. Wu Y. C., Tang S. J., Sun G. H., and Sun K. H. (2016) CXCR7 mediates TGFβ1-promoted EMT and tumor-initiating features in lung cancer. Oncogene 35, 2123–2132 [DOI] [PubMed] [Google Scholar]
- 44. Gonzalez D. M., and Medici D. (2014) Signaling mechanisms of the epithelial-mesenchymal transition. Sci. Signal. 7, re8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xie L., Law B. K., Chytil A. M., Brown K. A., Aakre M. E., and Moses H. L. (2004) Activation of the Erk pathway is required for TGF-β1-induced EMT in vitro. Neoplasia 6, 603–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gui T., Sun Y., Shimokado A., and Muragaki Y. (2012) The roles of mitogen-activated protein kinase pathways in TGF-β-induced epithelial-mesenchymal transition. J. Signal Transduct. 2012, 289243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Julien S., Puig I., Caretti E., Bonaventure J., Nelles L., van Roy F., Dargemont C., de Herreros A. G., Bellacosa A., and Larue L. (2007) Activation of NF-κB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene 26, 7445–7456 [DOI] [PubMed] [Google Scholar]
- 48. Zhou B. P., Deng J., Xia W., Xu J., Li Y. M., Gunduz M., and Hung M. C. (2004) Dual regulation of Snail by GSK-3β-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 6, 931–940 [DOI] [PubMed] [Google Scholar]
- 49. Zhou B. P., and Hung M. C. (2005) Wnt, hedgehog and snail: sister pathways that control by GSK-3β and β-Trcp in the regulation of metastasis. Cell Cycle 4, 772–776 [DOI] [PubMed] [Google Scholar]
- 50. Howe L. R., Watanabe O., Leonard J., and Brown A. M. (2003) Twist is up-regulated in response to Wnt1 and inhibits mouse mammary cell differentiation. Cancer Res. 63, 1906–1913 [PubMed] [Google Scholar]
- 51. Peinado H., Olmeda D., and Cano A. (2007) Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 52. Chen H., Zhu G., Li Y., Padia R. N., Dong Z., Pan Z. K., Liu K., and Huang S. (2009) Extracellular signal-regulated kinase signaling pathway regulates breast cancer cell migration by maintaining slug expression. Cancer Res. 69, 9228–9235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jiao X., Katiyar S., Willmarth N. E., Liu M., Ma X., Flomenberg N., Lisanti M. P., and Pestell R. G. (2010) c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. J. Biol. Chem. 285, 8218–8226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Y., Pu X., Shi M., Chen L., Song Y., Qian L., Yuan G., Zhang H., Yu M., Hu M., Shen B., and Guo N. (2007) Critical role of c-Jun overexpression in liver metastasis of human breast cancer xenograft model. BMC Cancer 7, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nateri A. S., Spencer-Dene B., and Behrens A. (2005) Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 437, 281–285 [DOI] [PubMed] [Google Scholar]
- 56. Gao M., Labuda T., Xia Y., Gallagher E., Fang D., Liu Y. C., and Karin M. (2004) Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306, 271–275 [DOI] [PubMed] [Google Scholar]
- 57. Nateri A. S., Riera-Sans L., Da Costa C., and Behrens A. (2004) The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 [DOI] [PubMed] [Google Scholar]
- 58. Migliorini D., Bogaerts S., Defever D., Vyas R., Denecker G., Radaelli E., Zwolinska A., Depaepe V., Hochepied T., Skarnes W. C., and Marine J. C. (2011) Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J. Clin. Investig. 121, 1329–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu J., Zhang W., Gao F., Liu Y. X., Chen Z. Y., Cheng L. Y., Xie S. F., and Zheng S. S. (2014) FBW7 increases chemosensitivity in hepatocellular carcinoma cells through suppression of epithelial-mesenchymal transition. Hepatobiliary Pancreat. Dis. Int. 13, 184–191 [DOI] [PubMed] [Google Scholar]
- 60. Ling C., Zuo D., Xue B., Muthuswamy S., and Muller W. J. (2010) A novel role for 14-3-3σ in regulating epithelial cell polarity. Genes Dev. 24, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hosing A. S. (2009) Regulation of the G2/M DNA Damage Checkpoint by 14-3-3 Proteins In Human Cells. Ph.D. Thesis, University of Mumbai [Google Scholar]
- 62. Kundu S. T., Gosavi P., Khapare N., Patel R., Hosing A. S., Maru G. B., Ingle A., Decaprio J. A., and Dalal S. N. (2008) Plakophilin3 downregulation leads to a decrease in cell adhesion and promotes metastasis. Int. J. Cancer 123, 2303–2314 [DOI] [PubMed] [Google Scholar]
- 63. Spector D. L., Goldman R. D., and Leinwand L. A. (eds) (1998) Cells: a Laboratory Manual, Vol. 2, pp. 86.2–86.4, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 64. Niessen K., Fu Y., Chang L., Hoodless P. A., McFadden D., and Karsan A. (2008) Slug is a direct Notch target required for initiation of cardiac cushion cellularization. J. Cell Biol. 182, 315–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mei Y., Yuan Z., Song B., Li D., Ma C., Hu C., Ching Y. P., and Li M. (2008) Activating transcription factor 3 up-regulated by c-Jun NH2-terminal kinase/c-Jun contributes to apoptosis induced by potassium deprivation in cerebellar granule neurons. Neuroscience 151, 771–779 [DOI] [PubMed] [Google Scholar]
- 66. Gosavi P., Kundu S. T., Khapare N., Sehgal L., Karkhanis M. S., and Dalal S. N. (2011) E-cadherin and plakoglobin recruit plakophilin3 to the cell border to initiate desmosome assembly. Cell. Mol. Life Sci. 68, 1439–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chaudhary N., and Maddika S. (2014) WWP2-WWP1 ubiquitin ligase complex coordinated by PPM1G maintains the balance between cellular p73 and ΔNp73 levels. Mol. Cell. Biol. 34, 3754–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chandra S., Priyadarshini R., Madhavan V., Tikoo S., Hussain M., Mudgal R., Modi P., Srivastava V., and Sengupta S. (2013) Enhancement of c-Myc degradation by BLM helicase leads to delayed tumor initiation. J. Cell Sci. 126, 3782–3795 [DOI] [PubMed] [Google Scholar]
- 69. Khapare N., Kundu S. T., Sehgal L., Sawant M., Priya R., Gosavi P., Gupta N., Alam H., Karkhanis M., Naik N., Vaidya M. M., and Dalal S. N. (2012) Plakophilin3 loss leads to an increase in PRL3 levels promoting K8 dephosphorylation, which is required for transformation and metastasis. PLoS One 7, e38561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sehgal L., Mukhopadhyay A., Rajan A., Khapare N., Sawant M., Vishal S. S., Bhatt K., Ambatipudi S., Antao N., Alam H., Gurjar M., Basu S., Mathur R., Borde L., Hosing A. S., et al. (2014) 14-3-3γ-mediated transport of plakoglobin to the cell border is required for the initiation of desmosome assembly in vitro and in vivo. J. Cell Sci. 127, 2174–2188 [DOI] [PubMed] [Google Scholar]
- 71. Telles E., Gurjar M., Ganti K., Gupta D., and Dalal S. N. (2011) Filamin A stimulates cdc25C function and promotes entry into mitosis. Cell Cycle 10, 776–782 [DOI] [PubMed] [Google Scholar]
- 72. Basu S., Thorat R., and Dalal S. N. (2015) MMP7 is required to mediate cell invasion and tumor formation upon Plakophilin3 loss. PLoS One 10, e0123979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Alam H., Kundu S. T., Dalal S. N., and Vaidya M. M. (2011) Loss of keratins 8 and 18 leads to alterations in α6β4-integrin-mediated signalling and decreased neoplastic progression in an oral-tumour-derived cell line. J. Cell Sci. 124, 2096–2106 [DOI] [PubMed] [Google Scholar]
- 74. Muslin A. J., Tanner J. W., Allen P. M., and Shaw A. S. (1996) Interaction of 14-3-3 with signaling proteins is mediated by recognition of phosphoserine. Cell 84, 889–897 [DOI] [PubMed] [Google Scholar]
- 75. Roberts B. J., Reddy R., and Wahl J. K. 3rd (2013) Stratifin (14-3-3σ) limits plakophilin-3 exchange with the desmosomal plaque. PLoS One 8, e77012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brunet A., Kanai F., Stehn J., Xu J., Sarbassova D., Frangioni J. V., Dalal S. N., DeCaprio J. A., Greenberg M. E., and Yaffe M. B. (2002) 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 156, 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dalal S. N., Schweitzer C. M., Gan J., and DeCaprio J. A. (1999) Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell. Biol. 19, 4465–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Yang H., Zhao R., and Lee M. H. (2006) 14-3-3σ, a p53 regulator, suppresses tumor growth of nasopharyngeal carcinoma. Mol. Cancer Ther. 5, 253–260 [DOI] [PubMed] [Google Scholar]
- 79. Brabletz T. (2012) EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell 22, 699–701 [DOI] [PubMed] [Google Scholar]
- 80. Hou Z., Peng H., White D. E., Wang P., Lieberman P. M., Halazonetis T., and Rauscher F. J. 3rd (2010) 14-3-3 binding sites in the snail protein are essential for snail-mediated transcriptional repression and epithelial-mesenchymal differentiation. Cancer Res. 70, 4385–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Liu T. A., Jan Y. J., Ko B. S., Liang S. M., Chen S. C., Wang J., Hsu C., Wu Y. M., and Liou J. Y. (2013) 14-3-3epsilon overexpression contributes to epithelial-mesenchymal transition of hepatocellular carcinoma. PLoS One 8, e57968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Dalal S. N., Yaffe M. B., and DeCaprio J. A. (2004) 14-3-3 family members act coordinately to regulate mitotic progression. Cell Cycle 3, 672–677 [PubMed] [Google Scholar]
- 83. Herron B. J., Liddell R. A., Parker A., Grant S., Kinne J., Fisher J. K., and Siracusa L. D. (2005) A mutation in stratifin is responsible for the repeated epilation (Er) phenotype in mice. Nat. Genet. 37, 1210–1212 [DOI] [PubMed] [Google Scholar]
- 84. Steinacker P., Schwarz P., Reim K., Brechlin P., Jahn O., Kratzin H., Aitken A., Wiltfang J., Aguzzi A., Bahn E., Baxter H. C., Brose N., and Otto M. (2005) Unchanged survival rates of 14-3-3γ knockout mice after inoculation with pathological prion protein. Mol. Cell. Biol. 25, 1339–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Toyo-oka K., Shionoya A., Gambello M. J., Cardoso C., Leventer R., Ward H. L., Ayala R., Tsai L. H., Dobyns W., Ledbetter D., Hirotsune S., and Wynshaw-Boris A. (2003) 14-3-3ϵ is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat. Genet. 34, 274–285 [DOI] [PubMed] [Google Scholar]
- 86. Graves P. R., Lovly C. M., Uy G. L., and Piwnica-Worms H. (2001) Localization of human Cdc25C is regulated both by nuclear export and 14-3-3 protein binding. Oncogene 20, 1839–1851 [DOI] [PubMed] [Google Scholar]
- 87. Lopez-Girona A., Furnari B., Mondesert O., and Russell P. (1999) Nuclear localization of Cdc25 is regulated by DNA damage and a 14-3-3 protein. Nature 397, 172–175 [DOI] [PubMed] [Google Scholar]
- 88. Su C. H., Zhao R., Velazquez-Torres G., Chen J., Gully C., Yeung S. C., and Lee M. H. (2010) Nuclear export regulation of COP1 by 14-3-3σ in response to DNA damage. Mol. Cancer 9, 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Waldmann I., Wälde S., and Kehlenbach R. H. (2007) Nuclear import of c-Jun is mediated by multiple transport receptors. J. Biol. Chem. 282, 27685–27692 [DOI] [PubMed] [Google Scholar]
- 90. Su C. H., Zhao R., Zhang F., Qu C., Chen B., Feng Y. H., Phan L., Chen J., Wang H., Wang H., Yeung S. C., and Lee M. H. (2011) 14-3-3σ exerts tumor-suppressor activity mediated by regulation of COP1 stability. Cancer Res. 71, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Phan L., Chou P. C., Velazquez-Torres G., Samudio I., Parreno K., Huang Y., Tseng C., Vu T., Gully C., Su C. H., Wang E., Chen J., Choi H. H., Fuentes-Mattei E., Shin J. H., et al. (2015) The cell cycle regulator 14-3-3σ opposes and reverses cancer metabolic reprogramming. Nat. Commun. 6, 7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wei W., Jin J., Schlisio S., Harper J. W., and Kaelin W. G. Jr. (2005) The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 93. Morton S., Davis R. J., McLaren A., and Cohen P. (2003) A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 22, 3876–3886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Taira N., Mimoto R., Kurata M., Yamaguchi T., Kitagawa M., Miki Y., and Yoshida K. (2012) DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J. Clin. Investig. 122, 859–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Yang H. W., Menon L. G., Black P. M., Carroll R. S., and Johnson M. D. (2010) SNAI2/Slug promotes growth and invasion in human gliomas. BMC Cancer 10, 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Luanpitpong S., Li J., Manke A., Brundage K., Ellis E., McLaughlin S. L., Angsutararux P., Chanthra N., Voronkova M., Chen Y. C., Wang L., Chanvorachote P., Pei M., Issaragrisil S., and Rojanasakul Y. (2015) SLUG is required for SOX9 stabilization and functions to promote cancer stem cells and metastasis in human lung carcinoma. Oncogene 22, 2824–2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Wang L., Sang Y., Tang J., Zhang R. H., Luo D., Chen M., Deng W. G., and Kang T. (2015) Down-regulation of prostate stem cell antigen (PSCA) by Slug promotes metastasis in nasopharyngeal carcinoma. J. Pathol. 237, 411–422 [DOI] [PubMed] [Google Scholar]
- 98. Tanno B., Sesti F., Cesi V., Bossi G., Ferrari-Amorotti G., Bussolari R., Tirindelli D., Calabretta B., and Raschellà G. (2010) Expression of Slug is regulated by c-Myb and is required for invasion and bone marrow homing of cancer cells of different origin. J. Biol. Chem. 285, 29434–29445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kusewitt D. F., Choi C., Newkirk K. M., Leroy P., Li Y., Chavez M. G., and Hudson L. G. (2009) Slug/Snai2 is a downstream mediator of epidermal growth factor receptor-stimulated reepithelialization. J. Invest. Dermatol. 129, 491–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ding X., Park S. I., McCauley L. K., and Wang C. Y. (2013) Signaling between transforming growth factor β (TGF-β) and transcription factor SNAI2 represses expression of microRNA miR-203 to promote epithelial-mesenchymal transition and tumor metastasis. J. Biol. Chem. 288, 10241–10253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. He Q., Zhou X., Li S., Jin Y., Chen Z., Chen D., Cai Y., Liu Z., Zhao T., and Wang A. (2013) MicroRNA-181a suppresses salivary adenoid cystic carcinoma metastasis by targeting MAPK-Snai2 pathway. Biochim. Biophys. Acta 1830, 5258–5266 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.