Abstract
Motivation: Moonlighting proteins (MPs) show multiple cellular functions within a single polypeptide chain. To understand the overall landscape of their functional diversity, it is important to establish a computational method that can identify MPs on a genome scale. Previously, we have systematically characterized MPs using functional and omics-scale information. In this work, we develop a computational prediction model for automatic identification of MPs using a diverse range of protein association information.
Results: We incorporated a diverse range of protein association information to extract characteristic features of MPs, which range from gene ontology (GO), protein–protein interactions, gene expression, phylogenetic profiles, genetic interactions and network-based graph properties to protein structural properties, i.e. intrinsically disordered regions in the protein chain. Then, we used machine learning classifiers using the broad feature space for predicting MPs. Because many known MPs lack some proteomic features, we developed an imputation technique to fill such missing features. Results on the control dataset show that MPs can be predicted with over 98% accuracy when GO terms are available. Furthermore, using only the omics-based features the method can still identify MPs with over 75% accuracy. Last, we applied the method on three genomes: Saccharomyces cerevisiae, Caenorhabditis elegans and Homo sapiens, and found that about 2–10% of proteins in the genomes are potential MPs.
Availability and Implementation: Code available at http://kiharalab.org/MPprediction
Contact: dkihara@purdue.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
As the last decade has witnessed a momentous expansion in the number of functionally well-characterized proteins as well as rapid progress in large-scale proteomics studies, protein multi-functionality has become a highly perceived phenomenon (Campbell and Scanes, 1995; Jeffery, 1999; Weaver, 1998). These multifunctional, ‘moonlighting’ proteins demonstrate multiple autonomous and usually unrelated functions within a single polypeptide chain, which cannot be individually assigned into separate domains. It has become evident that the functional diversity of these proteins is neither specific to genomes or certain protein families nor facilitated by common function-switching mechanisms. Many of the known moonlighting proteins (MPs) were originally recognized as enzymes, but there are also others that are known as receptors, channel proteins, chaperone proteins, ribosomal proteins or scaffold proteins (Jeffery, 1999, 2004). There are speculations that MPs evolved to broaden the functional aspects of a genome without expanding the genome size (Jeffery, 1999). Studies suggest significant impacts of MPs in diseases and disorders (Ovádi, 2011; Sriram et al., 2005) as well as roles in important biochemical pathways (Jeffery, 2004). Despite the potential abundance of MPs in various genomes and their important roles in pathways and disease developments, the size of existing databases (Hernández et al., 2014; Mani et al., 2014) that contain experimentally confirmed MPs is still too small to obtain a comprehensive picture of the cellular mechanisms underlying their functional diversity. This quantitative insufficiency is due in large part to the tendency for the additional function of these proteins to be found serendipitously in the course of unrelated experiments. Hence, a systematic bioinformatics approach could make substantial contributions in identifying novel MPs on a genome scale and also to an overall understanding of the underlying biology of their multi-functional nature.
The functional diversity of MPs poses a significant challenge to computational protein function annotation as current methods do not explicitly consider the possibility of dual functions for a protein. Conventional sequence-based functional annotation methods, based on the concept of homology (Altschul et al., 1997) or conserved motifs/domains (Bru et al., 2005; Finn et al., 2014; Hunter et al., 2012), will have problems identifying secondary functions because there are cases where a homolog of a MP does not possess the secondary function (Ozimek et al., 2006) or has a different secondary function (Banerjee et al., 2007; Chen et al., 2005). Due to these intrinsic computational challenges, systematic studies of MPs are still in an early stage for obtaining a comprehensive picture of proteins’ moonlighting functions or for developing computational methods for predicting MPs [review by (Khan and Kihara, 2014)]. Existing bioinformatics approaches for detection of MPs have two general shortcomings. First, they rely heavily on the existence of functional annotation of a protein (Chapple et al., 2015; Pritykin et al., 2015), which is a major bottleneck of the problem. Second, all the existing methods address different aspects of MPs’ functional diversity: sequence similarity (Gomez et al., 2003; Khan et al., 2012), motifs/domains, structural disorder (Hernández et al., 2011), or protein–protein interaction (PPI) patterns combined with existing gene ontology (GO) annotations (Chapple et al., 2015; Gómez et al., 2011; Pritykin et al., 2015). However, the diverse nature of MPs’ functions, cellular locations, function switching mechanisms, and the organisms in which they are found gives compelling evidence that in order to understand and identify the overall functional aspects of these proteins, one should characterize these proteins in a wider functional/proteomic space.
Previously, we have identified functional characteristics of MPs in different proteomic aspects using a computational framework (Khan et al., 2014). Here, we have constructed an automated prediction model to identify MPs based on features we characterized in our previous study. To address the diverse nature of MPs, we have used a wide feature space ranging from GO (Gene Ontology Consortium, 2013) and several omics-scale data, namely PPI, gene expression (GE), phylogenetic profiles (Phylo), genetic interactions (GIs) and network-based graph properties (such as node between-ness, degree centrality, closeness-centrality), to protein structural properties such as the number and the length of intrinsically disordered regions in the protein chain. Based on our computed GO and the omics-based protein feature space, we used machine learning classifiers as the framework for MP prediction and used an existing MP database to cross-validate our prediction model. Because a significant fraction of proteins do not have certain functional/network features in databases, we have additionally developed an imputation technique using random forest (RF) to predict missing features for proteins. Cross-validation results on the dataset of known moonlighting and non-MPs (control dataset) show that if GO information is available, MPs can be predicted with over 98% accuracy. More importantly, leveraging just the non-GO based features, our imputation-classification models can predict MPs with over 75% accuracy. The latter result is very important because it indicates that MPs without sufficient function annotations can be identified by analyzing available omics data, which is the first such development. Lastly, we have run our imputation-classification models with the best performing omics-based feature combinations on three genomes, Saccharomyces cerevisiae (yeast), Caenorhabditis elegans and Homo sapiens (human), and found that about 2–10% of the proteomes are potential MPs.
2 Methods
The overall computational prediction model, named MPFit (Moonlighting protein Prediction with missing Feature Imputation) undergoes four phases: data construction, feature computation, missing feature imputation (when needed) and classification into MP or non-MP. Each of the steps is discussed in detail below.
2.1 Data construction
We used a manually curated MP database, MoonProt (Mani et al., 2014), and extracted 268 proteins that had Uniprot ID mapping. Two hundred sixty-eight MPs include those from human (45 proteins, 16.8%), Escherichia coli (30 proteins, 11.19%), yeast (27 proteins, 10.1%) and mouse (11 proteins, 4.1%). In order for our model to train on negative examples of such proteins along with the positive examples, we used the following criteria to select negative examples of MPs (referred as non-MPs) from these four genomes as developed in our previous work (Khan et al., 2014). A protein was selected as a non-MP if it has (i) at least eight GO term annotations, (ii) when GO terms in the Biological Process (BP) category were clustered using the semantic similarity score (Schlicker et al., 2006) thresholds of 0.1 and 0.5, not more than one cluster was obtained at each threshold. We further added a criterion on Molecular Function (MF) category GO terms: (iii) not more than one cluster of MF GO terms at semantic similarity scores of 0.1 and 0.5. In essence, a non-MP is a protein that has a sufficient number of GO annotations but they are not functionally diverse. For this procedure, full GO annotations (including computationally predicted terms such as IEA) were taken from UniProt and parental propagation of GO terms was not applied, to be consistent with the criteria established in our previous work (Khan et al., 2014). Furthermore, we computed pairwise sequence similarity of the selected non-MPs from the above three conditions and further ruled out redundant proteins that had >25% sequence identity to other sequences. This process yielded 162 non-MPs, among which 60 are from human (37.0%), 52 from mouse (32.1%), 34 from yeast (20.9%) and 16 from E.coli (9.88%). The MP and non-MP datasets are made available at http://kiharalab.org/MPprediction/.
2.2 Feature computation and selection
As MPs have dual functions, intuitively they interact with more proteins with different functions compared with non-MPs. In our previous work (Khan et al., 2014), we have characterized MPs and non-MPs in terms of different omics-based features (including PPI, GE, Phylo, GIs) and showed that when the interacting partners are clustered based on their functional similarity, the number of clusters tend to be higher for MPs than non-MPs. Based on this analysis, we develop the MPFit model in this work that uses the number of functional clusters as the features to classify MPs and non-MPs.
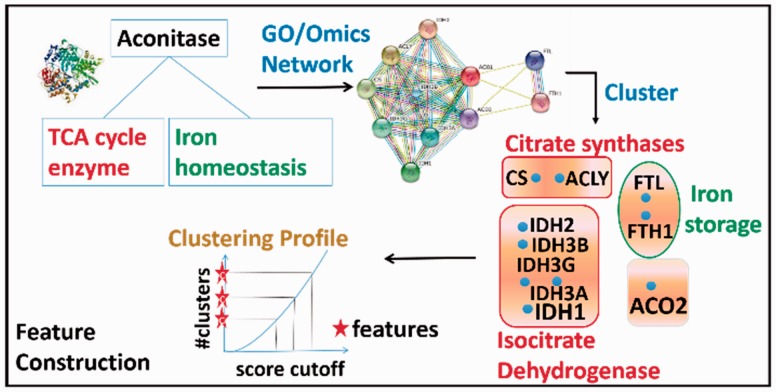
To characterize MP and non-MPs we selected a broad range of features, i.e. GO annotations, PPI network, GE profiles, Phylo, GIs, disordered protein regions (DOR), and the protein’s graph properties in the PPI network (NET). In order to extract the feature for a protein Pi in any information domain, we first extracted the GO terms or proteins associated with Pi in that domain and built a network Ni for Pi. Each node in Ni can be either a GO term (if the information domain is GO) or a protein (if the information domain is any of the omics-based information); edges in Ni represent association weights among nodes. Then we applied single linkage clustering on Ni and the number of clusters at several score thresholds were selected as features of Pi (Khan et al., 2014). Figure 1 illustrates the feature computation procedure for human aconitase (aco1), an MP, for the PPI network. First, we extracted interacting partners for aco1, then based on the GO annotation similarity score of the interacting partners, the PPI network was clustered and four clusters were obtained with a certain similarity cutoff i. Two of these clusters (circled in red) contain proteins related to the (tricarboxylic acid) TCAcycle and are associated to the first function of aco1 while another cluster (green) was relevant to the second function. Such clustering was performed with five different similarity cutoffs (from 0.1 to 0.9 with an interval of 0.2), which resulted in a clustering profile shown in the bottom of Figure 1. Finally, we extracted the number of clusters at multiple score cutoffs as the PPI network features. More details about the feature computation in the PPI network domain are provided in the Supplementary Figure S1.
Fig. 1.
Schematic diagram of MPFit. Feature construction of moonlighting protein Aconitase in PPI network
To construct the GE network, expression profiles were obtained from the COEXPRESdb database (Okamura et al., 2014). Gene pairs that have an absolute value of their Pearson correlation of expression levels within the top 2% among all the pairs were connected in the network. Phylo network was constructed using the STRING database (Szklarczyk et al., 2014). A protein pair was connected in the network if they have a sufficient score (>0.7 as recommended by STRING) at ‘neighborhood’, ‘co-occurrence’ or ‘gene-fusion’ in STRING. For the GI network, we used the BIOGRID database (Stark et al., 2006) and extracted gene pairs that had the ‘experiment type’ listed as ‘genetic’ to be associated in the GI network. For the NET feature, three graph properties of proteins, namely, degree centrality, closeness centrality and between-ness centrality, based on the PPI network were computed as features. For the DOR feature, using the D2P2 database (Oates et al., 2013), we computed three properties of protein’s intrinsically disordered regions, namely, the number and the total length of disordered regions as well as the proportion of disordered regions in the sequence.
2.3 Missing data imputation
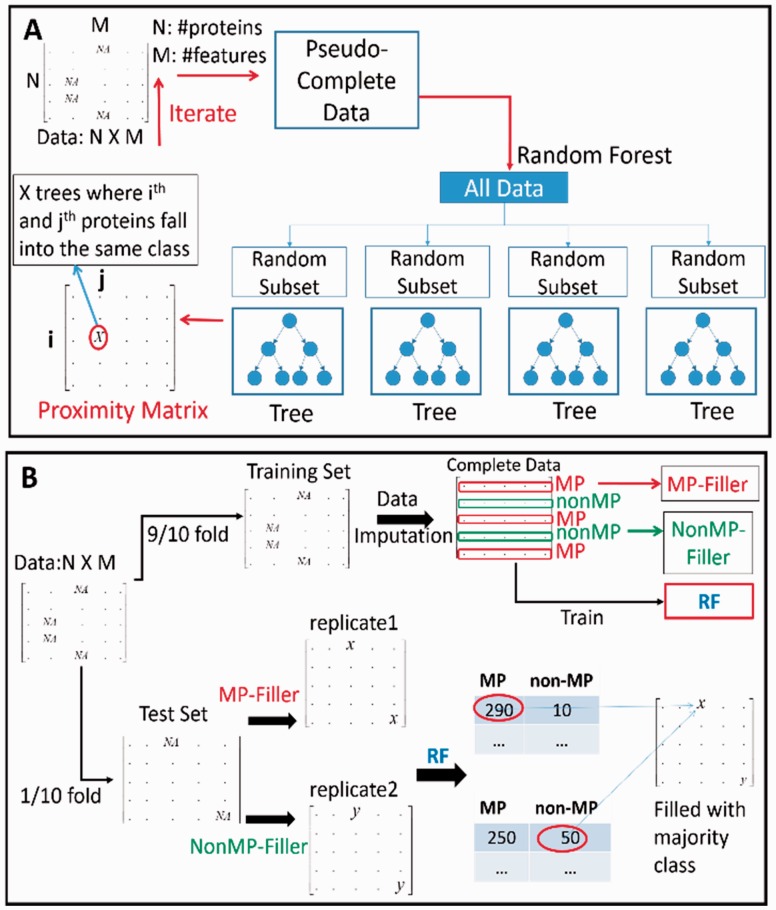
In order to deal with missing data, imputation is an approach that fills in the missing data rather than discarding the data points entirely and using only the complete subset of the data. Among known imputation approaches, there are methods that fill in the missing feature from mean or median of the known values of the same features in other instances (Little and Rubin, 1987; Zloba 2002). On the other hand, there are methods that perform partial imputation based on known features of small neighborhood of the incomplete data (Morin and Raeside, 1981; Zhang, 2008). In this work, we used a RF-based imputation technique (Breiman, 2001; Liaw, 2003). Figure 2A–B shows the procedure. In Figure 2A, the training dataset is represented as a matrix where rows are proteins and columns are features. Missing features in the dataset are represented by NAs. The algorithm starts by replacing NAs with the column medians. Then a RF was constructed using the temporally filled features in the previous step (pseudo-complete data in Fig. 2A). Next, the proximity matrix from the RF was used to update the imputed values of the NAs. The (i, j) element of the proximity matrix is the fraction of trees in which the proteins i and j fall in the same class. The imputed value for a feature is the weighted average of the non-missing features from other proteins, where weights are the proximities. The imputation was iterated until the proximity matrixes converged or the procedure is iterated 10 times. Finally, a RF RFtrain was computed with this imputed training data matrix.
Fig. 2.
Schematic diagram of MPFit. A-B: Missing feature imputation method. RF: Random Forest. See text for details
In order to impute missing features in the test set (Fig. 2B), the training dataset with missing values imputed was used to compute two filler vectors (referred to as MP-filler and non-MP-filler), one for each of the MP and non-MP classes. The ith element of the filler vector MP-filler (non-MP-filler) is the mean of the imputed features at the ith column of the training matrix with the MP (non-MP) class label. The test dataset was represented as a matrix similar to the training data (rows are proteins, columns are features). For the test data row ritest, since the label (MP/non-MP) is not known, two replicates were made: the missing features in the first replicate were filled with the MP-filler and the same for the second replicate was filled with the non-MP-filler vector. Now these two completed test replicates were run down through the previously trained RF RFtrain. Each protein receives tree votes of MP and non-MP in RFtrain from replicates 1 and 2, and the higher vote between the MP vote in replicate 1 and the non-MP vote in replicate 2 finally determines the MP/non-MP-fillers to be used in the missing features of the protein. In Figure 2B, the first protein received higher MP votes from replicate 1 (290 votes) over non-MP votes from replicate 2 (50 votes); thus, the missing features of the protein are filled with the MP-filler vector. Finally, proteins in the test set were predicted to be MP or non-MP using a classifier. When RF was used for the classifier, this voting was used as the final prediction. We have also used support vector machine (SVM) and naïve-Baiyes as the final classifier and compared the results.
Aside from this explicit RF-based imputation technique, an alternative imputation method (termed as ‘probabilistic imputation’) was used in this work where the splitting probabilities in the RF were learned from the subset of complete data and later used to classify the incomplete data. Detail of this method is discussed in Supplementary Figure S4 and its associated text.
3 Results
In this section, we present and discuss the performance of MPFit with different combinations of features. MPFit was run and evaluated with the GO term feature and all possible combinations of six omics feature domains (namely, PPI, GE, Phylo, GI, DOR and NET). There are 1+ (26 − 1) = 64 such combinations.
3.1 Imputation of missing features facilitates usage of omics data
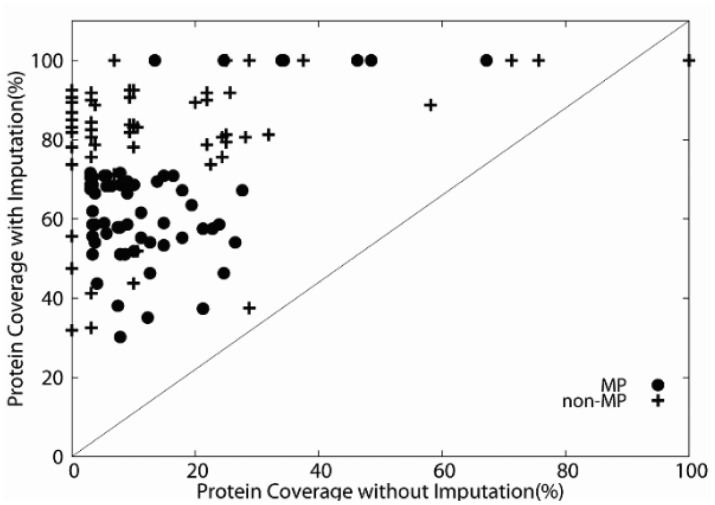
For a given combination of omics features, there are proteins which lack some of the feature data. One way to handle such missing data by a classifier is to impute the missing data so that a classifier trained on the full features can be applied. Figure 3 contrasts the number of target proteins that were predicted by MPFit before and after the imputation. A point represents one of the 64 feature combinations. For each feature combination considered, proteins that have at least one feature were subject to imputation and those that do not have any features are discarded (data points in Fig. 3 with under 100% protein coverage after imputation).
Fig. 3.
Impact of missing feature imputation. Comparison of the number of proteins used in classifiers without (x-axis) and with (y-axis) missing feature imputation. Each point represents % of proteins that were used in a classifier for a certain feature combination
It is evident that the imputation technique substantially increased the dataset coverage, which also consequently improved classifier performance as explained in later sections. For example, the number of MP proteins for a feature combination of (PPI, Phylo, GE, GI, DOR) was originally 8 (2.9%), which increased to 192 (71.7%) after imputation. The features with 100% coverage after imputation are seven single features, GO, GE, Phylo, PPI, GI, NET and DOR.
3.2 Prediction accuracy of MPs
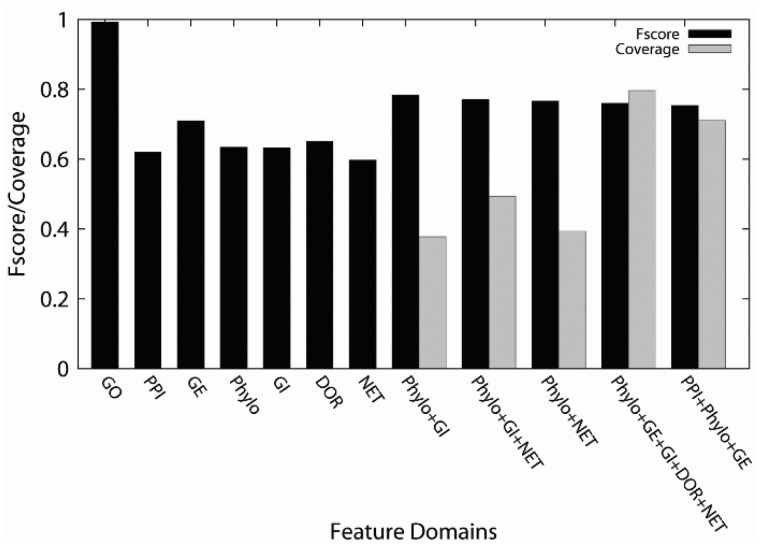
Next, we discuss prediction performance of MPFit using RF (Breiman, 2001) as the final classifier in the pipeline (Fig. 2B). Prediction performance was evaluated by a weighted class average F-score, where the F-score was computed separately for MP and non-MP protein classes and weighted by the number of proteins in the corresponding class. The F-score is defined as (2*precision*recall)/(precision + recall), where precision and recall are defined as (TP/(TP + FP)) and (TP/(FP + FN)), respectively. Here, TP, FP and FN stand for true positive, false positive and false negative, respectively. Figure 4 presents results with the seven single features as well as the five combinations of features that showed the highest F-score. Average F-score from a 5-fold cross-validation was reported.
Fig. 4.
Performance of MPFit with RF. Average of results from a 5-fold cross-validation are reported. Black bars show the F-score while the gray bars show the coverage (i.e. the fraction of the proteins in the dataset that were predicted). Feature legends on the x-axis: GO, gene ontology, PPI, protein–protein interactions; Phylo, phylogenetic profile; GE, gene expression; DOR, disordered regions; GI, genetic interactions; NET, 3 graph properties; i.e. between-ness, degree centrality, and closeness centrality. Coverage was computed as the mean protein coverage of MP and non-MP
When proteins have GO annotations, it is shown that prediction can be very accurate, with an F-score of 0.993. Among the six individual omics features, GE showed the best F-score of 0.710, and the rest of the features performed similarly (F-scores range from 0.597 to 0.651). Results of all the possible combinations of omics features are provided in Supplementary Figure S2. Their F-scores range from 0.784 to 0.571. Among the feature combinations, Phylo + GI showed highest accuracy (precision, recall and F-score are 0.799, 0.771 and 0.784, respectively), followed by Phylo + GI + NET and Phylo + NET. However, these three combinations have relatively low coverage (Fig. 4), while the fourth and fifth best performing feature combinations, Phylo + GE + GI + DOR + NET and PPI + Phylo + GE, have a high coverage with good F-scores that are close to the best value achieved by Phylo + GI (0.7964, 0.7602 for coverage and 0.7109, 0.7538, for F-score, respectively). For this reason we used the fourth and fifth feature combinations in the genome-scale prediction performed in the subsequent section. Among the proteins in MoonProt, there are five protein pairs from the same organism that have over 25% sequence identity. We removed five proteins, one from each of these similar protein pairs and recomputed the F-score with cross-validation for the two-feature combinations, Phylo + GE + GI + DOR + NET and PPI + Phylo + GE. The changes of F-score were marginal: an increase of 0.87 and 3.09 were observed for the former and the latter combinations, respectively.
Here, we discuss two cases where combinations of different omics-based features improved prediction over single feature. The first example is a MP in human, which is a ribosomal protein (part of the 60S subunit) (UniProt ID: P46777) (Horn and Vousden, 2008). The secondary function of this protein is inhibition of HDM2, an ubiquitin ligase, which results in stabilization of the p53 tumor suppressor protein. Using only the PPI features, this protein is incorrectly predicted as non-MP. This is because 63 interacting proteins in the PPI network have a relatively small number of functional clusters for MP. When clustered using functional similarity (funsim) scores for BP and MF combined (see Supplementary Figure S1B), the relative number of clusters stay below 0.32 at each clustering cutoff, which is significantly lower compared with the MP distribution. However, the protein was correctly predicted as MP with the PPI + Phylo + GE combination. Twenty-five associated proteins for this target in the Phylo network were clustered in to 2, 3, 3, 3 and 24 groups at similarity cutoffs 0.1, 0.3, 0.5, 0.7 and 0.9 of the funsim score, which are larger than the non-MP distribution (Supplementary Figure S5A). Thus for this protein, addition of Phylo features made the prediction correct.
The second example is DNA replication factor Cdt1 (UniProt ID: Q9H211) (Varma et al., 2012). Besides its primary function as DNA replication factor, this MP’s secondary function is a role in mitosis where it localizes to kinetochores through binding to the Hec1 component of the Ndc80 complex. Using PPI features only, this protein is incorrectly predicted as non-MP, because its 29 interacting proteins in the PPI network do not have sufficient number of functionally different groups. Clustering using the funsim BP + MF score, the relative number of clusters stays below 0.35, which is significantly lower than the MP distribution. However, the PPI + Phylo + NET feature combination made correct prediction as MP. This is partly because the NET feature of this protein has high values, e.g. a between-ness centrality of 0.267, which is high (above 75 percentile) for this feature’s distribution for MP (Supplementary Figure S5B).
We also ran MPFit with RF without imputation, i.e. only on proteins that do not have any missing feature in a feature combination. The results for all the feature combinations are shown in Supplementary Figure S3. Skipping imputation substantially lowers coverage (Fig. 4, and Supplementary Figures S2 and S3). Without imputation the coverage decreases as the number of features in a combination increases, which resulted in 0 coverage for 16 out of 64 cases (Supplementary Figure S3). Also, the data sizes of MP and non-MP classes become substantially different and imbalanced for several feature combinations. In contrast, imputation not only increases prediction coverage but also improves accuracy by increasing the size of the training set, as indicated by the cases that improved F-score by imputation.
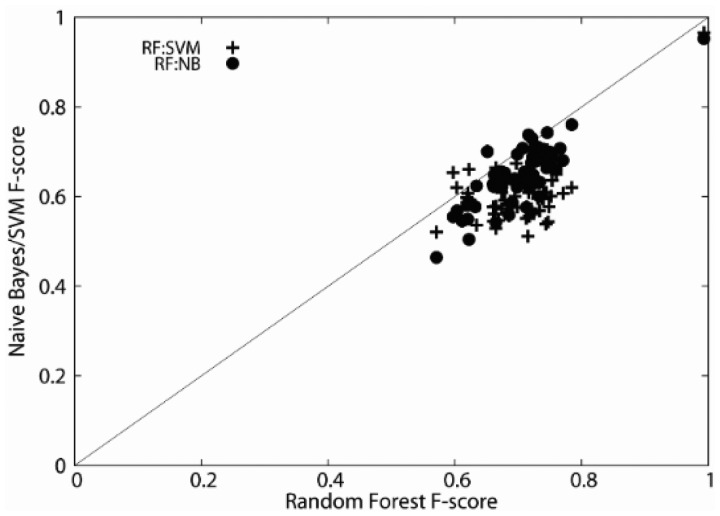
We examined prediction performance of MPFit when naïve Bayes (Andrew and Kamal, 1998) or SVM (Cortes and Vapnik, 1995), was used as the last classifier in the procedure. As explained with Figure 2, the missing data imputation was performed with RF, and naïve Bayes or SVM was applied as the final classifier to proteins with full imputed features. Results with all 64 feature combinations were shown in comparison with the results by RF in Figure 5. Results in the lower triangle in Figure 5 are the cases where RF performed better than the counterpart. It is apparent that RF performed better than SVM and naïve Bayes for the majority of the cases. Using the GO term features showed the highest F-score by all the classifiers (the upper right corner of Fig. 5). F-scores of feature combinations by the three classifiers correlated moderately. The correlation coefficient between RF and naïve Bayes was highest, 0.828, that for RF with SVM was 0.542, and between SVM and naïve Bayes it was 0.561. Our speculation for RF outperforming SVM is that the fairly low number of features used in this work is probably more suitable for RF than SVM, which is shown to perform well for a high dimensional feature space (Caruana et al., 2008).
Fig. 5.
Performance comparison of RF with two other classifiers. F-score using each of the different feature combinations by MPFit with RF was compared with SVM (cross) or naïve Bayes (filled circles). The imputed dataset was used. Results are the weighted class average F-score over 5-fold cross-validation
We also computed cross-validation F-score for the alternative imputation technique (termed as ‘probabilistic imputation’) and compared the result with the F-score shown in Figure 4 with explicit imputation. The result is discussed in Supplementary Figure S4 with the conclusion that explicit imputation outperforms the probabilistic imputation.
To summarize this section, MP and non-MP can be classified very accurately by MPFit when GO terms of the proteins are available. Encouragingly, prediction can be made with a sufficient accuracy even when no function annotation is available using proper combinations of omics-based features. Missing feature imputation increases the coverage of proteins that are subject to prediction and also helps to improve accuracy by increasing the training data of a classifier. Among the three classifiers tested, RF performed better than SVM and naïve Bayes.
3.3 Genome-wide prediction of MPs
In the last section of this work, we report genome-wide prediction of MPs performed with MPFit on three genomes, S.cerevisiae (yeast), C.elegans and human. We used two feature combinations that gave high performance in both F-score and coverage (Fig. 4): Phylo + GE + GI + DOR + NET and PPI + Phylo + GE. MPFit with the two feature combinations were run separately with explicit feature imputation and RF as the last classifier. Then, proteins that were predicted as MPs by consensus of both runs were taken as plausible MPs. Consensus was taken to only count highly plausible MPs and avoid over-estimation of the MP fraction in the genomes. For MPFit runs with a feature combination, proteins were discarded if they had no features in the combination. In the yeast genome, which has 6718 proteins in UniProt (UniProt Consortium, 2014), there were 4673 proteins (69.6%) that had at least one feature among PPI, Phylo or GE, and 5845 proteins (87.0%) that had at least one feature in Phylo, GE, GI, DOR or NET. The coverages for C.elegans are 79.8% and 89.5%, while that for the human genome are 68.1 and 82.4%, respectively for the PPI + Phylo + GE and Phylo + GE + GI+ DOR + NET feature combinations. The results are summarized in Table 1. A list of predicted MPs is available at http://kiharalab.org/MPprediction.
Table 1.
Genome-wide prediction of MPs
| Genome | No. Proteins | Cov. (%)a | Known MPs Predictedb | MPs (%) c |
|---|---|---|---|---|
| yeast | 6718 | 69.56 | 22/27 (81.4%) | 10.97 |
| C.elegans | 20 133 | 79.82 | 1/1 (100%) | 2.73 |
| human | 20 098 | 67.91 | 33/45 (73.3%) | 7.82 |
aThe fraction of proteins that were subject to the prediction among all the proteins in the genome.
bThe number of known MPs in MoonProt predicted as MPs.
cThe fraction of predicted MPs among the proteins in the genome.
First, we examined if known MPs listed in the MoonProt database in each genome were correctly predicted as MPs. The results in the second column from the right in Table 1 show that MPFit predicts known MPs reasonably well with recall of over 73% to each genome.
Next, we moved onto the blind genome-wide prediction to the three genomes. In yeast, MPFit with the two feature combinations Phylo + GE + GI + DOR + NET and PPI + Phylo + GE predicted 24.6 and 18.5% of the proteins as MPs, respectively, and among them, 10.9% of the proteins have a consensus prediction as MPs with the two feature sets. We note that this number of MPs in yeast is similar to the numbers obtained by a recent work by a different group (Pritykin et al., 2015). In human, 67.6% of the total genome was subject to MPFit by both feature combinations, and 7.8% of the total genome was predicted as MP by consensus of the two feature combinations.
In C.elegans, 79.8% of proteins were subject to prediction by the two feature combinations. For this genome, the two feature combinations showed difference in the number of proteins predicted as MPs. With the Phylo + GE + GI + DOR + NET combination, 15.4% of the proteins were predicted as MPs while the fraction was 4.0% using the PPI + Phylo + GE combination, which resulted in a consensus of 2.73% of the proteins predicted as MPs. The fraction of predicted MPs by the latter feature combination was particularly lower than the other mainly because 48.5% of the predicted MPs by Phylo + GE + GI + DOR + NET were not subject to prediction with the PPI + Phylo + GE combination due to missing features.
To date there are two methods that predict whether a protein is MP or not. A method by Chapple et al. (2015) considers a protein as MP if it is within an overlapping cluster in the PPI network and further passes a GO-based analysis. Out of the 45 known MPs in human in MoonProt, only 3 were predicted by this method (recall 0.0667). The method by Pritykin et al. 2015) uses GO-based multifunctional filtering criteria to predict MPs. Their method predicted 22 out of 45 known MPs in human (recall 0.4889) and 13 out of 27 known MPs in yeast (recall 0.4815) as MPs. Thus, as shown in Table 1, MPFit showed a larger recall (Table 1) in both human and yeast than the two methods.
3.4 Analysis of Genome-wide MP prediction
We examined functions of predicted MPs in the three genomes by considering GO and Kyoto encyclopedia of genes and genomes (KEGG) pathway association (Kanehisa and Goto, 2000). In order to assign a protein to GO categories, we first mapped its GO annotations onto the terms at the second depth in the GO hierarchy, and performed GO enrichment analysis (NaviGO at http://kiharalab.org/web/compare.php). Table 2 lists the enriched GO categories of the predicted MPs. This GO analysis covers 100%, 99.3%, and 99.9% of predicted MPs in yeast, C.elegans and human, respectively, which have GO annotations. Table 3 is a list of associations of the predicted MPs to KEGG pathways. Note that this analysis was based on the predicted MPs that exist in KEGG (Kanehisa and Goto, 2000) database (66.36, 35.21 and 51.92% in yeast, C.elegans and human genomes, respectively).
Table 2.
GO categories of the predicted MPs
| Genome | Enriched GO terms | MP (%) |
|---|---|---|
| yeast | enzyme (BP/MF) | 91.86 |
| GO:0005488 binding (MF) | 59.29 | |
| GO:0032991 macromolecular complex (CC) | 51.70 | |
| GO:0071840 cellular component organization or biogenesis (BP) | 42.61 | |
| GO:0031974 membrane enclosed lumen (CC) | 26.05 | |
| GO:0005198 structural molecule activity (MF) | 19.95 | |
| GO:0009295 nucleoid (CC) | 0.951 | |
| GO:0016209 antioxidant activity (MF) | 0.810 | |
| C.elegans | enzyme (BP/MF) | 73.67 |
| GO:0005198 structural molecule activity (MF) | 15.72 | |
| GO:0002376 immune system process (BP) | 3.47 | |
| GO:0060089 mol. transducer activity (MF) | 1.65 | |
| GO:0004872 receptor activity (MF) | 1.65 | |
| Human | enzyme (BP/MF) | 76.77 |
| GO:0005488 binding (MF) | 63.84 | |
| GO:0050896 response to stimulus (BP) | 45.51 | |
| GO:0032501 multicellular organismal process (BP) | 38.19 | |
| GO:0005576 extracellular region (CC) | 36.54 | |
| GO:0071840 cellular component organization or biogenesis (BP) | 33.23 | |
| GO:0051179 localization (BP) | 29.03 | |
| GO:0051704 multi-organism process (BP) | 15.15 | |
| GO:0040011 locomotion (BP) | 10.18 | |
| GO:0032991 macromolecular complex (CC) | 9.41 | |
| GO:0030054 cell junction (CC) | 7.51 | |
| GO:0000003 reproduction (BP) | 7.26 | |
| GO:0005198 structural molecule activity (MF) | 7.07 | |
| GO:0040007 growth (BP) | 4.58 | |
| GO:0031012 extracellular matrix (CC) | 3.95 | |
| GO:0009055 electron carrier activity (MF) | 1.15 |
GO category ‘Enzyme’ is upon membership of either GO:0008152 metabolic process or GO:0003824 catalytic activity. The percentage of GO terms will not sum to 100% for a genome because a protein can have multiple assigned GO terms.
Table 3.
KEGG pathway associations of predicted MPs
| Genome | Top 5 KEGG pathways | MP (%) |
|---|---|---|
| yeast | Metabolic pathways (KEGG ID 1100) | 29.17 |
| Ribosome (3010) | 15.33 | |
| Biosynthesis of secondary metabolites (1110) | 13.70 | |
| Carbon metabolism (1200) | 6.92 | |
| Biosynthesis of amino acids (1230) | 6.38 | |
| C.elegans | Ribosome (3010) | 13.79 |
| Metabolic pathways (1100) | 12.34 | |
| Purine metabolism (230) | 2.72 | |
| Pyrimidine metabolism (240) | 2.54 | |
| Oxidative phosphorylation (190) | 2.54 | |
| human | Metabolic pathways (1100) | 18.38 |
| Ribosome (3010) | 4.45 | |
| Olfactory transduction (4740) | 3.94 | |
| Purine metabolism (230) | 2.54 | |
| Cytokine-cytokine receptor interaction (4060) | 2.42 |
In Tables 2 and 3, the major proportion of MPs is enzymes. This observation is consistent with previous reports that many MPs were known primarily as enzymes when their secondary function was discovered (Hernández et al., 2014; Jeffery, 2003; Mani et al., 2014).
Ribosome was listed as a KEGG pathway for the three genomes. An example is 40S ribosomal protein S3 (Uniprot ID: P23396) in human, which functions primarily as a ribosomal protein (part of the 40S subunit), and has a second function of being a subunit of a DNA-binding complex involved in NF-kappaB-mediated transcription (Wan et al., 2007). The second example of MPs is glyceraldehyde-3-phosphate dehydrogenase (UniProt ID: P04406) in human. Besides its primary function as enzyme in the glycolysis pathway, this protein moonlights as interferon-gamma-activated inhibitor of translation that silences ceruloplasmin mRNA translation (Sampath et al., 2004). In a proteomics study (Prunotto et al., 2013), this protein was identified as one of the urinary exosome proteins, and thus contains GO:0070062 extracellular exosome, which is a child term of GO:0005576 extracellular region, and hence falls in the latter GO category in Table 2. Both are these examples are correctly predicted MPs in human by the two omics-based combinations Phylo + GE + GI + DOR + NET and PPI + Phylo + GE.
4 Discussion
We proposed a novel computational approach, MPFit, for detecting MPs from GO annotations or omics-based features. MPFit can be applied to a large fraction of proteins in a genome due to the use of several omics-based features and the implemented imputation protocol for filling missing features. As the mechanisms by which MPs exhibit multiple functions differ from case by case, using various feature types is reasonable to capture MPs of different nature.
Although there is a possibility that some predictions made by MPFit are not correct, the overall estimation is probably not too far from the truth and serves as workable hypotheses for future research projects. We believe that our work is an imperative step toward a systematic and integrative approach of studying MPs and it will open up new opportunities to investigate the multi-functional nature of proteins at a systems level.
Supplementary Material
Acknowledgements
The authors thank Jennifer Neville for useful discussion on the probabilistic imputation. Lenna X. Peterson for proofreading the article.
Funding
This work was partly supported by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM097528) and the National Science Foundation (IIS1319551, DBI1262189, IOS1127027).
Conflict of Interest: none declared.
References
- Altschul S.F. et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew M., Kamal N. (1998) A comparison of event models for Naive Bayes text classification In: AAAI-98 Workshop on Learning for Text Categorization, vol. 752, pp. 41–48. [Google Scholar]
- Banerjee S. et al. (2007) Iron-dependent RNA-binding activity of Mycobacterium tuberculosis aconitase. J. Bacteriol., 189, 4046–4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L. (2001) Random forests. Mach. Learn., 45, 5–32. [Google Scholar]
- Bru C. et al. (2005) The ProDom database of protein domain families: more emphasis on 3D. Nucleic acids research., 33, D212–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.M., Scanes C.G. (1995) Endocrine peptides ‘moonlighting’ as immune modulators: roles for somatostatin and GH-releasing factor. J. Endocrinol., 147, 383–396. [DOI] [PubMed] [Google Scholar]
- Caruana R. et al. (2008) An empirical evaluation of supervised learning in high dimensions In: Proceedings of the 25th international conference on Machine learning, pp. 96–103. [Google Scholar]
- Chapple C.E. et al. (2015) Extreme multifunctional proteins identified from a human protein interaction network. Nature communications., 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.J. et al. (2005) Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science, 307, 714–717. [DOI] [PubMed] [Google Scholar]
- Cortes C., Vapnik V. (1995) Support-vector network. Mach. Learn., 20, 273–297. [Google Scholar]
- Finn R.D. et al. (2014) The Pfam protein families database. Nucleic Acids Res., 42, D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium. (2013) Gene Ontology annotations and resources. Nucleic Acids Res., 41, D530–D535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez A. et al. (2011) Do protein-protein interaction databases identify moonlighting proteins? Mol. BioSyst., 7, 2379–2382. [DOI] [PubMed] [Google Scholar]
- Gomez A. et al. (2003) Do current sequence analysis algorithms disclose multifunctional (moonlighting) proteins? Bioinformatics, 19, 895–896. [DOI] [PubMed] [Google Scholar]
- Hernández S. et al. (2011) Do moonlighting proteins belong to the intrinsically disordered protein class? Proteomics Bioinformatics., 5, 262–264. [Google Scholar]
- Hernández S. et al. (2014) MultitaskProtDB: a database of multitasking proteins. Nucleic Acids Res., 42, D517–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn H.F., Vousden K.H. (2008) Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene, 27, 5774–5784. [DOI] [PubMed] [Google Scholar]
- Hunter S. et al. (2012) InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res., 40, D306–D312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery C.J. (2003) Moonlighting proteins: old proteins learning new tricks. Trends Genet., 19, 415–417. [DOI] [PubMed] [Google Scholar]
- Jeffery C. (1999) Moonlighting proteins. Trends Biochem. Sci., 24, 8–11. [DOI] [PubMed] [Google Scholar]
- Jeffery C. (2004) Moonlighting proteins: complications and implications for proteomics research. Drug Discov. Today, 3, 71–78. [Google Scholar]
- Kanehisa M., Goto S. (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res., 28, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I. et al. (2014) Genome-scale identification and characterization of moonlighting proteins. Biol. Direct., 9, 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I., Kihara D. (2014) Computational characterization of moonlighting proteins. Biochem. Soc. Trans., 42, 1780–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I. et al. (2012) Evaluation of function predictions by PFP, ESG, and PSI-BLAST for moonlighting proteins. BMC Proc., 6, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liaw A. (2003) Missing Value Imputations by randomForest, R Documentation. Available online at http://rss. acs. unt. edu/Rdoc/library/randomForest/html/rfImpute. html.
- Little R.J.A, Rubin D.B. (1987) Statistical Analysis with Missing Data. John Wiley & Sons. [Google Scholar]
- Mani M. et al. (2014) MoonProt: a database for proteins that are known to moonlight. Nucleic acids research, gku954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin R.L., Raeside D.E. (1981) A reappraisal of distance-weighted k-nearest neighbor classification for pattern recognition with missing data. IEEE Transactions on Systems, Man, and Cybernetics, 3, 241–243. [Google Scholar]
- Oates M.E. et al. (2012) D2P2: Database of Disordered Protein predictions. Nucleic acids research, gks1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura Y. et al. (2014) COXPRESdb in 2015: coexpression database for animal species by DNA-microarray and RNAseq-based expression data with multiple quality assessment systems. Nucleic acids research, gku1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovádi J. (2011) Moonlighting proteins in neurological disorders. IUBMB Life, 63, 453–457. [DOI] [PubMed] [Google Scholar]
- Ozimek P. et al. (2006) Hansenula polymorpha and Saccharomyces cerevisiae Pex5p’s recognize different, independent peroxisomal targeting signals in alcohol oxidase. FEBS Lett., 580, 46–50. [DOI] [PubMed] [Google Scholar]
- Pritykin Y. et al. (2015) Genome-Wide Detection and Analysis of Multifunctional Genes. PLoS Comput. Biol., 11, e1004467.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prunotto M. et al. (2013) Proteomic analysis of podocyte exosome-enriched fraction from normal human urine. J. Proteomics, 82, 193–229. [DOI] [PubMed] [Google Scholar]
- Sampath P. et al. (2004) Noncanonical function of glutamyl-prolyl-tRNA synthetase: gene-specific silencing of translation. Cell, 119, 195–208. [DOI] [PubMed] [Google Scholar]
- Schlicker A. et al. (2006) A new measure for functional similarity of gene products based on Gene Ontology. BMC Bioinformatics, 7, 302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram G. et al. (2005) Single-gene disorders: what role could moonlighting enzymes play? American journal of human genetics., 76, 911–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C. et al. (2006) BioGRID: a general repository for interaction datasets. Nucleic Acids Res., 34, D535–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D. et al. (2014) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic acids research, gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium. (2014) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res., 42, D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma D. et al. (2012) Recruitment of the human Cdt1 replication licensing protein by the loop domain of Hec1 is required for stable kinetochore-microtubule attachment. Nat. Cell. Biol., 14, 593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan F. et al. (2007) Ribosomal protein S3: a KH domain subunit in NF-kappaB complexes that mediates selective gene regulation. Cell, 131, 927–939. [DOI] [PubMed] [Google Scholar]
- Weaver D.T. (1998) Telomeres: moonlighting by DNA repair proteins. Curr. Biol., 8, R492–R494. [DOI] [PubMed] [Google Scholar]
- Zhang S. (2008) Parimputation: From imputation and null-imputation to partially imputation. IEEE Intel. Inform. Bull., 9, 32–38. [Google Scholar]
- Zloba E. (2002) Statistical methods of reproducing of missing data. J. Comp. Model. New Technol., 6, 51–61. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.