ABSTRACT
Glycans or carbohydrates attached to therapeutic glycoproteins can directly affect product quality, safety and efficacy, and therefore must be adequately analyzed and controlled throughout product life cycles. However, the complexity of protein glycosylation poses a daunting analytical challenge. In this study, we evaluated the utility of a lectin microarray for assessing protein glycans. Using commercial lectin chips, which contain 45 lectins toward distinct glycan structures, we were able to determine the lectin binding patterns of a panel of 15 therapeutic proteins, including 8 monoclonal antibodies. Lectin binding signals were analyzed to generate glycan profiles that were generally consistent with the known glycan patterns for these glycoproteins. In particular, the lectin-based microarray was found to be highly sensitive to variations in the terminal carbohydrate structures such as galactose versus sialic acid epitopes. These data suggest that lectin microarray could be used for screening glycan patterns of therapeutic glycoproteins.
KEYWORDS: Glycan analysis, lectin microarray, monoclonal antibodies, therapeutic glycoproteins
Introduction
Glycosylation of proteins is a complex post-translational modification that attaches carbohydrates or named glycans at specific sites on a protein backbone, most commonly at Asn (N-linked) or Ser/Thr (O-linked) residues.1 The N-linked glycosylation occurs at the consensus sequence of Asn-X-Ser/Thr (where X is any amino acid except proline), whereas O-linked glycans are usually attached to Ser or Thr residues. Both N- and O-glycosylation involve a series of enzymatic reactions catalyzed by glycan-processing enzymes, which are responsible for trimming and modifications of glycan epitopes, resulting in diverse N-glycan structures (e.g., high-mannose, complex, and hybrid glycans) and O-glycan variants containing up to 8 O-GalNAc glycan core structures. To add complexity, protein glycosylation is influenced by the type of host cells and fluctuations in fermentation conditions (e.g., media, pH, temperature, agitation).2 For instance, therapeutic glycoproteins produced by mammalian cells such as Chinese hamster ovary (CHO) cells usually contain human-like glycans. By contrast, proteins expressed by yeast strains usually contain high levels of mannose (up to 100 units). Other hosts including engineered plant cells and genetically modified animals may produce proteins with non-human glycan variants such as xylose, N-glycolylneuraminic acid (Neu5Gc) or terminal α-galactose (α-Gal), which are known to be immunogenic. As a result, a glycoprotein produced by living cell systems usually contains a mixture of different glycoforms. These protein variants share an identical peptide backbone, but may differ in glycosylation properties such as glycosylation site, glycan structure and content.
Glycans attached to a therapeutic protein can directly affect product quality, safety and efficacy. It is well documented that glycans attached to a protein affect protein solubility and stability,3-5 pharmacokinetics/pharmacodynamics (PK/PD),3,6-8 and immunogenicity.2,9 In the latter, non-human glycans attached onto a therapeutic protein such as Neu5Gc and terminal α-Gal epitopes could cause immunogenic responses. For many monoclonal antibodies (mAbs), proper glycosylation of the crystallizable fragment (Fc) is essential to IgG antibody effector functions.10 Therefore, glycan moieties of therapeutic proteins must be adequately characterized and controlled throughout product life cycle. The commonly used methods include high-performance liquid chromatography (HPLC), high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD), mass spectrometry (MS) and capillary electrophoresis (CE),11-16 which provide information on glycosylation sites, site occupancy, and contents of glycan variants attached to glycoproteins.
There is growing interest in the development of high throughput platforms for assessing protein glycan profiles. Lectins are glycan binding proteins (GBPs) that selectively recognize glycan epitopes of free carbohydrates or glycoproteins.17 Lectin-based microarrays have been used to analyze glycan profiles of purified glycoproteins or cell surface proteins.18-23 In this study, we evaluated the potential utility of a lectin microarray for characterization of therapeutic glycoproteins. Using commercial lectin chips containing 45 distinct lectins, we tested a panel of 15 therapeutic proteins for their glycan profiles. Our data show that the lectin microarray is robust in generating glycan profiles that are generally consistent with the known glycan characteristics of an individual glycoprotein.
Results
The utility of a lectin microarray in determining glycan profiles of therapeutic mAbs
We assessed the utility of a lectin microarray in profiling glycan variants of therapeutic proteins using commercial lectin microchips printed with 45 distinct lectin proteins (Fig. 1A). These include lectins that selectively bind core-fucose, sialic acids, N-acetyl-D-lactosamine (Galβ1-4GlcNAc), mannose, or N-acetylglucosamine (GlcNAc) oligomers, respectively (Table 1).18 These glycan structures are commonly found in recombinant glycoproteins. We tested a panel of 15 distinct proteins, including 8 therapeutic mAbs, one Fc-fusion protein, 4 recombinant therapeutic cytokines and enzymes, and 2 different versions of human transferrin proteins (Table 2). To facilitate microarray analysis, protein samples were fluorescently labeled with Cy3 followed by incubation onto lectin-coated chips. We first performed dose-titrations and identified the optimal concentrations of Cy3-labeled protein sample that fell within the linear response ranges for most lectin spots (Supplement I). We also tested the effect of incubation time on lectin binding signals, showing that overnight incubation is required to obtain optimal binding between a Cy3-labeled glycoprotein and the lectins printed on chips (Supplement II). In the following experiments, a fixed concentration of protein samples (125 ng/mL) was applied to lectin chips and incubated overnight then lectin binding signals were detected. Bound glycoprotein signals were determined using evanescent-field fluorescence scanner, which allows a direct measure of glycans bound onto lectins without the need of washing procedures to remove unbound species (Fig. 1B). Using lectin chips from different batches, we determined the assay reproducibility to be < 10% CV for most lectin-glycan binding signals of the samples tested.
Figure 1.
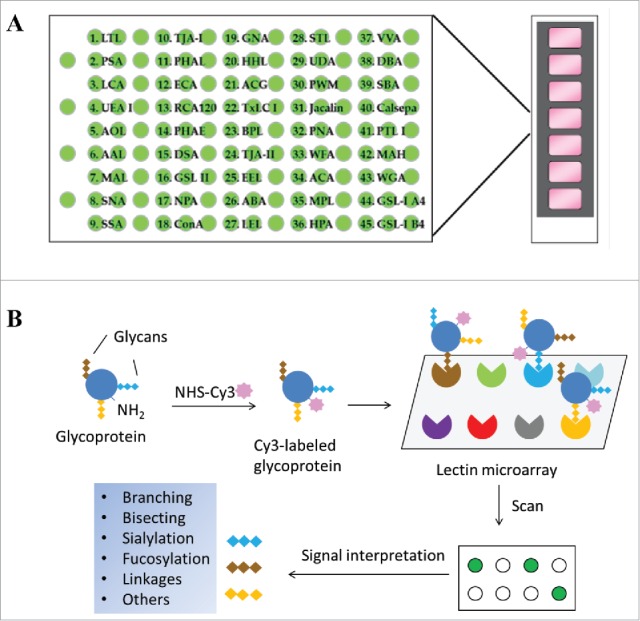
Schematic view of lectin microarray. (A) Lectin microchips used in this study consist of 45 distinct lectins that selectively bind structural variants of carbohydrates attached onto a protein. Each lectin is printed in triplicate. The lectin-printing layout of lectin chips was provided by the vendor (GlycoTechnica). (B) Protein samples are labeled with a fluorescent dye (e.g., Cy3) and then applied onto the lectin chips. The binding signals at each lectin spots are measured using an evanescent-field fluorescence scanner, detecting the presence or absence of glycan variants in the testing sample based on the known selectivity of lectins toward particular glycan structures.
Table 1.
Reported glycan selectivity of the 45 lectins used in the microarray assay.*16
| Lectin No. | Lectin (origin) | Reported glycan selectivity |
|---|---|---|
| 1 | LTL (Lotus tetragonolobus) | Fucα1-3(Galβ1-4)GlcNAc (Lewis x), Fucα1-2Galβ1-4GlcNAc (H-type 2) |
| 2 | PSA (Pisum sativum) | Fucα1-6GlcNAc (Core Fuc) , α-Man |
| 3 | LCA (Lens culinaris) | Fucα1-6GlcNAc (Core Fuc), α-Man |
| 4 | UEA-I (Ulex europaeus) | Fucα1-2Galβ1-4GlcNAc (H-type 2) |
| 5 | AOL (Aspergillus oryzae) | Fucα1-6GlcNAc (Core Fuc), Fucα1-2Galβ1-4GlcNAc (H-type 2) |
| 6 | AAL (Aleuria aurantia) | Fucα1-3(Galβ1-4)GlcNAc (Lewis x), Fucα1-6GlcNAc (Core Fuc) |
| 7 | MAL_I (Maackia amurensis) | Siaα2-3Galβ1-4GlcNAc |
| 8 | SNA (Sambucus nigra) | Siaα2-6Gal/GalNAc |
| 9 | SSA (Sambucus sieboldiana) | Siaα2-6Gal/GalNAc |
| 10 | TJA-I (Trichosanthes japonica) | Siaα2-6Gal/GalNAc, HSO3(-) -6Galβ1-4GlcNAc |
| 11 | PHAL (Phaseolus vulgaris) | tri/tetra-antennary complex-type N-glycan |
| 12 | ECA (Erythrina cristagalli) | Galβ1-4GlcNAc (up with increasing the number of terminal Gal), no affinity for fully sialylated N-type, fully agalactosylated N-type |
| 13 | RCA120 (Ricinus communis) | Galβ1-4GlcNAc (up with increasing the number of terminal Gal), Galβ1-3Gal (weak), no affinity for agalactosylated N-type |
| 14 | PHAE (Phaseolus vulgaris) | bi-antennary complex-type N-glycan with outer Gal and bisecting GlcNAc, no affinity for fully sialylated N-type |
| 15 | DSA (Datura stramonium) | (GlcNAcβ1-4)n, tri/tetra-antennary N-glycan |
| 16 | GSL-II (Griffonia simplicifolia) | agalactosylated tri/tetra antennary glycans, GlcNAc, no affinity for fully galactosylated or sialylated N-type |
| 17 | NPA (Narcissus pseudonarcissus) | High-Mannose including Manα1-6Man |
| 18 | ConA (Canavalia ensiformis) | High-Mannose including Manα1-6(Manα1-3)Man |
| 19 | GNA (Galanthus nivalis) | High-Mannose including Manα1-3Man |
| 20 | HHL (Hippeastrum hybrid) | High-Mannose including Manα1-3Man or Manα1-6Man |
| 21 | ACG (mushroom, Agrocybe cylindracea) | Galβ1-3Gal, Siaα2-3Galβ1-4GlcNAc |
| 22 | TxLCI (Tulipa gesneriana) | Manα1-3(Manα1-6)Man, bi/tri-antennary complex-type N-glycan, GalNAc |
| 23 | BPL (Bauhinia purpurea) | Galβ1-3GalNAc (up with Lewis x, down with Core Fuc), GalNAc |
| 24 | TJA-II (Tanthes japonica) | Fucα1-2Galβ1-> or GalNAcβ1-> groups at their non-reducing terminals |
| 25 | EEL (Euonymus europaeus) | Galα1-3Galβ1-4GlcNAc, Fucα1-2Galβ1-3GlcNAc (H antigen) |
| 26 | ABA (fungus, Agaricus bisporus) | Galβ1-3GalNAc, GlcNAc |
| 27 | LEL (tomato, Lycopersicon esculentum) | (GlcNAcβ1-4)n (Chitin), (Galβ1-4GlcNAc)n (polylactosamine) |
| 28 | STL (potato, Solanum tuberosum) | (GlcNAcβ1-4)n (Chitin), oligosaccharide containing GlcNAc and MurNAc |
| 29 | UDA (Urtica dioica) | GlcNAcβ1-4GlcNAc (Chitin), High-Mannose (3 to High, up with increasing the number of Man) |
| 30 | PWM (pokeweed, Phytolacca Americana) | (GlcNAcβ1-4)n (Chitin) |
| 31 | Jacalin (Artocarpus integrifolia) | GlcNAcβ1-3GalNAc (Core3), Siaα2-3Galβ1-3GalNAc (sialyl T), Galβ1-3GalNAc (T-antigen), α-GalNAc (Tn-antigen) |
| 32 | PNA (peanut, Arachis hypogaea) | Galβ1-3GalNAc |
| 33 | WFA (Wisteria floribunda) | GalNAcβ1-4GlcNAc (LacdiNAc), Galβ1-3(-6)GalNAc |
| 34 | ACA (Amaranthus caudatus) | Galβ1-3GalNAc (T-antigen), Siaα2-3Galβ1-3GalNAc (sialyl T) |
| 35 | MPA (Maclura pomifera) | α-GalNAc (Tn-antigen), Galβ1-3GalNAc (T-antigen), |
| 36 | HPA (snail, Helix pomatia) | α-GalNAc |
| 37 | VVA (Vicia villosa) | GalNAcβ1-4Gal, GalNAcβ1-3Gal, α-GalNAc |
| 38 | DBA (Dolichos biflorus) | Blood group A, GalNAcα1-3GalNAc, GalNAcβ1-4(Siaα2-3)Galβ1-4Glc (GM2) |
| 39 | SBA (soybean, Dolichos biflorus) | α- or β-linked GalNAc, Galα1-4Gal-Glc |
| 40 | Calsepa (Calystegia sepium) | Galactosylated bianntenary N-type with bisecting GlcNAc (galacto > agalacto, down with Core Fuc), High-Mannose (Man2-6) |
| 41 | PTL-I (Psophocarpus tetragonolobus) | α-GalNAc, Galα1-3(Fucα1-2)Gal (B-antigen) |
| 42 | MAH (Maackia amurensis) | Siaα2-3Galβ1-3(Siaα2-6)GalNAc (disialyl-T) |
| 43 | WGA (wheat germ, Triticum aestivum) | (GlcNAcβ1-4)n (Chitin), Hybrid type N-glycan, Sia |
| 44 | GSL-I A4 (Griffonia simplicifolia) | α-Gal, α-GalNAc |
| 45 | GSL-I B4 (Griffonia simplicifolia) | α-Gal, α-GalNAc |
LfDB database http://jcggdb.jp/rcmg/glycodb/LectinSearch
Table 2.
Information of protein samples used in lectin microarray assay.*
| Number | Proprietary name | USAN name | Class | Expression system |
|---|---|---|---|---|
| 1 | Avastin | Bevacizumab | mAb | CHO |
| 2 | Herceptin | Trastuzumab | mAb | CHO |
| 3 | Humira | Adalimumab | mAb | CHO |
| 4 | Remicade | Infliximab | mAb | Sp2/0 |
| 5 | Rituxan | Rituximab | mAb | CHO |
| 6 | Xolair | Omalizumab | mAb | CHO |
| 7 | Vectibix | Panitumumab | mAb | CHO |
| 8 | Erbitux | Cetuximab | mAb | Sp2/0 |
| 9 | Enbrel | Etanercept | Fc-fusion protein | CHO |
| 10 | Aranesp | Darbepoetin alfa | cytokine | CHO |
| 11 | Pulmozyme | Dornase alfa | enzyme | CHO |
| 12 | Elitek | Rasburicase | enzyme | S. cerevisiae |
| 13 | Recombinant human transferrin, expressed in rice | |||
| 14 | Transferrin purified from human blood plasma | |||
| 15 | Neupogen | Filgrastim | cytokine | E. coli |
Abbreviations used in this table: mAb, monoclonal antibody; CHO, Chinese Hamster Ovary cells; Sp2/0, murine myeloma cell line; S. cerevisiae, Saccharomyces cerevisiae; E. coli, Escherichia coli.
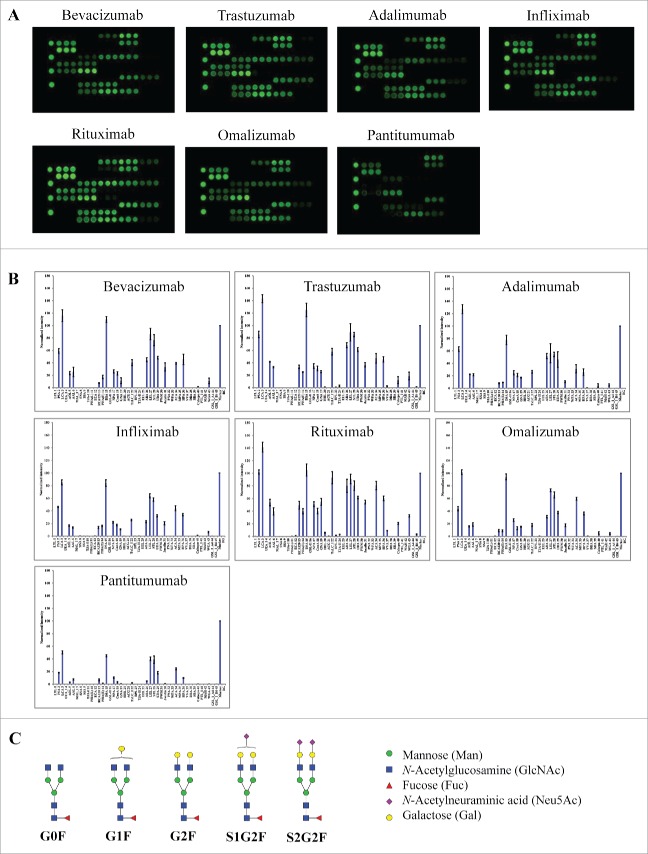
We first tested a panel of 6 therapeutic mAbs, including bevacizumab (Avastin®), trastuzumab (Herceptin®), adalimumab (Humira®), infliximab (Remicade®), rituximab (Rituxan®) and omalizumab (Xolair®). These products are known to be IgG1 isotypes with glycosylation exclusively occurring in the Fc.24-26 All these IgG1 mAbs showed similar lectin binding patterns (Fig. 2A & B) in which strong binding signals were detected at lectins with binding selectivity to core fucose (PSA, LCA, AOL, AAL), galactose (RCA120, PHAE), mannose (NPA, ConA and GNA), and GlcNAc oligomer (LEL, STL, UDA). No binding signals were detected at lectins for sialic acids (MAL_I, SNA, SSA, TJA-I) or tri/tetra-antennary complex-type N-glycan (PHAL). Based on the known selectivity of lectin-glycan interactions (Table 1), the lectin binding patterns indicate the presence of core fucose, outer galactose, mannose, and GlcNAc oligomer, and the absence of sialic acids and tri/tetra-antennary complex-type N-glycans. Overall, the glycan profiles derived from lectin microarray are consistent with the reported glycans in IgG1 Fc that are known to contain principally bi-antennary non-sialylated complex-type N-glycans with little or no high-mannose type or sialylation (Fig. 2C).27 Using the specified lectin chips, we detected similar lectin binding patterns for IgG1 mAbs containing only Fc glycosylation (Fig. 2B). Compared to the other 5 mAbs, rituximab appeared to display relative higher binding signals at AOL/AAL (core fucose), RCA120 (terminal galactose), and GNA (high mannose). Panitumumab (Vectibix®), an IgG2 isotype, displayed much weaker signal intensities across the lectin chip despite a similar lectin binding pattern as observed for IgG1 mAbs (Fig. 2A). This data is consistent with the reported lower level of glycan content in the IgG2 antibodies in comparison with IgG1 antibodies.28,29
Figure 2.
Lectin binding profiles of therapeutic IgG monoclonal antibodies. The indicated therapeutic mAbs, including bevacizumab, trastuzumab, adalimumab, infliximab, rituximab, omalizumab, and panitumumab were labeled with Cy3 and applied onto the lectin chips containing 45 distinct lectins with each being printed in triplicate. (A) Lectin binding images of the indicated samples. (B) Relative binding signals at specific lectin spots were derived from the images in A and normalized to protein markers on the same chip (mean ± SD). Shown are representatives of 3 independent experiments. The coefficient of variation (CV) was determined to be < 10% for most lectin-glycan binding signals of the samples tested. (C) Typical glycan structures present in the Fc portion of therapeutic IgG1 mAbs.27
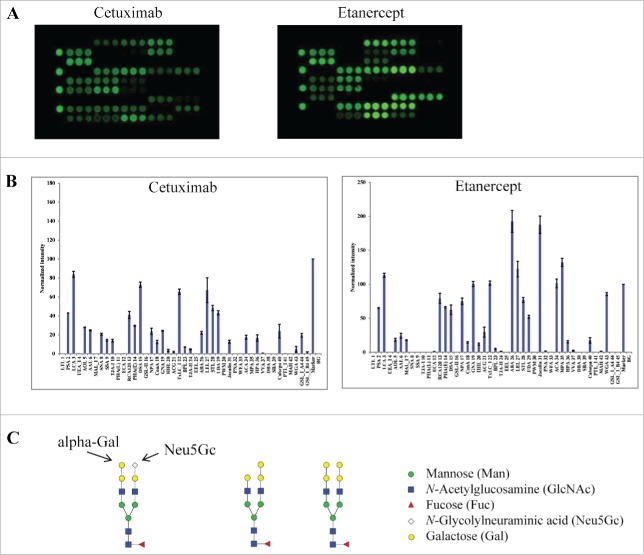
Next, we tested 2 other therapeutic glycoproteins that are associated with more complex glycosylation patterns. Cetuximab (Erbitux®) was chosen because it contains N-glycosylation sites in both the antigen-binding fragment (Fab) and Fc of the molecule.30 In contrast to glycan profiles for IgG1 mAbs (Fig. 2), cetuximab showed unique binding patterns at lectins (SNA, SSA and TJA-I), which are known to bind α2-6-linked sialic acids (Fig. 3A & B). A binding signal was also detected at the α-Gal binding lectin GSL-I-A4,31,32 suggesting the presence of α-Gal structures in cetuximab proteins. Such a glycan variant was not detected in other samples tested in this study. Overall, this data is consistent with the known glycan patterns of cetuximab Fab, which include an abundant N-linked sialic acid (Neu5Gc) and terminal α-Gal variants (Fig. 3C).30
Figure 3.
Lectin binding profiles of cetuximab and etanercept. Cy3-labeled samples were applied onto the lectin chips as in Figure 2. Shown are (A) representative lectin binding images, (B) Relative binding signals at specific lectin spots (mean ± SD), and (C) Typical glycan structures present in the Fab portion of cetuximab.30
Another sample tested was etanercept (Enbrel®), a homodimer of Fc-fusion protein consisting of TNF-α receptor and an IgG1 Fc portion, which was reported to contain 3 N-linked and 13 O-linked glycosylation sites.33 Etanercept displayed distinct lectin binding patterns compared to IgG1 mAbs. For example, strong binding signals were detected at MAL-I and ACG, which are known to selectively bind α2-3 linked sialic acid epitope. No signals were detected at lectins SNA, SSA or TJA-1, showing the lack of α2-6 sialylation in the protein sample.34 This data confirms the presence of complex glycans in etanercept, including α2-3-sialic acids and abundant bi-antennary neutral glycans. Compared to other IgG1 mAbs, etanercept displayed a strong and novel signal at MPA, a lectin that is known to selectively bind Galβ1-3GalNAc and αGalNAc.35 This data is consistent with the reported abundance of O-glycans onto etanercept.33
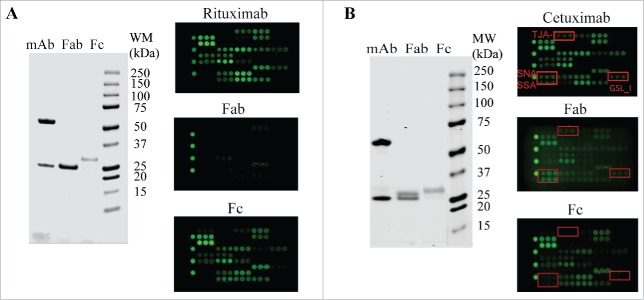
To support the selectivity of lectin binding signals, we tested the Fab and Fc purified from rituximab and cetuximab, respectively. Rituximab is known to contain only one N-glycosylation site in its Fc.26 In the lectin microarray, the isolated rituximab Fab showed little or no signals across the lectin chip whereas the rituximab Fc displayed a similar lectin profile as intact rituximab (Fig. 4A). By contrast, cetuximab contains 2 N-glycans, one located within its Fc portion and another in the Fab.30 The uncommon α-Gal and Neu5Gc epitopes were reported to be solely in the Fab (Fig. 3C).30 The cetuximab Fc expressed a typical lectin profile for IgG1 Fc glycans (bi-antennary G0F, G1F, and G2F) (Fig. 4B). The lectin signals of SNA, SSA and TJA-1, which indicate expression of α2-6-sialylation, were only present in intact cetuximab and cetuximab Fab profiles, but were absent in the Fc profile. GSL-I signal also indicated the presence of α-Gal structure in the Fab, but not in the Fc (Fig. 4B). These data not only confirm the proper locations of glycosylation sites in Fc or Fab portions, but also support the selectivity of lectin bindings toward particular glycans in a testing sample. As noted, the rituximab Cy3-labeled Fab showed no lectin-binding signals across the lectin chips, confirming no or little interaction between the protein backbone and the lectins.
Figure 4.
Glycan profiling of Fabs and Fcs. The Fabs and Fcs of rituximab (A) and cetuximab (B) were prepared as described in Materials and Methods. Purified Fab and Fc were analyzed by reducing SDS-PAGE (left panel) and lectin microarray (right panel). As noted, the dimeric Fcs (˜ 55 kDa) were reduced to monomeric products (˜ 30 kDa) on SDS-PAGE under reducing conditions.
The utility of lectin microarray in glycan profiling of proteins produced by different host cell systems
We assessed whether lectin microarray is capable of profiling glycan variants of therapeutic proteins that are produced by different host cell systems such as mammalian cells, yeast strains and bacterial strains. These cell systems are different in their glycosylation machinery, which produce proteins with distinct glycan patterns. For example, glycoproteins expressed by yeast strains usually contain high-mannose structures whereas Escherichia coli (E. coli) proteins are all non-glycosylated due to the lack of glycosylation machinery in natural bacterial.2 We evaluated a panel of 6 proteins, including 2 therapeutic proteins produced by CHO cells (darbepoetin alfa (Aranesp®) and dornase alfa (Pulmozyme®)), one therapeutic protein produced by yeast (rasburicase (Elitek®)), human transferrin proteins expressed by recombinant rice strain or isolated from human plasma, and filgrastim (Neupogen®) produced by E. coli (Fig. 5A & B). Darbepoetin alfa showed strong signals at MAL-I, demonstrating the presence of α2-3-sialylation structures. Moreover, darbepoetin alfa displayed strong signals at PHAL-coated spots, which are known to be selective for tri-/tetra-antennary N-glycan structures. Such a signal was not detected in other proteins, confirming the absence of tri-/tetra-antennary glycans in these samples. This pattern was consistent with the reported data that darbepoetin alfa contains high levels of sialylation and abundant tri- or tetra-antennary structures (Fig. 5C).36 Darbepoetin displayed relative week binding signals at other lectin spots compared to MAL-I and PHAL, raising a possibility that those other glycan species (e.g., Galβ1-4GlcNAc and mannose oligomers) might be “capped” by the outermost galactose and sialic acid.
Figure 5.
Glycan profiles of proteins produced by different host cell systems. The proteins tested include therapeutic proteins produced by CHO cells (darbepoetin alfa and dornase alfa), yeast strains (rasburicase), or E. coli (filgrastim), and human transferrin protein expressed by recombinant rice (transferrin-rice) or isolated from human plasma (transferrin-human). (A) Lectin binding images. (B) Relative binding signals at specific lectin spots (mean ± SD). (C) Typical glycan structures present in darbepoetin alfa.36
Dornase alfa (Pulmozyme®), a recombinant enzyme expressed by CHO cells, displayed a simpler lectin binding pattern compared to the above IgG1 and IgG2 mAbs (Fig. 5A and 5B). The sample tested showed unique binding signals at SNA/SSA for α2-6-sialylation, RCA120 for Galβ1-4GlcNAc, DSA for GlcNAc oligomer and/or Galβ1-4GlcNAc,37,38 ConA for mannose, and LEL/STL for GlcNAc oligomers. The spectrum of selective binding signals suggests the presence of complex-type glycans with α2-6-sialylation in dornase alfa molecules. By contrast, rasburicase (Elitek®), a therapeutic glycoprotein produced by yeast strains, displayed distinctively different lectin profiles compared to the above described products produced by mammalian cells. Rasburicase showed relatively weak binding signals across the lectin chips, which is consistent with its known low level of glycosylation.39 Despite the overall weak binding signals, rasburicase appeared to interact exclusively with mannose binding lectins (NPA, ConA, and GNA) and GlcNAc oligomer binding lectins (STL and UDA). This data confirms the presence of high-mannose carbohydrates that are mainly attached onto glycoproteins produced by yeast strains.40 No binding signals were detected at sialic acid-binding lectins (e.g., MAL_I, SNA, SSA, and TJA-I), fucose-binding lectins (e.g., PSA and LCA) or galactose-binding lectins (e.g., RCA 120 and PHAE), even when the protein concentration of rasburicase was enhanced to 500 ng/mL (data not shown), demonstrating the absence of the relevant glycan species in rasburicase. The two versions of human transferrin proteins also showed distinct glycan patterns in which the recombinant human transferrin expressed in rice (transferrin-rice) showed binding signals primarily at mannose-binding lectin (NPA) and GlcNAc oligomer-binding lectins (LEL, STL and UDA). The DSA signal indicated the presence of either GlcNAc oligomer or Galβ1-4GlcNAc. By contrast, transferrin proteins isolated from human plasma showed additional signals at α2-6-sialic acid-binding lectins (SNA, SSA, and TJA-I) and galactose-binding lectins (RCA120 and PHAE). As expected, no lectin binding signals were detected for filgrastim (Neupogen®) that is produced by E. coli as a non-glycosylated protein.41
The utility of lectin microarray in monitoring terminal galactosylation and sialylation of glycoproteins
To further evaluate the utility of lectin microarray in glycan profiling, we prepared protein variants with defined galactosylation and sialylation modifications. This was achieved through in vitro enzymatic glycoengineering of rituximab using commercially available galactosyltransferase and sialyltransferase. β1-4-galactosyltransferase (β1-4GalT) catalyzes the transfer of galactose from donor substrate UDP-galactose (UDP-Gal) to GlcNAcβ1-2Man units of glycoproteins to form a β1-4-galactosylation linkage, while α2-6-sialyltransferase (α2-6SiaT) facilitates sialylation by adding sialic acids to terminal Galβ1-4GlcNAc units. Modified rituximab protein variants were purified and then characterized using mass spectrometry (MS), revealing distinct deconvoluted MS spectra for the light chain and heavy chain (Fig. 6A). The light chain fragments resolved as a single species at an average mass of 23036 Da, corresponding to the theoretical mass of rituximab light chain.42,43 Consistent with the lack of glycosylation sites within the rituximab light chains, the mass of light chain remained unchanged after treatments of rituximab with β1-4GalT or further with α2-6SiaT. The other 3 major mass species at 50507, 50669, 50832 Da correspond to the heavy chains of rituximab containing G0F, G1F or G2F glycoforms, respectively (Fig. 2C).42,43 Treatment of rituximab with β1-4GalT resulted in a mass shift from G0F and G1F to G2F, indicating galactosylation reactions were effectively accomplished. When the galactosylated rituximab mixture was sequentially treated with α2-6SiaT, the final product showed a further mass shift from 50832 Da (G2F) to 51122 Da (+290) and 51414 Da (+582). These mass shifts corresponded to an addition of one or two Neu5Ac residues, indicating that G2F glycoform was effectively converted to primarily mono-sialylated species (S1G2F) and a small portion of di-sialylated species (S2G2F). These data indicate that the in vitro glycan engineering reactions produced rituximab variants containing the desired modifications (e.g., galactosylation and sialylation).
Figure 6.
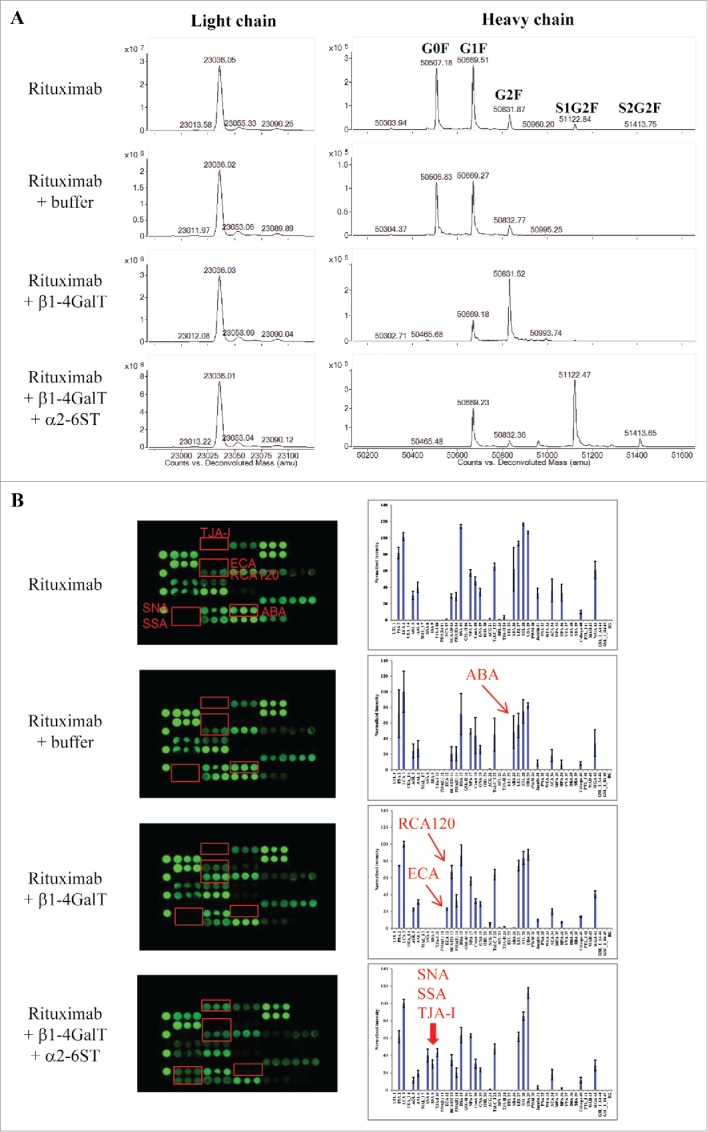
Assessing glycan variants in glycan-engineered rituximab protein samples. Rituximab was incubated with a reaction buffer alone (rituximab + buffer), β1-4-galactosyltransferase (rituximab + β1-4GalT), and β1-4-galactosyltransferase followed by α2-6-sialyltransferase (rituximab + β1-4GalT + α2-6SiaT) (see detail in Materials and Methods). After affinity purification, the resulting samples were analyzed using mass spectrometry and lectin microarray, respectively. Shown are representatives of deconvoluted mass spectra (A) and corresponding lectin binding profiles (B) for the samples produced under the indicated conditions.
The engineered rituximab samples with defined glycan variations were then analyzed by lectin microarray (Fig. 6B). The reaction buffer alone had no effect on lectin binding profile of rituximab. The sample resulting from β1-4GalT reaction (rituximab + β1-4GalT), which was confirmed by MS to majorly contain G2F galactosylation (Fig. 6A), displayed strong binding signals at positions of ECA and RCA120. Both lectins are known to bind N-glycan Galβ1-4GlcNAc, and therefore the incurred ECA lectin-binding signal and the significantly enhanced RCA120 signal were indicative of the increased galactose species in the samples. Notably, there was a concomitant decrease in the binding signals at ABA, a lectin with dual binding affinity toward Gal-exposed O-glycans and GlcNAc-exposed N-glycans.44 Because rituximab was not reported to undergo O-glycosylation, the ABA binding signal detected for native rituximab was likely due to GlcNAc-exposed N-glycans. Such GlcNAc-exposed N-glycans appeared to be occupied upon galactosylation catalyzed in the reactions with β1-4GalT. Samples of native rituximab and galactosylated species showed no detectable signals at the lectins SNA, SSA, TJA-I that are known to bind sialic acids. By contrast, strong binding signals at these lectins were detected for the samples derived from the sequential reactions with β1-4GalT and α2-6SiaT enzymes. No signal was detected at MAL-I (a lectin selective to α2-3-sialylation), demonstrating not only the specificity of glycan engineering but also the utility of the lectin microarray in distinguishing different terminal sialic acid linkages.
Discussion
The complexity of glycosylation poses an analytical challenge in the development of therapeutic glycoproteins. The methods most commonly used for analysis include MS, HPLC, HPAEC-PAD, and CE. Among these methods, MS remains a powerful tool in the characterization of glycosylation site(s) occupancy and carbohydrate structures. MS-based methods involve enzymatic digestion of a glycoprotein into peptide fragments and separation by liquid chromatography. HPLC-, HPAEC- and CE-based methods usually require the release of glycans from a glycoprotein through enzymatic or chemical reactions. As such, an accurate assessment of glycosylation requires a complete release of all glycans that are present in a glycoprotein being tested. By contrast, lectin microarray directly measures glycan profiles on an intact protein without the need for enzymatic digestion or clipping glycans from the protein backbone. Such a platform is unique in increasing the likelihood of full coverage of all glycan variants of a glycoprotein. Using the commercial lectin chips, we were able to determine glycan profiles for a panel of therapeutic proteins that were generally consistent with their known glycosylation properties (Figs. 2-5). Notably the lectin microarray was highly sensitive to alterations in the terminal glycan structures, i.e., galactosylation vs. sialyation (Fig. 6). Lectin microarray can effectively distinguish glycan isomers containing different sialic acid linkages. Our data demonstrate a usefulness of the lectin microarray in screening glycan patterns of protein samples. As noted, the commercial lectin chips employed in this study was not designed specifically for assessing therapeutic mAbs. In principle, assay performance could be further improved through the use of lectins with improved selectivity and binding affinity to distinct glycan species. To analyze a specific glycoprotein, lectin chips can be customized to include the lectins that are relevant to the glycan species that are possibly present in the testing sample. Upon optimization of lectin chips, lectin microarray platform could be adopted as a complementary tool for high throughput screening of glycan profiles of therapeutic glycoproteins.
Materials and methods
Therapeutic proteins and reagents
All therapeutic proteins were purchased from the Division of Veterinary Resources pharmacy, Office of Research Services, National Institute of Health, and handled according to respective package inserts. Recombinant human transferrin expressed by rice and transferrin purified from human blood plasma were purchased from Sigma-Aldrich. Cy3 mono-reactive dye was obtained from GE Healthcare. Zeba spin desalting columns (7K MWCO) were purchased from Thermo Scientific. Lectin chips coated with 45 distinct lectin proteins and the probing solutions were obtained from GlycoTechnica. Glycosyltransferases β1-4-galactosyltransferase (β1-4GalT) and α2-6-sialyltransferase (α2-6SiaT) were purchased from Prozyme. Micro BCA Protein Assay Kit, NAb Protein A Plus Spin Kit and Pierce Fab micro preparation kit were obtained from Thermo Scientific. NuPAGE 4-12% Bis-Tris gels and Coomassie stain (SimplyBlue SafeStain) were purchased from Life Technologies.
Lectin microarray analysis
Protein samples were diluted to 50 µg/mL in phosphate-buffered saline (PBS). Aliquots (20 µL, 1 µg protein) were mixed with 100 µg Cy3 mono-reactive dye, and incubated at room temperature for 2 h. The unbound Cy3 dye was removed using Zeba spin desalting columns (7K MWCO). Cy3-labeled protein samples were diluted to 125 ng/mL in probing solution, and an aliquot of 100 µL was applied onto a lectin chip, and incubated at room temperature on an orbital shaker for 18 hours in dark. The resulting lectin chips were scanned for fluorescence intensities on each lectin-coated spot using an evanescent-field fluorescence scanner GlycoStation Reader 1200 (GlycoTechnica, Japan).
Preparation of IgG Fab and Fc
The Fab and Fc of therapeutic mAbs were prepared using Pierce Fab micro preparation kit following the vendor's protocol (Thermo Scientific). Briefly, immobilized papain (settled resin) was located in a 0.8 mL spin column and equilibrated using Fab digestion buffer. Antibody samples were diluted to 0.4 mg/mL using Fab digestion buffer, and aliquot of 120 µL was added to the spin column containing equilibrated immobilized papain. The digestion reaction was incubated overnight at 37°C. Digested samples were collected by column centrifugation and applied onto NAb Protein A Plus Spin Column to separate the Fab and Fc. Protein concentrations of the resulting samples were determined using Micro BCA Protein Assay Kit (Thermo Scientific).
Enzymatic glycan engineering of rituximab
Rituximab was used to create glycan variants of a mAb through in vitro enzymatic reactions. Briefly, rituximab stock solution was diluted to 2 µg/µL using 1× galactosyltransferase (GalT) reaction buffer (10 mM MnCl2 with 100 mM MES, pH 6.5). Uridine-5’-diphosphogalactose disodium salt (UDP-Gal) solution (50 µg/µL) was also prepared using 1× GalT reaction buffer. Cytidine 5’-monophospho-N-acetylneuraminic acid disodium salt (CMP-Neu5Ac) solution (50 µg/µL) was prepared using 1× sialyltransferase (SiaT) reaction buffer (50 mM Tris-Acetate, pH 7.5). To facilitate galactosylation, 50 µL rituximab (2 µg/µL in solution) was mixed with 4 µL UDP-Gal (50 µg/µL in solution) and 1.5 µg β1-4-galactosyltransferase (β1-4GalT). The final reaction volume was adjusted to 100 µL using 1× GalT reaction buffer, and was left to incubate overnight at 37°C, resulting galactosylation of rituximab. To further add sialylation, the above galactosylated rituximab was mixed with 200 µg CMP-Neu5Ac (50 µg/µL in solution) and 10 µg α2-6-sialyltransferase (α2-6SiaT). After adjusting volume to 200 µL using 1× SiaT reaction buffer, the mixture was incubated at 37°C for 3 hours and then frozen at −20°C. The engineered antibodies containing different glycan variants were purified using NAb Protein A Plus Spin Kit by following the protocol instructions (Thermo Scientific).
Liquid chromatography–mass spectrometry analysis
Liquid chromatography–mass spectrometry analyses were conducted on an Agilent 1260 HPLC-Chip nano-electrospray-ionization 6520 Q-TOF MS system. Solvent-A was 0.1% formic acid in water and solvent-B was 0.1% formic acid in 95% acetonitrile. Mass correction was enabled during the run using internal reference ions with masses of 299.2945 and 1221.9906 Da. Intact protein mass measurement was performed using an Agilent 43 mm 300 Å C8 chip with a 40 nL trap column (G4240-63001 SPQ105). All rituximab samples were reduced in 10 mM DTT for 1 hour at 37°C, and then formic acid was added to reach a final concentration of 0.1% (V/V). The samples were diluted to 150 ng/μL in 0.1% formic acid, and 1 µL (˜1 pmol) or larger volume was injected onto the trap column in the C8 chip at a flow rate of 2.5 µL/min in 100% A, then eluted at 0.5 µL/min with a linear gradient of 10–100% B in 18 min and held for additional 4 min. Q-TOF VCap, fragmentor, and skimmer settings were 1,890 eV, 225 eV, and 65 eV, respectively. HPLC-Chip gas temperature and drying gas flow rate were 350°C and 9 L/min, respectively. The data were analyzed using Agilent MassHunter (version B.05.00) Qualitative Analysis software and deconvoluted with Agilent MassHunter Bioconfirm software.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem 1985; 54:631-64; PMID:3896128; http://dx.doi.org/ 10.1146/annurev.bi.54.070185.003215 [DOI] [PubMed] [Google Scholar]
- 2.Ghaderi D, Zhang M, Hurtado-Ziola N, Varki A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnol Genet Eng Rev 2012; 28:147-75; PMID:22616486; http://dx.doi.org/ 10.5661/bger-28-147 [DOI] [PubMed] [Google Scholar]
- 3.Sola RJ, Griebenow K. Glycosylation of therapeutic proteins: an effective strategy to optimize efficacy. BioDrugs 2010; 24:9-21; PMID:20055529; http://dx.doi.org/ 10.2165/11530550-000000000-00000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J Pharm Sci 2009; 98:1223-45; PMID:18661536; http://dx.doi.org/ 10.1002/jps.21504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narhi LO, Arakawa T, Aoki KH, Elmore R, Rohde MF, Boone T, Strickland TW. The effect of carbohydrate on the structure and stability of erythropoietin. J Biol Chem 1991; 266:23022-6; PMID:1744097 [PubMed] [Google Scholar]
- 6.Desnick RJ, Schuchman EH. Enzyme replacement therapy for lysosomal diseases: lessons from 20 years of experience and remaining challenges. Annu Rev Genomics Hum Genet 2012; 13:307-35; PMID:22970722; http://dx.doi.org/ 10.1146/annurev-genom-090711-163739 [DOI] [PubMed] [Google Scholar]
- 7.Tsuda E, Kawanishi G, Ueda M, Masuda S, Sasaki R. The role of carbohydrate in recombinant human erythropoietin. Eur J Biochem 1990; 188:405-11; PMID:2156701; http://dx.doi.org/ 10.1111/j.1432-1033.1990.tb15417.x [DOI] [PubMed] [Google Scholar]
- 8.Morell AG, Gregoriadis G, Scheinberg IH, Hickman J, Ashwell G. The role of sialic acid in determining the survival of glycoproteins in the circulation. J Biol Chem 1971; 246:1461-7; PMID:5545089 [PubMed] [Google Scholar]
- 9.Houdebine LM. Production of pharmaceutical proteins by transgenic animals. Comp Immunol Microbiol Infect Dis 2009; 32:107-21; PMID:18243312; http://dx.doi.org/ 10.1016/j.cimid.2007.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimmerjahn F, Ravetch JV. Translating basic mechanisms of IgG effector activity into next generation cancer therapies. Cancer Immun 2012; 12:13; PMID:22896758 [PMC free article] [PubMed] [Google Scholar]
- 11.Marino K, Bones J, Kattla JJ, Rudd PM. A systematic approach to protein glycosylation analysis: a path through the maze. Nat Chem Biol 2010; 6:713-23; PMID:20852609; http://dx.doi.org/ 10.1038/nchembio.437 [DOI] [PubMed] [Google Scholar]
- 12.Beck A, Wagner-Rousset E, Ayoub D, Van DA, Sanglier-Cianferani S. Characterization of therapeutic antibodies and related products. Anal Chem 2013; 85:715-36; PMID:23134362; http://dx.doi.org/ 10.1021/ac3032355 [DOI] [PubMed] [Google Scholar]
- 13.Higgins E. Carbohydrate analysis throughout the development of a protein therapeutic. Glycoconj J 2010; 27:211-25; PMID:19888650; http://dx.doi.org/ 10.1007/s10719-009-9261-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lingg N, Zhang P, Song Z, Bardor M. The sweet tooth of biopharmaceuticals: importance of recombinant protein glycosylation analysis. Biotechnol J 2012; 7:1462-72; PMID:22829536; http://dx.doi.org/ 10.1002/biot.201200078 [DOI] [PubMed] [Google Scholar]
- 15.Pabst M, Altmann F. Glycan analysis by modern instrumental methods. Proteomics 2011; 11:631-43; PMID:21241022; http://dx.doi.org/ 10.1002/pmic.201000517 [DOI] [PubMed] [Google Scholar]
- 16.Rohrer JS, Basumallick L, Hurum D. High-performance anion-exchange chromatography with pulsed amperometric detection for carbohydrate analysis of glycoproteins. Biochemistry (Mosc ) 2013; 78:697-709; PMID:24010833; http://dx.doi.org/ 10.1134/S000629791307002X [DOI] [PubMed] [Google Scholar]
- 17.Varki A, Etzler ME, Cummings RD, Esko JD. Discovery and Classification of Glycan-Binding Proteins. In: Varki A, Cummings RD, Esko JD, et al. Essentials of Glycobiology. 2nd edition. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 2009. Chapter 26 [PubMed] [Google Scholar]
- 18.Hirabayashi J, Yamada M, Kuno A, Tateno H. Lectin microarrays: concept, principle and applications. Chem Soc Rev 2013; 42:4443-58; PMID:23443201; http://dx.doi.org/ 10.1039/c3cs35419a [DOI] [PubMed] [Google Scholar]
- 19.Cook MC, Kaldas SJ, Muradia G, Rosu-Myles M, Kunkel JP. Comparison of orthogonal chromatographic and lectin-affinity microarray methods for glycan profiling of a therapeutic monoclonal antibody. J Chromatogr B 2015; 997:162-78; PMID:26114652; http://dx.doi.org/ 10.1016/j.jchromb.2015.05.035 [DOI] [PubMed] [Google Scholar]
- 20.Huang WL, Li YG, Lv YC, Guan XH, Ji HF, Chi BR. Use of lectin microarray to differentiate gastric cancer from gastric ulcer. World J Gastroenterol 2014; 20:5474-82; PMID:24833877; http://dx.doi.org/ 10.3748/wjg.v20.i18.5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi Y, Masuda K, Banno K, Kobayashi N, Umene K, Nogami Y, Tsuji K, Ueki A, Nomura H, Sato K, et al.. Glycan profiling of gestational choriocarcinoma using a lectin microarray. Oncol Rep 2014; 31:1121-6; PMID:24424471; http://dx.doi.org/10.3892/or.2014.2979 [DOI] [PubMed] [Google Scholar]
- 22.Xin AJ, Cheng L, Diao H, Wang P, Gu YH, Wu B, Wu YC, Chen GW, Zhou SM, Guo SJ, et al.. Comprehensive profiling of accessible surface glycans of mammalian sperm using a lectin microarray. Clin Proteomics 2014; 11:10; PMID:24629138; http://dx.doi.org/ 10.1186/1559-0275-11-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta G, Surolia A, Sampathkumar SG. Lectin microarrays for glycomic analysis. OMICS 2010; 14:419-36; PMID:20726799; http://dx.doi.org/ 10.1089/omi.2009.0150 [DOI] [PubMed] [Google Scholar]
- 24.Song T, Ozcan S, Becker A, Lebrilla CB. In-depth method for the characterization of glycosylation in manufactured recombinant monoclonal antibody drugs. Anal Chem 2014; 86:5661-6; PMID:24828102; http://dx.doi.org/ 10.1021/ac501102t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stadlmann J, Pabst M, Kolarich D, Kunert R, Altmann F. Analysis of immunoglobulin glycosylation by LC-ESI-MS of glycopeptides and oligosaccharides. Proteomics 2008; 8:2858-71; PMID:18655055; http://dx.doi.org/ 10.1002/pmic.200700968 [DOI] [PubMed] [Google Scholar]
- 26.Visser J, Feuerstein I, Stangler T, Schmiederer T, Fritsch C, Schiestl M. Physicochemical and functional comparability between the proposed biosimilar rituximab GP2013 and originator rituximab. BioDrugs 2013; 27:495-507; PMID:23649935; http://dx.doi.org/ 10.1007/s40259-013-0036-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huhn C, Selman MH, Ruhaak LR, Deelder AM, Wuhrer M. IgG glycosylation analysis. Proteomics 2009; 9:882-913; PMID:19212958; http://dx.doi.org/ 10.1002/pmic.200800715 [DOI] [PubMed] [Google Scholar]
- 28.Wuhrer M, Stam JC, van de Geijn FE, Koeleman CA, Verrips CT, Dolhain RJ, Hokke CH, Deelder AM. Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 2007; 7:4070-81; PMID:17994628; http://dx.doi.org/ 10.1002/pmic.200700289 [DOI] [PubMed] [Google Scholar]
- 29.Perdivara I, Peddada SD, Miller FW, Tomer KB, Deterding LJ. Mass spectrometric determination of IgG subclass-specific glycosylation profiles in siblings discordant for myositis syndromes. J Proteome Res 2011; 10:2969-78; PMID:21609021; http://dx.doi.org/ 10.1021/pr200397h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian J, Liu T, Yang L, Daus A, Crowley R, Zhou Q. Structural characterization of N-linked oligosaccharides on monoclonal antibody cetuximab by the combination of orthogonal matrix-assisted laser desorption/ionization hybrid quadrupole-quadrupole time-of-flight tandem mass spectrometry and sequential enzymatic digestion. Anal Biochem 2007; 364:8-18; PMID:17362871; http://dx.doi.org/ 10.1016/j.ab.2007.01.023 [DOI] [PubMed] [Google Scholar]
- 31.Murphy LA, Goldstein IJ. Five alpha-D-galactopyranosyl-binding isolectins from Bandeiraea simplicifolia seeds. J Biol Chem 1977; 252:4739-42; PMID:68957 [PubMed] [Google Scholar]
- 32.Lescar J, Loris R, Mitchell E, Gautier C, Chazalet V, Cox V, Wyns L, Perez S, Breton C, Imberty A. Isolectins I-A and I-B of Griffonia (Bandeiraea) simplicifolia. Crystal structure of metal-free GS I-B(4) and molecular basis for metal binding and monosaccharide specificity. J Biol Chem 2002; 277:6608-14; PMID:11714720; http://dx.doi.org/ 10.1074/jbc.M109867200 [DOI] [PubMed] [Google Scholar]
- 33.Tan Q, Guo Q, Fang C, Wang C, Li B, Wang H, Li J, Guo Y. Characterization and comparison of commercially available TNF receptor 2-Fc fusion protein products. MAbs 2012; 4:761-74; PMID:23032066; http://dx.doi.org/ 10.4161/mabs.22276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houel S, Hilliard M, Yu YQ, McLoughlin N, Martin SM, Rudd PM, Williams JP, Chen W. N- and O-glycosylation analysis of etanercept using liquid chromatography and quadrupole time-of-flight mass spectrometry equipped with electron-transfer dissociation functionality. Anal Chem 2014; 86:576-84; PMID:24308717; http://dx.doi.org/ 10.1021/ac402726h [DOI] [PubMed] [Google Scholar]
- 35.Huang J, Xu Z, Wang D, Ogata CM, Palczewski K, Lee X, Young NM. Characterization of the secondary binding sites of Maclura pomifera agglutinin by glycan array and crystallographic analyses. Glycobiology 2010; 20:1643-53; PMID:20826825; http://dx.doi.org/ 10.1093/glycob/cwq118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shahrokh Z, Royle L, Saldova R, Bones J, Abrahams JL, Artemenko NV, Flatman S, Davies M, Baycroft A, Sehgal S, et al.. Erythropoietin produced in a human cell line (Dynepo) has significant differences in glycosylation compared with erythropoietins produced in CHO cell lines. Mol Pharm 2011; 8:286-96; PMID:21138277; http://dx.doi.org/ 10.1021/mp100353a [DOI] [PubMed] [Google Scholar]
- 37.Crowley JF, Goldstein IJ, Arnarp J, Lonngren J. Carbohydrate binding studies on the lectin from Datura stramonium seeds. Arch Biochem Biophys 1984; 231:524-33; PMID:6203486; http://dx.doi.org/ 10.1016/0003-9861(84)90417-X [DOI] [PubMed] [Google Scholar]
- 38.Myllyharju J, Nokkala S. Glycoproteins with N-acetylglucosamine and mannose residues in Chinese hamster metaphase chromosomes. Hereditas 1996; 124:251-9; PMID:8931358; http://dx.doi.org/ 10.1111/j.1601-5223.1996.00251.x [DOI] [PubMed] [Google Scholar]
- 39.Higgins E. Carbohydrate analysis throughout the development of a protein therapeutic. Glycoconj J 2010; 27:211-25; PMID:19888650; http://dx.doi.org/ 10.1007/s10719-009-9261-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hossler P, Khattak SF, Li ZJ. Optimal and consistent protein glycosylation in mammalian cell culture. Glycobiology 2009; 19:936-49; PMID:19494347; http://dx.doi.org/ 10.1093/glycob/cwp079 [DOI] [PubMed] [Google Scholar]
- 41.Jaffe SR, Strutton B, Levarski Z, Pandhal J, Wright PC. Escherichia coli as a glycoprotein production host: recent developments and challenges. Curr Opin Biotechnol 2014; 30:205-10; PMID:25156401; http://dx.doi.org/ 10.1016/j.copbio.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 42.Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. Chemoenzymatic glycoengineering of intact IgG antibodies for gain of functions. J Am Chem Soc 2012; 134:12308-18; PMID:22747414; http://dx.doi.org/ 10.1021/ja3051266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Gucinski AC, Keire DA, Buhse LF, Boyne MT. Structural comparison of two anti-CD20 monoclonal antibody drug products using middle-down mass spectrometry. Analyst 2013; 138:3058-65; PMID:23579346; http://dx.doi.org/ 10.1039/c3an36524g [DOI] [PubMed] [Google Scholar]
- 44.Nakamura-Tsuruta S, Kominami J, Kuno A, Hirabayashi J. Evidence that Agaricus bisporus agglutinin (ABA) has dual sugar-binding specificity. Biochem Biophys Res Commun 2006; 347:215-20; PMID:16824489; http://dx.doi.org/ 10.1016/j.bbrc.2006.06.073 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.