Although significant progress has been made in understanding the mechanisms of nuclear movement, the functions of nuclear movement have proven more elusive. Properly positioned nuclei are necessary to build a functional sarcomere network during muscle development.
Abstract
Two defining characteristics of muscle cells are the many precisely positioned nuclei and the linearly arranged sarcomeres, yet the relationship between these two features is not known. We show that nuclear positioning precedes sarcomere formation. Furthermore, ZASP-GFP, a Z-line protein, colocalizes with F-actin in puncta at the cytoplasmic face of nuclei before sarcomere assembly. In embryos with mispositioned nuclei, ZASP-GFP is still recruited to the nuclei before its incorporation into sarcomeres. Furthermore, the first sarcomeres appear in positions close to the nuclei, regardless of nuclear position. These data suggest that the interaction between sarcomere proteins and nuclei is not dependent on properly positioned nuclei. Mechanistically, ZASP-GFP localization to the cytoplasmic face of the nucleus did require the linker of nucleoskeleton and cytoskeleton (LINC) complex. Muscle-specific depletion of klarsicht (nesprin) or klariod (SUN) blocked the recruitment of ZASP-GFP to the nucleus during the early stages of sarcomere assembly. As a result, sarcomeres were poorly formed and the general myofibril network was less stable, incomplete, and/or torn. These data suggest that the nucleus, through the LINC complex, is crucial for the proper assembly and stability of the sarcomere network.
INTRODUCTION
The myofiber—the syncytial cellular unit of muscle—forms from the continual fusion of mononucleted myoblasts (Capers, 1960). Each newly added nucleus is moved to the center of the myofiber (Cadot et al., 2012) and then to the periphery of the cell to maximize the distance between nuclei (Bruusgaard et al., 2004). Although the mechanisms of these movements are emerging and are conserved between Drosophila and mammals (Capers, 1960; Cadot et al., 2012; Folker et al., 2012, 2014; Metzger et al., 2012; Wilson and Holzbaur, 2012, 2015), how nuclear movement benefits the muscle is not known.
Another highly organized structure within the myofiber is the myofibril—a linear arrangement of sarcomeres that extends throughout the cell. Sarcomeres are composed of bipolar myosin II filaments (thick filaments) and pairs of associated actin filaments (thin filaments) flanked by a Z-disk. The Z-disk is composed of many proteins, including α-actinin (Puszkin et al., 1977), titin (Furst et al., 1988), nebulin (Pierobon-Bormioli et al., 1989), and Z-band alternatively spliced PDZ-motif protein (ZASP; Faulkner et al., 1999), that stabilize and anchor the barbed ends of the thin filaments (Huxley, 1957). The Z-disk is vital to sarcomere assembly, integrity, and contraction (Rui et al., 2010; Katzemich et al., 2013; Fernandes and Schöck, 2014; Lin et al., 2014).
Although the structure–function relationship of the sarcomere is established, sarcomere assembly is poorly understood. Current models suggest that the sarcomere develops from preassembled complexes of sarcomere proteins, but the nature of the complexes is not clear. Early sarcomeric complexes are believed to be either premyofibril structures formed in an integrin-dependent manner or latent complexes that appear uniformly throughout the muscle cell (Rhee et al., 1994; Rui et al., 2010; Weitkunat et al., 2014). The latter hypothesis is supported by the presence of puncta containing α-actinin and F-actin throughout developing muscles (Rui et al., 2010), termed I-Z-I complexes, which are precursors to sarcomeres (Holtzer et al., 1997; Golson et al., 2004). Furthermore, sarcomeres develop rapidly throughout the entire muscle (Weitkunat et al., 2014), suggesting that preformed assemblies exist throughout the muscle. The mechanism of latent complex assembly, however, is not known.
Here we demonstrate that the nucleus contributes to sarcomere assembly. ZASP, a Z-line component, and F-actin colocalize at the cytoplasmic face of nuclei before they are incorporated into myofibrils. Furthermore, when nuclear position is disrupted, the site of initial sarcomere assembly is similarly disrupted and is colocalized with nuclei. Finally, the linker of nucleoskeleton and cytoskeleton (LINC) complex is crucial to nucleus-mediated sarcomere assembly and overall myofibril integrity. Together these data link two defining characteristics of the muscle cell and provide a purpose for the movement of nuclei.
RESULTS AND DISCUSSION
Nuclear positioning precedes sarcomere assembly
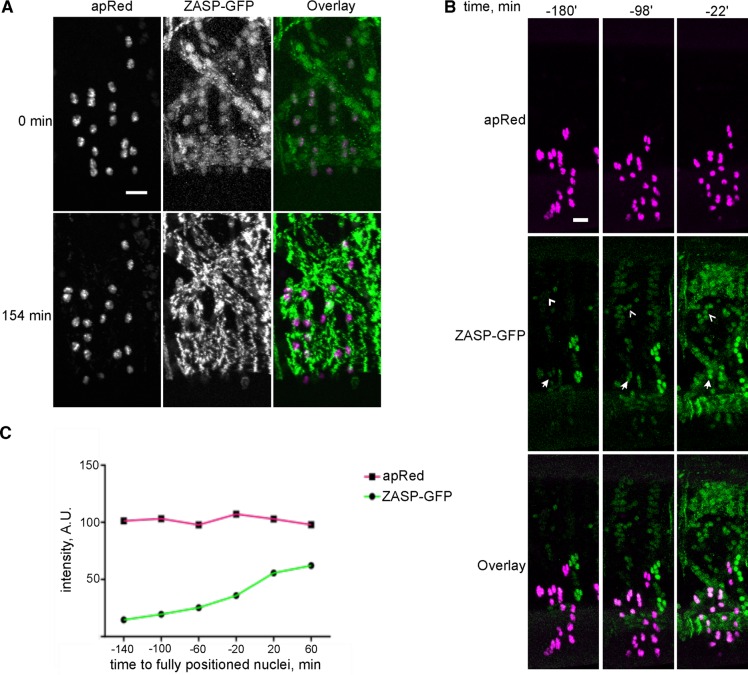
In Drosophila and mammals, myonuclei undergo a series of movements that leaves them spaced throughout the muscle fiber (Cadot et al., 2012; Metzger et al., 2012; Wilson and Holzbaur, 2012; Folker et al., 2014). However, the effect of that nuclear movement on muscle development is not known. To understand the spatiotemporal relationship between nuclear positioning and sarcomere assembly, we coimaged the myonuclei in the lateral transverse (LT) muscles of the Drosophila embryo and the assembly of sarcomeres. The nuclei of the LT muscles were labeled by the apRed reporter (Richardson et al., 2007), which is inserted into the genome that codes for a nuclear localization signal fused to the DsRed fluorescent protein. This reporter is downstream of the mesodermal enhancer for apterous, a transcription factor that is expressed only in the four lateral transverse muscles of the Drosophila embryo. Thus the nuclei of these four muscles can be monitored by live-embryo confocal microscopy (Folker et al., 2014). Sarcomeres were labeled with ZASP–green fluorescent protein (GFP), a Z-line component (Faulkner et al., 1999), that functions with α-actinin to stabilize F-actin within the sarcomere (Katzemich et al., 2013; Lin et al., 2014). ZASP-GFP results from the insertion of GFP into the endogenous ZASP coding locus and does not rely on overexpression of the reporter construct (Hudson et al., 2008; Katzemich et al., 2013). Nuclei in the LT muscles achieved their final position before the appearance of a regular sarcomere network (Figure 1A), indicating that the movement of nuclei is an early event in muscle cell development that can affect many aspects of muscle development, including sarcomere assembly.
FIGURE 1:
ZASP colocalizes with nuclei before sarcomere assembly. (A) The LT muscles in the Drosophila embryo from a time-lapse acquisition of ZASP-GFP (green) and NLS-DsRed (apRed; magenta). Here 0 min is the time at which nuclear movement is completed in the LT muscles. Defined Z-lines appear 154 min later. Scale bar, 10 μm. (B) Single hemisegment from an embryo that expressed ZASP-GFP (green) and apRed (magenta). Arrows indicate ZASP-GFP localized to apRed nuclei in the LT muscles. Arrowheads indicate ZASP-GFP that is colocalized with nuclei in the DO2 muscle, which does not express apRed. Scale bar, 10 μm. (C) Graph of the fluorescence intensity of apRed (magenta) and ZASP-GFP (green) on a single nucleus over time. Examination of many nuclei from different muscles produced similar results, although the precise levels varied.
The Z-line protein ZASP colocalizes with myonuclei
In LT muscles, ZASP-GFP accumulated in disks that colocalized with apRed, indicating that ZASP-GFP colocalized with nuclei before sarcomere assembly (Figure 1, A and B, and Supplemental Movie S1). Furthermore, the fluorescence intensity of ZASP-GFP that colocalized with nuclei increased over time, whereas the fluorescence intensity of the apRed was constant, suggesting that newly expressed ZASP-GFP was recruited to the nucleus before sarcomere assembly (Figure 1C). Further analysis revealed that before the appearance of sarcomeres, ZASP-GFP localized into disks in each muscle, including muscles that did not express apRed. These disks were the same size and shape as the nuclei and moved similarly to them, indicating that this localization pattern is not a consequence of apRed expression (Figure 1B). ZASP-GFP fluorescence that colocalized with nuclei increased earlier and more rapidly than the ZASP-GFP fluorescence throughout the muscle (Figure 2, A and B, and Supplemental Movie S1), suggesting that the colocalization with nuclei is not merely a consequence of increased ZASP levels.
FIGURE 2:
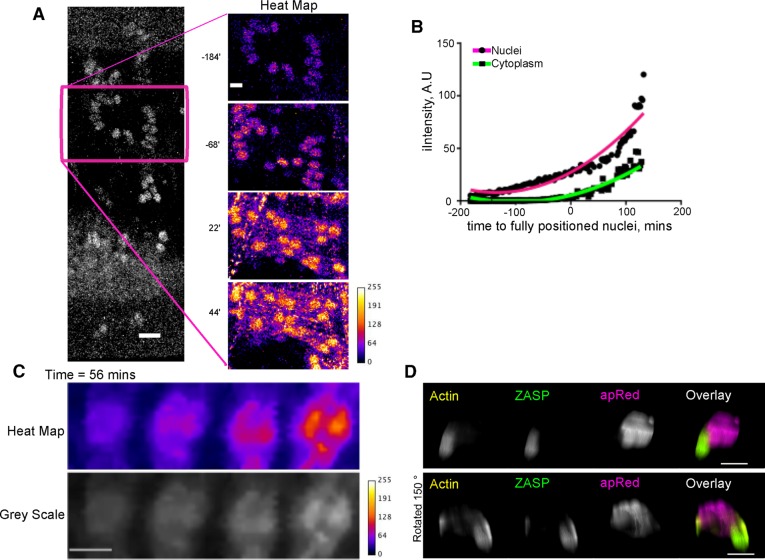
ZASP-GFP assemblies colocalize with the nuclei. (A) Left, ZASP-GFP fluorescence in a single hemisegment of a Drosophila embryo at stage 16 (18 h AEL); scale bar, 10 μm. Right, higher-magnification view of the ZASP-GFP signal in the DO2 muscle shown as a heat map. Time is relative to the completion of nuclear movement in the LT muscles. Scale bar, 5 μm. (B) Graph of the intensity of ZASP-GFP signal that colocalized with nuclei (magenta) and to regions of the muscle devoid of nuclei (green) in the DO2 muscle. Data are the mean of five individual nuclei and five regions of the muscle without nuclei in a single muscle. Specific values varied between muscles, but the trend was the same in all muscles. Time is relative to the completion of nuclear movement in the LT muscles. (C) Heat map and gray scale montage of a single myonucleus that accumulates ZASP-GFP puncta over a 56-min time period. Scale bar, 2 μm. (D) Three-dimensional projection of an individual nucleus in LT muscle from an embryo that was stained for apRed (magenta), ZASP-GFP (green), and F-actin (yellow). ZASP-GFP and actin are colocalized in puncta extending from the same myonucleus. Bottom is rotated 150° relative to the top. Scale bar, 2 μm.
ZASP-GFP assembly is colocalized with nuclei
The initial assembly of sarcomeres is hypothesized to occur through either a premyofibril templating mechanism in which sarcomeres emanate from the myotendinous junctions by an integrin-dependent mechanism (Volk et al., 1990) or assembly of nascent complexes of sarcomere proteins throughout the cytoplasm (Rhee et al., 1994; Rui et al., 2010; Weitkunat et al., 2014). To determine whether the localization of ZASP-GFP was related to the formation of nascent complexes, we analyzed the distribution of ZASP-GFP on individual nuclei. Initial ZASP-GFP localization to nuclei was uniform. ZASP-GFP then became punctate. Initial puncta colocalized with nuclei, and only later did puncta appear throughout the muscle (Figure 2, A and C). The latter pattern resembled previously observed α-actinin puncta, which were demonstrated to be precursors of sarcomere assembly (Rui et al., 2010). This suggested that ZASP-GFP was assembled into a similar nascent complex that colocalized with the nucleus before ZASP-GFP is organized throughout the muscle.
Imaging of fixed embryos indicated that ZASP-GFP puncta were localized to the cytoplasmic face of the nucleus and extended from the nucleus, without overlap between ZASP-GFP and apRed that is localized inside the nucleus (Figure 2D and Supplemental Movie S2). Furthermore, these puncta were identified by phalloidin, indicating that they contained F-actin (Figure 2D and Supplemental Movie S2). Thus a complex of F-actin and ZASP is colocalized at the cytoplasmic face of the nucleus. Similar clusters of Z-line proteins and F-actin were named I-Z-I complexes and shown to be precursors to sarcomere assembly (Rui et al., 2010). Therefore these data suggest that the assembly of I-Z-I complexes occurs on or near the nucleus.
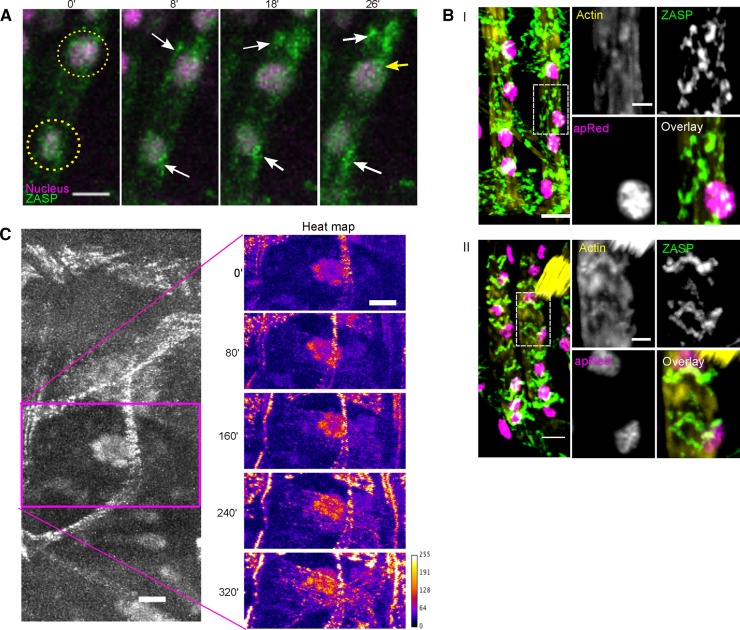
Although the first ZASP-GFP puncta colocalized with nuclei, puncta were later found throughout the muscle (Figure 2A), suggesting that each nucleus may assemble I-Z-I complexes at several different locations. To test this hypothesis, we performed live-embryo time-lapse microscopy in embryos that coexpressed the nuclear marker apRed and the Z-line marker ZASP-GFP. Often, after ZASP-GFP puncta formed on the nucleus, the nucleus moved to another location, whereas the ZASP-GFP puncta maintained its localization and continued to accumulate ZASP-GFP. After the nucleus moved, new ZASP-GFP puncta appeared on the nucleus (Figure 3A and Supplemental Movie S3). This dynamic association between ZASP-GFP and the nuclei occurred throughout the muscle. The observation that new puncta that did not previously exist continually appear at a position that is colocalized with the nucleus suggests that the nuclei are necessary for or dictate the position of the assembly of I-Z-I complexes.
FIGURE 3:
ZASP puncta are dictated by the position of the nucleus and contain F-actin. (A) Montage of two nuclei (magenta, indicated by yellow circles) in LT muscle from an embryo that expressed both apRed (magenta) and ZASP-GFP (green). White arrows indicate ZASP-GFP puncta that maintain their position after the nucleus moves away. Yellow arrows indicate newly formed ZASP-GFP puncta at the new position of the nucleus. Times are relative to the first frame of the time-lapse acquisition. Scale bar, 5 μm. (B) Image of two LT muscles before (I) and after (II) Z-line formation; scale bar, 5 μm. Right, higher-magnification images of individual signals and an overlay. Before Z-line formation (I), most actin is colocalized with the ZASP, but after Z-line formation (II), there are pools that colocalize and pools that do not colocalize with ZASP. Scale bar, 2 μm. (C) ZASP-GFP fluorescence in a single hemisegment of a stage 16 (18 h AEL) ensconsin-depleted embryo (left) and a higher-magnification view of the ZASP-GFP signal in the DO2 muscle shown as a heat map to indicate the locations of highest ZASP-GFP localization. Times are relative to the first frame of the time-lapse acquisition. Scale bar, 10 μm.
The puncta of ZASP and F-actin that previously colocalized with nuclei did mature into bona fide sarcomeres (Figure 3B), indicating that these puncta are productive precursors in the sarcomere assembly process. Furthermore, these data underscore that sarcomere assembly is a very dynamic process, with sarcomeric protein localization patterns changing, depending on the developmental stage of the muscle cell. (Rhee et al., 1994; Rui et al., 2010; Rosado et al., 2014; Weitkunat et al., 2014). Together these data suggest that nuclei contribute to the early stages of sarcomere assembly by facilitating the organization of ZASP and actin throughout the muscle.
Early sarcomere formation localizes to mispositioned nuclei
These data suggest that the changing position of the nuclei determines the position of sarcomere assembly. ZASP-GFP was expressed in Drosophila embryos that were depleted of the microtubule-associated protein ensconsin. In these embryos, nuclei were clustered in the center of the muscle (Figure 3C; Metzger et al., 2012). This defect in nuclear position occurs without other gross effects on the muscle, including effects on microtubule organization (Metzger et al., 2012). Ensconsin depletion did not affect the localization of ZASP-GFP to nuclei but did affect early sarcomere assembly. The majority of the ZASP-GFP puncta were clustered around the nuclei near the center of the muscle (Figure 3C and Supplemental Movie S4), and the first sarcomeres colocalized with the nuclei. Furthermore, as time progressed, sarcomeres appeared at increasing distances from the nuclei, giving the appearance that sarcomere assembly emanated from the clustered nuclei (Figure 3C and Supplemental Movie S4). These data demonstrate that the nuclei, independent of their position, recruit ZASP-GFP and instruct the position of initial sarcomere assembly.
The LINC complex is required for ZASP-GFP colocalization with the nucleus
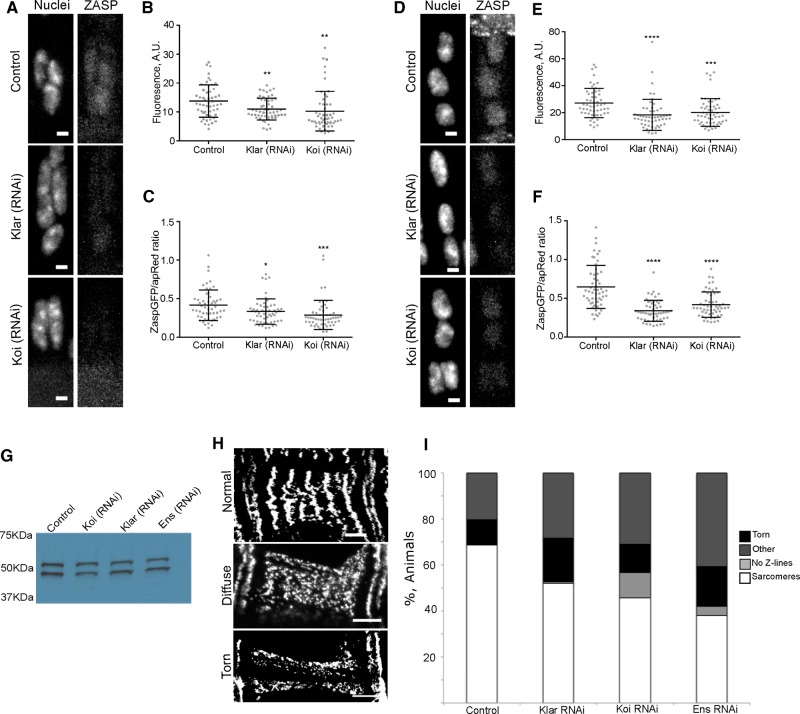
Colocalization of ZASP-GFP with the nucleus and nuclear-dependent assembly of sarcomeres require a molecular link to the nucleus. The LINC complex spans the nuclear envelope and regulates interactions between the nucleus and the cytoskeleton to move the nucleus (Starr, 2002; Luxton et al., 2010) and is therefore ideally localized to regulate nucleus-dependent sarcomere assembly. Muscle-specific depletion of the KASH domain–containing protein klarsicht (klar), which spans the outer nuclear envelope and directly links the nucleus to the cytoskeleton, or the SUN domain–containing protein klaroid (koi), which spans the inner nuclear membrane and is required for the localization of KASH-domain proteins to the outer nuclear envelope, blocked the early, uniform localization of ZASP-GFP to nuclei and the later punctate localization of ZASP-GFP (Figure 4, A and D). Quantitatively, the levels of ZASP-GFP that colocalized with nuclei were significantly lower in embryos depleted of either klar or koi both when nuclei began their movement back into the center of the muscle (Figure 4, B and C) and when nuclei approached their final position (Figure 4, E and F). Western blots indicated that ZASP-GFP was expressed at similar levels in all genotypes (Figure 4G). These data suggest that the expression of ZASP-GFP is not reduced but that the localization of ZASP-GFP to the nucleus requires the LINC complex. More broadly, this indicates that the structural interplay between the cytoskeleton and the nucleus is bidirectional. The LINC complex has been shown to regulate interactions between the nucleus and both the actin cytoskeleton and the microtubule cytoskeleton during muscle development and the Z-line specifically in muscles with a well-developed sarcomere network (Cadot et al., 2012; Elhanany-Tamir et al., 2012; Wilson and Holzbaur, 2012; Meinke et al., 2014). These data suggest that the LINC complex facilitates the interaction between the nucleus and the immature sarcomere and contributes to the assembly of the sarcomere.
FIGURE 4:
The LINC complex is required to recruit ZASP-GFP to Z-lines. (A) Images of apRed (nuclei) and ZASP-GFP in the LT muscle of embryos with indicated genotypes taken when nuclei are clustered but have begun movement into the center of the muscle (18 h AEL); scale bar, 2 μm. (B) Mean ZASP-GFP fluorescence on 54 nuclei from the stage described in A in indicated genotypes. (C) Mean ZASP-GFP fluorescence divided by the mean apRed fluorescence for the nuclei described in A. (D) Images of apRed (nuclei) and ZASP-GFP in the LT muscle of embryos with indicated genotypes taken when nuclei approach their final position (20 h AEL); scale bar, 2 μm. (E) Mean fluorescence across 54 nuclei described in D from each indicated genotype. (F) Mean ZASP-GFP fluorescence divided by the mean apRed fluorescence for the nuclei described in D. ****p < 0.0001, ***p < 0.001, **p < 0.01. (G) Western blot showing ZASP-GFP expression in embryos depleted of the indicated protein by RNAi. The same number of precisely staged embryos was used and the same fraction loaded for each condition. (H) Representative images of the classes of Z-line structure observed; scale bars, 10 μm. (I) Percentage of embryos/larva in each category of sarcomere structure shown in H. Data are from at least three independent experiments in which all animals (≥33) were counted.
Nucleus-dependent sarcomere assembly is crucial for stable sarcomeres
Although the localization of ZASP-GFP to the nucleus was lost in embryos depleted of the LINC complex, ZASP-GFP was often incorporated into structures that resembled Z-lines. Similarly, although the position of the first sarcomeres was disrupted in embryos depleted of ensconsin, Z-line–like structures did eventually assemble throughout the muscle. This likely represents the assembly of sarcomeric complexes through already described processes (Sparrow and Schöck, 2009; Rui et al., 2010; Sanger et al., 2010) and suggests that the contributions of the nucleus occur in parallel with already established mechanisms.
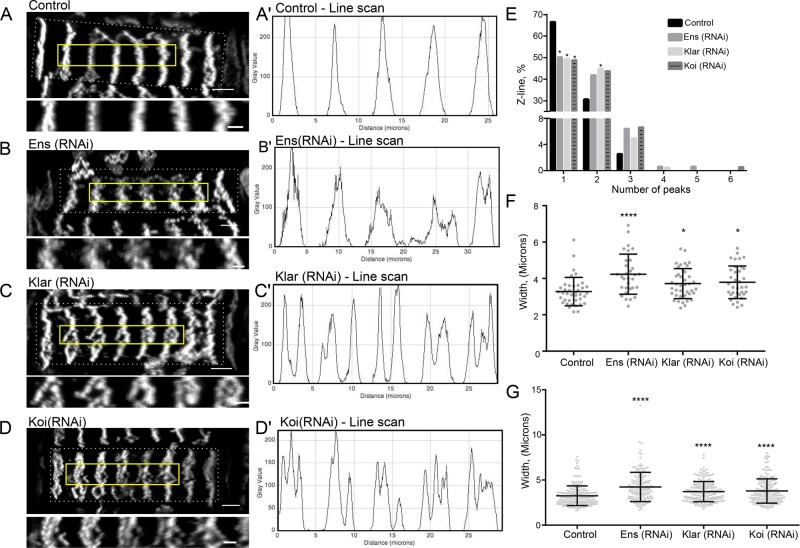
Characterization of the general muscle structure in animals that either had the position of early sarcomere assembly disrupted (ensconsin RNA interference [RNAi]) or lacked nucleus-dependent sarcomere assembly (klar RNAi and koi RNAi) revealed the critical role of the nucleus in sarcomere assembly and stability. In controls, >67% of muscles had defined Z-lines. In ensconsin-, klarsicht-, and klaroid-depleted embryos, <52% of muscles displayed a defined Z-line structure (Figure 4, H and I). The greatest increases were in animals with complex phenotypes that included torn muscles, muscles lacking Z-lines, and missing or detached muscles (Figure 4I). Furthermore, the Z-lines that did form were different in klarsicht-, klaroid-, and ensconsin-depleted animals than in control animals (Figure 5, A–D). Line scans performed perpendicular to the orientation of the sarcomeres in the ventral longitudinal 1 muscle (VL1; Figure 5, A′–D′) of these animals revealed an increase in the number of peaks (Figure 5E) and an increase in the width of the Z-line structure (Figure 5, F and G) compared with the controls. These additional peaks do not correspond to additional sarcomeres but to the splitting of individual sarcomeres, indicating that the ability of the nucleus to recruit and organize ZASP and the fact that this organization occurs throughout the cell rather than in a single cluster both are necessary for the integrity of the myofibril network. This finding is consistent with previous studies that indicated that the nucleus and the LINC complex contribute to the organization of the microtubule cytoskeleton (Stewart-Hutchinson et al., 2008) and the actin cytoskeleton (Khatau et al., 2009; Chambliss et al., 2013) in culture. Furthermore, the interaction between the nucleus and actin is necessary for the mechanical strength of the cell (Lombardi et al., 2011) in culture. Our data suggest that this is also true of muscle cells in vivo. In the absence of the LINC complex, the sarcomere network, which is sensitive to the mechanical state of the cell (Perkins et al., 2010; Pines et al., 2012; Weitkunat et al., 2014), is less stable. These data suggest that the nucleus and the LINC complex contribute to the initial assembly of specific cytoskeletal networks.
FIGURE 5:
Proper nuclear positioning and the LINC complex are required to establish a stable Z-line structure. (A–D) Images of ZASP-GFP in the VL1 muscle (indicated by the dashed white box) in L1 larva of indicated genotypes. Solid yellow boxes indicate the five Z-lines shown below each VL1 muscle. Scale bar, 5 μm (whole muscle), 2 μm (enlarged). (A′–D′) Line scans through the center of the enlarged images. (E) Number of peaks as a percentage of ≥150 Z-lines in L1 larva of the indicated genotypes. (F) Average width of Z-lines in ≥30 L1 larva per genotype. Each data point represents the average of five Z-lines within a single muscle from a single animal. (G) Graph showing individual Z-line widths from ≥150 Z-lines taken from first-instar larva. ****p < 0.0001, *p < 0.05.
This work begins an investigation into why the process of nuclear movement is conserved throughout eukaryotes. It demonstrated a role for the nucleus in sarcomere assembly, but similar actin–myosin contractile structures with less-exaggerated organizations are seen in processes such as cell migration and apical constriction (Fujiwara and Pollard, 1976; Young et al., 1991; Skwarek-Maruszewska et al., 2009). Furthermore, during each of these developmental processes, movement of the nucleus is an early and essential phenomenon. Thus, although the details regarding the nature by which the nucleus organizes the cytoskeleton may differ, that the nucleus is a crucial regulator of the cytoskeleton is likely conserved.
MATERIALS AND METHODS
Drosophila genetics
The following stocks were grown under standard conditions: apME-NLS::DsRed (Richardson et al., 2007), ZASP66-GFP (Katzemich et al., 2013), UAS-klar RNAi (36721; Bloomington Drosophila Stock Center [BDSC], Bloomington, IN), UAS-Koi RNAi (40924; BDSC), and UAS-ens RNAi (40825; BDSC). UAS-mCherry RNAi (35785; BDSC) UAS-RNAi constructs were driven specifically in the mesoderm using Twist-Gal4 (Greig and Akam, 1993). In all experiments, Gal4 was provided by the mother and UAS-RNAi by the father.
Live-embryo imaging
In all experiments, unless otherwise stated, control refers to Twist-Gal4, apRed; ZASP-GFP driving the expression of UAS-m-Cherry-RNAi. Embryos were washed in 50% bleach to remove the outer membrane, rinsed in water to remove all bleach, and mounted in halocarbon oil (H8898; Sigma-Aldrich). Stage 15/16 embryos were selected based on both the location of nuclei within the LT muscles and the morphology of the gut as previously described (Folker et al., 2014). Time-lapse imaging was performed on a Zeiss 700 LSM using a 40×/1.4 numerical aperture (NA) Apochromat objective. Z-stacks were acquired at a 1-μm step size and a rate of 2 min/stack for 13.3 h to ensure that embryo development was monitored until it hatched. Movies were assembled, and data were analyzed using ImageJ (National Institutes of Health, Bethesda, MD).
Fluorescence intensity quantification
Colocalization of ZASP-GFP and apRed was quantified by measuring the signal intensity of both across the same region of a given nucleus within LT muscle. For Figure 2B, ZASP-GFP signal intensity was measured across five nuclei within the DO2 muscles from when the signal was first visible until nuclei could no longer be distinguished, 132 min after nuclei were fully positioned. Similar fluorescence intensity measurements were also taken from five areas devoid of nuclei. Fluorescence measurements were taken across five nuclei, using ImageJ software. Examination of different muscles produced different raw values but had the same trend. All times are relative to nuclei in the LT muscles achieving their final position. Fluorescence measurements within the DO2 muscles were then plotted, fitting the data to a second-order polynomial. All indicated times are relative to the completion of nuclear movement within the LT muscles except for Figure 3, A and C.
Fixed-embryo imaging
To image nuclei at a higher resolution, embryos expressing ZASP-GFP and apRed were washed in 50% bleach to remove the outer membrane, washed with water, and then fixed in 50% Formalin (HT501128; Sigma-Aldrich, St. Louis, MO) diluted in 1:1 heptane for 20 min. Embryos were mounted in Prolong Gold (Invitrogen, Carlsbad, CA) and imaged with an Apochromat 40×/1.4 NA objective with a 3.3× optical zoom on a Zeiss 700 LSM for Figure 2D. Images were converted to three dimensions using ImageJ. Note that the imaging (except Figures 2D and 3B) was performed on the fluorescence of GFP and DsRed and was not indirect immunofluorescence. In Figures 2D and 3B, F-actin was identified with Acti-stain 670 Phalloidin (Cytoskeleton, Denver, CO), DsRed was identified with rabbit anti-DsRed (632496; Clontech, Mountain View, CA), and tropomyosin was identified with rat anti-tropomyosin (50567; Abcam, Cambridge, MA). Secondary antibodies were Alexa 488 anti-rat and Alexa 555 anti-rabbit (Life Technologies, Carlsbad, CA). Colocalization of ZASP-GFP and apRed was quantified by measuring the signal intensity of both apRed and ZASP-GFP across the same region of a given nucleus within LT muscle using ImageJ. All images were acquired with identical microscope settings on precisely staged embryos based on the morphology of the gut and the morphology of the trachea, as previously described (Folker et al., 2012). For the measurements in Figure 4, B, C, E, and F, ImageJ was used to measure the mean pixel intensity on six individual nuclei within the LT muscles of nine different embryos per genotype. Data are presented as both raw values and ratios relative to apRed signal, which served as an internal control and did not vary between embryos that expressed different RNAi constructs. More than 50 nuclei were analyzed and compared by Student’s t test using GraphPad Prism (GraphPad Software). p < 0.05 was considered significant.
Muscle phenotype analysis
Phenotype analysis was performed on embryos 20 h after egg laying (AEL) that were controlled for with timed lays and raised at 29°C. All genotypes were examined at the same age and under identical conditions. Embryos and larva were scored on whether ZASP-GFP had been taken up into the Z-line and normal muscle contraction was seen (normal), whether ZASP-GFP failed to be incorporated into the Z-line (no sarcomeres), whether torn muscles were present (Torn), and as complex phenotypes that included missing muscles, patterning defects, torn muscles, and muscles without sarcomeres (other).
Z-line structure analysis
Z-line analysis of L1 larva was performed by selecting L1 larva from laying pots raised at 25°C. Larva were placed on a coverslip using a fine brush, and another coverslip was placed on top to halt larval crawling. The entire VL1 muscle was then imaged using an Apochromat 40×/1.4 NA objective with Z-stacks acquired at a 0.5-μm step size. To normalize the differing depth of the muscle, 2.5-μm slices were taken from the top of each muscle. Line scans were then performed across the center of five separate Z-lines per muscle per animal, with the width of each Z-line taken from the raw data of the line scan. Statistical analysis was performed using GraphPad Prism 6. Measurements were evaluated in >30 animals using a Student’s t test using with p < 0.05 considered significant.
Western blot
Embryos were washed in 50% bleach to remove the outer membrane and then washed with water to remove bleach. Rather than using a standard loading control such as tubulin or actin, which is likely to respond to depletion of ensconsin, klarsicht, or klaroid, loading was controlled by using the same number of precisely staged embryos for each condition. Five embryos of each genotype (Twist-Gal4, apred; ZASP-GFP crossed to either w1118 [control], UAS-ens-RNAi, UAS-koi-RNAi, or UAS-klar-RNAi) were selected that were at stage 17 of embryo development using the morphology of the gut and appearance of the trachea, as previously described (Beckett and Baylies, 2007). Embryos were then crushed in an Eppendorf tube in 100 μl of standard 2× SDS sample buffer. This homogenate was then heated at 100°C in a boiling water bath for 5 min and cooled briefly before centrifuging of the sample at 20,000 × g for 5 min. The supernatant was then removed and placed in a fresh Eppendorf tube. One-fifth of each sample was loaded per lane for SDS–PAGE, with rabbit anti-GFP (TP-401; Torrey Pines Biolabs, Secaucus, NJ) at 1:10,000 and horseradish peroxidase goat anti-rabbit (656120; Life Technology) used for immunoblotting.
Supplementary Material
Acknowledgments
We thank members of the Folker lab for insightful discussions. E.S.F. is funded by The Charles H. Hood Foundation and by a Scientist Development Grant from the American Heart Association.
Abbreviations used:
- DO2
dorsal muscle 2
- GFP
green fluorescent protein
- KASH
Klarsicht, Anc-1 and Syne Homology
- klar
klarsicht
- koi
klaroid
- LINC complex
linker of nucleoskeleton and cytoskeleton
- LT
lateral transverse
- NLS
nuclear localization signal
- SUN
Sad1 and Unc84 homology domain
- VL1
ventral longitudinal muscle 1.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0021) on June 15, 2016.
REFERENCES
- Beckett K, Baylies MK. 3D analysis of founder cell and fusion competent myoblast arrangements outlines a new model of myoblast fusion. Dev Biol. 2007;309:113–125. doi: 10.1016/j.ydbio.2007.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard JC, Liestøl K, Ekmark M, Kollstad K, Gundersen K. Number and spatial distribution of nuclei in the muscle fibres of normal mice studied in vivo. J Physiol. 2004;551:467–478. doi: 10.1113/jphysiol.2003.045328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadot B, Gache V, Vasyutina E, Falcone S, Birchmeier C, Gomes ER. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 2012;13:741–749. doi: 10.1038/embor.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capers CR. Multinucleation of skeletal muscle in vitro. J Biophys Biochem Cytol. 1960;7:559–566. doi: 10.1083/jcb.7.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambliss AB, Khatau SB, Erdenberger N, Robinson DK, Hodzic D, Longmore GD, Wirtz D. The LINC-anchored actin cap connects the extracellular milieu to the nucleus for ultrafast mechanotransduction. Sci Rep. 2013;3:1087. doi: 10.1038/srep01087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanany-Tamir H, Yu YV, Shnayder M, Jain A, Welte M, Volk T. Organelle positioning in muscles requires cooperation between two KASH proteins and microtubules. J Cell Biol. 2012;198:833–846. doi: 10.1083/jcb.201204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G, Pallavicini A, Formentin E, Comelli A, Ievolella C, Trevisan S, Bortoletto G, Scannapieco P, Salamon M, Mouly V, et al. ZASP: a new Z-band alternatively spliced PDZ-motif protein. J Cell Biol. 1999;146:465–475. doi: 10.1083/jcb.146.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandes I, Schöck F. The nebulin repeat protein Lasp regulates I-band architecture and filament spacing in myofibrils. J Cell Biol. 2014;206:559–572. doi: 10.1083/jcb.201401094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Schulman VK, Baylies MK. Muscle length and myonuclear position are independently regulated by distinct Dynein pathways. Development. 2012;139:3827–3837. doi: 10.1242/dev.079178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Schulman VK, Baylies MK. Translocating myonuclei have distinct leading and lagging edges that require kinesin and dynein. Development. 2014;141:355–366. doi: 10.1242/dev.095612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara K, Pollard TD. Fluorescent antibody localization of myosin in the cytoplasm, cleavage furrow, and mitotic spindle of human cells. J Cell Biol. 1976;71:848–875. doi: 10.1083/jcb.71.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furst DO, Osborn M, Nave R, Weber K. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J Cell Biol. 1988;106:1563–1572. doi: 10.1083/jcb.106.5.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golson ML, Sanger JM, Sanger JW. Inhibitors arrest myofibrillogenesis in skeletal muscle cells at early stages of assembly. Cell Motil Cytoskeleton. 2004;59:1–16. doi: 10.1002/cm.20017. [DOI] [PubMed] [Google Scholar]
- Greig S, Akam M. Homeotic genes autonomously specify one aspect of pattern in the Drosophila mesoderm. Nature. 1993;362:630–632. doi: 10.1038/362630a0. [DOI] [PubMed] [Google Scholar]
- Holtzer H, Hijikata T, Lin ZX, Zhang ZQ, Holtzer S, Protasi F, Franzini-Armstrong C, Sweeney HL. Independent assembly of 1.6 microns long bipolar MHC filaments and I-Z-I bodies. Cell Struct Funct. 1997;22:83–93. doi: 10.1247/csf.22.83. [DOI] [PubMed] [Google Scholar]
- Hudson AM, Petrella LN, Tanaka AJ, Cooley L. Mononuclear muscle cells in Drosophila ovaries revealed by GFP protein traps. Dev Biol. 2008;314:329–340. doi: 10.1016/j.ydbio.2007.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. The double array of filaments in cross-striated muscle. J Biophys Biochem Cytol. 1957;3:631–648. doi: 10.1083/jcb.3.5.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzemich A, Liao KA, Czerniecki S, Schöck F. Alp/Enigma family proteins cooperate in Z-disc formation and myofibril assembly. PLoS Genet. 2013;9:e1003342. doi: 10.1371/journal.pgen.1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatau SB, Hale CM, Stewart-Hutchinson PJ, Patel MS, Stewart CL, Searson PC, Hodzic D, Wirtz D. A perinuclear actin cap regulates nuclear shape. Proc Natl Acad Sci USA. 2009;106:19017–19022. doi: 10.1073/pnas.0908686106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Ruiz J, Bajraktari I, Ohman R, Banerjee S, Gribble K, Kaufman JD, Wingfield PT, Griggs RC, Fischbeck KH, et al. Z-disc-associated, alternatively spliced, PDZ motif-containing protein (ZASP) mutations in the actin-binding domain cause disruption of skeletal muscle actin filaments in myofibrillar myopathy. J Biol Chem. 2014;289:13615–13626. doi: 10.1074/jbc.M114.550418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi ML, Jaalouk DE, Shanahan CM, Burke B, Roux KJ, Lammerding J. The interaction between nesprins and sun proteins at the nuclear envelope is critical for force transmission between the nucleus and cytoskeleton. J Biol Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GWG, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke P, Mattioli E, Haque F, Antoku S, Columbaro M, Straatman KR, Worman HJ, Gundersen GG, Lattanzi G, Wehnert M, et al. Muscular dystrophy-associated SUN1 and SUN2 variants disrupt nuclear-cytoskeletal connections and myonuclear organization. PLoS Genet. 2014;10:e1004605. doi: 10.1371/journal.pgen.1004605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger T, Gache V, Xu M, Cadot B, Folker ES, Richardson BE, Gomes ER, Baylies MK. MAP and kinesin-dependent nuclear positioning is required for skeletal muscle function. Nature. 2012;484:120–124. doi: 10.1038/nature10914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins AD, Ellis SJ, Asghari P, Shamsian A, Moore EDW, Tanentzapf G. Integrin-mediated adhesion maintains sarcomeric integrity. Dev Biol. 2010;338:15–27. doi: 10.1016/j.ydbio.2009.10.034. [DOI] [PubMed] [Google Scholar]
- Pierobon-Bormioli S, Betto R, Salviati G. The organization of titin (connectin) and nebulin in the sarcomeres: an immunocytolocalization study. J Muscle Res Cell Motil. 1989;10:446–456. doi: 10.1007/BF01771820. [DOI] [PubMed] [Google Scholar]
- Pines M, Das R, Ellis SJ, Morin A, Czerniecki S, Yuan L, Klose M, Coombs D, Tanentzapf G. Mechanical force regulates integrin turnover in Drosophila in vivo. Nat Cell Biol. 2012;14:935–943. doi: 10.1038/ncb2555. [DOI] [PubMed] [Google Scholar]
- Puszkin S, Puszkin E, Maimon J, Rouault C, Schook W, Ores C, Kochwa S, Rosenfield R. alpha-Actinin and tropomyosin interactions with a hybrid complex of erythrocyte-actin and muscle-myosin. J Biol Chem. 1977;252:5529–5537. [PubMed] [Google Scholar]
- Rhee D, Sanger JM, Sanger JW. The premyofibril: evidence for its role in myofibrillogenesis. Cell Motil Cytoskeleton. 1994;28:1–24. doi: 10.1002/cm.970280102. [DOI] [PubMed] [Google Scholar]
- Richardson BE, Beckett K, Nowak SJ, Baylies MK. SCAR/WAVE and Arp2/3 are crucial for cytoskeletal remodeling at the site of myoblast fusion. Development. 2007;134:4357–4367. doi: 10.1242/dev.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado M, Barber CF, Berciu C, Feldman S, Birren SJ, Nicastro D, Goode BL. Critical roles for multiple formins during cardiac myofibril development and repair. Mol Biol Cell. 2014;25:811–827. doi: 10.1091/mbc.E13-08-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Bai J, Perrimon N. Sarcomere formation occurs by the assembly of multiple latent protein complexes. PLoS Genet. 2010;6:e1001208. doi: 10.1371/journal.pgen.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger JW, Wang J, Fan Y, White J, Sanger JM. Assembly and dynamics of myofibrils. J Biomed Biotechnol. 2010;2010:1–8. doi: 10.1155/2010/858606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skwarek-Maruszewska A, Hotulainen P, Mattila PK, Lappalainen P. Contractility-dependent actin dynamics in cardiomyocyte sarcomeres. J Cell Sci. 2009;122:2119–2126. doi: 10.1242/jcs.046805. [DOI] [PubMed] [Google Scholar]
- Sparrow JC, Schöck F. The initial steps of myofibril assembly: integrins pave the way. Nat Rev Mol Cell Biol. 2009;10:293–298. doi: 10.1038/nrm2634. [DOI] [PubMed] [Google Scholar]
- Starr DA. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D. Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res. 2008;314:1892–1905. doi: 10.1016/j.yexcr.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volk T, Fessler LI, Fessler JH. A role for integrin in the formation of sarcomeric cytoarchitecture. Cell. 1990;63:525–536. doi: 10.1016/0092-8674(90)90449-o. [DOI] [PubMed] [Google Scholar]
- Weitkunat M, Kaya-Çopur A, Grill SW, Schnorrer F. Tension and force-resistant attachment are essential for myofibrillogenesis in Drosophila flight muscle. Curr Biol. 2014;24:705–716. doi: 10.1016/j.cub.2014.02.032. [DOI] [PubMed] [Google Scholar]
- Wilson MH, Holzbaur ELF. Opposing microtubule motors drive robust nuclear dynamics in developing muscle cells. J Cell Sci. 2012;125:4158–4169. doi: 10.1242/jcs.108688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MH, Holzbaur ELF. Nesprins anchor kinesin-1 motors to the nucleus to drive nuclear distribution in muscle cells. Development. 2015;142:218–228. doi: 10.1242/dev.114769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young PE, Pesacreta TC, Kiehart DP. Dynamic changes in the distribution of cytoplasmic myosin during Drosophila embryogenesis. Development. 1991;111:1–14. doi: 10.1242/dev.111.1.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.