Abstract
Single-molecule picometer resolution nanopore tweezers (SPRNT) is a new tool for analyzing the motion of nucleic acids through molecular motors. With SPRNT, individual enzymatic motions along DNA as small as 40 pm can be resolved on sub-millisecond time scales. Additionally, SPRNT reveals an enzyme’s exact location with respect to a DNA strand’s nucleotide sequence, enabling identification of sequence-specific behaviors. SPRNT is enabled by a mutant version of the biological nanopore formed by Mycobacterium smegmatis porin A (MspA). SPRNT is strongly rooted in nanopore sequencing and therefore requires a solid understanding of basic principles of nanopore sequencing. Furthermore, SPRNT shares tools developed for nanopore sequencing and extends them to analysis of single-molecule kinetics. As such, this review begins with a brief history of our work developing the nanopore MspA for nanopore sequencing. We then describe the underlying principles of SPRNT, how it works in detail, and propose some potential future uses. We close with a comparison of SPRNT to other techniques and we present the methods that will enable others to use SPRNT.
Keywords: Single-molecule, Helicase, Polymerase, Kinetics, Force spectroscopy
1. Introduction
Nanopores are emerging as a high-precision single-molecule tool. In its basic implementation, a nanopore device consists of a single pore within a membrane (Fig. 1). This membrane divides a salt solution into two wells called ‘cis’ and ‘trans.’ When a voltage is applied across this membrane, ion current flows through the pore. The magnitude of this ion current is the primary signal. Molecules of interest (i.e. DNA, RNA, peptides, nanoparticles, etc.) are drawn towards the pore and then through it by the electric field. As the molecules traverse the pore, they alter the ion current flowing through the pore. Depending on the degree to which the ion current is blocked, various properties of the molecule and its movement through the pore can be inferred. Generally, nanopores can be grouped into two categories: (1) ‘biological pores’ which are pore proteins such as alpha hemolysin (aHL) [1–4] or Mycobacterium smegmatis porin A (MspA) [5–7] embedded in a phospho-lipid bilayer and (2) solid-state pores, which are nanometer-scale holes drilled into thin membranes of silicon nitride [8–10], graphene [11–13], molybdenum disulfide [14], etc. Nanopores have been used to detect size and shape characteristics of proteins [15], separate polymers of different sizes and charges [16,17] or to separate two types of proteins [18].
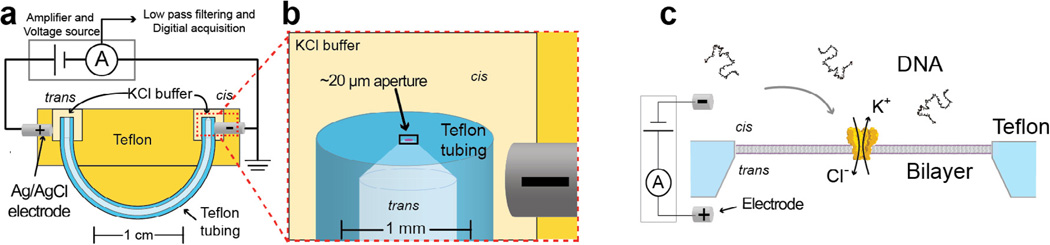
Fig. 1.
Schematic of the u-tube bilayer setup at various scales (originally described by Akeson et al. in 1999 [2], see Appendix A for detail on building and setting up such an experiment). The bilayer is established across the ~20 µm aperture. Once a bilayer is established, MspA is added to the cis solution until a single pore inserts. Once the operator detects an insertion, the remaining MspA is perfused from the cis well and the system is prepared for experiments. When a voltage is applied across the bilayer by two Ag/AgCl electrodes (as shown in c, not to scale), negatively charged DNA is drawn into and through the pore. While all experiments described below are performed using this single channel recording setup, it is possible to massively parallelize nanopore setups for higher throughput. Figure modified from [37].
In 1996, Kasianowicz et al. [1] proposed using a nanopore setup to sequence DNA. In this scheme, the applied voltage electrophoretically feeds single stranded DNA (ssDNA) into the pore. As the DNA passes through the pore, DNA bases within the pore block the ionic current. Because each base has differing size and charge distribution, it was proposed that each base has its own distinct ion current, allowing one to determine the DNA sequence by analyzing the current trace as DNA is drawn through the pore. Kasianowicz et al. found that they could translocate DNA strands through the pore and that the presence of DNA within the pore reduced the ion current. From these initial experiments, there emerged two clear challenges to the implementation of nanopore sequencing: researchers needed (1) a pore with the ability to distinguish between nucleotides with sufficient signal to noise and (2) a means of slowing the DNA translocation enough so that each base could be read for long enough [1].
In order to have single-nucleotide sensitivity, it is key that the size of the nanopore (diameter and length) is commensurate with the dimensions of the DNA bases [5,16]. Biological pores such as aHL [1,2,4,19–21] or MspA [5–7,22–27], with a geometry similar to the size of ssDNA nucleotides [5], have been used to study DNA.
2. MspA for sequencing DNA
MspA, in particular, proved nearly ideal for the analysis of single-stranded nucleic acid strands. This is because the pore constriction (the region of the pore with the highest ion current density and thereby highest sensitivity to nucleobase differences) is just 0.6 nm long and 1.2 nm wide [5] (Fig. 2). This compares well with the size and spacing of DNA nucleobases within ssDNA. In contrast, aHL has a long constriction resulting in several DNA sequence recognition sites spanning some ~12 nucleotides [20]. While it is possible to construct solid-state pores with dimensions similar to those of MspA, in graphene for instance [11–13], current fabrication techniques have not yet achieved the atomistic reproducibility inherent to biological systems and the efficacy of such solid-state pores for DNA sequencing has yet to be demonstrated.
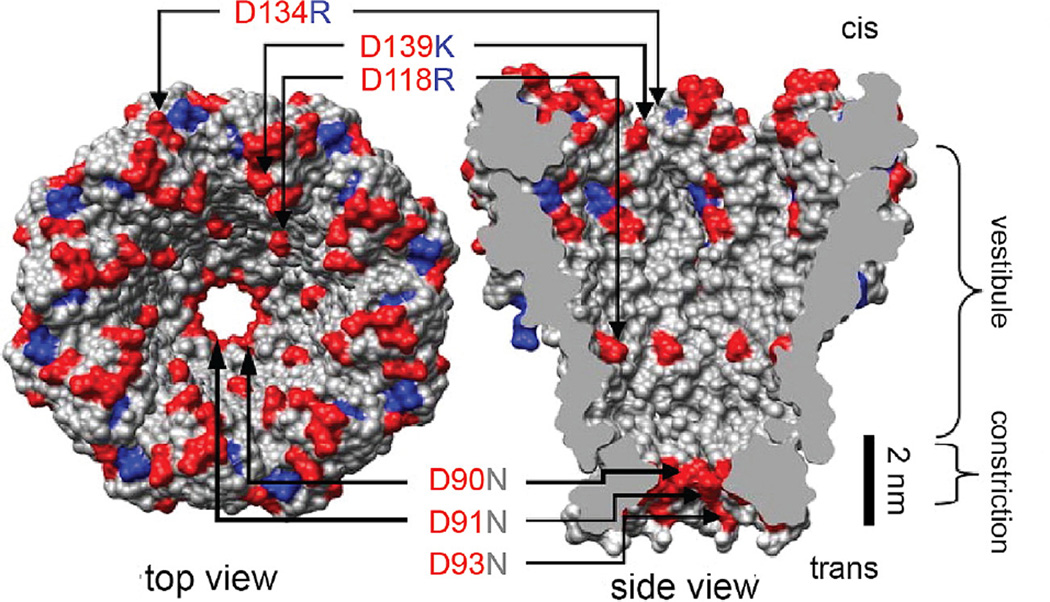
Fig. 2.
Mycobacterium smegmatis porin A (MspA). Wild type MspA contains negatively charged residues D90, D91, and D93. Butler et al. found that mutation of these residues to neutrally charged N allowed DNA to translocate through the MspA pore. Further mutation of the pore included positively charged residues within the vestibule (D118R, D134R, and E139K) and resulted in increased DNA interaction. Figure modified from [5].
2.1. Mycobacterium smegmatis porin A
MspA is an outer membrane protein that derives from Mycobacterium smegmatis. In 2008 Butler et al. proposed that its size and shape would make it a promising candidate for nanopore sequencing [5]. In initial experiments with wild-type MspA, DNA was unable to translocate through the pore. Butler et al. hypothesized that this was due to negatively charged aspartic acid residues that define the constriction of the pore (D90, D91, D93; Fig. 2). Mutating these residues to neutral asparagines resulted in clear ssDNA-induced current blockades. Further mutation of the pore to include positively charged residues within the vestibule (mutations D118R, D134R, and E139K; Fig. 2) significantly increased the rate of DNA capture by the pore allowing for a ~100-fold reduction in the minimum analyte concentration for the negatively charged DNA.
2.2. Base differentiation and single-nucleotide resolution
Early indications that DNA bases might actually create different blockage currents were shown with aHL [2–4,19,28]. In 1999 Akeson et al. translocated block homopolymer RNA strands consisting of 30 As followed by 70 C’s through an aHL pore and observed ssDNA translocation events that consisted of two distinct current levels, however they were unable to resolve individual bases. This confirmed what was apparent from the initial nanopore sequencing paper, that a method for slowing DNA translocation was needed [1]. In 2000 Henrickson et al. [19] tethered DNA to a large streptavidin molecule in order to hold DNA stationary within a nanopore. This technique was later used to demonstrate that each of the 4 bases created unique blockage currents within the aHL pore [3] however it was revealed that the region of sensitivity within the aHL pore was ~12 nt long [4,28], far too long to achieve the single-nucleotide resolution necessary for DNA sequencing.
As with aHL, ssDNA translocates through MspA far too quickly (>1 nt/µs) for individual bases to be resolved within the noise [5]. In 2010, Derrington et al. showed that MspA was capable of distinguishing between the four canonical bases when held within the pore by duplex DNA, which is too wide to fit through MspA’s constriction [6]. In 2011 Manrao et al. [7] showed that MspA was capable of distinguishing single bases using DNA nucleotides tethered to a streptavidin molecule (Fig. 3). By changing the location of a single nucleotide within a heterogeneous DNA strand held at different locations within the pore (Fig. 3a and c) Manrao et al. showed that MspA’s region of base sensitivity included ~4 nucleotides in and around the constriction and that the exchange of just a single DNA base could be detected. The ion current was even sensitive to the replacement of a C with methyl cytosine. These results showed that MspA’s ability to distinguish between bases was far better than observed in comparable experiments with aHL [4,20,21].
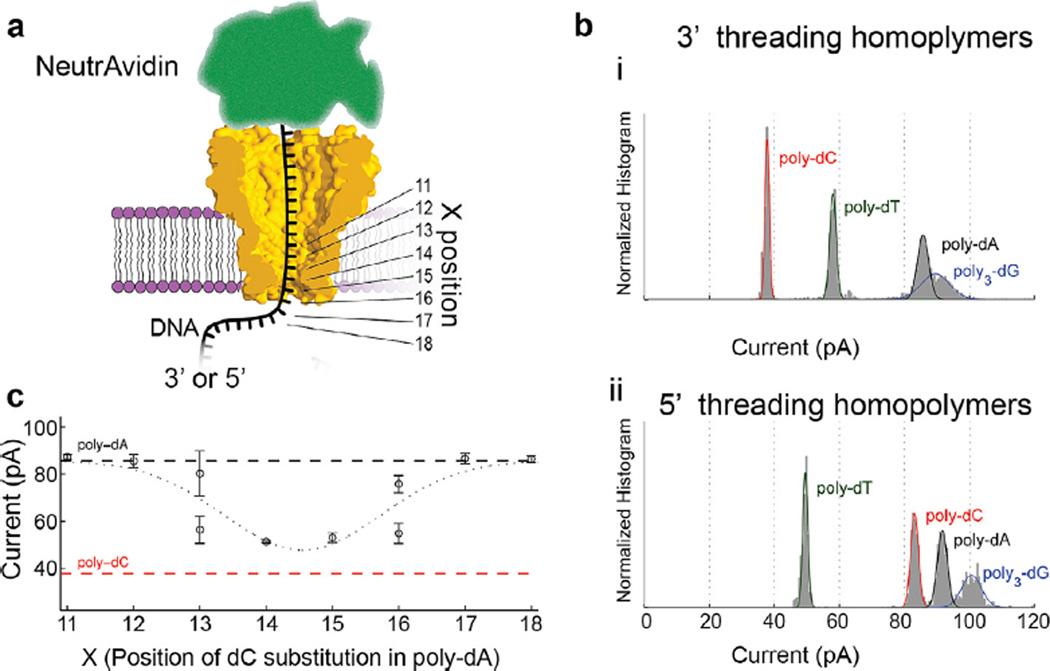
Fig. 3.
Single nucleotide sensitivity. (a) Schematic of initial experiments with DNA held stationary in the pore by tethering it to a large streptavidin molecule. (b) These experiments showed that homopolymer strands containing each base had its own unique current. (c) Single nucleotide substitutions of nucleotides held within the pore at various positions shown in (a) such as a single C replacing an A at various positions along a homopolymer A DNA strand showed that the region of sensitivity for MspA was ~4 nucleotides wide, and that the substitution of a single base held within the constriction was detectable. (b) and (c) modified from [7].
2.3. Enzyme control
All that was needed then for nanopore sequencing was a means of slowing the DNA translocation through the pore; with such control, it was possible that the superior base recognition of MspA could enable nanopore sequencing. Just as nature provided almost ideal biological nanopores, nature again offers a solution in the form of molecular motors that walk along DNA (polymerases, helicases, translocases). Gyarfas et al. [29] pioneered such techniques, using at first T7 and Klenow fragment. However, the most success was later found with phi29 DNA polymerase (phi29 DNAP) [30,31] a highly processive DNAP from Bacillus subtilis phage phi29.
Rather than attaching the molecular motor to the pore, it is much easier to load the molecular motor onto the DNA and then draw the DNA into the pore until the enzyme rests on the pore. A key challenge is preventing enzyme extension of the DNA primer in bulk, before the DNA-enzyme complex is loaded onto the pore. To solve this problem, Cherf et al. [31] developed the ‘blocking oligomer technique’ (Fig. 4) wherein the phi29 DNAP is prevented from extending a DNA primer strand by the presence of a third DNA strand: the non-extendable blocking oligomer. The blocking oligomer hybridizes with the template DNA strand immediately 3′ of the primer strand. In bulk, phi29 DNAP binds to the template strand near the 3′ end of the blocking oligomer but is unable to extend the blocker due to non-complementarity and chemical modification of the blocking oligomer’s 3′ end [31] (it turns out that non-complementarity of the last ~7 bases of the blocking oligomer to the template strand is sufficient to prevent bulk activity [22]). When the template strand is threaded into the pore and the phi29 DNAP comes into contact with the pore rim, the force of the pore rim on the enzyme pushes the phi29 DNAP in the 5′-3′ direction through the duplex region created by the blocking oligo (this is the reverse of the direction that phi29 DNAP usually walks during polymerase activity). The phi29 DNAP, being forced backwards along the template strand, unzips the blocking oligo from the template strand one nucleotide at a time, thereby feeding the template strand into the pore from cis to trans one nucleotide at a time until the blocking oligomer is fully dissociated and the primer’s 3′ end falls into the phi29 DNAP’s active site. In the presence of MgCl2 and deoxynucleotide triphosphates (dNTPs), the phi29 DNAP then goes to work extending the primer DNA strand and pulls the template back out of the pore from trans to cis (Fig. 4b). Cherf et al. found that as the DNA moves through an aHL pore, the ion current shows distinct current steps corresponding to each of the phi29 DNAP steps along the DNA strand, however they could not discern the DNA sequence.
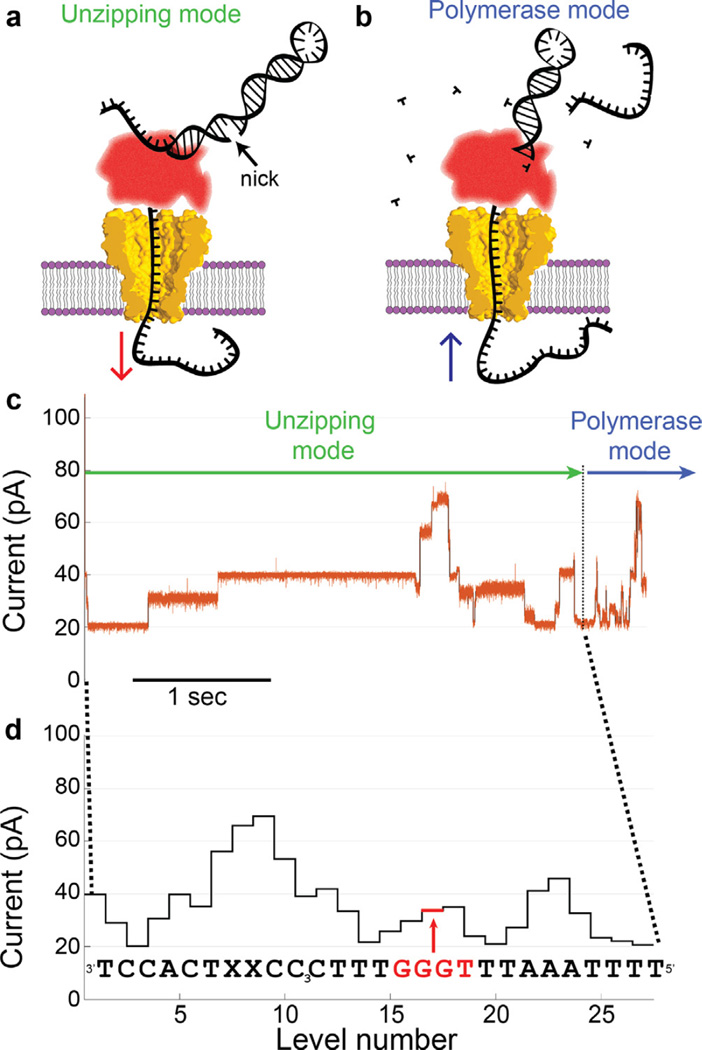
Fig. 4.
Phi29 DNAP controls DNA translocation via the blocking oligomer technique. (a) DNA initially translocates from cis to trans as the electric force on the DNA pulls it through the phi29 DNAP unzipping the frayed end of the duplex DNA like a zipper until it arrives at a nick between the blocking oligomer and the primer strand, at which point the blocking oligomer dissociates. (b) This exposes the extendable end of the primer formed by the hairpinned 3′ end of the template strand. If proper buffer conditions are present, the phi29 DNAP begins extending the primer thereby pulling the template strand back out of the pore from trans to cis. (c) As phi29 DNAP steps DNA through the pore, a series of current levels are observed. Green and blue arrows indicate the current levels that correspond to unzipping and polymerase mode, respectively. (d) Because the durations of the observed current levels have stochastic durations, it can be more clear to depict the observed current levels without duration information so it can be displayed with the corresponding DNA sequence. As in Manrao et al. 2011 [7], the current is sensitive to approximately 4 nucleotides held within the pore constriction. In (d) the four nucleotides centered on a given current level are those that contribute most to the observed current, for instance, the nucleotides GGGT contribute most to the level indicated in (d). We call these four nucleotides a ‘quadromer.’ (c) and (d) modified from [22].
Manrao et al. applied this technique to control DNA motion through MspA (Fig. 4c), and observed better-resolved current steps with durations from milliseconds to seconds. Because of the superior base-resolution of MspA, the series of current amplitudes of each step could be associated with DNA sequence [22]. As in Manrao et al. 2011 [7], single nucleotide substitutions could be detected and approximately four nucleotides within the constriction influenced each observed current level (Fig. 4d). This work laid the groundwork for nanopore strand sequencing [32]. Subsequent studies using the MspA-phi29 DNAP system found that MspA could be used to directly detect epigenetic base modifications [23,25,26] and unnatural bases dNaM and d5SICS [27] within single DNA strands.
2.4. Making sense of long DNA reads
The ability to make long reads of single DNA molecules is one of the key attractions of nanopore sequencing. Long reads of several thousand bases were achieved using the same MspA-phi29 DNAP system described above [24]. This can be done by linking genomic DNA from bacteriophage phi X 174 with adapters that facilitate the loading of phi29 DNAP as in Manrao et al. 2012 [22,24]. In Laszlo et al. 2014, the adapter on one end of a linearized phi X 174 genome, contained a frayed end and the adapter on the other end contained a hairpin with a nick to enable the polymerase mode.
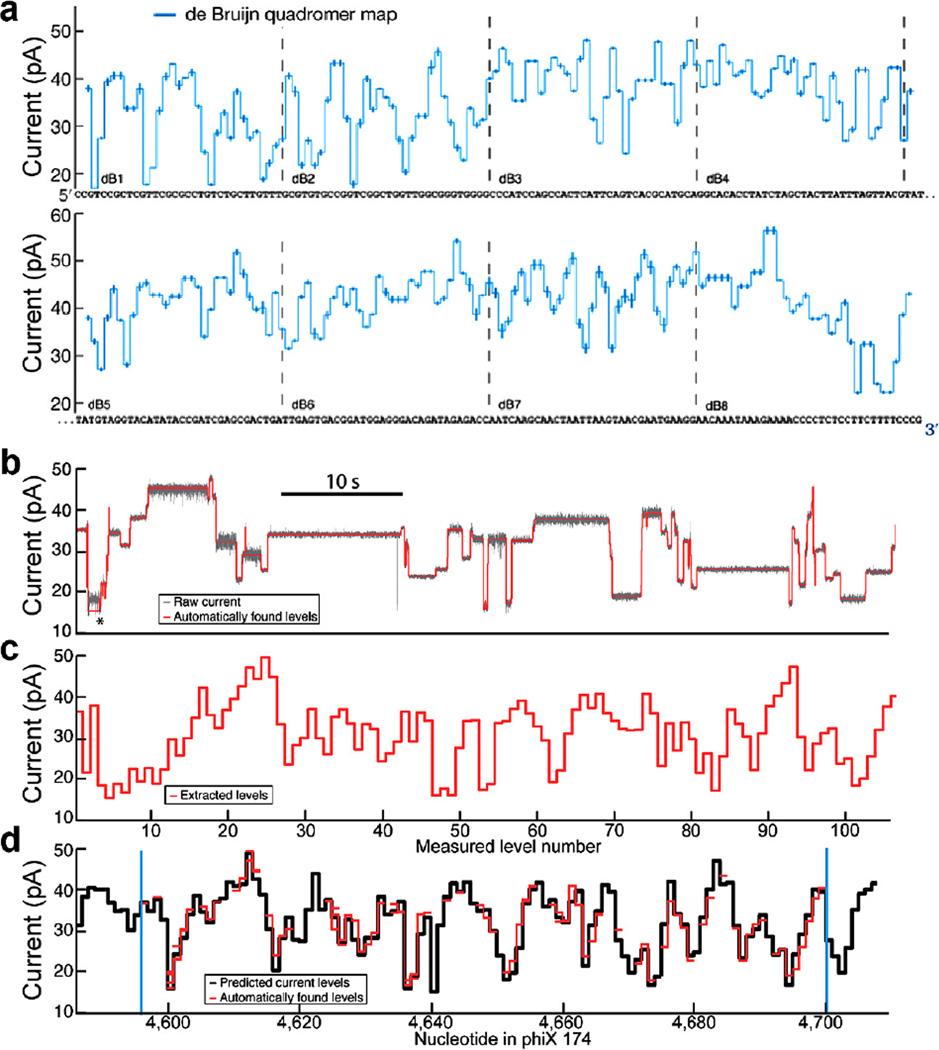
Because ~4 nucleotides within MspA’s constriction influence the observed current values, it was necessary to measure all 256 different sequence combinations of A, C, G, and T (we call each 4-nucleotide combination a quadromer, Fig. 5a). These measurements were conducted on a de Bruijn sequence [33,34] that contained 256 quadromers [24] (Fig. 5a).
Fig. 5.
Quadromer map and alignment to sequence. (a) Graphical representation of quadromer map values aligned to the de Bruijn sequence used to measure all 256 quadromers. Each measured current value is centered on the four nucleotides that contribute significantly to its current value. CCGT corresponds to the first current level displayed, CGTC corresponds to the next current level, etc. (b) Subset of a raw current read of phi X 174 DNA. Average current levels are automatically detected and displayed in (c). (d) These current reads can be aligned to quadromer map based predictions of the reference sequence (using methods described in Appendix C). The ability to perform such alignments allows for association of enzyme behaviors with DNA sequence. Figure modified from [24].
The quadromer map was able to predict the current values that would be observed for other previously unmeasured DNA strands. Translation of the known phi X 174 reference DNA sequence into a series of current levels enabled alignment of the long DNA reads to the reference sequence (Fig. 5b). To make sense of these long reads, it was no longer practical to align the DNA sequences to the measured current levels by hand. Automated alignment techniques were used to associate nanopore current readings to DNA sequence. Such alignments can be used in high-confidence species identification and hybrid genome assembly [24] (using a single ~3800 base nanopore read as a scaffold for assembly of 100 base long Illumina reads, Fig. 5c). While species identification and hybrid genome assembly are not necessary for SPRNT, we list them here to demonstrate the ability of an MspA-based system to identify DNA sequences. As we will see, the ability to accurately identify where an enzyme is along a long DNA strand is a valuable aspect of SPRNT, enabling direct study of DNA-sequence-specific enzyme behavior. (For a full description on how nanopore current reads are preprocessed for alignment see Appendix B, for a full description on how these reads are then aligned to DNA sequence see Appendix C).
2.5. Nanopore sequencing outlook
Nanopore sequencing is now entering commercialization and several companies are pursuing nanopore technologies. Nanopore sequencing has intrinsic advantages of long reads and high speed in addition to its ability to directly detect non-canonical bases [7,21,23,25–27]. Such abilities will allow nanopore sequencing to answer questions accessible by few, if any, currently available sequencing technologies. Nanopores are already being applied to problems of rapid species identification [24,35] and the long reads make them useful for hybrid genome assembly [24,32]. Nanopore devices approaching the market are also remarkably portable [32] enabling them to be deployed rapidly in the field [36]. Challenges still remain with regard to read accuracy, sample preparation (for example delivering small, unamplified genomic samples to the pore), and throughput (currently available nanopore sequencers do not have enough throughput to tackle a human-size genome). Such challenges are the subject of research in academic and industrial laboratories.
2.6. From sequencing to SPRNT
A remarkable consequence of MspA’s sensitivity to small changes within its constriction is that the current is sensitive to not only the nucleotides’ identity but also their precise position along the axis of the pore’s constriction. For instance, DNA held slightly more towards cis within the pore results in a significant change in the observed current. Specific nucleotide sequences can be so sensitive to the exact nucleotide positioning that displacements of the DNA as small as 40 pm can be resolved on one ms time scales [37].1 While such sensitivity to DNA registration within the pore may serve as a challenge to sequencing, it also enables use of MspA as a powerful new tool for analyzing the motion of nucleic acid processing enzymes at unprecedented resolution and therefore allows very detailed studies of how enzymes move along DNA. We cultivated this tool and called it Single-molecule Picometer Resolution Nanopore Tweezers (SPRNT) [37].
3. SPRNT
3.1. Historical use of nanopores for kinetics
Nanopores have been used to study single molecule kinetics for nearly two decades. In 2000, Henrickson et al. varied the applied voltage (thereby varying the applied force) to measure the varying dwell times of molecules held within an aHL pore at varying forces [19]. aHL was later used to measure the kinetics of DNA hairpin unzipping within aHL [39,40] and force dependent dissociation of DNA-exonuclease complexes [41]. Shortly thereafter, researchers used aHL pores to study the stepping kinetics of individual polymerases [30,42,43].
3.2. Introduction to SPRNT
While the use of nanopores to study single molecule kinetics is not new, the key advance that enabled SPRNT was the realization that the currents observed while DNA translocates through the pore can be directly related to the precise longitudinal position of DNA within the pore [37]. SPRNT takes advantage of this exquisite sensitivity to track the progression of DNA through enzymes.
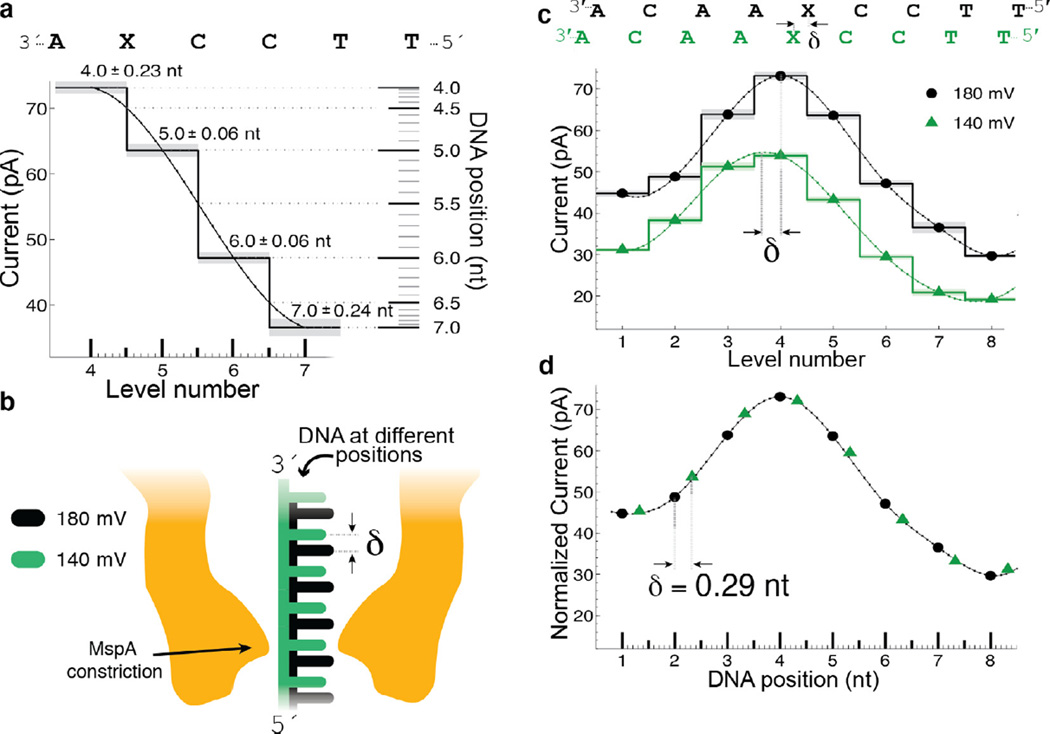
The high spatial resolution of MspA is readily apparent when studying the effect of variations in the applied voltage [37]. Fig. 6 shows a comparison of current levels observed at 180 mV and 140 mV while phi29 DNAP passes DNA containing an abasic residue through the pore. The observed pattern of current levels at 140 mV appears shifted when compared to current levels at 180 mV. In addition to decreasing the current amplitude because of the decreased applied voltage, the voltage change shifts the envelope of the current levels; this shift corresponds to the DNA being held further to the cis side of the pore at 140 mV compared to 180 mV due to the reduced electric force on the DNA. Interestingly, the current levels observed at 140 mV lie along a spline interpolation of the observed 180 mV current levels (Fig. 6d). In other words, measured current level amplitudes can be predicted using a spline interpolant and knowledge of the DNA displacement. Conversely, measurement of the current levels and knowledge of the underlying smooth curve can be used to gain information about the position of the DNA within the pore. In regions where the current levels change significantly for small displacements of the DNA, SPRNT provides a high-precision measurement of DNA position within the pore. In some sequence contexts that exhibit large current differences between adjacent levels, SPRNT has resolution at or below 40 pm on ms time scales.
Fig. 6.
Transduction of current to distance. (a) Regions of high current contrast can be used to measure DNA position precisely. Small uncertainties in measured current translate to small positional uncertainty. (b) Schematic depiction of DNA position within the pore at two different voltages; differences in the applied electric force result in different DNA extensions. (c) Current levels observed for phi29 DNAP controlled motion of DNA through MspA at 180 mV and 140 mV of applied potential. A cubic spline interpolant has been applied to each set of current steps. Note that, apart from a scaling factor, the shape of the spline is identical but the position of the spline has shifted a distance δ. (d) After a linear scale and offset is applied to the two splines, a horizontal displacement δ = 0.29 nt brings the two splines in line with one another. This experiment has two important results: (1) The current levels observed during single-nucleotide stepping of DNA through MspA lie along an underlying smooth curve that is well-approximated by a spline. (2) This spline provides a direct mapping from current to DNA position and we can use it to measure sub-nucleotide movement of DNA. Figure modified from [37].
Precision smaller than the diameter of a hydrogen atom may seem at first impossible or not meaningful given that the experiment takes place at room temperature in water. Naively, one might expect that Brownian motion of the DNA within the pore, Brownian hopping of the enzyme on top of the pore rim, and flopping of the bases along the axis of the pore may prohibit SPRNT’s exquisite spatial resolution. Even though the DNA’s position within the pore is fluctuating significantly more than the precision of the measurement, our measurements indicate that these fluctuations must occur on much faster time-scales than the measurement. This is evident in the experimental fact that the observed current follows a smooth curve and that, on the timescale of the measurement, the uncertainty in the observed current is relatively small compared to the change in current caused by a small translocation of the DNA. Thus, SPRNT is able to provide such a precise measurement of DNA position because it averages over these fluctuations.
3.3. Proof of principle application of SPRNT to another enzyme
In essence, SPRNT turns nanopore DNA sequencing on its head; instead of using enzymes to control the DNA’s motion through MspA in order to measure the DNA’s sequence, we are using our knowledge of the DNA sequence to measure the enzyme’s motions. To perform SPRNT, one needs precise knowledge of the underlying smooth curve that is associated with a given DNA sequence. Alignments of measured DNA levels using another enzyme can then be compared and aligned to this underlying smooth curve. Where along the curve aligned levels reside can then be translated to DNA position within the pore.
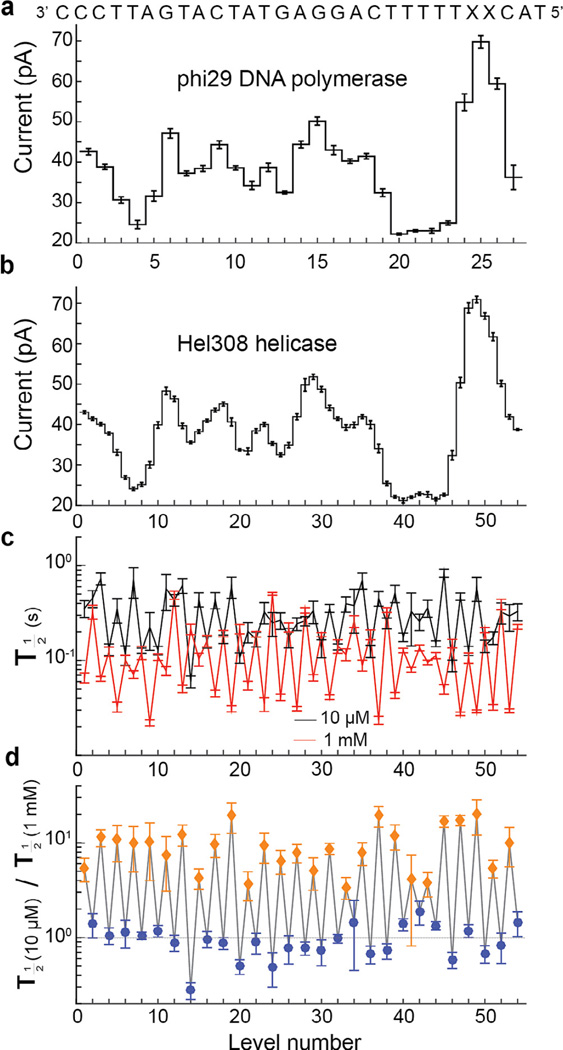
Derrington et al. [37] showed proof of principle demonstration of SPRNT to measure and compare the motions of phi29 DNAP and a Hel308 helicase from Thermococcus gammatolerans. Hel308 is a ski2-like superfamily II helicase/translocase that unwinds dsDNA in the 3° to 5° direction [44]. It is conserved in archaea and eukarya, including humans. Analysis of the crystal structure of Hel308 conjugated with a DNA substrate, in comparison with another superfamily II helicase VASA conjugated to an ATP analog, suggested that Hel308 walks along DNA via an ATP driven inchworm model [45].
SPRNT enabled the observation of this predicted [45], but previously unseen, two-step motion of Hel308. While the phi29 DNAP moves along the DNA strand in single-nucleotide steps via a Brownian ratchet model [31,43,46], Hel308 moves along the DNA strand in two steps per nucleotide, each about 0.5 nucleotides long (Fig. 7a and b; which seems to support a power stroke model proposed by Butner et al. [45]). By comparing these steps to phi29 DNAP, it became clear that the odd- and even-numbered steps correspond to DNA being held 0.14 ± 0.03 nt cis and 0.41 ± 0.03 nt trans of the closest observed phi29 DNAP level, respectively [37]. Because the spline interpolant is not a perfect approximation for the underlying smooth current curve, individual position measurements based off of this comparison to a spline through observed phi29 DNAP current levels carry systematic uncertainty. This systematic uncertainty can be corrected for by either averaging over several measurements at various positions along the DNA (as was done in Derrington et al.) or by improving the underlying curve through empirical measurements of the DNA at different positions within the pore. To achieve the highest spatial resolution with SPRNT, the shape of the underlying smooth current curve must be well understood. Even if this uncertainty makes it difficult to assign a precise position to the DNA, SPRNT’s ability to detect current changes associated with small conformational changes can still be used to gain insights into how enzymes work. For instance, while TIRF-FRET is capable of yielding distance measurements, it is commonly used only as an indicator of changes in enzymatic state [47].
Fig. 7.
Comparison of two enzymes. (a) Levels observed when stepping the indicated DNA strand through MspA with phi29 DNAP controlling the DNA motion and (b) with helicase Hel308 tga controlling the DNA motion. The observed current patterns are similar, however the helicase takes twice as many steps. (c) Comparison of the half-lives of each level at two different concentrations of ATP. (d) Ratio of observed durations from (c). Interestingly, the duration of odd-numbered levels (orange diamonds) varies as [ATP] changes, while even-numbered levels (blue circles) remain unchanged. With the ability to resolve such small substates and observe their chemical dependencies, SPRNT has the potential to shed light on how such motors actually do their work. Figure modified from [37].
Variation of [ATP] can be used to better understand the origin of the two steps observed in Hel308 translocation [37]. Fig. 7c shows the median level duration at 10 µM and 1 mM ATP for the levels shown in Fig. 7b. Taking the ratio of these durations at these two [ATP]’s (Fig. 7d) makes the effect of changing ATP clear: the stepping rate for one of the steps is [ATP] dependent while the other is unaffected by [ATP].
3.4. Generalizing SPRNT to new enzymes
Many of the analysis tools developed for using MspA to sequence DNA are applicable to SPRNT. Setup of a single MspA channel in a bilayer is identical for a SPRNT experiment as it is for a sequencing experiment. The quadromer map developed is directly applicable to interpretation of SPRNT results and the code for analysis of SPRNT data is based on code developed for nanopore sequencing. There are two additional key considerations in adapting SPRNT for use with other enzymes:
It is generally necessary to stall enzyme activity in bulk to ensure progression only begins when the DNA threads into the pore. Without preventing this activity, experiments will run short as enzymes in bulk consume reagents. Strategies to prevent bulk activity may use blocking oligomers, or sections of DNA that serve as temporary road-blocks such as known enzyme pause sites, or stretches of abasic residues or carbon spacers, etc. that impede progression of the enzyme until it comes into contact with the pore. Other schemes will likely arise as the technology develops further.
How a given enzyme sits on the pore will effect where DNA resides within the pore constriction; it may rest higher or lower on the pore rim resulting in current shifts similar to those observed in our voltage stretching experiments. Such registration shifts can be easily adjusted for by using a spline interpolant to shift predicted levels appropriately. Related to this, is the question of the number of steps we expect to observe for a particular enzyme and how we expect them to be spaced. Such questions can be easily answered by use of a standardized DNA sequences for SPRNT. For instance, comparison of Fig. 7a and b clearly suggests the presence of two separate steps per nucleotide. Automated alignments of nanopore data to a prediction based on the quadromer map, as was done in Laszlo et al. [24], will be invaluable.
3.5. Comparison to other techniques
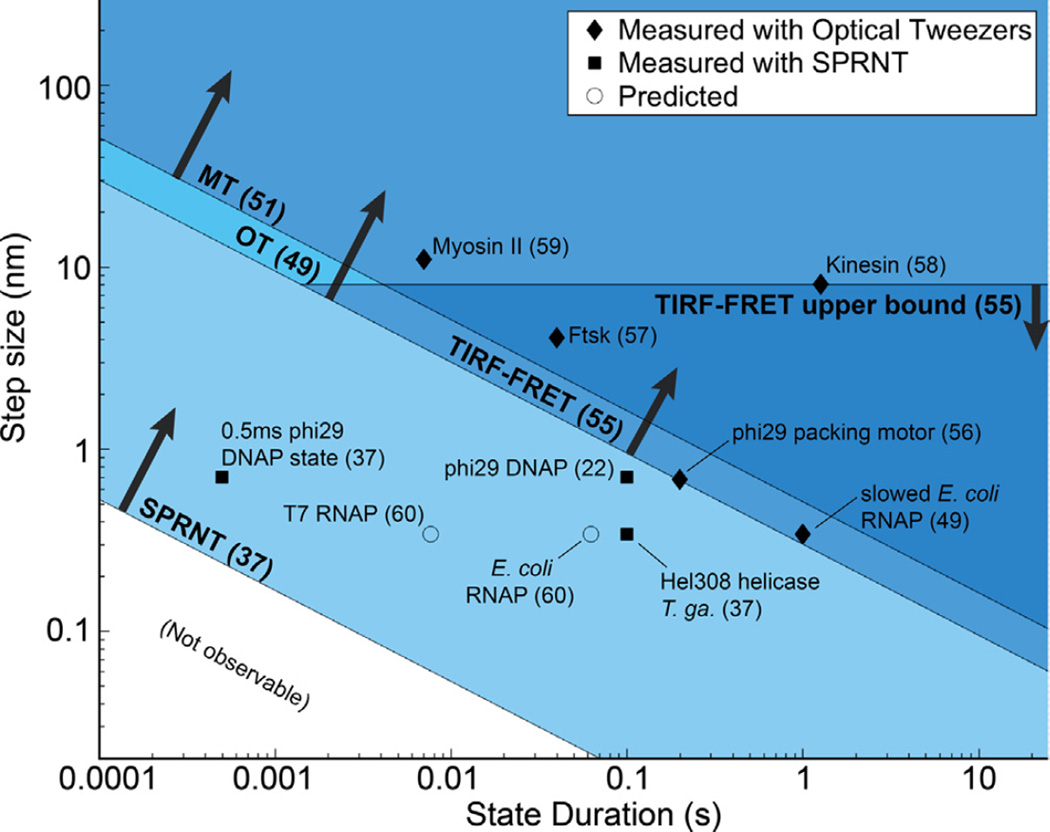
In Figs. 8 and 9 we compare the high spatiotemporal resolution of SPRNT to the spatiotemporal resolutions of other established single-molecule techniques used to study enzymes acting on DNA or RNA. With Gaussian-distributed noise, the spatiotemporal resolution of a given technique will follow a –½-power law relation between observable step-size and observable step duration [48] (Fig. 9). In other words, for a given technique there is a noise budget that is essentially a compromise between spatial and temporal resolution. By averaging for longer intervals, one can observe miniscule differences of state or by sacrificing spatial resolution, one can observe rapid single-molecule transitions. However, this also means some kinetic states are too small and fast to be observed with some techniques. Thus to observe some enzymes that take small steps, researchers have starved enzymes of reagents so that their steps were observable (Fig. 8d and “slowed E. coli RNAP” in Fig. 9) [49]. The spatiotemporal resolution of SPRNT should enable study of previously unobservable enzyme kinetics in conditions that more closely resemble those of the cell.
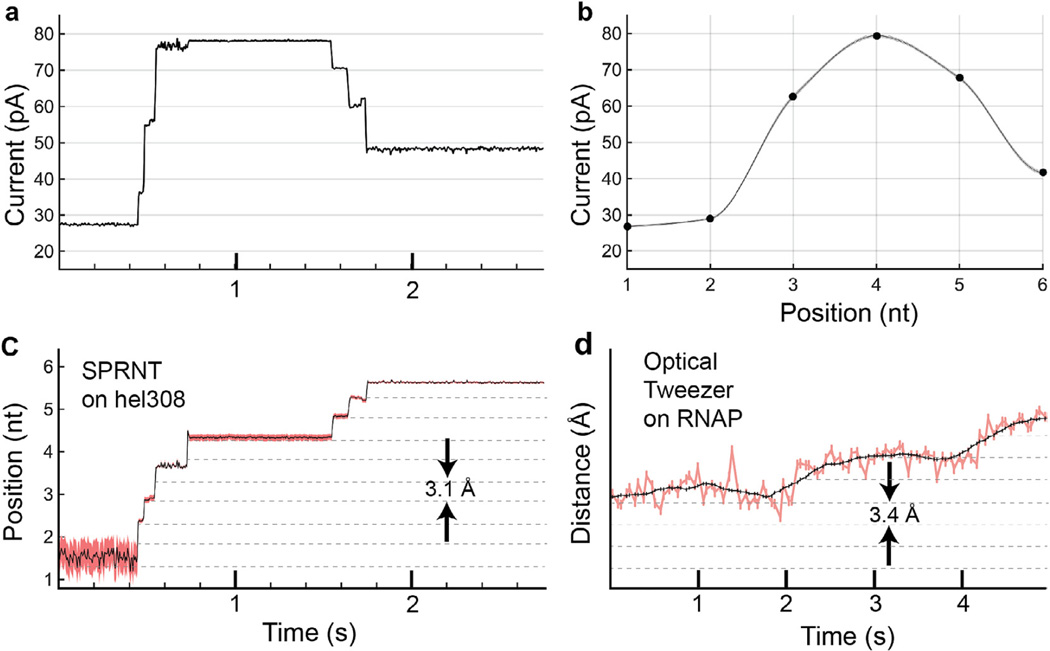
Fig. 8.
Conversion of current trace to DNA displacement and comparison to optical tweezers, OT. (a) Raw current trace from SPRNT measurement of Hel308 helicase median filtered at 200 Hz (5 ms). (b) A piecewise cubic hermite interpolating polynomial (PCHIP) interpolation of current values observed for translocation of the same DNA through an MspA pore using phi29 DNAP. Spline uncertainty, as determined by bootstrapping of measured phi29 current values, is depicted via gray shading and is, in most places, too small to be visible in this graph. (c) The interpolation in (b) is used to translate the currents observed in (a) to DNA position. Pink shading denotes positional uncertainty introduced by statistical variation in the measured phi29 current values. Note that the relatively uniform noise seen in the current trace is either enhanced or suppressed due to the slope of the interpolant. Regions of high slope yield precise measurements of position on short time-scales while regions of low slope yield less precise position measurements. Nanopore data is displayed at 200 Hz. (d) Example OT data of single nt RNAP steps from [49] for comparison. Note, OT data in pink and black is median filtered at 20 Hz (50 ms window) and 1.33 Hz (750 ms window) respectively while nanopore data is median filtered at 200 Hz (5 ms window).
Fig. 9.
Spatio-temporal resolution of SPRNT [37] as compared to optical tweezers, OT [48,49,54], magnetic tweezers, MT [51], and TIRF-FRET [55]. Many nucleic acid processing enzymes work on time-scales and step sizes that are too fast and too small to be resolved with current techniques. The spatiotemporal resolution provided by SPRNT may enable study of previously unobservable kinetic steps. SPRNT has already shown its ability to measure steps on time scales that exceed the spatiotemporal resolution of OT. To provide context, black diamonds indicate average step-sizes and durations for various enzyme steps observed with OT [48,49,56–59], black squares indicate enzymes measured with SPRNT [22,37], while circles represent estimated step-sizes and step durations for several other enzymes based on known stepping rates and suspected step size [60]. The square labeled “0.5 ms phi29 DNAP state” shows the spatiotemporal resolution required to resolve an observed single-step measurement from [37] that falls close to the noise limit. The enzyme and reference number from which it was taken are labeled.
It should be noted that spatiotemporal resolution is just one of several attributes that should be considered in single molecule experiments. For example, the ability to apply force [50] (or to apply no force) is often an important consideration. Table 1 compares other important aspects of various single molecule techniques individually [37,38,47,50,51]. Often single-molecule techniques are combined to observe different reaction coordinates in parallel [47].
Table 1.
In addition to spatiotemporal resolution, the displacement range, force range, ability to be parallelized, and ability to apply torque may be important considerations when planning single-molecule experiments.
| Technique | Displacement range (nm) | Force range (pN) | Massively parallelizable | Can apply torque? |
|---|---|---|---|---|
| SPRNT | 0.04–105 | 15–60 | Yes | No |
| Optical Tweezers | 0.1–105 | 0.1–100 | No | Yes |
| AFM single molecule probe | 0.5–105 | 10–10,000 | No | No |
| TIR-FRET | 2–10 | N/A | Yes | No |
| Magnetic Tweezers | 0.5–105 | 0.001–10,000 | Yes | Yes |
Similar to other single molecule techniques, performing SPRNT may affect the enzyme’s behavior compared to its in vivo behavior. In particular, forces applied to the enzyme by pulling on the DNA strand and the balancing force where the pore rim comes into contact with the enzyme may alter the enzyme’s function. This is similar to optical tweezers where attachment of an enzyme to a bead may affect its function.
Depending on the voltage applied, SPRNT applies forces from 15 to 60 pN in a force-clamped mode. We estimate that MspA applies a force of ~35 ± 10 pN to the DNA when 180 mV is applied across the membrane. More accurate force measurements with SPRNT will require direct and independent force calibration.
3.6. Discussion
The ability to detect DNA displacements as small as 40 pm on millisecond time scales establishes SPRNT as a new tool for probing nucleic acid-protein interactions with unprecedented spatiotemporal resolution. Such resolution enabled detection of two distinct substates within a single ATP hydrolysis cycle of a Hel308 helicase [37] that would have been unobservable with other techniques (Fig. 9). SPRNT’s high spatial precision makes it necessary to consider what exactly the DNA motion means: the measured DNA motion is a result of motion of the DNA relative to the active site of the enzyme plus the motion of the enzyme caused by conformational changes that move the enzyme relative to MspA. Interpretation of SPRNT data must take the origin of the DNA motion into account.
In addition to the ability to measure small DNA displacements, SPRNT provides a simultaneous measurement of an enzyme’s absolute location relative to the sequence of the DNA strand. This enables real-time study of sequence-dependence of enzyme activity. Hints of sequence dependence are already apparent in the SPRNT level duration data in Fig. 7c. Steps that are ATP-dependent or ATP-independent appear to come from statistically significant distributions depending on where along the DNA the enzyme is.
It’s worth noting that the MspA protein and phospholipid bilayer is remarkably robust to changes in experimental conditions. MspA is unchanged in a range of salt conditions from 0 M to 2 M and pH from 0 to 14 [52]. MspA retains pore-forming ability after extraction for 30 min at 100 °C and subsequent 15 min incubation at 80 °C in 2% SDS. While the presence of at least some salt is necessary to achieve an ionic current signal, the use of asymmetric salt conditions [53] should enable enzymatic studies at any desired condition.
SPRNT compares well with other single molecule techniques however it is less generalizable than other single-molecule tools. At present, force can only be applied between a single ssDNA strand and an attached molecule. Experiments that require pulling on microtubules or dsDNA are not yet feasible and complicated experimental geometries that require pulling on multiple DNA strands may be undoable without combining SPRNT with other single-molecule techniques. While SPRNT is currently limited to use with nucleic-acid processing enzymes, it is possible that strategic tethering of a ssDNA probe strand to particular locations on an enzyme could be used to reveal small conformational changes of those enzymes as they function.
SPRNT is at an early stage in its development and there is still work to do to make SPRNT more adaptable to new enzymes and experimental configurations. For example, DNA orientation within the pore affects the observed current (5′ threaded first or 3′ threaded first, see Fig. 3b vs. Fig. 3c). As such, a new quadromer map for 3′ feeding will be needed for certain experiments. Measurements of RNA motion through the pore are likely possible [2] but RNA will require its own set of quadromer maps. Development of DNA sequences that result in large current swings for small displacements will be necessary to maximize SPRNT’s resolution. We have already used MspA’s sensitivity to abasic residues to provide high precision position measurements and it is possible that current patterns caused by unnatural bases will yield even higher resolution. Improvements are also necessary in our understanding of the underlying smooth curve. While a cubic spline is a good interpolation to find the DNA position, it seems unlikely that a spline is the true mathematical form of the underlying curve. As such, the spline introduces small systematic error in places where the underlying curve has curvature (regimes that appear linear likely have less error). Theoretical work that accurately predicts nanopore ion current values with DNA held at various positions or empirical measurement of a known optimal DNA sequence held at different positions will reduce this systematic. Finally, massive parallelization will make SPRNT a high-throughput single-molecule technique, capable of obtaining thousands of single-molecule data-traces within minutes. As such, SPRNT may become a useful drug-discovery platform that enables exploration of how small molecules affect enzymatic reactions.
Acknowledgments
We would like to thank Dr. Richard Ebright, Dr. Paul Wiggins, and Dr. Charles Asbury for helpful discussions regarding Fig. 9. This work was supported by the National Institutes of Health, National Human Genome Research Institute (NHGRI) $1000 Genome Program Grant R01HG005115.
Abbreviations
- ssDNA
single stranded DNA
- dsDNA
double stranded DNA
- MspA
Mycobacterium smegmatis porin A. Octameric outer membrane protein from Mycobacterium smegmatis (accession no CAB56052.1) with mutations D90N, D91N, D93N, D118R, D134R, and E139K
- Phi29 DNAP
DNA polymerase from bacteriophage phi29. (Accession no P03680.1)
- Hel308
Helicase Hel308 from Thermococcus gammatolerans. (Accession no WP_015858487.1)
- SPRNT
Single-molecule Picometer Resolution Nanopore Tweezers
- OT
optical tweezers
- MT
Magnetic Tweezers
- TIRF-FRET
total internal reflection fluorescence forster resonance energy transfer
- dNTP
deoxynucleotide triphosphates
- ATP
adenosine triphosphate
- ADP
adenosine diphosphate
- nt
nucleotide
Appendix A. Single-channel experimental setup
A1. Setup description
In our setup (Fig. 1), originally described by Akeson et al. 1999 [2], two wells are established in a Teflon ‘puck’ and are connected by a Teflon heat-shrink ‘u-tube.’ The two ~50 µl wells are dubbed cis and trans. The u-tube volume is ~30 µl. On the cis well side, the u-tube narrows into a ~20 µm Teflon aperture across which the bilayer is painted. The aperture is formed by first heat shrinking a segment of tubing onto a sharp needle. The end of the tube is then shaved off carefully from the end until a ~20 µm opening is established at the end. Electrical connection to the wells is established using silver-silver chloride electrodes (A-M systems Part no 550010). Electrodes are assembled by attaching a connector pin to the far side and then covering the wire and electrode with Teflon heat shrink tubing (Component company part no SMDT-130-148). Coating the electrode provides electrical insulation and enables a water-tight fit for the electrode into the puck. Several different amplifiers are commercially available that will work for this kind of setup. In our laboratory we use Axopatch 200B amplifiers with a National Instruments PCI-6251 DAQ. The entire experiment takes place ontop of a copper plate and inside of a metal box which is attached to the amplifier ground. This box acts as a faraday cage and filters out electrical noise. The box need only be closed while acquiring data.
A2. Puck cleaning
The pucks are cleaned with Folch solution. Folch solution contains: 33.3 %methanol, 66.6 %chloroform protective gear includes: fume hood, lab coat, long pants, close toed shoes, nitrile gloves, and goggles.
procedure:
**steps 1–8 are conducted in a fume hood
Place pucks and stir-bar in 1 L beaker.
Place beaker on stir plate.
Pour chloroform over pucks until all pucks are covered, approximately 150 ml.
Pour methanol into beaker until desired ratio is achieved, approximately 75 ml.
Cover beaker with watch glass.
Turn on stir plate to 110 rpm and run for 25–40 min.
Decant Folch solution into waste container.
Remove Folch from u-tubes using a 2 ml NORM-JECT syringe.
Fill front well with deionized water and draw through the back well using a BD 3 ml syringe until the u-tube is half filled, remove water from the front well and suck water out of the u-tube.
Fill front well with 100% ethanol and draw through the back well using a BD 3 ml syringe until the u-tube is half filled, remove ethanol from the front well and suck ethanol out of the u-tube.
Fill front well with hexane using a Pasteur pipette and draw through the back well using a 2 ml NORM-JECT syringe until the u-tube is half filled, remove hexane from the front well and suck hexane out of the u-tube.
*if pucks are to be primed immediately, vacuum desiccate for 5 min.
A3. Bilayer formation
Pucks are prepared for an experiment by ‘priming.’ 20 µl of DphPC lipid at 10 mg/ml concentration in chloroform is dried down in the bottom of a test tube in a vacuum desiccator for 10 min (because chloroform attacks some plastics, we attach a glass capillary tube to the end of a pipet tip and suck up the chloroform only into the glass capillary tube). The dried down lipid is then resuspended in 0.066 g of hexane. One µl of the lipid suspended in hexanes is then applied to the Teflon aperture. After this, the operator uses a clean syringe to gently blow air through the trans side of the u-tube to clear any hexane from the aperture. We then let the puck dry for 15 min. After 15 min, a second µl of lipid-hexane suspension is placed onto the aperture in the same manner and let dry for 20 min. We prime several pucks this way at a time. Primed pucks are usable for experiments up to three days after priming but get worse over time; after three days bilayers formed on such pucks have a tendency to be short-lived.
Primed pucks are now ready for experiments. They are fitted with the electrodes and installed in the puck holder, then the wells are filled with buffer solution. It is usually easiest to avoid bubbles in the u-tube by first filling the u-tube, then filling the cis and trans wells. In experiments using asymmetric salt conditions, the bilayer is first established in symmetric salt conditions and then a new buffer is added to the cis well once the pore has been established in the membrane. Note that the u-tube restricts direct access to the trans side of the bilayer. Once the puck has been filled with experimental solution and electrical connection of the two wells has been verified the operator can begin ‘painting’ the bilayer.
The process of bilayer formation is performed under a dissection microscope. Lipid paint is made by taking lipid dried down on glass slides and mixing it with an oil. We have had success with both hexadecane and hexadecene. Mixing oil and lipid to the right consistency is key; if there is too little oil, then the bilayers will tend to be leaky or short-lived, while if there is too much oil, then the bilayers can instead turn into ‘clogs’ of oil within the aperture. The consistency of the paint should be a bit wetter than that of toothpaste. Once the lipid and oil have been mixed on the glass slide, the operator is ready to begin ‘painting’ the lipid onto the Teflon surface around the aperture. For this we often use small paint brushes with synthetic bristles available at an art supply store, that we have cut to have only one or a few bristles. Before use, brushes are cleaned with ethanol and water (note: residual ethanol on the brushes will kill bilayers). Once lipid paint has been applied around the aperture, one can attempt to make a bilayer. This is done by taking a 1–10 µl pipetter and forming a ~5 µl air bubble across the aperture. When the bubble is retracted slowly, often a bilayer is formed across the aperture indicated by zero current on the amplifier. To ensure that it is a bilayer and not a clog, try ‘zapping’ the bilayer by applying a 5 ms, 1 V pulse across the bilayer. If the measured current again rails, there was a bilayer (which is now broken and another bubble will establish it again). If the current does not rail after zapping, there is likely a clog. The clog can be cleared by forcing buffer into the trans side of the u-tube with a syringe. It is also important that the noise characteristics of the bilayer are ideal. Once a bilayer is established and the faraday cage closed, the rms noise at 5 kHz bandwidth should be above 0.45 pA and below 0.8 pA, indicating a good bilayer. While there are many sources of bilayer noise, bilayer noise can often be reduced by scraping the paint off the surface with a pipette tip and re-painting. Once a good bilayer is established, the operator can begin trying to insert nanopores into the bilayer.
A4. Nanopore insertion
We begin nanopore insertion by adding approximately half a µl of 1 µg/ml MspA into the cis well and waiting for several minutes. If a pore does not insert within a few minutes, more protein is added. The amount of additions and concentration of additions can be variable and titration may be necessary. Because MspA is diluted in a detergent (0.1% OPOE) too many additions of protein to the cis well (>2 µl of MspA in 0.1% OPOE) can disrupt bilayers. The concentration of MspA additions should be tuned so that an insertion can be acquired with minimal addition of detergent to the experimental volume. A pore is inserted into the bilayer once the current goes from zero to a characteristic single-pore current. The characteristic current varies depending on the conductance of the buffering solution, at 300 mM KCl a single MspA pore will have a current of ~115 pA, while a bilayer with two pores will have a current of ~230 pA. Once a single pore has inserted, the operator quickly perfuses new clean buffer into the cis well, rinsing out and reducing the MspA concentration within the cis well to reduce the chance of a second insertion. An operator perfuses the cis well with two syringes, one flowing in fresh buffer and another taking out excess buffer at the same rate so that the well does not overflow. Typically perfusing 1 ml of buffer through the cis well is sufficient to prevent further MspA insertions. It is important that the operator ground their syringes first by touching the buffer solution to a droplet on a grounded surface to avoid shocking and breaking the bilayer upon contact with the cis well. Once a single pore has been established in a bilayer, the operator can proceed with whichever experiment they would like to run. A well-trained operator can go from a primed puck to having a pore in as little as ten minutes.
We have found that evaporation can lead to conductivity changes of the solution over the course of tens of minutes. To prevent this, we place plastic well caps over the cis and trans well meniscuses. Well caps consist of the conical section of a 1.5 ml Eppendorf tube (carefully cut off with a razor blade). A droplet of buffer can be made to sit in the tip of the cone, held stationary by surface tension. When the well cap is placed over the well meniscus, the cap ensures that the small air volume over the well is saturated with water vapor and therefore minimizes evaporation. Without well caps, the conductivity of the buffer can vary by as much as 20%.
A5. Troubleshooting tips
To get good electrical connection between the two wells, it’s important to first fill the u-tube from the trans side and then fill the cis and trans wells. If there are bubbles in the u-tube, suction from the trans side with a syringe will remove them (albeit slowly). Applying pressure makes the bubbles shrink momentarily, but when the pressure is relieved, they will grow again and may again block the u-tube.
If buffer solution is leaking out around the electrode holes, one can get a better seal by applying some vacuum grease on the outside of the electrodes.
Over time, depletion of chloride ions from the electrodes can result in varying electrode voltage offsets that interfere with your amplifiers’ ability to maintain a constant voltage across the pore. Such offsets can be avoided by either rechloridizing your electrodes in bleach or by shaving off the electrode surface with a razor blade once every other week.
If bilayers are difficult to form, first check the consistency of the lipid paint. It may be too dry. Failing that, the puck priming may be bad; switching to another puck or priming a new puck with a different priming solution may be necessary. Lipid also goes bad after several weeks when exposed to oxygen. We’ve found that storing our lipid stocks under argon gas greatly improves its lifespan.
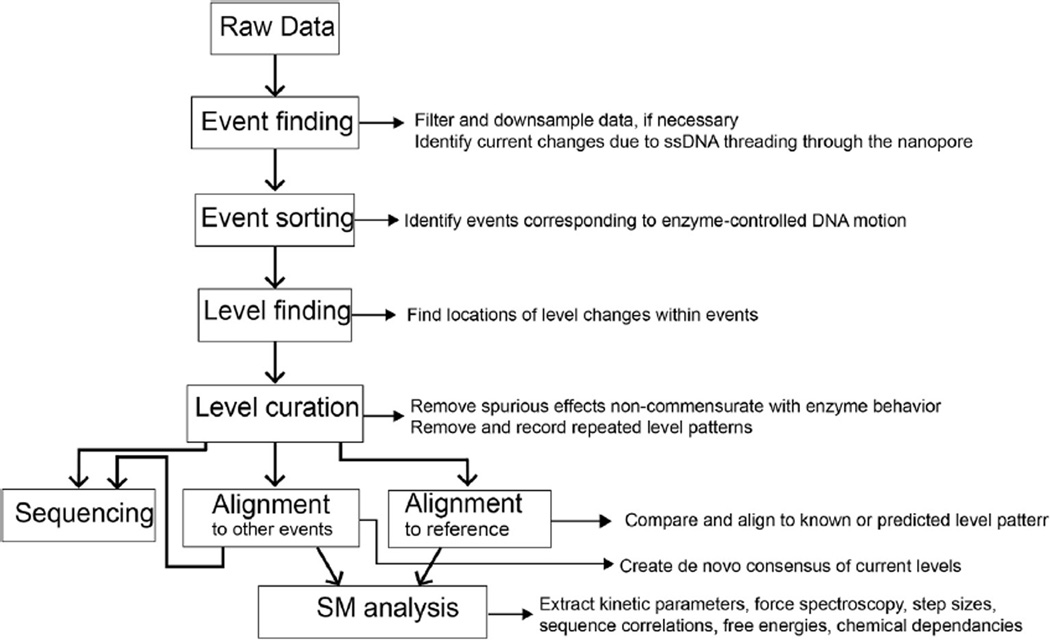
Appendix B. Data analysis pipeline
This section describes the data reduction and analysis as it occurs in the Gundlach Laboratory. Much of the preprocessing code used for SPRNT is available as supplemental information in ref [24]. Data analysis and reduction takes place over several stages (Fig. B1). Initial data is recorded into a raw data file at 500 kHz. Raw files are then preprocessed which consists of three stages. Initially, the file header is decoded and translated into a file containing meta data about the experiment. The data is next downsampled to 5000 Hz for use in later analysis. The next stage of preprocessing consists of searching the reduced data for DNA interaction events. This is performed via thresholding. First the open pore current is determined by finding the maximum in the current histogram above 90 pA and below 200 pA. Then an event is defined to begin when the current drops below 75% of the open pore current, while an event end is defined by current returning to above 94% of open pore current. The positions of these events and metadata about each of the events (average, standard deviation, duration, etc.) are stored for later analysis. Events matching certain criteria (usually events are longer than 1 s and an average between 10 and 70% of the open pore current) are automatically selected from the data to enable by-eye sorting.
After preprocessing, we classify the events based on their quality. In sequencing experiments using long DNA reads, it is sufficient to classify events lasting longer than a certain duration as good events and all others not worth using (a typical phi29 DNAP controlled unzipping event like those obtained in Laszlo et al. goes through DNA at ~1 base per second). When using short DNA strands, or testing a new enzyme with SPRNT it is often better to sort through events by eye as the events are harder to classify automatically. Classifying events by eye is done using a graphical user interface that allows the user to quickly scroll through flagged events and categorize them for later analysis.
We then use an automated level-finding algorithm to find current level transitions within good events. The output of this algorithm includes information on levels found within every ‘good’ event found in the original raw data file. The level finding algorithm consists of several steps: initial level finding, filtering, and removal of ‘toggles.’ Initial level finding proceeds iteratively through the data beginning with a window of size t points. It then evaluates points between 0 and t while querying whether the likelihood that the left and right consist of two separate distributions exceeds the likelihood that they are the same distribution by a set threshold. A transition point is called when this likelihood measure is maximal. The algorithm then continues iteratively through the left and right subsections of data seeing if it can further divide the data using the set threshold. The algorithm continues until it finds no more division points or the subsections are shorter than a set length L. This algorithm has two important parameters that will affect the result: the likelihood threshold and the minimum subsection length. This algorithm is prone to finding spurious spikes in the data that result in unnecessary division of the subsections. While these spikes are indeed statistically significant, tuning the algorithm to ignore them can result in too few levels being found. As such, we find it useful to filter the data after level-finding to remove such spurious levels and recombine levels that were unnecessarily split. This filtering step searches through found levels and cuts out levels that were shorter than a certain duration (the default value is 20 points for data at 5 kHz sample rate). Finally, the enzyme itself can sometimes introduce errors into the sequencing signal. In particular, it can step backwards before going forwards again. Such backsteps add extra levels to the series of current levels and can cause sequencing errors downstream. Thus our final toggle-filtering step removes and simplifies any levels that appear to take an ABABAB…C pattern of stepping and recombines them. Levels at all three stages of filtering (raw, filtered, and toggle filtered) are stored for future analysis. (When analyzing enzyme kinetics, these backsteps are important and are therefore not filtered however accurate sequencing requires removal of such errors.)
Fig. B1. Flow chart of data-processing pipeline. Squares linked by bold arrows outline individual processing steps while small arrows and text describe outputs at each stage of the analysis.
While the quadromer map is predictive, we have found that there are context dependent changes in observed current. For instance, a particular quadromer can measured in one sequence context can differ significantly that same quadromer in a different context. It appears that such differences are not simply due to additional nucleotides adjacent to the 4 that make up the quadromer. We propose that such inconsistencies are due to nucleotide-dependent differences in the stretchiness of DNA between where it is held by the enzyme and where it is read, in MspA’s constriction. Such differences could lead to small sequence-dependent positional shifts of the DNA within the pore constriction. We correct for this sequence-dependent systematic error by making an empirical consensus of observed current values. We do this automatically via iterative cycles of alignment of events to a predicted consensus followed by consensus correction. The procedure is described in detail in Derrington et al. 2015 [37].
Appendix C. Alignment of nanopore currents
Alignment of nanopore current levels to one another and to reference sequences is done via a dynamic programming algorithm such as Needleman-Wunch alignment [61,62]. There are many such alignment algorithms, each with their own virtues [62]. In this appendix we first describe basic Needleman-Wunch alignment and then describe a variation on it developed in our laboratory that is optimized for alignment of nanopore reads to a known reference.
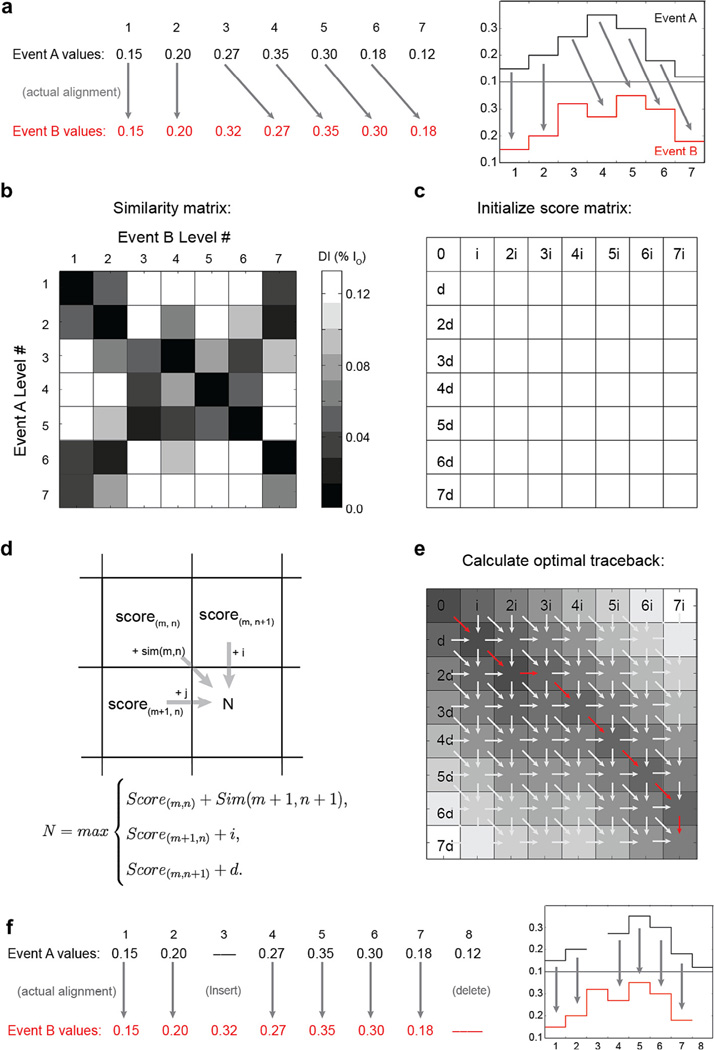
Lets begin by using Needleman-Wunch alignment to align two events A and B (Fig. C1). Events A and B have similar current patterns, however B has an extra level as the third level and event B ends prematurely and is missing the last level observed in A. Basic Needleman-Wunch alignment begins with a comparison of each level in event A to each level in event B (Fig. C1c). We first populate the “similarity matrix,” a matrix that is as tall as event A is long and as wide as event B is long. For simplicity, in this example we will use a simple scoring metric to decide if two levels are similar: the current difference between levels (in practice, more statistically realistic measures like a student’s T-test yield better results). Note that levels that should align have good match scores, indicated by dark squares in the similarity matrix. Next, we initialize the “score matrix.” Note the score matrix is one entry taller and wider than the similarity matrix. To initialize the score matrix we populate the first row and first column with multiples of the “insert penalty” i and “delete penalty” d respectively. Fig. C1d then demonstrates how to fill out the remainder of the score matrix. The score N of the box in position (m + 1,n + 1) is calculated using the equation in Fig. C1d. Where sim is the similarity matrix in Fig. C1c. Following this rule, the score matrix is then filled out iteratively from top to bottom, left to right. Fig. C1e shows a schematic for how the optimal alignment path is then determined. In this instance, we are trying to determine the optimal global alignment of event A with event B, that is we want to align all of event A to all of event B. The optimal alignment then is the path of steps taken that resulted in the score calculated in the bottom right element of the score matrix (red arrows in Fig. C1e). This path can be determined either by keeping track of all choices made while filling out the score matrix (in a separate matrix called the ‘traceback matrix’) or can be re-calculated by starting at the bottom right corner and tracing backwards using the rule in Fig. C1d to determine which choice was made to yield the final alignment score. How the trace-back proceeds through the score matrix determines how the two events best align to one another. Note that this technique is guaranteed to provide the optimal global alignment of event A to event B given the allowed steps and stepping penalties. When used in biological sequence alignment, values of the similarity matrix are log probabilities for how likely it is for two such elements of A and B to match. Similarly, i and d are the log probabilities of such types of errors taking place. Proper tuning of similarity scores and stepping penalties is necessary for proper alignment.
Fig. C1. Example Needleman-Wunch alignment. (a) Here we align two events, A and B, that each contain errors in the form of missing or extra levels. The correct alignment of the events is indicated by arrows. (b) A graphical representation of the similarity matrix shows a comparison of each level in event A with each level in event B. The dark square at position (3.4) indicates a good match between level 3 in A and level 4 in B, while the white square at (1.6) indicates that level 1 in A and level 6 in B differ significantly. Computation of the similarity matrix elements is the first step in Needleman-Wunch alignment. (c) Next the score matrix is initialized by filling the first row and column of the score matrix with iterative applications of the delete penalty or insert penalty, respectively. (d) Next, the remaining elements of the score matrix are calculated from left to right, top to bottom, using the rule shown in d. (e) Graphical representation of the score matrix, darker shades indicate better alignment scores. The traceback (red series of arrows) is determined by tracing the path chosen backwards at each square that resulted in the score calculated in the bottom right matrix element. (f) The traceback reveals the optimal alignment of event A to event B, given the allowed steps and error penalties. Properly tuned i and d result in more accurate alignments. Figure originally appeared in [63].
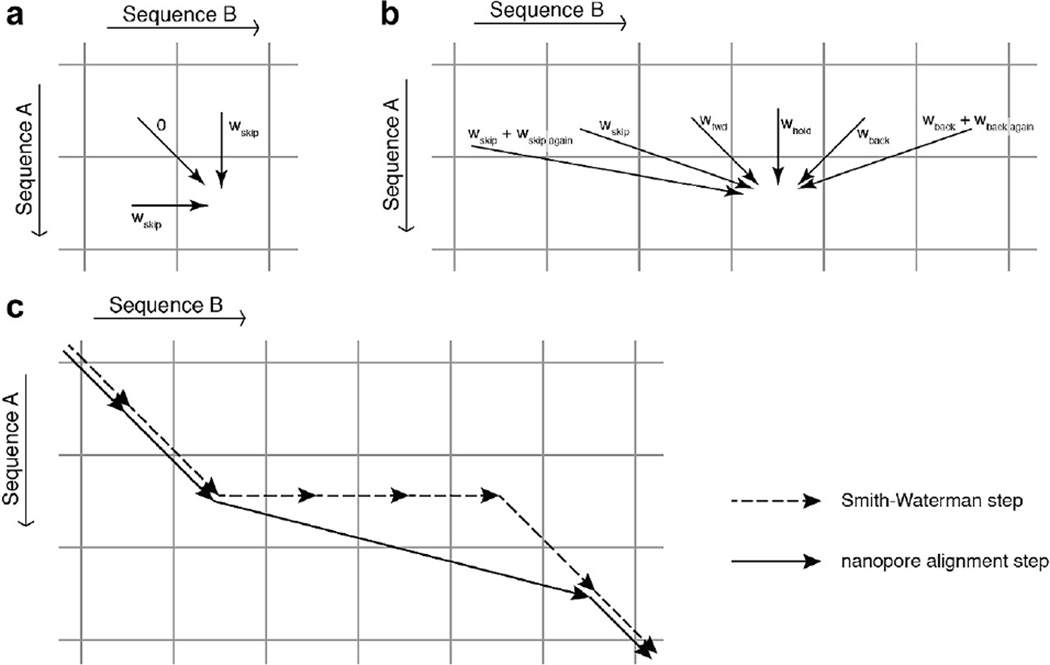
Many variations on Needleman-Wunch alignment exist [62]. By varying or changing the rules of such alignments one can do various kinds of alignment. For instance, if event A were far longer than event B and we suspected that event B started at the same position as event A but was a subset of the levels of event A, we would make the following change: instead of the starting our traceback at the lower right corner of the score matrix, we would allow the trace-back to begin anywhere along the right side of the matrix. We would select the start point as the point along the right side of the matrix with the best score.
Fig. C2. Schematic comparing the stepping rules allowed in (a) Needleman-Wunch alignment and (b) ‘Ross alignment.’ Ross alignment is designed for the alignment of nanopore current reads to predicted ion current levels based on a known consensus or quadromer map. In such an alignment we expect stepping ‘errors’ by the polymerase or helicase such as back-stepping or skipping forward in the sequence of levels. To allow for these errors, in Ross alignment we force the alignment to always go forward in level sequence A as in (b), this allows for the possibility of back-stepping. (c) All paths that are possible in Needleman-Wunch alignment are possible in Ross alignment. Figure from [24].
In our laboratory, we developed a variation on such alignment methods that is optimized for alignment of nanopore sequencing data to a reference sequence [24]. We call this variation “Ross alignment” for Brian Ross who invented it. In Ross alignment, we align an event containing nanopore sequencing errors like skips and backsteps to a reference sequence which is not expected to have errors. As such, the rules for filling out the score matrix are changed significantly. For this example, let event A be the measured nanopore read, and event B be the predicted current levels for the known reference sequence. Instead of allowing the alignment to either move forwards in both events, move forwards in A, or move forwards in B, we enforce that we must move forwards in A (Fig. C2b). Under this assumption, alignment of event A to event B can be interpreted as event A consisting of steps forwards and backwards along B. In Ross alignment the score matrix is filled out one row at a time element by element following the rules outlined in Fig. C2b. Matching element m of event A to element n of event B can be achieved via a ‘step forward’ from (m − 1, n − 1), a ‘hold’ from (m − 1, n), a ‘skip’ from (m − 1, n − 2), or a ‘backstep’ from (m − 1, n + 1). Additional numbers of skips and backsteps are also allowed via what is called an affine gap scoring method [62]. Note, that forward progression in the measured event is enforced by the fact that each step must come from the row above it. If the similarity score of two elements is below some threshold value wbad, the measured level can be considered to be a bad level so that it does not get included in the alignment. This allows the algorithm to skip over any spurious levels that may be introduced by overzealous level-finding, pore ‘gating,’ or sharp, brief current spikes caused by bilayer instabilities.
All paths that are possible in a Needleman-Wunch alignment are possible in a Ross alignment (Fig. C2c). However, unlike Needleman-Wunch alignment which is symmetric (when i = d) if events A and B are transposed, Ross alignment is asymmetric in its treatment of events A and B. As such, it should not be used for comparison of sequences that are on equal footing such as comparison of one event to another. It would yield different alignments depending on if event A were aligned to event B or if event B were aligned to event A.
Appendix D. Methods for constructing Fig. 9
Moffitt et al. [48] describe the inverse relationship between temporal and spatial resolution for optical tweezers:
where Δl is the length of the observable step, t is the observable step duration, SNR is the signal to noise ratio, and for a dual optical trap with identical beads, C is some constant dependent upon temperature, the number of measurements made, the drag coefficient of the beads, and the tension of the DNA tether attaching the two beads. This expression is generalizable to other techniques assuming that the noise in the signal is Gaussian distributed. It generally states that the minimum size of the step that one can observe is inversely proportional to the square root of the amount of time you are allowed to observe it. The optical tweezers line in Fig. 9 is exactly this relationship described in Moffitt et al. [48]. Positions for resolution limits of MT, TIRF-FRET, and SPRNT are based on standardized comparisons of signal to noise ratios of raw data from each technique to the best resolution achieved with optical tweezers [49].
We began by calculating benchmark signal to noise measurements for the highest resolution result that we could find in OT [49], MT [51], TIRF-FRET [55], and SPRNT [37]. Because each of these benchmark datasets are acquired at different sampling rates and different-sized steps, we normalize these SNRs by dividing the SNR by the step size and multiplying it by the square root of the sampling rate. The resulting ‘scaled SNR’ (sSNR) [54] can be interpreted as the SNR that would be observed with this technique for a 1 nm step at 1 Hz sampling rate [54]. The calculated sSNR values were sSNRSPRNT = 2360; sSNROT = 41.6; sSNRMT = 24.3; and sSNRTIRF-FRET= 41.6. Taking the ratio of sSNROT relative to each of the techniques’ sSNR gives a comparison of the resolving power for each of the techniques relative to OT. For instance sSNROT/sSNRMT = 1.7 tells us that OT has ~1.7 times the resolution available to MT, therefore, the line in Fig. 9 that represents MT’s resolution can be found 1.7× above the resolution limit of OT.
Datapoints that represent measured step-sizes and step durations and estimated step sizes and durations were pulled from the literature.
Footnotes
This has important implications for sequencing. For instance, changing electrode offsets caused by depletion of AgCl in the electrodes can cause the applied voltage (and thereby the applied force) to change resulting in repositioning of the DNA within the pore. Measured currents would be modified by both differences in voltage and registration shifts of the DNA within the pore, complicating the sequencing signal. Such a registration shift may be the cause of inaccuracies observed as the applied voltage changes as electrodes degrade in the Oxford MinION device [38].
References
- 1.Kasianowicz JJ, et al. Characterization of individual polynucleotide molecules using a membrane channel. Proc. Natl. Acad. Sci. USA. 1996;93(24):13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akeson M, et al. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 1999;77(6):3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purnell RF, Mehta KK, Schmidt JJ. Nucleotide identification and orientation discrimination of DNA homopolymers immobilized in a protein nanopore. Nano Lett. 2008;8(9):3029–3034. doi: 10.1021/nl802312f. [DOI] [PubMed] [Google Scholar]
- 4.Stoddart D, et al. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc. Natl. Acad. Sci. USA. 2009;106(19):7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler TZ, et al. Single-molecule DNA detection with an engineered MspA protein nanopore. Proc. Natl. Acad. Sci. USA. 2008;105(52):20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derrington IM, et al. Nanopore DNA sequencing with MspA. Proc. Natl. Acad. Sci. USA. 2010;107(37):16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manrao EA, et al. Nucleotide discrimination with DNA immobilized in the MspA nanopore. PLoS ONE. 2011;6(10):e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wanunu M, et al. Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors. Nat. Nanotechnol. 2010;5(11):807–814. doi: 10.1038/nnano.2010.202. [DOI] [PubMed] [Google Scholar]
- 9.Storm AJ, et al. Fabrication of solid-state nanopores with single-nanometre precision. Nat. Mater. 2003;2(8):537–540. doi: 10.1038/nmat941. [DOI] [PubMed] [Google Scholar]
- 10.Li J, et al. Ion-beam sculpting at nanometre length scales. Nature. 2001;412(6843):166–169. doi: 10.1038/35084037. [DOI] [PubMed] [Google Scholar]
- 11.Garaj S, et al. Graphene as a subnanometre trans-electrode membrane. Nature. 2010;467(7312):190–U73. doi: 10.1038/nature09379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merchant CA, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10(8):2915–2921. doi: 10.1021/nl101046t. [DOI] [PubMed] [Google Scholar]
- 13.Schneider GF, et al. DNA translocation through graphene nanopores. Nano Lett. 2010;10(8):3163–3167. doi: 10.1021/nl102069z. [DOI] [PubMed] [Google Scholar]
- 14.Liu K, et al. Atomically thin molybdenum disulfide nanopores with high sensitivity for DNA translocation. ACS Nano. 2014;8(3):2504–2511. doi: 10.1021/nn406102h. [DOI] [PubMed] [Google Scholar]
- 15.Yusko EC, et al. Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat. Nano. 2011;6(4):253–260. doi: 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reiner JE, et al. Theory for polymer analysis using nanopore-based single-molecule mass spectrometry. Proc. Natl. Acad. Sci. USA. 2010;107(27):12080–12085. doi: 10.1073/pnas.1002194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson JW, et al. Single-molecule mass spectrometry in solution using a solitary nanopore. Proc. Natl. Acad. Sci. USA. 2007;104(20):8207–8211. doi: 10.1073/pnas.0611085104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fologea D, et al. Electrical characterization of protein molecules by a solid-state nanopore. Appl. Phys. Lett. 2007;91(5):539011–539013. doi: 10.1063/1.2767206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henrickson SE, et al. Driven DNA transport into an asymmetric nanometer-scale pore. Phys. Rev. Lett. 2000;85(14):3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 20.Stoddart D, et al. Multiple base-recognition sites in a biological nanopore: two heads are better than one. Angew. Chem. Int. Ed. 2010;49(3):556–559. doi: 10.1002/anie.200905483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wallace EVB, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chem. Commun. 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat. Biotechnol. 2012;30(4):349–U174. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laszlo AH, et al. Detection and mapping of 5-methylcytosine and 5-hydroxymethylcytosine with nanopore MspA. Proc. Natl. Acad. Sci. USA. 2013;110(47):18904–18909. doi: 10.1073/pnas.1310240110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat. Biotechnol. 2014;32(8):829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schreiber J, et al. Error rates for nanopore discrimination among cytosine, methylcytosine, and hydroxymethylcytosine along individual DNA strands. Proc. Natl. Acad. Sci. USA. 2013;110(47):18910–18915. doi: 10.1073/pnas.1310615110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wescoe ZL, Schreiber J, Akeson M. Nanopores discriminate among five C5-cytosine variants in DNA. J. Am. Chem. Soc. 2014;136(47):16582–16587. doi: 10.1021/ja508527b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Craig JM, et al. Direct detection of unnatural DNA nucleotides dNaM and d5SICS using the MspA nanopore. PLoS ONE. 2015;10(11):e0143253. doi: 10.1371/journal.pone.0143253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purnell RF, Schmidt JJ. Discrimination of single base substitutions in a DNA strand immobilized inabiological nanopore. ACS Nano. 2009;3(9):2533–2538. doi: 10.1021/nn900441x. [DOI] [PubMed] [Google Scholar]
- 29.Gyarfas B, et al. Mapping the position of DNA polymerase-bound DNA templates in a nanopore at 5 Å resolution. ACS Nano. 2009;3(6):1457–1466. doi: 10.1021/nn900303g. [DOI] [PubMed] [Google Scholar]
- 30.Lieberman KR, et al. Processive replication of single DNA molecules in a nanopore catalyzed by phi29 DNA polymerase. J. Am. Chem. Soc. 2010;132(50):17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-angstrom precision. Nat. Biotechnol. 2012;30(4):344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ashton PM, et al. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat. Biotechnol. 2014;33:296–300. doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 33.de Bruijn NG. A combinatorial problem.. Koninklijke Netherlandse Akademie v. Wetenschappen. 1946;49:758–764. [Google Scholar]
- 34.Compeau PEC, Pevzner PA, Tesler G. How to apply de Bruijn graphs to genome assembly. Nat. Biotechnol. 2011;29(11):987–991. doi: 10.1038/nbt.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilianski A, et al. Bacterial and viral identification and differentiation by amplicon sequencing on the MinION nanopore sequencer. Gigascience. 2015;4:12. doi: 10.1186/s13742-015-0051-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quick J, et al. Real-time, portable genome sequencing for Ebola surveillance. Nature. 2016;530(7589):228–232. doi: 10.1038/nature16996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derrington IM, et al. Subangstrom single-molecule measurements of motor proteins using a nanopore. Nat. Biotechnol. 2015;33(10):1073–1075. doi: 10.1038/nbt.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ip CL, et al. MinION analysis and reference consortium, MinION analysis and reference consortium: phase 1 data release and analysis [version 1; referees: 2 approved] F1000 Res. 2015;4:1075. doi: 10.12688/f1000research.7201.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mathe J, et al. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 2004;87(5):3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sauer-Budge AF, et al. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 2003;90(23):238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 41.Hornblower B, et al. Single-molecule analysis of DNA-protein complexes using nanopores. Nat. Methods. 2007;4(4):315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 42.Cockroft SL, et al. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J. Am. Chem. Soc. 2008;130(3):818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieberman KR, et al. Kinetic mechanism of translocation and dNTP binding in individual DNA polymerase complexes. J. Am. Chem. Soc. 2013;135(24):9149–9155. doi: 10.1021/ja403640b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Woodman ILB, Wedward L. Winged helix domains with unknown functions in Hel308 and related helicases. Biochem. Soc. Trans. 2011;39:140–144. doi: 10.1042/BST0390140. [DOI] [PubMed] [Google Scholar]
- 45.Buttner K, Nehring S, -P K. Hopfner, Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007;14(7):647–652. doi: 10.1038/nsmb1246. [DOI] [PubMed] [Google Scholar]
- 46.Morin JA, et al. Mechano-chemical kinetics of DNA replication: identification of the translocation step of a replicative DNA polymerase. Nucl. Acids Res. 2015;43(7):3643–3652. doi: 10.1093/nar/gkv204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joo C, et al. Advances in single-molecule fluorescence methods for molecular biology. Ann. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 48.Moffitt JR, et al. Recent advances in optical tweezers. Ann. Rev. Biochem. 2008;77:205–228. doi: 10.1146/annurev.biochem.77.043007.090225. [DOI] [PubMed] [Google Scholar]
- 49.Abbondanzieri EA, et al. Direct observation of base-pair stepping by RNA polymerase. Nature. 2005;438(7067):460–465. doi: 10.1038/nature04268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neuman KC, Nagy A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods. 2008;5(6):491–505. doi: 10.1038/nmeth.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dulin D, et al. High spatiotemporal-resolution magnetic tweezers: calibration and applications for DNA dynamics. Biophys. J. 2015;109(10):2113–2125. doi: 10.1016/j.bpj.2015.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heinz C, Engelhardt H, Niederweis M. The core of the tetrameric mycobacterial porin MspA is an extremely stable b-sheet domain. J. Biol. Chem. 2003;278(10):8678–8685. doi: 10.1074/jbc.M212280200. [DOI] [PubMed] [Google Scholar]
- 53.Wanunu M, et al. Electrostatic focusing of unlabelled DNA into nanoscale pores using a salt gradient. Nat. Nano. 2010;5(2):160–165. doi: 10.1038/nnano.2009.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moffitt JR, et al. Differential detection of dual traps improves the spatial resolution of optical tweezers. Proc. Natl. Acad. Sci. USA. 2006;103(24):9006–9011. doi: 10.1073/pnas.0603342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holden SJ, et al. Defining the limits of single-molecule FRET resolution in TIRF microscopy. Biophys. J. 2010;99(9):3102–3111. doi: 10.1016/j.bpj.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chemla YR, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122(5):683–692. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 57.Saleh Fast OA, et al. DNA-sequence independent translocation by FtsK in a single-molecule experiment. EMBO J. 2004;23(12):2430–2439. doi: 10.1038/sj.emboj.7600242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Svoboda K, et al. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365(6448):721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 59.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: piconewton forces and nanometre steps. Nature. 1994;368(6467):113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 60.Seidel R, Dekker C. Single-molecule studies of nucleic acid motors. Curr. Opin. Struct. Biol. 2007;17(1):80–86. doi: 10.1016/j.sbi.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Needleman SB, Wunsch CD. A general method applicable to search for similarities in amino acid sequence of 2 proteins. J. Mol. Biol. 1970;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 62.Durbin R, et al. Biological Sequence Analysis. New York: C. Press; 2006. pp. 92–96. [Google Scholar]
- 63.Laszlo AH. Nanopores for DNA sequencing and epigenetic detection with a MspA nanopore ProQuest Dissertations And Theses. University of Washington: Department of Physics; 2014. p. 153. [Google Scholar]