SUMMARY
To provide a detailed analysis of the molecular components and underlying mechanisms associated with ovarian cancer, we performed a comprehensive mass spectrometry-based proteomic characterization of 174 ovarian tumors previously analyzed by The Cancer Genome Atlas (TCGA), of which 169 were high-grade serous carcinomas (HGSC). Integrating our proteomic measurements with the genomic data yielded a number of insights into disease such as how different copy number alternations influence the proteome, the proteins associated with chromosomal instability, the sets of signaling pathways that diverse genome rearrangements converge on, as well as the ones most associated with short overall survival. Specific protein acetylations associated with homologous recombination deficiency suggest a potential means for stratifying patients for therapy. In addition to providing a valuable resource, these findings provide a view of how the somatic genome drives the cancer proteome and associations between protein and post-translational modification levels and clinical outcomes in HGSC.
INTRODUCTION
A comprehensive molecular view of cancer is necessary for understanding the underlying mechanisms of disease, improving prognosis, and ultimately guiding treatment (Hanahan and Weinberg, 2011). The Cancer Genome Atlas (TCGA) conducted an extensive genomic and transcriptomic characterization of ovarian high-grade serous carcinoma (HGSC) aimed at defining the genomic landscape and aiding the development of targeted therapies for this highly lethal malignancy (TCGA Research Network, 2011). Key findings from TCGA were: 1) the prevalent role of TP53 mutations, 2) extensive DNA copy alterations, 3) preliminary transcriptional signatures associated with survival, 4) varied mechanisms of BRCA1/2 inactivation, and lastly, 5) CCNE1 aberrations. Subsequent analysis of genomic data from the TCGA consortium led to the refinement of the transcript-defined signatures, improving the statistical association with patient outcome (Yang et al., 2013), and integrating microRNA and mRNA expression profiles associated with HGSC to identify candidate microRNA targets (Creighton et al., 2012).
While the insights from genomic analyses are substantial, functions encoded in the genome are generally executed at the protein level, and are often further modulated by post-translational modifications (PTMs) (Vogel and Marcotte, 2012). For example, TCGA used reverse phase protein array (RPPA) analysis of 172 proteins (including 31 phosphoproteins with phospho-specific antibodies) to generate a signature associated with the risk of tumor recurrence (Yang et al., 2013). To obtain a more comprehensive assessment of the complex ovarian HGSC phenotype, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) conducted an extensive mass spectrometry (MS)-based proteomic and phosphoproteomic characterization of HGSC tumors characterized by TCGA, providing quantitative measurements for a combined total of 9,600 proteins from 174 tumors (an average of 7,952 proteins per tumor), and a total of 24,429 phosphosites from 6,769 phosphoproteins in a subset of 69 tumors (an average of 7,677 phosphosites per tumor). Our results provide insights on HGSC biology and correlate differences in protein and PTM levels with clinical outcome complementary to TCGA genomic analyses.
RESULTS
Proteomic analysis of TCGA HGSC samples
HGSC biospecimens and clinical data from 174 patients collected by TCGA were analyzed at two independent CPTAC Centers, Johns Hopkins University (JHU) and Pacific Northwest National Laboratory (PNNL); thirty-two samples were analyzed at both JHU and PNNL. Tumors were selected by examining the associated TCGA metadata to select tumors either 1) on the basis of putative homologous recombination deficiency (HRD), defined by the presence of germline or somatic BRCA1 or BRCA2 mutations, BRCA1 promoter methylation, or homozygous deletion of PTEN (Woodbine et al., 2014) (122 samples; 67 classified as HRD, 55 as non-HRD by the above criteria; JHU), or 2) to maximize differences in overall survival (84 samples; PNNL). For selection purposes, short survival was defined as overall survival of less than 3 years, and long survival as greater than 5 years. All but five tumors had somatic TP53 mutations, a characteristic feature of HGSC (TCGA Research Network, 2011); these five tumors were subsequently reclassified as other than HGSC (Vang et al., 2016), and removed from protein functional analyses (e.g., subtyping and survival analyses). The tumor selection criteria and the associated metadata are provided in the Supplemental Information Table S1, available online.
Proteomics measurements used isobaric tags for relative and absolute quantitation (iTRAQ; Ross et al., 2004) in conjunction with offline liquid chromatography fractionation via high-pH reversed phase liquid chromatography (RPLC) and online RPLC with high-resolution tandem MS to provide broad coverage for peptide and protein identification and quantification (Supplemental Information); this also alleviated quantitative interference potentially associated with the use of isobaric tags. We used the relative abundance measurements for each protein in the 32 patient samples analyzed at both JHU and PNNL to normalize across the two analysis sites, and then used clustering, principle component analysis (PCA), and statistical tests to identify any significant batch effects associated with the site of analysis (a detailed comparison of within site, between site, between sample measurement variability and the process used to merge the JHU and PNNL data is given in Supplemental Information, Figure S1). As shown in Figure S1C, the median coefficient of variation (CV) between measurements at the two sites was 16%.
A total of 9,600 proteins were identified with high confidence in all tumors, and the relative abundances in each tumor are given in Table S2. Functional analyses and proteome-transcriptome associations were restricted to 3,586 proteins observed and quantified in all 169 HGSC samples used for protein functional analyses and where sample variability (signal) exceeded technical variability (noise) in the merged data (Table S2), calculated as described in Supplemental Information. On average we identify peptides covering 29% of the amino acids in any of these 3,586 proteins in a given sample, with a range from 10% to 47%. In addition to protein abundance levels, phosphoproteomics data were acquired for 69 tumors with sufficient sample (Table S2). As with proteins, phosphopeptide abundances were calculated relative to the pooled reference sample. Because isobaric labeling was performed prior to splitting the samples for proteome and phosphoproteome analyses, phosphopeptide abundance could be corrected for changes in parent protein abundance to identify differences in the relative extent of phosphorylation at specific sites for each protein (Table S2). Overall, we achieved a protein dynamic range encompassing >4 orders of magnitude, ranging from low-level transcription factors to abundant structural proteins i.e., actin and tubulin.
Proteogenomic landscape of HGSC
The degree to which alterations observed at the genome and transcriptome levels are manifested at the protein level is variable, both qualitatively and quantitatively, and partially driven by multiple levels of post-transcriptional regulation (Zhang et al., 2014; Kislinger et al., 2006; Jovanovic et al., 2015; Mertins et al., 2016). To identify peptides encoded by single amino acid variants (SAAVs), splice variants, and novel exons documented by TCGA, mass spectra were searched against a custom graph database (Woo et al., 2014a) containing all peptide variations projected from the TCGA genomic analyses of the cohort using a multi-stage analysis pipeline (Supplemental Information). The most frequently observed variant peptides represented SAAVs and gene-level events. The evidence supporting each novel event is detailed in Table S2, including event type, genomic location, abundance information, and estimated false discovery rate (FDR). The validity of these variant peptides was further evaluated by the synthesis of twenty examples selected at random across the entire range of spectrum match scores. We obtained tandem mass spectra for all twenty synthetic peptides that matched spectra from our analyses, strongly supporting our observation of these variant peptides; three representative examples are given in Figure S1E. These results demonstrate the ability to confidently detect, identify and validate genome-level predictions at the protein level. More novel peptides would likely be observed if there was sufficient sample for more extensive fractionation and/or replicate analyses (Ruggles et al., 2015); these limitations preclude any conclusions about the biological significance of unobserved events. For example, only 2 mutant p53 peptides were identified despite the presence of p53 mutations in all tumors examined. Such low coverage can occur for reasons that include: excessively large or small tryptic fragments, inability to distinguish some amino acids, some possible biases against highly hydrophobic and hydrophilic peptides, or low abundance peptides co-eluting with very high abundance peptides.
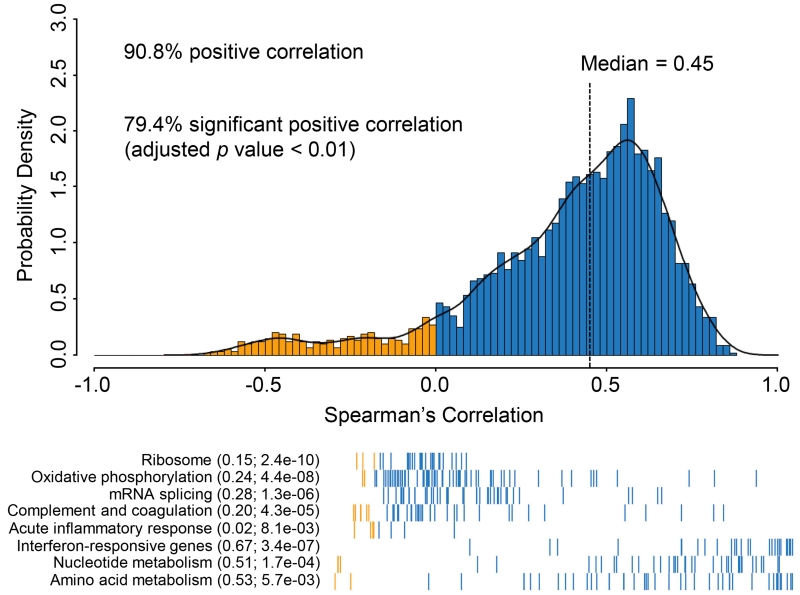
To assess the potential for post-transcriptional regulation (e.g., translational efficiency or protein degradation), we compared each protein to its corresponding transcript across all tumors, and correlation was assessed for those pairs with reliable measurements for both mRNA and protein, i.e., proteins with a corresponding mRNA measurement observed in all 169 HGSC tumors where sample variability exceeded technical variability (3,196 pairs). We excluded 391 proteins observed without a corresponding mRNA (e.g., HBA1) or discordant gene symbol annotation in the protein database (e.g., THOC4). Overall, 79.4% of the mRNA:protein pairs showed statistically significant positive Spearman correlations (Benjamini-Hochberg adjusted p value <0.01; Figure 1) when changes in mRNA abundance were compared to changes in relative protein abundance. The observed median r value of 0.45 for each mRNA-protein pair across all 169 tumors is similar to observations in colorectal cancer (Zhang et al., 2014), breast cancer (Mertins et al., 2016) and mouse tissues (Kislinger et al., 2006), although less than found for cell lines (Wu et al., 2013). In comparison, the median correlation of paired protein measurements from the same sample, but measured at two sites, was substantially higher, at 0.69 (Figure S1).
Figure 1. Correlations between mRNA and protein abundance in TCGA tumors.
mRNA and protein were correlated across all the samples, resulting in 90.6% positive correlations, and 79.4% were significantly correlated (Benjamini-Hochberg adjusted p value <0.01), with a mean Spearman’s correlation of 0.38 and a median value of 0.45 (top panel). Different biological pathways and processes showed significantly different levels of correlation (bottom panel). Metabolic pathways and the interferon response displayed high mRNA-protein correlation, and ribosome, mRNA splicing, oxidative phosphorylation, and complement and coagulation cascade were poorly correlated. The mean correlation is shown in parentheses followed by Benjamini-Hochberg adjusted p values calculated using a Kolmogorov–Smirnov test following the functional group names from MSIGDB. Blue bars indicate positive correlations and yellow indicates negative correlations; individual proteins (represented as bars on the x-axis) are sorted by correlation from low to high (bottom panel).
A wide range of mRNA:protein correlations was observed. In general, weaker correlations were observed for highly stable and abundant proteins associated with housekeeping or non-intrinsic functions (e.g., ribosomes, mRNA splicing, oxidative phosphorylation, complement cascade), while more dynamic proteins known to be transcriptionally regulated in response to nutrient demand or other perturbations (e.g., nucleotide metabolism, amino acid metabolism, acute inflammatory responses) were more highly correlated (Figure 1 and Table S3). This result is consistent with a previous colorectal cancer study (Zhang et al., 2014) and supports the hypothesis that while many biological functions are primarily regulated by mRNA abundance, post-transcriptional mechanisms likely have an important role in regulating certain house-keeping functions (Jovanovic et al., 2015; Komili and Silver, 2008; Lu et al., 2007; Marguerat et al., 2012). Additionally, we found that 23 mRNAs lacking polyA tails displayed lower correlation (mean −0.15) with their cognate proteins than did polyadenylated mRNAs, consistent with the decreased stability of mRNAs lacking polyA tails (Yang et al., 2011).
Clustering of tumors based on protein abundance
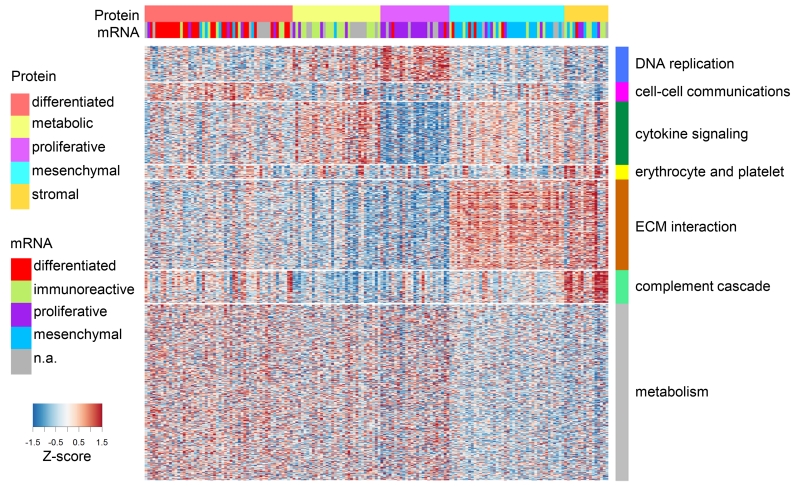
HGSC is the most common of the four major histological subtypes of epithelial ovarian cancer, and is characterized by distinct morphological features. Recent studies using mRNA abundance data have suggested four transcriptomic HGSC subtypes designated as differentiated, immunoreactive, mesenchymal, and proliferative (Yang et al., 2013). To build an unbiased molecular taxonomy of ovarian HGSC, we used protein abundance data from the 169 tumors to identify subtypes that might show biological differences that could be exploited in future studies. Figure 2 shows the resulting clustering analysis of individual tumors (vertical columns) by protein abundance (horizontal rows). The cluster assignment for each sample is provided in Table S4, and a consensus value matrix for the subtype comparisons is shown in Figure S2A. The results of a Weighted Gene Co-expression Network Analysis (WGCNA) (Langfelder and Horvath, 2008) of protein functional enrichment by subgroups are provided for the protein clusters, in Figure S2B and Table S4. The enrichment of KEGG and Reactome ontologies in the WGCNA-derived modules are shown in Figures S2C and S2D, respectively; membership of enriched pathways is provided in Table S3.
Figure 2. Proteomic subtypes and corresponding driving protein modules.
Global proteomics abundance is shown in a heatmap with TCGA samples represented by columns ordered by protein subtype where rows represent proteins. Color of each cell indicates z-score (log2 of relative abundance scaled by proteins’ standard deviation) of the protein in that sample; red is increased and blue is decreased (relative to the pooled reference). Transcriptome-based subtypes (Verhaak et al., 2013) and the proposed proteomic subtypes are indicated in color above the heatmap. WGCNA-derived modules are delineated with the row color panel and annotated according to the pathways based on enrichment of KEGG and Reactome ontologies.
Four of the proteomic clusters showed a clear correspondence to the mesenchymal, proliferative, immunoreactive, and differentiated subtypes defined by the TCGA transcriptome analysis (Figure 2 and Figure S2E). A relatively small fifth cluster of tumors significantly enriched in proteins associated with extracellular matrix interactions, erythrocyte and platelet functions, and the complement cascade was also observed using multiple approaches, including model based clustering with Bayesian information criteria, consensus clustering, and VISDA-based sub-phenotype clustering. This new group could not be attributed to tissue source site sampling bias or any other meta-data category, but may be related to tumor characteristics, including vascularization and tumor content, as the Tumor Purity score for this subtype (and the Mesenchymal subtype) was significantly lower than that of the other three subtypes (Figure S2F). The clinical relevance of these protein-based clusters will require validation in independent HGSC sample sets, particularly as no significant difference in survival was observed (Figure S2G), similar to mRNA-based clustering analysis (TCGA Research Network, 2011).
Proteomic analysis of CNA effects
Chromosomal instability, marked by extensive copy number alterations (CNAs) in each tumor, is a hallmark of HGSC, and a likely source of driver alterations in this disease (Cope et al., 2013; Kuo et al., 2009; Kobel et al., 2008). CNAs can affect the abundance of proteins at the same locus (cis effects), and may also act in trans, either directly or indirectly.
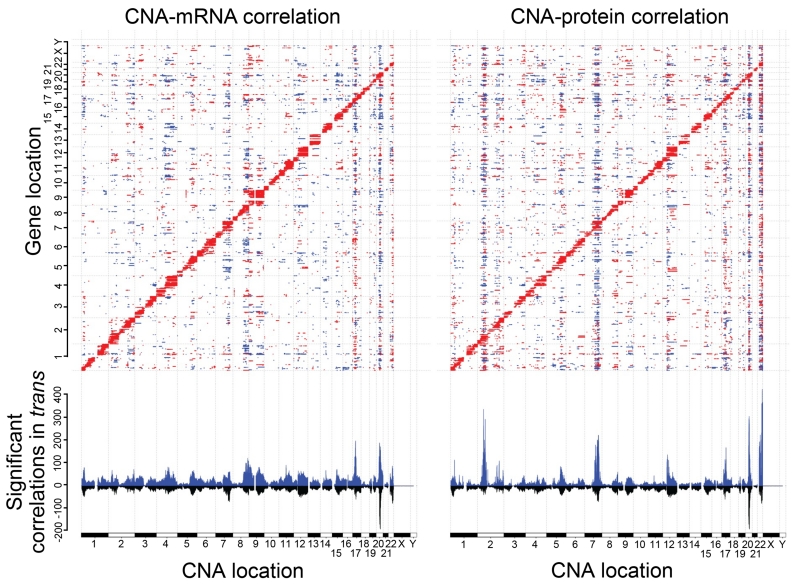
Hypothesizing that CNAs with strong trans effects are more likely to elicit a molecular phenotype and confer selective advantages, we sought to identify those CNA regions that have the broadest effect on protein expression. In all, 29,393 CNA loci (Table S5) were compared to our global proteomics data with 950,209 CNA/protein pairs (0.72% of the total) exhibiting significant association (Benjamini-Hochberg adjusted p value <0.01). We provide a complete list of the significantly associated proteins for each CNA in Table S5. The diagonal line evident in Figure 3 corresponds to cis effects, and vertical stripes correspond to trans effects where changes in copy number affect expression of numerous proteins across the genome. We performed the same analysis for mRNA to identify sites where associations are transcriptionally mediated. A similar number of CNA/mRNA pairs were found to be significantly associated (1,113,164 at Benjamini-Hochberg adjusted p value <0.01; Figure 3). This analysis revealed regions on chromosomes 2, 7, 20, and 22 correlated in trans with more than 200 proteins. In contrast to colorectal cancer where most of the trans-regulation of protein by CNA was accompanied by similar changes in mRNA (Zhang et al., 2014), we observed several loci associated with differences in protein abundance without a corresponding change in mRNA. For example, large regions on chromosome 2 have relatively little trans effect on mRNA levels, but are associated with more than 200 proteins in trans. Dissecting the mechanisms by which a specific CNA can alter protein levels in trans, without affecting the corresponding mRNA is difficult, given the extent of the amplified or deleted regions and the numerous genes affected at a given locus. Possible mechanisms include cis regulation of proteins associated with mRNA stability and translational efficiency, such as microRNAs and RNA binding proteins.
Figure 3. Functional impact of copy number alterations.
The top panel shows the correlation of CNAs to protein abundance (right) or mRNA (left) with significant positive correlations in red and negative correlations in blue (Benjamini-Hochberg adjusted p value <0.01, Spearman’s correlation coefficient). The x-axis plots the 29,393 CNAs obtained from TCGA. The y-axis plots 3,202 proteins. Genes are ordered by chromosomal location on both the x- and y-axes. The bottom panel shows the summed number of significantly correlated proteins (or mRNA) for each individual CNA. In blue is shown the total number, in black are those genes significantly correlated as both mRNA and protein. Where blue and black lines have similar magnitude, e.g., the hotspot on chromosome 20, the CNA associations are shared between protein and mRNA. Where a strong blue line has no mirrored black line, e.g., the protein hotspot on chromosome 2, the associations with CNA are largely unique.
Given the complex pattern of CNAs observed in HGSC, it has been difficult to identify a limited number of high impact genomic alterations that could function as drivers of the disease. We interrogated the trans-affected proteins associated with each putative CNA for common functions using pathways defined by the KEGG, NCI pathway interaction database (PID) and Reactome databases (Supplemental Information). Proteins associated with cell invasion and migration, and proteins related to immune function appeared to be enriched in association with multiple CNAs (Figure S3; Table S3). These observations suggest a convergence of multiple CNA targets on a common set of biological functions, namely motility/invasion and immune regulation, functions that are among the hallmarks of cancer (Hanahan and Weinberg, 2011).
Association of CNA trans-affected proteins with overall survival
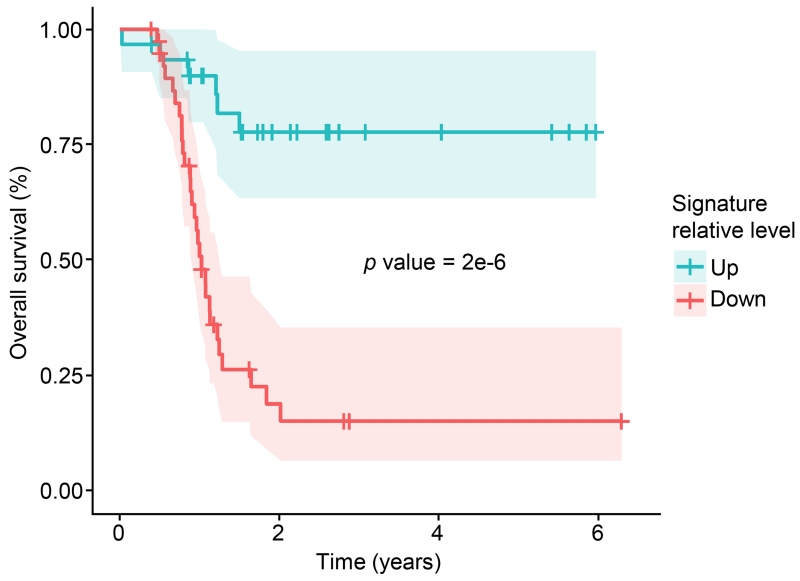
Availability of the TCGA survival data allowed us to use trans-affected protein data from the most influential CNAs (e.g., the four altered regions described on chromosomes 2, 7, 20, and 22; Figure 3) to build a model of overall survival. Because each CNA affects many proteins, we used a regression approach that identifies parsimonious Cox proportional hazards models with maximal predictive ability from the list of significantly correlated proteins for that CNA. We trained models on the proteomics data from PNNL (82 tumors) and tested the ability of the model to predict survival times in the data from JHU (87 tumors, not including the 32 overlapping). Each of the four most influential CNA regions produced models that were strongly associated with patient survival (p value <0.01, FDR <0.5% based on randomly selected proteins). A Kaplan-Meier plot illustrating the predictive value of each of the locus-specific models after validation is shown in Figure S4A; for the Kaplan-Meier plots, ‘high’ and ‘low’ expression were defined relative to the median for that signature across all samples, splitting patients into two equal groups of 45 (Table S6). We examined the overlap in predictions for patients for the four signatures and found that the predictions were unanimous (either high or low signature) for 62% of patients and 16% were evenly split, with the remaining having one dissenting signature. This suggested the utility of combining these signatures, using a voting method where the number of high or low calls was counted for each patient; this substantially improved prediction of survival time (p value 1.9e-6; Figure 4). We also examined the effects of tumor stage, tumor grade, patient age, surgical outcome, and platinum status, and found that our proteomic signatures were not improved by inclusion of these variables.
Figure 4. Kaplan-Meier plot of overall survival stratified by CNA-derived signatures.
A survival plot is shown for a consensus of the four best signatures (see Figure S4A and Table S6) identified from analysis of proteins affected in trans by CNAs. Models were trained using a lasso-based Cox proportional hazards model on the samples from PNNL, and shown are the survival curves from these models applied to the non-overlapping data from the JHU analysis, with the up (red; above the median) and down (blue, below the median) signatures each representing 45 patients. A vote was taken among the four most predictive signatures, and the results of this vote are shown. Probability of death is shown on the y-axis vs. survival time in years on the x-axis. Shaded ribbons denote 95% confidence intervals.
For each of these locus-specific models we analyzed the enrichment of genes and their regulatory sequences associated with outcome, and found all four models had genes significantly enriched (Benjamini-Hochberg adjusted p value <0.05) in binding sites for the proliferation-associated serum response factor (SRF), suggesting that SRF activity may be important in ovarian cancer outcome. Additionally, there was significant enrichment of proteins involved in the regulation of actin cytoskeleton, apoptosis, and adherens junction (Benjamini-Hochberg adjusted p value <0.05). We examined the protein abundance and phosphorylation status of SRF between short and long surviving groups and observed that they were higher in short surviving patients, but only slightly. Thus, although SRF alone was not predictive, the trends in SRF abundance and phosphorylation are consistent with the observation of enrichment for proteins potentially regulated by SRF binding. Details of the larger gene set analysis are provided in Supplemental Information.
Several proteins shared across all the signatures are known to be involved in cancer processes. Catenin B2 (CTNNA2) is a cell-cell adhesion protein and tumor suppressor (Fanjul-Fernández et al., 2013). The Rho GDP dissociation inhibitor (GDI) beta (ARHGDIB) is involved in invasion and migration in many cancers, and overexpression correlates with progression in pancreatic carcinoma (Yi et al., 2015). Protein kinase C and casein kinase substrate in neurons protein 2 (PACSIN2) is a repressor of cellular migration (Meng et al., 2011). Finally, the GTP-binding nuclear protein Ran, is a prognostic marker associated with increased survival in epithelial ovarian cancer (Barrès et al., 2010). The association of these previously described survival-related proteins with genes affected in trans by CNAs suggests a potential mechanism for the parallel activation of multiple pathways associated with poor prognosis in HGSC.
As a comparison we applied the previously described Provar signature (Yang et al., 2013), comprising five protein and four phosphoproteins that showed good survival prediction in the TCGA ovarian cancer dataset. We observed all proteins in the Provar signature in our proteomic data, but only one of the phosphosites (EGFR Y1173). Thus, we used the RPPA data from the original signature (Figure S4B). We found that the Provar signature was prognostic of survival (Benjamini-Hochberg adjusted p value = 0.11) in the 67 patients examined here (those with phosphoprotein data and had somatic TP53 mutations), but the statistical power of Provar is not as high as the statistical power of signatures derived from the CNA trans-affected proteins in this dataset. In addition, integrating the Provar signature with the CNA signatures did not improve survival prediction (Figure S4C).
Identification of proteins associated with chromosomal structural abnormality
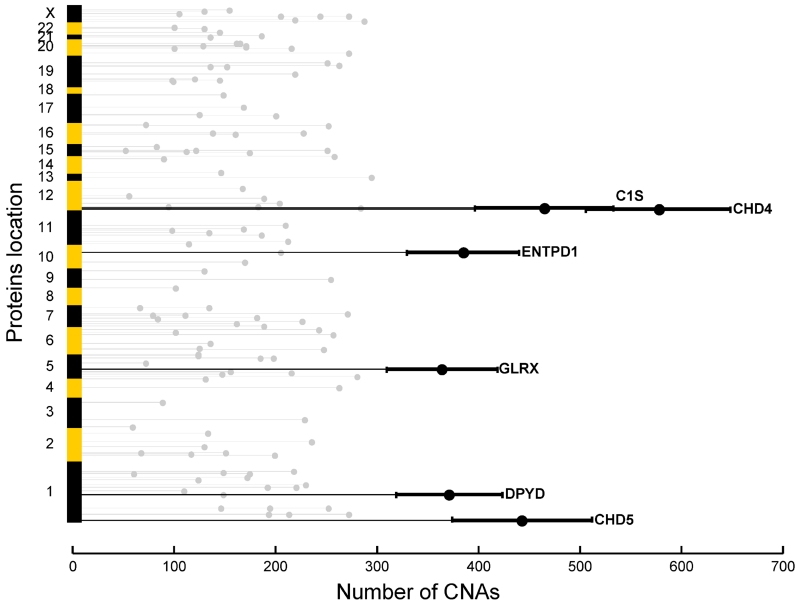
The degree of chromosomal instability exhibited by a tumor can be represented by a calculated chromosome instability (CIN) index, as described previously for lung cancer (TCGA Research Network, 2012). Identification of proteins associated with CIN may provide information on the processes contributing to chromosomal instability, while the analysis of trans-affected proteins described above is more closely related to the downstream consequences of specific CNAs. Using a bootstrapping method described in Supplemental Information, we identified a proteomic signature including 128 proteins (Table S7) showing significant correlation with CIN index (Benjamini-Hochberg adjusted p value <1e-4, Spearman correlation; Figure S5A). Functional annotation of these proteins (Figure S5B) showed that proteins up-regulated in tumors with high CIN index were preferentially involved in chromatin organization (p value 6.90e-5), whereas proteins up-regulated in tumors with low CIN index were more often associated with cell death (p value 2.13e-5). Correlation analysis of protein abundances and CIN indicated that a small number of proteins could account for the majority of the CIN (Figure 5); two of the most strongly associated proteins, CHD4 and CHD5, are known to be involved in chromatin organization (Liu and Matulonis, 2014).
Figure 5. Number of CNAs statistically explained by proteins significantly associated with CIN index.
A total of 128 proteins selected for their association with CIN were arranged along the chromosomal locations of their corresponding genes (y-axis). The length of the light grey horizontal lines indicates the number of CNAs significantly correlated with the protein across all chromosomes except for the protein’s corresponding gene coding region (x-axis). The top-ranked proteins are annotated and highlighted with dark lines to show the bootstrap-estimated 95% confidence intervals.
Identification of proteins associated with HRD status
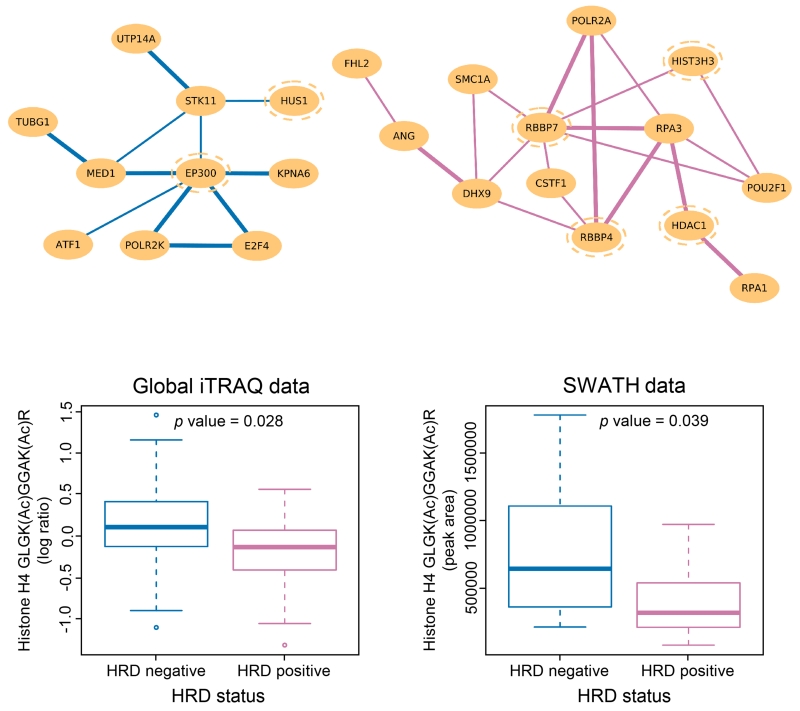
Since HRD is associated with susceptibility to PARP inhibitors and improved survival (Farmer et al., 2005; Liu and Matulonis, 2014), we sought to elucidate systemic changes associated with HRD and identify biomarkers that might be used to stratify patients for treatment. We defined tumors with HRD as having either germline or somatic mutations in BRCA1 or BRCA2, or BRCA1 promoter methylation, or homozygous deletion of PTEN (McEllin et al., 2010), with an overall survival >1.5 years, while non-HRD patients were defined as lacking these genomic aberrations and with a follow-up or time-to-death <2.5 years; additional selection criteria include available residual tissue volume and a tumor tissue contamination score estimated using copy number alterations (Yu et al., 2011). Applying differential dependency network (DDN) analysis (Zhang et al., 2009) on a set of 171 BRCA1- or BRCA2-related proteins curated from the literature and from the cBio portal, we identified a sub-network of 30 proteins that displayed co-expression patterns differentiating HRD from non-HRD patients (Table S7; Figure 6). Several of the proteins in these modules are known to be involved in histone acetylation or deacetylation, e.g., HDAC1, RBBP4, RBBP7, and EP300 (Mielcarek et al., 2015), and HUS1 (Cai et al., 2000).
Figure 6. DDN analysis and lysine-acetylation analysis between HRD and non-HRD patients.
DDN analysis revealed a sub-network of proteins that displayed distinct co-expression patterns between HRD and non-HRD patients, where purple connections indicate protein correlations that exist only in HRD samples and blue connections indicate protein correlations that exist only in non-HRD patients. The links between the nodes are drawn with two different thickness indicative of whether the connections meet the significance threshold of 0.05 (light lines) or 0.01 (bold lines). Proteins with blue dotted circles are known to be involved in histone acetylation or deacetylation. Further, identification and quantitation of lysine-acetylated peptides in global proteomics data showed that acetylation level at K12 and K16 of histone H4 are significantly different between HRD and non-HRD samples, suggesting a role of histone H4 acetylation (together with HDAC1) in modulating the choice of DSB repair mechanisms.
Although statistical association cannot distinguish between drivers and consequences of HRD status, the observed enrichment of proteins associated with histone acetylation motivated us to use an effective database search strategy (Supplemental Information) to identify and quantify acetylated peptides from the global proteomic data. Comparative analysis of 399 acetylated peptides identified 15 acetylated peptides with significant differences between the HRD and non-HRD samples (Table S7). Among these, dual acetylation at K12 and K16 of histone H4 showed a significant difference between HRD and non-HRD samples (Figure 6); differential acetylation of K12 and K16 was verified using synthetic peptides and targeted analysis using Sequential Windowed data independent Acquisition of the Total High-resolution Mass Spectra (SWATH-MS). In cell line data, acetylation of H4 has previously been reported to be involved in the choice of DNA double strand break (DSB) repair pathways (homologous recombination or non-homologous end joining (Gong and Miller, 2013; Tang et al., 2013). This relationship is regulated partially by HDAC1, a protein also identified in the DDN analysis. We observed a significant enrichment of HDAC1 and its co-regulated proteins in tumors with HRD and low H4 acetylation relative to non-HRD tumors with high H4 acetylation (DDN analysis: permutation tests, p value <0.05; differentially acetylated peptides: t-tests, p value <0.05 with an estimated FDR <0.5% by bootstrap/permutation tests). The combined observations of increased HDAC1 and associated proteins at the pathway level, together with decreased acetylation of H4 in HRD patients at the PTM level, provide insight regarding the potential role of HDAC1 in modulating the choice of DSB repair pathways.
Phosphoproteomic analysis of pathways associated with survival
An initial set of 24,464 different phosphopeptides (21,298 unique phosphorylation sites) contained within 4,420 proteins having data on net differences in phosphorylation (Table S2) was obtained. Since ischemia of the TCGA tumor samples may alter phosphopeptide abundance, we removed any phosphorylation sites previously shown to be altered by ischemia (Mertins et al., 2014). We also removed three tumor samples having substantially higher than average missing values and two tumor samples lacking somatic TP53 mutations (Supplemental Information). This yielded a final set of 7,675 different phosphopeptides (6,802 unique phosphorylation sites) from 2,324 proteins (Table S2) that were mapped to pathways via the NCI PID. The average net phosphorylation of all phosphopeptides mapping to a given pathway (i.e., the statistical average across all phosphoproteins known to be in the pathway, rather than the statistically significant individual proteins) was used as a measure of pathway activity and compared between short survivors (deceased surviving <3 years; N=17) and long survivors (patients surviving >5 years; N=19) using both proteomic and phosphoproteomic data.
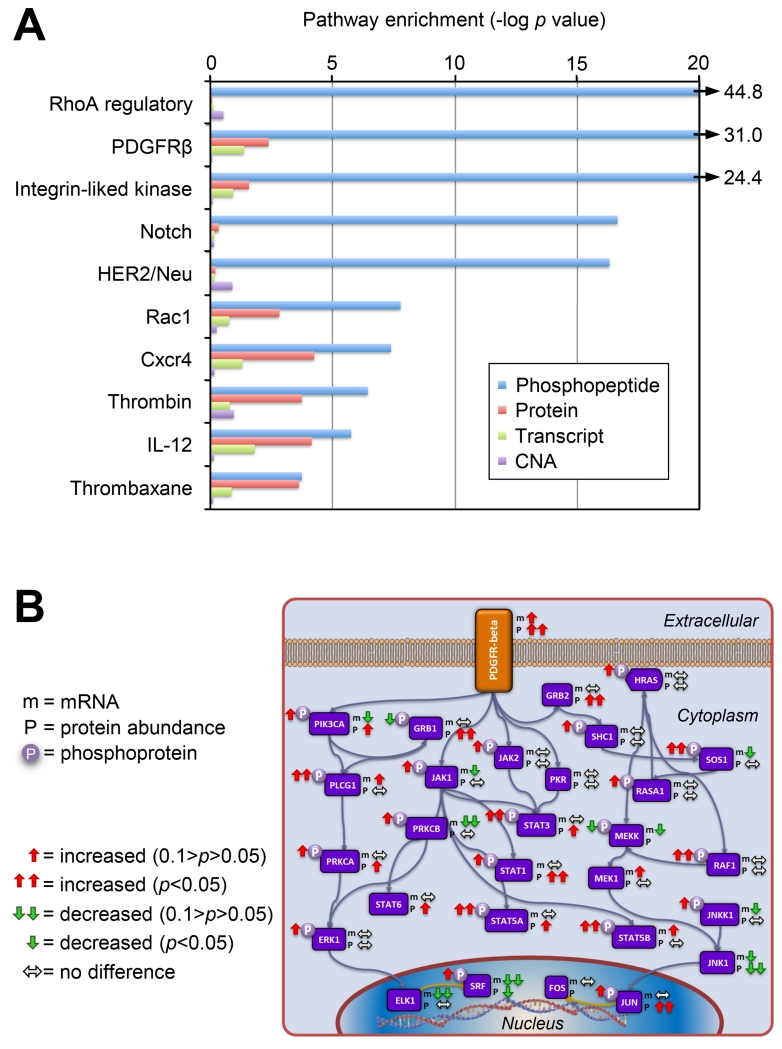
An issue in ranking the pathways is the distinction between phosphopeptides that are unique to a single pathway, such as the receptor peptides themselves, vs. peptides that map to proteins shared between pathways. In order to address this issue, we performed two analyses: one of all proteins associated with a specific pathway in the NCID database, and a second analysis in which proteins common to multiple pathways were excluded. Figure 7A shows a ranking of activated pathways, as inferred from the analysis of pathway specific components (i.e., differences in mRNA, protein and phosphoprotein levels) in short vs. long survivors. Fifteen pathways showed increased phosphorylation at Benjamini-Hochberg adjusted p values <0.01, in contrast with transcriptional data for the same samples which yielded only one statistically significantly increased pathway, androgen receptor signaling, which was also increased at the phosphoprotein level (Figure 7A and Supplemental Information). All pathways indicated in Figure 7A were increased in association with short survival relative to long survival, though some individual components within a pathway showed opposite changes (e.g. JNK1 transcript and protein levels, see Figure 7B). The RhoA-regulatory, PDGFRB, and integrin-like kinase pathways emerged as the most activated in this analysis (Figure 7A). Figure 7B shows the common and unique elements of the PDGFRB pathway, and the trends in mRNA, protein, and phosphoprotein levels in the comparison of short vs. long survivors, illustrating the benefit of combined protein and phosphoprotein measurements, as well as the substantial contribution of protein phosphorylation data to the overall analysis.
Figure 7. Pathway analysis associated with patient survival.
A) Pathway components were statistically analyzed using a two-sided t test between samples from patients surviving <3 years (short survival) and patients surviving >5 years (long survival) for differences in CNA, mRNA expression, protein or phosphorylation abundance. All significant pathways for mRNA and protein are shown (Benjamini-Hochberg adjusted p value <0.05) and the most significant pathways for phosphoproteomics are plotted on the x axis as the −log of the p value. Results show that phosphorylation provides additional information about the functional state of tumors. B) Simplified PDGFR-beta pathway showing differences between short and long survivor groups for protein abundance (p), transcript abundance (m), and phosphopeptide abundance (circled P).
We compared our results to the 31 phosphoproteins represented on the RPPA arrays used in the original TCGA ovarian cancer analysis (TCGA Research Network, 2011); RPPA identified differential phosphorylation of ERK1, RAF1, and STAT3 in the same patient samples analyzed by MS. For short surviving vs. long surviving patients, ERK1 showed statistically significantly increased phosphorylation by RPPA, on the same ERK1 phosphopeptides as found using MS, while AKT1, RAF1 and STAT3 showed increased phosphorylation by both RPPA and MS-based analyses, but based upon distinct phosphopeptides (Table S2). Good overlap was observed between RPPA and MS results for phosphoproteins on the RPPA array, but we did not observe three of the specific phosphosites from the Provar survival signature in our MS data, suggesting that proteomics is more sensitive to overall pathway activity than specific regulatory events in the pathway itself. However, many of the phosphopeptides identified by MS phosphoproteomics as components of the PDGFR-beta pathway were not represented in the RPPA, illustrating the ability to identify phosphorylation events not readily accessible with currently available antibodies (Figure 7B).
DISCUSSION
The addition of proteomic information, including post-translational modifications, to the rich genomic and transcriptomic data available from the ovarian HGSC samples analyzed by TCGA has provided additional insights into the biology of HGSC, including identification of changes at the level of pathway activation. Integration of genomic, proteomic, and phosphoproteomic measurements identified differentially regulated pathways and functional modules that displayed significant associations with patient outcomes, including survival and HRD status. Cox proportional hazard analysis of proteins associated with CNAs identified overall survival signatures enriched for targets of the proliferation-associated transcription factor SRF, and integrated proteomic, phosphoproteomic and transcriptomic data identified pathways that differentiated patients on the basis of survival, including the PDGFR-beta signaling pathway associated with angiogenesis, the RhoA regulatory and integrin-linked kinase pathways associated with cell mobility and invasion, and pathways associated with chemokine signaling and adaptive immunity. Proteins associated with cell invasion and motility emerged from both the integration of CNAs with protein abundance data and the phosphoproteomic investigation of up-regulated pathways in short versus long survivors. Despite the complexity of genomic alterations that characterize HGSC, these analyses suggest a functional convergence on a subset of key pathways. The association of increased invasiveness and motility with short overall survival in this study may help to explain more aggressive mechanisms of dissemination in ovarian cancer, including recently reported hematogenous metastasis (Pradeep et al., 2014), in addition to previously described lymphatic spread and known peritoneal spread by direct extension.
Focusing on functional modules has also revealed potential drivers of HGSC and more robust signatures for potentially stratifying ovarian HGSC patients into distinct cancer phenotypes to inform therapeutic management. For example, the identification of specific acetylation events associated with HRD, e.g., the simultaneous acetylation of K12 and K16 on histone H4, may provide an alternative biomarker of HRD and a rationale for the selection of patients in future clinical trials of HDAC inhibitors alone or in combination with PARP inhibition. This may help to resolve the current discrepancy between the initial observation of limited single agent activity of HDAC inhibitors in ovarian cancer (Mackay et al., 2010; Modesitt et al., 2008), and more recent findings of a >40% response rate when used in combination with cytotoxic chemotherapy in platinum-resistant patients (Dizon et al., 2012). Similarly, comprehensive interrogation of protein phosphorylation identified multiple pathways with significantly increased phosphorylation in patients with poor clinical outcomes. Specifically, the observed activation of PDGFR pathways in a subset of patients with short overall survival could potentially stratify patients for selective enrollment in trials of anti-angiogenic therapy, which is particularly relevant to current controversies over the use of bevacuzimab as a first-line therapy in ovarian cancer (Gadducci et al., 2015).
Overall, this work illustrates the ability of proteomics to complement genomics in providing additional insights into pathways and processes that drive ovarian cancer biology and how these pathways are altered in correspondence with clinical phenotypes. The comprehensive proteomic measurements for the HGSC tumor samples provide a public resource of information. Importantly, the ability to identify PTMs revealed a strong association between specific histone acetylation events and the HRD phenotype. In addition, the enhanced view of pathway activity provided by measurements of protein phosphorylation provides a foundation for linking genotype to proteotype, and ultimately to phenotype for understanding the molecular basis of cancer.
EXPERIMENTAL PROCEDURES
Full methods are available in Extended Experimental Procedures.
Tumor Samples
All tumor samples for the current study were obtained through the TCGA Biospecimen Core Resource as described previously (Zhang et al., 2014). Samples were selected based on overall survival, HRD status and availability.
Quantitative Proteomics
Tissue proteins were extracted and digested with trypsin; at each site, a portion of each resulting sample was also used to create a pooled reference in which each tumor sample contributed an equal percentage by peptide mass. The pooled reference sample was included in each multiplexed, isobaric labeling experiment to enable cross-experiment comparison in the entire sample cohort. The patient samples and the pooled reference sample were each labeled by different iTRAQ reagents (Sciex), combined, fractionated, and split for integrated, quantitative global proteome (10%) and phosphoproteome (90%) analysis using LC-MS/MS on an Orbitrap Velos mass spectrometer (Thermo Scientific). Raw data were processed for peptide identification by database searching and identified peptides were assembled as proteins and mapped to gene identifiers for proteogenomic comparisons. Quantitation was achieved by comparing the iTRAQ reporter ion intensities in each sample.
Comparison of mRNA and Protein Subtypes
A model-based clustering approach was used to model the protein abundance data as a mixture of subtypes. Bayesian information criteria, statistical resampling and VISDA-based sub-phenotype clustering approaches provided similar results with a stable optimization on five clusters as determined by consensus clustering. WGCNA analysis was performed to infer genes or gene networks that drive subtyping into 5 clusters, followed by correlation with subtype as a trait.
Integration of Proteomics and CNA data
Spearman correlation coefficients and corresponding adjusted p values were calculated for each protein/transcript by CNA locus, and gene set enrichment analysis was used to infer function for groups of proteins significantly correlated with a given CNA locus. Regression analysis was applied to the list of trans-affected protein correlated with each CNA to generate parsimonious Cox proportional hazards models with maximal predictive ability, using the PNNL data for training and JHU data for testing. Kaplan-Meier plots were used to visualize performance in predicting overall survival and progression-free survival. A CIN index (TCGA Research Network, 2012) was calculated for each sample as the mean absolute values of copy number measurements at the 29,393 selected loci. Bootstrap resampling was used to select proteins correlated with CIN index at high confidence.
Protein Acetylation and HRD
Acetylated peptides were identified by searching the global proteomics data for dynamic acetylation to lysines (+42 Da). Acetylation levels were compared between HRD and non-HRD cases by t-test. Targeted proteomics (SWATH; Collins et al., 2013) was used to orthogonally quantify the acetylated peptide with synthetic peptides as internal standard.
Survival-related Pathways Analysis
Phosphoproteome data from the short and long survivors were mapped to signaling pathways in the NCI pathway information database,(PID (http://pid.nci.nih.gov) using the gene names. For each signaling pathway in the PID, relative abundances for all phosphopeptides mapping to any pathway component were identified and separated into short and long survivor groups. The difference in distributions between the two sets of pathway-specific peptides, i.e., those associated with either short survivors or long long survivors, was then assessed using a two-tailed t test. Similar enrichment analyses were also performed using protein abundance, mRNA abundance, and CNA.
Data Repository
All of the primary mass spectrometry data on TCGA tumor samples are deposited at the CPTAC Data Coordinating Center as raw and mzML files and complete protein assembly data sets for public access (https://cptac-data-portal.georgetown.edu).
Supplementary Material
ARTICLE HIGHLIGHTS.
Comprehensive proteomic characterization of 174 ovarian tumors analyzed by TCGA
Copy number alterations affect the proteome in trans, converging on pathways
Acetylation of histone H4 correlates with homologous repair deficiency status
Protein and phosphoprotein abundance identifies pathways associated with survival
Acknowledgements
This work was supported by National Cancer Institute (NCI) CPTAC awards U24CA160019 and U24CA160036, and by National Institutes of Health grant P41GM103493. The PNNL proteomics work described herein was performed in the Environmental Molecular Sciences Laboratory, a U.S. Department of Energy (DOE) national scientific user facility located at PNNL in Richland, Washington. PNNL is a multi-program national laboratory operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL01830. Genomics data for this study were generated by TCGA pilot project established by the NCI and the National Human Genome Research Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Study conception and design: HZ, TL, ZZ, RDS, DWC, KDR. Investigation: performed experiment or data collection: TL, SS, FY, JZ, LijunC, PS, PA, YT, MAG, TRC, CC, ST, SN, RJM, HengZ. Computation and statistical analysis: ZZ, SHP, BaiZ, JEM, VAP, LiC, DR, MEM, SWC, SW, JW, DLT, DF, VB, YW, BingZ. Data interpretation and biological analysis: TL, HZ, SHP, ZZ, BaiZ, JEM, VAP, LiC, DR, SN, CW, IMS, AP, MPS, DAL, RDS, DWC, KDR. Writing/manuscript preparation and revision: HZ, TL, ZZ, SHP, JEM, DR, VAP, LeslieC, HR, IMS, AP, MPS, DAL, RDS, DWC, KDR. Supervision and administration: HR, ESB, TH, RCR, LS, RDS, DWC, KDR.
The authors declare no competing financial interests.
Supplemental information includes Extended Experimental Procedures, 5 figures, and 8 tables and can be found with this article online.
REFERENCES
- Barrès V, Ouellet V, Lafontaine J, Tonin PN, Provencher DM, Mes-Masson AM. An essential role for Ran GTPase in epithelial ovarian cancer cell survival. Mol. Cancer. 2010;9:272. doi: 10.1186/1476-4598-9-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai RL, Yan-Neale Y, Cueto MA, Xu H, Cohen D. HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J. Biol. Chem. 2000;275:27909–27916. doi: 10.1074/jbc.M000168200. [DOI] [PubMed] [Google Scholar]
- Collins BC, Gillet LC, Rosenberger G, Rost HL, Vichalkovski A, Gstaiger M, Aebersold R. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat. Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- Cope L, Wu RC, Shih Ie M, Wang TL. High level of chromosomal aberration in ovarian cancer genome correlates with poor clinical outcome. Gynecol. Oncol. 2013;128:500–505. doi: 10.1016/j.ygyno.2012.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creighton CJ, Hernandez-Herrera A, Jacobsen A, Levine DA, Mankoo P, Schultz N, Du Y, Zhang Y, Larsson E, Sheridan R, et al. Integrated analyses of microRNAs demonstrate their widespread influence on gene expression in high-grade serous ovarian carcinoma. PLoS One. 2012;7:e34546. doi: 10.1371/journal.pone.0034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dizon DS, Damstrup L, Finkler NJ, Lassen U, Celano P, Glasspool R, Crowley E, Lichenstein HS, Knoblach P, Penson RT. Phase II activity of belinostat (PXD-101), carboplatin, and paclitaxel in women with previously treated ovarian cancer. Int. J. Gynecol. Cancer. 2012;22:979–986. doi: 10.1097/IGC.0b013e31825736fd. [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fanjul-Fernández M, Quesada V, Cabanillas R, Cadiñanos J, Fontanil T, Obaya A, Ramsay AJ, Llorente JL, Astudillo A, Cal S, et al. Cell-cell adhesion genes CTNNA2 and CTNNA3 are tumour suppressors frequently mutated in laryngeal carcinomas. Nat. Commun. 2013;4:2531. doi: 10.1038/ncomms3531. [DOI] [PubMed] [Google Scholar]
- Gadducci A, Lanfredini N, Sergiampietri C. Antiangiogenic agents in gynecological cancer: State of art and perspectives of clinical research. Crit. Rev. Oncol. Hematol. 2015;96:113–28. doi: 10.1016/j.critrevonc.2015.05.009. [DOI] [PubMed] [Google Scholar]
- Gong F, Miller KM. Mammalian DNA repair: HATs and HDACs make their mark through histone acetylation. Mutat. Res. 2013;750:23–30. doi: 10.1016/j.mrfmmm.2013.07.002. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Jovanovic M, Rooney MS, Mertins P, Przybylski D, Chevrier N, Satija R, Rodriguez EH, Fields AP, Schwartz S, Raychowdhury R, et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science. 2015;347:1259038. doi: 10.1126/science.1259038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kislinger T, Cox B, Kannan A, Chung C, Hu P, Ignatchenko A, Scott MS, Gramolini AO, Morris Q, Hallett MT, et al. Global survey of organ and organelle protein expression in mouse: combined proteomic and transcriptomic profiling. Cell. 2006;125:173–186. doi: 10.1016/j.cell.2006.01.044. [DOI] [PubMed] [Google Scholar]
- Kobel M, Huntsman D, Gilks CB. Critical molecular abnormalities in high-grade serous carcinoma of the ovary. Expert Rev. Mol. Med. 2008;10:e22. doi: 10.1017/S146239940800077X. [DOI] [PubMed] [Google Scholar]
- Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat. Rev. Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- Kuo KT, Guan B, Feng Y, Mao TL, Chen X, Jinawath N, Wang Y, Kurman RJ, Shih Ie M, Wang TL. Analysis of DNA copy number alterations in ovarian serous tumors identifies new molecular genetic changes in low-grade and high-grade carcinomas. Cancer Res. 2009;69:4036–4042. doi: 10.1158/0008-5472.CAN-08-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Matulonis UA. New strategies in ovarian cancer: translating the molecular complexity of ovarian cancer into treatment advances. Clin. Cancer Res. 2014;20:5150–5156. doi: 10.1158/1078-0432.CCR-14-1312. [DOI] [PubMed] [Google Scholar]
- Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- Mackay HJ, Hirte H, Colgan T, Covens A, MacAlpine K, Grenci P, Wang L, Mason J, Pham PA, Tsao MS, et al. Phase II trial of the histone deacetylase inhibitor belinostat in women with platinum resistant epithelial ovarian cancer and micropapillary (LMP) ovarian tumours. Eur. J Cancer. 2010;46:1573–1579. doi: 10.1016/j.ejca.2010.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEllin B, Camacho CV, Mukherjee B, Hahm B, Tomimatsu N, Bachoo RM, Burma S. PTEN loss compromises homologous recombination repair in astrocytes: implications for glioblastoma therapy with temozolomide or poly(ADP-ribose) polymerase inhibitors. Cancer Res. 2010;70:5457–5464. doi: 10.1158/0008-5472.CAN-09-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Tian L, Zhou J, Li Z, Jiao X, Li WW, Plomann M, Xu Z, Lisanti MP, Wang C, Pestell RG. PACSIN 2 represses cellular migration through direct association with cyclin D1 but not its alternate splice form cyclin D1b. Cell Cycle. 2011;10:73–81. doi: 10.4161/cc.10.1.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, Wang X, Qiao JW, Cao S, Petralia F, et al. Proteogenomics connects somatic mutations to signaling in breast cancer. Nature. 2016 doi: 10.1038/nature18003. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, Clauser KR, Qiao JW, Gritsenko MA, Moore RJ, et al. Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol. Cell. Proteomics. 2014;13:1690–1704. doi: 10.1074/mcp.M113.036392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielcarek M, Zielonka D, Carnemolla A, Marcinkowski JT, Guidez F. HDAC4 as a potential therapeutic target in neurodegenerative diseases: a summary of recent achievements. Front. Cell. Neurosci. 2015;9:42. doi: 10.3389/fncel.2015.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modesitt SC, Sill M, Hoffman JS, Bender DP, Gynecologic Oncology Group A phase II study of vorinostat in the treatment of persistent or recurrent epithelial ovarian or primary peritoneal carcinoma: a Gynecologic Oncology Group study. Gynecol. Oncol. 2008;109:182–186. doi: 10.1016/j.ygyno.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Pradeep S, Kim SW, Wu SY, Nishimura M, Chaluvally-Raghavan P, Miyake T, Pecot CV, Kim SJ, Choi HJ, Bischoff FZ, et al. Hematogenous metastasis of ovarian cancer: rethinking mode of spread. Cancer Cell. 2014;26:77–91. doi: 10.1016/j.ccr.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- Ruggles KV, Tang Z, Wang X, Grover H, Askenazi M, Teubl J, Cao S, McLellan MD, Clauser KR, Tabb DL, et al. An analysis of the sensitivity of proteogenomic mapping of somatic mutations and novel splicing events in cancer. Mol. Cell. Proteomics. 2015 doi: 10.1074/mcp.M115.056226. M115.056226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Cho NW, Cui G, Manion EM, Shanbhag NM, Botuyan MV, Mer G, Greenberg RA. Acetylation limits 53BP1 association with damaged chromatin to promote homologous recombination. Nat. Struct. Mol. Biol. 2013;20:317–325. doi: 10.1038/nsmb.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vang R, Levine DA, Soslow RA, Zaloudek C, Shih Ie.M., Kurman RJ. Molecular Alterations of TP53 are a Defining Feature of Ovarian High-Grade Serous Carcinoma: A Rereview of Cases Lacking TP53 Mutations in The Cancer Genome Atlas Ovarian Study. Int. J. Gynecol. Pathol. 2016;35:48–55. doi: 10.1097/PGP.0000000000000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo S, Cha SW, Na S, Guest C, Liu T, Smith RD, Rodland KD, Payne S, Bafna V. Proteogenomic strategies for identification of aberrant cancer peptides using large-scale next-generation sequencing data. Proteomics. 2014a;14:2719–2730. doi: 10.1002/pmic.201400206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodbine L, Gennery AR, Jeggo PA. The clinical impact of deficiency in DNA non-homologous end-joining. DNA Repair. 2014;16:84–96. doi: 10.1016/j.dnarep.2014.02.011. [DOI] [PubMed] [Google Scholar]
- Wu L, Candille SI, Choi Y, Xie D, Jiang L, Li-Pook-Than J, Tang H, Snyder M. Variation and genetic control of protein abundance in humans. Nature. 2013;499:79–82. doi: 10.1038/nature12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JY, Yoshihara K, Tanaka K, Hatae M, Masuzaki H, Itamochi H, Takano M, Ushijima K, Tanyi JL, Coukos G, et al. Predicting time to ovarian carcinoma recurrence using protein markers. J. Clin. Invest. 2013;123:3740–3750. doi: 10.1172/JCI68509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi B, Zhang Y, Zhu D, Zhang L, Song S, He S, Zhang B, Li D, Zhou J. Overexpression of RhoGDI2 correlates with the progression and prognosis of pancreatic carcinoma. Oncol. Rep. 2015;33:1201–1206. doi: 10.3892/or.2015.3707. [DOI] [PubMed] [Google Scholar]
- Yu G, Zhang B, Bova GS, Xu J, Shih Ie M, Wang Y. BACOM: in silico detection of genomic deletion types and correction of normal cell contamination in copy number data. Bioinformatics. 2011;27:1473–1480. doi: 10.1093/bioinformatics/btr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Li H, Riggins RB, Zhan M, Xuan J, Zhang Z, Hoffman EP, Clarke R, Wang Y. Differential dependency network analysis to identify condition-specific topological changes in biological networks. Bioinformatics. 2009;25:526–532. doi: 10.1093/bioinformatics/btn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, Chambers MC, Zimmerman LJ, Shaddox KF, Kim S, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 2014;513:382–387. doi: 10.1038/nature13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.