Summary
The E7 oncoprotein of high-risk human papillomavirus (HPV) types induces DNA re-replication that contributes to carcinogenesis. Here, we demonstrated that Cdc6 plays an important role in E7-induced re-replication. These studies shed light on the mechanism by which HPV induces genomic instability.
Abstract
The E7 oncoprotein of high-risk human papillomavirus (HPV) types induces DNA re-replication that contributes to carcinogenesis; however, the mechanism is not fully understood. To better understand the mechanism by which E7 induces re-replication, we investigated the expression and function of cell division cycle 6 (Cdc6) in E7-expressing cells. Cdc6 is a DNA replication initiation factor and exhibits oncogenic activities when overexpressed. We found that in E7-expressing cells, the steady-state level of Cdc6 protein was upregulated and its half-life was increased. Cdc6 was localized to the nucleus and associated with chromatin, especially upon DNA damage. Importantly, downregulation of Cdc6 reduced E7-induced re-replication. Interestingly, the level of Cdc6 phosphorylation at serine 54 (S54P) was increased in E7-expressing cells. S54P was associated with an increase in the total amount of Cdc6 and chromatin-bound Cdc6. DNA damage-enhanced upregulation and chromatin binding of Cdc6 appeared to be due to downregulation of cyclin-dependent kinase 1 (Cdk1) as Cdk1 knockdown increased Cdc6 levels. Furthermore, Cdk1 knockdown or inhibition led to re-replication. These findings shed light on the mechanism by which HPV induces genomic instability and may help identify potential targets for drug development.
Introduction
Human papillomavirus (HPV) infection is one of the most common sexually transmitted infections (1) worldwide. To date, over 170 genotypes of HPV have been identified (1,2) and can be classified into two major groups: cutaneous and mucosal HPV. According to the clinical prognosis of the lesions they cause, mucosal (genital) HPV types can be categorized as either ‘high-risk’ or ‘low-risk’ subtypes. Approximately 12 HPV types, including types 16, 18, 31 and 45, are considered high-risk types because their infections can lead to the development of cancer (3). Cervical carcinoma is one of the leading causes of cancer death in women worldwide (4), and 99% of those cancer cases involve high-risk HPV types (5). Apart from uterine cervical cancer, HPV is etiologically associated with a subset of cancers of the head, neck, oropharynx, anus, penis, vagina and vulva (6). Although prophylactic vaccines are commercially available, they are type restricted. Therefore, understanding the pathogenesis of high-risk HPV types is still highly clinically important.
The primary targets of HPV infection are mucosal epithelial cells or cutaneous keratinocytes. Under physiological circumstances, epithelial cells exit from the cell cycle and undergo terminal differentiation. High-risk HPV encodes E6 and E7 genes, which interfere with critical cell cycle pathways and are consistently expressed in HPV-positive cervical cancers (7). The E6 and E7 genes induce DNA damage and genomic instability. The high-risk HPV E7 proteins bind to pRb family members, resulting in activation of the E2F transcription factors and entry of the cell into the S phase of the cell cycle. HPV DNA replication is dependent on host DNA replication machinery. Although E7 can efficiently immortalize keratinocytes in vitro, it is not sufficient for the induction of malignant progression (8). Clinically, HPV16 infections are very common in young sexually active women, but only a small percentage of HPV-infected people develop cancer. Moreover, progression from HPV infection to invasive cancer is slow and usually occurs over many years. These observations suggest that additional host genetic variations are needed for malignant progression.
The cell cycle is regulated by cyclins, cyclin-dependent kinases (Cdks) and their inhibitors. In each cell cycle, DNA replication must be tightly controlled to prevent the initiation of a second round of replication until mitosis is completed. Continuous DNA replication without cell division leads to the formation of abnormal polyploid cells. The components of the pre-replicative complex are considered key players in this regulation (9). Binding of the DNA replication factors Cdt1 and Cdc6 (cell division cycle protein 6) to the origin of replication is essential to form the pre-replicative complex because they both promote the loading of the license complex of minichromosome maintenance proteins (Mcms) (10,11). Overexpression of Cdc6 leads to re-replication in cancer cells, which is a form of endogenous DNA damage (12,13).
Cervical smear examination shows that the Cdc6 protein is expressed in most high-grade squamous intraepithelial lesions and is a potential marker for the diagnosis of high-grade and invasive cervical lesions (14,15). In cervical carcinoma cell lines, Cdc6 showed intense nuclear and cytoplasmic staining (15,16). Histological study demonstrated that Cdc6 staining was localized to the nucleus and was present in both cervical squamous carcinoma and adenocarcinoma. Interestingly, Cdc6-positive nuclei tended to be located lateral to or immediately beneath cells with high levels of HPV DNA, indicating a strong correlation between high-risk HPV infection and Cdc6 positivity (17). An increase in Cdc6 mRNA expression in CIN3 (cervical intraepithelial neoplasia) lesions and invasive cervical squamous cell carcinoma was also observed (18). Because Cdc6 is an E2F responsive gene (19), increased Cdc6 mRNA expression in cervical neoplasia may be a consequence of high-risk HPV oncoprotein E7 binding to pRb, resulting in the release of E2F inhibition.
The biological activities of Cdc6 are regulated by post-translational modifications. It is generally accepted that upon entry into the cell cycle, phosphorylation at Ser54 (S54P) by Cdk2 stabilizes Cdc6 by blocking Cdh1 from binding (20). After S-phase entry, Cdc6 is phosphorylated at Ser106 (S106) and translocated from chromatin to the cytoplasm before being degraded by ubiquitin-dependent proteolysis by the anaphase-promoting complex/cyclosome (21). Relocalization of Cdc6 to the cytoplasm prevents reinitiation of replication and is necessary for coupling the S phase to mitosis. However, some other studies have demonstrated that S-phase translocation occurs only with ectopically expressed Cdc6 and that endogenous Cdc6 persists in the nucleus bound to chromatin throughout the S phase (22,23).
We have recently demonstrated that HPV-16 E7 induces re-replication in response to DNA damage and that Cdt1 plays an important role in this process (24). However, as one of the key players in cell division, the role of Cdc6 in E7-induced re-replication has not been studied. The induction of genomic instability, polyploidy in particular, is an important step in cervical carcinogenesis (8). To further understand the mechanism by which E7 induces genomic instability, in the present study, we examined Cdc6 expression in HPV-16 E7-expressing cell lines and demonstrated its role and regulation in re-replication.
Materials and methods
Cell culture
Primary human keratinocytes (PHKs) derived from neonatal human foreskin epithelium were obtained from UMass Memorial Medical Center as described previously (25). Experiments were performed using PHKs within three passages. Cervical epithelial cells containing the HPV-16 genome (CE-HPV) (26) were provided by Aloysius Klingelhutz, University of Iowa. Spontaneously immortalized human keratinocytes (NIKS) were provided as described previously (25). These cells were cultured on mitomycin C-treated J2-3T3 feeder cells in E medium composed of three parts Dulbecco’s modified Eagle medium and one part Ham’s F-12 medium plus 5% fetal bovine serum, with all supplements as described previously (25). Human telomerase reverse transcriptase-expressing human retinal pigment epithelial (RPE1) cells were maintained in a 1:1 dilution of Dulbecco’s modified Eagle medium–Ham’s F-12 medium plus 10% fetal bovine serum.
The PHKs, NIKS and RPE1 cells expressing HPV-16 E7 were established using the pBabe retroviral system, as described previously (24). Populations of infected cells were pooled and expanded. The PHKs, NIKS and RPE1 cells were maintained in puromycin and used within 15 passages.
Cell proliferation was measured by the Cell Counting Kit-8 assay (CCK8; BosterBio) and performed according to the manufacturer’s instructions. Briefly, cells were seeded into 96-well plates at a density of 2×103 cells/well and incubated for 0, 12, 24, 36, 48 and 72h, respectively, after seeding. About 10 µl of CCK8 solution was added to each well, and then cells were incubated for 1h at 37°C. The absorbance of the colorimetric solution is determined at 450nm.
Immunoblotting
Protein extraction was performed in lysis buffer [10 mmol/l of Tris (pH 7.4), 1% sodium dodecyl sulfate and 1.0 mmol/l of sodium orthovanadate]. Equal amounts of protein from each cell lysate were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The membranes were blotted with antibodies against total Cdc6 (Santa Cruz, sc-9964), Cdc6 Ser54P (abcam, #75809), HPV-16 E7 (Santa Cruz, sc-6981), Cdk1 (BD Biosciences, #610038), Cdk2 (Santa Cruz, sc-6248), β-tubulin (Sigma, T-4026) and β-actin (Sigma, A-2066). HRP-conjugated goat anti-mouse or anti-rabbit was used as a secondary antibody. β-Tubulin, β-actin and Coomassie blue were used as loading controls. The half-life of Cdc6 was measured following cycloheximide (25 µg/ml) treatment and calculated using the Half Life Calculator (www.calculator.net).
Flow cytometry
For Cdk1 inhibitor-induction of polyploidy analysis, asynchronous cultures of RPE1 cells expressing HPV-16 E7 or vector alone were treated with RO-3306 (Alexis, 7.5 µM in dimethyl sulfoxide). During 48h of treatment, RO-3306 was replenished at 24h. For thymidine blocking analysis, RPE1 E7 cells were blocked with 2.5M thymidine for 16h and then released to regular media and collected at the indicated time points. All cells were fixed in 70% ethanol overnight, resuspended in phosphate-buffered saline (PBS), stained in propidium iodide (PI; Sigma, 50 µg/ml in PBS) staining solution supplemented with 70 µg/ml RNase A (Sigma) and analyzed by flow cytometry. Cell cycle analysis was conducted using FlowJo software (Tree Star, Ashland, OR).
For the bromodeoxyuridine (BrdU) labeling experiment, asynchronous cultures of RPE1 expressing HPV-16 E7 were transfected with 500 pM si-CDC6-001 for 72h. BrdU (final concentration: 20 µM) was added to the medium at 70h after transfection. After an additional 2h, cells were harvested and fixed in 70% ethanol overnight. The cells were permeablized with 2 N HCl–0.5% Triton X-100, neutralized with 0.1M sodium tetraborate and stained with monoclonal anti-BrdU (BD Biosciences), followed by anti-mouse IgG F(ab)2-FITC (Sigma), before being counterstained with PBS-PI-RNase A.
RNAi
All small interfering RNA (siRNA) oligonucleotides were purchased from Dharmacon. The siRNA duplexes were as follows: non-silencing control siRNA sense strand, 5′-UUCUCCGAACGUGUCACGU-3′; si-CDC6-001 sense strand, 5′-AAC UUC CCA CCU UAU ACC AGA-3′ (described previously in ref. 20); si-CDC6-002 sense strand, 5′-AAG AAU CUG CAU GUG UGA GAC-3′ (described previously in ref. 27); si-Cdk1 sense strand, 5′-GAU CAA CUC UUC AGG AUU U-3′ and si-Cdk2 sense strand, 5′-GCC AGA AAC AAG UUG ACG G-3′ (described previously in ref. 28). For cell cycle analysis, the cells were seeded onto 60mm dishes the day before transfection. Cells were transfected with 20nM or 500 pM siRNA for each target gene using Lipofectamine 2000 transfection reagent according to the manufacturer’s instructions (Invitrogen). After 24h, the cells were treated with 4–5 µg/ml bleomycin, incubated for an additional 48h and then harvested for fluorescence-activated cell sorting (FACS) analysis. For siRNA transfection into NIKS, 20nM siRNA for each target gene was transfected into the cells, which were collected for FACS analysis 72h after transfection. For protein knockdown analysis, the cells were seeded onto 60mm dishes the day before siRNA transfection. The cells were harvested 72h after transfection, and protein levels were analyzed by immunoblotting. For the Cdc6 immunofluorescence assay, cells were seeded onto 12-well plates the day before siRNA transfection. The cells were used for Cdc6 staining 72h after transfection, as described below.
Subcellular fractionation
Cytoplasmic, soluble and insoluble nuclear extracts were prepared as described (29,30), with some modifications. Briefly, an equal number of cells were lysed on ice for 10min with hypotonic buffer [20mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES), pH 8.0, 10mM KCl, 1mM MgCl2, 0.1% (vol/vol) Triton X-100 and 20% (vol/vol) glycerol]. The samples were centrifuged at 2300g for 2min, and the supernatants were collected and used as cytoplasmic fractions (CEs). The pellets were lysed for 20min on ice in hypertonic buffer [20mM HEPES, pH 8.0, 1mM ethylenediaminetetraacetic acid, 20% (vol/vol) glycerol, 0.1% (vol/vol) Triton X-100 and 400mM NaCl] with brief pipetting up and down. The samples were centrifuged at 18000g for 7min, and the supernatants were collected and used as soluble nuclear fractions (SNEs). The final chromatin pellet was resuspended in 1× Laemmli buffer without dithiothreitol and bromophenol blue for 10min at 70°C and sonicated for 15s in a 4710 Series Ultrasonic Homogenizer using a microtip at 25% amplitude (Cole-Parmer Instrument Co., Chicago, IL). The obtained lysates were used as insoluble chromatin-bound fractions (CBEs). The protein concentration was measured using a BCA protein assay kit. All fractions were boiled in 1× loading buffer for 10min at 70°C, and equal amounts of protein were used for immunoblotting. The purity of the obtained fractions was confirmed using anti-β-tubulin (Sigma, T-4026, for the CEs), anti-Sp1 (Cell Signaling #9389, for the SNEs) or anti-Histone H3 (Cell Signaling #3688, for the CBEs).
Immunofluorescence
For Cdc6 staining, 6×104 cells were seeded onto 12-well plates and grown on coverslips. The following day, the cells were treated with bleomycin (3 µg/ml). During 48h of treatment, bleomycin was replenished at 24h. The cells were fixed with cold methanol for 20min at room temperature and blocked with 5% normal goat serum in PBST (phosphate-buffered saline with 0.3% triton X 100) buffer for 30min at room temperature. The cells were incubated with an antibody against Cdc6 (Santa Cruz, sc-9964) or HPV-16 E7 (Santa Cruz, sc-1587) at 4°C overnight, followed by incubation with a fluorescein isothiocyanate-labeled anti-mouse secondary antibody. The cells were washed in PBS, counterstained with 4,5-diamidino-2-phenylindole dihydrochloride (Vector Laboratories) and analyzed using an Olympus BX51 epifluorescence microscope equipped with a multiband filter set. The two-color images were overlaid using Nikon NIS-Elements BR 3.10 imaging software.
Statistical analysis
All data are expressed as the mean ± SD. Student’s t-test was used to compare differences between means. Statistical significance was set at P ≤ 0.05.
Results
Upregulation of Cdc6 in HPV-16 E7-expressing cells
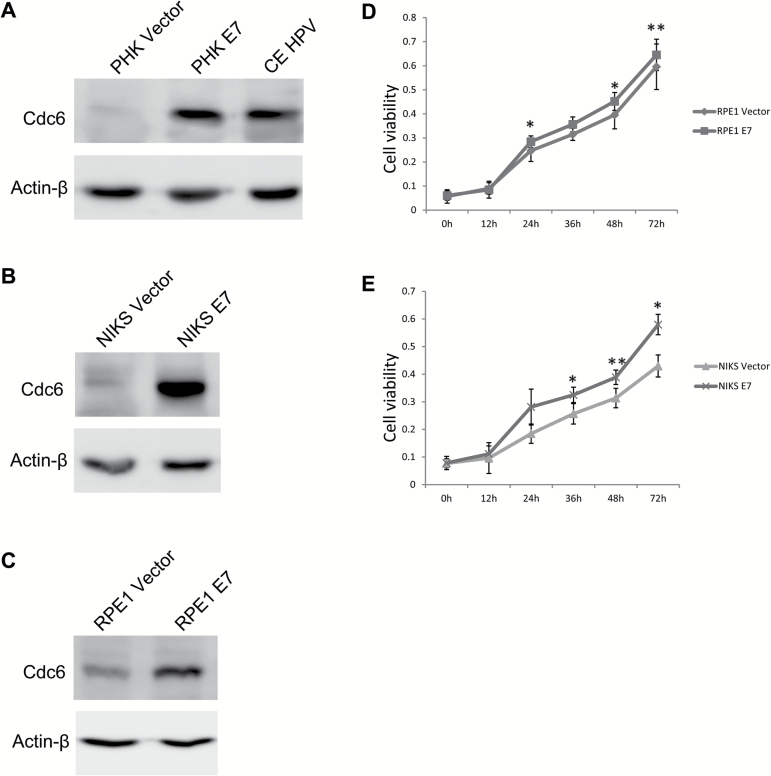
To understand the mechanism by which E7 induces re-replication, we examined the expression of Cdc6. Because Cdc6 is a target for E2F1 (19,31), it is expected to be transcriptionally upregulated in E7-expressing cells. We therefore examined Cdc6 protein expression. We normally initiate HPV studies using PHKs (24,25,32). However, as vector-containing PHKs do not proliferate efficiently in culture, we used human RPE1 cells (24) for experiments requiring high efficiency transfection or large amounts of cell extract. Alternatively, spontaneously immortalized human keratinocytes (NIKS) were also used. Accordingly, Cdc6 expression was examined in PHKs, NIKS and RPE1 cells. As shown in Figure 1A, the steady-state level of Cdc6 was significantly increased by more than 8-fold in E7-expressing PHKs compared with vector control PHKs. Similarly, Cdc6 levels were increased in cervical epithelial cells containing the HPV-16 genome (CE-HPV, Figure 1A). Upregulation of Cdc6 protein also occurred in NIKS E7 (Figure 1B) and RPE1 E7 cells (Figure 1C). In PHKs, it is well-established that E7-expressing cells proliferate more efficiently than the vector control cells. We measured the proliferation of RPE1 and NIKS cells expressing E7 and vector control. As shown in Figure 1D and E, E7-expressing cells proliferate more efficiently than the control cells.
Figure 1.
Upregulation of Cdc6 in E7-expressing cells. The steady-state levels of Cdc6 in PHKs and CE-HPV (A), NIKS (B) and RPE1 cells (C) expressing E7 or vector were examined by immunoblotting. Proliferation of E7-expressing RPE1 cells (D) and NIKS cells (E) was measured using a CCK8 assay. Cells were examined at 0, 12, 24, 36, 48 and 72h after seeding. Data are shown as the mean ± SD. A representative image of three experiments is shown. *P < 0.05, **P < 0.01.
Cdc6 plays an important role in E7-induced re-replication
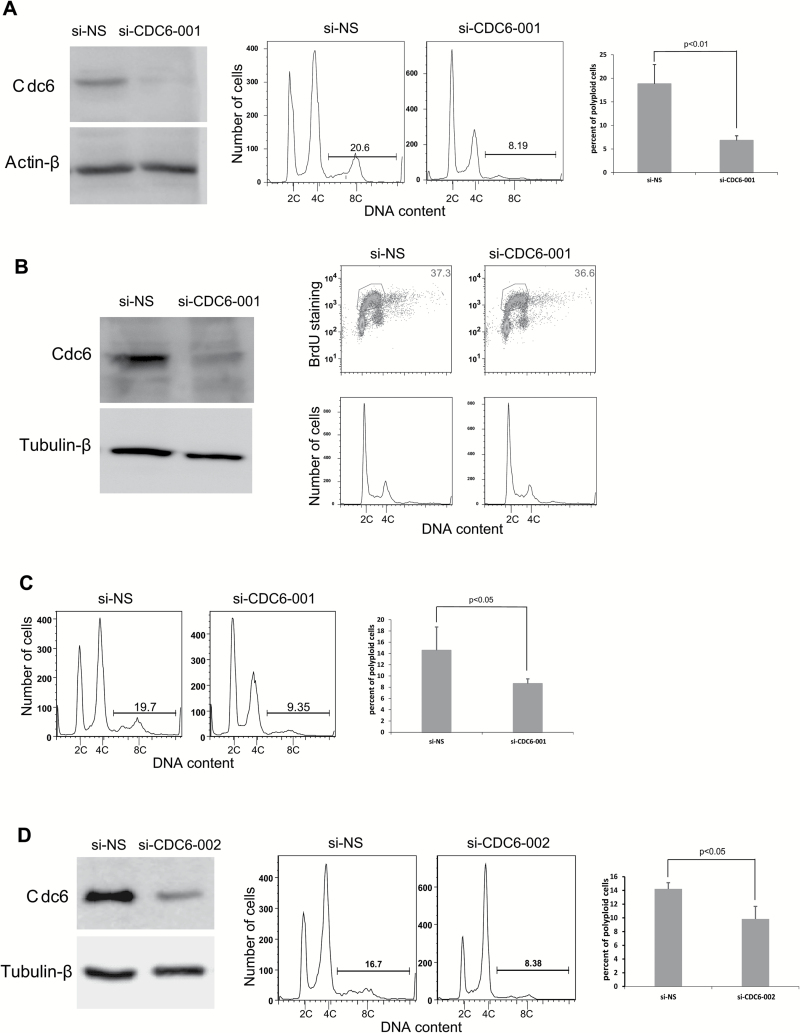
Because HPV-16 E7 induces re-replication (24) and is able to upregulate Cdc6 (Figure 1), whose overexpression leads to re-replication (12,13,33,34), it is reasonable to speculate that Cdc6 plays an important role in HPV E7-induced re-replication. To confirm this, we knocked down Cdc6 in HPV-16 E7-expressing RPE1 cells using siRNAs and examined the extent to which re-replication was affected. We used an siRNA (si-CDC6-001) that was reported to efficiently knockdown Cdc6 after transfection into cultured cells (20). Similar to the previous report, we found that the transfection of si-CDC6-001 efficiently reduced the steady-state levels of Cdc6 in RPE1 E7 cells to 5% of the original level (Figure 2A, left panel). Our recent study demonstrated that in response to DNA damage (bleomycin treatment), RPE1 E7 cells undergo re-replication, resulting in polyploidy formation (24). Accordingly, we transfected RPE1 E7 cells with siRNA targeting Cdc6 and treated the cells with bleomycin, followed by flow cytometry analysis. siRNA knockdown of Cdc6 significantly reduced the number of E7-expressing cells that underwent polyploidy formation by more than 60% (Figure 2A, middle and right panels), suggesting that upregulation of Cdc6 is important for E7 to induce re-replication.
Figure 2.
The downregulation of Cdc6 impairs the ability of E7 to induce polyploidy. RPE1 E7 cells were transfected with Cdc6 siRNAs, and DNA content/BrdU staining was analyzed by FACS. (A) Cdc6 protein levels were measured by immunoblotting 72h after si-CDC6-001 (20nM) transfection (left panel). For cell cycle analysis, bleomycin was added 24h post-transfection and incubated for an additional 48h. The cell cycle profiles are shown in the middle panel. The data are quantified and summarized in the right panel. (B) Regularly cultured RPE1 E7 cells were transfected with 500 pM si-CDC6-001 siRNA. Left panel, Cdc6 protein levels were measured by immunoblotting. A representative image from two independent experiments is shown. The cell cycle profiles are shown in the right panel; BrdU-positive cells are indicated. A histogram of the same profiles is shown below. (C) After si-CDC6-001 (500 pM) transfection, RPE1 E7 cells were treated with bleomycin. A representative image from two independent experiments of four determinants is shown (left panel). The data are summarized in the right panel. (D) After si-CDC6-002 transfection, the cells were treated with bleomycin and analyzed for Cdc6 expression (left panel) and cell cycle profiles (middle panel). A representative image from at least three independent experiments is shown, and the data are summarized (right panel). Error bars reflect the standard deviations of the mean. NS-siRNA, non-silencing siRNA.
Our typical transfection protocol uses siRNA at a final concentration of 20nM, which reduces the likelihood of siRNA-induced non-specific effects (35). However, there is concern that Cdc6 knockdown may disrupt normal DNA replication and cell cycle progression. Therefore, we reduced the amount of Cdc6 siRNA used for transfection to a final concentration of 500 pM and observed that si-CDC6-001 still efficiently knocked down Cdc6 (to 33%) (Figure 2B, left panel). Notably, the level of Cdc6 in E7-expressing RPE1 cells after siRNA knockdown was comparable with the level of Cdc6 in the vector control cells (data not shown). Furthermore, transfection of si-CDC6-001 at 500 pM did not affect the normal cell cycle profile (Figure 2B, right panel). Therefore, this concentration was used to examine the effect of si-CDC6-001 on E7-induced re-replication. As shown in Figure 2C, after siRNA knockdown of Cdc6 at the lower concentration, the number of polyploid cells was significantly reduced.
We observed that knockdown of Cdc6 by si-CDC6-001 reduced the number of E7-expressing cells at the G2/M phases upon DNA damage at both 20nM and 500 pM (Figure 2A and C). Our previous study showed that upon DNA damage, E7-expressing RPE1 cells are mainly at the G2 phase (24). Because re-replication in E7-expressing cells occurs at the G2 phase of the cell cycle (24), there is concern that the reduced number of polyploid cells may be the result of a reduced number of cells at the G2 stage. To alleviate this concern, we employed another siRNA targeting CDC6 (si-CDC6-002). At a final concentration of 20nM, si-CDC6-002 also efficiently reduced the steady-state level of Cdc6 to a level comparable with that in vector control cells (to 28%, Figure 2D, left panel). Notably, si-CDC6-002 transfection did not alter the number of cells at the G2 phase. Additionally, si-CDC6-002 knockdown of Cdc6 in E7-expressing RPE1 cells significantly reduced the number of polyploid cells (Figure 2D, middle and right panels). Taken together, our study demonstrates that upregulation of Cdc6 is important for E7 to induce re-replication.
DNA damage stabilizes the Cdc6 protein in E7-expressing cells
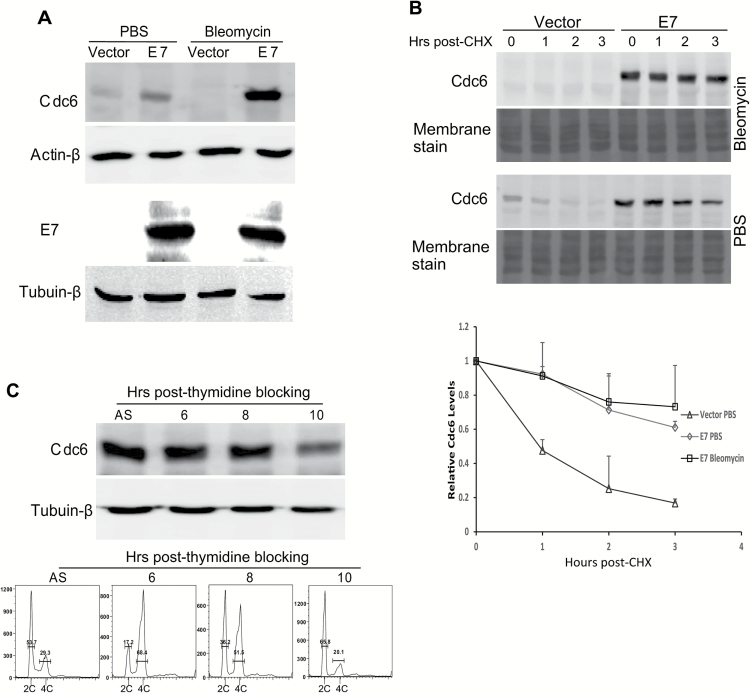
We were interested in understanding the mechanism by which Cdc6 induces re-replication upon DNA damage in E7-expressing cells. One possibility is that DNA damage arrests E7-expressing cells at the G2 stage, where re-replication occurs (24). An alternative possibility is that Cdc6 expression and/or localization may change upon DNA damage. In this experiment, we treated RPE1 cells expressing E7 with bleomycin and examined the steady-state level of Cdc6. Although bleomycin treatment led to a reduction of Cdc6 in vector control cells, a significant increase (3-fold) in the steady-state level of Cdc6 was seen in RPE1 E7 cells (Figure 3A). In contrast, the steady-state level of E7 remained unchanged with bleomycin treatment. We also determined the protein stability of Cdc6. The steady-state level of Cdc6 in vector-containing RPE1 cells was decreased by more than 80% 3h after cycloheximide treatment. In contrast, ~60% of the Cdc6 protein was maintained in E7-expressing cells. Cdc6 was undetectable in vector control cells after bleomycin treatment; however, more than 70% of Cdc6 remained in E7-expressing cells. Consistently, the half-life of Cdc6 in E7-expressing cells was longer than that in vector-containing cells (5.6 versus 1h; Figure 3B). The half-life of Cdc6 in vector-containing RPE1 cells upon DNA damage could not be measured due to the low levels of Cdc6 in these cells. In contrast, the half-life of Cdc6 in E7-expressing cells was further increased (to 6.4h; Figure 3B) after bleomycin treatment. These results demonstrate that the expression of Cdc6 in E7-expressing cells is regulated at the post-transcriptional level.
Figure 3.
Cdc6 in E7-expressing cells is stabilized with bleomycin treatment, and its abundance is not cell cycle related. RPE1 cells expressing E7 or vector were treated with PBS or bleomycin (5 µg/ml for 48h). (A) Cdc6 (upper panel) and E7 (lower panel) levels were examined by immunoblotting. (B) After bleomycin treatment, the cells were incubated with 25 µg/ml cycloheximide (CHX) and harvested at the indicated times. The stability of Cdc6 was monitored using immunoblotting. The data from at least of two independent experiments are shown in the upper panel and summarized in the lower panel. (C) RPE1 E7 cells were blocked with 2.5M thymidine for 16h and then released to regular media and collected at the time points indicated. Cdc6 levels were determined (upper panel), and the cell cycle profile was analyzed. The data from a representative of four independent experiments are shown. AS, asynchronous.
Our previous studies demonstrated that upon DNA damage, vector-containing RPE1 cells arrested at the G1 checkpoint, whereas E7-expressing RPE1 cells accumulated at the G2 phase (24). This observation provides a possible explanation for the increased half-life of Cdc6 in E7-expressing cells (i.e. cell cycle regulation: stabilized at G2 and destabilized at G1). To test this possibility, we arrested RPE1 E7 cells by thymidine blocking. The cells were then released and examined for cell cycle profile and Cdc6 steady-state level at several time points. We observed that 6h after release from thymidine blocking, the majority of cells were in the G2/M stage (Figure 3C). However, the steady-state level of Cdc6 was similar to that observed in unsynchronized cells. Additionally, 10h after thymidine blocking, the majority of cells entered the G1 phase, and the steady-state level of Cdc6 at the G1 phase was decreased (Figure 3C). Therefore, cell cycle-dependent Cdc6 fluctuations may explain the decrease in Cdc6 in vector control cells, but not in E7-expressing cells, upon DNA damage. These results indicate that a cell cycle-independent mechanism controls the stability of Cdc6 in E7-expressing cells.
Cellular localization of Cdc6 in E7-expressing cells
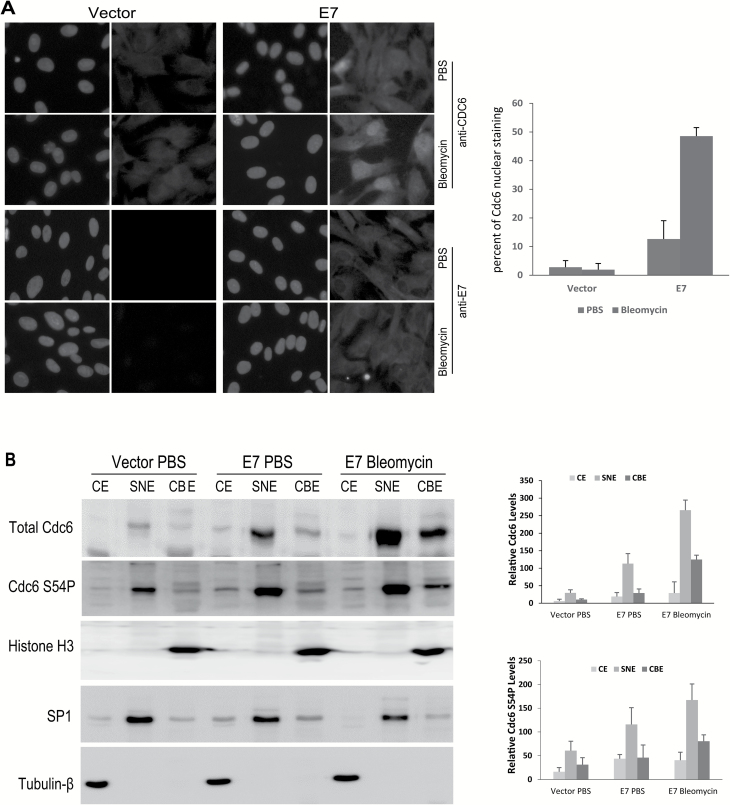
Next, we examined the cellular localization of Cdc6 in E7-expressing cells using immunofluorescence. Strong nuclear staining of Cdc6 protein was observed in a subset (~13%) of E7-expressing cells (Figure 4A). Upon DNA damage, increased nuclear localization of Cdc6 (~49%) was observed in E7-expressing cells (Figure 4A). In contrast, the Cdc6 immunofluorescence signal in regularly cultured RPE1 vector cells was too weak to capture at the same exposure time used in E7-expressing cells. Using a longer exposure time, cytoplasmic localization of Cdc6 was detected (Figure 4A). There was no change in cellular localization in vector control cells after bleomycin treatment. It is plausible that the difference in Cdc6 localization relates to different levels of E7 in the cells. We therefore examined E7 expression and localization by immunofluorescence. Our data show that E7 was uniformly expressed in all cells, indicating that nuclear localization of Cdc6 in a subset of E7-expressing cells is not related to different levels of E7 (Figure 4A).
Figure 4.
The localization and subcellular fractionation of Cdc6 in E7-expressing cells. RPE1 cells expressing E7 or vector were treated with bleomycin (3 µg/ml) for 48h. (A) Indirect immunofluorescence microscopy was performed to detect CDC6 and E7 expression and localization (green). 4,5-Diamidino-2-phenylindole dihydrochloride (blue) was used to counterstain the nucleus. Images were captured at ×40 magnification. The data from a representative of at least two independent experiments are shown. (B) The cells were fractionated as cytoplasmic extracts (CEs), soluble nuclear extracts (SNEs) and chromatin-bound fractions (CBEs). The total Cdc6 and Cdc6 S54P levels of each fraction were determined using immunoblotting. β-Tubulin, SP1 and histone H3 were used as loading controls for CE, SNE and CBE, respectively. The data from a representative of three independent experiments are shown in the left panel. The relative levels of total Cdc6 and Cdc6 S54P are summarized in the right panel.
We then examined Cdc6 localization by cellular fractionation to obtain more detailed and quantitative information. The protein extracts from the cells were fractionated into cytoplasmic (CE), soluble nuclear (SNE) and chromatin-bound (CBE) fractions and blotted with antibodies against Cdc6, β-tubulin (CE marker), SP1 (SNE marker) and histone H3 (CBE marker) (Figure 4B). Successful fractionation was confirmed by subcellular localization of cytoplasmic (β-tubulin), soluble nuclear (SP1) and chromatin-bound (Histone H3) protein markers. In the vector control RPE1 cells, a low amount of Cdc6 was detected in the SNE fraction (Figure 4B). Additionally, a faint band in the CBE fraction was also observed. However, little Cdc6 was detectable in the CE fraction (Figure 4B). This pattern was different from what was observed for immunofluorescence, where Cdc6 was detectable only in the cytoplasm (Figure 4A). In the E7-expressing cells, increased Cdc6 was detected in all three fractions. Upon DNA damage, the Cdc6 protein in the RPE1 vector cells became undetectable (data not shown). In contrast, increased Cdc6 was detected in the nucleus and chromatin fraction in E7-expressing cells (Figure 4B). These results demonstrate that in E7-expressing cells, Cdc6 mainly localizes in the nucleus and remains chromatin-bound. Upon DNA damage, Cdc6 localization increases in the nucleus and chromatin fractions, but not in the cytoplasm, in E7-expressing cells.
Phosphorylation of Cdc6 has been detected at several serine residues, including Ser54 (S54P) (36–38). In RPE1 cells, we detected Cdc6 phosphorylation at S54. Notably, Cdc6 S54P levels were higher in all three fractions in E7-expressing cells than in vector control cells (Figure 4B). Upon DNA damage in E7-expressing RPE1 cells, as the level of total Cdc6 increased and the localization of chromatin-bound Cdc6 protein increased in the nucleus, the level of Cdc6 S54P in the SNEs and CBEs also increased (Figure 4B). These observations indicate that Cdc6 S54P is associated with the increased stability, nuclear localization and chromatin binding of Cdc6.
Role of Cdk1 inactivation in Cdc6 stability and re-replication
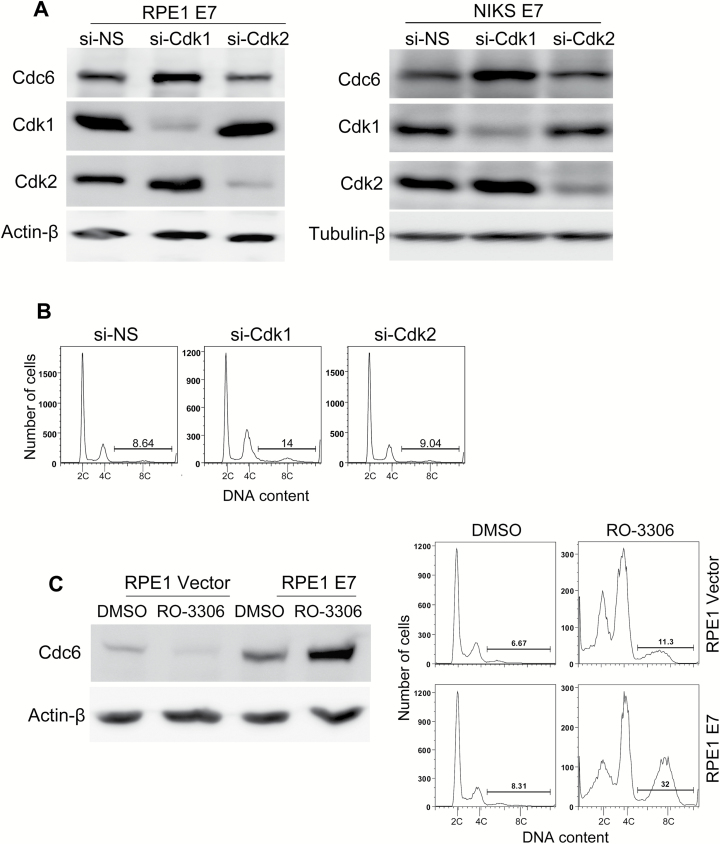
We aimed to determine how DNA damage increases the steady-state and S54P levels of Cdc6 in E7-expressing cells. It is known that Cdk2 is responsible for S54 phosphorylation (38). Therefore, we examined the effect of Cdk2 knockdown on the level of Cdc6. As previously shown, Cdk1 can compensate for Cdk2 activity in E7-expressing cells at the G1 checkpoint in response to DNA damage (39); as such, we included Cdk1 in our studies. Cdk2 knockdown in RPE1 E7 cells resulted in a reduction in the steady-state level of Cdc6 (30% reduction, Figure 5A, left panel). This result is consistent with a previous observation that Cdk2 phosphorylates S54 and stabilizes Cdc6 (35–37). Interestingly, siRNA knockdown of Cdk1 led to an increased steady-state level of Cdc6 (1.4-fold) (Figure 5A, left panel). Similar results were obtained in NIKS expressing HPV-16 E7 (Figure 5A, right panel).
Figure 5.
Cdk1 is involved in Cdc6 stability and polyploidy formation in E7-expressing cells. E7-expressing cells were transfected with 20nM si-Cdk1 or control siRNAs for 72h. (A) Downregulation of Cdk1 increased Cdc6 expression in E7-expressing RPE1 cells (left) or NIKS (right). Cdc6 levels were determined using immunoblotting. (B) Downregulation of Cdk1 induced polyploidy formation in E7-expressing NIKS. The cell cycle profile was analyzed using flow cytometry. The data from a representative experiment of two independent experiments are shown. (C) The Cdk1 inhibitor RO-3306 increased the Cdc6 level and efficiently induced polyploidy formation in RPE1 E7-expressing cells. RPE1 cells expressing E7 or vector were treated with 7.5 µM RO-3306 or dimethyl sulfoxide (DMSO) for 48h. Cdc6 levels were determined using immunoblotting, and the cell cycle profile was analyzed using flow cytometry. The data from at least three independent experiments are shown. NS-siRNA, non-silencing siRNA.
Consistent with the Western blotting results, immunofluorescence demonstrated that siRNA knockdown of Cdk1 in RPE1 E7 cells resulted in an increased number of cells with strong nuclear staining for Cdc6 (data not shown), which mimics the effect of bleomycin. If inhibition of Cdk1 is responsible for increased Cdc6 and bleomycin-triggered DNA re-replication in E7-expressing cells, knockdown of Cdk1 should lead to re-replication. We observed an increase in polyploidy formation in E7-expressing NIKS and RPE1 cells after Cdk1 siRNA transfection (Figure 5B and data not shown).
We also used RO-3306, a specific inhibitor of Cdk1 (Ki = 35nM for Cdk1/cyclin B1, Ki = 340nM for Cdk2/cyclin E and Ki > 2000nM for CDK4/cyclin D) (40), to treat the E7-expressing cells and examine the effect of RO-3306 on Cdc6 expression and polyploidy formation. As shown in Figure 5C, RO-3306 treatment resulted in a reduced level of Cdc6 expression in the control cells but an increased level of Cdc6 expression in the E7-expressing cells, a result similar to what was observed after bleomycin treatment and Cdk1 siRNA transfection. Furthermore, more polyploid cells formed in the E7-expressing RPE1 cells after treatment with the Cdk1 inhibitor (Figure 5C). These results suggest that bleomycin treatment induced Cdc6 upregulation and polyploidy formation in E7-expressing cells is due to an inhibitory effect on Cdk1. In conclusion, our data indicate that Cdk1 is a key molecule for Cdc6 stability and DNA re-replication.
Discussion
Our recent studies demonstrated that HPV-16 E7 induces re-replication in response to DNA damage and that the pre-replicative complex factor, Cdt1, plays an important role in this process (24). In this study, we showed that another DNA replication initiation factor, Cdc6, is increased in HPV-16 E7-expressing primary and immortalized cells and that upregulation of Cdc6 plays a role in DNA re-replication. DNA damage can stabilize Cdc6 and maintain its nuclear localization in E7-expressing cells through inhibition of Cdk1. We believe that DNA damage-induced inhibition of Cdk1 contributes to the status of CDC6 in E7 cells. The results from this study implicate Cdc6 in the development of HPV-associated carcinogenesis.
Similar to Cdt1, Cdc6 is an important protein in the initiation of DNA replication. Whereas previous studies have demonstrated overexpression of Cdc6 in cancer tissues, including cervical cancer (15,16), its expression in E7-expressing cells and its role in E7-mediated re-replication has not been examined. We found that Cdc6 was upregulated in E7-expressing cells. In addition, our results indicate that the half-life of Cdc6 was prolonged in E7-expressing cells. Therefore, additional mechanisms are responsible for the increased stability of Cdc6 in E7-expressing cells. Most current studies have focused on Cdk2’s regulation of Cdc6 stability. In E7-expressing cells, Cdk2 levels were increased, as described previously (39), and may contribute to upregulation of Cdc6. Furthermore, knockdown of Cdk2 reduced the steady-state level of Cdc6 (Figure 5A).
There is a concern that the effect Cdc6 siRNA on re-replication is due to a reduction in Cdc6’s normal function in DNA replication initiation. To alleviate this concern, we used siRNA to knockdown Cdc6 at a low concentration of 500 pM, which did not alter normal DNA replication but still reduced re-replication. Therefore, involvement of Cdc6 in re-replication is not due to interference with DNA replication but is a function obtained only upon overexpression.
Phosphorylation at Ser54 by Cdk2 stabilizes Cdc6 by preventing its association with the anaphase-promoting complex/cyclosome (20). Although ectopically expressed Cdc6 may behave differently, endogenous Cdc6 phosphorylated on S54 remains in the nucleus and bound to chromatin (23). However, a recent study found that S54P facilitates Cdc6 cytoplasmic translocation (41). In addition, another study noted Cdc6 phosphorylation at S54 in both cytoplasmic and nuclear insoluble compartments (21). Our results support the idea that S54P facilitates nuclear localization of Cdc6 in E7-expressing cells. Upon DNA damage, Cdc6 levels in vector control RPE1 cells were undetectable. Through induction of p21, the DNA damage-activated protein, p53, may lead to inhibition of cyclin E/Cdk2, which results in the loss of Cdc6 Ser54 phosphorylation and accelerated proteolysis of Cdc6 (42). However, in E7-expressing cells, treatment with bleomycin increased the total amount and nuclear localization of Cdc6 protein. The opposite effect was observed in vector control cells. One explanation for this observation is although Cdk2 activity in vector control cells is reduced, significant Cdk2 activity is maintained in E7-expressing cells (43).
Cdc6 levels and localization should also be affected by additional factors, such as Cdk1. Previous in vitro experiments showed that CycA/Cdk1 and CycB/Cdk1 can phosphorylate Cdc6, although CycB/Cdk1 is not as efficient as CycA/Cdk1 (16,44). A recent study suggested that although Cdc6 is exported to the cytoplasm by Cdk2 from the early S phase onwards, in the G2 phase, Cdc6 is exported to the cytoplasm via the increased activity of Cdk1 (45). Because phosphorylation at Ser106 (S106P) promotes Cdc6 cytoplasmic localization and degradation (21), Cdk1 is expected to negatively regulate the stability of Cdc6. We showed that Cdk1 regulates Cdc6 stability and localization in E7-expressing cells. DNA damage is thought to downregulate cyclins/Cdks, including Cdk1 and cyclin B activity (46–48). Additionally, downregulation of Cdk1 by either siRNA or the chemical inhibitor RO-3306 upregulated the total Cdc6 level and induced re-replication. This is consistent with the result from a previous study that showed that degradation of Cdc6 was substantially slower after RO-3306 treatment than during the normal cell cycle (49). Therefore, we predict that Cdk1 inhibition by DNA damage in E7-expressing cells is another major player in regulating Cdc6 stability. However, why Cdk1 inhibition by DNA damage in vector control cells does not lead to increased Cdc6 expression remains an unanswered question. We believe that this is due to a lack of sufficient Cdk2 activity in vector control cells upon DNA damage. The combination of Cdk2 and Cdk1 determined the stability of Cdc6. It has been proposed that cyclin E, which normally associates with Cdk2, opens a window of opportunity for replication complex assembly that is closed by cyclin A (50). Our data suggest that Cdk2 favors re-replication, whereas Cdk1, whose partners include cyclin A, inhibits re-replication.
There may be subtle feedback or crosstalk between Cdc6 and Cdk1 (51). Cdc6 may activate Cdk1 at the G1 stage, whereas during the G2 stage, Cdk1 may promote degradation of CDC6. The functional interaction between Cdc6 and Cdk1 contributes to re-replication in E7-expressing cells at the G2 stage. One may cast doubt on whether inhibition of Cdk1 caused by DNA damage can also arrest cells at the G1 stage. We believe that the checkpoint at the G1 stage is less stringent than the checkpoint at the G2 stage and that the degree of Cdk1 inhibition may not be strong enough to arrest cells in the G1 stage but is sufficient for promoting the cytoplasmic exportation and subsequent degradation of Cdc6. There is also a time difference between the G1 stage and re-replication, which occurs at the G2 stage. A recent study argues that phosphorylation of Cdc6 at Ser74 instead of Ser106 drives the translocation of Cdc6 to the cytoplasm (52). Due to the unavailability of an ideal anti-pS74P antibody, we cannot speculate on the relationship between S74 phosphorylation and Cdc6 stability. Further studies are needed to address these unanswered questions.
Funding
National Natural Science Foundation of China (81471944 to J.J.C.); National Cancer Institute (R01CA119134).
Acknowledgements
We thank Drs W.Jiang, Y.Gong and X.Wang for their helpful discussions and X.Zhang for technical assistance. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Conflict of Interest Statement: None declared.
Glossary
Abbreviations
- Cdk
cyclin-dependent kinase
- BrdU
bromodeoxyuridine
- HPV
human papillomavirus
- PBS
phosphate-buffered saline
- PHK
primary human keratinocyte
- RPE
retinal pigment epithelial
- siRNA
small interfering RNA
References
- 1. Van Doorslaer K., et al. (2013) The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res., 41, D571–D578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Villiers E.M. (2013) Cross-roads in the classification of papillomaviruses. Virology, 445, 2–10. [DOI] [PubMed] [Google Scholar]
- 3. Munoz N., et al. (2003) Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med., 348, 518–527. [DOI] [PubMed] [Google Scholar]
- 4. zur Hausen H. (2002) Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer, 2, 342–350. [DOI] [PubMed] [Google Scholar]
- 5. Walboomers J.M., et al. (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol., 189, 12–19. [DOI] [PubMed] [Google Scholar]
- 6. Chaturvedi A.K. (2010) Beyond cervical cancer: burden of other HPV-related cancers among men and women. J. Adolesc. Health, 46, S20–S26. [DOI] [PubMed] [Google Scholar]
- 7. Schwarz E., et al. (1985) Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature, 314, 111–114. [DOI] [PubMed] [Google Scholar]
- 8. Chen J.J. (2010) Genomic instability induced by human papillomavirus oncogenes. N. Am. J. Med. Sci., 3, 43–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sivaprasad U., et al. (2007) APC/C—the master controller of origin licensing? Cell Div., 2, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cook J.G., et al. (2002) Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc. Natl Acad. Sci. USA, 99, 1347–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xouri G., et al. (2007) Cdt1 interactions in the licensing process: a model for dynamic spatiotemporal control of licensing. Cell Cycle, 6, 1549–1552. [DOI] [PubMed] [Google Scholar]
- 12. Liontos M., et al. (2007) Deregulated overexpression of hCdt1 and hCdc6 promotes malignant behavior. Cancer Res., 67, 10899–10909. [DOI] [PubMed] [Google Scholar]
- 13. Vaziri C., et al. (2003) A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell, 11, 997–1008. [DOI] [PubMed] [Google Scholar]
- 14. Williams G.H., et al. (1998) Improved cervical smear assessment using antibodies against proteins that regulate DNA replication. Proc. Natl Acad. Sci. USA, 95, 14932–14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Murphy N., et al. (2005) p16INK4A, CDC6, and MCM5: predictive biomarkers in cervical preinvasive neoplasia and cervical cancer. J. Clin. Pathol., 58, 525–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fujita M., et al. (1999) Cell cycle regulation of human CDC6 protein. Intracellular localization, interaction with the human mcm complex, and CDC2 kinase-mediated hyperphosphorylation. J. Biol. Chem., 274, 25927–25932. [DOI] [PubMed] [Google Scholar]
- 17. Bonds L., et al. (2002) Immunohistochemical localization of cdc6 in squamous and glandular neoplasia of the uterine cervix. Arch. Pathol. Lab. Med., 126, 1164–1168. [DOI] [PubMed] [Google Scholar]
- 18. Murphy N., et al. (2005) Quantitation of CDC6 and MCM5 mRNA in cervical intraepithelial neoplasia and invasive squamous cell carcinoma of the cervix. Mod. Pathol., 18, 844–849. [DOI] [PubMed] [Google Scholar]
- 19. Hateboer G., et al. (1998) Cell cycle-regulated expression of mammalian CDC6 is dependent on E2F. Mol. Cell. Biol., 18, 6679–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mailand N., et al. (2005) CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell, 122, 915–926. [DOI] [PubMed] [Google Scholar]
- 21. Paolinelli R., et al. (2009) Acetylation by GCN5 regulates CDC6 phosphorylation in the S phase of the cell cycle. Nat. Struct. Mol. Biol., 16, 412–420. [DOI] [PubMed] [Google Scholar]
- 22. Coverley D., et al. (2000) Chromatin-bound Cdc6 persists in S and G2 phases in human cells, while soluble Cdc6 is destroyed in a cyclin A-cdk2 dependent process. J. Cell Sci., 113(Pt 11), 1929–1938. [DOI] [PubMed] [Google Scholar]
- 23. Alexandrow M.G., et al. (2004) Cdc6 chromatin affinity is unaffected by serine-54 phosphorylation, S-phase progression, and overexpression of cyclin A. Mol. Cell. Biol., 24, 1614–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Fan X., et al. (2013) Human papillomavirus E7 induces rereplication in response to DNA damage. J. Virol., 87, 1200–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu Y., et al. (2007) p53-independent abrogation of a postmitotic checkpoint contributes to HPV E6-induced polyploidy. Cancer Res., 67, 2603–2610. [DOI] [PubMed] [Google Scholar]
- 26. Berger K.L., et al. (2006) Cervical keratinocytes containing stably replicating extrachromosomal HPV-16 are refractory to transformation by oncogenic H-Ras. Virology, 356, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melixetian M., et al. (2004) Loss of geminin induces rereplication in the presence of functional p53. J. Cell Biol., 165, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. L’Italien L., et al. (2006) Unmasking the redundancy between Cdk1 and Cdk2 at G2 phase in human cancer cell lines. Cell Cycle, 5, 984–993. [DOI] [PubMed] [Google Scholar]
- 29. Tanaka T., et al. (2007) PDLIM2-mediated termination of transcription factor NF-kappaB activation by intranuclear sequestration and degradation of the p65 subunit. Nat. Immunol., 8, 584–591. [DOI] [PubMed] [Google Scholar]
- 30. Healy N.C., et al. (2009) Sequestration of PDLIM2 in the cytoplasm of monocytic/macrophage cells is associated with adhesion and increased nuclear activity of NF-kappaB. J. Leukoc. Biol., 85, 481–490. [DOI] [PubMed] [Google Scholar]
- 31. Yan Z., et al. (1998) Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc. Natl Acad. Sci. USA, 95, 3603–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Heilman S.A., et al. (2009) Abrogation of the postmitotic checkpoint contributes to polyploidization in human papillomavirus E7-expressing cells. J. Virol., 83, 2756–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mihaylov I.S., et al. (2002) Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol., 22, 1868–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Machida Y.J., et al. (2007) The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev., 21, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Persengiev S.P., et al. (2004) Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs). RNA, 10, 12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Petersen B.O., et al. (1999) Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J., 18, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jiang W., et al. (1999) Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc. Natl Acad. Sci. USA, 96, 6193–6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Delmolino L.M., et al. (2001) Multiple mechanisms regulate subcellular localization of human CDC6. J. Biol. Chem., 276, 26947–26954. [DOI] [PubMed] [Google Scholar]
- 39. Fan X., et al. (2014) Role of Cdk1 in DNA damage-induced G1 checkpoint abrogation by the human papillomavirus E7 oncogene. Cell Cycle, 13, 3249–3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vassilev L.T., et al. (2006) Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc. Natl Acad. Sci. USA, 103, 10660–10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hwang I.S., et al. (2014) Two nuclear export signals of Cdc6 are differentially associated with CDK-mediated phosphorylation residues for cytoplasmic translocation. Biochim. Biophys. Acta, 1843, 223–233. [DOI] [PubMed] [Google Scholar]
- 42. Duursma A., et al. (2005) p53-dependent regulation of Cdc6 protein stability controls cellular proliferation. Mol. Cell. Biol., 25, 6937–6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Funk J.O., et al. (1997) Inhibition of CDK activity and PCNA-dependent DNA replication by p21 is blocked by interaction with the HPV-16 E7 oncoprotein. Genes Dev., 11, 2090–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Herbig U., et al. (2000) Mutation of cyclin/cdk phosphorylation sites in HsCdc6 disrupts a late step in initiation of DNA replication in human cells. Mol. Biol. Cell, 11, 4117–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clijsters L., et al. (2014) PIP-box mediated degradation prohibits re-accumulation of Cdc6 during S phase. J Cell Sci., 127(Pt 6), 1336–1345. [DOI] [PubMed] [Google Scholar]
- 46. Tsao Y.P., et al. (1992) The involvement of active DNA synthesis in camptothecin-induced G2 arrest: altered regulation of p34cdc2/cyclin B. Cancer Res., 52, 1823–1829. [PubMed] [Google Scholar]
- 47. Herzinger T., et al. (1995) Ultraviolet B irradiation-induced G2 cell cycle arrest in human keratinocytes by inhibitory phosphorylation of the cdc2 cell cycle kinase. Oncogene, 11, 2151–2156. [PubMed] [Google Scholar]
- 48. Nakayama Y., et al. (2009) Bleomycin-induced over-replication involves sustained inhibition of mitotic entry through the ATM/ATR pathway. Exp. Cell Res., 315, 2515–2528. [DOI] [PubMed] [Google Scholar]
- 49. Ma H.T., et al. (2009) Cyclin A2-cyclin-dependent kinase 2 cooperates with the PLK1-SCFbeta-TrCP1-EMI1-anaphase-promoting complex/cyclosome axis to promote genome reduplication in the absence of mitosis. Mol. Cell. Biol., 29, 6500–6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coverley D., et al. (2002) Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol., 4, 523–528. [DOI] [PubMed] [Google Scholar]
- 51. Boronat S., et al. (2008) Linking mitosis with S-phase: Cdc6 at play. Cell Cycle, 7, 597–601. [DOI] [PubMed] [Google Scholar]
- 52. Yim H., et al. (2013) Phosphorylation of Cdc6 at serine 74, but not at serine 106, drives translocation of Cdc6 to the cytoplasm. J. Cell. Physiol., 228, 1221–1228. [DOI] [PubMed] [Google Scholar]