SUMMARY
Bacteria must maintain mechanical integrity to withstand the large osmotic pressure differential across the cell membrane and wall. Although maintaining mechanical integrity is critical for proper cellular function, a fact exploited by prominent cell wall-targeting antibiotics, the proteins that contribute to cellular mechanics remain unidentified. Here, we describe a high-throughput optical method for quantifying cell stiffness and apply this technique to a genome-wide collection of ~4000 Escherichia coli mutants. We identify genes with roles in diverse functional processes spanning cell wall synthesis, energy production, and DNA replication and repair that significantly change cell stiffness when deleted. We observe that proteins with biochemically redundant roles in cell wall synthesis exhibited different stiffness defects when deleted. Correlating our data with chemical screens reveals that reducing membrane potential generally increases cell stiffness. In total, our work demonstrates that bacterial cell stiffness is a property of both the cell wall and broader cell physiology and lays the groundwork for future systematic studies of mechanoregulation.
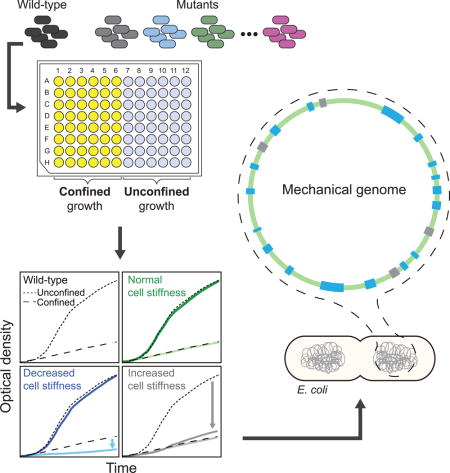
Graphical abstract
INTRODUCTION
Biological growth is as much about the creation of a physical body plan through structures such as membranes, cell walls, and extracellular matrix, as it is the synthesis of proteins, DNA, and RNA. The stiffness or rigidity of individual cells and tissues is the extent to which they resist deformation due to applied forces, either external or from within, and is a central property that directly impacts many critical aspects of physiology including cell viability and environmental sensing. Cell stiffness also governs sensing of mechanical stimuli, which are then transduced into chemical signals (Vogel and Sheetz, 2006). Changes in tissue stiffness can serve as a diagnostic readout for diseases such as cancer (Huang and Ingber, 2005). Moreover, the degree of stiffness in one cell can feedback on the behavior of other cells, with the mechanical properties of surrounding environment directing the commitment of mesenchymal stem cells (Engler et al., 2006). Cell mechanics are generally thought to be dictated by a combination of cytoskeletal polymers (Janmey and McCulloch, 2007) and exoskeletons such as the cell wall (Braybrook and Jonsson, 2016), although whether other cellular components play an important role is essentially unexamined.
Despite the fundamental importance of cell mechanics, there is a dearth of knowledge regarding the determinants of cell stiffness in any organism that stems in large part from measurement difficulties. There are several techniques available for directly measuring cell mechanics, including atomic force microscopy (Dufrene, 2014), tensile testing (Thwaites and Mendelson, 1989), and optical trapping (Wang et al., 2010). We previously reported a strategy using single-cell tracking of cell growth to extract a composite Young’s modulus of bacterial cells embedded in agarose gels of varying stiffness; the gels provide mechanical resistance to cell elongation (Tuson et al., 2012). However, all of these strategies, including our previous work (Tuson et al., 2012), have limited throughput and hence are not suitable for large-scale analyses. Thus, it remains unknown to what extent chemical and genetic perturbations can modulate cell stiffness, and whether cell stiffness is linked primarily to a specific cellular process or is a global emergent property of cell growth and metabolism.
In plant, fungal, and bacterial cells, the cell wall determines cell shape and provides mechanical stability by stretching to counter expansion due to large osmotic pressure differentials (turgor) on the order of atmospheres (Holtje, 1998). The cell wall is composed of peptidoglycan, a macromolecular network consisting of polysaccharides cross-linked by short peptides (Holtje, 1998). Enzymatic degradation of the peptidoglycan produces cells with altered shapes that are sensitive to osmotic shifts (Birdsell and Cota-Robles, 1967). The genes involved in peptidoglycan assembly code for a family of enzymes known as the penicillin binding proteins (PBPs).
The PBPs have been the subject of detailed biochemical analyses (Scheffers and Pinho, 2005) and are obvious candidate cell-stiffness modulators. However, chemical inhibition of an essential PBP does not impact peptidoglycan composition even at doses that alter cell shape and decrease growth rate (Lee et al., 2014). The relative contributions made by peptidoglycan composition, cell shape, and other aspects of growth to maintaining cellular stiffness are also unclear. More broadly, it is unknown whether perturbations to any part of the cell-wall synthesis and turnover machinery impact cellular mechanics. The only protein in E. coli that has been identified to affect cellular mechanics is the actin homolog MreB, which orchestrates the spatial organization of PBPs and is necessary for maintaining E. coli’s rod shape. Depolymerization of MreB using the small molecule A22 (Gitai et al., 2005) results in an immediate decrease in the bending stiffness of cells (Wang et al., 2010). This observation illustrates that changes to cell stiffness can be due to changes that are not connected to the composition of the cell envelope, which consists of the membrane(s) and cell wall.
More than 2000 of the ~4300 genes in E. coli K12 strains remain experimentally uncharacterized (Riley et al., 2006). This suggests that individual proteins with essential roles in maintaining or regulating cellular mechanics may remain unknown. Additionally, bacteria possess multiple systems to maintain essential cellular processes (Kitano, 2004). This apparent biochemical redundancy may mask specific functions related to cellular mechanics. However, genome-wide approaches that specifically score cell stiffness phenotypes have been lacking. Although perturbations to the membrane-biogenesis machinery can lead to more frequent cell lysis (Paradis-Bleau et al., 2014), which is generally accompanied by mechanical failure of the membranes and cell wall, lysis is not a faithful reporter of cell stiffness. A methodology for identifying proteins that modulate bacterial cell mechanics may provide fundamental insights into the biochemical, biophysical, and cell-biological aspects of cell growth.
Here, we describe a mechanical genomics assay, General Regulators Affecting Bacterial Stiffness (GRABS), which identifies mutants with altered cell stiffness. Screening the Keio collection of non-essential gene deletions in E. coli (Baba et al., 2006) using GRABS, we identified 46 candidate proteins that affect cell stiffness. These candidates have diverse functions that extend beyond cell-envelope assembly; the mechanical role of a subset of these proteins was validated using a microfluidic bending assay. By correlating our GRABS scores with chemical-genomics data (Nichols et al., 2011), we successfully predicted chemicals and/or environmental conditions that increased or decreased cell stiffness. Taken together, our methodology enables the rapid evaluation of how chemical perturbations or genetic disruptions, from single amino-acid changes to whole-gene deletions, alter the mechanical properties of cells.
RESULTS
A high-throughput screen for genes that affect bacterial cell mechanics
To identify modulators of bacterial cell stiffness, we developed a comprehensive, reproducible, and high-throughput methodology for quantifying the effects of genetic and chemical perturbations on cellular mechanics. We previously developed a methodology for determining bacterial cell stiffness using microscopy-based growth of single cells embedded in agarose hydrogels of tunable mechanical stiffness (Tuson et al., 2012). We observed a decrease in bacterial growth as we increased gel stiffness, which enabled us to estimate Young’s modulus of the cell envelope using a finite-element model of the growth of an elastic shell (Tuson et al., 2012). This method for determining cell stiffness, while quantitative and robust, is not amenable to high-throughput measurements at a genomic scale. We hypothesized that decreases in the growth of bacteria embedded in agarose could be measured optically using a standard microplate reader to measure light scattering. This strategy would enable rapid screens of a diverse collection of mutants, including genome-wide knockout libraries, for changes that specifically affect growth embedded in agarose.
To test this approach, we embedded cells of the Keio collection wild-type parent strain E. coli BW25113 in agarose gels (0.25% to 5%) that were infused with lysogeny broth (LB) and solidified in a 96-well plate (Experimental Procedures). We measured light scattering using a microplate reader (Figure 1A). The Keio collection contains deletions of virtually all non-essential genes in E. coli BW25113, and has served as a powerful resource for genome-wide screens for diverse phenotypes such as chemical sensitities (Nichols et al., 2011), membrane permeability (Paradis-Bleau et al., 2014), and phage infection (Qimron et al., 2006). As in our single-cell assay of cell stiffness (Tuson et al., 2012), we observed a monotonic decrease in the optical density (OD) (Figure 1A) and the maximum growth rate of wild-type cells as the concentration of agarose increased (Figure 1B). Based on these data, we inferred that a 1% (Young’s modulus 56 kPa) agarose gel is reasonable for detecting changes in cell growth, as wild-type cells growing in agarose had an OD decrease of ~80% relative to liquid medium (Figure 1B; determined at 8 h, which is approximately when wild-type cells enter stationary phase in liquid medium). The OD of cultures embedded in 1% agarose was linearly correlated with cell density over a broad range (Figure S1A). Gels with agarose concentrations below 1% are not suitable for our purposes because the pores have dimensions close to the diameter of cells (Narayanan et al., 2006).
Figure 1. GRABS methodology enables high-throughput measurements of cellular mechanical properties.
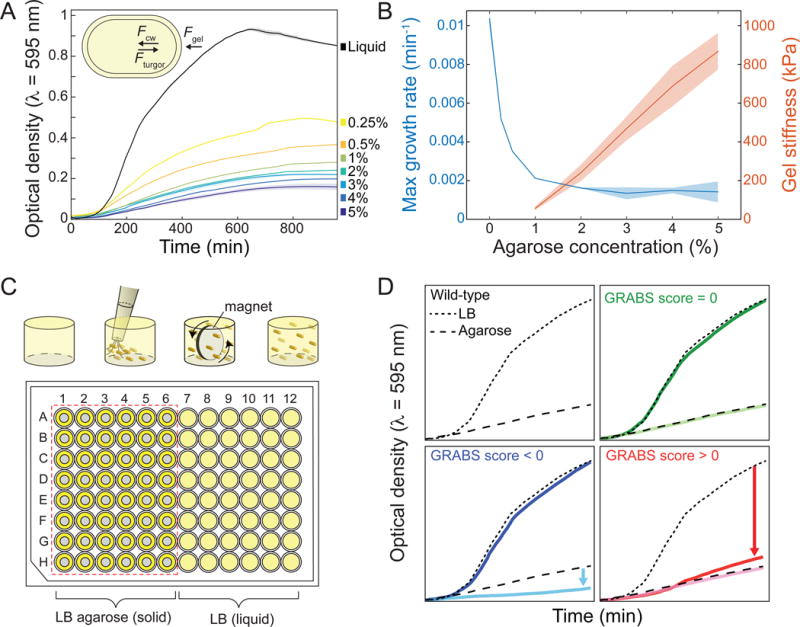
A) Growth curves of E. coli BW25113 embedded in 0.25%–5% agarose gels and liquid medium (n = 3) monitored with a plate reader; cell growth diminishes as gel stiffness increases. Solid lines and shaded areas represent the smoothed average and one standard deviation, respectively. Inset: schematic of a bacterial cell growing in agarose. Of the forces acting on the cell, Fgel is proportional to the stiffness of the agarose gel, Fcw is proportional to the composite stiffness of the cell envelope including the membranes and peptidoglycan layer, and Fturgor represents the turgor pressure across the cell membrane.
B) Maximal growth rate of an embedded E. coli culture and gel Young’s modulus (Tuson et al., 2012) as a function of gel stiffness. Solid lines and shaded areas represent the average and one standard deviation (n ≥ 3 for each concentration), respectively.
C) Schematic of the GRABS assay. A microwell is filled with 1% agarose or liquid medium, cells are pipetted into the well, and cells are mixed using a magnetic stirrer until homogenously dispersed. The stirrer is then removed and the agarose is left to solidify. In the 96-well microplate, wells in columns 1–6 are filled with 150 <image>L of 1% agarose and columns 7–12 are filled with liquid medium. Grey circles in columns 1–6 represent magnetic stirrers.
D) Example growth curves for wild-type E. coli BW25113 (black) and mutants with GRABS score = 0 (wild-type-like, green), > 0 (increased stiffness, red), and < 0 (decreased stiffness, blue). Wild-type growth curves in liquid are represented by dotted lines, and growth curves of cells in agarose are represented by dashed lines. Many mutants with a GRABS score < 0 have the same growth rate as wild-type cells in liquid and a reduced growth rate in 1% agarose. A GRABS score > 0 typically occurs when a mutant has a decreased growth rate in liquid and a growth rate similar to that of wild-type cells in agarose.
We applied GRABS (Figure 1C, D) to identify modulators of cell stiffness across the E. coli genome by screening 3,844 deletion mutants of non-essential genes from the Keio collection (Baba et al., 2006). To quantify changes in cell stiffness, we simultaneously measured the growth of 48 strains in a 1% agarose gel and liquid LB on each plate to control for variability in growth across 96-well plates (Figure 1C). While factors in addition to changes in cell stiffness could potentially affect the growth of embedded cells, we previously demonstrated that differences in the diffusion of nutrients, small molecules, or ions in agarose compared to liquid, changes in cell turgor due to osmolarity, and the secretion of enzymes that could alter the structure of the gel did not influence our microscopy-based measurements of E. coli’s Young’s modulus using agarose-embedded cells (Tuson et al., 2012). Additionally, agarose-embedded growth did not induce the SOS stress response (Figure S1B–I) or cause noticeable alterations in cell shape (Figure S2A), also suggesting that information from GRABS assays pertained to changes in cell stiffness per se.
To simplify growth curves to a scalar representation of the effects of agarose embedding on growing cells, we calculated the percent change in OD of each mutant (in agarose relative to liquid) compared to wild-type after 8 h (Figure S3A–C). We refer to this metric as the GRABS score (Extended Experimental Procedures), where a negative score indicates that the mutant exhibits less growth in agarose than expected based on its growth in liquid. By calculating GRABS scores across the Keio collection, we identified dozens of mutants that had a large decrease in growth rate in agarose and no significant change in growth rate in liquid relative to wild-type (Figure 1D, 2A), indicating a specific effect of the agarose on growth rate that is consistent with a reduction in cell stiffness. Interestingly, we also identified mutants that grew slowly in liquid, yet displayed growth rates in agarose that were comparable to the rate of wild-type cells. These mutants thus had a positive GRABS score (Figure 1D, 2A), although the large changes in liquid growth rate complicate the interpretation of these mutations as affecting cell stiffness alone.
Figure 2. Cell-stiffness modulators encompass a large range of cellular functions.
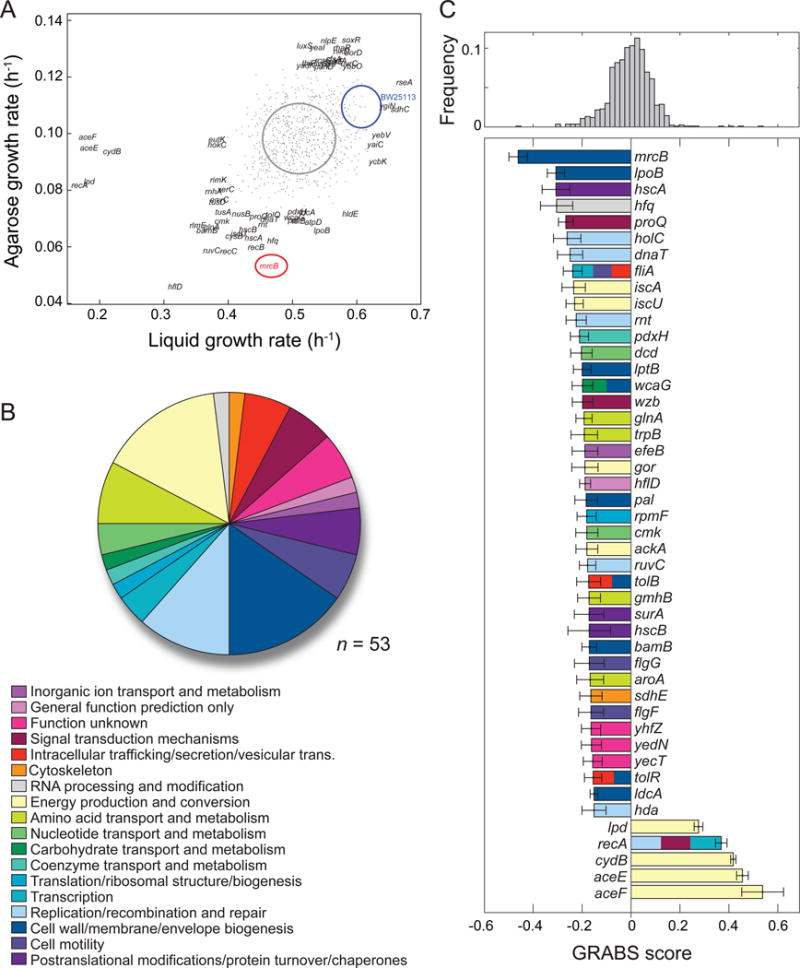
A) Maximal growth rates in liquid and agarose. Blue and red ovals represent the mean growth rates ± one standard deviation for BW25113 and ΔmrcB, respectively (n ≥ 5 independent experiments); the gray oval represents the distribution of values for the Keio collection.
B) The 46 genes with GRABS score < −0.15 (n = 41) or > 0.3 (n = 5) come from 18 Clusters of Orthologous Groups (COGs). Several genes (ΔrecA, ΔtolR, ΔtolB, ΔwcaG, ΔfliA) are assigned to two or more orthologous groups, which gives rise to n = 53 total assignments.
C) Top: distribution of GRABS scores. Bottom: GRABS hits in (B). The average GRABS score and standard deviation across ≥ 6 replicates from multiple independent experiments are shown. Genes are colored according to their COG assignment. Genes with ≥ 2 COG assignments are colored accordingly. E. coli BW25113 has a GRABS score of 0 by definition.
Cell-stiffness modulators span a wide range of functions
To exclude the possibility that kanamycin (the selective marker of the Keio collection) was directly responsible for changes in embedded growth, we rescreened ~25% of the Keio collection in the absence of kanamycin, focusing on mutants with the largest GRABS scores. Kanamycin had little effect on growth rates in liquid, but growth rates in 1% agarose were slightly higher in the absence of kanamycin. From this second screen, we identified 41 mutants (~1% of the genome) with a GRABS score < −0.15 and 5 mutants with a score > 0.3 (Figure 2C). We classified these hits using the Clusters of Orthologous Groups (COG) nomenclature (Galperin et al., 2015) and found that they represent a diverse set of functions (Figure 2B, 2C; Tables S1, S2). As anticipated, cell-wall and membrane biogenesis genes (nine mutants) constituted the largest subset of hits; these mutations likely affect cell stiffness by creating defects in the cell envelope through changes in peptidoglycan architecture or membrane structure. A mutant lacking mrcB, the gene encoding the bifunctional protein PBP1b, was the mutant with the largest negative GRABS score (Figure 2C). However, not all cell envelope-related deletions exhibited phenotypes, indicating that cell-wall synthesis activities can differentially impact mechanically confined growth. Moreover, 40/46 hits were from other functional categories (Table S1, S2; in addition to the 37 mutants not related to cell-envelope assembly, 3 of 9 cell-wall and membrane biogenesis genes had multiple assigned COG classifications), with the next-largest groups represented by energy production and conversion (eight mutants), replication/recombination and repair (six mutants), and amino-acid transport and metabolism (four mutants; Figure 2B, 2C). Cell-wall perturbations often result in changes in cell shape, which can affect the spatial distribution of stresses across the cell envelope (stresses are proportional to the radius in a idealized cylindrical thin shell under constant turgor pressure); however, we determined that all Keio mutants except ΔrodZ are rod-shaped (data not shown) and that GRABS scores did not correlate with average cell width or length (Figure S3D,E), indicating that mechanical changes identified in our screen could not be due to changes in cell geometry alone.
We also identified as modulators of cell stiffness a specific subset of genes known to have pleotropic effects. Cell stiffness decreased in strains harboring deletions of genes involved in homologous recombination (Figure 2B, 2C; Tables S1, S2). This is consistent with a recent report that mutations in these genes produce a phenotype in which cells exhibit higher rates of cell lysis and decreased cell envelope integrity (Paradis-Bleau et al., 2014). Both phenotypes could arise from defects in cell stiffness. Deletion of hfq (Figure 2C), which encodes the abundant small-RNA regulator Hfq (De Lay et al., 2013), also produces defects in cell stiffness. Hfq regulates the functions of 30 small RNAs in E. coli, and in turn these small RNAs have hundreds of putative direct or indirect target genes (Modi et al., 2011). These targets are associated with several of the functions we identified in our screen, including nucleotide transport and metabolism, cell motility, replication, and recombination and repair (Figure 2B, 2C; Tables S1, S2). Several mutants with decreased cell stiffness contained deletions of genes of unknown function, including yecT, yedN, and yhfZ (Figure 2C). yecT was previously found to display increased sensitivity to aztreonam, the antibiotic inhibitor of the division-specific peptidoglycan transpeptidase PBP3, as well as the outer membrane disruptor polymyxin B (Nichols et al., 2011). yedN was not included in a previous chemical genomics screen of the Keio collection (Nichols et al., 2011), so the reduced GRABS score defined here is the first known phenotype for this gene.
To confirm that our GRABS measurements were due to a specific gene deletion and not to polar effects on downstream genes or gene amplification during strain construction (Baba et al., 2006), we carried out complementation studies with four genes that yielded large decreases in GRABS score (mrcB, lpoB, hfq, and hscA) and two genes that produced essentially no stiffness change (mrcA and lpoA). We used a low-copy plasmid to express the gene of interest in trans, and observed 80–100% recovery of cell growth in agarose at 8 h for mrcB, lpoB, hfq, and hscA and no changes in the wild-type-like growth of mrcA and lpoA mutants (Figure S4A). Difficulties in fully complementing the in vivo function of Hfq using recombinant plasmids have been previously noted, as overexpression is toxic (Tsui et al., 1994). Our data, along with the lack of mechanical phenotypes in deletions of genes downstream of hfq in the operon containing hfq (Figure S4B), indicate that the stiffness phenotype we observe is unlikely to be due to polar effects. To further confirm the GRABS effects of these deletions, we constructed clean deletions in the well-characterized E. coli strain MG1655 (Extended Experimental Procedures). The phenotypes for E. coli MG1655 cells grown in agarose quantitatively matched the phenotypes of corresponding BW25113 mutants (Figure S4C), indicating that the role of these proteins in maintaining cellular mechanical properties is conserved in these E. coli strains.
GRABS scores are highly correlated with measurements of Young’s modulus
To confirm that our GRABS measurements reflect changes in cell stiffness, we directly measured Young’s modulus using two complementary microscopy-based methodologies: 1) a microfluidic-based bending assay (Amir et al., 2014), and 2) our previously developed single-cell assay of embedded growth (Tuson et al., 2012). In the bending assay, a shear fluid force is applied to filamentous cells (Figure 3A) and flexural (bending) rigidity is estimated by fitting the deflection (Figure S5A) of the cell body to a mechanical model (Extended Experimental Procedures); elastic-shell theory predicts that the magnitude of deflection of the cell tip under flow is proportional to l4, where l is the cell length exposed to flow (Amir et al., 2014).
Figure 3. Microfluidic bending assay and elongation rate measurements of embedded cells yield relative stiffness measurements consistent with GRABS scores.
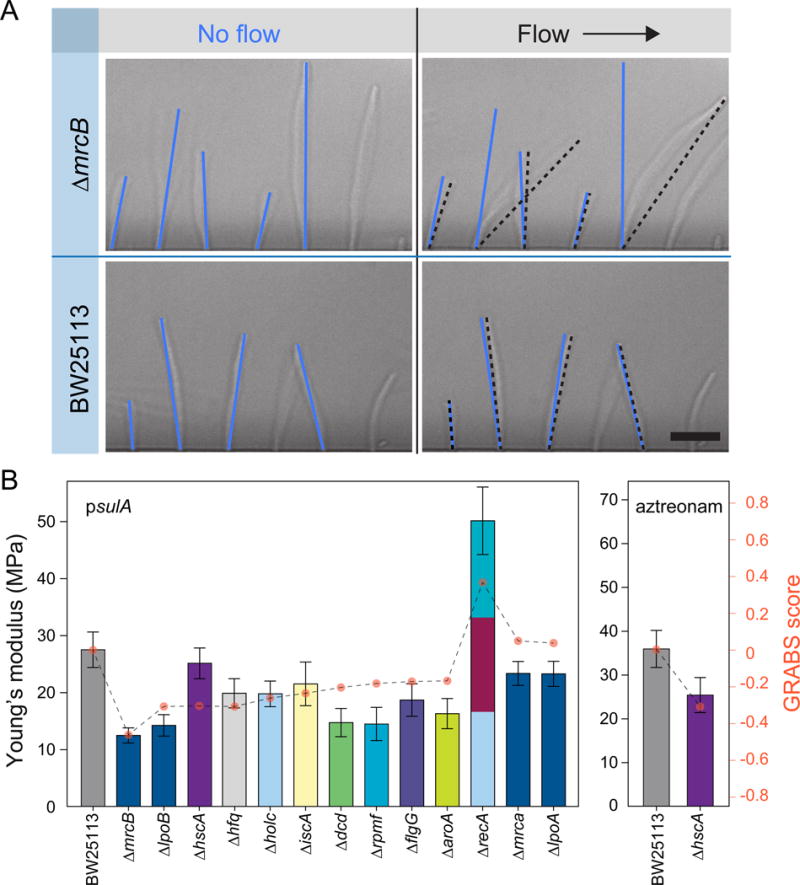
A) Representative images of filamentous wild-type cells and ΔmrcB cells in a microfluidic cell-bending device. Blue and black lines indicate the configuration of cells in the absence and presence of fluid flow, respectively. Deflection was measured as the difference between the position of the cell tip in the flow direction in the absence and presence of fluid flow. Scale bar: 10 μm.
B) Young’s moduli for sulA-induced (left) and aztreonam-treated (right) cells were determined by fitting deflection data (Extended Experimental Procedures) from a microfluidic-based bending assay. Error bars indicate 95% confidence interval to the fit (Extended Experimental Procedures). Data are colored according to each gene’s assignment to Clusters of Orthologous Groups, as in Figure 2B (n ≥ 130 cells, two independent experiments).
For a set of 13 mutants reflecting a broad range of cellular processes and spanning the complete range of GRABS scores, we induced filamentation through the production of SulA (Extended Experimental Procedures), which inhibits polymerization of the key division protein FtsZ (Mukherjee et al., 1998) and abolishes the recruitment of the division-specific cell-wall machinery to the division site (Chung et al., 2009). We obtained flexural rigidities of 8.9 × 10−20 N m2 for wild-type cells and 3.5 × 10−20 N m2 for ΔmrcB cells, the mutant with the largest magnitude GRABS score. Using an estimate of 4 nm for the thickness of the E. coli cell wall (Gan et al., 2008), we estimated the Young’s modulus of wild-type cells to be 28 MPa (Figure 3B, left; Extended Experimental Procedures), similar to previous studies (Tuson et al., 2012; Yao et al., 1999). By contrast, the Young’s modulus of ΔmrcB cells was 12 MPa (Figure 3B); the ~2-fold decrease relative to wild-type cells is in good agreement with our GRABS scores (Figure 2C), validating the importance of PBP1b for cell-stiffness determination. The additional 11 mutants had both positive and negative GRABS scores and displayed Young’s moduli in agreement with the GRABS scores (Figure 2C; Pearson correlation coefficient R = 0.83, Student’s t-test: p < 0.00025). We even observed a higher Young’s modulus in ΔrecA cells (Figure 3B, left), which had a positive GRABS score (Figure 2C) by virtue of the only slight defect in agarose-embedded growth relative to the substantial decrease in liquid growth (Figure S5B,C). Interestingly, the ΔhscA mutant, which had a large, negative GRABS score (Figure 2C), had a Young’s modulus close to that of wild-type cells (Figure 3B, left). Since HscA plays a role in cell division by affecting FtsZ localization at the division plane (Uehara et al., 2001), we hypothesized that the discrepancy between the Young’s modulus and the GRABS score for this mutant was due to inducing filamentation via SulA. To test this idea, we filamented ΔhscA cells using aztreonam, an inhibitor of the division-specific transpeptidase PBP3 (Arends and Weiss, 2004). Under aztreonam treatment, ΔhscA cells had a lower Young’s modulus than wild-type cells (Figure 3B, right), in agreement with our GRABS data (Figure 2C), indicating that the proper localization of FtsZ or division-specific cell-wall machinery is important for the cell stiffness decrease in ΔhscA cells.
To complement our microfluidics-based measure of radial cell stiffness, we also compared GRABS to our previous microscopy-based approach (Tuson et al., 2012), which measures longitudinal cell stiffness. We measured the elongation of wild-type (MG1655), ΔmrcB, ΔlpoB, Δhfq, ΔhscA, ΔmrcA, and ΔlpoA cells embedded in agarose gels of variable stiffness (Figure S5D–G). Our single-cell data was consistent with our GRABS scores, with wild-type, ΔmrcA, and ΔlpoA cells showing similar growth in gels from 1–4% agarose and ΔmrcB, ΔlpoB, Δhfq, and ΔhscA cells showing relatively less growth (Figure S5D–G). Using ultra performance liquid chromatography (Extended Experimental Procedures), we ascertained that the peptidoglycan composition in each of these mutants was similar to that in wild-type cells (Figure S6). We also measured cell viability within the gel using fluorescence microscopy (Extended Experimental Procedures) of wild-type cells and nine mutants in liquid and embedded in 1% agarose. There was no obvious difference in cell viability for wild-type cells or most of the mutants embedded in agarose. A few ΔmrcB and ΔlpoB cells were dead after 4 h, which is approximately the period of most rapid growth in 1% agarose; this level of cell death is insufficient to account for the decrease in embedded growth. Taken together, our results confirm that decreased cell stiffness is a major cause of the increased growth inhibition of mutant cells embedded in agarose, and validates GRABS as a methodology for identifying changes in Young’s modulus in bacterial cells.
Differential embedded growth among cell-wall synthesis mutants indicates a potential mechanism for reduced cell stiffness
Given that cell-wall assembly is of paramount importance for the mechanical integrity of bacterial cells and yet deletion of key enzymes had variable mechanical impacts in our experiments, as a case study, we interrogated how genetic perturbations to the biochemical activities of two of these enzymes affected cell stiffness. A major fraction of cell-wall synthesis in E. coli is carried out by the high molecular weight PBPs 1a and 1b (Scheffers and Pinho, 2005), which are encoded by the genes mrcA and mrcB, respectively. PBP1a and PBP1b are bifunctional enzymes that contain both glycosyltransferase and transpeptidase activities for glycan-strand polymerization and peptide crosslinking, respectively. The prevailing hypothesis for the functions of these proteins based on their spatial localization is that PBP1a is primarily responsible for peptidoglycan construction along the elongating peripheral cell wall and PBP1b controls peptidoglycan assembly at the division site (Typas et al., 2012). ΔmrcA ΔmrcB double mutants are synthetically lethal (Kato et al., 1985; Yousif et al., 1985), but deletion of mrcA or mrcB alone does not substantially affect growth (Figure S2B) or cell shape (Figure S2C, S2D), indicating partial compensation for each other’s in vivo activities (Paradis-Bleau et al., 2010). However, the two enzymes are not biochemically identical (Bertsche et al., 2005; Born et al., 2006), and thus it remains unclear to what extent their functions are redundant in vivo. For example, in contrast to ΔmrcA cells, ΔmrcB cells are outcompeted by wild-type cells in stationary phase (Pepper et al., 2006), have increased sensitivity to β-lactam antibiotics (Garcia del Portillo and de Pedro, 1990; Schmidt et al., 1981; Yousif et al., 1985), and cannot revert into walled rods from spheroplasts due to their inability to divide (Ranjit and Young, 2013). In our GRABS screen (Figure 2C) and bending measurements (Figure 3B), ΔmrcB was the mutant with the strongest phenotype, while ΔmrcA was phenotypically wild-type (Figure S2E), demonstrating that PBP1a and PBP1b make dramatically different contributions to cell stiffness.
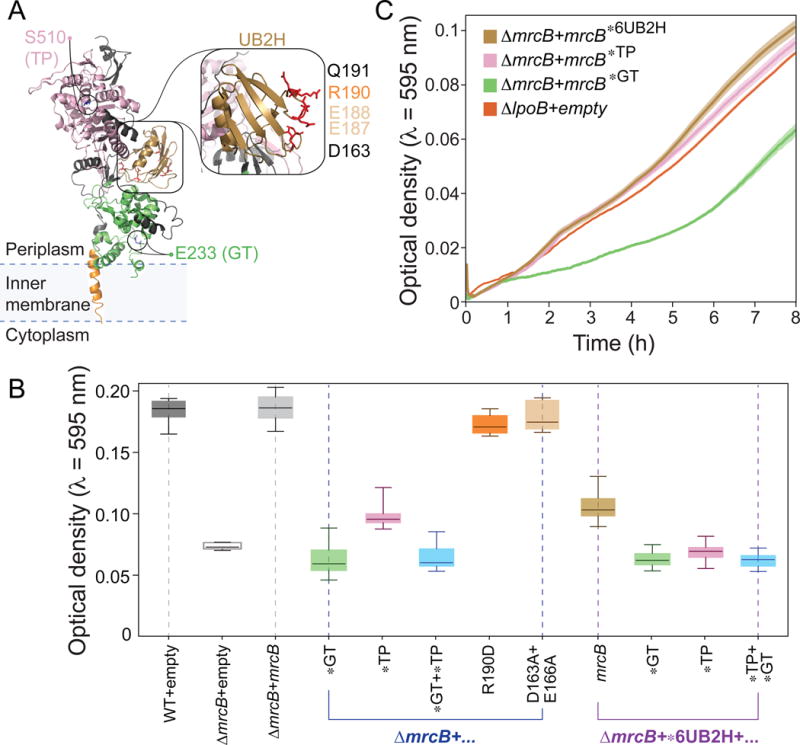
PBP1b contains a glycosyltransferase domain, a transpeptidase domain, and a UB2H domain (Figure 4A). To determine which domains and biochemical function(s) of PBP1b were responsible for the decrease in GRABS score (Figure 2C) and bending stiffness (Figure 3B), we complemented ΔmrcB cells with mutant forms of mrcB expressed in trans from a plasmid and measured cell growth in liquid and embedded in 1% agarose. Inactivation of the glycosyltransferase domain via the mutation E233Q (Meisel et al., 2003; Terrak et al., 1999) resulted in a decrease in the OD of agarose-embedded cells that was similar to that of ΔmrcB (Figure 4B), indicating that glycosyltransferase activity is necessary for the modulation of cell stiffness by PBP1b. By contrast, a mutant with inactivated transpeptidase activity (S510A) (Meisel et al., 2003; Terrak et al., 1999) (Figure 4A) partially restored the decrease in OD of embedded ΔmrcB cells (Figure 4B).
Figure 4. GRABS analysis of cell-wall synthesis mutants relates biochemical activities in vitro to changes in embedded growth in vivo.
A) Crystal structure of PBP1b (PDB ID: 3VMA) highlighting residues used for mutational analysis via GRABS. N-terminal trans-membrane α-helix, residues 64–87 (orange); UvrB Domain 2 Homolog (UB2H) domain, residues 109–200 (gold); glycosyltransferase (GT) domain, residues 203–367 (green); transpeptidase (TP) domain, residues 444–736 (pink). Other residues are colored in black. Inset: mutated residues in the UB2H domain.
B) OD at t = 8 h of agarose-embedded E. coli BW25113 cells and ΔmrcB cells expressing various mrcB alleles in trans. WT, wild-type. Error bars represent one standard deviation above and below the mean. * indicates a point mutation in the corresponding domain of PBP1b (UB2H, glycosyltransferase (GT), and transpeptidase (TP)). n ≥ 5 independent experiments.
C) Growth curves of agarose-embedded ΔlpoB cells and cells harboring mutations in the 6UB2H domain, E233Q (glycosyltransferase), and S510A (transpeptidase) of mrcB. Shaded regions represent one standard deviation above and below mean growth curves. * indicates a point mutation in the corresponding domain of mrcB. n ≥ 5 independent experiments.
The lipoprotein cofactor LpoB is important for the activation and enhancement of the transpeptidase and glycosyltransferase activities of PBP1b through the UB2H domain over an extended interface (Egan et al., 2014; Lupoli et al., 2014). It was previously shown that the D163A/E166A double mutation (Figure 4A) resulted in two-fold lower binding affinity of the PBP1b UB2H domain to LpoB in vitro (Egan et al., 2014); however, this mutation did not noticeably affect embedded OD relative to wild-type cells (Figure 4B). Although the R190D mutation decreased activation by LpoB ~16-fold (Egan et al., 2014), we observed only a slight decrease (~6%) in the ability of PBP1bR190D to fully complement embedded cell growth (Figure 4B). A PBP1b mutant with six amino-acid substitutions in the UB2H domain (6UB2H) displayed a 50-fold decrease in binding to LpoB (Egan et al., 2014); in our GRABS assay, this mutant exhibited a substantial decrease in OD when grown in agarose that was intermediate between the decreases in OD for BW25113 and ΔmrcB (Figure 4B). To confirm whether the 6UB2H mutant lost all interaction with LpoB, we compared its growth curve for embedded growth to that of the ΔlpoB mutant. The two curves did not qualitatively match (Figure 4C), indicating that there may be a minor interaction between LpoB and the UB2H domain that persists in the 6UB2H mutant.
The quantitative difference between the ΔmrcB and ΔlpoB phenotypes suggests a basal level of PBP1b functionality in the absence of LpoB. Interestingly, the growth curve of the embedded ΔlpoB mutant was qualitatively similar to that of an S510A transpeptidase mutant (Figure 4C), illustrating that in vivo activation of PBP1b’s glycosyltransferase domain may be independent of its interaction with LpoB. It is possible that these mutations may alter the stability or expression of PBP1b. Regardless, using the GRABS assay, we were able to disentangle nuances of the biochemical activities of PBP1b and its activator LpoB and link transglycosylase activity to changes in E. coli cell stiffness, thereby translating in vitro measurements to in vivo phenotypes; this strategy could be used to dissect the biochemical mechanism by which any protein modulates cell stiffness.
Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) increases cell stiffness
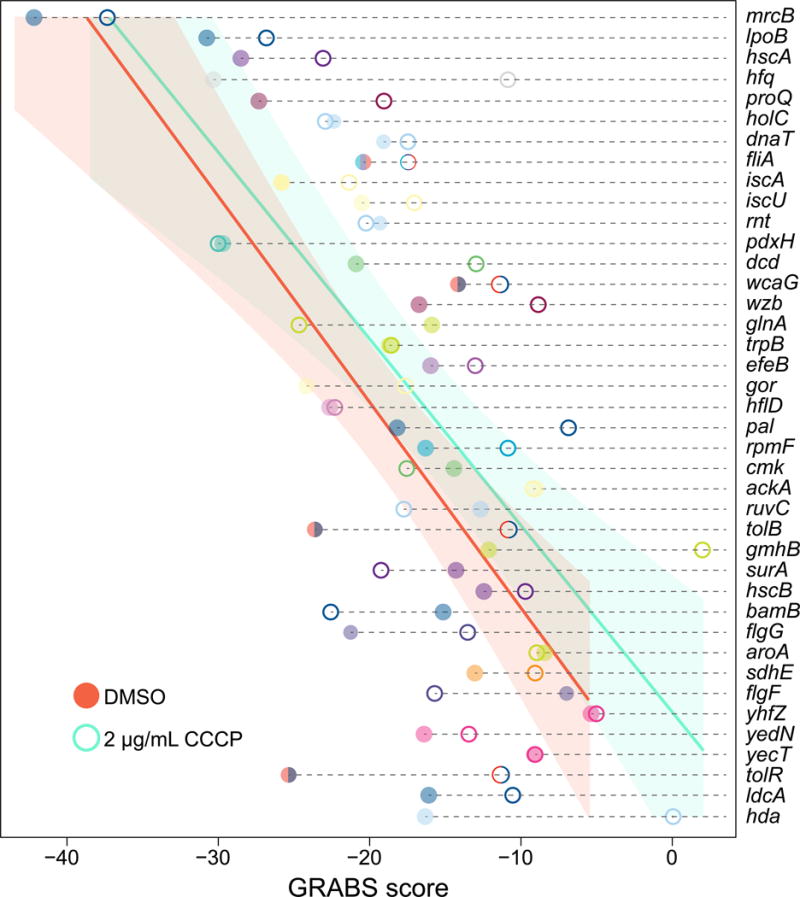
The genomic scale of our stiffness measurements empowered the use of statistical correlations to identify chemicals that generally affect cell stiffness across the strains in the Keio collection. We correlated our GRABS scores in the presence of kanamycin with a previously published chemical-genomics dataset of colony growth under hundreds of conditions (Nichols et al., 2011). The 20 conditions with the largest positive Pearson correlation coefficient, indicating that reductions in GRABS score coincide with negative S-score (reduced growth), included known envelope perturbations that are expected to disrupt cell mechanics (Table S3), such as the detergents sodium dodecyl sulfate and Triton X-100, the small molecule A22 (which inhibits the MreB cytoskeleton) (Gitai et al., 2005), and the antibiotic cefsulodin (which targets PBP1a and PBP1b) (Curtis et al., 1979). Intriguingly, there were also conditions associated with a negative correlation of a similar magnitude to the highest positive correlations in Table S3, indicating conditions in which GRABS score generally increased in strains with negative S-scores (Table S4). Thus, we hypothesized that some of these conditions may result in a general increase in cell stiffness.
We focused on the proton ionophore CCCP, which rapidly eliminates membrane potential by dissipating the proton motive force (Strahl et al., 2014); CCCP treatment at multiple concentrations had a significant negative correlation with GRABS scores (Table S4). We performed GRABS on wild-type cells and on 40 strains with the largest reduction in GRABS scores in the presence of 2 μg/mL CCCP. As expected, CCCP reduced growth rates in liquid (Figure S7A). However, embedded growth rates were reduced to a lesser extent (Figure S7B), and GRABS scores generally increased as predicted (Figure 5). Thus, our assay can be used to reveal conditions that modulate cell stiffness in both directions and to predict chemicals that will display synergy or antagonism with genetic perturbations that affect cell stiffness.
Figure 5. GRABS score generally increases under CCCP treatment.
GRABS score with and without CCCP, a proton ionophore that reduces membrane potential. Solid circles, representing GRABS score for the dimethyl sulfoxide (DMSO) control, are generally at lower values than the open circles, which represent GRABS score in the presence of 2 μg/mL CCCP. Genes are colored according to their assignment to Clusters of Orthologous Groups. Solid lines and shaded areas represent the average and 95% confidence interval to a linear fit, respectively, of n ≥ 4 independent experiments.
DISCUSSION
Here, we report a straightforward, high-throughput technology for rapid characterization of bacterial stiffness that should be widely applicable to the discovery of the mechanical effects of genetic or chemical perturbations to cellular functions. GRABS requires only standard lab equipment. Since absorbance-based optical measurements are sufficient for identifying genes connected to cell stiffness, as demonstrated here, GRABS is the first mechanical measurement technology that can be applied to genome-scale libraries. Because GRABS is based on a direct readout of proliferation, it can be easily applied to bacteria that display a variety of morphological phenotypes, such as filamentation, rounding, or branching. Embedding cells in an agarose hydrogel confines them and introduces a defined and tunable force that opposes growth while ensuring a constant supply of nutrients by diffusion. We previously demonstrated that agarose does not significantly alter the diffusion of growth factors, nor does it trigger contact-dependent phenotypes (Tuson et al., 2012). To normalize measurements of growth on agarose to growth in liquid, we also generated growth curves for Keio mutants in liquid LB (Table S5), a data-mining resource for the microbiology community. The GRABS score is correlated with the maximal growth rate of agarose growth curves (Figure S7C), potentially further simplifying future screens of cell mechanics. The GRABS score used here does not necessarily directly translate to quantitation of Young’s modulus, a standard measure of cellular stiffness. Nonetheless, GRABS scores were generally consistent with Young’s moduli determined from microfluidic-based bending assays (Figure 3) and single-cell embedded growth measurements (Figure S5D–G), suggesting that GRABS enables rapid screening of genomic-scale mutant collections for changes in cell stiffness (Figure 2). When computing Young’s modulus, we assumed a common value for wall thickness in all mutants; changes to this parameter would affect our modulus estimates. Cases such as ΔhscA, in which GRABS and other mechanical measurements appear to make conflicting estimates of cell stiffness (Figure 3B), can in fact reveal subtle feedback between biochemical functions of the affecting protein and cellular stiffness. Regardless, our identification of stiffness modulators related to numerous functions in E. coli (Figure 2B, 2C) indicates that cell mechanics is a global property connected to many intracellular pathways.
Future agarose growth-based studies of cells and mutants with morphological perturbations will be empowered by modeling the effects of mechanical perturbations on cellular dimensions and viability to disentangle changes in shape from changes in intrinsic mechanical properties. Changes in the anisotropy in the wall (Tropini et al., 2014) or the cell’s propensity for lysis can also alter cell mechanics. GRABS also enables secondary screens for genetic and chemical discovery, for example the identification of small molecules that affect cell stiffness individually or in combination with nonessential genes. Additionally, a recent study demonstrated that many E. coli genes increase the permeability of the cell envelope in responses to changes in temperature and salt concentration (among other conditions), which was sometimes accompanied by morphological changes (Paradis-Bleau et al., 2014). Investigating the effects of these environmental variables through growth in agarose may reveal genes that act synergistically or antagonistically with changes in the cell envelope.
The natural environments of bacterial cells often intrinsically involve mechanical stimuli or perturbations. Rapid transitions between a host and fresh water for pathogens such as Vibrio cholerae result in osmotic shocks that mechanically challenge the integrity of the cell envelope (Rowe et al., 2015). The intestine contains mucus layers that can embed nearby bacteria (Earle et al., 2015) and thereby potentially inhibit growth. The many avenues for genetic modulation of cell stiffness that we have revealed may provide the flexibility to adapt to new environments by balancing physical expansion with mechanical integrity, on both short and evolutionary time scales. Complementary to how cells respond to their environment, embedded growth and genetic perturbations that affect cell stiffness may also reorganize the interior of the cell and the membrane; any differences relative to non-embedded, wild-type cells would point to a potential role for cell mechanics in the underlying mechanisms of localization. Future applications of our technique to explore the growing number of genome-wide gene deletion libraries (Noble et al., 2010; Porwollik et al., 2014) should reveal whether mechanisms of mechanical modulation are conserved across species or selected by specific environmental contexts. The expansion of our knowledge base regarding cell stiffness determination should generate new opportunities for rewiring cellular mechanical properties and novel mechanisms for antibiotic chemotherapeutics.
EXPERIMENTAL PROCEDURES
Full experimental procedures can be found in the Extended Experimental Procedures. Briefly, bacterial cells were cultured before being transferred to liquid medium or embedded in 1% agarose in 96-well GRABS plates. Growth data were collected over 16 h for Keio mutants in liquid medium and agarose and GRABS scores were calculated from these data. Details about strain/plasmid construction, bacterial growth, the GRABS assay, data analysis, correlation with chemical-genomics data, complementation and deletion assays, assays of growth in the presence of chemicals, microscopy-based stiffness measurements, and microfluidics-based stiffness measurements appear in the Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
The authors thank the Huang and Weibel labs for useful discussions, Andrew Gray, Carol Gross, and Waldemar Vollmer for helpful feedback, Suresh Kannan and Kevin Young for bacterial strains, and the National BioResource Project for the Keio Collection. This work was supported by NIH Director’s New Innovator Awards DP2OD006466 (to K.C.H.) and DP2OD008735 (to D.W.), NSF CAREER Award MCB-1149328 (to K.C.H.), NSF Award DMR-1121288 (to U. Wisconsin at Madison), the Stanford Systems Biology Center funded by NIH grant P50 GM107615 (to K.C.H.), and a Wisconsin Alumni Research Foundation Distinguished Graduate Fellowship (to M.R.). This work was also supported in part by the National Science Foundation under Grant PHYS-1066293 and the hospitality of the Aspen Center for Physics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
G.K.A., K.C.H., and D.B.W. conceived of the methodology. G.K.A, M.R., K.C.H., and D.B.W. designed experiments. M.R. constructed mutants and plasmids associated with this study. T.K.L. analyzed data, performed analysis of Keio mutant morphologies, and built chemical genomics correlations. S.C. and A.M. performed and analyzed UPLC data. G.K.A., M.R., T.K.L., S.C., A.M., K.C.H. and D.B.W. interpreted data. G.K.A, K.C.H, D.B.W., M.R., T.K.L., S.C., and A.M. wrote the manuscript.
References
- Amir A, Babaeipour F, McIntosh DB, Nelson DR, Jun S. Bending forces plastically deform growing bacterial cell walls. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:5778–5783. doi: 10.1073/pnas.1317497111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends SJ, Weiss DS. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. Journal of bacteriology. 2004;186:880–884. doi: 10.1128/JB.186.3.880-884.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Breukink E, Kast T, Vollmer W. In vitro murein peptidoglycan synthesis by dimers of the bifunctional transglycosylase-transpeptidase PBP1B from Escherichia coli. The Journal of biological chemistry. 2005;280:38096–38101. doi: 10.1074/jbc.M508646200. [DOI] [PubMed] [Google Scholar]
- Birdsell DC, Cota-Robles EH. Production and ultrastructure of lysozyme and ethylenediaminetetraacetate-lysozyme spheroplasts of Escherichia coli. Journal of bacteriology. 1967;93:427–437. doi: 10.1128/jb.93.1.427-437.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born P, Breukink E, Vollmer W. In vitro synthesis of cross-linked murein and its attachment to sacculi by PBP1A from Escherichia coli. The Journal of biological chemistry. 2006;281:26985–26993. doi: 10.1074/jbc.M604083200. [DOI] [PubMed] [Google Scholar]
- Braybrook SA, Jonsson H. Shifting foundations: the mechanical cell wall and development. Current opinion in plant biology. 2016;29:115–120. doi: 10.1016/j.pbi.2015.12.009. [DOI] [PubMed] [Google Scholar]
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis NA, Orr D, Ross GW, Boulton MG. Affinities of penicillins and cephalosporins for the penicillin-binding proteins of Escherichia coli K-12 and their antibacterial activity. Antimicrobial agents and chemotherapy. 1979;16:533–539. doi: 10.1128/aac.16.5.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. The Journal of biological chemistry. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufrene YF. Atomic force microscopy in microbiology: new structural and functional insights into the microbial cell surface. MBio. 2014;5:e01363–01314. doi: 10.1128/mBio.01363-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE, Amieva MR, Huang KC, Sonnenburg JL. Quantitative Imaging of Gut Microbiota Spatial Organization. Cell host & microbe. 2015;18:478–488. doi: 10.1016/j.chom.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJ, Jean NL, Koumoutsi A, Bougault CM, Biboy J, Sassine J, Solovyova AS, Breukink E, Typas A, Vollmer W, et al. Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:8197–8202. doi: 10.1073/pnas.1400376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Francius G, Domenech O, Mingeot-Leclercq MP, Dufrene YF. Direct observation of Staphylococcus aureus cell wall digestion by lysostaphin. Journal of bacteriology. 2008;190:7904–7909. doi: 10.1128/JB.01116-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic acids research. 2015;43:D261–269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia del Portillo F, de Pedro MA. Differential effect of mutational impairment of penicillin-binding proteins 1A and 1B on Escherichia coli strains harboring thermosensitive mutations in the cell division genes ftsA, ftsQ, ftsZ, and pbpB. Journal of bacteriology. 1990;172:5863–5870. doi: 10.1128/jb.172.10.5863-5870.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gitai Z, Dye NA, Reisenauer A, Wachi M, Shapiro L. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell. 2005;120:329–341. doi: 10.1016/j.cell.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Ingber DE. Cell tension, matrix mechanics, and cancer development. Cancer cell. 2005;8:175–176. doi: 10.1016/j.ccr.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Janmey PA, McCulloch CA. Cell mechanics: integrating cell responses to mechanical stimuli. Annual review of biomedical engineering. 2007;9:1–34. doi: 10.1146/annurev.bioeng.9.060906.151927. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Sjovall H, Hansson GC. The gastrointestinal mucus system in health and disease. Nature reviews Gastroenterology & hepatology. 2013;10:352–361. doi: 10.1038/nrgastro.2013.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J, Suzuki H, Hirota Y. Dispensability of either penicillin-binding protein-1a or -1b involved in the essential process for cell elongation in Escherichia coli. Mol Gen Genet. 1985;200:272–277. doi: 10.1007/BF00425435. [DOI] [PubMed] [Google Scholar]
- Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–837. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- Lee TK, Tropini C, Hsin J, Desmarais SM, Ursell TS, Gong E, Gitai Z, Monds RD, Huang KC. A dynamically assembled cell wall synthesis machinery buffers cell growth. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:4554–4559. doi: 10.1073/pnas.1313826111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupoli TJ, Lebar MD, Markovski M, Bernhardt T, Kahne D, Walker S. Lipoprotein activators stimulate Escherichia coli penicillin-binding proteins by different mechanisms. J Am Chem Soc. 2014;136:52–55. doi: 10.1021/ja410813j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nature reviews Microbiology. 2011;9:265–278. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- Meisel U, Holtje JV, Vollmer W. Overproduction of inactive variants of the murein synthase PBP1B causes lysis in Escherichia coli. Journal of bacteriology. 2003;185:5342–5348. doi: 10.1128/JB.185.18.5342-5348.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mille Y, Beney L, Gervais P. Viability of Escherichia coli after combined osmotic and thermal treatment: a plasma membrane implication. Biochim Biophys Acta. 2002;1567:41–48. doi: 10.1016/s0005-2736(02)00565-5. [DOI] [PubMed] [Google Scholar]
- Modi SR, Camacho DM, Kohanski MA, Walker GC, Collins JJ. Functional characterization of bacterial sRNAs using a network biology approach. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15522–15527. doi: 10.1073/pnas.1104318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A, Cao C, Lutkenhaus J. Inhibition of FtsZ polymerization by SulA, an inhibitor of septation in Escherichia coli. Proceedings of the National Academy of Sciences. 1998;95:2885–2890. doi: 10.1073/pnas.95.6.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan J, Xiong JY, Liu XY. Determination of agarose gel pore size: Absorbance measurements vis a vis other techniques. J Phys Conf Ser. 2006;28:83–86. [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. Phenotypic landscape of a bacterial cell. Cell. 2011;144:143–156. doi: 10.1016/j.cell.2010.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet. 2010;42:590–598. doi: 10.1038/ng.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Kritikos G, Orlova K, Typas A, Bernhardt TG. A genome-wide screen for bacterial envelope biogenesis mutants identifies a novel factor involved in cell wall precursor metabolism. PLoS Genet. 2014;10:e1004056. doi: 10.1371/journal.pgen.1004056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE, Bernhardt TG. Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell. 2010;143:1110–1120. doi: 10.1016/j.cell.2010.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper ED, Farrell MJ, Finkel SE. Role of penicillin-binding protein 1b in competitive stationary-phase survival of Escherichia coli. FEMS microbiology letters. 2006;263:61–67. doi: 10.1111/j.1574-6968.2006.00418.x. [DOI] [PubMed] [Google Scholar]
- Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, et al. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One. 2014;9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats R, de Pedro MA. Normal growth and division of Escherichia coli with a reduced amount of murein. Journal of bacteriology. 1989;171:3740–3745. doi: 10.1128/jb.171.7.3740-3745.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qimron U, Marintcheva B, Tabor S, Richardson CC. Genomewide screens for Escherichia coli genes affecting growth of T7 bacteriophage. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19039–19044. doi: 10.1073/pnas.0609428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjit DK, Young KD. The Rcs stress response and accessory envelope proteins are required for de novo generation of cell shape in Escherichia coli. Journal of bacteriology. 2013;195:2452–2462. doi: 10.1128/JB.00160-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley M, Abe T, Arnaud MB, Berlyn MK, Blattner FR, Chaudhuri RR, Glasner JD, Horiuchi T, Keseler IM, Kosuge T, et al. Escherichia coli K-12: a cooperatively developed annotation snapshot–2005. Nucleic acids research. 2006;34:1–9. doi: 10.1093/nar/gkj405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe I, Patange S, Belyy V, Yasmann A, Sukharev S. The Tension-Activated Channels in the Cytoplasmic Membrane of Vibrio Cholerae. Biophysical Journal. 2015;108:564a. [Google Scholar]
- Scheffers DJ, Pinho MG. Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt LS, Botta G, Park JT. Effects of furazlocillin, a beta-lactam antibiotic which binds selectively to penicillin-binding protein 3, on Escherichia coli mutants deficient in other penicillin-binding proteins. Journal of bacteriology. 1981;145:632–637. doi: 10.1128/jb.145.1.632-637.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl H, Burmann F, Hamoen LW. The actin homologue MreB organizes the bacterial cell membrane. Nat Commun. 2014;5:3442. doi: 10.1038/ncomms4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrak M, Ghosh TK, van Heijenoort J, Van Beeumen J, Lampilas M, Aszodi J, Ayala JA, Ghuysen JM, Nguyen-Disteche M. The catalytic, glycosyl transferase and acyl transferase modules of the cell wall peptidoglycan-polymerizing penicillin-binding protein 1b of Escherichia coli. Mol Microbiol. 1999;34:350–364. doi: 10.1046/j.1365-2958.1999.01612.x. [DOI] [PubMed] [Google Scholar]
- Thwaites JJ, Mendelson NH. Mechanical properties of peptidoglycan as determined from bacterial thread. Int J Biol Macromol. 1989;11:201–206. doi: 10.1016/0141-8130(89)90069-x. [DOI] [PubMed] [Google Scholar]
- Tropini C, Lee TK, Hsin J, Desmarais SM, Ursell T, Monds RD, Huang KC. Principles of bacterial cell-size determination revealed by cell-wall synthesis perturbations. Cell Rep. 2014;9:1520–1527. doi: 10.1016/j.celrep.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Leung HC, Winkler ME. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Tuson HH, Auer GK, Renner LD, Hasebe M, Tropini C, Salick M, Crone WC, Gopinathan A, Huang KC, Weibel DB. Measuring the stiffness of bacterial cells from growth rates in hydrogels of tunable elasticity. Mol Microbiol. 2012;84:874–891. doi: 10.1111/j.1365-2958.2012.08063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A, Banzhaf M, Gross CA, Vollmer W. From the regulation of peptidoglycan synthesis to bacterial growth and morphology. Nat Rev Microbiol. 2012;10:123–136. doi: 10.1038/nrmicro2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Matsuzawa H, Nishimura A. HscA is involved in the dynamics of FtsZ-ring formation in Escherichia coli K12. Genes to cells : devoted to molecular & cellular mechanisms. 2001;6:803–814. doi: 10.1046/j.1365-2443.2001.00463.x. [DOI] [PubMed] [Google Scholar]
- Vogel V, Sheetz M. Local force and geometry sensing regulate cell functions. Nature reviews Molecular cell biology. 2006;7:265–275. doi: 10.1038/nrm1890. [DOI] [PubMed] [Google Scholar]
- Wang S, Arellano-Santoyo H, Combs PA, Shaevitz JW. Actin-like cytoskeleton filaments contribute to cell mechanics in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:9182–9185. doi: 10.1073/pnas.0911517107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Jericho M, Pink D, Beveridge T. Thickness and elasticity of gram-negative murein sacculi measured by atomic force microscopy. Journal of bacteriology. 1999;181:6865–6875. doi: 10.1128/jb.181.22.6865-6875.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousif SY, Broome-Smith JK, Spratt BG. Lysis of Escherichia coli by beta-lactam antibiotics: deletion analysis of the role of penicillin-binding proteins 1A and 1B. J Gen Microbiol. 1985;131:2839–2845. doi: 10.1099/00221287-131-10-2839. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


