Abstract
Unbiased estimation of causal parameters from marginal structural models (MSMs) requires a fundamental assumption of no unmeasured confounding. Unfortunately, the time-varying covariates used to obtain inverse probability weights are often error-prone. Although substantial measurement error in important confounders is known to undermine control of confounders in conventional unweighted regression models, this issue has received comparatively limited attention in the MSM literature. Here we propose a novel application of the simulation-extrapolation (SIMEX) procedure to address measurement error in time-varying covariates, and we compare 2 approaches. The direct approach to SIMEX-based correction targets outcome model parameters, while the indirect approach corrects the weights estimated using the exposure model. We assess the performance of the proposed methods in simulations under different clinically plausible assumptions. The simulations demonstrate that measurement errors in time-dependent covariates may induce substantial bias in MSM estimators of causal effects of time-varying exposures, and that both proposed SIMEX approaches yield practically unbiased estimates in scenarios featuring low-to-moderate degrees of error. We illustrate the proposed approach in a simple analysis of the relationship between sustained virological response and liver fibrosis progression among persons infected with hepatitis C virus, while accounting for measurement error in γ-glutamyltransferase, using data collected in the Canadian Co-infection Cohort Study from 2003 to 2014.
Keywords: causal inference, marginal structural models, measurement error, SIMEX, simulations, time-varying covariates
In longitudinal analyses of observational data, control of confounding is complicated whenever time-varying confounders are also intermediates on the causal pathway between the exposure and outcome. Adjustment for such covariates in standard regression models leads to biased estimators of exposure effects (1–3).
Marginal structural models (MSMs) represent an important improvement over conventional regression-based adjustment for time-varying covariates. The most common form of estimation for these models involves weighting by the inverse probability of exposure, or inverse probability weighting (IPW) (4). A fundamental assumption of MSMs is that there is no unmeasured confounding (1). In conventional, unweighted regression models, mismeasurement of confounders and mismodeling of functional forms in covariate-exposure relationships may undermine confounding adjustment (5–7). To date, the effect of measurement errors in confounders on inverse probability-weighted estimators has not been extensively studied (8, 9), even though MSMs are increasingly used in settings where measurement error is likely. For example, in analyses of human immunodeficiency virus (HIV) data, time-varying CD4+ cell counts are often used in IPW estimation (10). Like most laboratory measurements, CD4+ cell count is subject to high variability that is well-characterized following decades of use (11–13).
The behavior of exposure effect estimators in the context of misclassified or mismeasured confounders has received considerable attention in the literature. Greenland (14, 15) illustrated that adjustment for misclassified covariates impairs confounding control, and that nondifferential misclassification biases estimates in the direction of the crude exposure-outcome association. Ogburn and VanderWeele (16) demonstrated that adjusting for nondifferentially mismeasured ordinal covariates may increase bias—for example, if their effects differ qualitatively across exposure strata. In the context of IPW, covariate mismeasurement may result in over- or underestimation of the true parameters, and occasionally changes in the sign of their estimates (8). Overall, prior research suggests that the consequences of covariate mismeasurement may be unpredictable.
While most published IPW analyses do not implement modeling strategies to address measurement error, we note 2 important exceptions. To estimate the effect of HIV treatment on incident acquired immunodeficiency syndrome or death, Cole et al. (17) applied regression calibration to address exposure measurement error in a Cox MSM. McCaffrey et al. (9) suggested a propensity score-based approach to handle errors in covariates, relying partially on simulations, using the observed data to generate a weighting function. Their method performs well, but its implementation may be challenging for epidemiologists.
Here we consider a novel application of the simulation-extrapolation (SIMEX) procedure, originally proposed by Cook and Stefanski (18), to address measurement error in continuous time-varying covariates used for IPW. Our proposed adaptation of the SIMEX procedure to MSMs is straightforward and may be implemented with several statistical software packages. The method is both intuitive and easily explained. We validate the proposed approaches via simulation and conduct a simple analysis to illustrate a practical application.
SIMEX CORRECTION OF MEASUREMENT ERROR IN CONFOUNDING VARIABLES
Overview of the SIMEX procedure
Consider the classical measurement error model where is the observed jth measure for individual i of covariate Li, Lij is the corresponding true value, and ϵij represents a normally distributed mean-zero error term (19). Note that SIMEX procedures for Berkson error are also available (20, 21). For simplicity, we begin by describing the effect of a dichotomous exposure X in a single time interval:
| (1) |
Denote the target parameter by θ, where θ could be an average exposure effect or a vector of regression coefficients (β0, β1).
Given a regression model, the SIMEX procedure approximates the functional relationship between model parameters and the variance of measurement error using a 2-step simulation-based approach, assuming that the variance is known or accurately estimated (14). First, additional (“artificial”) random error is added to the original values of the error-prone covariate. The degree of artificial error is incremented over a set of user-defined multipliers λ. The number of λ values chosen need not be large, though Monte Carlo error is reduced as their quantity increases (18). For each value of λ, B data sets are simulated. The original regression model is refitted to each of B data sets to obtain Finally, the mean of the resulting parameter estimates is calculated: In the second step, is regressed on λ, often assuming a quadratic relationship. Extrapolating the function back to the hypothetical case with no measurement error, which corresponds to λ = −1, yields the SIMEX estimate of the parameter of interest (19).
Proposed adaptation of SIMEX to MSMs
In equation 1, the exposure parameter is β1, and the dependent variable Y represents the average difference between the counterfactuals YX=1 and YX=0, assuming all individuals had been exposed or not exposed, respectively. To obtain unbiased causal estimates of the effect of X while addressing confounding by L in equation 1, we may fit a weighted outcome model by inverting probabilities of exposure estimated from the following exposure model:
| (2) |
We consider 2 alternative applications of the SIMEX procedure to address errors in L. Both approaches employ the same error variance information, but the approaches differ in the choice of which mean estimates are regressed on λ (see Web Table 1, available at http://aje.oxfordjournals.org/). The direct correction approach targets the estimated parameter β1 in equation 1 while using naive weights obtained from the original, uncorrected exposure model. In denoting the SIMEX estimate for the bth (b = 1, …, B) data set as we obtain the direct SIMEX estimator by extrapolating the function to λ = −1.
In the second approach, termed indirect correction, SIMEX is applied to the exposure model, and the IPW weights are corrected. Here, the correction is targeted at the estimated parameters α0 and α1 in equation 2; extrapolating to λ = −1 yields the SIMEX-corrected estimators for exposure model coefficients (Web Appendix 1). The outcome model is fitted using the SIMEX-corrected IPW weights. The indirect SIMEX procedure is easily implemented in most statistical environments. In contrast, no software packages support direct correction, as existing implementations were not designed for 2-stage estimation procedures.
SIMULATION STUDIES
In simulations, we assessed bias in naive MSMs, which do not correct for measurement error in a time-varying covariate, and the potential of the 2 SIMEX approaches to reduce the bias. We considered different combinations of relevant parameter values, and we simulated 300 data sets for each combination. We performed analyses in R, version 3.1.1 (R Development Core Team, Vienna, Austria), using our own SIMEX implementation (Web Appendix 2) and the R sprint package to efficiently apply the nonparametric bootstrap (22, 23). In all analyses, we specified 200 SIMEX replicates and used a quadratic function to perform the extrapolation. To account for clustering and the 2-stage estimation procedure, we obtained 95% confidence intervals for the exposure effect by means of the nonparametric bootstrap percentiles approach, using 1,000 bootstrap replicates for each parameter combination (24).
Methods
Single-time-interval simulation study: data generation
We first considered a single time interval setting to examine SIMEX performance in a simplified context. True (i.e., error-free) values of a continuous confounder L were generated from a uniform [−4, 4] distribution. A dichotomous exposure X, conditional on L, was then generated, assuming
We fixed α0 at 0.25. To minimize the risk of positivity violations, we simulated exposure probabilities bounded between 0.1 and 0.9 by setting α1 at 0.25, 0.50, or 0.75. The error-prone covariate L* was generated by adding random error drawn from an N(0, σδ) distribution, with standard deviations σδ of 0.25, 0.75, and 1.25, representing scenarios with increasing measurement error.
The outcome, Y, was normally distributed with the expected value dependent on the exposure X and the “true” value of confounder L:
where ϵ ∼ N(0, 1). Figure 1A illustrates the data generation scheme using a directed acyclic graph. In all, we examined 27 simulated scenarios corresponding to various combinations of sample size (n = 100, n = 500, n = 1,000), measurement error magnitude (controlled by σδ), and strength of confounding due to L (controlled by β1).
Figure 1.
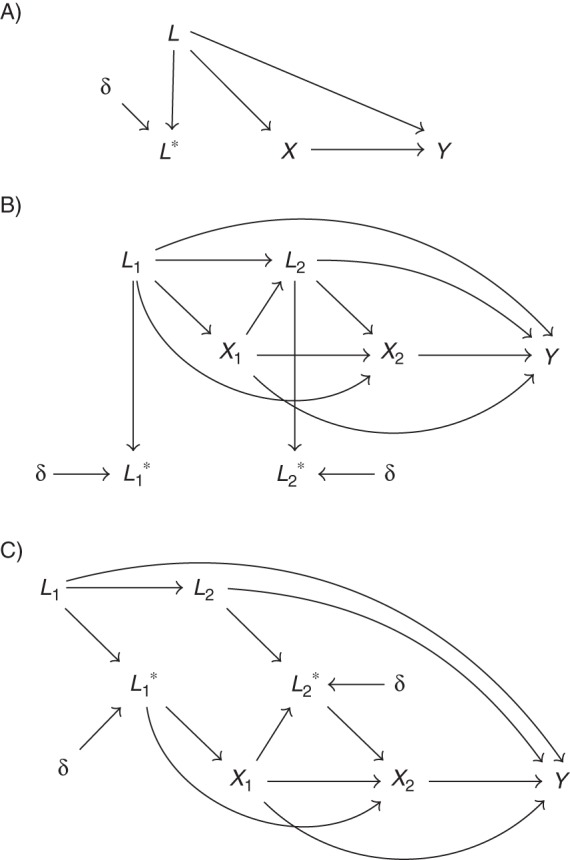
A) Data generation scenario for single-time-interval initial simulation studies, in which exposure is conditional on error-free covariate L. Measurement error is represented by δ. B) Data generation scenario for 2-interval simulation studies, in which exposure depends on error-free covariate Lj. Measurement error is represented by δ. C) Data generation scenario for secondary 2-interval simulation studies, in which exposure depends upon mismeasured covariate . Measurement error is represented by δ.
Two-time-interval simulation study: data generation
Initial error-free values of a continuous covariate L1 were drawn from a Uniform distribution with range −2 to 3. We generated first-interval exposure status X1 from a Bernoulli distribution, conditional on L1 with probability p = logit−1(0.25 + α1L1), with α1 equal to 0.50 or 0.75. The second-interval value of L was then generated from L2 = 0.3L1 + 1.25X1 + ϵ, where ϵ ∼ N(0, 1). We generated second-interval exposure status X2 given a Bernoulli distribution with probability p = logit−1(0.25 + α1L2 – 1.25X1). The outcome Y, at the second interval, was generated as
where η ∼ N(0, 1). Figure 1B depicts the 2-interval data generation scenario. Error-prone values of Lj (j = 1, 2) were obtained by adding random error, drawn from N(0, σδ), to the respective error-free values. We varied the strength of measurement error by specifying σδ to be 0.25 or 1.25.
Single-time-interval study: analysis
In each simulated data set, we fitted 2 alternative logistic models to predict exposure probabilities conditional on the error-free L or the mismeasured L*. Inverting the predicted probabilities from each of these models generated weights, which were used to fit the 2 corresponding MSMs for a linear regression of Y on X. We compared the resulting regression coefficients to assess the influence of measurement error.
Secondary single-time-interval study: application of SIMEX-corrected MSMs to actual clinical data contaminated with additional error
Web Appendix 3 provides results for secondary simulations designed to more closely mimic real data and the example considered in the case study, in which we systematically increase error in a time-varying covariate using a clinical data set (Web Figure 1).
Two-time-interval simulation study: analysis
The 2-time-interval simulations were analyzed similarly to the single-time-interval study, using separate logistic models to respectively regress X1 on L1 and regress X2 on X1 and L1. For the 2-time-interval MSMs, we stabilized the weights to reduce variance (1). Further details are available in Web Table 1.
Secondary 2-interval study: exposure status conditional on the mismeasured covariate
In our previous simulations, only the error-prone covariate was available to the analyst, while exposure was allocated using the true, error-free covariate. However, decisions to initiate, modify, or discontinue treatment may depend on the error-prone covariate, if no other relevant information is available to clinicians. In a sensitivity analysis, we assumed that exposure was conditional on the mismeasured covariate values of and as illustrated in Figure 1C. The simulated data were analyzed using the same methods as in the primary 2-interval study.
Sensitivity analysis: misspecification of measurement error variance
In our analyses of simulated data using the SIMEX procedure, we implicitly assumed that the measurement error variance is known precisely. In practice, this assumption may be violated to some degree. Consequently, we reanalyzed data in the 2-interval setting (Figure 1B), with α1 = 0.50, to assess the influence of measurement error variance misspecification on SIMEX performance. We varied the ratio of specified measurement error variance to the true variance (used to generate L*) across the range [0.25, 0.50, 0.75, 1.00, 1.25, 1.50, 1.75], with α1 = 0.50 and all other parameters identical to those from the primary 2-interval simulations.
Comparative assessment of SIMEX performance
In Web Appendix 3, we present pairwise correlations for both SIMEX approaches and the naive MSM across all single-time-interval simulations (Web Figure 2) and consider the accuracy of the quadratic extrapolation in further simulations (Web Figure 3).
Simulation results
Single-time-interval study
Web Table 2 presents estimates from the naive MSM, which does not correct for measurement error, while Table 1 shows estimates from the 2 error-corrected SIMEX models.
Table 1.
Results From a Single-Time-Interval Simulation Study With 27 Scenarios (Rows) in Which Sample Size (N), the Magnitude of the Confounder L (α1), and the Standard Deviation of Measurement Error σδ are Varied in Marginal Structural Models Including an Error-Free L and an Uncorrected Error-Prone L*
| α1, N, and Scenario | Error Deviation (σδ) | Model Using SIMEX-Corrected L* |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Direct Correction |
Indirect Correction |
||||||||
| Bias (β1) | Relative MSEa | MCSE | 95% Bootstrap Coverageb | Bias (β1) | Relative MSEa | MCSE | 95% Bootstrap Coverageb | ||
| 0.25 | |||||||||
| 100 | |||||||||
| A | 0.25 | −0.006 | 1.000 | 0.219 | 0.950 | −0.007 | 1.000 | 0.220 | 0.953 |
| B | 0.75 | 0.008 | 0.933 | 0.288 | 0.963 | 0.004 | 0.944 | 0.290 | 0.960 |
| C | 1.25 | 0.099 | 0.780 | 0.355 | 0.950 | 0.089 | 0.792 | 0.360 | 0.957 |
| 500 | |||||||||
| D | 0.25 | 0.008 | 1.000 | 0.099 | 0.943 | 0.008 | 1.000 | 0.099 | 0.943 |
| E | 0.75 | 0.030 | 0.472 | 0.127 | 0.943 | 0.026 | 0.472 | 0.128 | 0.943 |
| F | 1.25 | 0.127 | 0.385 | 0.156 | 0.890 | 0.120 | 0.375 | 0.158 | 0.900 |
| 1,000 | |||||||||
| G | 0.25 | 0.002 | 1.000 | 0.076 | 0.943 | 0.002 | 1.000 | 0.076 | 0.943 |
| H | 0.75 | 0.022 | 0.370 | 0.099 | 0.920 | 0.018 | 0.370 | 0.100 | 0.920 |
| I | 1.25 | 0.115 | 0.329 | 0.120 | 0.787 | 0.110 | 0.318 | 0.122 | 0.817 |
| 0.50 | |||||||||
| 100 | |||||||||
| J | 0.25 | −0.005 | 1.016 | 0.251 | 0.963 | −0.008 | 1.016 | 0.251 | 0.967 |
| K | 0.75 | 0.028 | 0.723 | 0.326 | 0.963 | 0.002 | 0.764 | 0.337 | 0.960 |
| L | 1.25 | 0.214 | 0.506 | 0.388 | 0.937 | 0.170 | 0.525 | 0.417 | 0.960 |
| 500 | |||||||||
| M | 0.25 | 0.005 | 0.933 | 0.117 | 0.937 | 0.001 | 0.933 | 0.118 | 0.937 |
| N | 0.75 | 0.035 | 0.271 | 0.146 | 0.930 | 0.008 | 0.271 | 0.152 | 0.950 |
| O | 1.25 | 0.222 | 0.256 | 0.173 | 0.730 | 0.181 | 0.217 | 0.186 | 0.830 |
| 1,000 | |||||||||
| P | 0.25 | −0.003 | 0.875 | 0.085 | 0.917 | −0.007 | 0.875 | 0.085 | 0.920 |
| Q | 0.75 | 0.025 | 0.178 | 0.111 | 0.907 | −0.002 | 0.178 | 0.115 | 0.927 |
| R | 1.25 | 0.211 | 0.216 | 0.132 | 0.603 | 0.170 | 0.171 | 0.141 | 0.750 |
| 0.75 | |||||||||
| 100 | |||||||||
| S | 0.25 | 0.025 | 1.009 | 0.335 | 0.943 | 0.015 | 1.009 | 0.335 | 0.943 |
| T | 0.75 | 0.055 | 0.755 | 0.452 | 0.947 | −0.013 | 0.792 | 0.467 | 0.967 |
| U | 1.25 | 0.327 | 0.479 | 0.499 | 0.890 | 0.220 | 0.486 | 0.559 | 0.930 |
| 500 | |||||||||
| V | 0.25 | −0.002 | 0.955 | 0.147 | 0.937 | −0.015 | 1.000 | 0.148 | 0.943 |
| W | 0.75 | 0.025 | 0.245 | 0.190 | 0.920 | −0.056 | 0.298 | 0.205 | 0.947 |
| X | 1.25 | 0.304 | 0.231 | 0.215 | 0.647 | 0.186 | 0.168 | 0.256 | 0.860 |
| 1,000 | |||||||||
| Y | 0.25 | 0.000 | 0.833 | 0.102 | 0.937 | −0.014 | 0.917 | 0.103 | 0.923 |
| Z | 0.75 | 0.026 | 0.139 | 0.135 | 0.920 | −0.056 | 0.182 | 0.147 | 0.907 |
| AA | 1.25 | 0.302 | 0.199 | 0.155 | 0.443 | 0.184 | 0.116 | 0.183 | 0.787 |
Abbreviations: MCSE, Monte Carlo standard error; MSE, mean squared error; SIMEX, simulation-extrapolation.
a MSEs are relative to those computed using a naive model including an error-prone L* with an identical parameter set.
b Bootstrap coverage was computed using the nonparametric bootstrap percentiles approach.
Bias in the naive estimators worsened systematically as measurement error increased (Web Table 2). For example, for a sample size of n = 1,000, with α1 = 0.25, a 3-fold increase in σδ (from 0.25 to 0.75) resulted in a 7-fold increase in bias. The bias in the naive model grew as the strength of the association between the error-prone covariate L* and exposure X increased. For n = 1,000 and σδ = 0.25, a 3-fold increase in α1 (from 0.25 to 0.75) resulted in a 2-fold increase in bias. Due to bias, coverage was systematically low; the lowest coverage corresponded to settings with the highest σδ and stronger L-X associations.
Table 1 compares both SIMEX-based approaches. When measurement error was lowest (σδ = 0.25), SIMEX-corrected models yielded nearly unbiased estimates comparable to those from the error-free model (Web Table 2). For σδ = 0.75, median bias in β1 was 99% and 88% lower for the indirect and direct SIMEX models, respectively, relative to the naive model. Due to a small degree of remaining bias, coverage rates were over 90% for both direct and indirect approaches, in contrast to poor coverage for the naive model (Table 1). Given high measurement error (σδ = 0.75), the indirect and direct SIMEX-corrected estimates were slightly biased, but they still reduced bias by 68% and 59% relative to the naive model estimates.
As anticipated, standard deviations of exposure effect estimates were slightly higher for both SIMEX-based models relative to naive estimates. The degree of variance inflation was minimal when the specified association between L and X was weakest (α1 = 0.25). For α1 = 0.50 and n = 1,000, average standard deviations were 23% greater for the indirect approach and 14% greater for the direct approach, in comparison with the naive model (data not shown). Given α1 = 0.75, σδ = 1.25, and n = 1,000, mean standard deviations for the direct and indirect approaches were 28% and 50% greater than those for the naive model, respectively. However, mean squared errors (MSEs) for the SIMEX-corrected models were systematically lower than those for the naive model, reflecting a good trade-off in variance for bias, particularly when α1 and σδ were largest (Table 1).
Given α1 = 0.25, both SIMEX approaches produced highly comparable estimates across varying sample sizes and measurement error magnitudes. When the influence of L on X was stronger (α1 = 0.50 or 0.75) and σδ = 1.25, the indirect SIMEX model (Table 1, right side) exhibited lower bias and MSE compared with the direct SIMEX model (Table 1, left side).
Both approaches yielded similar coverage for low-to-moderate confounding and smaller σδ. With increased N, coverage tended to decrease, as narrow intervals imply that even a relatively small bias results in suboptimal coverage. Coverage for the SIMEX-corrected models was noticeably poorer for higher levels of measurement error. Inadequate fit of the quadratic extrapolant is one possible explanation (25, 26). Kukush et al. (21) suggest that at larger error magnitudes, the naive ordinary likelihood estimator no longer conforms sufficiently to a quadratic relationship, and they propose alternative estimators to address this issue. An alternate hypothesis is that severe mismeasurement of L limits the amount of useful information available for adjustment using L*. Overall, estimates from both SIMEX approaches remained practically unbiased and very similar for low-to-moderate degrees of measurement error. When measurement error was more extreme, the indirect SIMEX approach yielded better coverage, lower bias, and lower MSE.
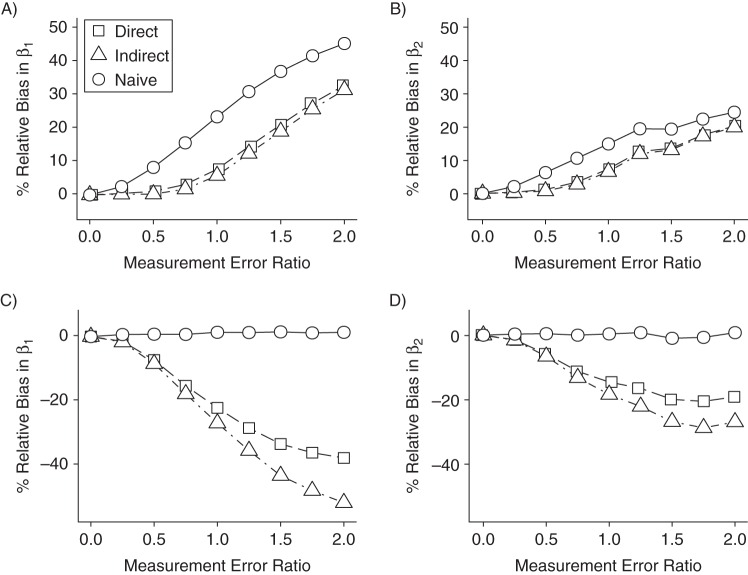
The 2-time-interval study
As in the 1-interval setting, SIMEX-corrected estimates were uniformly less biased and featured lower MSEs than the naive model (Table 2, panels A and B of Figure 2, and Web Table 3). Given higher measurement error and stronger confounding (scenario 4), the biases of direct and indirect SIMEX exposure effect estimates for the first interval were 56% and 71% smaller, respectively, than the bias from the naive model. In the second interval, biases in the direct and indirect estimates were 33% and 41% smaller, respectively.
Table 2.
Resultsa From a 2-Time-Interval Simulation Study With 4 Scenarios (Rows) in Which the Magnitude of the Confounder L (α1) and the Standard Deviation of Measurement Error σδ are Varied in Marginal Structural Models Including an Error-Free L and an Uncorrected Error-Prone L*
| α1 and Scenario | Error Deviation (σδ) | Model Using SIMEX-Corrected , |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Direct Correction |
Indirect Correction |
||||||||||||||
| Bias |
Relative MSEb |
MCSE |
95% Bootstrap Coveragec | Bias |
Relative MSEb |
MCSE |
95% Bootstrap Coveragec | ||||||||
| β1 | β2 | β1 | β2 | β1 | β2 | (β1, β2) | β1 | β2 | β1 | β2 | β1 | β2 | (β1, β2) | ||
| 0.50 | |||||||||||||||
| 1 | 0.25 | −0.003 | 0.009 | 1.000 | 1.000 | 0.097 | 0.107 | 0.923, 0.970 | −0.006 | 0.007 | 1.000 | 1.000 | 0.097 | 0.107 | 0.927, 0.970 |
| 2 | 1.25 | 0.187 | 0.154 | 0.263 | 0.552 | 0.112 | 0.114 | 0.600, 0.760 | 0.158 | 0.145 | 0.218 | 0.507 | 0.118 | 0.117 | 0.717, 0.793 |
| 0.75 | |||||||||||||||
| 3 | 0.25 | 0.008 | 0.013 | 1.000 | 0.950 | 0.126 | 0.137 | 0.907, 0.933 | −0.001 | 0.008 | 1.000 | 0.950 | 0.127 | 0.138 | 0.913, 0.933 |
| 4 | 1.25 | 0.249 | 0.213 | 0.240 | 0.565 | 0.127 | 0.141 | 0.527, 0.650 | 0.162 | 0.189 | 0.151 | 0.522 | 0.150 | 0.158 | 0.783, 0.720 |
Abbreviations: MCSE, Monte Carlo standard error; MSE, mean squared error; SIMEX, simulation-extrapolation.
a A sample size of n = 1,000 was used for all simulations.
b MSEs are relative to those computed using a naive model including an error-prone L* with an identical parameter set.
c Bootstrap coverage was computed using the nonparametric bootstrap percentiles approach.
Figure 2.
Relative bias in β1 estimates from naive and simulation-extrapolation (SIMEX)-corrected marginal structural models in 2-interval simulation studies, given varying ratios of covariate measurement error to residual error in Y. Parts A and B correspond to data generated according to Figure 1B, and parts C and D correspond to data generated according to Figure 1C. The left- and right-hand plots (parts A and C and parts B and D, respectively) represent first- and second-interval exposure effect estimates, respectively.
Both SIMEX models exhibited nearly identical coverage for α1 = [0.50 or 0.75] and σδ = 0.25 and improved coverage versus the naive model given σδ = 1.25. While coverage of the direct and indirect SIMEX-corrected intervals was as low as 53% and 78%, it was 0% for the naive model. In scenarios 1–3 from the primary study, where exposure depended on the error-free covariate, the two approaches produced nearly identical estimates. In scenario 4, given high measurement error and a strong L-X relationship, the indirect approach appeared more accurate (Table 2).
In contrast, in our secondary simulations, where exposure depended on error-prone covariates , both SIMEX approaches introduced bias, which grew as measurement error increased (Figure 2, parts C and D), while the naive MSM yielded unbiased estimates with the lowest MSE. This finding was not surprising; Figure 1C illustrates that inclusion of in the exposure (weighting) model effectively blocks the causal path between Xj and . Using IPW, we impose conditional independence between Xj and (27, 28).
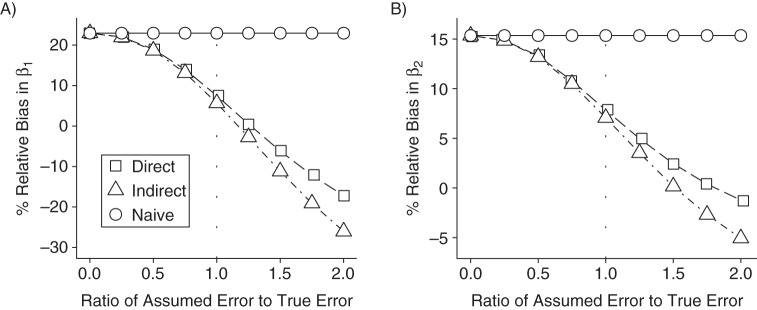
Figure 3 presents the results of a sensitivity analysis. It appears that when exposure depends on the error-free covariate, SIMEX-corrected estimates are less biased than the naive estimates, even when the variance of the error distribution is misspecified by as much as 50%.
Figure 3.
Influence of misspecification of measurement error variance on relative bias in exposure effect estimates from 2-interval simulations using naive, direct simulation-extrapolation (SIMEX)-corrected, and indirect SIMEX-corrected marginal structural models. A) first-interval estimates; B) second-interval estimates. The dotted line at X = 1.0 is used to indicate the scenario with correctly specified measurement error variance. Data were generated according to Figure 1B. Note that the axis scales differ between plots.
CASE STUDY: THE CANADIAN CO-INFECTION COHORT STUDY
To demonstrate an application of SIMEX-corrected MSMs, we analyzed data from the Canadian Co-infection Cohort Study (CCC). The CCC is an ongoing study of over 1,400 individuals coinfected with HIV and hepatitis C virus (HCV) (29). Persons aged 16 years or older may participate, given documented HIV infection (enzyme-linked immunosorbent assay confirmed by Western blot) and chronic HCV infection or laboratory evidence of HCV exposure (30). Study visits generally occur every 6 months (±1 month) and include a liver test profile. A key objective of the CCC is to explore predictors of liver disease progression among persons living with HIV/HCV. We analyzed data from a subset of 258 individuals treated for HCV, for whom at least 1 visit occurred after completing therapy (Web Figure 4, Web Table 4).
We investigated the relationship between HCV treatment and progression of liver disease. The exposure of interest was sustained virological response (SVR) to therapy, a clinically relevant measure of HCV treatment effectiveness and cure. Ghany et al. (31) define SVR as an undetectable HCV RNA result in a sensitive polymerase chain reaction assay at a specified number of weeks following discontinuation of therapy. We assessed SVR at 12 weeks posttherapy, which correlates well with SVR at 24 weeks and has recently been recommended as an endpoint for future HCV trials (32–34). The outcome variable was the aspartate aminotransferase:platelet ratio index (APRI), a continuous, noninvasive surrogate marker of liver fibrosis severity (35). APRI has been validated among persons coinfected with HIV and HCV (36, 37). We initially intended to conduct a 2-interval study but chose to consider a single time interval after identifying collinearity when modeling subsequent SVR measures.
The outcome model included the following covariates: female sex, age, duration of HCV infection, and HCV genotype. In the exposure model, we included continuous measures of current γ-glutamyltransferase (GGT) level, body mass index (weight (kg)/height (m)2), HIV viral load, and time since discontinuation of HCV therapy, as well as indicator variables for any reported alcohol consumption or injection drug use in the past 6 months and HIV therapy status. We applied both SIMEX approaches to correct measurement error in GGT. Although GGT is strongly correlated with APRI, recent research suggests that these measures reflect different disease features. Everhart and Wright (38) argue that GGT is associated with disease activity, while platelet counts (the denominator of APRI) capture disease severity; they further conclude that GGT may be more important prognostically than either aspartate aminotransferase or alanine aminotransferase for persons with advanced liver disease. In a more recent study, Mandorfer et al. (39) determined that GGT was an independent predictor of failure of HCV treatment to produce SVR. Note that the exposure variable represents a (biological) response, rather than a clinician-driven decision to modify or initiate treatment, and it is therefore affected by the error-free GGT value rather than the error-prone measurement available to the clinician. Our observation period began at the first study visit after completion of HCV therapy and concluded at the third posttherapy visit.
We use the following notation:
at first (baseline) visit after completing HCV therapy.
X1 = 1 if SVR is attained between the first and second study visits; otherwise X1 = 0.
Y = log10(APRI) at the third visit after completing HCV therapy.
Note that logarithmic transformation of the APRI score in a standard linear regression model produces estimates of the median APRI rather than the mean (40). Prior to analyses, we log-transformed values. For SIMEX, the standard deviation of the test error on was specified as 0.247, based on a recent systematic review (41). We addressed missing values in covariates using multiple imputation and the mice package in R (42). Because current HCV therapy may influence APRI measures, we censored individuals who reinitiated therapy, and we estimated inverse-probability-of-censoring weights for those who remained uncensored. We also censored persons with missing information on SVR status at the first or second study visit. To obtain estimates of the percent decrease in APRI reported below, we computed 1 − 10β. Additional details on the analysis are available in Web Tables 5 and 6.
RESULTS
Estimates from the naive model (Table 3) ignoring measurement error in GGT suggested a 45% decrease in median APRI among persons attaining SVR, relative to those not experiencing SVR (β = −0.26, 95% confidence interval (CI): −0.40, −0.11).
Table 3.
Association Between a Sustained Virological Response to Hepatitis C Virus Treatment at 12 Weeks Posttherapy and Progression of Liver Disease, as Measured by the Aspartate Aminotransferase:Platelet Ratio Index, Canadian Co-Infection Cohort Study, 2003–2014
| Variable | Model |
|||||
|---|---|---|---|---|---|---|
| Naive MSMa |
Direct SIMEX Correction |
Indirect SIMEX Correction |
||||
| 95% CIb | 95% CI | 95% CI | ||||
| SVR at third posttreatment visit | −0.26 | −0.40, −0.11 | −0.19 | −0.38, −0.01 | −0.17 | −0.36, 0.01 |
| Female sex | −0.09 | −0.25, 0.09 | −0.12 | −0.32, 0.07 | −0.08 | −0.29, 0.10 |
| Age, per 10 years | −0.07 | −0.18, 0.03 | −0.09 | −0.21, 0.03 | −0.08 | −0.21, 0.04 |
| Duration of HCV infection, per 10 years | 0.03 | −0.04, 0.09 | 0.02 | −0.07, 0.11 | 0.03 | −0.07, 0.12 |
| HCV genotype of 2/3/4 | 0.00 | −0.19, 0.19 | 0.02 | −0.24, 0.28 | −0.03 | −0.26, 0.25 |
Abbreviations: CI, confidence interval; HCV, hepatitis C virus; MSM, marginal structural model; SIMEX, simulation-extrapolation; SVR, sustained virological response.
a The naive MSM does not correct for measurement error in log10(γ-glutamyltransferase).
b All 95% CIs were computed by means of the nonparametric bootstrap percentiles approach.
The error-corrected models suggested a more modest relationship between SVR and APRI. The direct SIMEX-corrected model estimated a 35% decrease in median APRI relative to persons not experiencing SVR (β = −0.19, 95% CI: −0.38, −0.01). Similarly, the indirect SIMEX model estimated a 32% decrease in median APRI associated with SVR (β = −0.17, 95% CI: −0.36, 0.01). These results appear to support the hypothesis that successful treatment for HCV infection is associated with a meaningful improvement in liver health relative to persons who are treated but do not attain SVR.
DISCUSSION
We proposed, assessed, and validated 2 approaches to correcting measurement error in time-varying covariates in MSMs, by adapting Cook and Stefanski's SIMEX procedure (18) to this specific setting. Our simulation results suggest that both the direct and indirect SIMEX approaches largely reduce bias when exposure depends on the true value of mismeasured time-varying covariates. This finding is of practical importance, as indirect correction is supported in several statistical software packages (43, 44).
SIMEX-based estimators exhibited some bias with increasing measurement error variance and increasing strength of the covariate-outcome association; however, they provided uniformly better coverage than the naive estimators. We observed only minor differences between the 2 SIMEX approaches, which produced systematically more accurate estimators than those from the naive model. These findings and the easier implementation of the indirect approach suggest that the latter may be recommended for most applications.
Most estimators to which SIMEX has been applied do not have closed-form solutions, and therefore theoretical proofs are infeasible; this is true of our proposed SIMEX-based estimators also (45–48). Thus, we followed the previous literature by demonstrating performance by simulation, including the SIMEX-based estimators’ capacity to reduce bias under differing assumptions about the “true” data structure.
Our secondary simulations confirmed that ignoring measurement error was a reasonable choice if treatment assignment depended on the error-prone covariate; in this case, correcting for error introduced bias. However, MSMs are increasingly being used to estimate the causal effects of exposures that are neither treatments nor treatment decisions, where measurement error in time-varying covariates should be considered, and a reliable estimate of the error variance is available (49–51). The case study we have presented is one such example. Our findings support the application of measurement error models to time-varying covariates within this context.
We acknowledge several limitations of our study. MSMs are frequently applied to data collected over many points in time, including larger numbers of covariates, several of which may exhibit measurement errors. Due mostly to computational burden, the scenarios we examined were limited to a simplified 2-interval setting. However, the consistency of simulation results across a range of relevant parameters suggests that the potential advantages of SIMEX-corrected MSMs extend to multiple time points. Furthermore, practical application of the SIMEX procedure requires that the measurement error variance is known or can be estimated relatively well. We find it encouraging that sensitivity analyses demonstrated that a small degree of variance misspecification (<25%) did not markedly bias our corrected estimates, which remained less biased than the naive estimates even when we under- or overspecified the error variance by as much as 50%. This finding should be interpreted cautiously, as our investigations were limited to simple models fitted to simulated data (52).
Measurement error in time-varying covariates remains an important issue that must be carefully considered in any analysis. When the exposure of interest is based on the true (not mismeasured) covariate and the measurement error variance may be reasonably well estimated, the SIMEX procedure is a useful method with which to address error in time-varying covariates. Moreover, while several methods for measurement error correction exist, few are implemented in commonly used statistical packages; SIMEX is a notable exception. We have shown how analysts can take advantage of this approach to correct measurement error in time-varying covariates within the MSM framework.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Department of Epidemiology, Biostatistics and Occupational Health, Faculty of Medicine, McGill University, Montréal, Québec, Canada (Ryan P. Kyle, Erica E. M. Moodie, Marina B. Klein, Michał Abrahamowicz); Chronic Viral Illness Service, McGill University Health Centre, Montréal, Québec, Canada (Marina B. Klein); and Division of Clinical Epidemiology, McGill University Health Centre, Montréal, Québec, Canada (Michał Abrahamowicz).
This project was supported by Canadian Institutes of Health Research (CIHR) grants MOP-81275 and MOP-130402. E.E.M.M. was supported by CIHR grant MOP-130402 and a Chercheurs-Boursier career award from the Fonds de Recherche du Québec–Santé (FRQS) and is a William Dawson Scholar at McGill University. M.B.K. was supported by a Chercheur-National career award from the FRQS. M.A. is a James McGill Professor at McGill University. Statistical computations were performed on the Guillimin and Colosse supercomputers at McGill University and Université Laval, managed by Calcul Québec (Montréal, Québec, Canada) and Compute Canada (Toronto, Ontario, Canada). Operation of these computer clusters is funded by the Canada Foundation for Innovation, NanoQuébec (Montréal, Québec, Canada), the Reseau de Médecine Génétique Appliquée, and the Fonds de Recherche du Québec–Nature et Technologies. The Canadian Co-Infection Cohort Study (study CTN222) was funded by the Réseau SIDA/Maladies Infectieuses of the FRQS, CIHR grant MOP-79529, and the Canadian HIV Trials Network.
We thank Dr. Aman D. Verma and Jeremy Labrecque for their thoughtful suggestions regarding implementation of the simulations.
This project was conducted as part of Ryan P. Kyle's doctoral thesis research at McGill University, co-supervised by Drs. Michał Abrahamowicz and Erica E. M. Moodie.
Site investigators for the Canadian Co-infection Cohort Study: Jeff Cohen (Windsor Regional Hospital, Metropolitan Campus, Windsor, Ontario); Brian Conway (Vancouver Infectious Diseases Research and Care Centre, Vancouver, British Columbia); Curtis Cooper (The Ottawa Hospital Research Institute, Ottawa, Ontario); Pierre Côté (Clinique du Quartier Latin, Montréal, Québec); Joseph Cox (Montréal General Hospital, Montréal, Québec); John Gill (Southern Alberta HIV Clinic, Calgary, Alberta); Shariq Haider (McMaster University, Hamilton, Ontario); Marianne Harris (St. Paul's Hospital, Vancouver, British Columbia); David Haase (Capital District Health Authority, Halifax, Nova Scotia); Mark Hull (BC Centre for Excellence in HIV/AIDS, Vancouver, British Columbia); Julio Montaner (St. Paul's Hospital, Vancouver, British Columbia); Neora Pick (Oak Tree Clinic, Children's and Women's Health Centre of British Columbia, University of British Columbia, Vancouver, British Columbia); Anita Rachlis (Sunnybrook & Women's College Health Sciences Centre, Toronto, Ontario); Danielle Rouleau (Centre Hospitalier de l'Université de Montréal, Montréal, Québec); Roger Sandre (HAVEN Program, Sudbury, Ontario); Joseph Mark Tyndall (Department of Medicine, Infectious Diseases Division, University of Ottawa, Ottawa, Ontario); Marie-Louise Vachon (Centre Hospitalier Universitaire de Québec, Québec City, Québec); Sharon Walmsley (University Health Network, Toronto, Ontario); and David Wong (University Health Network, Toronto, Ontario).
Conflict of interest: none declared.
REFERENCES
- 1.Robins JM, Hernán MA, Brumback B. Marginal structural models and causal inference in epidemiology. Epidemiology. 2000;115:550–560. [DOI] [PubMed] [Google Scholar]
- 2.Cole SR, Hernán MA. Constructing inverse probability weights for marginal structural models. Am J Epidemiol. 2008;1686:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moodie EEM, Stephens DA. Using directed acyclic graphs to detect limitations of traditional regression in longitudinal studies. Int J Public Health. 2010;556:701–703. [DOI] [PubMed] [Google Scholar]
- 4.Mortimer KM, Neugebauer R, van der Laan M et al. . An application of model-fitting procedures for marginal structural models. Am J Epidemiol. 2005;1624:382–388. [DOI] [PubMed] [Google Scholar]
- 5.Brenner H, Blettner M. Controlling for continuous confounders in epidemiologic research. Epidemiology. 1997;84:429–434. [PubMed] [Google Scholar]
- 6.Royston P, Altman DG, Sauerbrei W. Dichotomizing continuous predictors in multiple regression: a bad idea. Stat Med. 2006;251:127–141. [DOI] [PubMed] [Google Scholar]
- 7.Fewell Z, Smith GD, Sterne JAC. The impact of residual and unmeasured confounding in epidemiologic studies: a simulation study. Am J Epidemiol. 2007;1666:646–655. [DOI] [PubMed] [Google Scholar]
- 8.Regier MD, Moodie EEM, Platt RW. The effect of error-in-confounders on the estimation of the causal parameter when using marginal structural models and inverse probability-of-treatment weights: a simulation study. Int J Biostat. 2014;101:1–15. [DOI] [PubMed] [Google Scholar]
- 9.McCaffrey DF, Lockwood JR, Setodji CM. Inverse probability weighting with error-prone covariates. Biometrika. 2013;1003:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernán MA, Brumback B, Robins JM. Marginal structural models to estimate the causal effect of zidovudine on the survival of HIV-positive men. Epidemiology. 2000;115:561–570. [DOI] [PubMed] [Google Scholar]
- 11.Wulfsohn MS, Tsiatis AA. A joint model for survival and longitudinal data measured with error. Biometrics. 1997;531:330–339. [PubMed] [Google Scholar]
- 12.Hoover DR, Graham NMH, Chen B et al. . Effect of CD4+ cell count measurement variability on staging HIV-1 infection. J Acquir Immune Defic Syndr. 1992;58:794–802. [PubMed] [Google Scholar]
- 13.Tsiatis AA, DeGruttola V, Wulfsohn MS. Modeling the relationship of survival to longitudinal data measured with error. Applications to survival and CD4 counts in patients with AIDS. J Am Stat Assoc. 1995;90429:27–37. [Google Scholar]
- 14.Greenland S. The effect of misclassification in the presence of covariates. Am J Epidemiol. 1980;1124:564–569. [DOI] [PubMed] [Google Scholar]
- 15.Greenland S. Intuitions, simulations, theorems: the role and limits of methodology. Epidemiology. 2012;233:440–442. [DOI] [PubMed] [Google Scholar]
- 16.Ogburn EL, VanderWeele TJ. Bias attenuation results for nondifferentially mismeasured ordinal and coarsened confounders. Biometrika. 2013;1001:241–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole SR, Jacobson LP, Tien PC et al. . Using marginal structural measurement-error models to estimate the long-term effect of antiretroviral therapy on incident AIDS or death. Am J Epidemiol. 2010;1711:113–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cook JR, Stefanski LA. Simulation-extrapolation estimation in parametric measurement error models. J Am Stat Assoc. 1994;89428:1314–1328. [Google Scholar]
- 19.Carroll RJ, Ruppert D, Stefanski LA et al. . Measurement Error in Nonlinear Models: A Modern Perspective. Boca Raton, FL: Taylor & Francis Group, LLC; 2006. [Google Scholar]
- 20.Althubaiti A, Donev A. Non-Gaussian Berkson errors in bioassay. Stat Methods Med Res. 2016;251:430–445. [DOI] [PubMed] [Google Scholar]
- 21.Kukush A, Shklyar S, Masiuk S et al. . Methods for estimation of radiation risk in epidemiological studies accounting for classical and Berkson errors in doses. Int J Biostat. 2011;71:Article 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill J, Hambley M, Forster T et al. . SPRINT: a new parallel framework for R. BMC Bioinformatics. 2008;9:558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 24.Efron B. Bootstrap methods: another look at the jackknife. Ann Stat. 1979;71:1–26. [Google Scholar]
- 25.Wang X-F, Fan Z, Wang B. Estimating smooth distribution function in the presence of heteroscedastic measurement errors. Comput Stat Data Anal. 2010;541:25–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He W, Yi GY, Xiong J. Accelerated failure time models with covariates subject to measurement error. Stat Med. 2007;2626:4817–4832. [DOI] [PubMed] [Google Scholar]
- 27.Pearl J. Causality: Models, Reasoning, and Inference. New York, NY: Cambridge University Press; 2009. [Google Scholar]
- 28.Rothman KJ, Greenland S, Lash TL. Modern Epidemiology. 3rd ed Philadelphia, PA: Lippincott Williams & Wilkins; 2012. [Google Scholar]
- 29.Cooper C, Rollet-Kurhajec KC, Young J et al. . HIV virological rebounds but not blips predict liver fibrosis progression in antiretroviral-treated HIV/hepatitis C virus-coinfected patients. HIV Med. 2015;161:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein MB, Saeed S, Yang H et al. . Cohort profile: the Canadian HIV–Hepatitis C Co-infection Cohort Study. Int J Epidemiol. 2010;395:1162–1169. [DOI] [PubMed] [Google Scholar]
- 31.Ghany MG, Strader DB, Thomas DL et al. . Diagnosis, management, and treatment of hepatitis C: an update. Hepatology. 2009;494:1335–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen J, Florian J, Carter W et al. . Earlier sustained virologic response end points for regulatory approval and dose selection of hepatitis C therapies. Gastroenterology. 2013;1447:1450–1455.e2. [DOI] [PubMed] [Google Scholar]
- 33.Martinot-Peignoux M, Stern C, Maylin S et al. . Twelve weeks posttreatment follow-up is as relevant as 24 weeks to determine the sustained virologic response in patients with hepatitis C virus receiving pegylated interferon and ribavirin. Hepatology. 2010;514:1122–1126. [DOI] [PubMed] [Google Scholar]
- 34.Rivero-Juárez A, Mira JA, Pérez-Camacho I et al. . Twelve week post-treatment follow-up predicts sustained virological response to pegylated interferon and ribavirin therapy in HIV/hepatitis C virus co-infected patients. J Antimicrob Chemother. 2011;666:1351–1353. [DOI] [PubMed] [Google Scholar]
- 35.Wai C-T, Greenson JK, Fontana RJ et al. . A simple noninvasive index can predict both significant fibrosis and cirrhosis in patients with chronic hepatitis C. Hepatology. 2003;382:518–526. [DOI] [PubMed] [Google Scholar]
- 36.Al-Mohri H, Cooper C, Murphy T et al. . Validation of a simple model for predicting liver fibrosis in HIV/hepatitis C virus-coinfected patients. HIV Med. 2005;66:375–378. [DOI] [PubMed] [Google Scholar]
- 37.Nunes D, Fleming C, Offner G et al. . HIV infection does not affect the performance of noninvasive markers of fibrosis for the diagnosis of hepatitis C virus-related liver disease. J Acquir Immune Defic Syndr. 2005;405:538–544. [DOI] [PubMed] [Google Scholar]
- 38.Everhart JE, Wright EC. Association of γ-glutamyl transferase (GGT) activity with treatment and clinical outcomes in chronic hepatitis C (HCV). Hepatology. 2013;575:1725–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandorfer M, Reiberger T, Payer BA et al. . Revisiting predictors of virologic response to PEGIFN + RBV therapy in HIV-/HCV-coinfected patients: the role of metabolic factors and elevated GGT levels. J Viral Hepat. 2014;211:33–41. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery DC, Peck EA, Vining GG. Introduction to Linear Regression Analysis. Hoboken, NJ: John Wiley & Sons, Inc.; 2012. [Google Scholar]
- 41.Carobene A, Braga F, Roraas T et al. . A systematic review of data on biological variation for alanine aminotransferase, aspartate aminotransferase and γ-glutamyl transferase. Clin Chem Lab Med. 2013;5110:1997–2007. [DOI] [PubMed] [Google Scholar]
- 42.van Buuren S, Groothuis-Oudshoorn K. Multivariate imputation by chained equations in R. J Stat Softw. 2011;453:1–67. [Google Scholar]
- 43.Hardin JW, Schmiediche H, Carroll RJ. The simulation extrapolation method for fitting generalized linear models with additive measurement error. Stata J. 2003;34:373–385. [Google Scholar]
- 44.Lederer W, Küchenhoff H. A short introduction to the SIMEX and MCSIMEX. R News. 2006;64:26–31. [Google Scholar]
- 45.Wang N, Lin X, Gutierrez RG et al. . Bias analysis and SIMEX approach in generalized linear mixed measurement error models. J Am Stat Assoc. 1998;93441:249–261. [Google Scholar]
- 46.Li Y, Lin X. Functional inference in frailty measurement error models for clustered survival data using the SIMEX approach. J Am Stat Assoc. 2003;98461:191–203. [Google Scholar]
- 47.Kim J, Gleser LJ. SIMEX approaches to measurement error in ROC studies. Commun Stat Theory Methods. 2000;2911:2473–2491. [Google Scholar]
- 48.Shang Y. Measurement error adjustment using the SIMEX method: an application to student growth percentiles. J Educ Meas. 2012;494:446–465. [Google Scholar]
- 49.Henegar CE, Westreich DJ, Maskew M et al. . Effect of pregnancy and the postpartum period on adherence to antiretroviral therapy among HIV-infected women established on treatment. J Acquir Immune Defic Syndr. 2015;684:477–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson AH, Yang W, Townsend RR et al. . Time-updated systolic blood pressure and the progression of chronic kidney disease: a cohort study. Ann Intern Med. 2015;1624:258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Howe CJ, Napravnik S, Cole SR et al. . African American race and HIV virological suppression: beyond disparities in clinic attendance. Am J Epidemiol. 2014;17912:1484–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maldonado G, Greenland S. The importance of critically interpreting simulation studies. Epidemiology. 1997;84:453–456. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.