Abstract
Organisms from all domains of life depend on filaments of the protein actin to provide structure and to support internal movements. Many eukaryotic cells use forces produced by actin polymerization for their motility, and myosin motor proteins use ATP hydrolysis to produce force on actin filaments. Actin polymerizes spontaneously, followed by hydrolysis of a bound adenosine triphosphate (ATP). Dissociation of the γ-phosphate prepares the polymer for disassembly. This review provides an overview of the properties of actin and shows how dozens of proteins control both the assembly and disassembly of actin filaments. These players catalyze nucleotide exchange on actin monomers, initiate polymerization, promote phosphate dissociation, cap the ends of polymers, cross-link filaments to each other and other cellular components, and sever filaments.
All life-forms depend on actin for structure and the support of internal movements. Dozens of proteins control actin dynamics, participating in the polymerization, depolymerization, capping, and cross-linking of filaments.
1. INTRODUCTION
The evolutionarily ancient, highly conserved actin molecule assembles reversibly into filaments that constitute one of the three major cytoskeletal polymers. This review explains how the structure of the actin molecule accounts for its functional properties, including polymerization dynamics and regulation by a suite of actin-binding proteins. Other contributions to this collection explain how actin participates in many cellular functions, including interactions with myosin motor proteins (Sweeney and Holzbaur 2016), intracellular transport (Titus 2016), cellular structure and motility (Svitkina 2016), muscle contraction (Sweeney 2016), and cytokinesis (Glotzer 2016).
2. GENES, SEQUENCE CONSERVATION, DISTRIBUTION, AND ABUNDANCE
The actin gene originated in the common ancestor of all life on Earth, as evidenced by the fact that bacteria, archaea, and eukaryotes all have actin molecules related structurally and functionally to each other (Gunning et al. 2015). Even earlier, the genes for actin and the glycolytic enzyme hexokinase might have had a common ancestor as their folds are similar and both bind ATP in a central cleft. Bacteria have genes for three types of actins called MreB, FtsA, and ParM. Polymers of each protein have different functions: MreB influences cell wall synthesis and shape, FtsA participates in cell division, and ParM separates large plasmids. In addition, bacterial plasmids and bacteriophages contain genes for more than 30 additional actin homologs (Derman et al. 2009). A subset of archaea has a gene for MreB, and organisms in the so-called TACK (Thaumarchaeota, Aigarchaeota, Crenarchaeota, and Korarchaeota) branch have another actin gene more closely related to eukaryotic actin in sequence and structure. A nearly complete genome sequence from a deep-sea environmental sample came from the prokaryote most closely related to eukaryotes. This “unseen” (not cultured) archaeal cell called Lokiarchaeota has a genuine actin gene and other eukaryotic genes such as Ras-family GTPases (Spang et al. 2015). An ancient relative of Lokiarchaeota is presumed to have founded the eukaryotes when it took on a bacterial symbiont that evolved into the mitochondria. Thus, the eukaryotic actin gene was present in the founding organism.
All eukaryotes have one or more genes for actin, and sequence comparisons have established that they are one of the most conserved gene families, varying by only a few amino acids between algae, amoeba, fungi, and animals. This conservation is attributed to constraints imposed by the interactions of actin with itself to polymerize, with motors and with a large number of regulatory proteins (Gunning et al. 2015). The early branching flagellate Giardia has the most divergent actin eukaryotic gene (Paredez et al. 2011).
Many eukaryotes, including budding yeast, fission yeast, and the green alga Chlamydomonas, get by with a single actin gene and protein to make all of the cytoskeletal structures required for life, but many species, including humans, have multiple actin genes expressed in different tissues (Herman 1993). Humans have three genes for α-actin (muscles), one gene for β-actin (nonmuscle cells), and two genes for γ-actin (one in some smooth muscles and one in nonmuscle cells). Plants have 10 or more actin genes; some are specialized for reproductive tissues and others for vegetative tissues.
Early during the evolution of eukaryotes, the primordial actin gene was duplicated multiple times and, through divergent evolution, gave rise to genes for actin-related proteins—so-called “Arps” (Muller et al. 2005). The Arp genes diversified into multiple families with distinct functions more than 1 billion years ago when the common ancestor of animals, fungi, and amoebas diverged from the large clade of organisms, including algae, plants, ciliates, and a diversity of other single-cell organisms. Arps share between 17% and 52% sequence identity with actin and are numbered Arp1–Arp11 according to their divergence from actin. Arp1 and Arp11 are part of the dynactin complex (Barlan and Gelfand 2016; Goodson and Jonasson 2016), Arp2 and Arp3 are part of the Arp2/3 complex (see below), and several Arps (Arp4–Arp9) participate in chromatin-remodeling complexes and other nuclear functions (Oma and Harata 2011).
Actin is one of the most abundant proteins on Earth and the most abundant protein in many cells, from amoebas to human, often accounting for 10% or more of total protein. Its abundance is topped only by tubulin in brain and keratins in skin. Actin molecules in cells turn over very slowly, on the order of weeks in muscle cells.
3. STRUCTURES OF ACTIN AND ACTIN FILAMENTS
The strong tendency of actin to polymerize into filaments thwarted efforts for decades to grow crystals, but eventually, in the early 1990s, cocrystallization with DNase I (Kabsch et al. 1990) or profilin (Schutt et al. 1993) allowed for high-resolution structures. Now dozens of crystal structures are available for eukaryotic and prokaryotic actins (Dominguez and Holmes 2011).
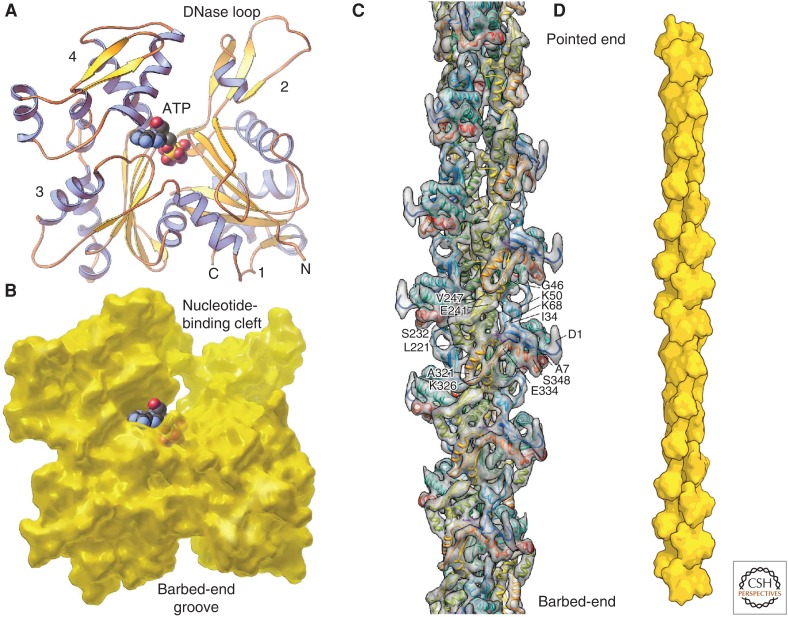
The eukaryotic actin polypeptide of 375 residues folds into a flat protein with a deep medial cleft that binds ATP (Fig. 1A,B). Actin is described as having four subdomains. The polypeptide winds from the amino terminus in subdomain 1 to subdomains 2, 3, and 4 and back to subdomain 1 at the carboxyl terminus. ATP binds in a deep cleft, interacting more strongly with subdomains 3 and 4, but also with residues in subdomains 1 and 2. Several proteins bind in a prominent groove between subdomains 1 and 3—and, hence, some call it the “target-binding groove” (Dominguez 2004).
Figure 1.
Structures of the actin molecule and actin filament. (A) Ribbon diagram of the actin molecule with space-filling ATP (protein data bank [PDB]: 1ATN). N, amino terminus; C, carboxyl terminus. Numbers 1, 2, 3, and 4 label the four subdomains. (B) Space-filling model of actin showing the nucleotide-binding cleft with ATP in situ and barbed-end groove. (C) Reconstruction of the actin filament from cryo-electron micrographs. The labels are single-letter abbreviations for selected amino acids. (D) Cartoon of the actin filament showing the position of the pointed and barbed ends. (A,B, Reprinted, with permission, from Pollard and Earnshaw 2007; C, reprinted, with permission from Macmillan Publishers Ltd., from Fujii et al. 2010; D, adapted, with permission, from Pollard and Earnshaw 2007.)
The two halves of the protein flanking the nucleotide-binding cleft have similar folds but no sequence similarity, suggesting that a very ancient duplication formed the original actin gene, followed by divergence of the two halves of the gene. Arps have the same fold as actin, including all of the atoms required to bind to ATP (Robinson et al. 2001), but their surfaces differ extensively from those of actin, with insertion of one or more surface loops and many amino acid substitutions that participate in forming unique macromolecular assemblies.
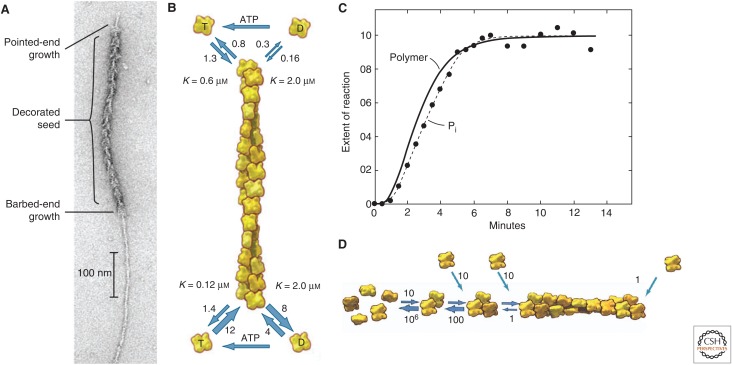
Early X-ray diffraction studies of live muscle and electron microscopy of isolated filaments (Huxley 1963) revealed that actin filaments consist of two strands of subunits in right-handed helices staggered by half the length of an actin monomer (2.7 nm) (Fig. 1C,D). This structure can also be described as a single-stranded left-hand helix that encompasses all of the subunits of the filament. The polarity of the filament was revealed by electron micrographs of negatively stained preparations of filaments saturated with myosin heads, which form an arrowhead-shaped complex with each turn of the helix (Fig. 2A). These myosin arrowheads define the “barbed” and “pointed” ends of a filament. The target-binding groove is at the barbed end, and the nucleotide-binding cleft is at the pointed end of the actin subunit.
Figure 2.
Actin polymerization. (A) Electron micrograph of a negatively stained actin filament. A seed was first decorated with myosin heads and then allowed to grow bare extensions. Elongation was faster at the barbed end than at the pointed end. (B) Diagram showing the rate constants for actin association and dissociation at the two ends of an actin filament. The pointed end is at the top and the barbed end is at the bottom. Unit of association rate constants, µm−1 sec−1; unit of dissociation rate constants, sec−1. The K values are the ratios of dissociation rate constants to association rate constants, the critical concentrations for each of the four reactions. The horizontal arrows indicate the exchange of adenosine diphosphate (ADP) for ATP. (C) Time course of spontaneous polymerization of Mg-ATP–actin monomers. The solid line is the polymer concentration measured by the fluorescence of pyrene-labeled actin. The initial lag comes from slow spontaneous nucleation. The reaction reaches a steady state when the free actin monomer concentration reaches the overall critical concentration. Filled circles are the extent of hydrolysis of the bound ATP, which lags behind polymerization by a few seconds. (D) Mechanism of nucleation, showing monomers, a dimer, a trimer, and a filament, with estimates of the rate constants for each step. Unit of association rate constants, µm−1 sec−1; unit of dissociation rate constants, sec−1. (A,B,D, Adapted, with permission, from Pollard and Earnshaw 2007; C, reprinted from Pollard and Weeds 1984.)
The best available actin filament structures are reconstructions of electron micrographs of frozen-hydrated filaments (Fig. 1C) (Fujii et al. 2010; von der Ecken et al. 2014) and models from X-ray fiber diffraction of aligned filaments (Oda et al. 2009). These models revealed extensive contacts along the short-pitch helix and between subdomain 2 of each subunit and the barbed-end groove of the next subunit along the long-pitch helix. Compared with crystal structures, the subunits in filaments are “flatter” because of a scissors-like rotation between subdomains 1 and 3.
4. NUCLEOTIDE BINDING AND POLYMERIZATION
The availability of large quantities (hundreds of milligrams) of purified actin from muscle and other sources (see Box 1) has enabled a half-century of quantitative mechanistic experiments on nucleotide binding and hydrolysis and their roles in polymerization.
BOX 1. PURIFICATION AND HANDLING OF ACTIN
The classic preparation of actin from skeletal muscle starts by extracting myosin from homogenized tissue with a high concentration of salt, precipitating the residual proteins with acetone, and drying to make an acetone powder. Actin monomers are extracted from this dry powder with a dilute buffer containing ATP and polymerized by adding salt. The filaments are pelleted by ultracentrifugation and depolymerized by dialysis against low-salt buffer. Gel filtration removes actin oligomers, capping protein and other minor contaminants, yielding monomers suitable for quantitative assembly experiments (McLean-Fletcher and Pollard 1980). Purification from other cells usually requires a preliminary step to concentrate the actin by ion-exchange chromatography (Gordon et al. 1976) or affinity chromatography with an actin-binding protein such as DNase I (Schafer et al. 1998) or gelsolin (Ohki et al. 2009). One or more cycles of polymerization and depolymerization, followed by gel filtration, complete the purification. Actin can be stored for days at 4°C in low-salt buffer with ATP, a sulfhydryl reducing agent, 0.1 mm CaCl2, and sodium azide to prevent bacterial growth. Freezing is not recommended.
Actin monomers bind ATP or adenosine diphosphate (ADP) tightly, provided that either Ca2+ or Mg2+ is present in the buffer. One of these divalent cations associates with the β- and γ-phosphates of ATP, stabilizing its interaction with the protein (Kabsch et al. 1990). Ca2+ is used during the purification of actin, but Mg2+ is bound under physiological conditions and is assumed to be present in the following discussion of polymerization. ATP binds nucleotide-free actin monomers rapidly, with a rate constant of 6 µm−1 sec−1 and dissociates at ∼10−2 sec−1, and so the Kd is in the nanomolar range (De La Cruz and Pollard 1995). Chelation of free divalent cations increases the dissociation rate of ATP 20-fold. Monomers without a bound nucleotide denature in seconds unless stabilized by high sucrose concentrations.
Actin monomers polymerize spontaneously under physiological salt conditions with either or both monovalent and divalent cations in the buffer. Cations bind specific sites that promote interactions between subunits in the filament (Kang et al. 2013). Spontaneous polymerization begins with a lag period that depends very strongly on the concentration of the actin monomers. Already, in the 1960s, the lag was correctly interpreted as a slow nucleation step that forms small oligomers suitable for elongation (Oosawa and Asakura 1975). Computer modeling of the complete time course of polymerization of a wide range of actin monomer concentrations established that nucleation consists of two unfavorable steps: formation of a dimer and addition of a third subunit to form a trimer (Sept and McCammon 2001). The association reactions are fast, but dimers and trimers are very unstable—dimers dissociate at ∼106 sec−1, and trimers dissociate a subunit at 100 sec−1 (Cooper et al. 1983; Frieden 1983). Thus, dimers and trimers are present at exceedingly low concentrations in a polymerization reaction. The oligomer is more stable after adding a fourth subunit, presumably because the subunits have a full complement of intermolecular contacts. Larger oligomers elongate at the same rate as long filaments, with the rate of elongation depending on the monomer concentration.
Actin filament elongation is understood much better than nucleation because one can measure the elongation rates in bulk solution or by observing single filaments by electron (Pollard 1986) or light microscopy (Kuhn and Pollard 2005). Barbed ends grow much faster than pointed ends, with a diffusion-limited association rate constant of ∼10 µm−1 sec−1 for ATP–actin and slow dissociation at ∼1 sec−1 (Fig. 2D). The ratio of the dissociation and association rate constants gives the “critical concentration” for polymerization at the barbed end of ∼0.1 µm. Association and dissociation of ATP–actin is much slower at pointed ends. A bound nucleotide stabilizes monomers but is not required for polymerization. Free energy from association of actin subunits with the barbed end can be used to produce piconewton forces (Kovar and Pollard 2004).
The conformational changes associated with actin polymerization increase the rate of hydrolysis of bound Mg-ATP from 7 × 10−6 sec−1 by actin monomers to 0.3 sec−1 in filaments (Fig. 3) (Blanchoin and Pollard 2002) by repositioning the protein and water molecules around the γ-phosphate (McCullagh et al. 2014). Hydrolysis is irreversible (Carlier and Pantaloni 1986). The hydrolysis reaction is “concerted” in the sense that association of the attacking water with the γ-phosphate is coupled with dissociation of the bond between the β- and γ-phosphates (McCullagh et al. 2014). Most evidence favors the hypothesis that hydrolysis occurs randomly on polymerized Mg-ATP–actin subunits (Jégou et al. 2011), although the nucleotide state of neighboring subunits might influence the rate. One version of such influence was a proposal that hydrolysis occurs in a zipper-like fashion along the polymer (Korn et al. 1987).
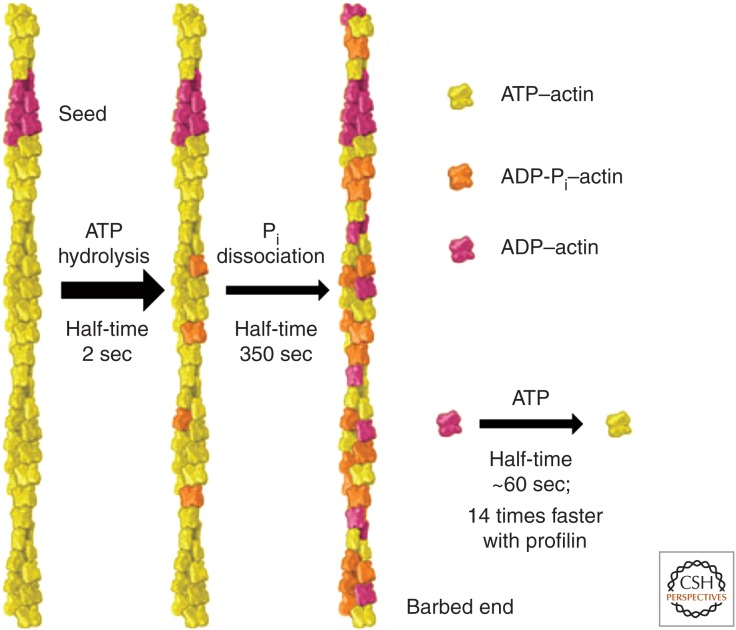
Figure 3.
Nucleotide reactions of actin. The cartoon shows an actin filament that has grown from both ends of an ADP–actin seed. Over time, ATP bound to the polymerized subunits is hydrolyzed randomly to ADP and phosphate (Pi), followed by slow dissociation of the phosphate, leaving ADP–actin. ADP dissociates from ADP–actin monomers and is rapidly replaced by ATP. Profilin speeds ADP dissociation. (Adapted, with permission, from Pollard and Earnshaw 2007.)
After ATP hydrolysis, the γ-phosphate dissociates very slowly from polymerized actin, with a half-time of ∼6 min (dissociation rate constant ∼ 0.003 sec−1) (Carlier and Pantaloni 1986), and so ADP-Pi–actin is a long-lived intermediate in the polymerization process (Fig. 3). The polymerization properties of ADP-Pi–actin are close to those of ATP–actin (Fujiwara et al. 2007), but once the γ-phosphate dissociates, the ADP–actin subunit behaves quite differently. Most notably, ADP–actin subunits dissociate faster from both ends than ATP–actin subunits (Fig. 2). The critical concentration for polymerization of Mg-ADP–actin is the same at both ends, 1.8 µm (Pollard 1986).
With ATP in the buffer and bound to actin monomers, the critical concentrations for polymerization differ at the two ends of filaments (Fig. 2B). Consequently, at steady state, net addition occurs at the barbed end balanced by net loss of subunits at the pointed end—so-called treadmilling. The treadmilling rate of <1 subunit/sec is so slow that it contributes little to actin filament turnover in cells, but it has fascinated the field since its discovery in the 1970s (Wegner 1976).
Filaments trap ADP irreversibly, but they exchange phosphate with the medium. The affinity of polymerized ADP–actin for phosphate is very low because phosphate binding is very slow, with an association rate constant of only ∼2 m−1 sec−1 (Carlier and Pantaloni 1986). Thus, the Kd for phosphate-binding ADP–actin filaments is on the order of 1 mm, depending on the pH.
Differences in phosphate binding explain the polymerization asymmetry between the ends (Fujiwara et al. 2007). Subunits at the ends of filaments bind and dissociate phosphate faster than interior subunits, and the affinity of pointed ends for phosphate is 10 times weaker than that of barbed ends. Given this difference and given that subunits associate and dissociate slower at pointed ends, pointed ends are much more likely than barbed ends to hydrolyze bound ATP and dissociate γ-phosphate before being buried in the filament. This exposes ADP–actin with its higher critical concentration for dissociation from the pointed end.
Cycles of actin assembly in cells occur in an environment with millimolar concentrations of Mg-ATP and phosphate. Most of the cytoplasmic actin monomers have bound Mg-ATP (Rosenblatt et al. 1995), which is hydrolyzed on polymerized subunits, followed by phosphate dissociation. Depolymerization releases Mg-ADP–actin monomers into the cytoplasm, where ADP exchanges for ATP, restarting the cycle (Fig. 3). Regulatory proteins, discussed below, promote phosphate dissociation from filaments, disassembly, and nucleotide exchange, but none has been shown to influence ATP hydrolysis.
Actins from species as divergent as mammals and amoeba polymerize (they even copolymerize) and handle the bound ATP similarly, but one must be alert for exceptions. For example, polymerized yeast actins hydrolyze bound ATP and dissociate the γ-phosphate much faster than other actins (Harris et al. 2004; Ti and Pollard 2011).
5. OVERVIEW OF ACTIN-BINDING PROTEINS
The behavior of actin in cells differs dramatically from that of the purified protein in a test tube. At the total concentrations found in cells (50–200 µm), >99% of purified actin would polymerize in seconds, and subunits would exchange on and off the barbed ends roughly once per second and at the pointed end would exchange more slowly. In contrast to this largely static situation, approximately half of total actin in cells is unpolymerized at concentrations in the range ∼25–100 µm, orders of magnitude higher than the critical concentration. Furthermore, filaments assemble and turn over on timescales of tens of seconds—far faster than relatively inert actin filaments in a test tube.
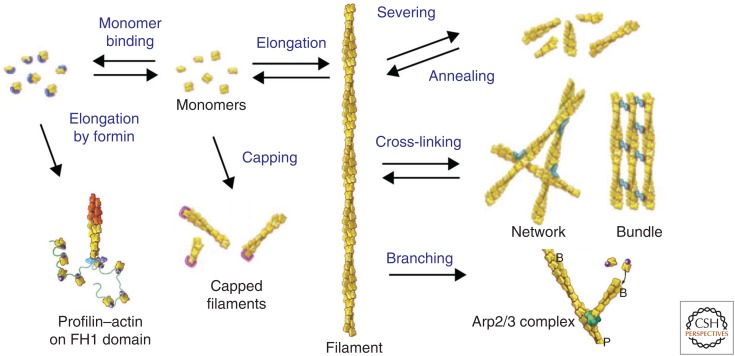
Actin-binding proteins account for these differences by regulating virtually every aspect of actin assembly (Fig. 4). Collectively, these proteins maintain a large pool of actin monomers available for polymerization, nucleate assembly of new filaments, promote elongation, cap barbed or pointed ends to terminate elongation, sever filaments, and cross-link filaments.
Figure 4.
Overview of families of actin-binding proteins, including monomer binding, polymerases such as formins, capping proteins, severing proteins, cross-linking proteins, and branching protein Arp2/3 complex. Filaments can anneal end to end, but no proteins are known to facilitate this reaction. The drawing does not include tropomyosin and myosin motors, which bind to the sides of filaments. (Adapted, with permission, from Pollard and Earnshaw 2007.)
Most actin-binding proteins are widely distributed in eukaryotes, and so they arose in an ancient common ancestor. Giardia is an exception as it lacks genes encoding many actin-binding proteins, including myosin, cofilin, formins, and the Arp2/3 complex (Paredez et al. 2011). It might have branched before the actin system was fully developed or lost these genes. The following sections explain the properties of proteins in each of these families, most of which have subtle mechanisms of action that contribute to the dynamics of the actin system in cells.
6. ACTIN-MONOMER-BINDING PROTEINS
The small (∼13–14-kDa), actin-monomer-binding protein profilin is essential for the viability of most eukaryotes. Given its affinity (Kd = 0.1 µm) for ATP–actin monomers and a cellular concentration in the range 50–100 µm, most of the unpolymerized actin in the cytoplasm is bound to profilin, except for mammalian cells that express thymosin-β4 (see below). Profilin bound to the barbed end of an actin monomer (Fig. 5C) sterically inhibits nucleation and elongation at pointed ends, but not elongation at barbed ends. Profilin binds weakly to ATP–actin on the barbed end of filaments (Kd > 20 µm), so profilin dissociates rapidly after a profilin–actin complex binds, freeing the end for further elongation. (Profilin has much higher affinity for the barbed ends of ADP–actin filaments [Courtemanche and Pollard 2013].) However, high concentrations of free profilin can slow elongation and even promote dissociation of the terminal subunit (Jégou et al. 2011; Courtemanche and Pollard 2013).
Figure 5.
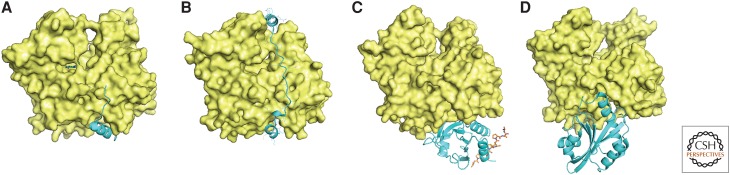
Proteins that bind actin monomers. Space-filling models of the actin monomer with ribbon diagrams of bound proteins. This is the standard view of actin (see Fig. 1), with the ATP-binding cleft at the top and the barbed-end groove at the bottom. (A) The WH2 helix binds in the barbed-end groove (PDB: 3M1F, from Vibrio parahaemolyticus Vopl). (B) Thymosin-β4 helices bind in both the barbed-end groove and across the pointed-end cleft (PDB: 4PL7). (C) Profilin can bind simultaneously to the barbed end of actin and to polyproline helices such as that from vasodilator-stimulated phosphoprotein (VASP), shown here as a red stick figure (PDB: 2PBD). (D) The carboxy-terminal cofilin domain from twinfilin binds on the barbed end of the actin molecule (PDB: 3DAW).
Two other activities of profilin are essential for viability (Lu and Pollard 2001). Bound profilin reduces the affinity of actin monomers for ATP or ADP, so profilin catalyzes nucleotide exchange (Mockrin and Korn 1980) by rapidly dissociating ADP from newly depolymerized actin monomers and allowing ATP to bind (Fig. 3) (Vinson et al. 1998). Profilin also binds polyproline sequences at a site physically separated from the actin-binding site (Archer et al. 1994; Ferron et al. 2007). As explained below, this interaction allows profilin to deliver actin to polyproline sequences in elongation factors, such as formins and Ena/VASP, and to promote elongation of actin filament barbed ends. Phosphatidylinositol (4,5)-bisphosphate (PtdIns(4,5)P2, commonly known as PIP2) binds profilin and competes for binding actin.
Thymosin-β4 is a peptide of 43 residues originally described as a thymic hormone, but it is also the most abundant actin-monomer-binding protein in some cells, including leukocytes and platelets (Safer et al. 1991). The amino terminus of thymosin-β4 forms a short helix that binds in the barbed-end groove, and the rest of the peptide consists of an extended region that binds the front surface of actin and a second helix that caps the pointed end at the top of the nucleotide-binding cleft (Fig. 5B) (Xue et al. 2014). Given concentrations of >100 µm in the cytoplasm and a micromolar affinity for Mg-ATP–actin, thymosin-β4 can sequester a large pool of actin monomers, preventing them from engaging in any of the polymerization reactions because of steric interference with all of the interactions required for polymerization (Fig. 2D).
Profilin competes with thymosin-β4 for binding actin monomers, offering a pathway for actin monomers sequestered by thymosin-β4 to participate in elongation (Pantaloni and Carlier 1993). This shuttling process is possible because both proteins exchange rapidly on and off actin monomers. Given the physiological concentrations of all three proteins, most of the actin monomers are bound to either profilin or thymosin-β4, leaving a low (submicromolar) concentration of free actin monomers.
Other proteins have one or more sequences homologous to the amino-terminal half of thymosin-β4, including the helix that binds in the barbed-end groove of actin (Fig. 5A) (Chereau et al. 2005; Rebowski et al. 2010). These sequences are called WH2 (WASp-homology 2) motifs after their discovery in Wiskott–Aldrich syndrome protein (WASp; see Sec. 8). WH2 motifs can deliver an actin subunit to the barbed end of a filament such as those nucleated by the Arp2/3 complex (see Sec. 8). Tandem WH2 motifs allow proteins in the Ena/VASP family to promote actin filament elongation (see Sec. 8) and the proteins Lmod (Chereau et al. 2008) and Spire (Quinlan et al. 2005) to promote nucleation by bringing together multiple actin monomers.
7. SEVERING PROTEINS
The two main families of actin filament–severing proteins differ greatly in size—tiny 15-kDa cofilin and large multidomain proteins of the gelsolin family. In addition, at least two formins, FRL-α (Harris et al. 2004) and INF-2 (Gurel et al. 2014), can sever actin filaments.
Most eukaryotes express high concentrations of cofilin, a small protein that binds in the barbed-end groove of actin monomers and to actin filaments (Fig. 5D). Cofilin bound to actin monomers inhibits nucleotide exchange (Nishida 1985), but profilin overcomes this effect (Blanchoin and Pollard 1998).
The main function of cofilin is to sever actin filaments. Cofilin binds cooperatively to the sides of actin filaments, with a higher affinity for ADP–actin subunits than ATP- or ADP-Pi subunits (Cao et al. 2006). Thus, ATP hydrolysis and phosphate dissociation act as a timer for cofilin binding. Nevertheless, weak binding of cofilin to ADP-Pi subunits in filaments promotes dissociation of the γ-phosphate, producing ADP–actin polymers in seconds rather than in minutes (Blanchoin and Pollard 1999), a timescale reasonable for the rapid turnover of filaments in cells.
A crystal structure showed that a cofilin domain from the actin-monomer-binding protein twinfilin binds in the barbed-end groove of monomeric actin (Fig. 5D) (Paavilainen et al. 2008). This interaction is maintained when cofilin binds an actin filament, whereas other parts of cofilin contact subdomain 2 at the pointed end of the adjacent actin subunit along the long-pitch helix (Galkin et al. 2011). These interactions force actin subdomains 1 and 2 (the outer domain) to rotate ∼30° from the flattened conformation in the filament to become even more skewed than monomers. To avoid steric clashes, the twist between successive subunits along the short-pitch helix is reduced from 167° to 162°, enough to reduce the repeat of the long-pitch helices from 36 nm to 27 nm and make the filaments more flexible (McCullough et al. 2011).
Cellular processes dependent on actin filament severing by cofilin include motility and cytokinesis. The mechanism of severing is remarkable. Filaments saturated with cofilin are very stable, but binding of small numbers of cofilins promotes severing, most likely at interfaces between flexible decorated sites and stiffer bare segments (Elam et al. 2013; Ngo et al. 2015). Therefore, steady state severing is optimal at concentrations of cofilin far below the Kd (Andrianantoandro and Pollard 2006). However, high concentrations of cofilin sever transiently as the first few cofilins bind to a bare filament or if other proteins compete with cofilin for binding.
Cofilin was originally called actin-depolymerizing factor (ADF) (Bamburg et al. 1980), because it reduced pelleting of actin filaments in the ultracentrifuge. Severing explains this behavior. Severing also creates free ends for depolymerization, but cofilin does not promote dissociation of subunits from either end of filaments (Andrianantoandro and Pollard 2006), contrary to a widely cited theory (Carlier et al. 1997).
Cells use multiple strategies to regulate cofilin (Mizuno 2013). Binding of cofilin to PtdIns(4,5)P2 or phosphorylation of Ser3 of cofilin interferes with cofilin binding to actin, so either inactivates all functions. LIM kinases phosphorylate Ser3, and phosphatases, including Slingshot and chronophin, remove the phosphate. The extent of cofilin phosphorylation changes during many cellular activities, developmental processes, and diseases because many signaling pathways impinge on LIM kinase. These include bone morphogenetic protein signaling through receptor serine kinases, inflammatory mediators operating through Rho and ROCK, guidance molecules through Rac, Cdc42, and PAK, and vascular endothelial growth factor through the mitogen-activated protein kinase pathway. Chemotactic signals also influence the activity of Slingshot. In addition to disassembling actin filaments, severing produces a free barbed end that can stimulate actin filament assembly (Bravo-Cordero et al. 2013). Actin-interacting protein Aip1 cooperates with cofilin to promote actin filament turnover in cells and actin filament severing in vitro (Chen et al. 2015).
Many organisms, from yeasts to flies, have a single cofilin gene that is essential for their viability, but mammals have three isoforms (cofilin-1, muscle-specific cofilin-2, and ADF) (Poukkula et al. 2011). Cofilin-1 is required for viability of mice, whereas ADF-null mutant mice are viable but have ocular defects.
Other members of the cofilin family are widespread from fungi to humans, but none of them is as essential to viability as cofilin itself (Poukkula et al. 2011). Glia maturation factor (GMF) shares the cofilin fold, but, rather than interacting with actin, it binds Arp2/3 complex and dissociates actin filament branches (Ydenberg et al. 2013). Twinfilin consists of tandem cofilin domains that bind actin monomers and filaments, suppress polymerization (Paavilainen et al. 2008), and promote subunit dissociation from both filament ends in cooperation with cyclase associate protein (Johnston et al. 2015).
Members of the gelsolin family comprise two to six homologous gelsolin domains. The fold of the gelsolin domain resembles that of cofilin (Nag et al. 2013), although no strong evidence exists that gelsolin and cofilin have a common ancestor. Mammalian family members consist of three or six gelsolin domains plus other domains, whereas homologous proteins in other species have two, four, or five gelsolin domains.
Gelsolin and related proteins sever actin filaments and cap their barbed ends. Calcium binding regulates most family members by releasing gelsolin from an autoinhibited inactive state and stabilizing the gelsolin domains into a conformation that permits binding to the actin filament. The affinities of the calcium-binding sites range from Kd = 0.2 µm, which is physiologically relevant, to Kd > 100 µm.
The long α-helices of gelsolin domains 1, 2, and 4 bind in the barbed-end groove of actin, similar to the binding of cofilin. This interaction with a filament results in severing, leaving gelsolin associated with the barbed end of the severed filament. Polyphosphoinositides (e.g., PtdIns(4,5)P2) can compete the gelsolin cap from the end of the filament (Janmey and Stossel 1987). Mice with a deletion mutation of the gelsolin gene are viable, but they have defects in cellular motility and platelet function during blood clotting.
One splicing isoform of gelsolin has an amino-terminal signal sequence that directs the protein into the secretory pathway and eventually into the blood. The high calcium concentration in the blood keeps gelsolin active so that, together with vitamin D–binding protein, it forms a “scavenger” system to depolymerize actin filaments released into the bloodstream from damaged cells (Nag et al. 2013). A mutation in patients with one form of familial amyloidosis makes gelsolin susceptible to proteolysis in the Golgi apparatus, and the resulting peptides form amyloid deposits that damage multiple organs (Solomon et al. 2012).
The eight members of the gelsolin gene family in mammals have specific expression patterns (Nag et al. 2013). For example, adseverin helps secretory granules reach the plasma membrane in endocrine organs, and villin has an extra domain that allows it to bundle actin filaments in microvilli. Macrophages express CapG, a family member with three gelsolin domains that cap, but do not sever, actin filaments.
8. NUCLEATION PROTEINS
Given that actin filament nucleation is intrinsically unfavorable and suppressed by profilin and thymosin-β4, cells rely on regulatory proteins to initiate actin filament polymerization in a controlled manner. They use Arp2/3 complex to produce actin filament branches, formins to initiate unbranched filaments, and proteins with tandem WH-2 domains to form other filaments.
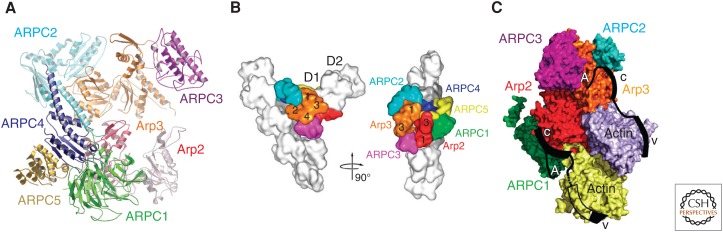
Arp2/3 complex is an ancient (basal eukaryote) assembly of seven subunits, including Arp2 and Arp3 (Fig. 6). The complex is intrinsically inactive because the other subunits hold the two Arp moieties apart (Robinson et al. 2001). When Arp2/3 complex binds the side of an actin filament, Arp2 and Arp3 move closer together and form the base for growth of a branch (Rouiller et al. 2008). The free barbed end of the daughter filament elongates, whereas the Arp2/3 complex anchors the pointed end of the filament rigidly to the side of the mother filament.
Figure 6.
The Arp2/3 complex. (A) Ribbon diagram showing the two actin-related proteins (Arps) and the five novel subunits. Subdomains 1 and 2 of Arp2 were disordered in this structure (PDB: 1K8K). (B) Model of the branch junction from reconstructions of electron micrographs (Rouiller et al. 2008). The Arp2/3 complex anchors a branch represented by daughter subunits D1 and D2 on the side of the mother filament. The numbers indicate the subdomains of Arp2 (red) and Arp3 (orange). (C) Model (Padrick et al. 2011) for the interaction of Arp2/3 complex with two verprolin-cofilin-acidic (VCA) motifs (black), each with an actin subunit bound to the V (WH2) motif (Chereau et al. 2005). The location of the A motif on Arp3 was determined by X-ray crystallography (Ti et al. 2011). Other aspects of the model were inferred from binding and cross-linking experiments (Padrick et al. 2011). (A, Reprinted, with permission, from Pollard and Earnshaw 2007; B, reprinted from Rouiller et al. 2008; C, adapted, with permission, from Padrick et al. 2011.)
A variety of nucleation-promoting factors activate Arp2/3 complex (Rottner et al. 2010), each in a particular cellular context, including at the leading edge of motile cells (WASp, N-WASP [neural-WASp], Scar [suppressor of cAMP activator]/WAVE [WASP family verprolin homologous protein]), at sites of endocytosis (WASp), and for internal membrane traffic (Wiskott–Aldrich syndrome protein and Scar homolog [WASH]) (Rotty et al. 2013). All of these nucleation-promoting factors are intrinsically inactive because of sequestration of their binding sites for actin monomers and Arp2/3 complex. Intramolecular interactions inhibit WASp and N-WASP. Rho-family GTPases, polyphosphoinositides, and proteins with Src-homology 3 domains overcome these autoinhibitory interactions, allowing WASp and N-WASP to interact with actin monomers and Arp2/3 complex. The WAVE regulatory complex, consisting of five subunits, inhibits the Scar/WAVE family (Chen et al. 2010). The Rac GTPase binds the WAVE regulatory complex and frees Scar/WAVE to interact with Arp2/3 complex. A similar protein complex regulates WASH (Campellone and Welch 2010).
Binding of Arp2/3 complex to the side of an actin filament is unfavorable because it requires conformational changes in both partners (Rouiller et al. 2008). These high-energy states are poorly populated, so stable binding is slow (Beltzner and Pollard 2008) because most interactions result in dissociation (Smith et al. 2013). Association of two molecules of nucleation-promoting factor prepares Arp2/3 complex for binding actin filaments (Padrick et al. 2011; Ti et al. 2011), which completes the activation process. Each nucleation-promoting factor also brings along an actin monomer; together, they become the first two subunits in the daughter filament. Binding to the mother filament is thought to drive the conformational changes that position the Arps to initiate the branch (Ti et al. 2011), but one nucleation-promoting factor (Dip1) can activate Arp2/3 complex to nucleate a filament independent of a preexisting filament (Wagner et al. 2013). In contrast, a protein named arpin inhibits nucleation of actin filament branches by Arp2/3 complex (Dang et al. 2013).
Actin filament branches are quite rigid and stable for tens of seconds. The protein cortactin stabilizes branches (Weaver et al. 2001), but both cofilin (Chan et al. 2009) and the related protein GMF (Ydenberg et al. 2013) promote dissociation of branches. A drug-like molecule called CK-666 is available to inhibit branch formation (Nolen et al. 2009).
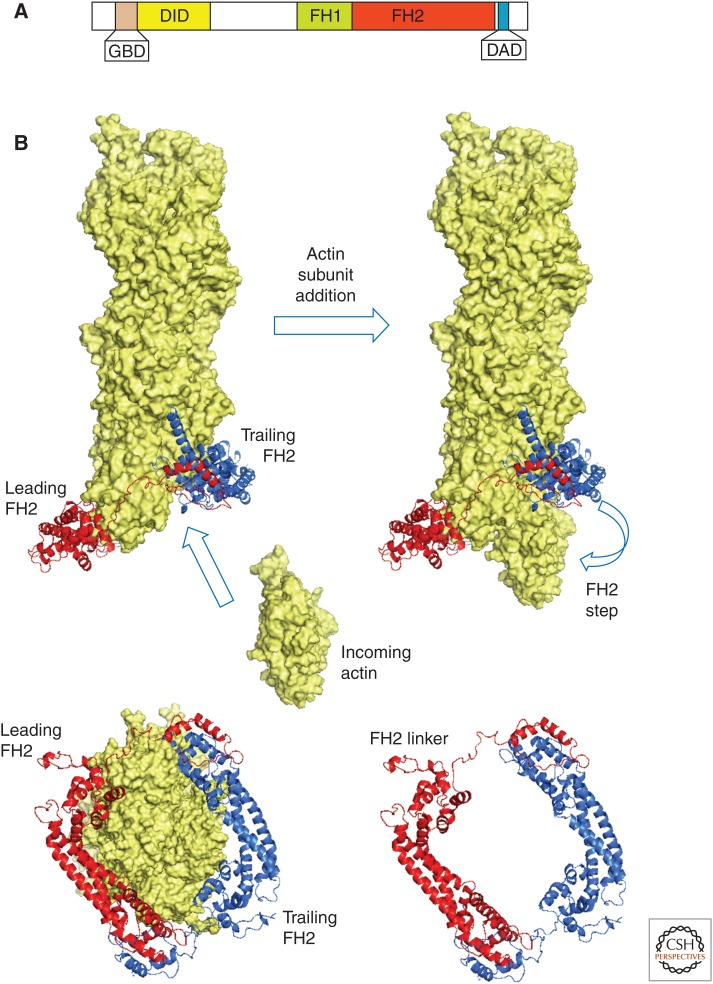
Formins are multidomain, homodimeric proteins characterized by a formin homology 2 (FH2) domain that interacts with the barbed end of an actin filament (Fig. 7A) (Goode and Eck 2007; Paul and Pollard 2009). FH2 domains form head-to-tail dimers (Xu et al. 2004). When free of actin, an α-helical link holds the two halves of the dimer close together, but the linker can stretch into an extended chain to allow the FH2 dimer to wrap around an actin filament (Fig. 7B) (Otomo et al. 2005). The actin in cocrystals with FH2 forms a polymer with twofold symmetry rather than the 167° helical twist of actin filaments (Otomo et al. 2005). A refined model of an FH2 dimer on the end of an actin filament (Fig. 7B) has extensive contacts between the FH2 and actin (Baker et al. 2015). Next to the FH2 domain, most formins have an FH1 domain with multiple proline-rich sequences that bind profilin. Many formins are autoinhibited by interactions between a diaphanous autoinhibitory domain (DAD) near the amino terminus and a DAD-interacting domain (DID) near the carboxyl terminus (Alberts 2001). Binding of Rho-family GTPases near the DAD overcomes this autoinhibition.
Figure 7.
Structure and role of formins. (A) Domain organization of a diaphanous (Dia)-type formin. DAD, diaphanous autoinhibitory domain; DID, DAD-interacting domain; GBD, GTPase-binding domain; FH1, proline-rich formin homology 1 domain; FH2, formin homology 2 domain. (B) Model of the actin filament elongation mechanism, with actin subunits shown by a space-filling model and the formin Bni1 FH2 dimer shown as red and blue ribbon diagrams. The model is based on the crystal structure (Otomo et al. 2005) docked on the barbed end of the actin filament model from fiber diffraction (Oda et al. 2009) and refined by molecular-dynamics simulation (Baker et al. 2015). An actin subunit binds from solution, creating a binding site for the trailing FH2 domain (blue) to “step” toward the barbed end. The lower diagrams show end-on views, including (left) the actin filament and (right) the FH2 dimer alone.
Most formins nucleate actin filaments, presumably by stabilizing actin dimers (Pring et al. 2003). Only free actin monomers appear to participate in this nucleation process (Paul and Pollard 2009). In fission yeast, each of the three formins nucleates unbranched filaments for specific structures, such as the cytokinetic contractile ring or interphase actin filament cables, but with some overlap. The situation is much more complicated in mammals, which have 15 isoforms with partially overlapping functions (Higgs and Peterson 2005; Campellone and Welch 2010), and so assignment of biological functions is still in progress. These formins also nucleate unbranched filaments for the contractile ring, stress fibers, and filopodia. Some formins are associated with the plasma membrane or intracellular membranes, such as the endoplasmic reticulum, and others interact with microtubules. Transmembrane helices anchor some plant formins to membranes. Most formins can either inhibit or promote actin filament elongation, as explained in Section 9.
Proteins with tandem WH2 domains promote nucleation, presumably by favoring the association of dimers or trimers (Dominguez and Holmes 2011). This family includes proteins called spire, cordon bleu, and JMY that have been implicated in the development of the nervous system and other tissues (Campellone and Welch 2010). A protein with one WH2 domain, called leiomodin, participates in actin polymerization in muscle (Chereau et al. 2008). Some bacteria express proteins with WH2 domains that subvert cellular actin assembly.
Two other mechanisms can create barbed ends. As noted above, severing by cofilin produces a free barbed end and a free pointed end, a reaction shown to stimulate polymerization in live cells (Bravo-Cordero et al. 2013). Removing a cap from the end of a filament has the theoretical capacity to open a site for elongation (see Sec. 10).
9. ACTIN FILAMENT POLYMERASES
In addition to nucleating actin filaments, formins both inhibit and promote the elongation of actin filament barbed ends by interacting processively with the growing end (Paul and Pollard 2009). On their own, FH2 domains from all formins tested slow barbed-end elongation. A simple explanation is that the complex of the FH2 domain and the end of the filament has two conformations: Actin monomers can bind to the open state but not the closed state (Vavylonis et al. 2006). Depending on the formin, barbed ends are open between 5% and 90% of the time. Profilin overcomes this inhibition and can bias polymerization toward filaments with formins, providing that the formin construct has an FH1 domain in addition to the dimer of FH2 domains. FH1 domains are flexible “tentacles,” located amino-terminal to the FH2 domain, with one to 14 polyproline tracks that bind profilin–actin complexes. After rate-limiting binding of profilin–actin to multiple sites in the FH1 domain, diffusion of the FH1 domains delivers profilin very rapidly to the end of the filament, allowing rapid elongation in spite of the fact that the end is in the closed state part of the time. In favorable cases, such as formin mDia1, which is open 90% of the time and has an FH1 domain with 14 potential profilin–actin-binding sites, elongation can be five times faster than for a free barbed end (Kovar et al. 2006). In spite of the rapid elongation, all formins tested are remarkably processive, “stepping” onto the newly added subunit for thousands of cycles without failure. This polymerase activity inhibits capping by capping protein and allows actin filaments associated with a formin to grow very quickly and persistently in the cell. For example, formin mDia1 grows filaments at 700 subunits/sec in fibroblasts (Higashida et al. 2004).
Similar to formins, tetramers of Ena/VASP associate with growing actin filament barbed ends, promote elongation, and inhibit capping (Edwards et al. 2014). In contrast to formins, Ena/VASP proteins do not seem to nucleate polymerization. VASP can deliver either free actin monomers or profilin–actin to the barbed end of the filament, using either an actin-monomer-binding site related to a WH2 domain (Ferron et al. 2007) or polyproline tracks that bind profilin (Hansen and Mullins 2010). VASP is much less processive than formins, with a dwell time on barbed ends of only 1.5 sec. Mice need at least one of their three Ena/VASP genes for viability. These proteins concentrate at the leading edge of motile cells and the tips of filopodia, where they contribute to the growth of the filaments.
10. CAPPING PROTEINS
Capping protein (Edwards et al. 2014) is a heterodimer of structurally similar α- and β-subunits that binds tightly to actin filament barbed ends (Fig. 8). Virtually all eukaryotic cells express capping protein. Given micromolar concentrations of capping protein in cells, barbed ends are capped in seconds—and remain capped as the half-time for dissociation is 30 min. Capping protein cooperates with profilin to maintain the actin monomer pool, limit the number of barbed ends available for growth during actin-based protrusion of the leading edge, and stabilize the barbed ends of filaments in the Z-disk of striated muscles.
Figure 8.
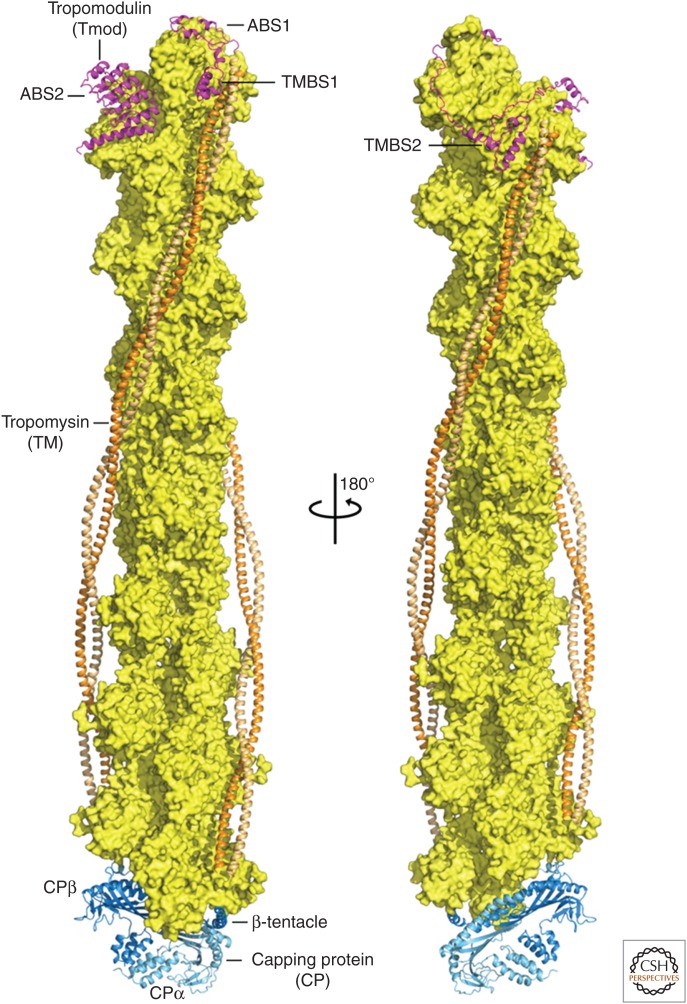
Capping of the two ends of the actin filament. Depicted is a space-filling model of a short filament with two laterally aligned tropomyosin molecules (orange), terminating with tropomodulin at the filament pointed end (magenta) (Rao et al. 2014) and heterodimeric capping protein (CP; cyan and blue) at the filament barbed end (Urnavicius et al. 2015). (Figure prepared by Roberto Dominguez of the University of Pennsylvania.)
Capping protein is constitutively active, but is regulated allosterically by proteins that are generally unrelated but contain a capping protein interaction motif, including “capping protein Arp2/3 myosin I linker” (CARMIL). Other molecules that regulate capping protein do so by sterically inhibiting its binding to the barbed end (Edwards et al. 2014). Polyphosphoinositides bind capping protein and block its interaction with barbed ends but do not dissociate capping protein from the end of a filament. V-1/myotrophin sequesters capping protein in the cytoplasm of animal cells, whereas CARMIL allows capping protein to associate weakly with barbed ends and slow elongation.
As discussed above, members of the gelsolin family are calcium-regulated proteins that not only sever filaments but also bind to barbed ends (Nag et al. 2013). This capping activity allows gelsolin and capping protein to nucleate filaments that grow from their pointed ends, as observed in skeletal muscles (Littlefield et al. 2001).
Tropomodulin is exclusively a pointed-end capping protein (Rao et al. 2014). The protein wraps around the three terminal subunits at the pointed end and blocks subunit addition and loss (Fig. 8). Interactions with the amino-terminal ends of two tropomyosin molecules strengthen the capping activity. These interactions stabilize the pointed end of the thin filaments in muscle but still allow slow exchange of actin subunits. A shorter isoform of tropomodulin caps the pointed ends of the tiny actin filaments in the spectrin–actin network of red blood cells.
Arp2/3 complex also caps pointed ends in the act of nucleating actin filament branches that grow at their free barbed ends. When a branch dissociates from the side of a mother filament (spontaneously or promoted by cofilin), Arp2/3 complex caps the pointed end of the former daughter filament.
11. CROSS-LINKING PROTEINS
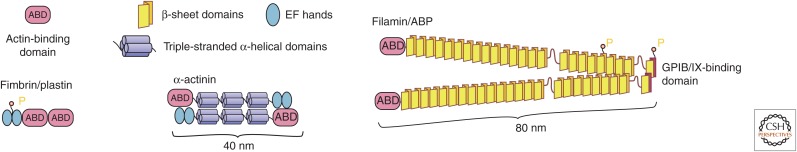
Physical connections between actin filaments made by a large family of cross-linking proteins (Matsudaira 1994) stabilize higher-order structures, such as bundles of filaments in microvilli, filopodia, and cytoplasmic cables, as well as networks of actin filaments (see Svitkina 2016). Two actin-binding sites in the same polypeptide or in two subunits of oligomeric proteins are required to connect two filaments (Fig. 9). The actin-binding domains (ABDs) of many of these proteins consist of two calponin-homology domains (Borrego-Diaz et al. 2006), but the distance between the pairs of ABDs varies considerably. The tandem ABDs of fimbrin and the twofold symmetry of fascin promote the formation of actin filament bundles, whereas the widely separated ABDs of filamin (ABP) cross-link less-organized networks of filaments, such as those at the leading edge of motile cells (Matsudaira 1994; see Svitkina 2016).
Figure 9.
Cross-linking proteins. Illustrations depicting the size, domains, and organization of the actin filament cross-linking proteins fimbrin, α-actinin, and filamin (ABP). The different patterns of distribution of the actin-binding domains lend the proteins distinct cross-linking capabilities. ABD, actin-binding domain; EF hand, calcium-binding motif. (Adapted from Pollard and Earnshaw 2007, with permission from Elsevier.)
ABDs typically have relatively low affinity for actin filaments (Kd ∼ 10 µm), so they exchange on and off filaments on a subsecond timescale. Rapid exchange of these linking proteins makes the mechanical properties of actin filament networks much different from those of covalently cross-linked synthetic polymers (Yao et al. 2011). When deformed rapidly, cross-linked actin networks are stiff, but the rapidly rearranging cross-links do not resist slow deformation (Xu et al. 1998). This explains why cells are stiff and elastic on fast timescales but deformable on timescales of tens of seconds.
12. FILAMENT-BINDING PROTEINS
The coiled-coil protein tropomyosin binds along each of the two long-pitch helices of the actin filament (Fig. 8) (von der Ecken et al. 2014). Genes for one or more tropomyosins are present in fungi and animals but not amoebas, plants, or other types of eukaryotes. Tropomyosin protects filaments from severing by cofilin (Maciver et al. 1991) and influences which myosins interact with a filament (Pollard and Lord 2014). In yeast, particular tropomyosin isoforms associate with filaments produced by different formins. In striated muscles, tropomyosin and troponin comprise the calcium-sensitive switch that regulates contraction (see Sweeney 2016).
13. CONCLUDING REMARKS
Evolution has produced a system of proteins that use actin subunits to build a rich array of different structures in prokaryotes and eukaryotes. These range from the stable sarcomeres of striated muscles (Sweeney 2016) to force-producing branched networks at the leading edges of motile cells that turn over in seconds (Svitkina 2016). Appreciating the mechanisms of the modest number of regulatory proteins covered in this review will allow the reader to understand actin filament dynamics in all of the systems described in this collection.
ACKNOWLEDGMENTS
The author’s research reported in this publication was supported by National Institute of General Medical Sciences of the National Institutes of Health under awards number R01GM026132, R01GM026338, and PO1GM066311. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health. The author thanks Roberto Dominguez for many valuable suggestions and Figure 8, and Qian Chen, Naomi Courtemanche, and Shalini Nag for comments on a draft of this review.
Footnotes
Editors: Thomas D. Pollard and Robert D. Goldman
Additional Perspectives on The Cytoskeleton available at www.cshperspectives.org
REFERENCES
*Reference is in this collection.
- Alberts AS. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J Biol Chem 276: 2824–2830. [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E, Pollard TD. 2006. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell 24: 13–23. [DOI] [PubMed] [Google Scholar]
- Archer SJ, Vinson VK, Pollard TD, Torchia DA. 1994. Elucidation of the poly-l-proline binding site in Acanthamoeba profilin-I by NMR spectroscopy. FEBS Lett 337: 145–151. [DOI] [PubMed] [Google Scholar]
- Baker JL, Courtemanche N, Parton DL, McCullagh M, Pollard TD, Voth GA. 2015. Electrostatic interactions between the Bni1p formin FH2 domain and actin influence actin filament nucleation. Structure 23: 68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamburg JR, Harris HE, Weeds AG. 1980. Partial purification and characterization of an actin depolymerizing factor from brain. FEBS Lett 121: 178–182. [DOI] [PubMed] [Google Scholar]
- *.Barlan K, Gelfand VI. 2016. Microtubule-based transport and the distribution, tethering, and organization of organelles. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a025817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. 2008. Pathway of actin filament branch formation by Arp2/3 complex. J Biol Chem 283: 7135–7144. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. 1998. Interaction of actin monomers with Acanthamoeba actophorin (ADF/cofilin) and profilin. J Biol Chem 273: 25106–25111. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. 1999. Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J Biol Chem 274: 15538–15546. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Pollard TD. 2002. Hydrolysis of ATP by polymerized actin depends on the bound divalent cation but not profilin. Biochemistry 41: 597–602. [DOI] [PubMed] [Google Scholar]
- Borrego-Diaz E, Kerff F, Lee SH, Ferron F, Li Y, Dominguez R. 2006. Crystal structure of the actin-binding domain of α-actinin 1: Evaluating two competing actin-binding models. J Struct Biol 155: 230–238. [DOI] [PubMed] [Google Scholar]
- Bravo-Cordero JJ, Magalhaes MA, Eddy RJ, Hodgson L, Condeelis J. 2013. Functions of cofilin in cell locomotion and invasion. Nat Rev Mol Cell Biol 14: 405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. 2010. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol 11: 237–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Goodarzi JP, De La Cruz EM. 2006. Energetics and kinetics of cooperative cofilin–actin filament interactions. J Mol Biol 361: 257–267. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. 1986. Direct evidence for ADP-Pi-F-actin as the major intermediate in ATP-actin polymerization. Rate of dissociation of Pi from actin filaments. Biochemistry 25: 7789–7792. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Laurent V, Santolini J, Melki R, Didry D, Xia GX, Hong Y, Chua NH, Pantaloni D. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: Implication in actin-based motility. J Cell Biol 136: 1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C, Beltzner CC, Pollard TD. 2009. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Curr Biol 19: 537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK. 2010. Structure and control of the actin regulatory WAVE complex. Nature 468: 533–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Courtemanche N, Pollard TD. 2015. Aip1 promotes actin filament severing by cofilin and regulates the constriction of the cytokinetic contractile ring. J Biol Chem 290: 2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. 2005. Actin-bound structures of Wiskott–Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci 102: 16644–16649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D, Boczkowska M, Skwarek-Maruszewska A, Fujiwara I, Hayes DB, Rebowski G, Lappalainen P, Pollard TD, Dominguez R. 2008. Leiomodin is an actin filament nucleator in muscle cells. Science 320: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JA, Buhle EL Jr, Walker SB, Tsong TY, Pollard TD. 1983. Kinetic evidence for a monomer activation step in actin polymerization. Biochemistry 22: 2193–2202. [DOI] [PubMed] [Google Scholar]
- Courtemanche N, Pollard TD. 2013. Interaction of profilin with the barbed end of actin filaments. Biochemistry 52: 6456–6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang I, Gorelik R, Sousa-Blin C, Derivery E, Guérin C, Linkner J, Nemethova M, Dumortier JG, Giger FA, Chipysheva TA, et al. 2013. Inhibitory signalling to the Arp2/3 complex steers cell migration. Nature 503: 281–284. [DOI] [PubMed] [Google Scholar]
- De La Cruz EM, Pollard TD. 1995. Nucleotide-free actin: Stabilization by sucrose and nucleotide binding kinetics. Biochemistry 34: 5452–5461. [DOI] [PubMed] [Google Scholar]
- Derman AI, Becker EC, Truong BD, Fujioka A, Tucey TM, Erb ML, Patterson PC, Pogliano J. 2009. Phylogenetic analysis identifies many uncharacterized actin-like proteins (Alps) in bacteria: Regulated polymerization, dynamic instability and treadmilling in Alp7A. Mol Microbiol 73: 534–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. 2004. Actin-binding proteins—A unifying hypothesis. Trends Biochem Sci 29: 572–578. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC. 2011. Actin structure and function. Annu Rev Biophys 40: 169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. 2014. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol 15: 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam WA, Kang H, De La Cruz EM. 2013. Biophysics of actin filament severing by cofilin. FEBS Lett 587: 1215–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. 2007. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J 26: 4597–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieden C. 1983. Polymerization of actin: Mechanism of the Mg2+-induced process at pH 8 and 20°C. Proc Natl Acad Sci 80: 6513–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Iwane AH, Yanagida T, Namba K. 2010. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature 467: 724–728. [DOI] [PubMed] [Google Scholar]
- Fujiwara I, Vavylonis D, Pollard TD. 2007. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc Natl Acad Sci 104: 8827–8832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Kudryashov DS, Solodukhin A, Reisler E, Schröder GF, Egelman EH. 2011. Remodeling of actin filaments by ADF/cofilin proteins. Proc Natl Acad Sci 108: 20568–20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Glotzer M. 2016. Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode BL, Eck MJ. 2007. Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 76: 593–627. [DOI] [PubMed] [Google Scholar]
- *.Goodson HV, Jonasson EM. 2016. Microtubules and MAPs. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DJ, Eisenberg E, Korn ED. 1976. Characterization of cytoplasmic actin isolated from Acanthamoeba castellanii by a new method. J Biol Chem 251: 4778–4786. [PubMed] [Google Scholar]
- Gunning PW, Ghoshdastider U, Whitaker S, Popp D, Robinson RC. 2015. The evolution of compositionally and functionally distinct actin filaments. J Cell Sci 128: 2009–2019. [DOI] [PubMed] [Google Scholar]
- Gurel PS, Ge P, Grintsevich EE, Shu R, Blanchoin L, Zhou ZH, Reisler E, Higgs HN. 2014. INF2-mediated severing through actin filament encirclement and disruption. Curr Biol 24: 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. 2010. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol 191: 571–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris ES, Li F, Higgs HN. 2004. The mouse formin, FRLα, slows actin filament barbed end elongation, competes with capping protein, accelerates polymerization from monomers, and severs filaments. J Biol Chem 279: 20076–20087. [DOI] [PubMed] [Google Scholar]
- Herman IM. 1993. Actin isoforms. Curr Opin Cell Biol 5: 48–55. [DOI] [PubMed] [Google Scholar]
- Higashida C, Miyoshi T, Fujita A, Oceguera-Yanez F, Monypenny J, Andou Y, Narumiya S, Watanabe N. 2004. Actin polymerization-driven molecular movement of mDia1 in living cells. Science 303: 2007–2010. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Peterson KJ. 2005. Phylogenetic analysis of the formin homology 2 domain. Mol Biol Cell 16: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley HE. 1963. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol 7: 281–308. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Stossel TP. 1987. Modulation of gelsolin function by phosphatidylinositol 4,5-biphosphate. Nature 325: 362–364. [DOI] [PubMed] [Google Scholar]
- Jégou A, Niedermayer T, Orbán J, Didry D, Lipowsky R, Carlier MF, Romet-Lemonne G. 2011. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol 9: e1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AB, Collins A, Goode BL. 2015. High-speed depolymerization at actin filament ends jointly catalysed by Twinfilin and Srv2/CAP. Nat Cell Biol 17: 1504–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. 1990. Atomic structure of the actin:DNase I complex. Nature 347: 37–44. [DOI] [PubMed] [Google Scholar]
- Kang H, Bradley MJ, Elam WA, De La Cruz EM. 2013. Regulation of actin by ion-linked equilibria. Biophys J 105: 2621–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn ED, Carlier MF, Pantaloni D. 1987. Actin polymerization and ATP hydrolysis. Science 238: 638–644. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Pollard TD. 2004. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci 101: 14725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. 2006. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell 124: 423–435. [DOI] [PubMed] [Google Scholar]
- Kuhn JR, Pollard TD. 2005. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J 88: 1387–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R, Almenar-Queralt A, Fowler VM. 2001. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat Cell Biol 3: 544–551. [DOI] [PubMed] [Google Scholar]
- Lu J, Pollard TD. 2001. Profilin binding to poly-l-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell 12: 1161–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciver SK, Zot HG, Pollard TD. 1991. Characterization of actin filament severing by actophorin from Acanthamoeba castellanii. J Cell Biol 115: 1611–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsudaira P. 1994. Actin crosslinking proteins at the leading edge. Semin Cell Biol 5: 165–174. [DOI] [PubMed] [Google Scholar]
- McCullagh M, Saunders MG, Voth GA. 2014. Unraveling the mystery of ATP hydrolysis in actin filaments. J Am Chem Soc 136: 13053–13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough BR, Grintsevich EE, Chen CK, Kang H, Hutchison AL, Henn A, Cao W, Suarez C, Martiel JL, Blanchoin L, et al. 2011. Cofilin-linked changes in actin filament flexibility promote severing. Biophys J 101: 151–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean-Fletcher S, Pollard TD. 1980. Identification of a factor in conventional muscle actin preparations which inhibits actin filament self association. Biochem Biophys Res Commun 96: 18–27. [DOI] [PubMed] [Google Scholar]
- Mizuno K. 2013. Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation. Cell Signal 25: 457–469. [DOI] [PubMed] [Google Scholar]
- Mockrin SC, Korn RD. 1980. Acanthamoeba profilin interacts with G-actin to increase the rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry 19: 5359–5362. [DOI] [PubMed] [Google Scholar]
- Muller J, Oma Y, Vallar L, Friederich E, Poch O, Winsor B. 2005. Sequence and comparative genomic analysis of actin-related proteins. Mol Biol Cell 16: 5736–5748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, Larsson M, Robinson RC, Burtnick LD. 2013. Gelsolin: The tail of a molecular gymnast. Cytoskeleton 70: 360–384. [DOI] [PubMed] [Google Scholar]
- Ngo KX, Kodera N, Katayama E, Ando T, Uyeda TQ. 2015. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. Elife 4: e04806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida E. 1985. Opposite effects of cofilin and profilin from porcine brain on rate of exchange of actin-bound adenosine 5′-triphosphate. Biochemistry 24: 1160–1164. [DOI] [PubMed] [Google Scholar]
- Nolen BJ, Tomasevic N, Russell A, Pierce DW, Jia Z, McCormick CD, Hartman J, Sakowicz R, Pollard TD. 2009. Characterization of two classes of small molecule inhibitors of Arp2/3 complex. Nature 460: 1031–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. 2009. Nature of the globular to fibrous actin transition. Nature 457: 441–445. [DOI] [PubMed] [Google Scholar]
- Ohki T, Ohno C, Oyama K, Mikhailenko SV, Ishiwata S. 2009. Purification of cytoplasmic actin by affinity chromatography using the C-terminal half of gelsolin. Biochem Biophys Res Commun 383: 146–150. [DOI] [PubMed] [Google Scholar]
- Oma Y, Harata M. 2011. Actin-related proteins localized in the nucleus: From discovery to novel roles in nuclear organization. Nucleus 2: 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosawa F, Asakura S. 1975. Thermodynamics of the polymerization of protein. Academic, New York. [Google Scholar]
- Otomo T, Tomchick DR, Otomo C, Panchal SC, Machius M, Rosen MK. 2005. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433: 488–494. [DOI] [PubMed] [Google Scholar]
- Paavilainen VO, Oksanen E, Goldman A, Lappalainen P. 2008. Structure of the actin-depolymerizing factor homology domain in complex with actin. J Cell Biol 182: 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK. 2011. Arp2/3 complex is bound and activated by two WASP proteins. Proc Natl Acad Sci 108: E472–E479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaloni D, Carlier MF. 1993. How profilin promotes actin filament assembly in the presence of thymosin β4. Cell 75: 1007–1014. [DOI] [PubMed] [Google Scholar]
- Paredez AR, Assaf ZJ, Sept D, Timofejeva L, Dawson SC, Wang CJ, Cande WZ. 2011. An actin cytoskeleton with evolutionarily conserved functions in the absence of canonical actin-binding proteins. Proc Natl Acad Sci 108: 6151–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul A, Pollard TD. 2009. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton 66: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD. 1986. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J Cell Biol 103: 2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Earnshaw WC. 2007. Cell biology, 2nd ed Saunders, New York. [Google Scholar]
- Pollard LW, Lord M. 2014. Getting myosin-V on the right track: Tropomyosin sorts transport in yeast. Bioarchitecture 4: 35–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Weeds AG. 1984. The rate constant for ATP hydrolysis by polymerized actin. FEBS Lett 170: 94–98. [DOI] [PubMed] [Google Scholar]
- Poukkula M, Kremneva E, Serlachius M, Lappalainen P. 2011. Actin-depolymerizing factor homology domain: A conserved fold performing diverse roles in cytoskeletal dynamics. Cytoskeleton 68: 471–490. [DOI] [PubMed] [Google Scholar]
- Pring M, Evangelista M, Boone C, Yang C, Zigmond SH. 2003. Mechanism of formin-induced nucleation of actin filaments. Biochemistry 42: 486–496. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. 2005. Drosophila Spire is an actin nucleation factor. Nature 433: 382–388. [DOI] [PubMed] [Google Scholar]
- Rao JN, Madasu Y, Dominguez R. 2014. Mechanism of actin filament pointed-end capping by tropomodulin. Science 345: 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebowski G, Namgoong S, Boczkowska M, Leavis PC, Navaza J, Dominguez R. 2010. Structure of a longitudinal actin dimer assembled by tandem W domains: Implications for actin filament nucleation. J Mol Biol 403: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. 2001. Crystal structure of Arp2/3 complex. Science 294: 1679–1684. [DOI] [PubMed] [Google Scholar]
- Rosenblatt J, Peluso P, Mitchison TJ. 1995. The bulk of unpolymerized actin in Xenopus egg extracts is ATP-bound. Mol Biol Cell 6: 227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottner K, Hanisch J, Campellone KG. 2010. WASH, WHAMM, and JMY: Regulation of Arp2/3 complex and beyond. Trends Cell Biol 20: 650–661. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Biol 14: 7–12. [DOI] [PubMed] [Google Scholar]
- Rouiller I, Xu XP, Amann KJ, Egile C, Nickell S, Nicastro D, Li R, Pollard TD, Volkmann N, Hanein D. 2008. The structural basis of actin filament branching by the Arp2/3 complex. J Cell Biol 180: 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safer D, Elzinga M, Nachmias VT. 1991. Thymosin β4 and Fx, an actin-sequestering peptide, are indistinguishable. J Biol Chem 266: 4029–4032. [PubMed] [Google Scholar]
- Schafer DA, Jennings PB, Cooper JA. 1998. Rapid and efficient purification of actin from nonmuscle sources. Cell Motil Cytoskeleton 39: 166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. 1993. The structure of crystalline profilin–β-actin. Nature 365: 810–816. [DOI] [PubMed] [Google Scholar]
- Sept D, McCammon JA. 2001. Thermodynamics and kinetics of actin filament nucleation. Biophys J 81: 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BA, Padrick SB, Doolittle LK, Daugherty-Clarke K, Corrêa IR Jr, Xu MQ, Goode BL, Rosen MK, Gelles J. 2013. Three-color single molecule imaging shows WASP detachment from Arp2/3 complex triggers actin filament branch formation. Elife 2: e01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon JP, Page LJ, Balch WE, Kelly JW. 2012. Gelsolin amyloidosis: Genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit Rev Biochem Mol Biol 47: 282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Saw JH, Jørgensen SL, Zaremba-Niedzwiedzka K, Martijn J, Lind AE, van Eijk R, Schleper C, Guy L, Ettema TJ. 2015. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature 521: 173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Svitkina TM. 2016. Actin cytoskeleton and actin-based motility. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney L. 2016. Muscle contraction. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a023200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sweeney L, Holzbaur E. 2016. Motor proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S-C, Pollard TD. 2011. Purification of actin from fission yeast S. pombe and characterization of functional differences from muscle actin. J Biol Chem 286: 5784–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S-C, Jurgenson C, Nolen BJ, Pollard TD. 2011. Structural and biochemical characterization of two binding sites for nucleation promoting factor WASp-VCA on Arp2/3 complex. Proc Natl Acad Sci 108: E463–E471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Titus MA. 2016. Myosin-driven intracellular transport. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a021972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnavicius L, Zhang, Diamant AG, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. 2015. The K structure of the dynactin complex and its interaction with dynein. Science 347: 1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. 2006. Model of formin-associated actin filament elongation. Mol Cell 21: 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson VK, De La Cruz EM, Higgs HN, Pollard TD. 1998. Interactions of Acanthamoeba profilin with actin and nucleotides bound to actin. Biochemistry 37: 10871–10880. [DOI] [PubMed] [Google Scholar]
- von der Ecken J, Müller M, Lehman W, Manstein DJ, Penczek PA, Raunser S. 2014. Structure of the F-actin-tropomyosin complex. Nature 519: 114–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner AR, Luan Q, Liu SL, Nolen BJ. 2013. Dip1 defines a class of Arp2/3 complex activators that function without preformed actin filaments. Curr Biol 23: 1990–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Karginov AV, Kinley AW, Weed SA, Li Y, Parsons JT, Cooper JA. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr Biol 11: 370–374. [DOI] [PubMed] [Google Scholar]
- Wegner A. 1976. Head to tail polymerization of actin. J Mol Biol 108: 139–150. [DOI] [PubMed] [Google Scholar]
- Xu J, Wirtz D, Pollard TD. 1998. Dynamic cross-linking by α-actinin determines the mechanical properties of actin filament networks. J Biol Chem 273: 9570–9576. [DOI] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, Poy F, Pellman D, Goode BL, Eck MJ. 2004. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell 116: 711–723. [DOI] [PubMed] [Google Scholar]
- Xue B, Leyrat C, Grimes JM, Robinson RC. 2014. Structural basis of thymosin-β4/profilin exchange leading to actin filament polymerization. Proc Natl Acad Sci 111: E4596–E4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao NY, Becker DJ, Broedersz CP, Depken M, Mackintosh FC, Pollak MR, Weitz DA. 2011. Nonlinear viscoelasticity of actin transiently cross-linked with mutant α-actinin-4. J Mol Biol 411: 1062–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ydenberg CA, Padrick SB, Sweeney MO, Gandhi M, Sokolova O, Goode BL. 2013. GMF severs actin-Arp2/3 complex branch junctions by a cofilin-like mechanism. Curr Biol 23: 1037–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]