Abstract
The p53 pathway is perturbed in the majority of human cancers. Although this most frequently occurs through the direct mutation or deletion of p53 itself, there are a number of other alterations that can attenuate the pathway and contribute to tumorigenesis. For example, amplification of important negative regulators, MDM2 and MDM4, occurs in a number of cancers. In this work, we will review both the normal regulation of the p53 pathway and the different mechanisms of pathway inhibition in cancer, discuss these alterations in the context of the global genomic analyses that have been conducted across tumor types, and highlight the translational implications for cancer diagnosis and treatment.
The p53 tumor suppressor pathway is inactivated in many human cancers. This is usually caused by mutations in p53 itself, but other mechanisms (e.g., overexpression of the negative regulators MDM2 and MDM4) also contribute.
Inactivation of the p53 pathway is a defining feature of human cancers, with nearly all cancers evolving a way to circumvent this essential tumor-suppressive mechanism. Although a large number of human cancers directly inactivate p53 through mutations or deletions of the TP53 (p53) locus (Hainaut and Pfeifer 2016), there are a vast number of other molecular alterations that can functionally serve to attenuate the pathway. Through studies using mouse models, it has become clear that even small changes that perturb p53 levels or activity can have profound implications for tumor development (Eischen and Lozano 2014). A thorough understanding of the mechanisms of p53-pathway regulation and the biological outcomes downstream from p53 activation combined with the recent cancer genomic studies is needed to appreciate how extensively this pathway is perturbed in human cancers. Additionally, this knowledge has and continues to provide novel opportunities to develop therapeutic strategies to overcome the inactivation of p53 in both prevention and treatment of human malignancies.
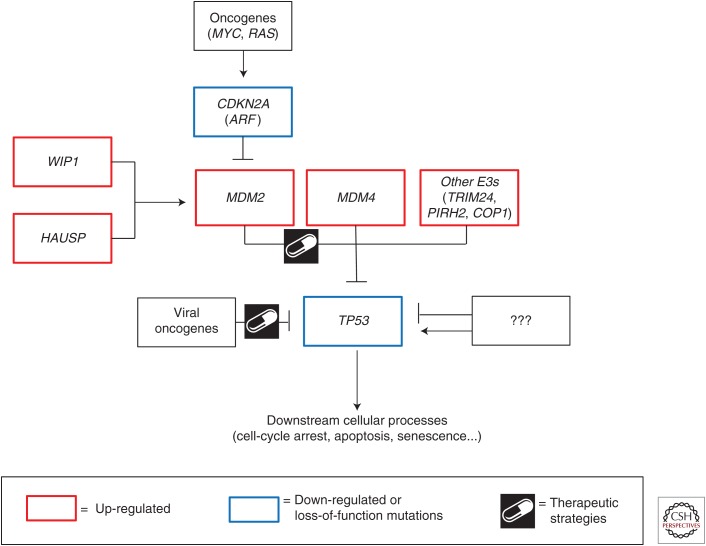
As an extensive discussion of the p53 pathway and the downstream cellular consequences is provided in other articles within this collection, only a brief recap is required to provide the contextual framework for the discussions that follow (Fig. 1). p53 is widely acknowledged as the guardian of the genome (Lane 1992). In normal, unstressed cells, p53 levels and transcriptional activity are kept in check by important negative regulators. In response to a variety of cellular stresses, including DNA damage, oncogene activation, and oxidative stress, posttranslational mechanisms stabilize and activate p53. As a transcription factor, p53 can then bind specific promoters and regulate the expression of genes that drive cell-cycle arrest, apoptosis, senescence, and several other cellular functions discussed in this collection. Ultimately, these activities preserve the fidelity and integrity of the cell, and are important tumor-suppressive mechanisms.
Figure 1.
The p53 pathway. Diagrammatic representation of effectors (positive and negative) of the p53 pathway discussed in this review.
ALTERATIONS IN DIRECT p53 REGULATORS
MDM2
One effective mechanism to attenuate p53 activity is through overexpression of important negative regulators. The most prominent and well studied of these negative regulators is MDM2. Mdm2 was first identified from the murine tumorigenic cell line 3T3-DM. This cell line shows an amplified genomic region in the form of double minute chromosomes, and Mdm2 (murine double minute 2) was found to be the transforming gene contained within this amplicon (Cahilly-Snyder et al. 1987; Fakharzadeh et al. 1991). Mechanistically, MDM2 contributes to cellular transformation through interaction with p53 and inhibition of its transcriptional activity (Momand et al. 1992; Oliner et al. 1993; Wu et al. 1993; Zauberman et al. 1993). Further studies showed that MDM2 is an E3 ubiquitin ligase that directly ubiquitinates and targets the p53 protein for proteosomal degradation (Haupt et al. 1997; Honda et al. 1997; Kubbutat et al. 1997).
The essential role of Mdm2 as a negative regulator of p53 was highlighted in mouse models. Mdm2-null mice are early embryonic lethal, losing viability preimplantation (Jones et al. 1995; Montes de Oca Luna et al. 1995; Chavez-Reyes et al. 2003). High levels of apoptosis, as measured by TUNEL staining in blastocysts, indicate that loss of Mdm2 drives inappropriate p53 activation and subsequent cell death. Importantly, the Mdm2-null embryonic lethality is rescued by the concomitant deletion of p53, demonstrating that the cell death phenotype is a result of deregulated p53 activity. This relationship has also been investigated in adult tissues of the mouse. Tissue-specific loss of Mdm2 in the smooth muscle and epithelial cells of the gastrointestinal system (Boesten et al. 2006; Valentin-Vega et al. 2009), cardiomyocytes (Grier et al. 2006), neuronal progenitor cells (Francoz et al. 2006; Xiong et al. 2006), or hepatocytes (Kodama et al. 2011) all lead to p53-dependent cell death. Two different models have been used to evaluate the effect of unrestricted p53 activity (via Mdm2 loss) throughout the adult mouse, both demonstrating p53-dependent apoptosis and tissue atrophy in classically radiosensitive tissues (spleen, bone marrow, and intestine), as well as generally radioinsensitive tissues (kidney, liver, heart, retina, and hippocampus) (Ringshausen et al. 2006; Zhang et al. 2014). Further, Mdm2 hypomorphic mice show p53-dependent pathologies (Mendrysa et al. 2003). Combined, these data clearly establish MDM2 as an essential negative regulator of the p53 pathway in multiple tissues in vivo.
Physiologically, the p53 pathway is important in response to stress and one of the primary mechanisms of p53 stabilization is through dissociation from MDM2. Specifically, the DNA damage-dependent phosphorylation of p53 and MDM2 inhibits the interaction between these two proteins allowing for p53-dependent activation of downstream transcriptional targets (Canman et al. 1998; Khosravi et al. 1999; Chehab et al. 2000; Hay and Meek 2000). Further, an important safety mechanism built into the system prevents the negative consequences of prolonged p53 activation. In addition to the genes that drive cell-cycle arrest and apoptosis, p53 also transcriptionally activates the expression of MDM2 by binding two p53-response elements within the P2 promoter, subsequently ensuring its own degradation (Barak et al. 1993; Juven et al. 1993; Wu et al. 1993; Wu and Levine 1997). A mouse model with mutations that prevent p53 binding and transactivation of the Mdm2 promoter is viable under normal physiological conditions (Pant et al. 2013). However, mice show enhanced radiosensitivity, and die in response to sublethal doses of radiation caused by hematopoietic failure as a result of prolonged p53 activity in the bone marrow. Combined, all of the above-described genetic experiments have established MDM2 as an essential negative regulator of p53 during development, normal tissue homeostasis, and DNA damage response.
Mouse models have also been generated to evaluate the physiological and pathological consequences of Mdm2 overexpression. A transgenic mouse model was established, in which two- to fourfold increased levels of Mdm2 were expressed under the control of the endogenous Mdm2 promoter (Jones et al. 1998). Transgene expression is sufficient to drive tumorigenesis in mice, with half of animals hemizygous for the transgene developing tumors by 84 weeks of age. Similar to p53+/− or p53−/−, Mdm2Tg mice primarily develop lymphomas and sarcomas. It is, however, interesting to note that there is a marked increase in the number of sarcomas formed in Mdm2Tgp53−/− mice compared with p53−/−, suggesting that there may be p53-independent roles for Mdm2 in promoting transformation. Mdm2Tg mice have not yet been characterized on a p53+/− background to evaluate the impact of the Mdm2Tg on loss of heterozygosity (LOH) of the p53 locus or to further interrogate potential p53-independent mechanisms contributing to tumorigenesis.
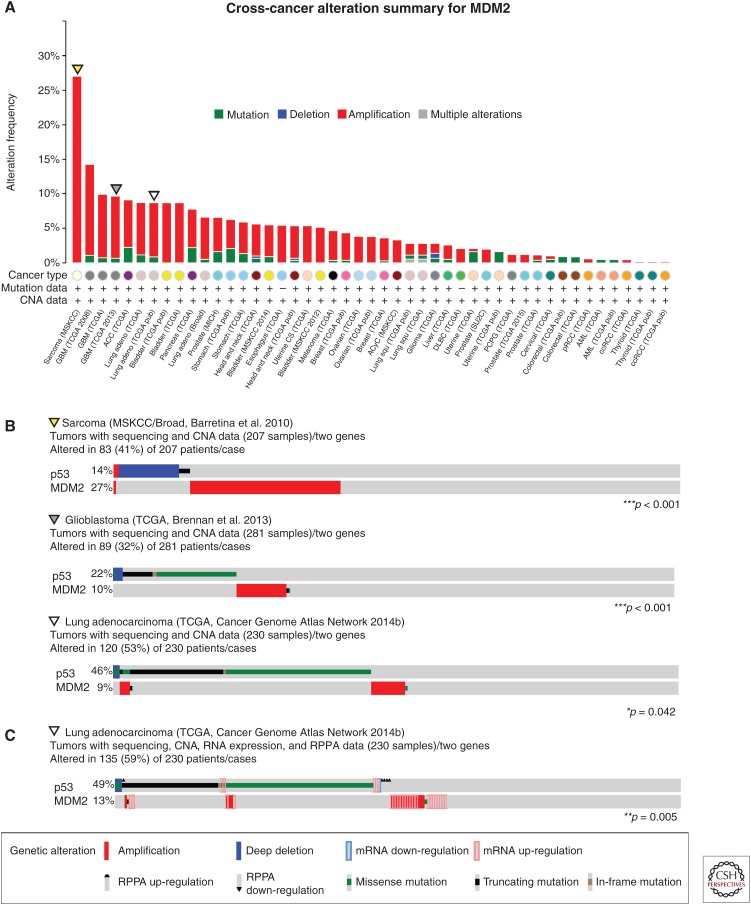
Given the above studies, it is therefore not surprising that one common mechanism by which human cancers abrogate p53 is through overexpression of MDM2. Soon after the identification of MDM2, it was appreciated that a large proportion of human sarcomas amplify the MDM2 locus (Oliner et al. 1992). In recent years, large-scale cancer genome profiling has provided a catalog of all of the somatic alterations that occur across a large number and broad range of cancer types, and the cBioPortal for Cancer Genomics (www.cbioportal.org) is a valuable tool for visualizing and analyzing these cancer genomics data sets (Cerami et al. 2012; Gao et al. 2013). Combined, these studies have highlighted the high levels of MDM2 amplification in sarcomas and revealed several other tumor types that show frequent amplification of the MDM2 locus (Fig. 2A). Specifically, MDM2 is most frequently amplified in sarcomas (27.1%) (Barretina et al. 2010), glioblastoma (13.2% and 8.9%, in two different data sets) (Cancer Genome Atlas Network 2008; Brennan et al. 2013), bladder urothelial carcinoma (8.7%) (Cancer Genome Atlas Network 2014a), and lung adenocarcinoma (7.8%) (Cancer Genome Atlas Network 2014b). Importantly, MDM2 amplification tends to be mutually exclusive with p53 mutations, suggesting that both serve the same purpose to attenuate the p53 pathway. Specifically, the data available through the cBioPortal reveal a statistically significant mutually exclusive relationship between genetic alterations in p53 and MDM2 in the data sets mentioned above. Examples from sarcomas, glioblastomas, and lung adenocarcinomas are presented in Figure 2B. In addition, high levels of MDM2 can be achieved through transcriptional and posttranslational mechanisms. More recent genomic studies, specifically those being conducted through The Cancer Genome Atlas (TCGA), have incorporated expression studies, including RNA-sequencing and reverse phase protein arrays (RPPAs), which give a more comprehensive analysis of gene and protein expression levels in cancers. For example, adding expression data both increases the number of tumors with alterations in these two p53-pathway components in lung adenocarcinoma (52% vs. 59%) and improves the statistical significance of the mutually exclusive relationship between p53 and MDM2 lesions (p = 0.042 vs. 0.005) in these cancers (Fig. 2C) (Cancer Genome Atlas Network 2014b). Further dissection of the mechanisms that drive increased levels of MDM2 independent of gene amplification will also be important for realizing the full impact of MDM2 in tumorigenesis.
Figure 2.
MDM2 alterations in cancer. Analyses of genomic data of tumors with p53 mutation or deletion and high levels of MDM2 from data sets accessed and prepared using the cBioPortal for Cancer Genetics (www.cbioportal.org) (Cerami et al. 2012; Gao et al. 2013). (A) Frequency of MDM2 copy number alterations and mutations across tumor types. Data sets derived from cell lines or xenografts, as well as studies without copy number data, were excluded. (B) Detailed analyses of MDM2 amplification and p53 mutation of the three tumors (sarcoma, glioblastoma, and lung adenocarcinoma) with high levels of MDM2 amplification in A. Statistically significant mutually exclusive relationships between p53 and MDM2 alterations are observed in all three of these data sets. (C) Combined analysis of MDM2 amplification and RNA and protein expression data with p53 mutation in lung adenocarcinoma. CNA, Copy number alterations; RPPA, reverse phase protein array. *p < 0.05, **p < 0.01, ***p < 0.001. Data were accessed on July 16, 2015.
Single-nucleotide polymorphisms (SNPs) in the MDM2 promoter also contribute to cancer risk (Bond et al. 2005). A high-frequency SNP has been identified within the second and p53-activated P2 promoter of MDM2 at position 309. This T to G polymorphism is present in the population in the heterozygous (TG) state at a frequency of 40% and in the homozygous (GG) state at 14% (Bond et al. 2004). The G allele creates a preferential binding site for the SP1 transcription factor and subsequently results in increased levels of MDM2 (Bond et al. 2004; Knappskog and Lonning 2011). Genome-wide association studies identified significant associations between the G allele and increased tumor risk (Bond and Levine 2007; Grochola et al. 2010). To directly test this association, mouse models with homozygous SNP309 T and G alleles were developed (Post et al. 2010). When crossed to a genetic model of cancer, Mdm2SNP309G/G mice show reduced survival and an increased number with multiple tumors when compared with the Mdm2SNP309T/T animals. This SNP and potentially other germline variants, both alone and in combination, are likely to play a role in attenuating the p53 pathway in cancers.
MDM4
MDM4 is a closely related protein to MDM2 and is also an important in vivo regulator of the p53 pathway. MDM4 (originally and often referred to as MDMX) was identified in a screen for p53 interacting proteins (Shvarts et al. 1996). Unlike MDM2, however, MDM4 does not have an enzymatically active RING domain and is unable to directly target p53 for ubiquitination. It can, however, bind to and inhibit the activity of the p53 transactivation domain (Shvarts et al. 1997). Genetically engineered mouse models have served to establish the essential role for Mdm4 in regulating p53 during development, as Mdm4-null mice also show p53-dependent embryonic lethality (Parant et al. 2001). There is additional evidence to suggest that MDM4 can interact with and enhance the E3 ligase activity of MDM2. MDM4 interacts with MDM2 through its RING domain (Sharp et al. 1999; Tanimura et al. 1999), and indeed, mice with deletion of or a point mutation in the RING domain of Mdm4 are also embryonic lethal because of increased p53 activity, suggesting that this domain and its interaction with Mdm2 are essential for negatively regulating p53 activity during embryonic development (Huang et al. 2011; Pant et al. 2011).
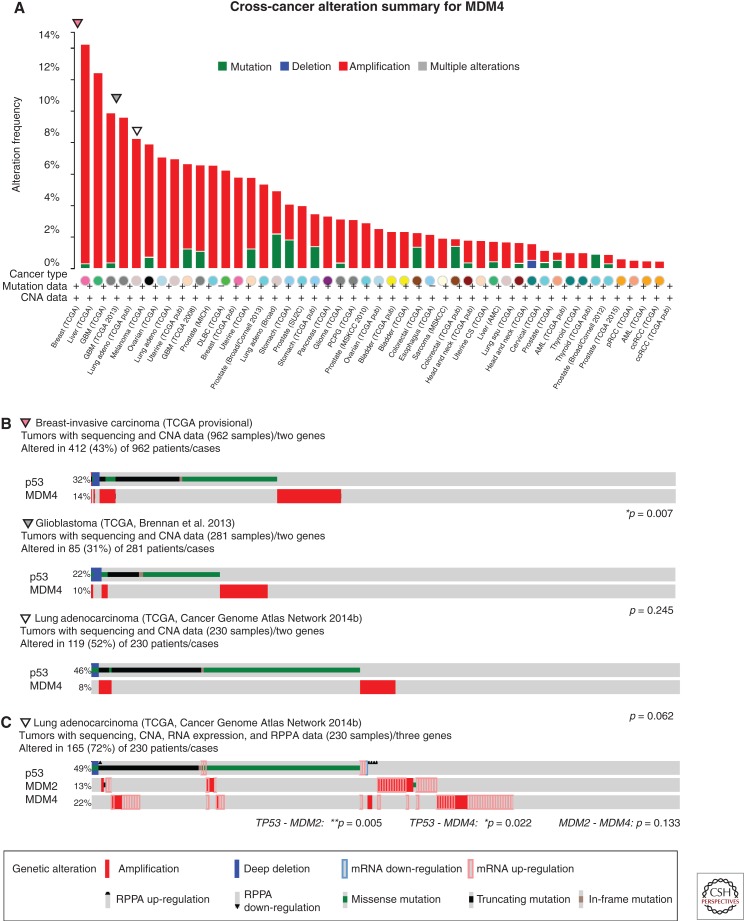
Similar to MDM2, the pathological consequences of MDM4 overexpression were modeled using transgenic mice. Although epitope-tagged Mdm4 did not promote tumorigenesis in vivo (De Clercq et al. 2010), two independent untagged transgenic lines develop spontaneous tumors, with sarcomas being the most frequent (Xiong et al. 2010). Elevated expression of MDM4 is also seen in a number of cancers. Using the cBioPortal (Cerami et al. 2012; Gao et al. 2013) to evaluate MDM4 status across available data sets (Fig. 3A) reveals the highest levels of MDM4 amplification in invasive breast carcinoma (14.2%, TCGA provisional), liver hepatocellular carcinoma (12.4%, TCGA provisional), glioblastoma (9.6%) (Brennan et al. 2013), and lung adenocarcinoma (8.3%) (Cancer Genome Atlas Network 2014b). Additionally, MDM4 amplifications occur in 65% of retinoblastomas (Laurie et al. 2006). Although MDM4 amplification or mRNA overexpression are seen in 12% of skin cutaneous melanomas from TCGA profiling (provisional), a recent study has found elevated protein expression of MDM4 in ∼65% of stage II–V melanomas (Gembarska et al. 2012). These results suggest that, although current genomic studies are very informative, they likely underestimate the fraction of tumors that have deregulated MDM4 protein expression.
Figure 3.
MDM4 alterations in cancer. Analyses of genomic data of tumors with high levels of MDM4 and p53 mutation or deletion from data sets accessed and prepared using the cBioPortal for Cancer Genetics (www.cbioportal.org) (Cerami et al. 2012; Gao et al. 2013). (A) Frequency of MDM4 copy number alterations and mutations across tumor types. Data sets derived from cells lines or xenografts, as well as studies without copy number data, were excluded. (B) Detailed analyses of MDM4 amplification and p53 mutation of the three tumors (breast, glioblastoma, and lung adenocarcinoma) with high levels of MDM4 in A. MDM4 and p53 alterations show statistically significant mutual exclusivity in breast cancer, and a noted trend in glioblastoma and lung adenocarcinoma. (C) Combined analysis of MDM2 and MDM4 amplification and overexpression data with p53 mutation status. CNA, Copy number alteration; RPPA, reverse phase protein array. *p < 0.05, **p < 0.01. Data were accessed on July 16, 2015.
Similar to the results obtained from the comparison of p53 and MDM2 lesions in cancers, p53 and MDM4 alterations show significant evidence of mutual exclusivity in invasive breast carcinoma (TCGA provisional), and clear trends in lung adenocarcinoma (Cancer Genome Atlas Network 2014b) and glioblastoma (Fig. 3B) (Brennan et al. 2013). Not all tumor types, however, show strong evidence of mutual exclusivity. In the hepatocellular carcinoma data set (TCGA provisional), approximately one-third of the tumors with MDM4 amplification also have deletions or mutations in p53. These findings suggest that MDM4 may have p53-independent functions that also contribute to tumorigenesis, or that these tumors may retain a wild-type p53 allele and the combination of p53 heterozygosity and amplified MDM4 further dampens the p53 pathway (Eischen and Lozano 2014). Again, the data are further enriched when RNA and protein expression are also considered. The relationship between the status of p53, MDM2, and MDM4 in lung adenocarcinoma (Cancer Genome Atlas Network 2014b) is shown in Figure 3C, with strong mutual exclusivity being shown among all genes. Combined, these data show that overexpression of MDM4 is another mechanism through which tumors can inactivate the p53 pathway.
Other E3 Ubiquitin Ligases
Although mouse models have provided definitive in vivo evidence that MDM2 and MDM4 are essential negative regulators of p53, a number of other E3 ubiquitin ligases have been identified to regulate p53 in vitro. Several reviews have been written describing these other regulators (Jain and Barton 2010; Lee and Gu 2010; Hock and Vousden 2014; Pant and Lozano 2014). As small changes to levels or activity of p53 impact tumor suppression in mice, increased expression of these enzymes has the potential to attenuate p53 and contribute to tumorigenesis. Herein, we will focus on the tumor-specific up-regulation of three of the more prominent E3 enzymes: PIRH2, COP1, and TRIM24.
Before discussion of each of these regulators individually, however, there are a few salient points relevant to all of them. First, it is important to consider that these enzymes likely all have tissue, stimulus, and context-specific activities. Although p53 is the dominant substrate of MDM2, the other E3 enzymes are all known to have multiple substrates, including some oncogenes. Copy number alterations or mutations, therefore, are likely to be influenced by a balance of the effects on all substrates in a given cellular context. Second, although knockout mouse models have not provided convincing evidence of p53 regulation during development (Hakem et al. 2011; Migliorini et al. 2011; Jiang et al. 2015), studies have shown strong evidence of regulation in vitro. The in vivo models to date have modeled loss of these enzymes, not overexpression, which would be expected to better mimic the pathological state for a negative regulator of p53 during tumorigenesis. Thus, as even partial inhibition of p53 can impact tumorigenesis, it is therefore important to consider the potential impact of the overexpression of these enzymes.
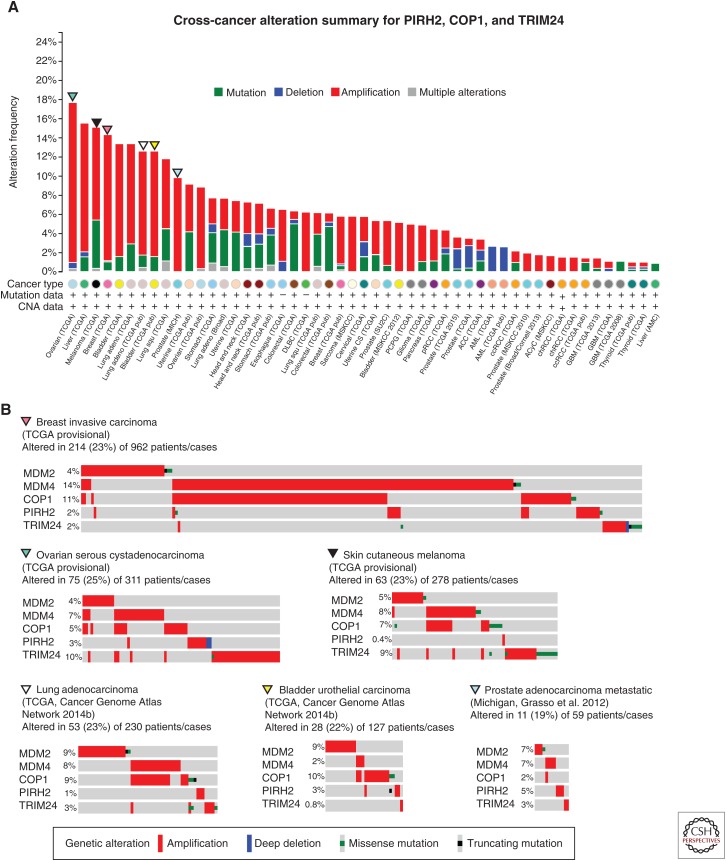
PIRH2 (official gene name, RCHY1) was first identified as a p53-activated target gene and was subsequently shown to interact with and target p53 for degradation (Leng et al. 2003). PIRH2 amplifications are seen in some tumors, albeit less frequently than MDM2 or MDM4. Specifically, the most frequent lesions occur in metastatic prostate adenocarcinoma (amplified in 4.9% of cases) (Grasso et al. 2012), lung squamous cell carcinoma (3.4%, TCGA provisional), and ovarian serous cystadenocarcinoma (2.6%, TCGA provisional).
COP1 (official gene name, RFWD2) is another p53-induced E3 ubiquitin ligase. The association between COP1 and p53 was first identified from affinity purification of ectopic COP1 from cell lysates followed by mass spectrometry analysis to identify interacting proteins (Dornan et al. 2004). This interaction was confirmed on endogenous proteins, and COP1 was shown to increase p53 protein turnover and target it for ubiquitin-dependent proteosomal degradation (Dornan et al. 2004). Interestingly, the pattern of COP1 copy number alterations and mutations in the cBioPortal resembles those of MDM2 and MDM4, with frequent amplification in a number of cancers: liver hepatocellular carcinoma (13%, TCGA provisional), breast-invasive carcinoma (10.8%, TCGA provisional), bladder urothelial carcinoma (10.2%) (Cancer Genome Atlas Network 2014a), and lung adenocarcinoma (7.8%) (Cancer Genome Atlas Network 2014b). In all four of these data sets, there is also a subset of COP1-amplified tumors that retains wild-type p53: 60% of the liver hepatocellular carcinomas, 73% of the breast-invasive carcinomas, 27% of the bladder urothelial carcinomas, and 61% of the lung adenocarcinomas.
TRIM24 is a p53-interacting protein identified through the affinity purification of a TAP-tagged p53, from mouse embryonic stem cells (Allton et al. 2009). TRIM24 also promotes the ubiquitination and protein turnover of p53. The most frequent amplifications of TRIM24 are seen in ovarian serous cystadenocarcinoma (9.6%, TCGA provisional), which is a tumor type in which p53 mutations are a frequent lesion (87% of cases in this data set), and skin cutaneous melanoma (5.4%, TCGA provisional) in which 87% of these cases have wild-type p53.
While considering any one of these other E3 ligases alone does not necessarily show a pronounced influence on human cancers, when the combined amplification from all three are evaluated together the impact is much stronger (Fig. 4A). Frequent amplifications are observed in ovarian serous adenocarcinoma (16.7%, TCGA provisional), liver hepatocellular carcinoma (13.5%, TCGA provisional), breast-invasive carcinoma (13.5%, TCGA provisional), bladder urothelial carcinoma (11%) (Cancer Genome Atlas Network 2014a), and lung adenocarcinoma (10.9%) (Cancer Genome Atlas Network 2014b). It is also interesting to consider amplifications of these E3 enzymes in addition to MDM2 and MDM4. Although the frequencies of any of the individual lesions is quite low, making the statistical analyses largely underpowered, a clear trend is present when you compare all five of these negative regulators across several data sets, with tumors frequently amplifying only one (Fig. 4B). The one noted exception to this is the large proportion of tumors that amplify both MDM4 and COP1; however, these two genes are both located on the long arm of chromosome 1, making them likely candidates for coamplification. Taken together, these data suggest that the up-regulation of p53-negative regulators is a common mechanism through which cancers may inactivate the p53 pathway.
Figure 4.
PIRH2, COP1, and TRIM24 alterations in cancer. Analyses of genomic data of tumors with high levels of p53 inhibitors COP1, PIRH2, and TIRM24 from data sets accessed and prepared using the cBioPortal for Cancer Genetics (www.cbioportal.org) (Cerami et al. 2012; Gao et al. 2013). (A) Frequency of COP1, PIRH2, and TRIM24 alterations across tumor types. (B) Detailed analyses of amplification data for five p53 inhibitors (MDM2, MDM4, COP1, PIRH2, and TRIM24) in tumors with high levels of amplification in A (breast-invasive carcinoma, glioblastoma, ovarian, melanoma, and lung adenocarcinoma). For B, because of the relatively low frequency of alterations in each regulator, only tumors with alterations were included. CNA, Copy number alteration; RPPA, reverse phase protein array. Data were accessed on July 16, 2015.
REGULATORS OF MDM2
Given the profound impact of MDM2 regulation on p53, another mechanism to inhibit this tumor suppressor pathway is through alterations of regulators of MDM2.
ARF
Upstream of MDM2, ARF is part of an important regulatory mechanism that allows for p53 activation in response to deregulated expression or activation of potent oncogenes (Fig. 1). Specifically, ARF is activated downstream from oncogenes, such as MYC and RAS (Palmero et al. 1998; Zindy et al. 1998), and binds and sequesters MDM2, thereby stabilizing p53 (Zhang et al. 1998; Honda and Yasuda 1999; Tao and Levine 1999; Weber et al. 1999; Llanos et al. 2001). As a result, tumors with amplified MYC frequently evolve mechanisms to inactive the p53 pathway and evade the downstream induction of apoptosis. Experimentally, mice expressing the Eμ-Myc transgene develop B-cell lymphomas at a much shorter latency when combined with either Arf or p53 loss (Eischen et al. 1999). This study highlights the strong cooperation between Myc overexpression and Arf loss in lymphomagenesis. Consistent with being a positive regulator of p53, Arf-null mice develop spontaneous tumors early in life (Kamijo et al. 1997). ARF is expressed from an alternate reading frame from the CDKN2A locus, which also codes for another important tumor suppressor protein, p16. The CDKN2A locus is mutated or deleted in a large proportion of human cancers. Specifically, frequent alterations are seen in malignant nerve sheath tumors (73.3%) (Lee et al. 2014), glioblastoma (60.0% and 46.2%) (Cancer Genome Atlas Network 2008; Brennan et al. 2013), head and neck squamous cell carcinoma (48.4%, TCGA in revision), and several others. Deletions of CDKN2A and, therefore, ARF may dampen the p53 pathway by releasing MDM2 and allowing it to inhibit p53.
HAUSP
HAUSP (official gene name, USP7) is a ubiquitin-specific protease, or deubiquinating enzyme (DUB), that is able to remove ubiquitin from target proteins and prevent their degradation. MDM2 is one of the physiological targets of HAUSP, resulting in MDM2 stabilization and negative regulation of the p53 pathway (Cummins et al. 2004; Li et al. 2004). The Hausp-null mouse is embryonic lethal and shows evidence of p53 activation, but is only partially rescued by loss of p53 (Kon et al. 2010). Given that HAUSP stabilizes MDM2, HAUSP levels are expected to be up-regulated in cancers. Data available through the cBioPortal show amplifications in bladder urothelial carcinoma (6.2%) (Iyer et al. 2013) and invasive breast cancers (4.7%, TCGA provisional). Interestingly, in the data set of invasive breast cancers, RNA expression levels of HAUSP are elevated in an additional 18% of cases, suggesting that the primary mode of deregulation of HAUSP is not through gene amplifications in these tumors. There are trends toward mutual exclusivity in genomic alterations (copy number and mutation) between both HAUSP and p53 and HAUSP and MDM2 in invasive breast cancer (TCGA provisional, data not shown). These associations are difficult to make in bladder urothelial carcinoma because of the relatively small proportion of tumors with alterations.
WIP1
WIP1 (official gene name, PPM1D) is a protein phosphatase that plays important roles in regulating the p53 pathway. Like many of the factors already discussed, WIP1 is a transcriptional target of p53, and is also part of an important feedback mechanism (Fiscella et al. 1997). As discussed previously, MDM2 is phosphorylated in response to DNA damage, and dissociates from p53. WIP1 dephosphorylates MDM2, restores the interaction between MDM2 and p53, and thereby prevents the deleterious effects of prolonged p53-pathway activation (Fiscella et al. 1997; Lu et al. 2007). Pancreatic neuroendocrine tumors (PanNETs) generally do not have mutations in p53, but show frequent amplifications of MDM2 (22%), MDM4 (45%), and WIP1 (51%) (Hu et al. 2010; Jiao et al. 2011). WIP1 amplifications are also evident in invasive breast carcinoma (9.3%) (Cancer Genome Atlas Network 2012b) and liver hepatocellular carcinoma (6.2%, TCGA provisional). Interestingly, WIP1 amplification in invasive breast cancer shows a significant (p = 0.002) mutual exclusivity with p53 mutations (Cancer Genome Atlas Network 2012b). Insufficient numbers of tumors or tumors with alterations makes this analysis challenging in other tumor types.
NONENDOGENOUS MECHANISMS OF p53-PATHWAY INACTIVATION: VIRAL ONCOGENES
p53 was originally identified through its association with a polypeptide, the large T antigen, produced by a small DNA virus, Simian virus 40 (SV40) (Lane and Crawford 1979; Linzer and Levine 1979). Subsequently, a number of human tumor viruses were shown to express proteins that bind and inhibit the p53 pathway. Probably, the most prominent example is the high-risk subtypes of human papilloma virus (HPV), specifically HPV16 and 18. More than 90% of cervical cancers are positive for HPV (Walboomers et al. 1999). Mechanistically, two viral proteins, E6 and E7, are essential for the transforming capabilities of the virus as they bind to and inhibit important tumor suppressor proteins in the cell. Specifically, the viral E6 protein complexes with E6AP (E6-associated protein or UBE3A) within the host cell and subsequently binds p53 and targets it for ubiquitination and proteosomal degradation (Scheffner et al. 1990; Werness et al. 1990; Huibregtse et al. 1991). As HPV infection is involved in the etiology of the vast majority of cervical cancers, and leads to the expression of a viral oncoprotein that can attenuate the p53 pathway, the majority of cervical cancers have wild-type p53 (Crook et al. 1992). In the infrequent HPV-negative cases, however, p53 mutations are observed, suggesting that loss of the p53 pathway is an essential and characterizing feature of these cancers (Crook et al. 1992). Understanding the molecular basis of cervical cancer has led to the development of a vaccine against the high-risk types of HPV for the prevention of malignancy (Cutts et al. 2007; Anderson 2012). Although the global impact of HPV vaccines remains to be determined, they represent an important advance in the area of cancer prevention.
Other oncogenic DNA viruses, including Epstein–Barr virus (EBV), and Kaposi’s sarcoma–associated herpesvirus (KHSV), also express viral oncoproteins that similarly promote the inhibition of p53 (Collot-Teixeira et al. 2004; Sato and Tsurumi 2013). EBV has been associated with Burkitt’s lymphoma, nasopharyngeal cancers, Hodgkin’s lymphoma and T-cell lymphomas, and KHSV with Karposi’s sarcomas, which are commonly diagnosed in AIDS patients. To the best of our knowledge, a comprehensive evaluation of p53 mutation status in cancers positive for EBV and/or KHSV has not yet been completed, although studies in a small number of samples have suggested that the majority of tumors (with the exception of Burkitt’s lymphomas) do retain wild-type p53 (Farrell et al. 1991; Effert et al. 1992; Lo et al. 1992; Edwards and Raab-Traub 1994; Nador et al. 1996; Katano et al. 2001; Petre et al. 2007).
TUMOR SPECIFICITY IN p53-PATHWAY INACTIVATION
Tumors with Significant p53-Pathway Inactivation
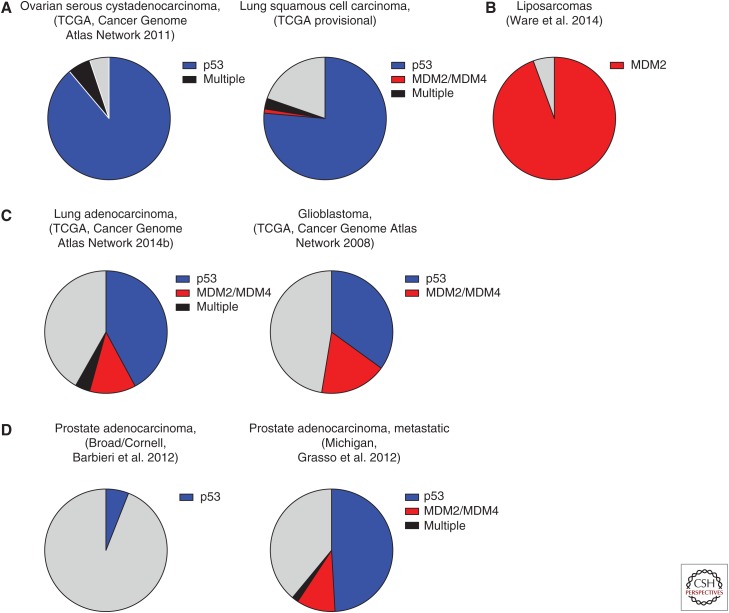
A number of interesting trends emerge when the scope of p53 alterations across different cancer types is considered. There are some tumors that inactivate the pathway, almost exclusively, through direct deletion or mutation of the p53 locus (Fig. 5A). For example, this is a characteristic lesion in ovarian serous cystadenocarcinomas (95%) (Cancer Genome Atlas Network 2011), small-cell lung cancer (90%) (Peifer et al. 2012; Rudin et al. 2012), esophageal squamous cell carcinoma (84%) (Song et al. 2014), and lung squamous cell carcinoma (79%, TCGA provisional). Alternatively, other tumor types are characterized by other p53-pathway lesions (Fig. 5B). Liposarcomas have near complete penetrance of MDM2 amplification (Ware et al. 2014), and MDM4 overexpression is observed in the majority of melanomas and retinoblastomas (Laurie et al. 2006; Gembarska et al. 2012). A third group of cancers show alterations in multiple known components of the p53 pathway (Fig. 5C, limited to p53, MDM2, and MDM4). For example, lung adenocarcinomas show pathway inactivation in greater than 65% of cases, with only approximately two-thirds being a direct mutation at the p53 locus (Cancer Genome Atlas Network 2014b). These observations may hold some valuable insights into the molecular mechanisms contributing to tumorigenesis in these cancers. For example, evidence from both in vitro and in vivo studies has shown the missense mutations in p53 both eliminate the normal functions of the protein, but also have additional gain-of-function (GOF) properties that contribute to tumorigenesis (Lang et al. 2004; Olive et al. 2004; Oren and Rotter 2010; Shetzer et al. 2016). In tumors that show high proportions of missense mutations in p53, this GOF might be an important contributing factor in tumor development. Similarly, MDM2 has also been shown to have p53-independent roles, which may be important in the development of the tumors that are characterized by MDM2 amplification (Ganguli and Wasylyk 2003; Bouska and Eischen 2009). Finally, some tumors, such as glioblastomas or lung adenocarcinomas (Fig. 5C), inactivate known components of the p53 pathway ∼50% of the time, suggesting that unknown factors that impinge on p53 activity might be responsible for tumor development in the other cases. Genome-wide analyses to look for mutual exclusivity with other genes are worthwhile, and might identify novel regulators of the p53 pathway.
Figure 5.
Summary of p53-pathway alterations in cancer. Pie charts summarizing the alteration spectrums in p53-pathway components seen across different cancer types. For simplicity, only p53, MDM2, and MDM4 are included in this representation. Blue, p53 mutation/deletion; red, Mdm2/Mdm4 amplification; black, alterations in more than one of p53, MDM2, and MDM4; gray, tumors with no alterations in these components of the p53 pathway. (A) Tumors with frequent alterations in p53. (B) Tumor with frequent alterations in a p53 regulator. (C) Tumors with alterations in multiple pathway components, as well as a subset with no evidence of pathway attenuation. (D) Comparison of prostate cancer data sets taken from primary and metastatic tumors, suggesting that p53-pathway attenuation occurs late in the pathogenesis of this cancer. Data were accessed on July 16, 2015.
Tumors with Little Evidence of p53-Pathway Inactivation
Despite the large number of p53-pathway alterations that occur across different cancer types, the genomic analyses available in the cBioPortal suggest that there are a number of tumor types that have only infrequent alterations in the pathway. A number of hypotheses arise from this observation and will be discussed.
Insufficient Data on Known Pathway Components
To date, large-scale global cancer genome projects, such as the TCGA and International Cancer Genome Consortium (ICGC) have provided a wealth of knowledge about the recurrent somatic lesions and copy number aberrations found across a large number and wide variety of human cancer types. These studies have since expanded to include mRNA and protein expression, and more recently epigenetic profiling. As these data sets continue to expand and include expression and epigenetic data, the proportion of tumors with alterations in known pathway components will likely increase. For example, melanoma protein analysis has revealed MDM4 overexpression in 65% of cases (Gembarska et al. 2012).
Timing in Tumorigenesis and Progression
The frequency of alterations in a given gene may be used to provide suggestive evidence as to which lesions are important for the initiation of those tumors, and which may be associated with progression. This idea that a specific order of genetic lesions underlies tumor development has been well established for colorectal cancer, in which mutations in the tumor suppressor APC occur early and transform the normal epithelia to adenomas, and subsequent mutations in KRAS followed by p53 result in progression to adenocarcinoma (Fearon and Vogelstein 1990). Indeed, in one study, 72% of colorectal cancers show mutations in APC, and 42% and 52% have mutations in KRAS and p53, respectively (Cancer Genome Atlas Network 2012a). It is interesting to consider the data available in the cBioPortal for prostate cancer in this context. Specifically, the different data sets clearly reflect the understanding that p53 lesions occur late in the pathogenesis and progression of prostate cancers. The majority of these data sets are from primary tumors isolated from patients, and these show infrequent lesions in the p53 pathway (Fig. 5D) (Taylor et al. 2010; Barbieri et al. 2012; Baca et al. 2013). One data set, on the other hand, profiled metastatic and high-grade localized tumors and found a much higher frequency of p53-pathway alterations, including mutations or deletions in p53 in 52% of cases (Fig. 5D) (Grasso et al. 2012). There is additional evidence to suggest that p53 mutations arise late in the pathogenesis of thyroid tumors, occurring more frequently in poorly differentiated and anaplastic tumors (Shahedian et al. 2001; Malaguarnera et al. 2007). These examples clearly illustrate the value of comparing the genomic landscape of early primary lesions to that of late-stage and metastatic tumors to better understand the events that are essential for initiation and those that contribute to tumor progression.
Unknown Regulators of the p53 Pathway
Despite being one of the most extensively studied proteins and pathways in human cancer, the possibility remains that there are important disease-relevant mechanisms of p53-pathway regulation to be uncovered (indicated by “?” in Fig. 1). TCGA analyses from the cBioPortal show little evidence of p53-pathway inactivation in clear cell and papillary renal cell carcinoma (RCC). Interestingly, in one study, four cell lines derived from RCCs with wild-type p53 were unable to transactivate p53 downstream target genes (Gurova et al. 2004). This was shown to be independent of MDM2, MDM4, and ARF, and could only be overcome by superphysiological expression of p53 from a lentivirus. Further, cell hybrids with these RCC lines resulted in dominant pathway inhibition when fused to cells with a functional p53 response. Additional work suggested that activation of the NF-κB pathway was responsible for the attenuated p53 response (Gurova et al. 2005). It should also be noted that, like the other tumor types discussed previously, there is also evidence to suggest that p53 mutations can also occur late in the pathogenesis of RCCs (Zigeuner et al. 2004; Noon et al. 2010). Nonetheless, this example highlights the notion that there may be novel, tumor-specific mechanisms of p53-pathway inactivation still to be identified.
Further, in addition to the potential to uncover novel regulators of the p53 pathway, there is still a lot to be understood about many of the currently known regulators. With the exception of MDM2 and MDM4, which have clear roles in regulating p53 during development and have pathological consequences when overexpressed, we do not yet have a full appreciation for the physiological roles of many of the other identified regulators, including the three E3 ubiquitin ligases we have discussed in detail. Specifically, a better understanding of the in vivo cellular contexts in which these proteins regulate p53 will be valuable.
TRANSLATING THIS KNOWLEDGE: IMPLICATIONS OF THERAPY
The understanding of both p53 function and regulation has not only brought an expanded appreciation of the breadth of pathway inactivation across human cancers, but it has also provided opportunities for therapeutic intervention.
MDM2:p53 Inhibitors
In tumors with wild-type p53, in which pathway inactivation occurs through the overexpression of important negative regulators, the inhibition of the p53:inhibitor interaction serves as a promising therapeutic approach to restore the tumor-suppressive activity of the pathway. This strategy is the most advanced for inhibitors of the p53:MDM2 interaction. The Nutlin class of small molecules was identified as potent inhibitors of this interaction by competitively binding the p53-interaction domain of MDM2. Treatment of cells expressing wild-type p53 with Nutlin results in the induction of downstream p53 target genes, such as p21, and a subsequent reduction in cell viability (Vassilev et al. 2004). Importantly, oral administration of Nutlin also slows xenograft tumor growth in vivo (Vassilev et al. 2004). The success of the preclinical studies of Nutlin has led to the development of related compounds with more favorable pharmacological properties, which have been evaluated in early clinical trials for both hematological malignancies and solid tumors. Nutlins, both alone and in combination with either cytotoxic chemotherapy or other targeted agents, show promise for the treatment of cancers with wild-type p53 (Vassilev et al. 2004; Li and Lozano 2013). Nutlins, however, represent only one of several approaches that have been developed to inhibit the MDM2:p53 interaction. For example, the small molecule RITA binds to the amino terminus of p53, and prevents association with MDM2 (Issaeva et al. 2004). We direct readers to several reviews that have been written on this subject (Vassilev 2007; Shangary and Wang 2009; Li and Lozano 2013) and to articles in this collection (Cheok and Lane 2016; Wang et al. 2016). Although elevated levels of MDM2 in the context of wild-type p53 are largely used to predict therapeutic benefit of these inhibitors, WIP1 and HAUSP-overexpressing tumors and those with deletions of ARF may also be sensitive to p53:MDM2 inhibition.
Other Pathway Inhibitors
We have described a number of other interactors and regulators of p53 that could also be exploited for the treatment of cancers. Specifically, MDM4 shows the strongest levels of amplification and overexpression in cancers. The development and advancement of specific p53:MDM4 inhibitors have clinical potential, with a number of small molecules already identified (Reed et al. 2010; Wang et al. 2011; Li and Lozano 2013). As our understanding and appreciation for the biological contexts in which other p53 regulators act, it is likely that inhibitors of these interactions will be developed.
SUMMARY AND FUTURE PERSPECTIVES
Inactivation of the p53 pathway plays a prominent role across numerous malignancies. Although mutations and deletions in the p53 locus are most common, a number of other important mechanisms attenuate this pathway. MDM2 and MDM4 amplification show clear mutual exclusivity with p53-mutation status in several tumor types. Additionally, a number of other direct p53-negative regulators have been identified in vitro; however, the pathological significance of overexpression of these genes, for example, through transgenic mouse models, has not yet been evaluated.
Interestingly, some tumors rarely alter known components of the p53 pathway, suggesting that our understanding of the pathway remains incomplete and additional regulatory mechanisms are still to be uncovered. The existing genomic data could provide an opportunity to identify genes with mutually exclusive alterations with p53 as potential candidate regulators.
In summary, the current understanding of both the normal regulation of the pathway and subsequent mechanisms of pathway deregulation has important implications for human cancers. The development of p53:MDM2 inhibitors is a strong example of how understanding fundamental biological mechanisms can lead to novel therapeutic approaches. Additionally, HPV infection and the subsequent inhibition of p53 by the viral E6 oncoprotein are essential to the etiology of the majority of cervical cancers, and this knowledge has led to the development of vaccines to prevent these cancers. Combined, understanding the p53 pathway as a whole has had and will continue to have tremendous significance to the prevention, diagnosis, and treatment of human cancers.
ACKNOWLEDGMENTS
We thank Drs. Vinod Pant and Yun Zhang for critical reading of this manuscript. We additionally acknowledge and thank the work of all of our colleagues and apologize for those studies we were unable to include because of space constraints. This work is supported by National Cancer Institute Grant CA47296 (G.L.) and a fellowship from the Canadian Institutes of Health Research (A.R.W.).
Footnotes
Editors: Guillermina Lozano and Arnold J. Levine
Additional Perspectives on The p53 Protein available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Allton K, Jain AK, Herz HM, Tsai WW, Jung SY, Qin J, Bergmann A, Johnson RL, Barton MC. 2009. Trim24 targets endogenous p53 for degradation. Proc Natl Acad Sci 106: 11612–11616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson LA. 2012. Prophylactic human papillomavirus vaccines: Past, present and future. Pathology 44: 1–6. [DOI] [PubMed] [Google Scholar]
- Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, et al. 2013. Punctuated evolution of prostate cancer genomes. Cell 153: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M. 1993. mdm2 expression is induced by wild type p53 activity. EMBO J 12: 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat JP, White TA, Stojanov P, Van Allen E, Stransky N, et al. 2012. Exome sequencing identifies recurrent SPOP, FOXA1 and MED12 mutations in prostate cancer. Nat Genet 44: 685–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, et al. 2010. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet 42: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesten LS, Zadelaar SM, De Clercq S, Francoz S, van Nieuwkoop A, Biessen EA, Hofmann F, Feil S, Feil R, Jochemsen AG, et al. 2006. Mdm2, but not Mdm4, protects terminally differentiated smooth muscle cells from p53-mediated caspase-3-independent cell death. Cell Death Differ 13: 2089–2098. [DOI] [PubMed] [Google Scholar]
- Bond GL, Levine AJ. 2007. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene 26: 1317–1323. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. 2004. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 119: 591–602. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Levine A. 2005. A single nucleotide polymorphism in the MDM2 gene: From a molecular and cellular explanation to clinical effect. Cancer Res 65: 5481–5484. [DOI] [PubMed] [Google Scholar]
- Bouska A, Eischen CM. 2009. Murine double minute 2: p53–independent roads lead to genome instability or death. Trends Biochem Sci 34: 279–286. [DOI] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. 2013. The somatic genomic landscape of glioblastoma. Cell 155: 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahilly-Snyder L, Yang-Feng T, Francke U, George DL. 1987. Molecular analysis and chromosomal mapping of amplified genes isolated from a transformed mouse 3T3 cell line. Somat Cell Mol Genet 13: 235–244. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2011. Integrated genomic analyses of ovarian carcinoma. Nature 474: 609–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2012a. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2012b. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2014a. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507: 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. 2014b. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. 1998. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science 281: 1677–1679. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Reyes A, Parant JM, Amelse LL, de Oca Luna RM, Korsmeyer SJ, Lozano G. 2003. Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res 63: 8664–8669. [PubMed] [Google Scholar]
- Chehab NH, Malikzay A, Appel M, Halazonetis TD. 2000. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev 14: 278–288. [PMC free article] [PubMed] [Google Scholar]
- *.Cheok CF, Lane DP. 2016. Exploiting the p53 pathway for therapy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collot-Teixeira S, Bass J, Denis F, Ranger-Rogez S. 2004. Human tumor suppressor p53 and DNA viruses. Rev Med Virol 14: 301–319. [DOI] [PubMed] [Google Scholar]
- Crook T, Wrede D, Tidy JA, Mason WP, Evans DJ, Vousden KH. 1992. Clonal p53 mutation in primary cervical cancer: Association with human-papillomavirus-negative tumours. Lancet 339: 1070–1073. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. 2004. Tumour suppression: Disruption of HAUSP gene stabilizes p53. Nature 428: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- Cutts FT, Franceschi S, Goldie S, Castellsague X, de Sanjose S, Garnett G, Edmunds WJ, Claeys P, Goldenthal KL, Harper DM, et al. 2007. Human papillomavirus and HPV vaccines: A review. Bull World Health Organ 85: 719–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq S, Gembarska A, Denecker G, Maetens M, Naessens M, Haigh K, Haigh JJ, Marine JC. 2010. Widespread overexpression of epitope-tagged Mdm4 does not accelerate tumor formation in vivo. Mol Cell Biol 30: 5394–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, O’Rourke K, Koeppen H, Dixit VM. 2004. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 429: 86–92. [DOI] [PubMed] [Google Scholar]
- Edwards RH, Raab-Traub N. 1994. Alterations of the p53 gene in Epstein–Barr virus–associated immunodeficiency-related lymphomas. J Virol 68: 1309–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effert P, McCoy R, Abdel-Hamid M, Flynn K, Zhang Q, Busson P, Tursz T, Liu E, Raab-Traub N. 1992. Alterations of the p53 gene in nasopharyngeal carcinoma. J Virol 66: 3768–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Lozano G. 2014. The Mdm network and its regulation of p53 activities: A rheostat of cancer risk. Hum Mutat 35: 728–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. 1999. Disruption of the ARF–Mdm2–p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev 13: 2658–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakharzadeh SS, Trusko SP, George DL. 1991. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J 10: 1565–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell PJ, Allan GJ, Shanahan F, Vousden KH, Crook T. 1991. p53 is frequently mutated in Burkitt’s lymphoma cell lines. EMBO J 10: 2879–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767. [DOI] [PubMed] [Google Scholar]
- Fiscella M, Zhang H, Fan S, Sakaguchi K, Shen S, Mercer WE, Vande Woude GF, O’Connor PM, Appella E. 1997. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci 94: 6048–6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francoz S, Froment P, Bogaerts S, De Clercq S, Maetens M, Doumont G, Bellefroid E, Marine JC. 2006. Mdm4 and Mdm2 cooperate to inhibit p53 activity in proliferating and quiescent cells in vivo. Proc Natl Acad Sci 103: 3232–3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli G, Wasylyk B. 2003. p53-independent functions of MDM2. Mol Cancer Res 1: 1027–1035. [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gembarska A, Luciani F, Fedele C, Russell EA, Dewaele M, Villar S, Zwolinska A, Haupt S, de Lange J, Yip D, et al. 2012. MDM4 is a key therapeutic target in cutaneous melanoma. Nat Med 18: 1239–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC, et al. 2012. The mutational landscape of lethal castration-resistant prostate cancer. Nature 487: 239–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. 2006. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol 26: 192–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochola LF, Zeron-Medina J, Meriaux S, Bond GL. 2010. Single-nucleotide polymorphisms in the p53 signaling pathway. Cold Spring Harb Perspect Biol 2: a001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurova KV, Hill JE, Razorenova OV, Chumakov PM, Gudkov AV. 2004. p53 pathway in renal cell carcinoma is repressed by a dominant mechanism. Cancer Res 64: 1951–1958. [DOI] [PubMed] [Google Scholar]
- Gurova KV, Hill JE, Guo C, Prokvolit A, Burdelya LG, Samoylova E, Khodyakova AV, Ganapathi R, Ganapathi M, Tararova ND, et al. 2005. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-κB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci 102: 17448–17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hainaut P, Pfeifer GP. 2016. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakem A, Bohgaki M, Lemmers B, Tai E, Salmena L, Matysiak-Zablocki E, Jung YS, Karaskova J, Kaustov L, Duan S, et al. 2011. Role of Pirh2 in mediating the regulation of p53 and c-Myc. PLoS Genet 7: e1002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299. [DOI] [PubMed] [Google Scholar]
- Hay TJ, Meek DW. 2000. Multiple sites of in vivo phosphorylation in the MDM2 oncoprotein cluster within two important functional domains. FEBS Lett 478: 183–186. [DOI] [PubMed] [Google Scholar]
- Hock AK, Vousden KH. 2014. The role of ubiquitin modification in the regulation of p53. Biochim Biophys Acta 1843: 137–149. [DOI] [PubMed] [Google Scholar]
- Honda R, Yasuda H. 1999. Association of p19ARF with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J 18: 22–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R, Tanaka H, Yasuda H. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett 420: 25–27. [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Modica I, Klimstra DS, Song L, Allen PJ, Brennan MF, Levine AJ, Tang LH. 2010. Gene amplifications in well-differentiated pancreatic neuroendocrine tumors inactivate the p53 pathway. Genes Cancer 1: 360–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Yan Z, Liao X, Li Y, Yang J, Wang ZG, Zuo Y, Kawai H, Shadfan M, Ganapathy S, et al. 2011. The p53 inhibitors MDM2/MDMX complex is required for control of p53 activity in vivo. Proc Natl Acad Sci 108: 12001–12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huibregtse JM, Scheffner M, Howley PM. 1991. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J 10: 4129–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. 2004. Small molecule RITA binds to p53, blocks p53–HDM-2 interaction and activates p53 function in tumors. Nat Med 10: 1321–1328. [DOI] [PubMed] [Google Scholar]
- Iyer G, Al-Ahmadie H, Schultz N, Hanrahan AJ, Ostrovnaya I, Balar AV, Kim PH, Lin O, Weinhold N, Sander C, et al. 2013. Prevalence and co-occurrence of actionable genomic alterations in high-grade bladder cancer. J Clin Oncol 31: 3133–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain AK, Barton MC. 2010. Making sense of ubiquitin ligases that regulate p53. Cancer Biol Ther 10: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S, Minter LC, Stratton SA, Yang P, Abbas HA, Akdemir ZC, Pant V, Post S, Gagea M, Lee RG, et al. 2015. TRIM24 suppresses development of spontaneous hepatic lipid accumulation and hepatocellular carcinoma in mice. J Hepatol 62: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331: 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208. [DOI] [PubMed] [Google Scholar]
- Jones SN, Hancock AR, Vogel H, Donehower LA, Bradley A. 1998. Overexpression of Mdm2 in mice reveals a p53-independent role for Mdm2 in tumorigenesis. Proc Natl Acad Sci 95: 15608–15612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven T, Barak Y, Zauberman A, George DL, Oren M. 1993. Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8: 3411–3416. [PubMed] [Google Scholar]
- Kamijo T, Zindy F, Roussel MF, Quelle DE, Downing JR, Ashmun RA, Grosveld G, Sherr CJ. 1997. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell 91: 649–659. [DOI] [PubMed] [Google Scholar]
- Katano H, Sato Y, Sata T. 2001. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92: 3076–3084. [DOI] [PubMed] [Google Scholar]
- Khosravi R, Maya R, Gottlieb T, Oren M, Shiloh Y, Shkedy D. 1999. Rapid ATM-dependent phosphorylation of MDM2 precedes p53 accumulation in response to DNA damage. Proc Natl Acad Sci 96: 14973–14977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knappskog S, Lonning PE. 2011. Effects of the MDM2 promoter SNP285 and SNP309 on Sp1 transcription factor binding and cancer risk. Transcription 2: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama T, Takehara T, Hikita H, Shimizu S, Shigekawa M, Tsunematsu H, Li W, Miyagi T, Hosui A, Tatsumi T, et al. 2011. Increases in p53 expression induce CTGF synthesis by mouse and human hepatocytes and result in liver fibrosis in mice. J Clin Invest 121: 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kon N, Kobayashi Y, Li M, Brooks CL, Ludwig T, Gu W. 2010. Inactivation of HAUSP in vivo modulates p53 function. Oncogene 29: 1270–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303. [DOI] [PubMed] [Google Scholar]
- Lane DP. 1992. Cancer. p53, guardian of the genome. Nature 358: 15–16. [DOI] [PubMed] [Google Scholar]
- Lane DP, Crawford LV. 1979. T antigen is bound to a host protein in SV40-transformed cells. Nature 278: 261–263. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. 2004. Gain of function of a p53 hot spot mutation in a mouse model of Li–Fraumeni syndrome. Cell 119: 861–872. [DOI] [PubMed] [Google Scholar]
- Laurie NA, Donovan SL, Shih CS, Zhang J, Mills N, Fuller C, Teunisse A, Lam S, Ramos Y, Mohan A, et al. 2006. Inactivation of the p53 pathway in retinoblastoma. Nature 444: 61–66. [DOI] [PubMed] [Google Scholar]
- Lee JT, Gu W. 2010. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 17: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Teckie S, Wiesner T, Ran L, Prieto Granada CN, Lin M, Zhu S, Cao Z, Liang Y, Sboner A, et al. 2014. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet 46: 1227–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, Parant JM, Lozano G, Hakem R, Benchimol S. 2003. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell 112: 779–791. [DOI] [PubMed] [Google Scholar]
- Li Q, Lozano G. 2013. Molecular pathways: Targeting Mdm2 and Mdm4 in cancer therapy. Clin Cancer Res 19: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. 2004. A dynamic role of HAUSP in the p53–Mdm2 pathway. Mol Cell 13: 879–886. [DOI] [PubMed] [Google Scholar]
- Linzer DI, Levine AJ. 1979. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17: 43–52. [DOI] [PubMed] [Google Scholar]
- Llanos S, Clark PA, Rowe J, Peters G. 2001. Stabilization of p53 by p14ARF without relocation of MDM2 to the nucleolus. Nat Cell Biol 3: 445–452. [DOI] [PubMed] [Google Scholar]
- Lo KW, Mok CH, Huang DP, Liu YX, Choi PH, Lee JC, Tsao SW. 1992. p53 mutation in human nasopharyngeal carcinomas. Anticancer Res 12: 1957–1963. [PubMed] [Google Scholar]
- Lu X, Ma O, Nguyen TA, Jones SN, Oren M, Donehower LA. 2007. The Wip1 phosphatase acts as a gatekeeper in the p53–Mdm2 autoregulatory loop. Cancer Cell 12: 342–354. [DOI] [PubMed] [Google Scholar]
- Malaguarnera R, Vella V, Vigneri R, Frasca F. 2007. p53 family proteins in thyroid cancer. Endocr Relat Cancer 14: 43–60. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. 2003. mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol 23: 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliorini D, Bogaerts S, Defever D, Vyas R, Denecker G, Radaelli E, Zwolinska A, Depaepe V, Hochepied T, Skarnes WC, et al. 2011. Cop1 constitutively regulates c-Jun protein stability and functions as a tumor suppressor in mice. J Clin Invest 121: 1329–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George D, Levine AJ. 1992. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell 69: 1237–1245. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206. [DOI] [PubMed] [Google Scholar]
- Nador RG, Cesarman E, Chadburn A, Dawson DB, Ansari MQ, Sald J, Knowles DM. 1996. Primary effusion lymphoma: A distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 88: 645–656. [PubMed] [Google Scholar]
- Noon AP, Vlatkovic N, Polanski R, Maguire M, Shawki H, Parsons K, Boyd MT. 2010. p53 and MDM2 in renal cell carcinoma: Biomarkers for disease progression and future therapeutic targets? Cancer 116: 780–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. 1992. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358: 80–83. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Pietenpol JA, Thiagalingam S, Gyuris J, Kinzler KW, Vogelstein B. 1993. Oncoprotein MDM2 conceals the activation domain of tumour suppressor p53. Nature 362: 857–860. [DOI] [PubMed] [Google Scholar]
- Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, Crowley D, Jacks T. 2004. Mutant p53 gain of function in two mouse models of Li–Fraumeni syndrome. Cell 119: 847–860. [DOI] [PubMed] [Google Scholar]
- Oren M, Rotter V. 2010. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol 2: a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmero I, Pantoja C, Serrano M. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395: 125–126. [DOI] [PubMed] [Google Scholar]
- Pant V, Lozano G. 2014. Limiting the power of p53 through the ubiquitin proteasome pathway. Genes Dev 28: 1739–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Xiong S, Iwakuma T, Quintas-Cardama A, Lozano G. 2011. Heterodimerization of Mdm2 and Mdm4 is critical for regulating p53 activity during embryogenesis but dispensable for p53 and Mdm2 stability. Proc Natl Acad Sci 108: 11995–12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V, Xiong S, Jackson JG, Post SM, Abbas HA, Quintas-Cardama A, Hamir AN, Lozano G. 2013. The p53–Mdm2 feedback loop protects against DNA damage by inhibiting p53 activity but is dispensable for p53 stability, development, and longevity. Genes Dev 27: 1857–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J, Chavez-Reyes A, Little NA, Yan W, Reinke V, Jochemsen AG, Lozano G. 2001. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet 29: 92–95. [DOI] [PubMed] [Google Scholar]
- Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. 2012. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44: 1104–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petre CE, Sin SH, Dittmer DP. 2007. Functional p53 signaling in Kaposi’s sarcoma-associated herpesvirus lymphomas: Implications for therapy. J Virol 81: 1912–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Pant V, Iwakuma T, Hamir A, Jackson JG, Maccio DR, Bond GL, Johnson DG, Levine AJ, et al. 2010. A high-frequency regulatory polymorphism in the p53 pathway accelerates tumor development. Cancer Cell 18: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, et al. 2010. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem 285: 10786–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringshausen I, O’Shea CC, Finch AJ, Swigart LB, Evan GI. 2006. Mdm2 is critically and continuously required to suppress lethal p53 activity in vivo. Cancer Cell 10: 501–514. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. 2012. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44: 1111–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y, Tsurumi T. 2013. Genome guardian p53 and viral infections. Rev Med Virol 23: 213–220. [DOI] [PubMed] [Google Scholar]
- Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63: 1129–1136. [DOI] [PubMed] [Google Scholar]
- Shahedian B, Shi Y, Zou M, Farid NR. 2001. Thyroid carcinoma is characterized by genomic instability: Evidence from p53 mutations. Mol Genet Metab 72: 155–163. [DOI] [PubMed] [Google Scholar]
- Shangary S, Wang S. 2009. Small-molecule inhibitors of the MDM2–p53 protein–protein interaction to reactivate p53 function: A novel approach for cancer therapy. Annu Rev Pharmacol Toxicol 49: 223–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp DA, Kratowicz SA, Sank MJ, George DL. 1999. Stabilization of the MDM2 oncoprotein by interaction with the structurally related MDMX protein. J Biol Chem 274: 38189–38196. [DOI] [PubMed] [Google Scholar]
- *.Shetzer Y, Molchadsky A, Rotter V. 2016. Oncogenic mutant p53 gain of function nourishes the vicious cycle of tumor development and cancer stem cell formation. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Steegenga WT, Riteco N, van Laar T, Dekker P, Bazuine M, van Ham RC, van der Houven van Oordt W, Hateboer G, van der Eb AJ, et al. 1996. MDMX: A novel p53-binding protein with some functional properties of MDM2. EMBO J 15: 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Shvarts A, Bazuine M, Dekker P, Ramos YF, Steegenga WT, Merckx G, van Ham RC, van der Houven van Oordt W, van der Eb AJ, Jochemsen AG. 1997. Isolation and identification of the human homolog of a new p53-binding protein, Mdmx. Genomics 43: 34–42. [DOI] [PubMed] [Google Scholar]
- Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al. 2014. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 509: 91–95. [DOI] [PubMed] [Google Scholar]
- Tanimura S, Ohtsuka S, Mitsui K, Shirouzu K, Yoshimura A, Ohtsubo M. 1999. MDM2 interacts with MDMX through their RING finger domains. FEBS Lett 447: 5–9. [DOI] [PubMed] [Google Scholar]
- Tao W, Levine AJ. 1999. p19ARF stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc Natl Acad Sci 96: 6937–6941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Box N, Terzian T, Lozano G. 2009. Mdm4 loss in the intestinal epithelium leads to compartmentalized cell death but no tissue abnormalities. Differentiation 77: 442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT. 2007. MDM2 inhibitors for cancer therapy. Trends Mol Med 13: 23–31. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. 2004. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303: 844–848. [DOI] [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189: 12–19. [DOI] [PubMed] [Google Scholar]
- Wang H, Ma X, Ren S, Buolamwini JK, Yan C. 2011. A small-molecule inhibitor of MDMX activates p53 and induces apoptosis. Mol Cancer Ther 10: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wang S, Zhao Y, Aguilar A, Bernard D, Yang C-Y. 2016. Targeting the MDM2–p53 protein–protein interaction for new cancer therapy: Progress and challenges. Cold Spring Harb Perspect Med 10.1101/cshperspect.a026245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware PL, Snow AN, Gvalani M, Pettenati MJ, Qasem SA. 2014. MDM2 copy numbers in well-differentiated and dedifferentiated liposarcoma: Characterizing progression to high-grade tumors. Am J Clin Pathol 141: 334–341. [DOI] [PubMed] [Google Scholar]
- Weber JD, Taylor LJ, Roussel MF, Sherr CJ, Bar-Sagi D. 1999. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol 1: 20–26. [DOI] [PubMed] [Google Scholar]
- Werness BA, Levine AJ, Howley PM. 1990. Association of human papillomavirus types 16 and 18 E6 proteins with p53. Science 248: 76–79. [DOI] [PubMed] [Google Scholar]
- Wu L, Levine AJ. 1997. Differential regulation of the p21/WAF-1 and mdm2 genes after high-dose UV irradiation: p53-dependent and p53-independent regulation of the mdm2 gene. Mol Med 3: 441–451. [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ. 1993. The p53–mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132. [DOI] [PubMed] [Google Scholar]
- Xiong S, Van Pelt CS, Elizondo-Fraire AC, Liu G, Lozano G. 2006. Synergistic roles of Mdm2 and Mdm4 for p53 inhibition in central nervous system development. Proc Natl Acad Sci 103: 3226–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong S, Pant V, Suh YA, Van Pelt CS, Wang Y, Valentin-Vega YA, Post SM, Lozano G. 2010. Spontaneous tumorigenesis in mice overexpressing the p53-negative regulator Mdm4. Cancer Res 70: 7148–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. 1993. Sequence-specific DNA binding by p53: Identification of target sites and lack of binding to p53–MDM2 complexes. EMBO J 12: 2799–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xiong S, Li Q, Hu S, Tashakori M, Van Pelt C, You MJ, Pageon L, Lozano G. 2014. Tissue-specific and age-dependent effects of global Mdm2 loss. J Pathol 233: 380–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigeuner R, Ratschek M, Rehak P, Schips L, Langner C. 2004. Value of p53 as a prognostic marker in histologic subtypes of renal cell carcinoma: A systematic analysis of primary and metastatic tumor tissue. Urology 63: 651–655. [DOI] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, Roussel MF. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev 12: 2424–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]