Abstract
Light chain amyloidosis is a protein misfolding disease in which immunoglobulin light chains aggregate as insoluble fibrils that accumulate in extracellular deposits. Amyloid fibril formation in vitro has been described as a nucleation–polymerization, autocatalytic reaction in which nascent fibrils catalyze formation of new fibrils, recruiting soluble protein into the fibril. In this context, it is also established that preformed fibrils or “seeds” accelerate fibril formation. In some cases, seeds with a substantially different sequence are able to accelerate the reaction, albeit with a lower efficiency. In this work, we studied the recruitment and addition of monomers in the presence of seeds of five immunoglobulin light chain proteins, covering a broad range of protein stabilities and amyloidogenic properties. Our data reveal that in the presence of homologous or heterologous seeds, the fibril formation reactions become less stochastic than de novo reactions. The kinetics of the most amyloidogenic proteins (AL-T05 and AL-09) do not present significant changes in the presence of seeds. Amyloidogenic protein AL-103 presented fairly consistent acceleration with all seeds. In contrast, the less amyloidogenic proteins (AL-12 and κI) presented dramatic differential effects that are dependent on the kind of seed used. κI had a poor efficiency to elongate preformed fibrils. Together, these results indicate that fibril formation is kinetically determined by the conformation of the amyloidogenic precursor and modulated by the differential ability of each protein to either nucleate or elongate fibrils. We observe morphological and conformational properties of some seeds that do not favor elongation with some proteins, resulting in a delay in the reaction.
Graphical abstract
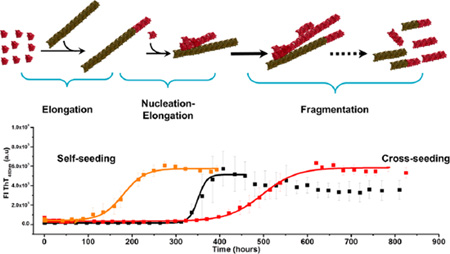
Light chain (AL) amyloidosis is a progressive and lethal disease characterized by the anomalous aggregation of monoclonal immunoglobulin light chains into amyloid fibrils and tissue deposition, which results in organ dysfunction.1,2 It is widely accepted that in vitro, amyloid fibrils are formed following the nucleation–polymerization model.3–8 The reaction can be described in two phases: the so-called nucleation/lag phase, in which the partially folded, aggregation-prone conformation of monomers associates into a multimolecular nucleus, and the polymerization/elongation phase, in which the nucleus acts as a “seed”, to elongate into fibrils by the recruitment and addition of monomers and/or multimolecular oligomers.
The ability to self-propagate is a fundamental property of all amyloid fibrils and proceeds as a continuous autocatalytic process via the recruitment of additional protein molecules from the surrounding solution.1,8,9 In fact, the sigmoidal nature of the amyloid fibril formation kinetics is characteristic of autocatalytic reactions,4,7,8,10 where the rate increases as the materials react. Autocatalytic reactions proceed slowly in the beginning because there is little catalyst present. As the reaction continues, the rate of the reaction accelerates with an increased amount of catalyst and then slows as the reactant is consumed.
The characteristic basic steps of amyloid formation are primary nucleation and elongation of fibrils. Primary nucleation starts from monomers in solution, while elongation describes new growth that is dependent on already formed fibrils.
It is well established that the rate of fibril formation is accelerated by the addition of preformed fibrils (a phenomenon called “seeding” because of its analogy with the crystallization process). Recent experimental evidence from a number of laboratories11–13 supports the idea that in vitro, the recruitment of the monomers to the nascent fibril includes elongation and secondary nucleation components. Elongation is the recruitment of monomers into the ends of a nascent fibril. Secondary nucleation is a complementary mechanism that generates new recruitment competent fibrils at a rate that is dependent on the concentration of existing fibrils.
Three different mechanisms for the secondary nucleation have been proposed by Ruschak et al.14 (a) Fragmentation breaks fibrils to produce new ends suitable for growth with a rate depending only upon the concentration of the existing fibrils and results in fibrils with a long and linear morphology.15 (b) “Branching” allows a new fibril to grow from within an existing fibril. Finally, (c) lateral interactions occur when addition of a monomer facilitates side-to-side interactions with another fibril, with the rate of lateral interactions being dependent on both the concentration of monomers and that of existing fibrils.11 Branching and lateral interactions are equivalent processes of secondary nucleation.
Two distinct types of seeding reactions are known. Addition of soluble protein identical to the fibril seed or nucleus involves homologous seeding or self-seeding. Addition of a different soluble protein induces heterologous seeding or cross-seeding. Heterogeneity of amyloid protein composition within a single patient has been clinically demonstrated16,17 and has been observed in murine amyloidosis models.18,19 It reinforces the critical importance of understanding the recruitment of soluble proteins in both types of seeding reactions in the context of human diseases.
All processes (primary nucleation, elongation, and secondary nucleation) involved in fibril formation are active during all phases of the reaction; in other words, none of the three phases seen in the overall kinetics (lag phase, elongation phase, and the final plateau) can be related to a single microscopic process (primary nucleation, elongation, and secondary nucleation).3 Thus, modifications of each microscopic process influence the overall kinetic curve; i.e., changes in the elongation rate constant have large effects on the duration of the lag phase with respect to similar changes in the primary nucleation constant.
The seeded reactions circumvent these problems by bypassing the de novo formation of nuclei from monomers,11 resulting, in the ideal cases, in exponential traces in which fibril elongation is the dominant process. The presence of a lag phase usually implies a secondary nucleation mechanism in seeded reactions.11
In the context of AL amyloidosis, previous work by our group and others20–27 has shown a direct correlation between low protein stability and the propensity for amyloid formation for some AL proteins. However, we have also found that proteins with similar thermodynamic parameters present different fibril formation kinetics.23–25,27,28 This suggests that, within a certain thermodynamic stability regime, there are additional factors that play a role in determining the amyloidogenic potential of AL proteins.
Here, we have investigated the mechanism of in vitro fibril formation within the context of AL amyloidosis by studying the aggregation kinetics of four immunoglobulin light chains and the wild-type, κI germline protein. The κI patient-derived, variable domain proteins share substantial sequence identity with the κI germline sequence: AL-09, 94%; AL-12, 91%; and AL-103, 96%. AL-T05, from the λ1b subgroup, is 48% identical with respect to the κI germline. The ability of AL proteins to form an “amyloidogenic nucleus” and to self-propagate was assessed using de novo fibril formation assays. We then studied the differential ability of AL fibrils to self-seed (homologous) and cross-seed (heterologous) to investigate the conformational specificity and the kinetic dominance of elongation in the amyloid reaction. Self-seeding reactions allow the determination of the elongation properties of amyloid fibrils and the efficiency of each protein to elongate the nascent fibril with preformed fibrils from the same protein. Cross-seeding experiments evaluate the ability and efficiency of a protein for sequence and conformational specificity.
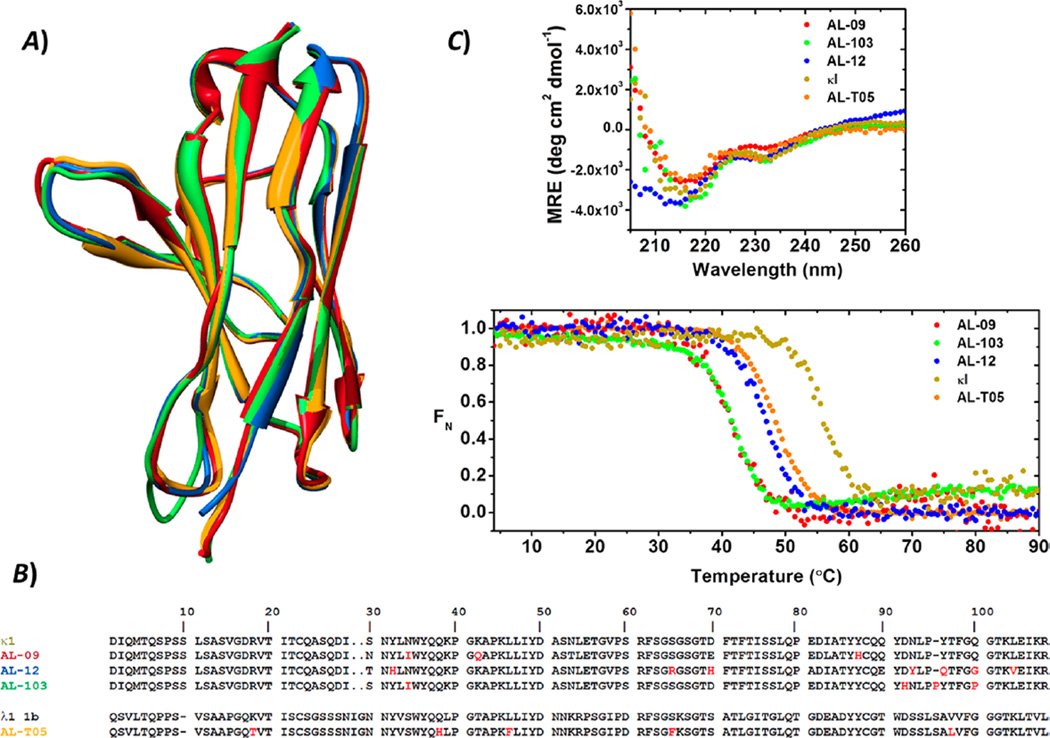
Of the κI subgroup proteins, AL-09 is quite amyloidogenic.29 AL-09 forms fibrils quickly and over a broad pH range (pH 2.0–9.0). The stability of the AL-09 protein has been extensively characterized, presenting a fully reversible, two-state chemical and thermal unfolding.23,30 AL-103 presents thermodynamic stability parameters comparable to those of AL-09 (Figure 1B) but shows strong kinetic control of protein folding with delayed and restricted kinetics of amyloid formation with a strong pH dependence (pH 2.0–7.0), because of the presence of the somatic mutation/insertion P95aIns.24,27,29 AL-12 is slightly more stable than AL-09 and AL-103 and presents delayed amyloid formation kinetics with respect to its germline protein κI.28 AL-12 amyloid fibril formation kinetics show a weak pH dependence, as it is able to form fibrils within the pH range of 2.0–8.0. κI germline protein (WT), used as control, is known to exhibit higher stability and delayed fibril formation compared to those of the patient proteins AL-09 and AL-103.27 κI is able to form amyloid fibrils only at pH 2.0; hence, the fibril formation experiments were conducted at this pH value. To further probe the relationship between the seeding specificity and the sequence, AL-T05 protein was included as a heterologous protein. Despite having a sequence only 48% identical to that of the κI proteins, AL-T05 shares their monomeric structure and has a thermodynamic stability comparable to that of AL-12. AL-T05 presents the fastest kinetics of fibril formation studied by our group to date.31
Figure 1.
Structural and stability properties of the VL proteins prior to amyloid formation reactions. (A) Structural and (B) sequence alignments of the VL immunoglobulin proteins. The sequence of the λ1 1b germline VL was included for the purpose of comparison. Nonconservative somatic mutations are colored red. (C) Far-UV CD spectra and thermal unfolding analysis of AL-09 (red), AL-103 (green), AL-12 (blue), AL-T05 (black), and κI (olive green). All proteins display β-sheet structure with the two characteristic minima (235 and ~217 nm) for these proteins. Experimental conditions were as follows: 20 µM protein in 10 mM Tris-HCl (pH 7.4). Far UV-CD spectra were recorded at 4 °C. Thermal denaturation experiments were performed from 4 to 90 °C at a rate of 0.5 °C min−1.
In this work, we have shown that the efficiency of recruitment between proteins with different sequences differs depending on the amyloidogenic nature of the protein, with the most amyloidogenic proteins showing very little effect and the least amyloidogenic proteins showing a preference for specific seeds to accelerate amyloid formation.
EXPERIMENTAL PROCEDURES
Chemicals
Water was Milli-Q grade. Yeast extract and tryptone were from Difco. Other reagents were from Sigma-Aldrich.
Cloning, Expression, Extraction, and Purification of Recombinant VL AL-09, AL-12, AL-103, and κI
AL-09, AL-12, and AL-103 are patient-derived variable domain proteins belonging to the germline gene product, κI O18/O8 (also known as IGKV 1–33); their sequences were obtained from patients with cardiac involvement.32 Germline κI O18/O8 will be called κI herein for the sake of simplicity. We also omitted VL from the name of all proteins herein for the sake of simplicity.
Protein expression was performed as reported previously.25,30,33 Briefly, all plasmids were transformed into Escherichia coli BL21(DE3) Gold competent cells (Stratagene, La Jolla, CA), and protein expression was induced overnight with 0.8 mM isopropyl β-d-1-thiogalactopyranoside.
Proteins were extracted and then purified by size-exclusion chromatography with a HiLoad 16/60 Superdex 75 column on an AKTA FPLC (GE Healthcare, Piscataway, NJ) system, in 10 mM Tris-HCl (pH 7.4).
Protein purity and homogeneity were determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis and analytical size-exclusion chromatography (see below). Protein concentration was determined by UV absorption at 280 nm using an extinction coefficient calculated from the amino acid sequence as follows: ε = 14890 M−1 cm−1 for κI and AL-103, ε = 13610 M−1 cm−1 for AL-09 and AL-12, and ε = 18020 M−1 cm−1 for AL-T05. Pure proteins were frozen and stored at −80 °C.
Circular Dichroism Spectroscopy
As a quality control measurement, far-UV circular dichroism (CD) spectroscopy was used to confirm that the proteins retained their native secondary structure at pH 7.4 and at the beginning of the fibril formation reaction at pH 2.0. Far-UV CD spectra from 260 to 200 nm (1 nm bandwidth) were acquired at 4 °C, on a Jasco model 810 spectropolarimeter (JASCO, Inc., Easton, MD) using a 0.2 cm path-length quartz cuvette. All samples (20 µM) were prepared in 10 mM Tris-HCl (pH 7.4) or 10 mM acetate borate citrate (ABC) buffer and 150 mM NaCl (pH 2.0). Thermal unfolding/refolding experiments were conducted following the ellipticity at 217 nm over a temperature range of 4–90 °C to determine the melting temperature (Tm) and to ensure the quality of the preparation, as reported previously.24 Temperature was regulated within ±0.002 °C using a Peltier system.
Sample Preparation for the in Vitro Fibril Formation Assay
Proteins were thawed at 4 °C, filtered using 0.45 µm membranes, and equilibrated for 24 h at 4 °C. Filtered, equilibrated proteins were ultracentrifuged at a speed of 90000 rpm (645000g) for 3.3 h in an NVT-90 rotor on an Optima L-100 XP centrifuge (Beckman Coulter). This step was performed to remove any preformed aggregates formed during the thawing process of the soluble protein, as reported previously.24 After ultracentrifugation, and prior to initiation of the fibril formation reaction, the global structure and oligomeric state (monomer–dimer equilibrium) of the proteins were confirmed via far-UV CD spectra, thermal unfolding, and analytical size-exclusion chromatography to ensure the integrity and homogeneity of the proteins before the initiation of the fibril formation reaction.
Size-Exclusion Chromatography
Analytical size-exclusion chromatography was conducted at 4 °C using a BioSil 125-5 high-performance liquid chromatography (HPLC) (Bio-Rad) size-exclusion column on an AKTA FPLC system (GE Healthcare). The column was equilibrated with 50 mM Na2HPO4, 50 mM NaH2PO4, 150 mM NaCl, and 0.02% NaN3 (pH 6.8). Chromatographic analyses were conducted at 0.2 mL/min. κ light chain variable domains have been previously characterized as weak homodimers at relatively high concentrations (~700 µM), with dissociation constants that range between 300 and 0.2 µM as calculated by NMR diffusion experiments.34 To re-establish the dimer–monomer equilibrium, protein samples (200 µL of pure proteins diluted to 20 µM) were incubated for 24 h at 4 °C prior to injection. Chromatographic peaks were detected by UV absorbance at 280 nm. Molecular weight and oligomeric states were estimated from elution volumes using a molecular weight calibration curve as reported previously.25 All proteins studied under these experimental conditions were monomeric when they were injected at 20 µM, demonstrating the monodispersity of the protein before the beginning of the reaction.
In Vitro Fibril Formation Assay
De Novo Nucleation (nonseeded) Experiments
Samples of each filtered and ultracentrifuged protein were prepared, on ice, at a final concentration of 20 µM in 10 mM ABC buffer (pH 2.0) containing 150 mM NaCl, 10 µM thioflavin T (ThT), and 0.02% NaN3, in 1.5 mL low-binding microcentrifuge tubes (total volume of 1.0 mL).
In vitro fibrillogenesis reactions were conducted by monitoring the fluorescence emission enhancement of thioflavin T (ThT) that occurs when ThT binds to amyloid fibrils. All fibril formation assays were performed in triplicate (260 µL per well) using black 96-well polystyrene plates (Greiner, Monroe, NC) sealed with plate sealers (Nunc, Roskilde, Denmark), covered with a black polystyrene cover, and sealed with tape to reduce evaporation. Samples were incubated at 37 °C while being continuously orbitally shaken (300 rpm) in a New Brunswick Scientific Innova40 incubator shaker. Fibril formation was monitored daily for 1 month (~750 h) following fluorescence on a plate reader (Analyst AD, Molecular Devices, Sunnyvale, CA). The excitation wavelength used was 440 nm, and the emission wavelength was 480 nm. The plate sealer and the tape sealing the cover were replaced daily.
We considered that a fibril formation reaction had occurred when we observed an at least 4-fold ThT fluorescence enhancement (~200000 AU in our system). The presence of amyloid fibrils was confirmed by electron microscopy.
Preparation of Amyloid Seeds
For the seeding experiments, fresh amyloid seeds were prepared as follows. A 800 µL volume of an amyloid fibril solution (20 µM soluble monomer at the beginning of the reaction) from de novo experiments was collected and transferred into low-binding microcentrifuge tubes. Samples were sonicated at room temperature for 10 s using a Branson model 8510 water bath sonicator (Branson Ultrasonics Corp., Danbury, CT).
The yield of the de novo fibril formation reaction for each protein was verified by an reverse phase HPLC assay following the method used by O’Nuallain et al.,35 measuring the free monomer concentration remaining in solution when the plateau of the de novo reactions was reached. The reaction yields, relative to the initial monomer concentration, were as follows: AL-09, 67.2% (13.4 µM); AL-103, 86.5% (17.3 µM); AL-12, 89.1% (17.8 µM); κI, 93.4% (18.7 µM); AL-T05, 94.3% (18.8 µM). The values in parentheses are the equivalent monomer concentrations in the fibrils.
Fibril Elongation [self-seeding (homologous) and cross-seeding (heterologous)] Experiments
Fresh solutions of each AL protein at 20 µM were prepared as we prepared them for de novo reactions. Ten microliters of freshly prepared seeds [1% (v/v)] was sonicated and added to the reaction mixture. On the basis of the fibril reaction yield, we added seeds at concentrations between 0.13 and 0.18 µM (monomer concentration in the fibrils). ThT fluorescence was monitored in the plate reader as previously described for the de novo experiments. In all cases, the same batch of freshly prepared seeds was used to avoid variability within the seeded reactions. Samples were kept on ice until the beginning of the reaction.
De novo control reactions as well as control wells containing only ThT without protein were performed in parallel, to ensure that any differences in fibril formation kinetics (measured as t50 values or the time it takes to complete 50% of the fibril formation reaction as well as elongation rates) could be attributed directly to the presence of the specific seed, while ensuring the reproducibility of the observations.
Analysis of Experimental Kinetic Data
The t50 value was obtained by fitting each independent kinetic trace to a sigmoidal function (defined as a Boltzmann function by the Origin software package http://www.originlab.com/www/helponline/Origin/en/UserGuide/Boltzmann.html) as previously reported.27,28,36
| (1) |
where A1 is the initial fluorescence value, A2 is the final fluorescence value, x0 is the midpoint (or t50 value), and dx is defined as the time constant.
A shorter t50 value indicates a faster reaction to form fibrils. The rates of fibril elongation were calculated from the slope of the linear fitting of the growth phase. The comparison and statistical analysis of the effects of homologous and heterologous seeding over the fibril formation were based on a paired Student’s t test. Significance was reported at the 95% (p < 0.05) confidence level.
Electron Microscopy
A 3 µL fibril sample was placed on a 300 mesh copper Formvar/carbon grid (Electron Microscopy Science, Hatfield, PA), and excess liquid was removed. The samples were negatively stained with 2% uranyl acetate, washed twice with H2O, and air-dried. Grids were analyzed on a Philips Tecnai T12 transmission electron microscope at 80 kV (FEI, Hillsboro, OR).
RESULTS
Sequence and Biophysical Characteristics of the Proteins Used in This Study
In this work, four variable domain proteins (VL) derived from AL amyloidosis patients have been selected, to study the effect of preformed fibrils or “seeds” on the kinetics of fibril formation of these proteins. By using this set of proteins (our group has extensively studied their biophysical and amyloidogenic properties), we cover a broad range of protein stabilities and amyloidogenic properties (Figure 1). To ensure that all the fibril formation reactions start with the proteins in the native conformation, far-UV CD spectral and thermal unfolding experiments were conducted. All proteins presented the characteristic far-UV CD spectrum at pH 7.4 (Figure 1C, top panel), observed in VL proteins, as previously reported.20–22
At pH 2.0 in the presence of 150 mM NaCl, AL-09 shows stable β-sheet structure. AL-12, κI, and AL-T05 also retain their secondary structure. AL-103 presented an increment in random coil content (Figure S3).
The thermal unfolding transitions of all samples were sigmoidal (Figure 1C, bottom panel) and fully reversible at both pH values tested. Thermodynamic parameters at pH 7.4 were in agreement with the previously reported values.23–25,27,36
At pH 2.0 in the presence of 150 mM NaCl, AL-T05, AL-09, and AL-103 showed a destabilization in thermal unfolding, while κI and AL-12 have similar Tm values at pH 2.0 and 7.4. AL-T05 showed the smallest change in ellipticity during the thermal unfolding at pH 2.0 (Figure S3).
Preformed Seeds Have Differential Effects in the Fibrillogenesis of AL Proteins
Preformed fibrils or “seeds” of each of the five proteins were screened for their ability to accelerate amyloid fibril formation in the presence of soluble AL proteins at pH 2.0 [10 mM ABC buffer (pH 2.0) and 150 mM NaCl (37 °C)], conditions under which all proteins studied are able to form fibrils.
In the absence of preformed seeds, nucleation and elongation processes drive the fibril formation reaction, and the kinetics present the classical sigmoidal transitions.3 For amyloid reactions with a high concentration of seeds, the overall reaction is dominated by fibril elongation processes (elongation and secondary nucleation) and the lag phase is abolished, resulting in exponential kinetic traces with slopes for the reaction different from those of de novo reactions.3,13 In the case of amyloid kinetics seeded with a very low concentration of preformed fibrils, we expect that some seeds will shorten the lag phase without affecting the slope of the exponential phase, allowing for the comparison among different proteins without confounding the analysis.3,13
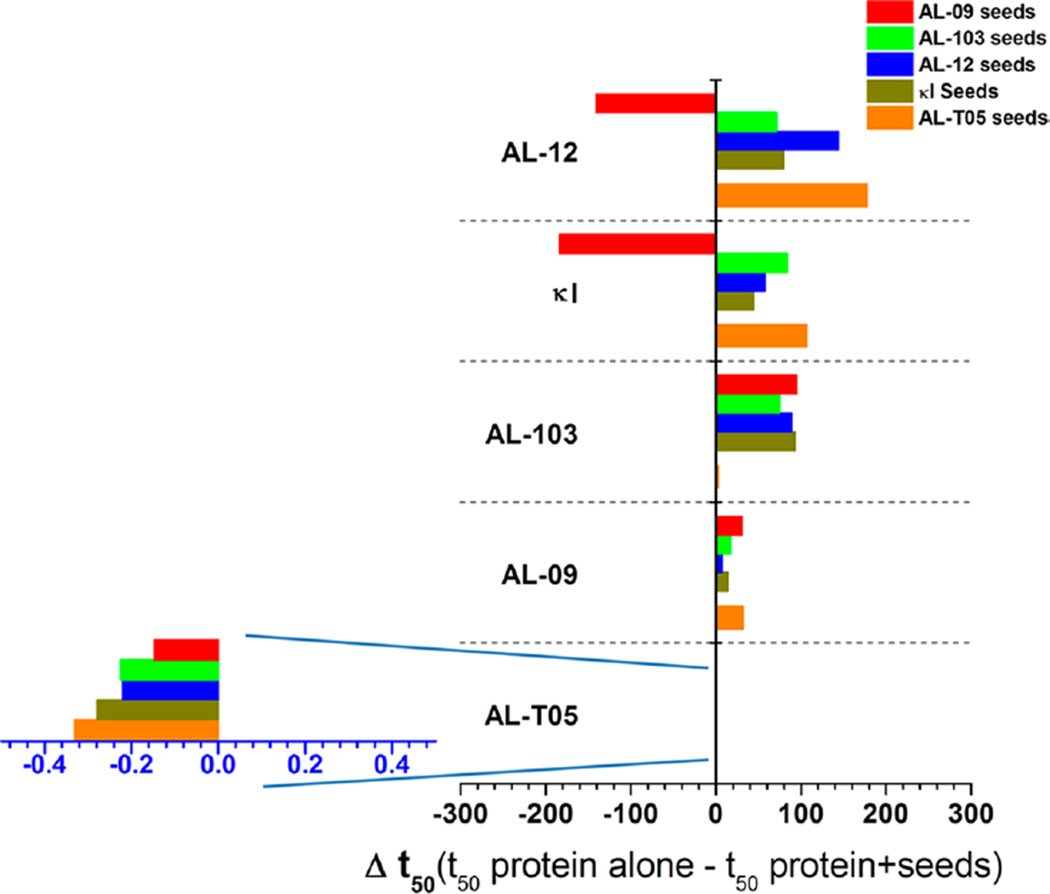
Figure 2 shows a summary of the effect of the homologous and heterologous seeding. We are showing the difference in the time it takes to complete 50% of the fibril formation reaction (t50 values) between de novo reactions and the reactions in the presence of each seed (Δt50). We will highlight only the most important results for our comparative analysis between AL proteins and their mutants (see Figures 3–7 for all the amyloid fibril formation kinetic traces).
Figure 2.
Differential effect of seeding on the t50 value of fibril formation kinetics of VL proteins. Comparison of Δt50 values as a function of the seed employed. Data are from fibril formation reactions conducted in triplicate. A reaction was considered positive when the ThT fluorescence increased 4-fold (>200000 AU). All proteins tested were able to form fibrils at pH 2.0. Note the difference in the x-axis for AL-T05.
Figure 3.
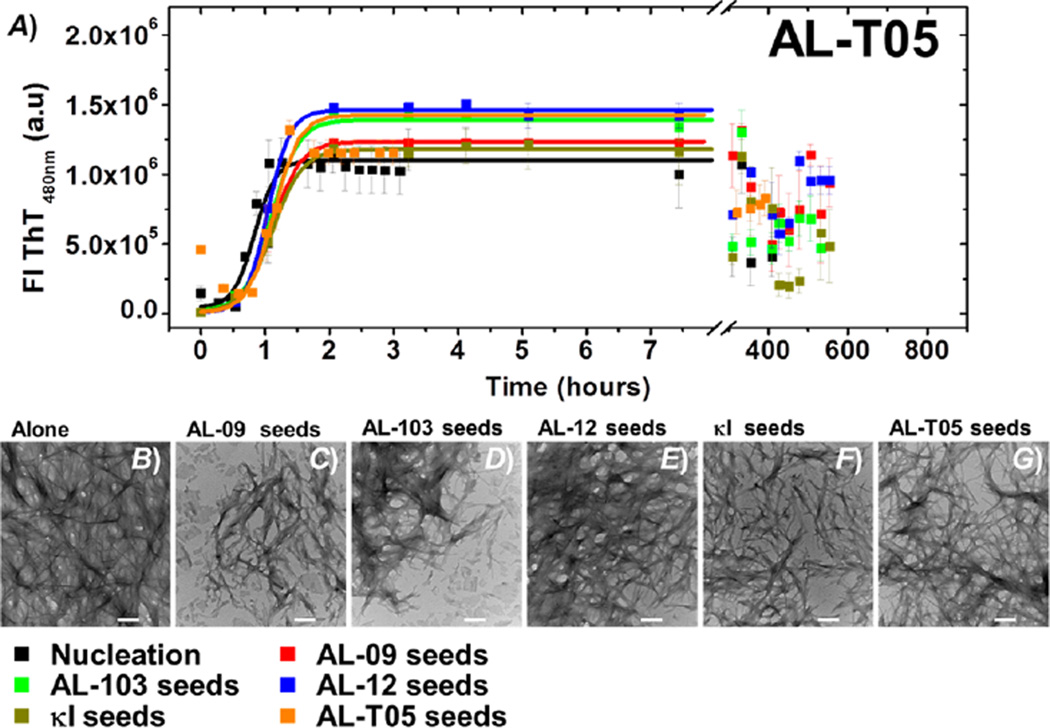
(A) Self-seeding and cross-seeding experiments with AL-T05: de novo at pH 2.0 (black), AL-09 seeds (red), AL-103 seeds (green), AL-12 seeds (blue), κI seeds (brown), and AL-T05 seeds (orange). All reactions were performed with 20 mM protein in the presence of 1% seeds. Transmission electron microscopy (TEM) images of AL-09 at the end point of the reaction. (B) De novo experiment at pH 2.0 in the presence of (C) AL-09 seeds, (D) AL-103 seeds, (E) AL-12 seeds, (F) κI seeds, and (G) AL-T05 seeds. The scale bar represents 200 nm.
Figure 7.
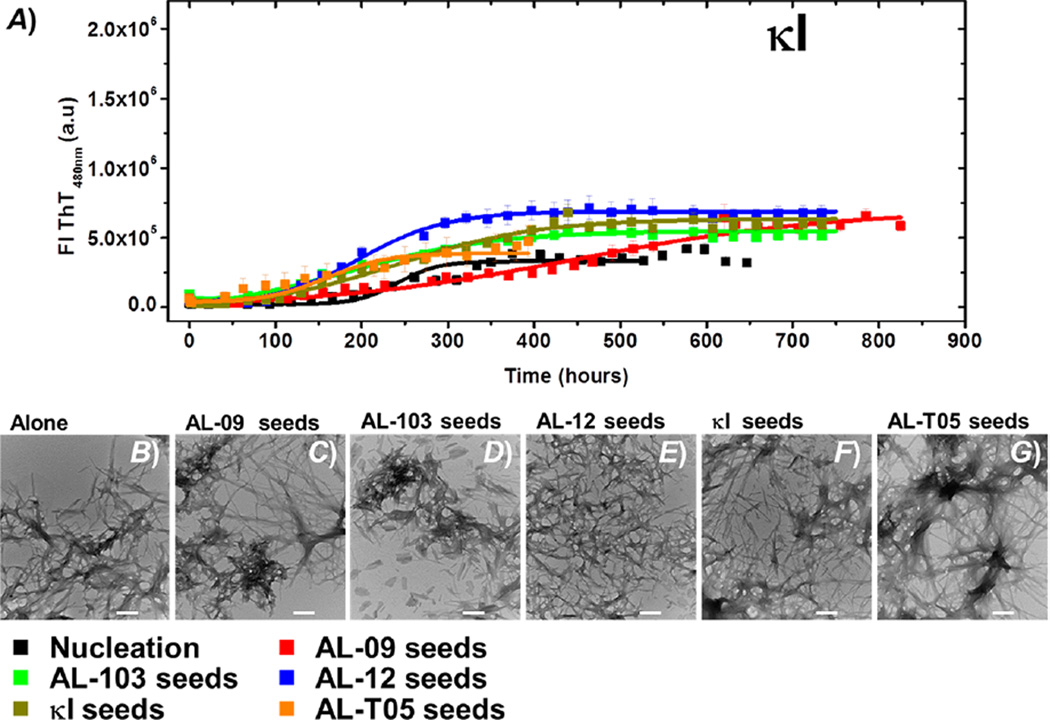
(A) Self- and cross-seeding experiments with κI: de novo at pH 2.0 (black), AL-09 seeds (red), AL-103 seeds (green), AL-12 seeds (blue), κI seeds (brown), and AL-T05 seeds (orange). All reactions were performed with 20 µM protein in the presence of 1% seeds. Transmission electron microscopy (TEM) images of AL-09 at the end point of the reaction. (B) De novo experiment at pH 2.0 in the presence of (C) AL-09 seeds, (D) AL-103 seeds, (E) AL-12 seeds, (F) κI seeds, and (G) AL-T05 seeds. The scale bar represents 200 nm.
The proteins AL-T05 (t50 de novo = 0.8 ± 0.1 h) and AL-09 (t50 de novo = 54.5 ± 0.8 h) have the fastest de novo kinetics of all proteins tested. For AL-T05 (Figure 3), neither homologous nor heterologous seeding affected the kinetics of fibril formation significantly (see Figure 2 and Table 1).
Table 1.
t50 (in hours) Values of Fibril Formation Kinetics of AL Proteins at pH 2.0
| AL-09 | AL-103 | AL-12 | κI | AL-T05 | |
|---|---|---|---|---|---|
| de novo (pH 7.4) | 244.1 ± 14.2a | 172.1 ± 36.7b | 430.0 ± 20.0c | − | 15.8 ± 0.9d |
| de novo | 54.5 ± 0.8 | 107.1 ± 0.9 | 357.0 ± 1.7 | 266.2 ± 10.9 | 0.8 ± 0.1 |
| AL-09 seeds | 24.1 ± 0.8 | 12.4 ± 0.2 | 497.4 ± 2.7 | 450.0 ± 14.3 | 1.0 ± 0.1 |
| AL-103 seeds | 37.6 ± 0.5 | 32.7 ± 0.6 | 285.8 ± 1.8 | 182.4 ± 6.1 | 1.1 ± 0.1 |
| AL-12 seeds | 47.3 ± 1.2 | 18.3 ± 0.9 | 213.2 ± 27.6 | 208.6 ± 3.7 | 1.0 ± 0.1 |
| κI seeds | 40.6 ± 0.6 | 14.1 ± 0.4 | 278.2 ± 2.4 | 221.8 ± 6.1 | 1.1 ± 0.1 |
| AL-T05 seeds | 23.3 ± 1.8 | 104.6 ± 5.2 | 179.8 ± 2.6 | 159.7 ± 46.6 | 1.2 ± 0.2 |
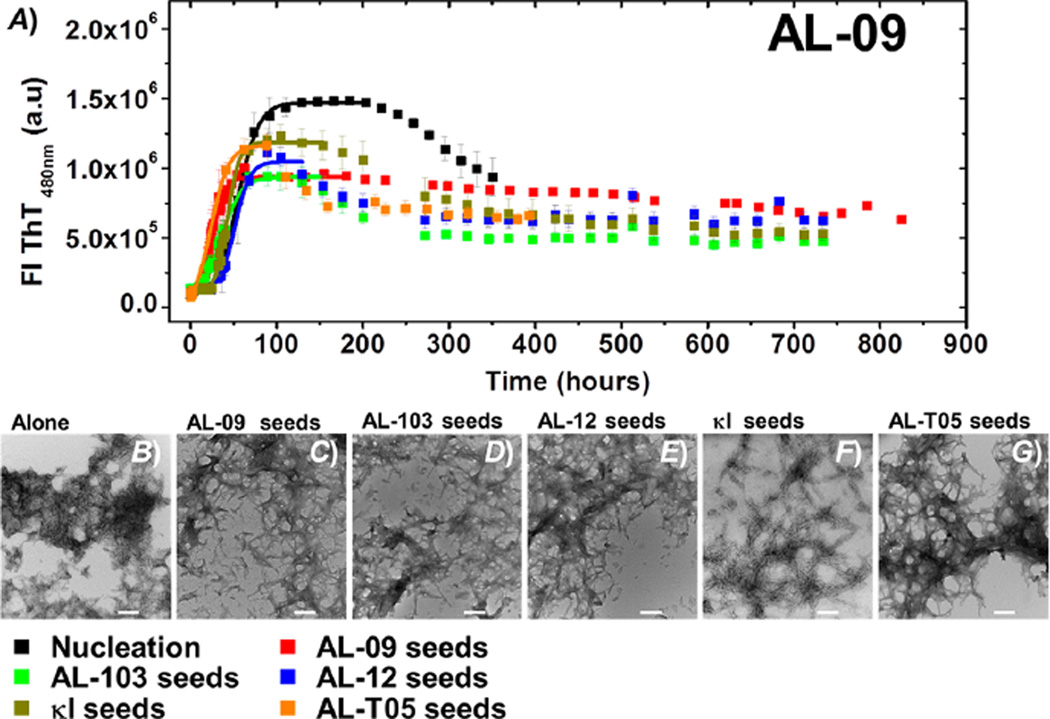
In the case of AL-09 (Figure 4), a slight acceleration can be observed in all seeded reactions, including with AL-T05 seeds and the homologous (self) seeding experiment.
Figure 4.
(A) Self- and cross-seeding experiments with AL-09: de novo at pH 2.0 (black), AL-09 seeds (red), AL-103 seeds (green), AL-12 seeds (blue), κI seeds (brown), and AL-T05 seeds (orange). All reactions were performed with 20 µM protein in the presence of 1% seeds. Transmission electron microscopy (TEM) images of AL-09 at the end point of the reaction. (B) De novo experiment at pH 2.0 in the presence of (C) AL-09 seeds, (D) AL-103 seeds, (E) AL-12 seeds, (F) κI seeds, and (G) AL-T05 seeds. The scale bar represents 200 nm.
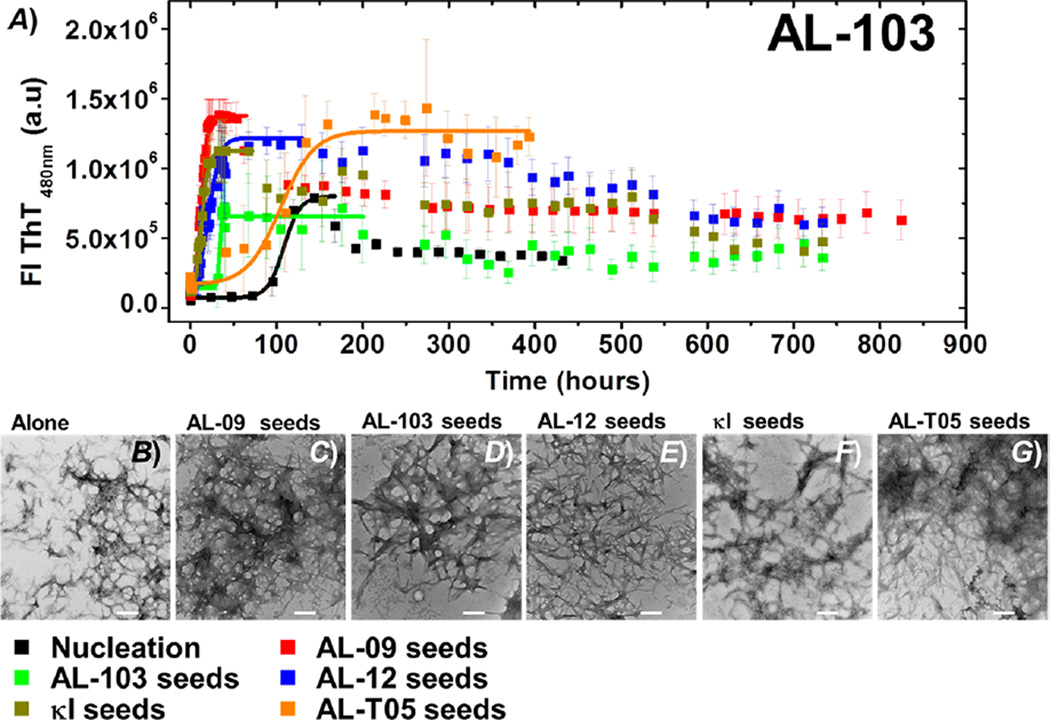
AL-103 (t50 de novo = 107.1 ± 0.9 h) shows that homologous seeding has a dramatic accelerating effect on the AL-103 kinetics. Heterologous seeding reactions of proteins of the same subtype (κI) also accelerate the reaction (Figure 2 and Table 1). Interestingly, AL-T05 seeds were also able to recruit AL-103 monomers (Figure 5G), but less efficiently (t50 = 104.6 ± 5.2 h).
Figure 5.
(A) Self- and cross-seeding experiments with AL-103: de novo at pH 2.0 (black), AL-09 seeds (red), AL-103 seeds (green), AL-12 seeds (blue), κI seeds (brown), and AL-T05 seeds (orange). All reactions were performed with 20 µM protein in the presence of 1% seeds. Transmission electron microscopy (TEM) images of AL-09 at the end point of the reaction. (B) De novo experiment at pH 2.0 in the presence of (C) AL-09 seeds, (D) AL-103 seeds, (E) AL-12 seeds, (F) κI seeds, and (G) AL-T05 seeds. The scale bar represents 200 nm.
In addition, it is worth noting that the AL-103 de novo amyloid formation kinetics is highly stochastic; this stochastic behavior is reduced to the point of being eliminated in most of the seeded reactions.
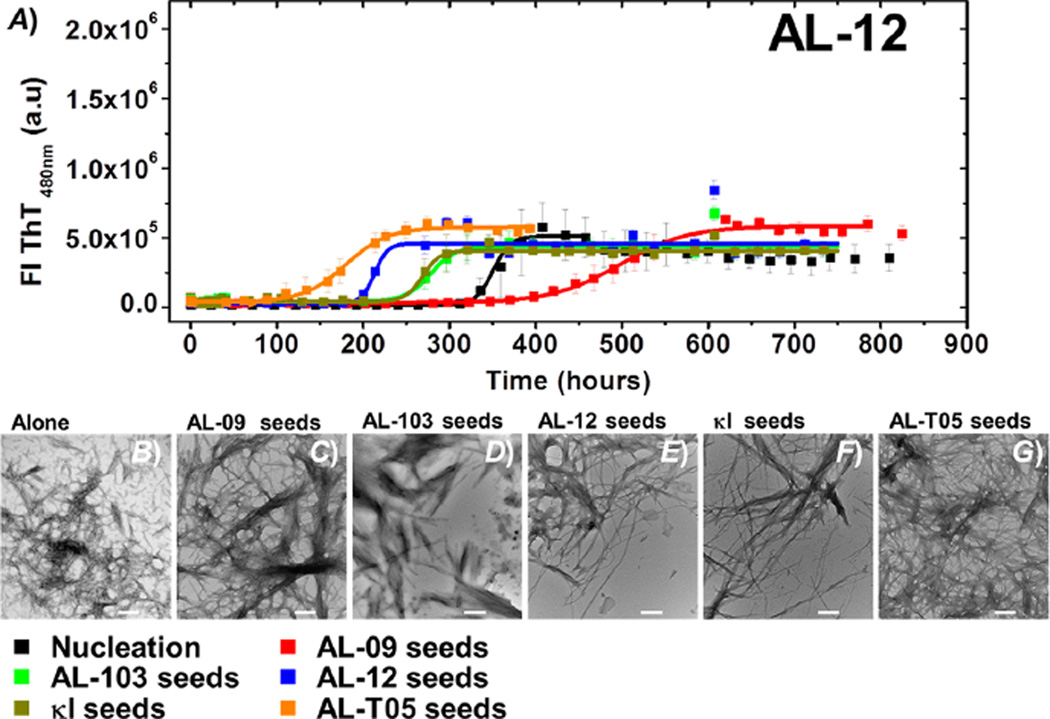
AL-12 shows the most remarkable results as its de novo amyloid formation kinetics are the slowest at pH 2.0 (t50 de novo = 356.96 ± 1.70 h). The most striking results are the dramatic acceleration of AL-12 seeded kinetics in the presence of AL-T05 seeds (t50 = 179.8 ± 2.6 h) and the delay observed when the reaction is seeded with AL-09 fibrils (t50 = 497.4 ± 2.7 h), opposite to what we expected according to the seeding sequence–homology specificity observed for AL-103 (Figure 6).
Figure 6.
(A) Self- and cross-seeding experiments with AL-12: de novo at pH 2.0 (black), AL-09 seeds (red), AL-103 seeds (green), AL-12 seeds (blue), κI seeds (brown), and AL-T05 seeds (orange). All reactions were performed with 20 µM protein in the presence of 1% seeds. Transmission electron microscopy (TEM) images of AL-09 at the end point of the reaction. (B) De novo experiment at pH 2.0 in the presence of (C) AL-09 seeds, (D) AL-103 seeds, (E) AL-12 seeds, (F) κI seeds, and (G) AL-T05 seeds. The scale bar represents 200 nm.
When κI or AL-103 seeds are used to seed AL-12 monomeric protein, comparable accelerations are observed (t50 value of 278.2 ± 2.4 or 285.8 ± 1.8 h, respectively).
Analogous to AL-12, the wild-type control κI presents interesting results. De novo amyloid fibril kinetics of κI is among the slowest reactions (t50 de novo = 266.2 ± 10.9 h). Homologous and heterologous seeding kinetics of κI present a slight acceleration with respect to de novo kinetics (Figure 7). Like that of AL-12, the fibril formation reaction in the presence of AL-09 seeds is delayed; AL-T05 seeds accelerate the reaction. Although we observe a differential effect in the κI reactions depending on the type of seed employed, the effect is not as dramatic as that observed for the AL-12 seeded reactions. Additionally, we noted that, contrary to the sharp transitions observed for the other proteins tested, κI seeded reactions present smooth and steady transitions, suggesting low cooperativity. This is probably due to κI’s poor nucleation and recruitment efficiency.
Consistent with what we observed for t50 values, the elongation rates calculated from the seeding reactions are faster for the amyloidogenic proteins (AL-T05, AL-09, and AL-103) than for the nonamyloidogenic proteins (AL-12 and κI) (Figure S1). AL-103 has overall elongation rates faster than those of AL-09 in the presence of seeds.
For many years, it has been considered that VL corresponds to the central region undergoing misfolding and aggregation in AL amyloidosis.1,2 However, recent proteomic studies with amyloid deposits from fat aspirates37 found full-length (FL) light chains as part of the amyloid deposits. This was later confirmed by a proteomics study using biopsy samples from affected tissues.38 These results suggest that AL amyloid deposits in patients could be formed by FL, VL, or mixtures of FL, VL, and/or other fragments.
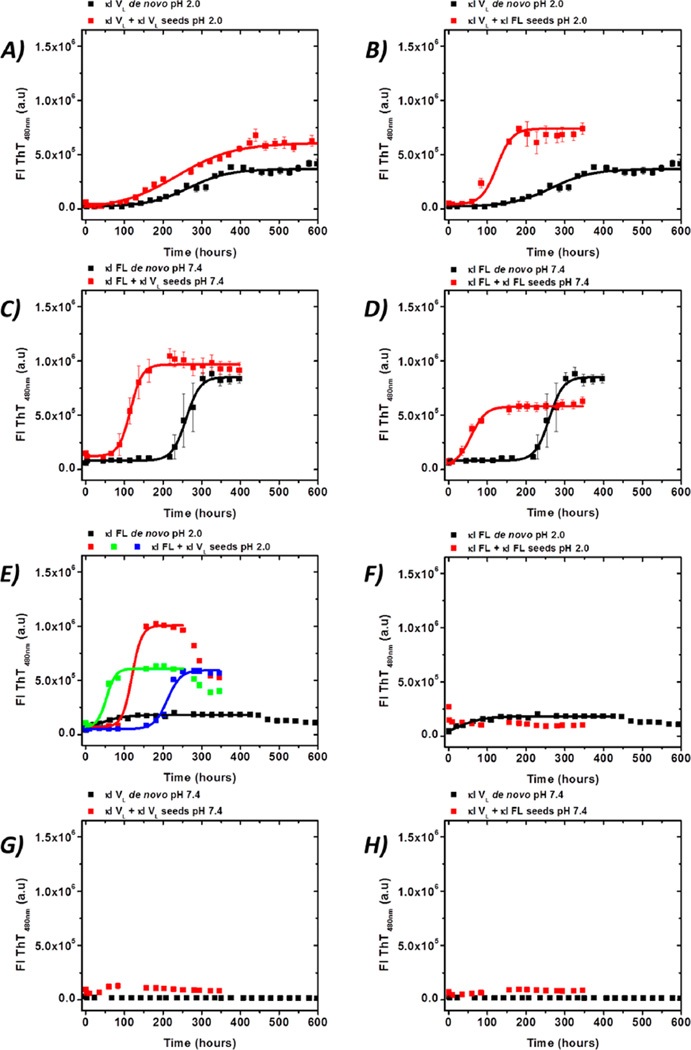
To test whether the VL domain could promote the fibril formation of full-length (FL) proteins, a proof-of-principle cross-seeding experiment using κI VL and FL was conducted. We chose κI because this was the protein that presented the lowest seeding efficiency. For this particular experiment, we added an experimental condition at pH 7.4 because κI FL is able to form fibrils at neutral pH.39 Comparing the reactions at pH 2.0 and 7.4 will allow us to determine if the different solution conditions play a role in the elongation of κI VL and FL fibril formation.
The presence of κI VL seeds shows a large increase in the rate of acceleration of the κI VL reaction at pH 2.0 (Figure 8A). However, the effect of κI VL seeds on soluble κI VL at pH 2.0 (Figure 8A) is not as dramatic as the effect we observe with κI FL seeds at pH 2.0 (Figure 8B).
Figure 8.
Self- and cross-seeding experiments with κI FL in the presence of κI VL seeds and κI VL in the presence of κI FL seeds. De novo fibril formation (black) and seeded (red) reactions were conducted with 20 µM protein: κI VL seeds (left) and κI FL seeds (right). For the seeded reaction, 1% (v/v, monomer concentration in the fibrils of 0.18 µM) seeds were used in all cases.
Homologous seeding of κI FL with κI FL seeds is accelerated at pH 7.4 and more favorable than in the presence of κI VL seeds (Figure 8C,D).
Preformed κI VL seeds have a highly stochastic accelerating effect on the κI FL fibril formation reaction at pH 2.0, with each well showing a different kinetic profile (Figure 8E).
By contrast, κI FL preformed seeds cannot initiate or elongate the fibril formation of κI FL at pH 2.0 (Figure 8F) or κI VL at 7.4 (Figure 8H). As expected, κI VL seeds are unable to initiate or elongate fibril formation at pH 7.4, with a behavior consistent with de novo reactions (Figure 8G)
Our results for κI VL/FL seeding are consistent with the idea of the variable domain being the main component of the in vivo amyloid aggregates, and the constant domain playing a modulating role in both the misfolding and fibril formation processes.39 It is interesting to note that for κI, the presence of FL seeds appears to have an accelerating effect, suggesting that the constant domain can accelerate the reaction.
Preformed Seeds Have Minor Effects on the Morphology of Mature Fibrils of AL Proteins
To further test whether the sequence-dependent seeding effects correlate with morphological characteristics of amyloid fibrils, we have performed transmission electron microscopy (TEM). As illustrated in Figures 3–7 (see Figure S2 for a compilation of all the TEM images), TEM images of fibrils at the end of the reaction present different morphologies and degrees of clustering as a function of the protein forming the aggregates and the kind of seed used in the reaction.
We observe in all de novo experiments at pH 2.0 (Figure S2 and Figures 3–7B) that the AL proteins form amyloid fibrillar aggregates consistent with the length and morphology of AL protein mature fibrils previously characterized in our laboratory.24,27,28,31
The most dramatic effect of seeding on fibril morphology was observed for AL-12. We observe that in the de novo experiments, AL-12 forms networks of straight fibrils (Figure S2 and Figure 6B). However, the presence of AL-09 seeds promotes the formation of the classical morphology we observe for this protein [long straight fibrils (Figure S2 and Figure 6C)], possibly as a consequence of the delay observed in the kinetics of fibril formation, where a slow reaction allows the formation of fewer fibril nuclei and longer fibrils.
In contrast, samples from kinetics seeded with AL-103 seeds form large clusters of long straight fibrils, laterally stacked together (Figure S2 and Figure 6D), whereas the self-seeded reaction, as well as the presence of κI and AL-T05 seeds, results in classical long straight fibrils (Figure 6F,G), consistent with the observed acceleration of fibril formation. It is interesting to note that AL-T05 seeds, with the heterologous amino acid sequence, induce the formation of AL-12 fibrils without affecting the fibril morphology significantly.
De novo AL-T05 fibril formation forms a dense network of clustered fibrils in a totally random arrangement (Figure S2 and Figure 3B), where individual amyloid fibril segments are difficult to observe. This is a typical behavior for in vitro AL amyloid fibril samples. In the self- and cross-seeded samples, we observe fewer dense clusters of individual straight fibrils laterally stacked together, consistent with the seeding of fibril formation (Figure S2 and Figure 3C–G). Individual fibrils showed the same morphological features that were observed in AL-T05 de novo kinetics under all conditions. In the presence of AL-09 seeds or AL-103 seeds, we also observe the presence of short, ribbonlike fibrils laterally stacked together (Figure S2 and Figure 3C,D), along with the typical long fibrils of AL-T05.
AL-09 fibrils obtained from de novo kinetics look like short rods, clustered in a random arrangement (Figure S2 and Figure 4B). By contrast, samples of AL-09 from self-seeded kinetics present slightly longer fibrils, forming clusters of entangled fibrils. A similar fibril morphology was found when AL-103 seeds or AL-12 seeds were used; however, we observed strong lateral interactions (Figure S2 and Figure 4D,E). Fibrils formed in the presence of κI seeds are long and thin straight fibrils. In the presence of AL-T05, the fibrils aggregate as large bundles of long fibrils (Figure S2 and Figure 4G). Fibrils obtained from AL-103 de novo kinetics did not present any significant difference in the fibril morphology of self- and cross-seeds samples (Figure S2 and Figure 5C–G) and were identical to previous reports: ribbonlike individual fibrils, with a strong tendency to form dense tangles. We speculate that AL-T05 and AL-09 form a large number of fibril nuclei, resulting in a large number of short mature fibrils.
With regard to the cross-seeding experiment of κI FL, fibrils obtained from κI FL de novo kinetics did not present a significant morphological difference compared to the crossseeded samples (Figure S2) and were identical to the results of previous reports.39
DISCUSSION
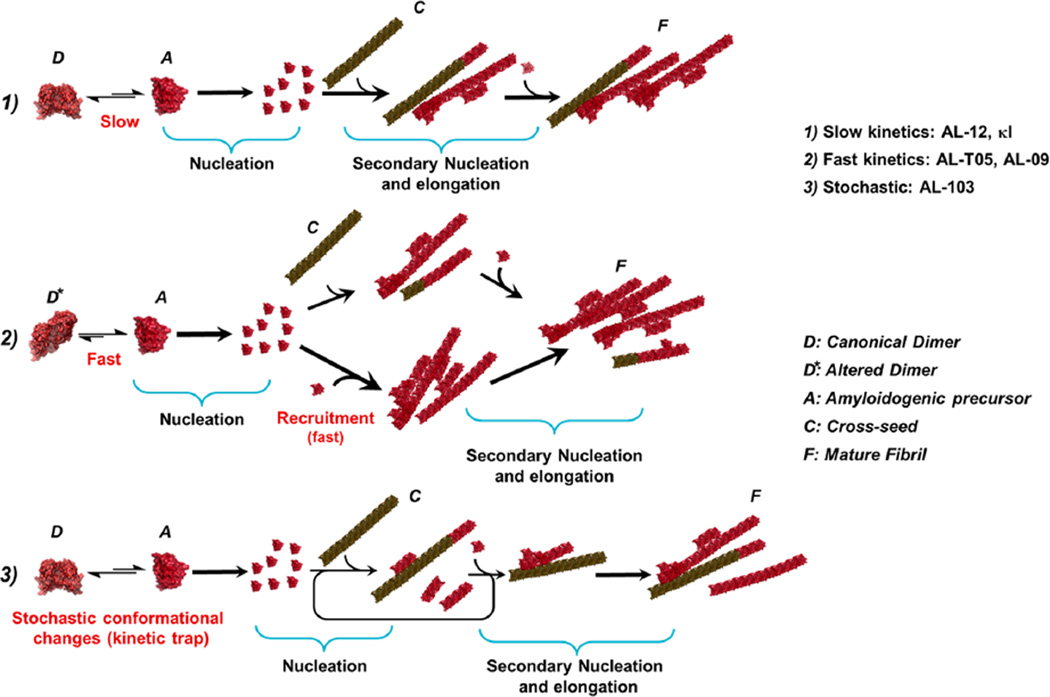
The seeded fibril formation of the AL proteins studied here shows differential effects that can be broadly grouped into three different behaviors (see Scheme 1).
Scheme 1.
Proposed Mechanism of Cross-Seeding.
(1) AL-09 and AL-T05 presented the fastest kinetics of fibril formation (de novo) and do not benefit significantly from the presence of seeds in conducting fibril formation. AL-09 has an amyloidogenic, altered dimer interface30 rotated 90° with respect to the canonical dimer interface. In the case of AL-T05 [a protein with a key mutation in the dimer interface (Q38H)], it presents a dimeric interface that is rotated 180°, similar to κI Y87H from ref 34 (unpublished observations).
We propose that the high efficiency of AL-09 and AL-T05 to form and populate amyloid nuclei is due to the mutations these proteins have in the dimer interface that shift the equilibrium toward the altered dimer, which facilitates misfolding events that trigger amyloid formation. The amyloid formation rates do not really increase with seeding because the nucleation reaction is already close to the maximal rate of fibril formation for those proteins.
(2) The proteins with the most delayed de novo kinetics, AL-12 and κI, presented a differential acceleration depending on the seed used. AL-12 exhibited the strongest differential effect of the kind of seeds on the fibril formation reaction. AL-12 de novo fibril formation kinetics is slow and highly independent of the pH,28 possibly experiencing a higher energy barrier of amyloid formation compared to those of AL-09 and AL-103. This barrier is reduced significantly in the presence of certain seeds or other accessory molecules, such as glycosaminoglycans.36
Unexpectedly, cross-seeded kinetics of AL-12 with AL-T05 seeds present the highest rate of acceleration observed. We speculate that the high efficiency of cross-seeding with AL-T05 is associated a conformational similarity rather than the degree of similarity of the amino acid sequence. κI presents a promiscuity of the dimer interface.23,30,34 κI has slow de novo fibril formation kinetics and is able to form fibrils at only pH 2.0.27 Although a similar differential acceleration as a function of the seed can be observed, it is not as dramatic as in the case of AL-12.
We speculate that κI is not efficient in populating the amyloidogenic conformation even though the dimer interface is promiscuous, and therefore, the recruitment of monomers is less efficient than that of the rest of the proteins. This supports the idea that seeds do not modify the amyloid formation pathway; it is only modulated by increasing or decreasing the rate of the secondary nucleation process.
(3) During de novo fibril formation, AL-103 undergoes stochastic conformational fluctuations to populate an amyloidogenic state.27,29,36
It is important to reiterate that AL-103 stability is not only thermodynamically but also kinetically determined, by the insertion of proline 95a cis–trans prolyl isomerization, which can enhance the population of a transient kinetic intermediate prone to fibril formation.27
The dramatic acceleration of the seeded reactions of AL-103 is in agreement with an autocatalytic reaction. We propose that the recruitment of soluble AL-103 monomers occurs preferentially via secondary nucleation, as described by Andersen and co-workers in their study of glucagon fibrils.11
Interestingly, AL-103 homologous seeding is less efficient than the cross-seeding. We propose that the conformational features of the AL-103 fibrils are slightly different from the incoming AL-103 monomer, decreasing the efficiency of incorporation of AL-103 protein. For cross-seeded reactions, the conformation of the AL-103 amyloidogenic precursor resembles the structures of heterologous seeds, and the incorporation into mature fibrils is more efficient. This could be a special case linked to the morphologies of AL-103 amyloid fibrils. Further studies are still needed.
The presence of preformed fibrils or seeds does not change the species populated in the amyloid formation pathway; it only modulates it by increasing the rate of the secondary nucleation process and the rate of elongation for AL-103. This is consistent with a kinetic process in which elongation and secondary nucleation occur, where seeding shortens the lag phase but does not affect the growth rate, and a lag phase is still present. This is because the surfaces of existing fibrils catalyze the nucleation of new fibrils from the monomeric state, with a rate that is dependent on both the concentration of monomers and that of existing fibrils.11
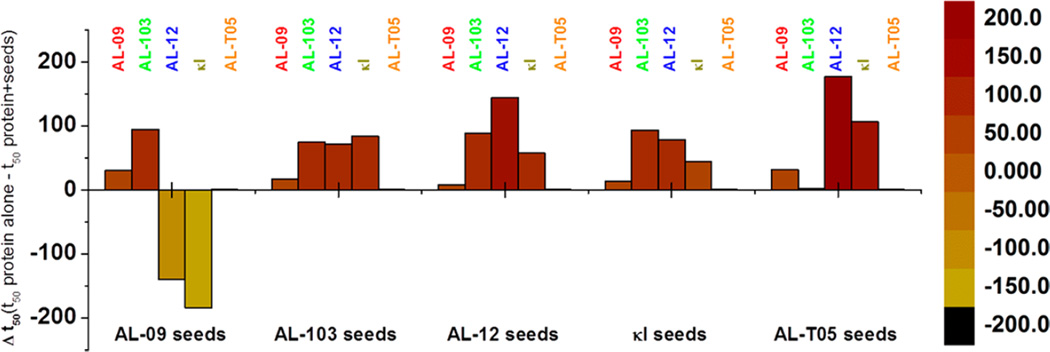
The efficiency of preformed fibril to cross-seed fibril formation of the proteins studied does not follow any specific pattern per seed used (Figure 9), suggesting that the efficiency to cross seed is more related to the efficiency of the soluble protein in populating amyloidogenic conformations rather than to the properties of the seed; however, this response is variable depending on the seed.
Figure 9.
Comparison of Δt50 values as a function of the seeds employed. Data are from fibril formation reactions that were conducted in triplicate. The reactions were considered positive when ThT fluorescence increased 4-fold (>200000 AU). All proteins tested were able to form fibrils at pH 2.0.
An interesting and surprising delay was observed in cross-seeded reactions with respect to de novo reactions for a few reactions. The most notorious delay was observed for the AL-12 fibril formation cross-seeded with AL-09, followed by κI reactions seeded with the AL-09 seeds. We speculate that the morphological and conformational properties of AL-09 seeds are not as favorable for elongation of the species populated by delaying the fibril formation pathway.
We propose that a combination of both the biophysical properties of the amyloid precursors and the topology of the nuclei plays an important role in the progression of the reaction, probably due to steric effects, consistent with a lateral or surface secondary nucleation process. This could be one of the explanations for why there are cases of in vivo amyloid plaques that have more than one type of amyloid precursor protein.2,17 Lateral secondary nucleation is a process similar to the one described by Ferrone et al. and Cohen et al.9,13 and has been revisited in recent reviews.3,6,40
Finally, the dramatic acceleration of κI FL kinetics caused by the presence of κI variable domain seeds stands in contrast with the low efficiency of the κI VL seeds to accelerate the κI VL soluble protein in the self-seeding experiments. We propose that κI FL protein populates to a major extent an amyloidogenic misfolded conformation compared to κI VL, because the κI FL folding pathway is kinetically controlled, resulting in a sort of kinetically trapped conformation prone to aggregation. De novo κI FL fibril formation is pH-dependent, with a stochastic behavior at pH <4.0,39 in contrast with that of κI VL, for which the de novo kinetics are highly homogeneous albeit restricted to pH 2.0. The presence of κI VL preformed seeds not only accelerates κI FL fibril formation at pH 7.4 but also results in a more homogeneous kinetic behavior without affecting the morphology of the fibrils, compared to the kinetics at pH 2.0. This suggests that preformed seeds of κI VL accelerate fibril formation via direct interaction with a kinetic misfolded intermediate of κI FL, whose stability is determined by the constant domain.39
Preformed homologous and heterologous seeds can accelerate reactions only under conditions under which de novo reactions occur, indicating that the κI species populated during the fibril formation pathway remain unchanged in the presence of seeds, suggesting that the energy barrier to populating the critical species needed for amyloid formation remains unchanged.
Future studies of the growing fibrils by real-time observations at a single-fibril level, in addition to the determination of the atomic-level structure of AL light chain fibrils, would be advantageous for elucidating the molecular basis of the autocatalytic process and the recruitment of soluble protein in AL amyloidosis disease.
Collectively, our results lead us to conclude that for AL proteins, fibril formation proceeds by the recruitment of soluble protein into fibrils via a lateral secondary nucleation mechanism. The interaction between the soluble protein and the nascent fibril is a conformation-dependent process, modulated by the ability of each protein to populate amyloidogenic conformations. It is clear that in addition to protein stability, disruptions in the quaternary structure could significantly increase the protein amyloidogenic propensity.
Supplementary Material
Acknowledgments
Funding
This work was supported by National Institutes of Health Grant R01 GM 071514. We are also thankful for the financial support offered by the Mayo Foundation and the generosity of amyloidosis patients and their families.
We thank Alex Tischer, the Ramirez-Alvarado lab members, Jon Wall, and Steve Kennel for helpful comments regarding the manuscript.
ABBREVIATIONS
- ThT
thioflavin T
- CD
circular dichroism
- VL
variable domain of immunoglobulin light chain
- FL
immunoglobulin light chain
- FI
fluorescence intensity
- T50
time at which 50% of the fibril formation reaction is complete
Footnotes
ASSOCIATED CONTENT
Supporting Information
- Supplemental Figures 1–4 (PDF).
Author Contributions
L.M.B.-M. and M.R.-A. designed the study. L.M.B.-M. conducted the experiments. L.M.B.-M. and M.R.-A. analyzed the data. L.M.B.-M. and M.R.-A. wrote and edited the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Blancas-Mejía LM, Ramirez-Alvarado M. Systemic Amyloidoses. Annu. Rev. Biochem. 2013;82:745–774. doi: 10.1146/annurev-biochem-072611-130030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buxbaum JN, Linke RP. A Molecular History of the Amyloidoses. J. Mol. Biol. 2012;421:142–159. doi: 10.1016/j.jmb.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Arosio P, Knowles TP, Linse S. On the lag phase in amyloid fibril formation. Phys. Chem. Chem. Phys. 2015;17:7606–7618. doi: 10.1039/c4cp05563b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jarrett JT, Lansbury PT., Jr Seeding ″one-dimensional crystallization″ of amyloid: a pathogenic mechanism in Alzheimer’s disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 5.Lomakin A, Teplow DB, Kirschner DA, Benedek GB. Kinetic theory of fibrillogenesis of amyloid beta-protein. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7942–7947. doi: 10.1073/pnas.94.15.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales R, Moreno-Gonzalez I, Soto C. Cross-seeding of misfolded proteins: implications for etiology and pathogenesis of protein misfolding diseases. PLoS Pathog. 2013;9:e1003537. doi: 10.1371/journal.ppat.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Naiki H, Gejyo F. Kinetic analysis of amyloid fibril formation. Methods Enzymol. 1999;309:305–318. doi: 10.1016/s0076-6879(99)09022-9. [DOI] [PubMed] [Google Scholar]
- 8.Harper JD, Lansbury PT., Jr Models of amyloid seeding in Alzheimer’s disease and scrapie: mechanistic truths and physiological consequences of the time-dependent solubility of amyloid proteins. Annu. Rev. Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 9.Ferrone FA, Hofrichter J, Sunshine HR, Eaton WA. Kinetic studies on photolysis-induced gelation of sickle cell hemoglobin suggest a new mechanism. Biophys. J. 1980;32:361–380. doi: 10.1016/S0006-3495(80)84962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofrichter J, Ross PD, Eaton WA. Kinetics and mechanism of deoxyhemoglobin S gelation: a new approach to understanding sickle cell disease. Proc. Natl. Acad. Sci. U. S. A. 1974;71:4864–4868. doi: 10.1073/pnas.71.12.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen CB, Yagi H, Manno M, Martorana V, Ban T, Christiansen G, Otzen DE, Goto Y, Rischel C. Branching in amyloid fibril growth. Biophys. J. 2009;96:1529–1536. doi: 10.1016/j.bpj.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yanagi K, Sakurai K, Yoshimura Y, Konuma T, Lee YH, Sugase K, Ikegami T, Naiki H, Goto Y. The monomerseed interaction mechanism in the formation of the beta2-microglobulin amyloid fibril clarified by solution NMR techniques. J. Mol. Biol. 2012;422:390–402. doi: 10.1016/j.jmb.2012.05.034. [DOI] [PubMed] [Google Scholar]
- 13.Cohen SI, Linse S, Luheshi LM, Hellstrand E, White DA, Rajah L, Otzen DE, Vendruscolo M, Dobson CM, Knowles TP. Proliferation of amyloid-beta42 aggregates occurs through a secondary nucleation mechanism. Proc. Natl. Acad. Sci. U. S. A. 2013;110:9758–9763. doi: 10.1073/pnas.1218402110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ruschak AM, Miranker AD. Fiber-dependent amyloid formation as catalysis of an existing reaction pathway. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12341–12346. doi: 10.1073/pnas.0703306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ban T, Hoshino M, Takahashi S, Hamada D, Hasegawa K, Naiki H, Goto Y. Direct observation of Abeta amyloid fibril growth and inhibition. J. Mol. Biol. 2004;344:757–767. doi: 10.1016/j.jmb.2004.09.078. [DOI] [PubMed] [Google Scholar]
- 16.Ihse E, Suhr OB, Hellman U, Westermark P. Variation in amount of wild-type transthyretin in different fibril and tissue types in ATTR amyloidosis. J. Mol. Med. (Heidelberg, Ger.) 2011;89:171–180. doi: 10.1007/s00109-010-0695-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson K, Barisone GA, Diaz E, Jin LW, DeCarli C, Despa F. Amylin deposition in the brain: A second amyloid in Alzheimer disease? Ann. Neurol. 2013;74:517–526. doi: 10.1002/ana.23956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardt F. Transfer amyloidosis. I. Studies on the transfer of various lymphoid cells from amyloidotic mice to syngeneic nonamyloidotic recipients. II. Induction of amyloidosis in mice with spleen, thymus and lymph node tissue from casein-sensitized syngeneic donors. Am. J. Pathol. 1971;65:411–424. [PMC free article] [PubMed] [Google Scholar]
- 19.Hardt F, Hellung-Larsen P. Transfer amyloidosis. Studies on the nature of the amyloid inducing factor in a murine transfer system. Clin. Exp. Immunol. 1972;10:487–492. [PMC free article] [PubMed] [Google Scholar]
- 20.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40:3525–3535. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 21.Hu D, Qin Z, Xue B, Fink AL, Uversky VN. Effect of Methionine Oxidation on the Structural Properties, Conformational Stability, and Aggregation of Immunoglobulin Light Chain LEN†. Biochemistry. 2008;47:8665–8677. doi: 10.1021/bi800806d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blancas-Mejia LM, Tellez LA, del Pozo-Yauner L, Becerril B, Sanchez-Ruiz JM, Fernandez-Velasco DA. Thermodynamic and Kinetic Characterization of a Germ Line Human λ6 Light-Chain Protein: The Relation between Unfolding and Fibrillogenesis. J. Mol. Biol. 2009;386:1153–1166. doi: 10.1016/j.jmb.2008.12.069. [DOI] [PubMed] [Google Scholar]
- 23.Baden EM, Randles EG, Aboagye AK, Thompson JR, Ramirez-Alvarado M. Structural insights into the role of mutations in amyloidogenesis. J. Biol. Chem. 2008;283:30950–30956. doi: 10.1074/jbc.M804822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DiCostanzo AC, Thompson JR, Peterson FC, Volkman BF, Ramirez-Alvarado M. Tyrosine residues mediate fibril formation in a dynamic light chain dimer interface. J. Biol. Chem. 2012;287:27997–28006. doi: 10.1074/jbc.M112.362921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Randles EG, Thompson JR, Martin DJ, Ramirez-Alvarado M. Structural alterations within native amyloidogenic immunoglobulin light chains. J. Mol. Biol. 2009;389:199–210. doi: 10.1016/j.jmb.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wall J, Murphy CL, Solomon A. In vitro immunoglobulin light chain fibrillogenesis. Methods Enzymol. 1999;309:204–217. doi: 10.1016/s0076-6879(99)09016-3. [DOI] [PubMed] [Google Scholar]
- 27.Blancas-Mejia LM, Tischer A, Thompson JR, Tai J, Wang L, Auton M, Ramirez-Alvarado M. Kinetic control in protein folding for light chain amyloidosis and the differential effects of somatic mutations. J. Mol. Biol. 2014;426:347–361. doi: 10.1016/j.jmb.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marin-Argany M, Güell-Bosch J, Blancas-Mejia LM, Villegas S, Ramirez-Alvarado M. Mutations can cause light chains to be too stable or too unstable to form amyloid fibrils. Protein Sci. 2015;24:1829–1840. doi: 10.1002/pro.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin DJ, Ramirez-Alvarado M. Comparison of amyloid fibril formation by two closely related immunoglobulin light chain variable domains. Amyloid. 2010;17:129–136. doi: 10.3109/13506129.2010.530081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baden EM, Owen BA, Peterson FC, Volkman BF, Ramirez-Alvarado M, Thompson JR. Altered dimer interface decreases stability in an amyloidogenic protein. J. Biol. Chem. 2008;283:15853–15860. doi: 10.1074/jbc.M705347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poshusta TL, Katoh N, Gertz MA, Dispenzieri A, Ramirez-Alvarado M. Thermal stability threshold for amyloid formation in light chain amyloidosis. Int. J. Mol. Sci. 2013;14:22604–22617. doi: 10.3390/ijms141122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraham RS, Geyer SM, Price-Troska TL, Allmer C, Kyle RA, Gertz MA, Fonseca R. Immunoglobulin light chain variable (V) region genes influence clinical presentation and outcome in light chain-associated amyloidosis (AL) Blood. 2003;101:3801–3807. doi: 10.1182/blood-2002-09-2707. [DOI] [PubMed] [Google Scholar]
- 33.Sikkink LA, Ramirez-Alvarado M. Salts enhance both protein stability and amyloid formation of an immunoglobulin light chain. Biophys. Chem. 2008;135:25–31. doi: 10.1016/j.bpc.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson FC, Baden EM, Owen BA, Volkman BF, Ramirez-Alvarado M. A single mutation promotes amyloidogenicity through a highly promiscuous dimer interface. Structure. 2010;18:563–570. doi: 10.1016/j.str.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Nuallain B, Thakur AK, Williams AD, Bhattacharyya AM, Chen S, Thiagarajan G, Wetzel R. Methods in Enzymology. New York: Academic Press; 2006. Kinetics and Thermodynamics of Amyloid Assembly Using a High-Performance Liquid Chromatography-Based Sedimentation Assay; pp. 34–74. [DOI] [PubMed] [Google Scholar]
- 36.Blancas-Mejia LM, Hammernik J, Marin-Argany M, Ramirez-Alvarado M. Differential effects on light chain amyloid formation depend on mutations and type of glycosaminoglycans. J. Biol. Chem. 2015;290:4953–4965. doi: 10.1074/jbc.M114.615401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lavatelli F, Perlman DH, Spencer B, Prokaeva T, McComb ME, Theberge R, Connors LH, Bellotti V, Seldin DC, Merlini G, Skinner M, Costello CE. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Mol. Cell. Proteomics. 2008;7:1570–1583. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vrana JA, Gamez JD, Madden BJ, Theis JD, Bergen HR, 3rd, Dogan A. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood. 2009;114:4957–4959. doi: 10.1182/blood-2009-07-230722. [DOI] [PubMed] [Google Scholar]
- 39.Blancas-Mejia LM, Horn TJ, Marin-Argany M, Auton M, Tischer A, Ramirez-Alvarado M. Thermodynamic and fibril formation studies of full length immunoglobulin light chain AL-09 and its germline protein using scan rate dependent thermal unfolding. Biophys. Chem. 2015;207:13–20. doi: 10.1016/j.bpc.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen SI, Vendruscolo M, Dobson CM, Knowles TP. From macroscopic measurements to microscopic mechanisms of protein aggregation. J. Mol. Biol. 2012;421:160–171. doi: 10.1016/j.jmb.2012.02.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.