Abstract
Localization of the Ca2+/calmodulin-dependent protein kinase II (CaMKII) to dendritic spine synapses is determined in part by the actin cytoskeleton. We determined binding of GFP-tagged CaMKII to tag-RFP-labeled actin cytoskeleton within live cells using total internal reflection fluorescence microscopy and single-molecule tracking. Stepwise photobleaching showed that CaMKII formed oligomeric complexes. Photoactivation experiments demonstrated that diffusion out of the evanescent field determined the track lifetimes. Latrunculin treatment triggered a coupled loss of actin stress fibers and the colocalized, long-lived CaMKII tracks. The CaMKIIα (α) isoform, which was previously thought to lack F-actin interactions, also showed binding, but this was threefold weaker than that observed for CaMKIIβ (β). The βE′ splice variant bound more weakly than α, showing that binding by β depends critically on the interdomain linker. The mutations βT287D and αT286D, which mimic autophosphorylation states, also abolished F-actin binding. Autophosphorylation triggers autonomous CaMKII activity, but does not impair GluN2B binding, another important synaptic protein interaction of CaMKII. The CaMKII inhibitor tatCN21 or CaMKII mutations that inhibit GluN2B association by blocking binding of ATP (βK43R and αK42M) or Ca2+/calmodulin (βA303R) had no effect on the interaction with F-actin. These results provide the first rationale for the reduced synaptic spine localization of the αT286D mutant, indicating that transient F-actin binding contributes to the synaptic localization of the CaMKIIα isoform. The track lifetime distributions had a stretched exponential form consistent with a heterogeneously diffusing population. This heterogeneity suggests that CaMKII adopts different F-actin binding modes, which is most easily rationalized by multiple subunit contacts between the CaMKII dodecamer and the F-actin cytoskeleton that stabilize the initial weak (micromolar) monovalent interaction.
Introduction
The calcium calmodulin-dependent kinase (CaMKII) is a multifunctional kinase that has a prominent role in long-term potentiation (LTP) (1, 2, 3). The four major isoforms of vertebrate CaMKII have ∼40 splice variants and are expressed in diverse tissues (3). Two isoforms, CaMKIIα (<α>) and CaMKIIβ (<β>), are dominant in the brain and their relative expression levels vary among different regions of the brain as well as during development (4). Their relative levels also vary within individual neurons between the cell body and dendritic/axonal processes (2). CaMKII has a prominent structural role in hippocampal dendritic spines, the postsynaptic computational units for LTP. CaMKII concentrations in spines are high (5), consistent with its structural role. The <β> isoform targets αβ hetero-oligomers to dendritic spines by binding to the spine actin cytoskeleton (6). Synaptic stimulation triggers CaMKII sequestration to dendritic spines and the postsynaptic density (PSD) within a few seconds of stimulation (7, 8, 9, 10). This rapid sequestration is coupled to actin polymerization and expansion of the stimulated spine (11). Expansion is due to the direct effects of CaMKII on the actin cytoskeleton (12, 13) as well as to indirect effects mediated by the activation of other kinases (14). The increase in spine size persists after termination of the stimulus-induced calcium transient. CaMKII levels in stimulated spines are also increased due to association with the PSD, in particular, the NMDA receptor GluN2B subunit (15) and the enlarged actin cytoskeleton (16). In the longer term, CaMKII promotes axonal branching and outgrowth (17).
The neuronal isoforms have highly homologous kinase and association domains, but the linker that connects these two domains is variable in sequence and length (1). The individual subunits assemble into homo- or hetero-oligomers of variable isoform compositions, and the atomic structure of the dodecameric enzyme has been previously described (18). The <α> and <β> isoforms form 12 subunit homo-oligomers of similar size, with one study reporting a slightly smaller <β> oligomer (19). Calmodulin binding to the regulatory segment relieves inhibition, and transphosphorylation activates the enzyme at <α>T286 (T287 in the other isoforms), which confers autonomous activity to the enzyme.
Binding of <β>, but not <α>, to the actin cytoskeleton has been shown by various approaches, including colocalization, fluorescence photobleaching, and pharmacological manipulations in neuronal and non-neuronal cell cultures (6, 20, 21, 22, 23). In vitro sedimentation assays and electron microscopy have demonstrated the <β>-dependent formation of F-actin bundles (12, 22, 23, 24). Activation of <β> by both autophosphorylation and the phosphomimetic T287D mutation (22) abolishes actin bundling activity. Furthermore, an alternative splice variant, <βE′>, which has a short linker similar to that of <α>, does not bind or bundle F-actin in pull-down assays. The differences observed with the mutants in pull-down assays are consistent with colocalization in neuronal cell cultures. Pyrene fluorescence measurements (12) have shown that both <α> and <β> isoforms bind globular (G-) actin, and <β> binds with 2.4 μM affinity and a stoichiometry of 12 actin monomers per oligomer (24). However, quantitative estimates of the affinity of <β> or <α> for F-actin, or of modulation via activation through stimulation or mutation, are not available.
Here, we characterized the association of CaMKII with labeled F-actin in live human umbilical vein endothelial cells (HUVECs) (25) by using total internal reflection fluorescence microscopy (TIRFM) to image and track single molecules (26, 27). We previously exploited this approach to study motor proteins, ion channels, and G-protein-coupled receptors (27, 28, 29, 30). Here, we extended the method to measure the association of enhanced green fluorescent protein (eGFP)-tagged CaMKII native and mutant proteins with red fluorescent protein (RFP)-tagged actin to mark the cytoskeletal structures. Single-molecule tracking experiments have shown that actin depolymerization increases CaMKII mobility in dendritic spines, and revealed different, heterogeneous mobility distributions for stimulated versus unstimulated states (16). We used HUVECs as a model system because they are ideal for TIRF imaging, have a defined cytoskeletal architecture, and are amenable to transient transfection methods. Our measurements show that both neuronal CaMKII isoforms bind cytoskeletal actin, but with affinities that differ by threefold over the first decade range of a log-normal binding curve. Our results explain why association of <α> may have been overlooked in earlier studies, and have implications for CaMKII transport and cytoskeletal remodeling within neurons.
Materials and Methods
All biochemicals were sourced from Sigma-Aldrich (Poole Dorset, UK) unless noted otherwise.
TIRFM
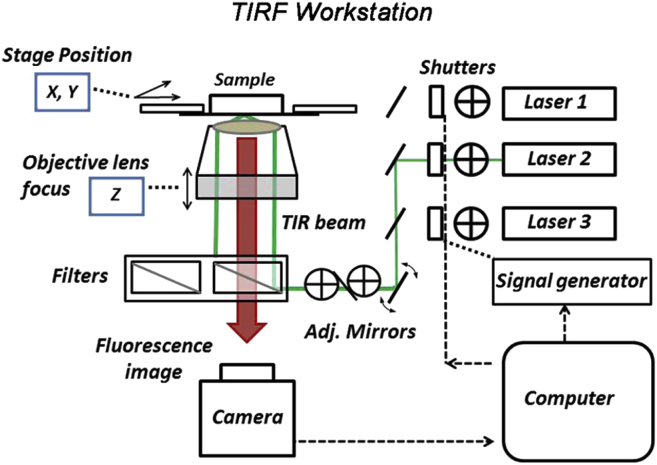
We used a custom-built TIRF microscope workstation based on an inverted microscope (Nikon Eclipse, TE 2000U; Nikon, Kingston-upon-Thames, UK) (Fig. 1). Complete details are provided in Supporting Materials and Methods in the Supporting Material.
Figure 1.
TIRF workstation. The choice of the laser excitation wavelength (laser 1 = 561 nm; laser 2 = 488 nm) was computer controlled; excitation (green line) and fluorescence emission (red arrow) light paths are shown. The TIRF incident angle was adjusted by an external mirror. The microscope stage and objective lens employed piezo-positioners to control specimen position and image focus. Images were acquired with an EMCCD camera. A waveform generator set the duration, delay, and frequency of photoactivation pulses (laser 3 = 405 nm), also in TIRF mode. Separate, exchangeable filter cassettes were used for GFP and tRFP fluorescence.
In vitro assays
For use as a single fluorophore calibration specimen, we immobilized GFP molecules on the surface of a microscope flow cell with a GFP antibody by first filling the flow cell with a phosphate-buffered saline (PBS) solution (pH 7.4) containing 5 μg/mL (3 nM) polyclonal anti-GFP antibody (Abcam, Cambridge, UK) as described previously (27). This solution was left to incubate in the flow cell for 5 min and then washed with PBS supplemented with 0.5 mg/mL bovine serum albumin to block regions of bare coverglass. The solution was then replaced with PBS containing 10 ng/mL (0.37 nM) GFP (Clontech, Palo Alto, CA) for 5 min, and unbound protein was washed out of the flow cell by several washes with assay buffer (AB− (20 mM imidazole (pH 7.4), 50 mM KCl, 2 mM EGTA, 4 mM MgCl2)) before it was viewed by TIRFM. The molecules were imaged in degassed and argon-purged AB− supplemented with an oxygen-scavenger system consisting of 3 mg/mL glucose, 0.5 mg/mL catalase, 0.2 mg/mL glucose oxidase, and 20 mM dithiothreitol. Using the antibody-immobilized GFP molecules as a control sample, we measured the single fluorophore intensity as a function of excitation power. The average value measured over several hundred fluorophores was linear with the laser power. The mean single fluorophore intensity could therefore be used as an independent internal check of excitation power in subsequent experiments.
Cell culture
CaMKII fusion proteins tagged with monomeric eGFP (GFP) or photoactivatable eGFP (PaGFP) carrying the A206K mutation have been described previously (22, 31, 32, 33, 34). The GFP tag does not interfere with kinase activity or holoenzyme assembly (24), and immunoelectron microscopy has shown that native CaMKII sequesters to the PSD of dendritic spines (35) with kinetics similar to those reported by the tagged proteins (22, 36). We studied the following tagged actin fusion proteins: mCherry-actin (37), tagRFP-actin (38), and mTurquoise2-actin (39). We chose tagRFP-actin (tRFP-actin) for its brightness, photostability, and expression level (40). The plasmids encoding GFP-CaMKII and tRFP-actin constructs were mixed and cotransfected into primary HUVECs or Cos7 cells at 70–80% confluence, primarily by nucleofection (Nucleofector Model 2b; Lonza, Blackley, UK). Alternatively, Lipofectamine-2000 (Life Technologies, Paisley, UK) transfection was used as previously described (41). With either method, the transfection efficiency was typically >50%. The cells were plated on poly-lysine-coated dishes (Lab-Tek chambered borosilicate, #1 coverglass; Nunc, Rochester, NY) in Dulbecco’s modified Eagle’s medium with added 10% fetal bovine serum and streptomycin (50 μg/mL). Cell culture dishes were removed from the CO2 incubator (Galaxy R; Scientific Laboratory Supplies, East Riding of Yorkshire, UK) 24–36 h after transfection. These incubation times were optimal for visualizing single GFP-CaMKII molecules. TIRF imaging was conducted at 25°C within an hour after the samples were removed from the incubator.
The HUVECs we chose as a model system for most of our TIRF imaging experiments attach firmly to the culture dish substrate and have long ventral stress fibers (42) that form oriented arrays. Other cytoskeletal substructures (i.e., arcs (43) and filopodia) are also present. Although HUVECs express a variant CaMKIIδ6 isoform (44), they do not natively express the <α> or <β> isoforms found in neurons. Expression was monitored by epifluorescence, and cell morphology was determined by phase contrast. In addition to morphology, we checked the integrity of the physiological state by noting an absence of CaMKII aggregation caused by high pH or calcium (36).
Single-molecule image analysis
A typical experiment involved a set of cotransfections of the plasmid encoding tRFP-actin with a plasmid encoding a GFP-CaMKII fusion (two dishes per CaMKII construct; up to four constructs per experiment). Control dishes cotransfected with plasmids encoding tRFP-actin and GFPCaMKIIβ were included in each experiment to assess the viability of the primary culture. First, tRFP-actin fluorescence was used to identify transfected cells, and then GFP-fluorescence was recorded. Many thousands of single-particle tracks were obtained for each construct using >12 cells from four different culture dishes and two separate experiments. Details regarding the single-particle tracking algorithm and the analytical measures used are provided in Supporting Materials and Methods.
Multiple analysis of variance (ANOVA) and simultaneous pairwise t-tests were conducted in R (https://www.r-project.org/) as detailed in (45). The variance was the sum of the variation within and between groups normalized by their degrees of freedom. The probability (p-value) that differences between populations were significant was then computed from the F-value (F). Significant differences reported by ANOVA were then tested by means of simultaneous, pairwise t-tests with default Holm correction for multiple testing.
Results
Our experimental study consisted of two parts, First, we used dual-color TIRFM to visualize and track (27) individual GFP-tagged <α> and <β> isoforms in HUVECS, and derived their properties from population statistics and spatial colocalization with F-actin cytoskeletal structures. We then studied different mutants and pharmacological agents to understand the structural basis of CaMKII association with F-actin.
Assay development
Visualization of GFP molecules in control specimens and live cells
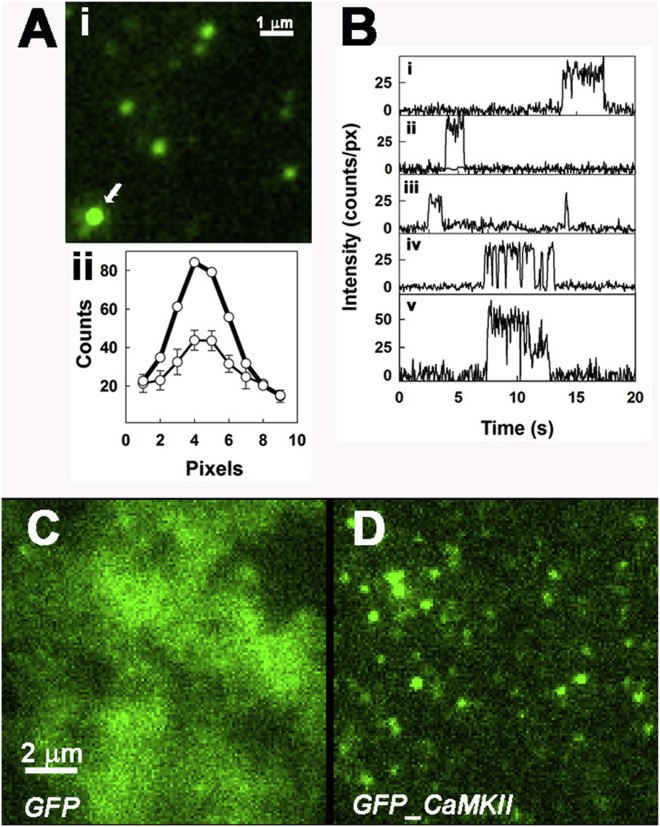
We visualized antibody-immobilized GFPs at low surface density (<1 μm−2) using TIRFM to establish the emission intensity of individual GFP molecules under our standard imaging conditions. Individual GFPs were readily identified as discrete fluorescent spots that had a diffraction-limited point spread function (PSF) with a characteristic spot intensity (Fig. 2 A). The spots had a mean duration of 2.0 ± 0.4 s and exhibited single-step photobleaching with a unitary intensity of 27.4 ± 2.2 counts/pixel. Brighter spots with twofold greater intensity exhibited two-step photobleaching (Fig. 2 B). Next, we obtained TIRFM video recordings of cultured HUVECs and Cos7 cells that were expressing GFP. In contrast to the video recordings of antibody-immobilized GFP molecules, the GFP fluorophores within cells could not be resolved (Fig. 2 C). This was because rapid diffusive motion within the cytosol caused image blurring during the frame acquisition period, as explained below.
Figure 2.
TIRFM visualization of GFP in vitro and in living cells versus GFP-CaMKII. (A) i: Antibody-immobilized GFP molecules (10-frame averaged image)). ii: Line intensity profiles of the four spots in field center, top and right (± standard error (SE), thin line), and of the brighter spot (arrow) show the diffraction-limited size. (B) Intensity-versus-time records of spots shown in A(i), illustrating single-step photobleaching (i–iii), blinking behavior (iv), and double-step photobleaching of the brighter spot (v).The single-step modal value was 27.5 ± 2.5 counts/pixel (doubling and tripling occurs when fluorophore PSFs overlap). (C) Single video frame (50 ms exposure) of a HUVEC expressing GFP alone, showing that motion blurring prevents single fluorophore observation. (D) Single video frame of a HUVEC expressing GFP-CaMKIIβ, showing that discrete fluorescent spots are now visible.
Visualization of homomeric GFP-CaMKIIβ complexes in the cellular cortex
In marked contrast to cell cultures expressing GFP molecules alone (see above), single fluorescent particles were visualized by TIRFM in cell cortices when GFP-tagged <β> (henceforth termed β) was expressed (Fig. 2 D). This discrepancy can be explained by attenuation of the spot intensity by motion blurring during the 50 ms frame acquisition period (δt). The attenuation factor of the computed centroid is given by the ratio of the area covered by the diffusing particle during a single video frame = π(δx)2 (where δx = (4D.δt)1/2, and D is the diffusion coefficient) and the area that captures 90% of the object’s PSF (here a 3 × 3 pixel region on the camera) = 0.9 μm2.
The expected lateral diffusion coefficients, DStokes, for the relevant species were computed from the diffusion equation:
| (1) |
where Stokes radius as = (3M/4πAσ)1/3, M is the molecular mass (kDa), A is Avogadro’s number, σ is the protein density (1300 kg/m3) (46), and η is the cortical viscosity (0.0032 Pa.s) (47, 48). This gives an estimated DStokes for GFP (M = 27 kDa; as = ∼2 nm) of ∼30 μm2s−1 and diffusive motion blurring during a 50 ms video frame of ∼20 μm2. Therefore, the expected reduction in fluorescence intensity (per pixel) is 20/0.9, ∼23-fold. This explains why freely diffusing GFP molecules were not resolved at the video imaging rates. Since DStokes varies inversely as the cube root of the molecular mass, we were also unable to resolve the GFP-<a> (henceforth α) mutant, which is monomeric due to deletion of the association domain (αΔ316) (M = 62 kDa) and tRFP-G-actin (M = 70 kDa), as both exhibit a >15-fold estimated attenuation of spot intensity due to motion blurring. We were able to satisfactorily visualize β molecules because they form dodecameric complexes (M = 87 × 12 = 1044 kDa) (18, 49). Thus, the intensity attenuation by motion blurring (∼7-fold) is more than compensated for by the 12-fold increase in intensity due to the increased number of GFPs.
Decoration of actin stress fibers with single GFP-CaMKIIβ holoenzymes
We used two-color TIRFM to image β (excited at 488 nm) and tRFP-actin (excited at 561 nm) to characterize β complexes interacting with F-actin cortical structures. TIRFM of HUVECs transfected with t-RFP actin revealed long linear fibers in the actin cortex. The morphology was consistent with ventral stress fibers (42), and these structures will henceforth be referred to as such. Approximately 100 video frames were averaged to enhance the relatively static fibers above the background of rapidly diffusing G-actin monomers. The averaged tRFP-actin image was then overlaid onto TIRFM video recordings of β molecules to reveal their movement within the cytosol and their association/dissociation with the tRFP-tagged F-actin structures (Movie S1).
Individual GFP-fluorescent spots were identified and tracked in the video sequences to yield spatiotemporal trajectories (300–3000 per record) of individual objects. Tracks were generated by linking centroids for successive frames. Apparent diffusion coefficients (Dlat) were computed from the centroid frame-to-frame displacements (Δx):
| (2) |
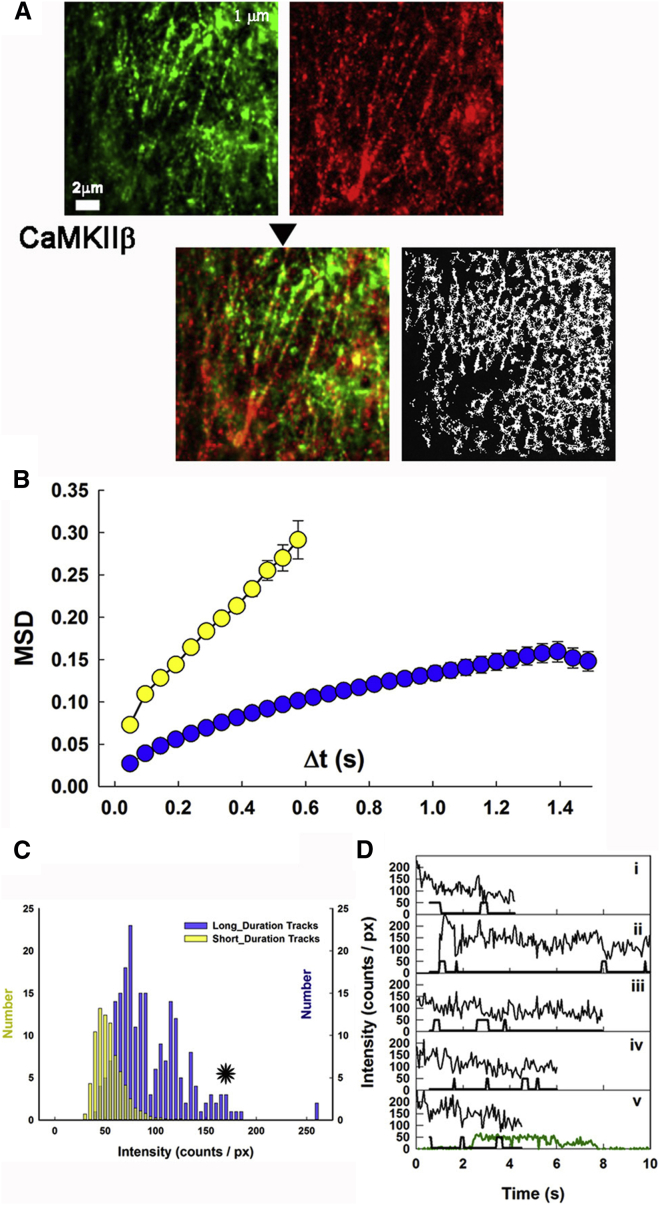
For free diffusion, c = 1. For confined diffusion, c = >1 and the denominator preexponent = <4. Individual tracks terminated when the object intensity dropped below the detection threshold due to diffusion from the excitation region (evanescent field), photobleaching, or tracking errors (considered below). Superposition of the image showing all of the particle tracks obtained over one video recording (lasting 25 s) onto the averaged tRFP-actin image provided a measure of colocalization (Pearson’s correlation coefficient, Ppix), as described in Supporting Materials and Methods. It was clear that β associated with the cortical actin fibers (Fig. 3 A).
Figure 3.
CaMKIIβ decoration of the actin cytoskeleton visualized by two-color TIRFM. (A) Averaged images of β (left panel, green, 400 frames) and tRFP-actin (right panel, red, 100 frames). Mean tRFP intensity = 119 ± 9 counts/pixel. The bottom panels (arrow) show the two frames superimposed (Ppix = 0.27, Prand = 0.09 ± 0.07) (left) and the single-particle tracks (right) accumulated over 10 s of video (Movie S1). (B) MSD-versus-time interval (Δt) for the total population of tracks (white circles) and short-lived (yellow circles) and long-lived (blue circles) track subpopulations (± standard deviation (σ)). The initial gradient of the short-lived track data gives Dlat = 0.28 μm2/s, whereas that of the long-lived tracks gives 0.04 μm2/s. Total number of tracks, n = 12,723. (C) Intensity histograms for the short-lived track subpopulation (yellow bars) and long-lived track subpopulation (blue bars). The asterisk (black) marks the region of the histogram that was used to analyze photobleaching. (D) Sample intensity-versus-time plots for some of the objects from the asterisk-marked region. Stepwise intensity changes as detected by Student’s t-test (Fig. S1) are marked immediately below each trace to indicate sudden intensity transitions. The starting intensity for each spot was >170 counts/pixel, which is ∼8-fold greater than the unitary GFP intensity. The green line (in the lowest panel), is the two-step immobilized GFP photobleaching, redrawn from Fig. 2B(v), shown for reference.
Dynamics of the interaction between CaMKIIβ and the actin cytoskeleton
Automated single-particle tracking (27) was used to identify and track individual β complexes. The object tracks were characterized with the measures defined in Materials and Methods. Short-lived particle trajectories (t < 0.58 s; Fig. 3 B, yellow symbols) closely approximated Brownian motion. In contrast, the plot for longer-lived trajectories (t > 2.5 s; Fig. 3 B, blue symbols) was nonlinear, with little increase in the mean-square deviation (MSD) beyond Δt > 1.2 s. Further analysis showed that the binned subpopulation of short-lived tracks had a unimodal intensity distribution with a lower mean relative to the parent population, whereas the subpopulation of longer-lived tracks had higher intensity relative to the parent population and the intensity distribution was greatly skewed toward higher values (Fig. 3 C). The different subpopulation characteristics are consistent with the notion that tracks from weakly bound, more mobile molecules have a short duration and dominate the <0.5 s subpopulation. In contrast, more strongly bound molecules dominate the >2.5 s subpopulation, with lower average Dlat. The modal intensities for both subpopulations are lower than expected for the multimeric (10–12 subunits) tagged β holoenzymes. Thus, although at first it may seem that the correlation between intensity and mobility differences is simply due to a difference in aggregate size, it is better explained by an intensity attenuation due to motion blurring (see above). To ascertain whether this was the case, we examined single spots and tracks.
To test for multisubunit states, we measured stepwise changes in fluorescence intensity. Spots immobilized on actin stress fibers had the highest intensities, but they exhibited PSF-limited spatial profiles similar to those obtained for single GFP fluorophores (Fig. 2 A). A small subset of such spots was analyzed (Fig. 3 D). The average initial intensity was ∼8-fold greater (171.6 ± 11.5 counts/pixel) than that measured for individual GFP molecules in vitro (∼27 counts/pixel). We used a running Student’s t-test to detect significant jumps in local mean intensity over adjacent sections of data (Fig. S1 in the Supporting Material). The mean intensity drop for each stepwise change in intensity was 22.1 ± 2.0 counts/pixel and the mean step duration was 2.6 ± 0.4 s, which are similar to the values obtained for single GFP molecules immobilized in vitro (27.4 ± 2.2 counts/pixel and 2.0 ± 0.4 s). Many of these spots showed a severalfold greater final intensity drop (e.g., 90–0 counts/pixel (spot v)) relative to the 27.4 ± 2.2 counts/pixel drops obtained for single GFP photobleaching. Simultaneous photobleaching of multiple (three for spot v) GFP fluorophores is not likely. Instead, the final intensity drops presumably report the dissociation of β holoenzymes from the fibers and diffusion out of the evanescent field before all their fluorophores have bleached.
Sample tracks and their MSD versus Δt plots were analyzed next (Fig. S2). In addition to high intensities, the long-lived tracks had highly nonlinear MSD versus Δt plots, and the MSD and Δt correlation was abolished for intervals greater than a few frames, consistent with immobilization as validated by an examination of the single tracks. Centroid intensity inversely correlated with mobility in the short-lived tracks of diffusing spots, with values consistent with the motion-induced sevenfold attenuation relative to the intensity of immobilized holoenzymes. The slopes (MSD versus Δt) of these short tracks correlated with the fraction of time during which they were mobile. Analyses of the single-spot photobleaching and single tracks show that motion blurring is responsible for the observed mobility-intensity correlations in the subpopulation distributions. We conclude that the rapid decrease in the track population with time is governed predominantly by the diffusion of unbound molecules out of the evanescent field.
Filopodia kymographs support the tracking analysis
Cultured HUVECs exhibit numerous filopodia, which are actin-rich tubular extensions >2 μm long and ∼150 nm in diameter. Some of the filopodia protruded close to the coverslip and were visualized in our video recordings by the evanescent field excitation. This gave us the opportunity to track GFP-tagged molecules that were essentially constrained to a single dimension independently of the depth, z, of the evanescent field. The molecule movements were suitable for kymograph analysis. We straightened the image data by using spline fits to the overall filopodial shape, and then extracted a linear strip of image pixels to form the abscissa in the kymograph time-series image (Fig. S3).
The β complexes produced punctate images on each video frame, and their motion within the filopodium then created a pattern of vertical trajectories (i.e., along the ordinate, time axis). The trajectories consisted of linear, bright segments that were tilted slightly toward the cell body (at ∼1.5 μm/min), consistent with complexes binding tightly to actin and reporting the slow rearward flow of the central F-actin bundle of the filopodium (50). These events were interspersed with haphazard, dim trajectories as the particles dissociated from actin and diffused within the body of the filopodium. Both types of trajectories were observed for closely adjacent objects within the same filopodium over the same time window, indicating that dim trajectories result from mobility of the β complexes within the filopodium rather than movement of the filopodium relative to the glass coverslip.
Our initial goal was to achieve a time-resolved characterization of bound and free episodes of β molecules constrained within the evanescent field by the filopodia. However, to our surprise, the kymographs also revealed that both α and β associated with filopodial F-actin.
F-actin dependence of CaMKII α and β lifetime distributions by evanescent field fluorescence photoactivation microscopy
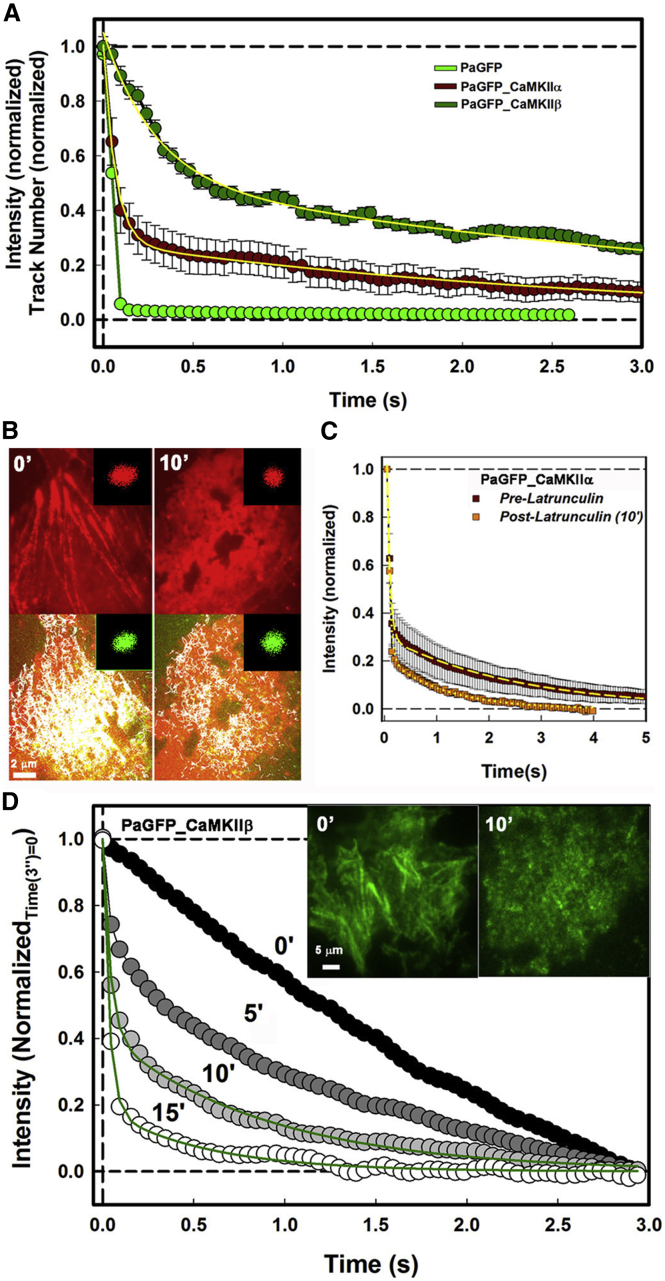
To follow up the finding that both α and β isoforms bind actin within filopodia, we examined the kinetics of fluorescence decay after photoactivation of PaGFP fusion constructs within the cell cortex. A brief flash of TIR laser light at 405 nm was used to activate PaGFP, and continuous illumination at 488 nm allowed the activated fluorescence to be visualized. The fluorescence of PaGFP alone decayed rapidly, reaching half its initial value within a single video frame (<50 ms; Fig. 4 A). The decay was two orders of magnitude more rapid than the photobleaching rate estimated from photobleaching of immobilized GFP molecules or photoactivation of fixed cells (see Materials and Methods). Therefore, the decay must reflect diffusion of the photoactivated PaGFP molecules out of the evanescent field.
Figure 4.
Results from photoactivated localization microscopy TIRFM and latrunculin treatment show that both CaMKII isoforms associate with the actin cytoskeleton. (A) Normalized fluorescence decay curves of PaGFP and PaGFP-CaMKII fusion proteins after photoactivation by a 405 nm laser (at t = 0). Decay was measured as single-molecule track lifetimes. Time 0 is the time required to exceed the five-frame track duration threshold (0.24 s). The data were least-square fitted to two exponentials (yellow lines): PaGFP-α = 0.72 ± 0.01(e(−15.2±0.5t)) + 0.28 ± 0.01(e(−0.35±0.07t)), n = 4306; PaGFP-β = 0.51 ± 0.01(e(−3.56±0.08t)) + 0.49 ± 0.01(e(−0.24±0.07t)), n = 11,160. In contrast, photoactivated PaGFP fluorescence intensity measured over the image field decayed by >50% within 0.1 s (two frames). (B) HUVEC stress fibers after 10 min (10’) treatment with latrunculin (5 μM). The top panels (red) show the averaged tRFP-actin images: although there is little change in the total fluorescence (97 ± 17 counts/pixel (before latrunculin treatment); 100 ± 23 counts/pixel (after latrunculin treatment)), the fibers disappear after treatment. The bottom panels show PaGFP-CaMKIIα (green) and single-particle tracks (white lines (n = 3777 (0’) and 1573 (10’)) superimposed on actin (red). Insets: FT spectra (tRFP-actin (red); GFP (green)). (C) PaGFP-CaMKIIα fluorescence decay before and after latrunculin treatment. Dual exponential fits: 0.71 ± 0.01(e(−18.1±0.5t)) + 0.29 ± 0.01(e(−0.39±0.01t)) (dashed yellow line) (0’); 0.8 ± 0.01(e(−19.9±0.8t)) + 0.21 ± 0.01(e(−0.92±0.04t)) (dotted yellow line) (10’). (D) Fluorescence intensity decay curves of PaGFP-β at various times (in minutes) after addition of latrunculin (5 μM) to a Cos7 cell culture. Intensity was normalized to unity at t = 0 s (t0) and zero at t = 3 s (t3); (t3/t0) ∼50%. Control fit (unnormalized): (0.26 ± 0.01) + (0.74 ± 0.01(e(−0.28±0.01t)). Fits after latrunculin treatment (green lines): 0.58(e(−24.3t)) + 0.42(e(−1.2)) (10’); 0.81(e(−29.3t)) + 0.19(e(−1.8)) (15’). Inset: Filamentous structures visualized when PaGFP-β was photoactivated in the absence of latrunculin (0’) were not observed (10’) after addition of latrunculin. Correlation coefficient R2 > 0.99 for all fits.
The PaGFP-CaMKII fusion constructs (PaGFP-α and PaGFP-β) showed slower and more complex kinetics (Fig. 4 A), although it was still rapid relative to photobleaching. Their fluorescence decay could be followed by single-molecule tracking. The decay profiles were approximated by dual-exponential fits with a 0.24 s offset relative to the PaGFP intensity decay due to the five-frame lifetime tracking filter. Direct image field intensity measurements, analogous to those used for PaGFP but corrected for the offset, showed a twofold difference in the fast-component, but not the slow-component, decay. Tracks may terminate for reasons other than fluorescence loss, specifically crossover of tracks of unbound particles and imperfections of the tracking algorithm (Supporting Materials and Methods), that could account for the modest discrepancy.
The slow components for PaGFP-α (1.9 s) and PaGFP-β (3.0 s) were incompatible with free diffusion. Therefore, we used latrunculin B (latrunculin) (51) to test whether disruption of the actin cytoskeleton affected the mobility of PaGFP-CaMKII fusion proteins. The effect of latrunculin on HUVEC stress fibers was evident within a few minutes (Fig. 4 B). Before latrunculin treatment, PaGFP-α colocalized weakly, generating an anisotropic pattern that aligned with the stress-fiber arrays as revealed by the elliptical Fourier transform (FT) spectra of the red/green images (red FT (R(maj/min) (major/minor axial ratio)) = 1.35, angle = 16° ± 5°); green FT (R(maj/min) = 1.33, angle = 24° ± 5°). After incubation (10 min) with latrunculin, the pattern had disappeared (FT R(maj/min) = ∼1 for both channels; Fig. 4 B, insets). We measured the photoactivated fluorescence decay kinetics at 0 and 10 min after latrunculin treatment. Dual-exponential fits to the fluorescence decrease showed that the amplitude and rate of the fast-decay component increased with time after treatment, consistent with a reduced F-actin-immobilized fraction (Fig. 4 C). We repeated the experiment with PaGFP-β. Photoactivated PaGFP-β formed brightly fluorescent filamentous substructures that disappeared after latrunculin treatment. The kinetics of PaGFP-β fluorescence decay also changed (Fig. 4 D) concomitantly with the observed structural changes. The fluorescence decay after photoactivation revealed a substantial fast-decay component for pulses applied 5 min after latrunculin treatment. The fast component increased with incubation time, so for photoactivation pulses 15 min after latrunculin treatment, the decay was similar to that seen for photoactivated PaGFP-α 10 min after latrunculin treatment. The fast-component decay was consistent with the formation of a PaGFP-like inert species.
Structural determinants of the CaMKII F-actin interaction
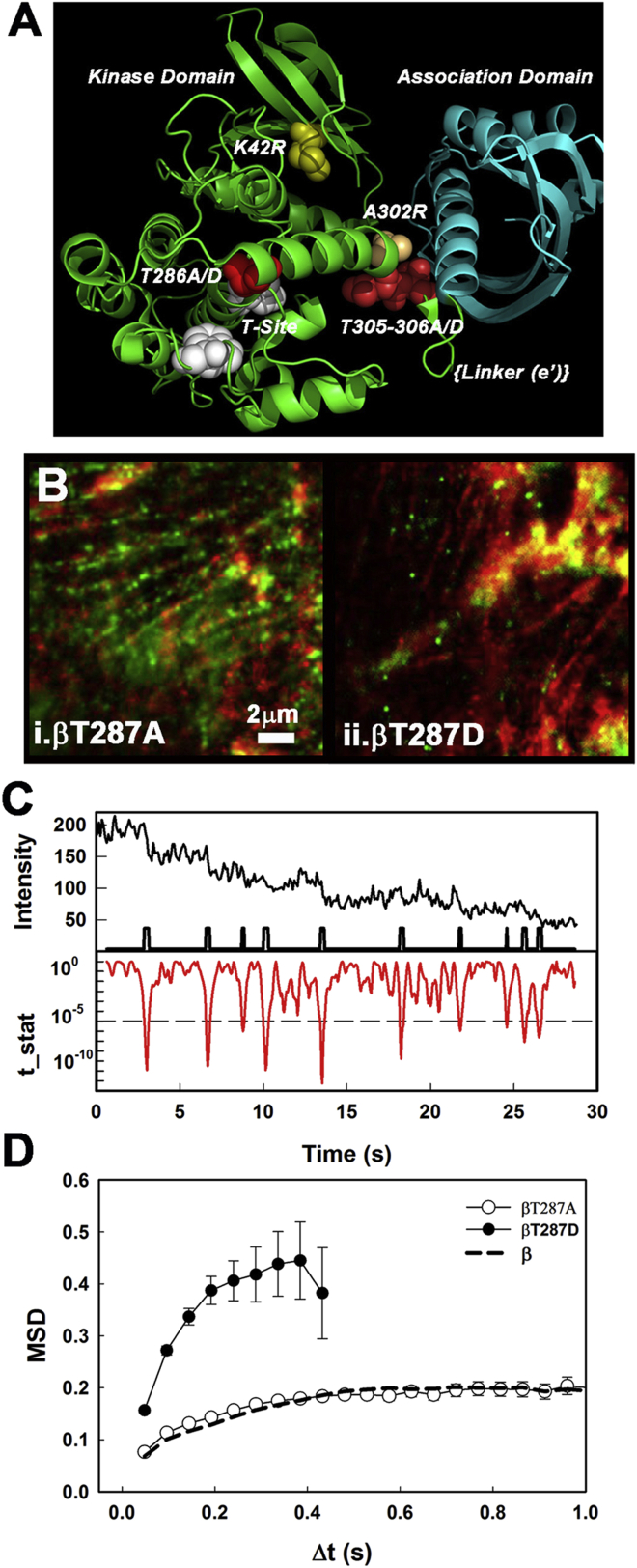
Having established single-molecule imaging techniques using native α and β isoforms, we next examined the GFP fusions of a panel of functionally significant CaMKII mutants. The mutations are mapped onto the CaMKII structure in Fig. 5 A (residue positions are incremented by one in the corresponding β sequence). The primary phosphorylation site, αT286, is important for long-term depression (LTD) as well as LTP since these functions are impaired in <αT286A> mutant mice (52, 53) and are affected or abolished, respectively, by overexpression of a constitutively active<αT286D> (54). To explore its role in single-molecule binding to cytoskeletal actin, we studied the homologous βT287A and βT287D mutants (1). Phosphorylation of the secondary sites αT305 and αT306 is known to inhibit kinase activity (33). We compared differences among the αT286D/T305/T306 triple mutants with both secondary sites mutated to either aspartate or alanine. Other mutations/lesions of interest were αK42M, which blocks ATP binding necessary for CaMKII activation, LTP, and spine enlargement (55); αA302R, which disrupts calmodulin binding and translocation to the PSD (9); and the βE′ splice variant, which lacks linker sequences encoded by exons I and IV (56). Finally, we used the tatCN21 inhibitor, which competes with the NR2B NMDA receptor subunit for binding to the T-site (57), to see whether CaMKII binding targets elicit structural changes (58) that affect F-actin association.
Figure 5.
The T287D point mutation downregulates association with the actin cytoskeleton. (A) Atomic structure (PDB: 3SOA) of rat CaMKII (18). The positions of the mutated sites (K42 (yellow), T286 (red), A302 (peach), and T305/T306 (magenta)) studied; the junction of kinase (green) and association (blue) domains where the splice E′ linker segment would be located; and the substrate-binding T-site (white) are shown in relation to the secondary-structure elements (cartoon representation). (B) Superimposed averaged images, processed as in Fig. 3A, show localization of single molecules (Fig. S4; Movies S2 and S3) of (i) the dephosphorylated mimic, β287A (550 frames, Ppix = 0.29, Prand = 0.16 ± 0.07, n = 6511) and (ii) the phosphomimic, β287D (475 frames (Ppix = 0.08, Prand = 0.0 ± 0.08, n = 1020) with tRFP-actin (red, 100 frames). The mean tRFP intensities were 105 ± 6 (βT287A) and 213 ± 75 (βT287D) counts/pixel. (C) Photobleaching profile of a long-lived β287A-GFP track (upper black line plot) with the corresponding t-test statistic (t-stat) based on a rolling, nonoverlapping 12-frame window (lower red line plot). The t-stat axis denotes the probability that successive 12-frame segments have the same mean. The probability threshold was set to 10−5 for detection of a step change. The bars on the time axis of the upper plot mark the 10 steps identified by the t-test. The mean lifetime and intensity decrease per step were 2.6 ± 0.4 s and 15.5 counts/pixel, respectively. (D) Average track MSD-versus-Δt plots for the β287D and β287A populations. The dashed line is the plot for the native β population redrawn from Fig. 3C.
The primary phosphorylation site mutants have dramatically different effects on F-actin binding
Averaged images show that the phosphorylation-incompetent βT287A mutant decorates cortical actin structures (Figs. 5 B(i) and S4). In contrast, the βT287D videos (Movies S2 and S3) show an isotropic distribution of fast-moving spots in the cell cortex that did not map onto the stress fibers (Figs. 5 B(ii) and S4). As for β, the tracks of immobilized βT287A spots have initial intensities that are several multiples of individual GFP fluorophores and show multistep photobleaching time courses. A rare example of a long-lived track reveals 10 steps (Fig. 5 C), consistent with the intensity ratio of the immobilized βT287A spot relative to single GFP fluorophores. In contrast, averaged images of the phosphomimetic βT287D mutant show no evidence of actin colocalization.
The difference between the two mutant proteins was emphasized by an analysis of MSD versus Δt plots (Fig. 5 D). For βT287A, the initial slope and subsequent behavior were superimposable with results obtained using native CaMKIIβ. The addition of tatCN21 (1 μM) had no effect on the association of βT287A with F-actin (two different cultures, >10,000 tracks). In contrast, the initial (MSD versus Δt) slope for βT287D was much greater than that for β and βT287A, with virtually no (<7%) tracks of duration longer than 0.4s (+0.24 s offset), consistent with fast-moving objects that diffused rapidly out of the evanescent field.
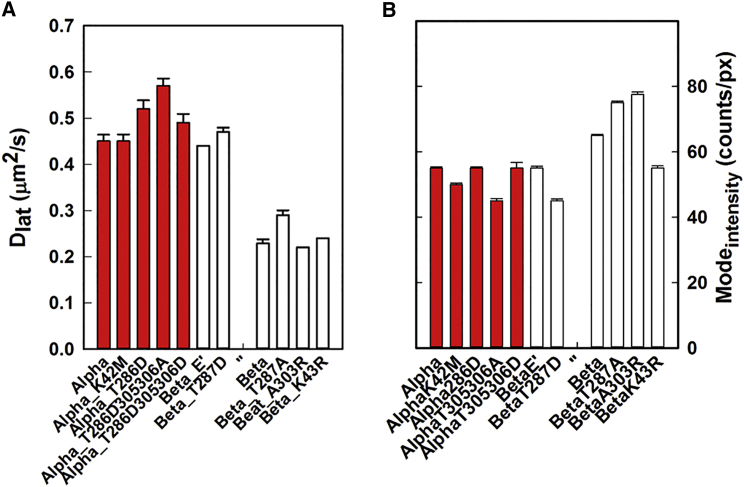
All of the mutant isoforms showed similar single-object intensities but formed two distinct mobility groups
ANOVA was used to test for significant differences in values for the panel of CaMKII mutants (Fig. 6 A) based on estimates of variance within and between data sets (Table S1). Variances were normalized for different degrees of freedom, and the probability, p, that differences between populations were significant (p < 0.05) was computed. Consistent with a visual inspection of the data (Fig. 6 A), the results showed two distinct groups: a low-mobility group comprised of the β proteins (native β, βK43R, βA303R, and βT287A) and a high-mobility group comprised of all the α isoform mutants together with βT287D and βE′. A similar pattern was obtained when instantaneous velocities were compared (Fig. S5). The modal, single-spot intensity values (Fig. 6 B) obtained across all proteins are similar and vary between 50 and 80 counts/pixel, which is four- to sixfold lower than the anticipated value for the CaMKII holoenzyme and twofold greater than for single GFP fluorophores. The similar values rule out oligomer aggregation as a possible cause of the mobility differences between species. The brighter spots seen decorating stress fibers in some video frames are due to PSF overlap of closely opposed spots; however, their tracks can be separated provided the spots are not stationary (27). Spot intensity measurements suggest that the expression level affects only the holoenzyme number and not the subunit stoichiometry (Fig. S5 B). Disassembly is also not the cause of the interspecies mobility differences, since the distributions lack peaks for the single GFP intensity and monomeric α could not be tracked (see “Visualization of homomeric GFP-CaMKIIβ complexes in the cellular cortex” above). The intensity histograms of β, βT287D, and α (Fig. S5 C) are differentiated by their skewness rather than their modes. The skewness reflects long-lived track lifetimes and results from oligomer immobilization on actin stress fibers (as shown in Fig. 3, A and B).
Figure 6.
Characterization of the mutant proteins. (A) Mobility (Dlat (mean ± SE)) values for the protein populations. Red bars indicate α-isoforms; white bars indicate β-isoforms. βT287D and βE′ have mobility similar to that of the α proteins. (B) Mode (± SE) intensities for the native and mutant GFP-CaMKII fusion protein populations. The bar colors indicate isoforms as in (A).
CaMKII dissociation from cytoskeletal actin
Thus far, our analysis indicates that the track lifetimes for both β and α are biphasic, with MSD versus Δt plots of the short-lived population being consistent with diffusion out of the evanescent field (e.g., Figs. 3 B and 5 D). The photoactivation experiments in the presence and absence of latrunculin demonstrate that the population track lifetime is dramatically reduced coincidently with stress-fiber disassembly. The reduction is mainly due to loss of the long-lived population, implying that these population lifetimes are limited by dissociation from the actin cytoskeleton (Fig. 4). With this in mind, we used track lifetime histograms to estimate the bound fraction and the F-actin dissociation rate for different CaMKII mutants.
The lifetime of the phosphomimic βT287D was taken as representative of unbound molecules, based on βT287D’s failure to decorate cytoskeletal structures (Fig. 5 B; Movie S3) and its high mobility (Fig. 6 A). Consistent with this idea, the βT287D track lifetime data were also fairly monotonic with single exponential decay (rate constant = 6.85 s−1 (1–0.05, R2 = 0.99); Fig. S6). We then fitted all of the other track distributions over this range to a function that assumed there was a nonbinding fraction (i.e., like βT287D) and another longer-lived fraction (Ao) that represented actin-binding complexes with an unknown but slower dissociation rate (k):
| (3) |
The cytoskeletal actin content was assumed to be the same for all experiments, consistent with the modest variation (127 ± 49 counts/pixel) in the mean tRFP-actin intensities in the images (Figs. 3, 4, and 5). The additional information obtained from Eq. 3 is the estimate of the bound () to freely diffusing pools () of molecules and of the dissociation rate, , of molecules from the actin cytoskeleton. The bound fraction, Ao, was 0.22 ± 0.02 for all strong-binding β fusion proteins (minus βE′). A0 was ∼2-fold lower for (α) proteins. The overall group pattern was similar to the pattern observed in the Dlat analysis. The -values were 2.9 s−1 and 1.3 s−1, respectively, for native (α) and (β) (Fig. S6).
Equation 3 would be valid over the complete (1–0) range only for homogeneous populations that follow single-parameter Poisson probability time distributions. This is not the case for the two populations. For the unbound population, as represented by the βT287D proteins, the Dlat value for the most mobile among them (∼0.5 μm2/s; Fig. 6 A) was ∼18-fold lower than the DStokes value calculated for free-diffusing (β) holoenzymes (∼10 μm2/s; Eq. 1). This discrepancy, as well as the deviation of the βT287D distribution from the single exponential fit (Fig. S4), indicates hindered diffusion, although bias introduced by exclusion of rapidly diffusing objects by the five-frame (0.24 s) track filter would also contribute. Power-law distributions due to hindered diffusion have been characterized for F-actin gels in vitro (59), as well as in vivo for membrane proteins confined by the actin cortex (60, 61). The tRFP-actin labeling does not resolve F-actin single filaments in the dense cortex or F-actin spacing in stress fibers, but limits on physical entrapment may be estimated (Supporting Materials and Methods) to rule out this scenario for stress-fiber decoration. For the bound population, a single will obtain only if the dissociation of CaMKII from F-actin subunits does not depend on neighboring subunits. This is not the case, since the detachment probability of a subunit will be lower if neighboring subunits participate in binding together the CaMKII holoenzymes and F-actin.
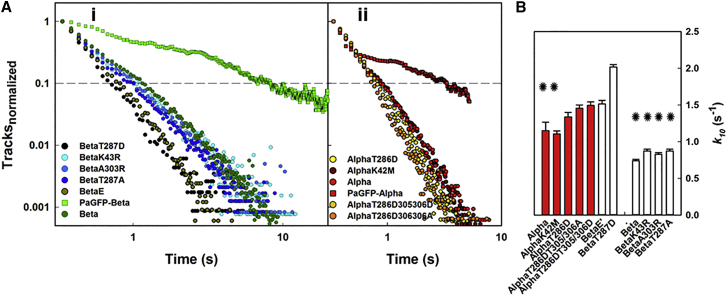
Therefore, we replotted all of our data on log-log axes. We found that they deviated markedly from a dual-exponential process once the population fraction was <5% (Fig. 7 A). All distributions showed the same convex log-log relation, consistent with a multiexponential, log-normal distribution of dissociation times. The initial phase of the log plots over which Eq. 3 is valid provides important estimates of the major binding modes. Nevertheless, it was clear that at longer times the data deviated from a single-parameter, two-population model, and this observation was consistent across all data sets.
Figure 7.
Track lifetimes show log-normal decay. (A) Log-log plots of the CaMKII track lifetime distributions deviate from dual exponential fits and show a downward curvature that is most evident at longer times (i, β proteins; ii, α proteins). (B) Histogram of rates (k10) computed from decay times to 10% amplitude. Asterisks mark species that associate with F-actin.
We compared the times required to reach 10% of the initial amplitude (t1/10) between data sets (Fig. 7 B) to better represent the log relations. We found the same grouping of different mutants as observed in the Dlat analysis. ANOVA (Table S2.I) did not reveal significant differences between the grouped (α) proteins, but did so when these were grouped with βT287D, βE′.
As expected, t1/10 was lowest for βT287D (1.14 + 0.02 s), the reference unbound state, and highest for native β (3.1 ± 0.08 s). We further analyzed differences between the data sets by conducting pairwise t-tests against the βT287D reference (Table S2.II) to parse out differences between group members that were not revealed by the ANOVA. The t-tests revealed α and αK2M as outliers within the weak-binding group, whereas the t1/10 values measured for βE′ and the αT286D proteins with and without secondary phosphorylation site mutations were not significantly different from those obtained for βT287D (Fig. 7 B).
We used the photoactivation data to estimate the dissociation constants (KD)app of CaMKII for actin. These data provide a more valid estimate of the actin dissociation rate, koff, since locally activated PaGFP-tagged molecules essentially only leave the evanescent field, whereas GFP-tagged molecules can both exit and enter from the bulk cytoplasm (Supporting Materials and Methods), resulting in a sevenfold difference in the observed decay (Fig. 7). Our t1/10 decay rates for PaGFP-α (3.4 ± 0.4 s) and PaGFP-β (9.4 + 0.2 s) give koff (= k10 ((log (10))/t1/10) estimates of 0.68 s−1 and 0.23 s−1, respectively. If we assume that the rate of actin association (kon) is in the middle (5× 105 M−1s−1) of the narrow (105–106 M−1s−1) diffusion-controlled range (62) applicable to high-ionic strength media such as cell cytoplasm (63), (KD)app (= koff/kon) is 0.5 μM for β and 1.4 μM for α. The estimate for β is comparable to its measured 2.4 μM affinity for G-actin (24). It is consistent with the simplest scenario of a common binding surface for both G- and F-actin, although more complex scenarios are possible (64, 65).
Discussion
In this work, we used TIRFM-based single-molecule imaging experiments, based on dual-color and photoactivation techniques, to study the dynamics of the interactions of CaMKII isotypes with F-actin within live cultured cells. Our ability to detect micromolar-affinity, weak-binding interactions at subsecond resolution provides information that complements classical sedimentation and gel chromatography assays, and leads to important new, to our knowledge, insights.
CaMKII binding to cytoskeletal actin
We conducted mutant analyses to characterize CaMKII binding to cytoskeletal actin. Substitution of the primary phosphorylated threonine residue by aspartate (βT287D and αT286D) abolished F-actin association for both isoforms, whereas substitution with alanine had no effect. The αT286D mutation abrogated affinity for actin and this effect was independent of mutations at the secondary phosphorylation sites. The βT287A and βT287D data are consistent with the idea that primary-site phosphorylation acts as a single-stage toggle switch, in line with some activation scenarios (66), to control F-actin binding affinity. Consistent with this idea, the βK43R and αK42M mutations that abolish ATP binding had no effect on F-actin association. Both β and α isoforms should bind ATP in HUVECs, since the CaMKII Michaelis constant for ATP is ∼40 μM (58) and the cytoplasmic ATP concentration is typically 2–5 mM (67). In addition, the βK43R/αK42M data show that, in contrast to association with the receptor subunit GluN2B (68), the association with CaMKII F-actin is insensitive to ATP binding and subsequent hydrolysis per se. Elimination of calmodulin binding by the βA303R mutation or use of the peptide inhibitor tatCN21 had no effect on F-actin association. These observations are most simply consistent with low basal Ca2+ and CaMKII activity within HUVECs. Finally, the splice segment that is absent in βE′ is essential for actin binding by the β isoform, consistent with sedimentation assays (22).
Using single-molecule live-cell imaging, we built upon the initial report of stress-fiber decoration in fixed cells (20), which established the CaMKII-F-actin interaction. Our direct observation of actin stress-fiber decoration by immobilized GFP-CaMKII holoenzymes in the presence of a mobile background fraction is consistent with specific binding to F-actin and is incompatible with nonspecific entrapment based on the known stress-fiber architecture. This is also the case for mobility distributions in other actin-rich regions of the cell, based on the known cortical F-actin density and calculated filament mesh size (Supporting Materials and Methods). We found no evidence for higher-order clustering of holoenzymes into larger aggregates that could become either entrapped within or excluded from the actin cortical network or stress fibers. The multistep photobleaching behavior of static spots, along with our histogram analysis of single-object fluorescence intensities, establishes that holoenzymes of CaMKII were the predominant species analyzed in our assays. The presence of larger aggregates is further ruled out by the fact that mutations that abolish Ca2+/CaM binding (A303R) or nucleotide binding (K42M/K43R) required for aggregate formation (36, 69) did not alter the native α/β mobility and lifetime distributions. Thus, the mobility differences between the β287D/αT286D proteins and other CaMKII species, as well as the differences between weak- and strong-binding groups analyzed in this study, can only be explained by differences in F-actin binding affinity.
Although the possibility of CaMKII association with other stress-fiber actin-binding proteins (ABPs) cannot be eliminated, three considerations argue for direct association with F-actin. First, the ABP would need to be abundantly and uniformly distributed along the fibers to be consistent with our images (Figs. 3 A and 4 A; Movies S1 and S2). Other stress-fiber structural ABPs (α-actinin and nonmuscle myosin II) display periodic banding (42). CaMKII binds to α-actinin (70), but this binding is not affected by primary-site phosphorylation (71) and thus may be ruled out. Second, although activated CaMKII has multiple binding targets, there are few binding partners for inactive CaMKII (2), which, as argued above, may be the dominant form in our HUVEC cultures. Third, the relative binding strengths of the CaMKIIβ mutant proteins in our measurements correlate well with results obtained with synthetic F-actin filaments in bundling assays (22).
In neuronal cultures, differences in dendritic arborization (31) and mobility (32) between native and mutant (A303R and K43R) β GFP fusion proteins have been reported, but these differences are probably due to spontaneous neuronal activity that triggers Ca2+.CAM binding for CaMKII activation. Hence, although we do not think that multiple binding partners play a role in our assays, they likely do so in dendritic spines. The reported multiple CaMKII kinetic spine subpopulations (16) may also be due, in part, to multimodal interactions with the F-actin network documented in this study.
Mechanisms for the log-normal bound lifetime distribution
The estimated dissociation constants, (KD)app, for actin are on the order of micromolar for both isoforms. The weak (micromolar) binding of the major β mode is in the ballpark of its reported G-actin affinity. The log-normal distribution of track lifetimes indicates that stronger binding modes exist in addition to the dominant initial mode. These modes may arise from engagement of a variable number of CaMKII subunits with one or more actin filaments (Fig. S7). The fact that the log-normal relation holds for both isoforms and their mutant variants is consistent with the notion that both have a common F-actin binding determinant that is more accessible in β due to its longer, more flexible linker. The alternatively spliced linker region encoded by exons I and IV may increase flexibility between subunits comprising the multimeric complex, thereby ameliorating the geometrical mismatch between CaMKII subunits (72) and binding sites on actin (73). The increased flexibility would optimize contact at the CaMKII and F-actin binding interface and facilitate simultaneous binding at two or more sites, thereby increasing binding avidity. This flexibility could also contribute to heterogeneous binding kinetics, as single-molecule studies indicate that proteins may exist as fluctuating conformational ensembles that lead to power-law distributions in enzyme-turnover experiments over the 10−3 to 10 s timescale (74). Phosphorylation or substitution of serine/threonine residues within the linker may also attenuate flexibility, based on differences in residue size and charge, to regulate persistent CaMKII association with F-actin (13). The possibility that the two isoforms have distinct binding determinants for F-actin cannot be ruled out, but we favor the idea of a common determinant as an explanation based on linker length, which would also account for the difference seen between β and βE′.
Physiological implications of the log-normal binding curve
The log-normal binding curve extends the concentration range for interaction with the actin cytoskeleton. It has two consequences, as described below:
First, it explains why the temporal resolution of the assay determines the detection sensitivity (Fig. S7). Our ability to detect weak-binding species will decrease as the time resolution of the assay increases. The likely explanation for the inability of classical assays to resolve the weak F-actin binding of <α> is that they only detect long-lived, tight-binding states. The binding of <α> to F-actin, thus established, must have a physiological rationale. Variations in isoform expression ratios occur as neurons develop. The binding ensures that CaMKII holoenzymes, predominantly composed of α subunits, can also target the actin cytoskeleton. Although the noted functional effects of αT286D mutation are thought to be mediated by α kinase activity, F-actin association may also play a role. <αT286D> has reduced synaptic localization (75, 76) even though it binds GluN2B in vitro, and loss of F-actin association could account for this effect.
Second, the curve has implications for the transport of <β> down neuronal processes. in contrast to <α>, <β> is expressed only in neuronal cell bodies. Increased avidity due to binding of multiple subunits to F-actin would depend on both the concentration and geometry of F-actin, as well as the multimeric state and flexibility of CaMKII. We estimate expression levels of 0.2–0.4 μM in our assays, based on the density of fluorescent spots in the videos (2000/20 μm2 image field area) and the 100 nm effective depth of the evanescent field. CaMKII concentrations in neurons are severalfold higher: above ∼10 μM holoenzyme in dendritic spines and ∼2–5 μM holoenzyme in dendritic processes (5). In regions of the cell where the actin cytoskeleton is sparse (i.e., dendritic and axonal branches (77)), the low (micromolar) affinity of monovalent <β> binding would minimize its association with cytoskeletal actin during transport along the long neuronal processes (78). However, in regions where the actin cytoskeleton forms a dense three-dimensional meshwork (i.e., dendritic spines), binding via multiple subunits would be favored and <β> would be immobilized. Thus, the extended binding range would facilitate unhindered transport of CaMKII along neuronal processes and sequestration at dendritic spines.
Within dendritic spines, the avidity difference between the two isoforms and between <β> and <βE′> would increase, with qualitatively different effects on the spine actin cytoskeleton. High avidity mediated by a few β subunits in the αβ hetero-oligomers might be sufficient to stabilize the dynamic actin cytoskeleton. The <βE′> splice variant is expressed in immature neurons (21, 31) when affinity for F-actin, which is not required for structural remodeling of synaptic sites, would only hinder the transport needed for targeted kinase activity. Our results provide a quantitative rationale for the fact that the expression of <βE′> has different physiological effects compared with that of <β>.
In conclusion, using single-molecule assays, we were able to resolve CaMKII F-actin binding events on the subsecond-to-second timescale in live mammalian cells. We documented the binding of both neuronal CaMKII isoforms and measured the effect of mutations that act at different points in the CaMKII activation cycle. The behavior of the mutants establishes that binding of CaMKII to actin only occurs when CaMKII is inactive (specifically, when it is not phosphorylated at the primary phosphorylation site). This is in contrast to binding of CaMKII to GluNB, which is triggered only when the kinase is active. This new, to our knowledge, information should be valuable for modeling the role of the actin cytoskeleton in CaMKII transport and synaptic localization.
Author Contributions
S.K. designed and performed research, analyzed data, and wrote the manuscript. I.C. performed research. T.C. contributed reagents. K.U.B. designed research, contributed reagents, and wrote the manuscript. J.E.M. designed research, analyzed data, and wrote the manuscript.
Acknowledgments
We thank Dr. Margaret Stratton for comments on the manuscript, and Dr. Gregory Mashanov for help with computer interfacing and data processing.
This work was supported by grants from the Royal Society Collaborative Exchange (grant U1175), National Institutes of Health (grant R01-NS081248 to K.U.B.), Molecular Biology Consortium (S.K.), Francis Crick Institute which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust (J.E.M.). The University of Colorado holds the patent rights for tatCN21, its derivatives, and its uses (PCT/US08/077934, Compositions and Methods for Improved CaMKII Inhibitors and Uses Thereof). K.U.B. is the owner of Neurexus Therapeutics, LLC.
Editor: Cecile Sykes.
Footnotes
Supporting Materials and Methods, seven figures, two tables, and three movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30440-4.
Supporting Citations
References (79, 80, 81, 82, 83, 84, 85) appear in the Supporting Material.
Supporting Material
The first section of the video shows tRFP-actin (red) and gfp-CaMKII-β (green), played back in real time; the middle section shows the averaged intensity for both channels overlaid as a static image; and the final section shows a section of the separate averaged images after local background subtraction. Colocalization of β and F-actin is seen at actin stress fibers (long white arrows) as well as in regions that are expected to be rich in F-actin (membrane ruffles, short yellow arrows). CaMKII/stress-fiber colocalization was quantified.
The first section of the video shows tRFP-actin (red) and βT287A (green), played back in real time, and the middle section shows the averaged intensity data for both channels overlaid as a static image. The single-particle tracks are overlaid and finally the averaged tRFP-actin image is shown.
The first section of the video shows tRFP-actin (red) and βT287D (green), played back in real time, and the middle section shows the averaged intensity data for both channels overlaid as a static image. The single-particle tracks are overlaid and finally the averaged tRFP-actin image is shown.
References
- 1.Coultrap S.J., Bayer K.U. CaMKII regulation in information processing and storage. Trends Neurosci. 2012;35:607–618. doi: 10.1016/j.tins.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hell J.W. CaMKII: claiming center stage in postsynaptic function and organization. Neuron. 2014;81:249–265. doi: 10.1016/j.neuron.2013.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hudmon A., Schulman H. Neuronal CA2+/calmodulin-dependent protein kinase II: the role of structure and autoregulation in cellular function. Annu. Rev. Biochem. 2002;71:473–510. doi: 10.1146/annurev.biochem.71.110601.135410. [DOI] [PubMed] [Google Scholar]
- 4.Bayer K.U., Löhler J., Harbers K. Developmental expression of the CaM kinase II isoforms: ubiquitous gamma- and delta-CaM kinase II are the early isoforms and most abundant in the developing nervous system. Brain Res. Mol. Brain Res. 1999;70:147–154. doi: 10.1016/s0169-328x(99)00131-x. [DOI] [PubMed] [Google Scholar]
- 5.Otmakhov N., Lisman J. Measuring CaMKII concentration in dendritic spines. J. Neurosci. Methods. 2012;203:106–114. doi: 10.1016/j.jneumeth.2011.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borgesius N.Z., van Woerden G.M., Elgersma Y. βCaMKII plays a nonenzymatic role in hippocampal synaptic plasticity and learning by targeting αCaMKII to synapses. J. Neurosci. 2011;31:10141–10148. doi: 10.1523/JNEUROSCI.5105-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dosemeci A., Tao-Cheng J.H., Reese T.S. Glutamate-induced transient modification of the postsynaptic density. Proc. Natl. Acad. Sci. USA. 2001;98:10428–10432. doi: 10.1073/pnas.181336998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen K., Teruel M.N., Meyer T. Molecular memory by reversible translocation of calcium/calmodulin-dependent protein kinase II. Nat. Neurosci. 2000;3:881–886. doi: 10.1038/78783. [DOI] [PubMed] [Google Scholar]
- 9.Shen K., Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science. 1999;284:162–166. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- 10.Bayer K.U., Schulman H. Regulation of signal transduction by protein targeting: the case for CaMKII. Biochem. Biophys. Res. Commun. 2001;289:917–923. doi: 10.1006/bbrc.2001.6063. [DOI] [PubMed] [Google Scholar]
- 11.Okamoto K., Nagai T., Hayashi Y. Rapid and persistent modulation of actin dynamics regulates postsynaptic reorganization underlying bidirectional plasticity. Nat. Neurosci. 2004;7:1104–1112. doi: 10.1038/nn1311. [DOI] [PubMed] [Google Scholar]
- 12.Hoffman L., Farley M.M., Waxham M.N. Calcium-calmodulin-dependent protein kinase II isoforms differentially impact the dynamics and structure of the actin cytoskeleton. Biochemistry. 2013;52:1198–1207. doi: 10.1021/bi3016586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim K., Lakhanpal G., Okamoto K. A temporary gating of actin remodeling during synaptic plasticity consists of the interplay between the kinase and structural functions of CaMKII. Neuron. 2015;87:813–826. doi: 10.1016/j.neuron.2015.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakoshi H., Yasuda R. Postsynaptic signaling during plasticity of dendritic spines. Trends Neurosci. 2012;35:135–143. doi: 10.1016/j.tins.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayer K.U., De Koninck P., Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- 16.Lu H.E., MacGillavry H.D., Blanpied T.A. Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J. Neurosci. 2014;34:7600–7610. doi: 10.1523/JNEUROSCI.4364-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang F., Kalil K. Netrin-1 induces axon branching in developing cortical neurons by frequency-dependent calcium signaling pathways. J. Neurosci. 2005;25:6702–6715. doi: 10.1523/JNEUROSCI.0871-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chao L.H., Stratton M.M., Kuriyan J. A mechanism for tunable autoinhibition in the structure of a human Ca2+/calmodulin- dependent kinase II holoenzyme. Cell. 2011;146:732–745. doi: 10.1016/j.cell.2011.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanaseki T., Ikeuchi Y., Yamauchi T. Structural features of Ca2+/calmodulin-dependent protein kinase II revealed by electron microscopy. J. Cell Biol. 1991;115:1049–1060. doi: 10.1083/jcb.115.4.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen K., Teruel M.N., Meyer T. CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron. 1998;21:593–606. doi: 10.1016/s0896-6273(00)80569-3. [DOI] [PubMed] [Google Scholar]
- 21.Lin Y.C., Redmond L. CaMKIIbeta binding to stable F-actin in vivo regulates F-actin filament stability. Proc. Natl. Acad. Sci. USA. 2008;105:15791–15796. doi: 10.1073/pnas.0804399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O’Leary H., Lasda E., Bayer K.U. CaMKIIbeta association with the actin cytoskeleton is regulated by alternative splicing. Mol. Biol. Cell. 2006;17:4656–4665. doi: 10.1091/mbc.E06-03-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okamoto K., Narayanan R., Hayashi Y. The role of CaMKII as an F-actin-bundling protein crucial for maintenance of dendritic spine structure. Proc. Natl. Acad. Sci. USA. 2007;104:6418–6423. doi: 10.1073/pnas.0701656104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanabria H., Swulius M.T., Waxham M.N. betaCaMKII regulates actin assembly and structure. J. Biol. Chem. 2009;284:9770–9780. doi: 10.1074/jbc.M809518200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong A.J., Pollard T.D., Herman I.M. Actin filament stress fibers in vascular endothelial cells in vivo. Science. 1983;219:867–869. doi: 10.1126/science.6681677. [DOI] [PubMed] [Google Scholar]
- 26.Mashanov G.I., Tacon D., Molloy J.E. Visualizing single molecules inside living cells using total internal reflection fluorescence microscopy. Methods. 2003;29:142–152. doi: 10.1016/s1046-2023(02)00305-5. [DOI] [PubMed] [Google Scholar]
- 27.Mashanov G.I., Molloy J.E. Automatic detection of single fluorophores in live cells. Biophys. J. 2007;92:2199–2211. doi: 10.1529/biophysj.106.081117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hern J.A., Baig A.H., Birdsall N.J. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc. Natl. Acad. Sci. USA. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nenasheva T.A., Mashanov G.I., Molloy J.E. Imaging individual myosin molecules within living cells. Methods Mol. Biol. 2011;778:123–142. doi: 10.1007/978-1-61779-261-8_9. [DOI] [PubMed] [Google Scholar]
- 30.Nenasheva T.A., Neary M., Molloy J.E. Abundance, distribution, mobility and oligomeric state of M2 muscarinic acetylcholine receptors in live cardiac muscle. J. Mol. Cell. Cardiol. 2013;57:129–136. doi: 10.1016/j.yjmcc.2013.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fink C.C., Bayer K.U., Meyer T. Selective regulation of neurite extension and synapse formation by the beta but not the alpha isoform of CaMKII. Neuron. 2003;39:283–297. doi: 10.1016/s0896-6273(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 32.Khan S., Reese T.S., Shabbir A. Spatiotemporal maps of CaMKII in dendritic spines. J. Comput. Neurosci. 2012;33:123–139. doi: 10.1007/s10827-011-0377-1. [DOI] [PubMed] [Google Scholar]
- 33.Pi H.J., Otmakhov N., Lisman J. Autonomous CaMKII can promote either long-term potentiation or long-term depression, depending on the state of T305/T306 phosphorylation. J. Neurosci. 2010;30:8704–8709. doi: 10.1523/JNEUROSCI.0133-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bayer K.U., LeBel E., De Koninck P. Transition from reversible to persistent binding of CaMKII to postsynaptic sites and NR2B. J. Neurosci. 2006;26:1164–1174. doi: 10.1523/JNEUROSCI.3116-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otmakhov N., Tao-Cheng J.H., Lisman J. Persistent accumulation of calcium/calmodulin-dependent protein kinase II in dendritic spines after induction of NMDA receptor-dependent chemical long-term potentiation. J. Neurosci. 2004;24:9324–9331. doi: 10.1523/JNEUROSCI.2350-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hudmon A., Lebel E., De Koninck P. A mechanism for Ca2+/calmodulin-dependent protein kinase II clustering at synaptic and nonsynaptic sites based on self-association. J. Neurosci. 2005;25:6971–6983. doi: 10.1523/JNEUROSCI.4698-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Subach F.V., Patterson G.H., Verkhusha V.V. Photoactivatable mCherry for high-resolution two-color fluorescence microscopy. Nat. Methods. 2009;6:153–159. doi: 10.1038/nmeth.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merzlyak E.M., Goedhart J., Chudakov D.M. Bright monomeric red fluorescent protein with an extended fluorescence lifetime. Nat. Methods. 2007;4:555–557. doi: 10.1038/nmeth1062. [DOI] [PubMed] [Google Scholar]
- 39.Goedhart J., von Stetten D., Royant A. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat. Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conte I.L., Hellen N., Carter T. Interaction between MyRIP and the actin cytoskeleton regulates Weibel-Palade body trafficking and exocytosis. J. Cell Sci. 2016;129:592–603. doi: 10.1242/jcs.178285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khan S., Zou Y., Reese T.S. Sequestration of CaMKII in dendritic spines in silico. J. Comput. Neurosci. 2011;31:581–594. doi: 10.1007/s10827-011-0323-2. [DOI] [PubMed] [Google Scholar]
- 42.Pellegrin S., Mellor H. Actin stress fibres. J. Cell Sci. 2007;120:3491–3499. doi: 10.1242/jcs.018473. [DOI] [PubMed] [Google Scholar]
- 43.Burnette D.T., Manley S., Lippincott-Schwartz J. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 2011;13:371–381. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Z., Ginnan R., Singer H.A. Calcium/calmodulin-dependent protein kinase II delta 6 (CaMKIIdelta6) and RhoA involvement in thrombin-induced endothelial barrier dysfunction. J. Biol. Chem. 2010;285:21303–21312. doi: 10.1074/jbc.M110.120790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalgaard P. 2nd ed. Springer; New York: 2008. Introductory Statistics with R. [Google Scholar]
- 46.Fischer H., Polikarpov I., Craievich A.F. Average protein density is a molecular-weight-dependent function. Protein Sci. 2004;13:2825–2828. doi: 10.1110/ps.04688204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swaminathan R., Hoang C.P., Verkman A.S. Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys. J. 1997;72:1900–1907. doi: 10.1016/S0006-3495(97)78835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swaminathan R., Bicknese S., Verkman A.S. Cytoplasmic viscosity near the cell plasma membrane: translational diffusion of a small fluorescent solute measured by total internal reflection-fluorescence photobleaching recovery. Biophys. J. 1996;71:1140–1151. doi: 10.1016/S0006-3495(96)79316-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rellos P., Pike A.C., Knapp S. Structure of the CaMKIIdelta/calmodulin complex reveals the molecular mechanism of CaMKII kinase activation. PLoS Biol. 2010;8:e1000426. doi: 10.1371/journal.pbio.1000426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kerber M.L., Jacobs D.T., Cheney R.E. A novel form of motility in filopodia revealed by imaging myosin-X at the single-molecule level. Curr. Biol. 2009;19:967–973. doi: 10.1016/j.cub.2009.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coué M., Brenner S.L., Korn E.D. Inhibition of actin polymerization by latrunculin A. FEBS Lett. 1987;213:316–318. doi: 10.1016/0014-5793(87)81513-2. [DOI] [PubMed] [Google Scholar]
- 52.Giese K.P., Fedorov N.B., Silva A.J. Autophosphorylation at Thr286 of the alpha calcium-calmodulin kinase II in LTP and learning. Science. 1998;279:870–873. doi: 10.1126/science.279.5352.870. [DOI] [PubMed] [Google Scholar]
- 53.Coultrap S.J., Freund R.K., Bayer K.U. Autonomous CaMKII mediates both LTP and LTD using a mechanism for differential substrate site selection. Cell Reports. 2014;6:431–437. doi: 10.1016/j.celrep.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mayford M., Bach M.E., Kandel E.R. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- 55.Yamagata Y., Kobayashi S., Okabe S. Kinase-dead knock-in mouse reveals an essential role of kinase activity of Ca2+/calmodulin-dependent protein kinase IIalpha in dendritic spine enlargement, long-term potentiation, and learning. J. Neurosci. 2009;29:7607–7618. doi: 10.1523/JNEUROSCI.0707-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brocke L., Srinivasan M., Schulman H. Developmental and regional expression of multifunctional Ca2+/calmodulin-dependent protein kinase isoforms in rat brain. J. Neurosci. 1995;15:6797–6808. doi: 10.1523/JNEUROSCI.15-10-06797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vest R.S., Davies K.D., Bayer K.U. Dual mechanism of a natural CaMKII inhibitor. Mol. Biol. Cell. 2007;18:5024–5033. doi: 10.1091/mbc.E07-02-0185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Raveendran R., Devi Suma Priya S., Omkumar R.V. Phosphorylation status of the NR2B subunit of NMDA receptor regulates its interaction with calcium/calmodulin-dependent protein kinase II. J. Neurochem. 2009;110:92–105. doi: 10.1111/j.1471-4159.2009.06108.x. [DOI] [PubMed] [Google Scholar]
- 59.Schmidt C.F., Barmann M., Sackmann E. Chain dynamics, mesh size and diffusive transport in networks of polymerized actin. A quasielastic light scattering and microfluorescence study. Macromolecules. 1989;22:3638–3649. [Google Scholar]
- 60.Heinemann F., Vogel S.K., Schwille P. Lateral membrane diffusion modulated by a minimal actin cortex. Biophys. J. 2013;104:1465–1475. doi: 10.1016/j.bpj.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Umemura Y.M., Vrljic M., Kusumi A. Both MHC class II and its GPI-anchored form undergo hop diffusion as observed by single-molecule tracking. Biophys. J. 2008;95:435–450. doi: 10.1529/biophysj.107.123018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlosshauer M., Baker D. Realistic protein-protein association rates from a simple diffusional model neglecting long-range interactions, free energy barriers, and landscape ruggedness. Protein Sci. 2004;13:1660–1669. doi: 10.1110/ps.03517304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fersht A. W.H. Freeman & Co.; New York: 1999. Structure and Mechanism in Protein Science. [Google Scholar]
- 64.Dominguez R. Actin-binding proteins—a unifying hypothesis. Trends Biochem. Sci. 2004;29:572–578. doi: 10.1016/j.tibs.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 65.dos Remedios C.G., Chhabra D., Nosworthy N.J. Actin binding proteins: regulation of cytoskeletal microfilaments. Physiol. Rev. 2003;83:433–473. doi: 10.1152/physrev.00026.2002. [DOI] [PubMed] [Google Scholar]
- 66.Thaler C., Koushik S.V., Vogel S.S. Structural rearrangement of CaMKIIalpha catalytic domains encodes activation. Proc. Natl. Acad. Sci. USA. 2009;106:6369–6374. doi: 10.1073/pnas.0901913106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sachs J.R. Internal potassium stimulates the sodium-potassium pump by increasing cell ATP concentration. J. Physiol. 1981;319:515–528. doi: 10.1113/jphysiol.1981.sp013923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Leary H., Liu W.H., Bayer K.U. Nucleotides and phosphorylation bi-directionally modulate Ca2+/calmodulin-dependent protein kinase II (CaMKII) binding to the N-methyl-D-aspartate (NMDA) receptor subunit GluN2B. J. Biol. Chem. 2011;286:31272–31281. doi: 10.1074/jbc.M111.233668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vest R.S., O’Leary H., Bayer K.U. Differential regulation by ATP versus ADP further links CaMKII aggregation to ischemic conditions. FEBS Lett. 2009;583:3577–3581. doi: 10.1016/j.febslet.2009.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Walikonis R.S., Oguni A., Kennedy M.B. Densin-180 forms a ternary complex with the (alpha)-subunit of Ca2+/calmodulin-dependent protein kinase II and (alpha)-actinin. J. Neurosci. 2001;21:423–433. doi: 10.1523/JNEUROSCI.21-02-00423.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robison A.J., Bartlett R.K., Colbran R.J. Differential modulation of Ca2+/calmodulin-dependent protein kinase II activity by regulated interactions with N-methyl-D-aspartate receptor NR2B subunits and alpha-actinin. J. Biol. Chem. 2005;280:39316–39323. doi: 10.1074/jbc.M508189200. [DOI] [PubMed] [Google Scholar]
- 72.Rosenberg O.S., Deindl S., Kuriyan J. Oligomerization states of the association domain and the holoenyzme of Ca2+/CaM kinase II. FEBS J. 2006;273:682–694. doi: 10.1111/j.1742-4658.2005.05088.x. [DOI] [PubMed] [Google Scholar]
- 73.Wu Y., Ma J. Refinement of F-actin model against fiber diffraction data by long-range normal modes. Biophys. J. 2004;86:116–124. doi: 10.1016/S0006-3495(04)74089-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Min W., Xie X.S. Kramers model with a power-law friction kernel: dispersed kinetics and dynamic disorder of biochemical reactions. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2006;73:010902. doi: 10.1103/PhysRevE.73.010902. [DOI] [PubMed] [Google Scholar]
- 75.Marsden K.C., Shemesh A., Carroll R.C. Selective translocation of Ca2+/calmodulin protein kinase IIalpha (CaMKIIalpha) to inhibitory synapses. Proc. Natl. Acad. Sci. USA. 2010;107:20559–20564. doi: 10.1073/pnas.1010346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barcomb K., Buard I., Bayer K.U. Autonomous CaMKII requires further stimulation by Ca2+/calmodulin for enhancing synaptic strength. FASEB J. 2014;28:3810–3819. doi: 10.1096/fj.14-250407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu K., Zhong G., Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adamec R., Hebert M., Mervis R.F. Dendritic morphology of amygdala and hippocampal neurons in more and less predator stress responsive rats and more and less spontaneously anxious handled controls. Behav. Brain Res. 2012;226:133–146. doi: 10.1016/j.bbr.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patterson G.H. Photobleaching and photoactivation of fluorescent proteins for studies in cell biology. Microsc. Microanal. 2007;13:294–295. [Google Scholar]
- 80.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Manders E.M., Verbeek F.J., Aten J.A. Measurement of co-localization of objects in dual-colour confocal images. J. Microsc. 1993;169:375–382. doi: 10.1111/j.1365-2818.1993.tb03313.x. [DOI] [PubMed] [Google Scholar]
- 82.Costes S.V., Daelemans D., Lockett S. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys. J. 2004;86:3993–4003. doi: 10.1529/biophysj.103.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Qian H., Sheetz M.P., Elson E.L. Single particle tracking. Analysis of diffusion and flow in two-dimensional systems. Biophys. J. 1991;60:910–921. doi: 10.1016/S0006-3495(91)82125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Meyer R.K., Aebi U. Bundling of actin filaments by alpha-actinin depends on its molecular length. J. Cell Biol. 1990;110:2013–2024. doi: 10.1083/jcb.110.6.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Koestler S.A., Rottner K., Small J.V. F- and G-actin concentrations in lamellipodia of moving cells. PLoS One. 2009;4:e4810. doi: 10.1371/journal.pone.0004810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The first section of the video shows tRFP-actin (red) and gfp-CaMKII-β (green), played back in real time; the middle section shows the averaged intensity for both channels overlaid as a static image; and the final section shows a section of the separate averaged images after local background subtraction. Colocalization of β and F-actin is seen at actin stress fibers (long white arrows) as well as in regions that are expected to be rich in F-actin (membrane ruffles, short yellow arrows). CaMKII/stress-fiber colocalization was quantified.
The first section of the video shows tRFP-actin (red) and βT287A (green), played back in real time, and the middle section shows the averaged intensity data for both channels overlaid as a static image. The single-particle tracks are overlaid and finally the averaged tRFP-actin image is shown.
The first section of the video shows tRFP-actin (red) and βT287D (green), played back in real time, and the middle section shows the averaged intensity data for both channels overlaid as a static image. The single-particle tracks are overlaid and finally the averaged tRFP-actin image is shown.