Abstract
Many drugs and other xenobiotics may reach systemic concentrations where they interact not only with the proteins that are their therapeutic targets but also modify the physicochemical properties of the cell membrane, which may lead to altered function of many transmembrane proteins beyond the intended targets. These changes in bilayer properties may contribute to nonspecific, promiscuous changes in membrane protein and cell function because membrane proteins are energetically coupled to their host lipid bilayer. It is thus important, for both pharmaceutical and biophysical reasons, to understand the bilayer-modifying effect of amphiphiles (including therapeutic agents). Here we use atomic force microscopy topography imaging and nanomechanical mapping to monitor the effect of statins, a family of hypolipidemic drugs, on synthetic lipid membranes. Our results reveal that statins alter the nanomechanical stability of the bilayers and increase their elastic moduli depending on the lipid bilayer order. Our results also suggest that statins increase bilayer heterogeneity, which may indicate that statins form nanometer-sized aggregates in the membrane. This is further evidence that changes in bilayer nanoscale mechanical properties may be a signature of lipid bilayer-mediated effects of amphiphilic drugs.
Introduction
Statins are a class of cholesterol (Chol) lowering drugs that were discovered in the 1970s as a result of a search for 3-hydroxy-3-methyl-glutaryl-Coenzyme A (HMG-CoA) reductase (HMG-CoA reductase) inhibitors (1). HMG-CoA reductase is an integral membrane protein in the endoplasmic reticulum that catalyzes the rate-limiting step in the endogenous synthesis of Chol, the acetylation of HMG-CoA to CoA and mevalonate. HMG-CoA reaction substrates are nontoxic, making HMG-CoA reductase a suitable target for the development of hypolipidemic drugs, and the first statins were put in clinical use in the 1980s (2). Since then they have become first-line drugs for the reduction of morbidity and mortality related to cardiovascular diseases—the primary cause of disability and premature death worldwide (3)—and one of the most consumed classes of drugs in the world (4).
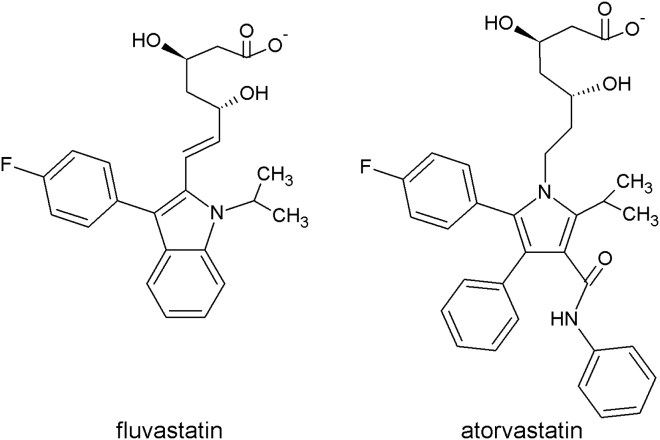
Statins inhibit HMG-CoA reductase activity by blocking access of HMG-CoA to its hydrophobic binding site (5). The key molecular feature of statins is a β-hydroxy-δ-lactone, which is open in most statins in current use and resembles HMG (1) (which binds in the active site) and rigid, hydrophobic groups that fit in a nonpolar pocket of the enzyme. First-generation statins had a decaline moiety, which was considered important for the interaction with the accessory hydrophobic zone of the receptor, but it is not essential for the activity. Examples of novel statins are fluvastatin and atorvastatin, where the decaline portion was substituted by a heteroaromatic group (Fig. 1).
Figure 1.
Chemical structures of fluvastatin (left) and atorvastatin (right). Pharmacokinetic data show plasma statins concentration in the nanomolar range. In vitro experiments are usually performed in the micromolar range (74).
Statins interact with the HMG-CoA reductase with high specificity, but they also exert effects that appear unrelated to the lowering of blood plasma Chol and the concentration of low-density lipoproteins in blood (6)—some being beneficial (7, 8, 9, 10), others not (11, 12, 13, 14).
Such pleiotropic effects could arise for a variety of reasons—some being due to the inhibition of HMG-CoA reductase because mevalonate is a precursor in the synthesis of a variety of isoprenes (6), others being less well understood. Among the proteins that are modified by statins are a variety of membrane-spanning proteins (15, 16), whose function may be altered through statin-induced changes in protein prenylation (6), membrane cholesterol concentration (17, 18), or by statin-dependent changes in lipid bilayer properties. The latter arises because statins are amphiphiles that will partition reversibly in the bilayer/solution interface (19, 20) and thereby alter membrane protein function (21, 22). Any of these mechanisms, alone or in combination, could lead to drug promiscuity. In the nanomolar range, the drug may interact primarily with the desired target; in the micromolar range, amphiphiles may alter lipid bilayer properties sufficiently to modulate the function of a wide range of membrane proteins (20, 23, 24).
Such alterations arise because membrane proteins undergo conformational changes that involve the proteins’ bilayer-spanning domains (25) and therefore will alter the lipid packing adjacent to the protein, which has an associated energetic cost, the bilayer deformation energy , which will be a contribution to the free energy difference between different protein conformations. Changes in lipid bilayer elasticity may alter the conformational preference of membrane receptors and transmembrane proteins and thereby alter their function (21, 26). The effect(s) of the incorporation of small molecules on membrane mechanics has been widely studied; for example, short-chain alcohols (27, 28), bile acids (29), salicylates (30), etc. In general, small surfactants change the bilayer lateral pressure profile, elasticity, and thickness (31), and modify the thermodynamic parameters of the lipid membrane (32). The gramicidin (gA) channels have proven useful as probes to detect changes in bilayer properties, as gA monomers in each membrane leaflet form dimeric bilayer-spanning channels, hence sensing lipid bilayer physical-chemical alterations (20, 33, 34, 35, 36, 37). gA has also been used to examine the effect of statins on lipid-bilayer properties (38).
We measured the elasticity of synthetic supported lipid bilayers (SLBs) by means of atomic force microscopy (AFM) topography imaging and quantitative nanomechanical mapping (QNM), in the absence and presence of statins. This imaging technique is based on short-range approach-and-retract cycles between the AFM tip and the sample surface. This process is accomplished on every location (pixel) of the image, thereby resulting in topographical (point of contact) and mechanical (slope of indentation) information simultaneously. Upon the addition of two different statins, fluvastatin and atorvastatin, as well as the commonly used detergent Triton X-100 (24), we observed an increased Young’s modulus (E) of the SLBs, for the lipid domains that are in a solidlike or gel phase with much more modest changes in the ld domains. Moreover, the statins increased the dispersity of the elasticity values, which might indicate heterogeneous miscibility of the drug with the lipid. Based on these measurements, we also estimated the bending stiffness, stretch modulus, and the hydrophobic mismatch elastic stretching energy at domain interfaces. Our nanomechanical approach using AFM-based measurements is, to our knowledge, novel for characterizing the effect of statins and other amphiphiles on membranes, and provides a different perspective on the bilayer-mediated effect of amphipathic drugs.
Materials and Methods
Sample preparation
SLBs were prepared as described in Mingeot-Leclercq et al. (39). In brief, phospholipids, DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) and DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), were dissolved in chloroform/methanol (3:1) to a final phospholipid concentration of 3 mM. An aliquot was poured into a glass vial and evaporated to dryness under a nitrogen stream. The resulting lipid film was kept under reduced pressure overnight, to ensure the absence of organic solvent traces, and hydrated with 10 mM Tris-HCl, 150 mM KCl, pH 7.4 buffer (measurement buffer) to form a 500 μM multilamellar lipid suspension, which was extruded through a 100-nm polycarbonate filter to produce large unilamellar vesicles (LUVs). 30 μL of the LUV suspension were deposited onto freshly cleaved mica disks (area 25 mm2) preincubated with 10 μL of 10 mM Tris-HCl, 150 mM KCl, 25 mM MgCl2, pH 7.4 (adsorption buffer). The mica disks were mounted on a disk of Teflon (Chemours, Wilmington, DE) and incubated for 1 h at 60°C, leading to the formation of SLBs. The SLBs were carefully rinsed with measurement buffer before imaging and were always kept under aqueous environment.
AFM measurements
AFM measurements were carried out in a fluid cell containing 100 μL measurement buffer at room temperature and ambient pressure on a Nanoscope-V AFM (Bruker, Santa Barbara, CA) equipped with Nanoscope-8 control software, in Peak-Force-Quantitative-Nano-Mechanics (PF-QNM) mode. Time-lapse experiments were performed by introducing the statin (fluvastatin or atorvastatin) solution through the inlet of the AFM fluid cell to give a final nominal statin concentration of ∼10 μM (the free statin concentration will be less than the nominal concentration due to drug partitioning into the SLB, but the depletion should be minimal). We used Si3N4 cantilevers with a nominal spring constant of either 100 pN/nm or 500 pN/nm and silicon tips with a 2-nm nominal radius (MSNL; Bruker). The actual spring constant of the cantilevers was determined using the thermal fluctuation method, and the cantilever sensitivity calibrated through force curve cycles on solid support before each measurement (40). Images were obtained at a resolution of 256 × 256 pixels at a line scan rate of 2 Hz. In the PF-QNM imaging mode, the sample support oscillates at constant rate (2 kHz) and amplitude (between 15 and 25 nm). During each oscillation cycle, the deflection (force) of the cantilever was monitored to obtain a short force-distance curve. The vertical amplitude of the piezoelectric displacement was set to allow the tip to completely separate out of contact from the sample surface, allowing accurate determination of the zero force and the applied indentation peak force in each cycle. The approach trace was used to control the maximum force applied (∼300 pN). The Young’s modulus (E) was calculated at each pixel of the image from the force F applied in the contact part of the retraction trace of each oscillation cycle by fitting the Hertz model for a spherical tip of radius R indenting an elastic half-space (41, 42) (Fig. S1 in the Supporting Material):
| (1) |
where ν is the Poisson ratio (assumed to be 0.5, the value for a perfectly elastic uncompressed material), and δ is the indentation. The tip radius (R) was assumed 2 nm, the nominal value. Even if the approximation is not valid, our main interest is the relative changes in elasticity upon statin addition. Using the Hertz model to describe the indentation of thin films introduces uncertainties in the determination of E (∼25% at 20% relative indentation (43)). The results reported here were acquired at low forces and small deformations (∼1 nm; see Fig. S1), conditions under which the Hertz approximation should be valid.
To avoid contributions from long-range electrostatic forces and van der Waals interactions, we restricted the Hertz-model fit to a range between 30 and 90% of the maximum indentation force F (peak-force set point). The E values were obtained through averaging and distribution analysis of the 65,536 measured values per image on micron-sized lipid areas from at least three independent measurements.
AFM image processing
Image and data processing was performed using the Nanoscope Analysis Software (Bruker) and Gwyddion 2.38 open software (gwyddion.net). In the topographical maps, the heights of the SLBs were measured from topographical maps displaying domain edges, measuring the height difference between the SLB and the bare mica. The nanomechanical parameter values were obtained after histogram analysis of the height and E maps from at least three different samples for each composition, without application of plane fitting, and applying a mask after domain edge detection (41, 44). The elasticity maps are log-normal distributed and were fitted with a Gaussian in the log scale. The reported mean values are the exponentiated center of the Gaussians; the errors are calculated from the SDs in the logarithmically transformed distribution.
Results
DPPC/DOPC model system
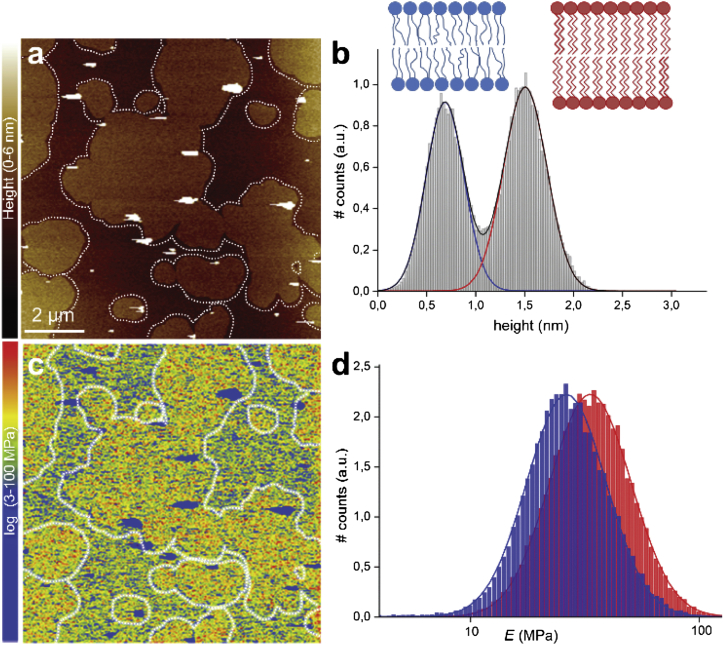
To study the mechanical effects of statins in lipid bilayers, we chose a binary lipid mixture DPPC/DOPC at a 1:1 molar ratio. At room temperature, this mixture shows coexistence of a liquidlike domain (liquid-disordered domain, ld) and a solidlike domain (solid-ordered domain, so). As would be expected for the chosen lipid mixture (45, 46), the lipid domains in the SLBs are large and directly visible (Fig. 2 a) with clearly distinguishable heights (∼0.8 nm height difference; Fig. 2 b). Nanomechanical mappings (Fig. 2 c) are comparable with E values in the literature, characterized mechanically by AFM (41, 47, 48, 49, 50) and micropipette aspiration (51, 52). (For an overview of experimental approaches used to assess the elasticity of membranes, see Dimova (53).) The mean E values are MPa for the ld domain and MPa for the so domain, respectively (Fig. 2 d). The sub- and superscripts denote the mean – and + the standard deviation from the fit to the log-normal transformed data.
Figure 2.
PF-QNM AFM topography and elasticity mapping of DOPC/DPPC (1:1) SLBs. (a) Topography image (nm) and (b) height histogram analysis of the topography (a). The dashed lines in (a) are drawn to represent the domain edges defined by edge detection. The solid lines in (b) are Gaussian fits to the height distributions, indicating an average height difference between the DOPC ld (blue line) and DPPC so (red line) domains of 0.82 nm. (c) The corresponding stiffness map (log scale, MPa) and (d) Young’s modulus values histograms (log scale, MPa) of the regions outlined in the topography (a) and stiffness (c) maps, corresponding to the ld (blue) and so (red) domains. The solid lines are Gaussian fits to the distributions peaking at MPa (ld) and MPa (so). To see this figure in color, go online.
Statins alter the bilayer elasticity
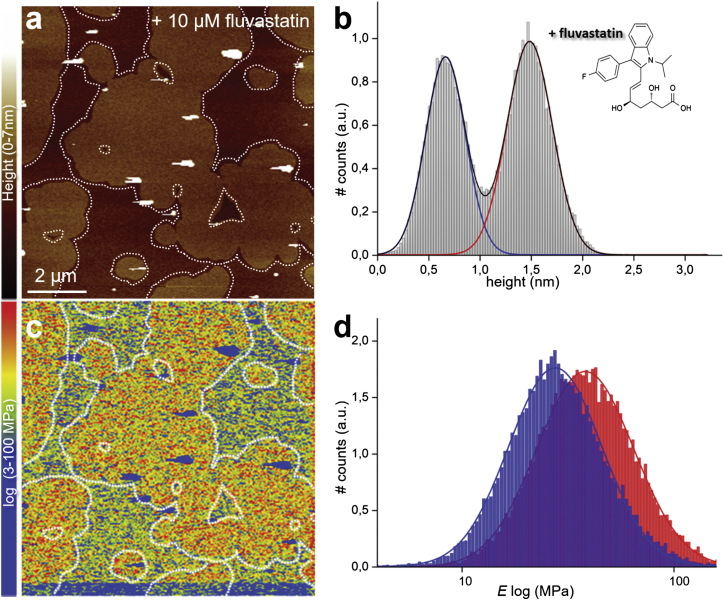
After addition of fluvastatin (∼10 μM total concentration) to the SLB characterized in Fig. 2, and equilibrating for 1 h, we repeated the topography imaging and mechanical mapping, which is possible because of the AFM’s unique capability of buffer exchange during operation. The topography image (Fig. 3 a) has a similar topography to the control image, and the height difference between the ld and so domains varied little (being 0.86 vs. 0.82 nm before statin addition; Fig. 3 b).
Figure 3.
PF-QNM AFM topography and elasticity mapping of DOPC/DPPC (1:1) SLBs after addition of fluvastatin. (a) Representative topography image (nm) and (b) height histogram analysis of topography (a) of the same DOPC/DPPC (1:1) SLBs as shown in Fig. 2a after ∼1 h of incubation with ∼10 μM fluvastatin. The dashed lines in (a) represent the domain edges defined by edge detection. The solid lines in (b) are Gaussian fits to height distribution of the whole height image, indicating an average height difference between the ld (blue line) and so (red line) domains of 0.86 nm. (c) Corresponding stiffness map (log scale, MPa) and (d) Young’s modulus values histograms (log scale, MPa) of the regions outlined in the topography (a) and stiffness (c) maps, corresponding to ld (blue) and so (red) domains. The solid lines are Gaussian fits to the distributions peaking at MPa (ld) and MPa (so). To see this figure in color, go online.
The stiffness of the so domain increased to MPa, whereas the ld domain remained almost unchanged, with a modulus of MPa. The stiffness increase of the so domain is accompanied by only minor morphological changes of the domains when comparing the topography images in Figs. 2 a and 3 a. Yet, the relative areas of the ld and so domains changed from ∼43 and ∼57%, respectively, to ∼33 and ∼67%; i.e., the so domains expanded when comparing Figs. 2 a and 3 a, suggesting a preferential insertion of fluvastatin into the so domains. To assess changes in the membrane elasticity, results from at least three different samples for each composition were averaged. After fluvastatin addition, the stiffness of the so domain increased to 50 ± 30 MPa (from 40 ± 10 MPa), while the ld domain changed to 31 ± 14 MPa (from 33 ± 7 MPa; mean and SD from three different experiments).
When atorvastatin was added to the SLB, the topography images similarly did not reveal significant morphological or height differences and, as observed with fluvastatin, the Young’s modulus of the so domain increased with respect to the untreated SLB (Fig. S2). After atorvastatin addition, the stiffness of the so domain increased to 81 ± 37 MPa (from 40 ± 10 MPa), while the ld domain increased to 36 ± 9 MPa (from 33 ± 7 MPa) (mean and SD from three different experiments).
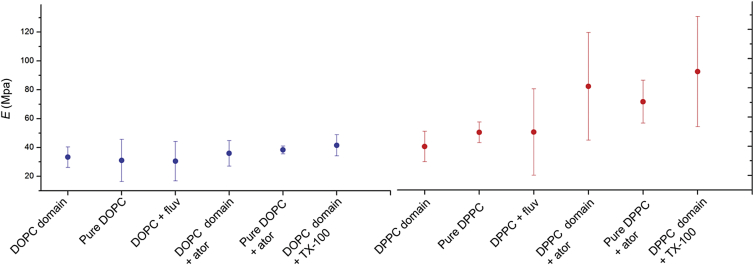
Addition of the statins thus increases the stiffness of the solidlike so domains (presumably enriched in DPPC), with minimal changes in the liquidlike ld domains (presumably enriched in DOPC). Atorvastatin has a higher impact on the Young’s modulus than fluvastatin (Fig. 4; Table 1). In addition to the changes in E of the so domains, the statins also caused a three- to fourfold increase of the SD of the log-normal transformed results, probably emerging from heterogeneity of the bilayer stiffness upon statin addition, although other variables may also contribute to this heterogeneity—notably heterogeneity in the boundary of the domains (54).
Figure 4.
Graphical representation of Young’s modulus for both fluidlike ld (DOPC) and solidlike so (DPPC) domains in the absence and presence of fluvastatin, atorvastatin, and TX-100. Data are shown as the mean ± SD from the peak position of the log-normal distributions from at least three different samples and measurements for each composition. To see this figure in color, go online.
Table 1.
Experimental Values of Bilayer Thickness h and E Obtained from the Topography and Nanomechanical Measurement of the Lipid Domains and Estimated Values for KA and kC
| Bilayer thickness |
Young’s modulus |
Stretch modulus |
Bending stiffness |
|
|---|---|---|---|---|
| h (nm) | E (MPa) | KA (pN/nm) | kC (kBT) | |
| ld DOPC domain | 4.6 ± 0.2 | 33 ± 7 | 202.0 | 23.8 |
| ld DOPC domain + fluvastatin | 4.5 ± 0.2 | 31 ± 14 | 183.4 | 21.3 |
| ld DOPC domain + atorvastatin | 4.4 ± 0.2 | 36 ± 9 | 212.0 | 23.7 |
| so DPPC domain | 5.5 ± 0.2 | 40 ± 10 | 289.2 | 48.9 |
| so DPPC domain + fluvastatin | 5.4 ± 0.2 | 50 ± 30 | 355.9 | 58.5 |
| so DPPC domain + atorvastatin | 5.7 ± 0.3 | 81 ± 37 | 615.6 | 114.1 |
Data are averaged from the means of the log-normal distributions of at least three different samples for each composition.
Estimation of the elastic energetic cost of mixing statins into lipid bilayers
Using the Young’s modulus for each domain of the DOPC/DPPC mixture together with the experimental bilayer thickness (h), we can use thin shell theory to calculate the area compression-expansion modulus and bending modulus as (47):
The results are summarized in Table 1. Our estimates for and (columns 3 and 4) are in quantitative agreement with values reported in the literature, based on micropipette aspiration methods (47) and AFM-based methods (41). We similarly estimated and for the fluidlike ld and the solidlike so domains in the presence of fluvastatin and atorvastatin, and found minimal changes in the stretch and bending stiffness of the ld domain upon addition of statins. In contrast, for the so domains, the stretch and bending moduli were higher in the presence of the statins, with the stronger effect, doubled and , for the more bulky (Fig. 1) atorvastatin.
When lipid domains separate in lipid mixtures, or when lipids surround membrane-embedded proteins, there may be a hydrophobic mismatch at the domain interfaces (55) or protein/bilayer interfaces (56, 57, 58), with an ensuing compression or stretching of the hydrocarbon chains in the vicinity of the interface. This hydrocarbon tail stretching or compression will influence the distribution of domain sizes and the kinetics of domain separation (59), as well as membrane protein function. We can estimate the compression/stretching of the acyl chains of the so and ld domains by minimizing the elastic energy of the system, . We can express assuming volume conservation (55) as
| (2) |
where and are the stretch moduli for so and ld, respectively. The values and are the lipid area per molecule for so and ld, respectively. The A values are well documented in literature (notably through x-ray diffraction and Langmuir methods), being 0.72 nm2 for ld DOPC and 0.49 nm2 for so DPPC at room temperature (60). We assume the variation of and upon addition of statins does not qualitatively change estimation. The values and are the measured bilayer thickness of so and ld, respectively. Experimental values for bilayer thicknesses and stretch moduli are shown in Table 1.
We wish to determine the value of x when has its minimum, therefore
| (3) |
for simplification, we replace
| (4) |
and solve Eq. 2 for x:
| (5) |
The estimated values for the ld and so domains are 0.27 kBT in the absence of statins, and 0.31 kBT and 0.88 kBT in the presence of fluvastatin and atorvastatin, respectively. This results in line tensions of ∼0.35 and 1.14 kBT∙nm in the absence and presence of statins, respectively; meaning that the presence of statins increase the energetic penalty for transferring a bilayer-spanning protein (with a diameter of a few nanometers) from ld to so domains. Furthermore, as the addition of statins changes both the stretch modulus and the bilayer thickness h initially adopted by the lipid, the energetic cost to domain-separate and/or surround membrane-embedded components changes following statin addition.
As a control experiment, to ascertain that the effect of the statins on the different domains in the binary mixture is not influenced by the mixing itself, we measured the topography and Young’s modulus in absence and presence of atorvastatin on single-component lipid bilayers (Table 2). The measured E for the one-component SLBs in the absence and presence of atorvastatin agree with the measured E for the correspondent ld-enriched and so-enriched domains for the binary mixtures, excluding a significant effect of lipid mixing.
Table 2.
Experimental Values of Young’s Modulus Obtained from the Nanomechanical Measurements of Single-Component Lipid Bilayers
| Bilayer thickness |
Young’s modulus |
|
|---|---|---|
| h (nm) | E (MPa) | |
| ld DOPC | 4.6 ± 0.2 | 31 ± 15 |
| ld DOPC + atorvastatin | 4.9 ± 0.2 | 36 ± 12 |
| ld DOPC + TX-100 | 4.5 ± 0.2 | 42 ± 7 |
| so DPPC | 5.1 ± 0.2 | 50 ± 7 |
| so DPPC + atorvastatin | 5.9 ± 0.3 | 71 ± 15 |
| so DPPC + TX-100 | 5.5 ± 0.3 | 91 ± 38 |
Data are averaged from at least three different samples for each composition.
To test whether the statin effects represent a general amphiphile effect, we also added Triton X-100 (TX-100) to the SLBs. TX-100 has been widely used to study detergent-resistant membrane domains or rafts. The resistance of membranes to surfactants is directly related to the melting temperature (Tm) of the lipid and the aggressiveness (critical micellar concentration, CMC) of the detergent (61). Detergent-resistant membrane domains are known to be enriched in gel-domain phospholipids (62). In the case of SLBs, it has been shown that TX-100, below the CMC (0.2–0.25 mM), does not solubilize the lipid membranes. However, it inserts into the bilayer and has an eroding effect on so domains, which was attributed to the modification of the molecular packing of the gel-domain induced by the TX-100 (63). In analogy to the experiments with the statins, we therefore examined the effect of TX-100 on SLBs at concentrations below the CMC. The nanomechanical effects of TX-100 in pure ld DOPC SLBs, pure so DPPC SLBs, and DOPC/DPPC (1:1) SLBs, were similar to what was used for the statin experiments, and we compared it to the effect of the statins (Table 2). For the DPPC and DPPC/DOPC experiments, we used 10 μM TX-100, which is far below the CMC; for the DOPC experiments we needed to decrease the TX-100 concentration to ∼2 μM to maintain bilayer integrity. As was the case for the statins, TX-100 increased the bilayer stiffness, with the major effect being on the so DPPC domains, again accompanied by an increase in the dispersion of the Young’s moduli.
Discussion
The incorporation of amphiphiles into lipid bilayers alters bilayer properties, such as the intrinsic curvature, thickness, elasticity, roughness, fluidity, etc. (20, 64). These alterations of bilayer properties are not expected to be isolated; any change in one property usually will be associated with changes in other bilayer properties, e.g., Bruno et al. (20). It is a priori difficult to discern which is the key modification that may explain the overall effect of an amphiphile on a lipid bilayer (20), however the impact of the alterations of the physical-chemical properties on membrane protein function is unquestioned (for reviews, see Lundbaek et al. (21) and Phillips et al. (26)).
We examined how lipid bilayer modifiers, i.e., statins and TX-100, alter the nanomechanical properties of SLBs. The statins and TX-100 increase the membrane’s Young’s modulus E of the so, with little effect on the ld domains, and with little effect on the thickness of the different domains or the domain morphology. Yet, despite the almost unchanged domain morphology, the relative area of the so domains increased by ∼15%, suggesting a preferential insertion of the statins into the so domain.
Along with increased rigidity, the dispersion of the E values is increased, whether the bilayer modifier was a statin or TX-100, suggesting that the amphiphiles induce bilayer mechanical heterogeneity at nanometer-scale but also on large length scales. The increase in E occurs preferentially in the so domain and would indicate preferential partitioning into the so lipids, a notion that is difficult to rationalize in the framework of earlier observations that amphiphiles tend to prefer ld over so domains (65, 66).
When comparing the AFM-based elasticity mapping with published bilayer stiffness measurements using gA channels as force transducers (25, 67), we note a qualitative difference between the results obtained in SLBs and results obtained in LUVs or planar lipid bilayers (20, 22, 68, 69). Gramicidin A is a cation-selective channel composed of two head-to-head-associated half-channels, each formed by β-strands coiled into a helical barrel (70); it has been widely used to study the effects of amphiphiles on the mechanics of the lipid bilayer, as its channel formation depends on the interaction of the two half-channels in each leaflet and hence on the elastic properties of the membrane. Channel formation leads to ion flow, i.e., current measurement. Importantly, the associated gA channel provokes a membrane deformation when formed in a bilayer with a thicker hydrophobic core. Changes in the elasticity of the lipid bilayer alter the bilayer disjoining force to the gA channel, and this was observable in single-channel current traces performed by means of patch-clamp measurements (37).
Based on the nanomechanical experiments presented here, the amphiphiles tested increase bilayer stiffness (Young’s modulus, as estimated using Eq. 1) of the so domains in SLBs. In contrast, the bilayer stiffness decreases when recorded using the gA channels as force transducers (38, 68), as would be expected for thermodynamic reasons (19, 20, 29). This difference between the results in SLBs and unsupported bilayers seems to be general: Khadka et al. (71), for example, found that the anticancer drug tamoxifen increases the bilayer thickness and Young’s modulus of SLBs, whereas tamoxifen shifts the gramicidin monomer-to-dimer equilibrium toward the conducting dimers (R.L.S. and O.S.A., unpublished results). The different results somehow reflect differences between the SLBs used in the AFM experiments and the unsupported bilayers used in the gramicidin experiments, either because of different responses to the partitioning of amphiphiles into membranes or because of the different nature of the deformations produced by the different approaches.
With respect to the former possibility, it is well established in micromechanical measurements that the membrane area increases when amphiphiles partition into the bilayer/solution interface (19, 28, 29, 30). This area increase is likely to have little consequence in unsupported bilayers, but could produce additional stresses in SLBs due to the constraints imposed by collective movements of the lipids in the SLB relative to the rigid support, which would tend to decrease the compressibility of the SLB. Related to this, the topographical profiles are remarkably invariant over an hour (after the addition of the amphiphile), which suggests that the ability of the SLB to respond to the insertion/removal of amphiphile molecules, which constitutes the basis for the amphiphile-induced increase in elasticity, may be reduced relative to the unsupported bilayers.
With respect to the latter possibility, lipid bilayers have anisotropic elastic properties, meaning that several elasticity moduli are required to fully describe the bilayer properties (i.e., the transverse elasticity, area expansion, volume compressibility, and elastic moduli) (72). When the bilayer is compressed, as is the case when a gA channel forms, the vertical deformation is imposed on the hydrophobic region of the lipid bilayer, leading to a local membrane thinning. In the case of AFM nanoindentations, the stress is applied perpendicular to the membrane plane from the top of the hydrophilic headgroups but the deformation will also have a radial component (72). Thus, considering the lipid membrane as an anisotropic element, the different approaches for measuring changes in the elasticity of the lipid bilayer may simply be different, complementary descriptors for the influence of amphiphiles on the lipid membrane. Statins may interact with the bilayer isotropically, maybe with a vertical intercalation of the heteroaromatic groups between the lipids. In this context, the ability of an amphiphile to induce changes in the cohesion of lipid bilayers has been shown to depend on the nature of the lipid molecules: cholesterol increases the molecular order only in lipid bilayers that have saturated hydrocarbon chains (73). It could be argued that statins have a similar interaction with lipids. Finally, whereas the gA experiments report about the compressibility of the hydrophobic membrane core, the AFM measures the compressibility of the entire membrane, including the headgroups.
Conclusions
Here we report a nanomechanical characterization and mapping of SLBs, using PF-QNM AFM topography, in the absence and presence of statins. Adding the statins to the buffer solution increases the force required to deform the bilayer (and thus the elastic modulus E), especially in the gel domain. The dispersion of the elasticity values is also higher in the presence of statins, suggesting an increase of heterogeneity of the lipid bilayers at the nanoscopic level and in general. The experimental estimate of E allowed us to estimate the stretch modulus and the bending stiffness of the lipid membrane, as well as the energetic cost of the lipid domains to accommodate to different thickness. In the presence of statins, the values are increased, probably due to changes in the phospholipid order, in particular in the so domain.
We propose that AFM-based nanomechanical mapping may be an alternative way to study the physical-chemical modification of amphiphilic drugs in host lipid bilayers, because nanoscale mechanics is a relevant signature of a lipid bilayer as this is the length-scale of membrane protein size and function. In the case of statins, this can provide a novel, to our knowledge, angle to analyze how bilayer properties are modified and maybe shed insight on the pleiotropic effects of statins that are not related to the inhibition of their target, HMGCo-A reductase.
Author Contributions
All authors designed the initial project; L.R.-M. performed the research; L.R.-M. and S.S. analyzed the data; and all authors wrote the article.
Acknowledgments
We thank Dr. F. Rico for helpful discussions.
This research was supported by the French Agence National de la Recherche grants ANR-Nano (No. ANR-12-BS10-009-01) and ANR-BBMS (No. ANR-12-BSV8-0006-01); the European Research Council grant (No. 310080) to S.S.; and National Institutes of Health grant (No. GM0021342) to O.S.A.
Editor: Klaus Gawrisch.
Footnotes
Two figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)30462-3.
Supporting Material
References
- 1.Tobert J.A. Lovastatin and beyond: the history of the HMG-CoA reductase inhibitors. Nat. Rev. Drug Discov. 2003;2:517–526. doi: 10.1038/nrd1112. [DOI] [PubMed] [Google Scholar]
- 2.Endo A. The discovery and development of HMG-CoA reductase inhibitors. J. Lipid Res. 1992;33:1569–1582. [PubMed] [Google Scholar]
- 3.World Health Organization . WHO; Geneva, Switzerland: 2012. World Health Statistics 2012. [Google Scholar]
- 4.Aitken M. IMS Institute for Healthcare Informatics; Parsippany, NJ: 2011. The Global Use of Medicines: Outlook Through 2015. [Google Scholar]
- 5.Istvan E.S., Deisenhofer J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science. 2001;292:1160–1164. doi: 10.1126/science.1059344. [DOI] [PubMed] [Google Scholar]
- 6.Liao J.K., Laufs U. Pleiotropic effects of statins. Annu. Rev. Pharmacol. Toxicol. 2005;45:89–118. doi: 10.1146/annurev.pharmtox.45.120403.095748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altwairgi A.K. Statins are potential anticancerous agents (review) Oncol. Rep. 2015;33:1019–1039. doi: 10.3892/or.2015.3741. [DOI] [PubMed] [Google Scholar]
- 8.Krishna R.K., Issa O., Santana O. Pleiotropic effects of the 3-hydroxy-3-methylglutaryl-CoA reductase inhibitors in pulmonary diseases: a comprehensive review. Pulm. Pharmacol. Ther. 2015;30:134–140. doi: 10.1016/j.pupt.2014.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Bonfrate L., Procino G., Portincasa P. A novel therapeutic effect of statins on nephrogenic diabetes insipidus. J. Cell. Mol. Med. 2015;19:265–282. doi: 10.1111/jcmm.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pon D., Abe A., Gupta E.K. A review of statin use and prostate cancer. Curr. Atheroscler. Rep. 2015;17:474. doi: 10.1007/s11883-014-0474-5. [DOI] [PubMed] [Google Scholar]
- 11.Dobrzynski J.M., Kostis J.B. Statins and cataracts—a visual insight. Curr. Atheroscler. Rep. 2015;17:477. doi: 10.1007/s11883-014-0477-2. [DOI] [PubMed] [Google Scholar]
- 12.Moosmann B., Behl C. Selenoprotein synthesis and side-effects of statins. Lancet. 2004;363:892–894. doi: 10.1016/S0140-6736(04)15739-5. [DOI] [PubMed] [Google Scholar]
- 13.Vaklavas C., Chatzizisis Y.S., Giannoglou G.D. Molecular basis of statin-associated myopathy. Atherosclerosis. 2009;202:18–28. doi: 10.1016/j.atherosclerosis.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Bełtowski J., Wójcicka G., Jamroz-Wiśniewska A. Adverse effects of statins—mechanisms and consequences. Curr. Drug Saf. 2009;4:209–228. doi: 10.2174/157488609789006949. [DOI] [PubMed] [Google Scholar]
- 15.Malenda A., Skrobanska A., Nowis D.A. Statins impair glucose uptake in tumor cells. Neoplasia. 2012;14:311–323. doi: 10.1593/neo.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimitroulakos J., Yeger H. HMG-CoA reductase mediates the biological effects of retinoic acid on human neuroblastoma cells: lovastatin specifically targets P-glycoprotein-expressing cells. Nat. Med. 1996;2:326–333. doi: 10.1038/nm0396-326. [DOI] [PubMed] [Google Scholar]
- 17.Lundbæk J.A., Andersen O.S. John Wiley; Hoboken, NJ: 2012. Cholesterol Regulation of Membrane Protein Function by Changes in Bilayer Physical Properties—An Energetic Perspective. [Google Scholar]
- 18.Dopico A.M., Bukiya A.N. Lipid regulation of BK channel function. Front. Physiol. 2014;5:312. doi: 10.3389/fphys.2014.00312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhelev D.V. Material property characteristics for lipid bilayers containing lysolipid. Biophys. J. 1998;75:321–330. doi: 10.1016/S0006-3495(98)77516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruno M.J., Rusinova R., Andersen O.S. Interactions of drugs and amphiphiles with membranes: modulation of lipid bilayer elastic properties by changes in acyl chain unsaturation and protonation. Faraday Discuss. 2013;161:461–480. doi: 10.1039/c2fd20092a. discussion 563–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lundbaek J.A., Birn P., Andersen O.S. Capsaicin regulates voltage-dependent sodium channels by altering lipid bilayer elasticity. Mol. Pharmacol. 2005;68:680–689. doi: 10.1124/mol.105.013573. [DOI] [PubMed] [Google Scholar]
- 22.Rusinova R., Herold K.F., Andersen O.S. Thiazolidinedione insulin sensitizers alter lipid bilayer properties and voltage-dependent sodium channel function: implications for drug discovery. J. Gen. Physiol. 2011;138:249–270. doi: 10.1085/jgp.201010529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornecki E., Ehrlich Y.H., Niewiarowski S. Granulocyte-platelet interactions and platelet fibrinogen receptor exposure. Am. J. Physiol. 1988;255:H651–H658. doi: 10.1152/ajpheart.1988.255.3.H651. [DOI] [PubMed] [Google Scholar]
- 24.Sawyer D.B., Koeppe R.E., 2nd, Andersen O.S. Induction of conductance heterogeneity in gramicidin channels. Biochemistry. 1989;28:6571–6583. doi: 10.1021/bi00442a007. [DOI] [PubMed] [Google Scholar]
- 25.Lundbaek J.A., Collingwood S.A., Andersen O.S. Lipid bilayer regulation of membrane protein function: gramicidin channels as molecular force probes. J. R. Soc. Interface. 2010;7:373–395. doi: 10.1098/rsif.2009.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips R., Ursell T., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ly H.V., Block D.E., Longo M.L. Interfacial tension effect of ethanol on lipid bilayer rigidity, stability, and area/molecule: a micropipet aspiration approach. Langmuir. 2002;18:8988–8995. [Google Scholar]
- 28.Ly H.V., Longo M.L. The influence of short-chain alcohols on interfacial tension, mechanical properties, area/molecule, and permeability of fluid lipid bilayers. Biophys. J. 2004;87:1013–1033. doi: 10.1529/biophysj.103.034280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans E., Rawicz W., Hofmann A.F. Lipid bilayer expansion and mechanical disruption in solutions of water-soluble bile acid. Bile Acids Gastroenterol. Basic Clin. Adv. 1995;80:59–68. [Google Scholar]
- 30.Zhou Y., Raphael R.M. Effect of salicylate on the elasticity, bending stiffness, and strength of SOPC membranes. Biophys. J. 2005;89:1789–1801. doi: 10.1529/biophysj.104.054510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cantor R.S. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Theodoropoulou E., Marsh D. Effect of angiotensin II non-peptide AT1 antagonist losartan on phosphatidylethanolamine membranes. Biochim. Biophys. Acta Biomembr. 2000;1509:346–360. doi: 10.1016/s0005-2736(00)00317-5. [DOI] [PubMed] [Google Scholar]
- 33.Beaven A.H., Pastor R.W., Im W. Exploring protein-lipid interactions using gramicidin a as a model system. Biophys. J. 2013;104:432A. [Google Scholar]
- 34.Goforth R.L., Chi A.K., Andersen O.S. Hydrophobic coupling of lipid bilayer energetics to channel function. J. Gen. Physiol. 2003;121:477–493. doi: 10.1085/jgp.200308797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim T., Lee K.I., Im W. Influence of hydrophobic mismatch on structures and dynamics of gramicidin a and lipid bilayers. Biophys. J. 2012;102:1551–1560. doi: 10.1016/j.bpj.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen O.S., Bruno M.J., Koeppe R.E., 2nd Single-molecule methods for monitoring changes in bilayer elastic properties. Methods Mol. Biol. 2007;400:543–570. doi: 10.1007/978-1-59745-519-0_37. [DOI] [PubMed] [Google Scholar]
- 37.Ingólfsson H.I., Andersen O.S. Screening for small molecules’ bilayer-modifying potential using a gramicidin-based fluorescence assay. Assay Drug Dev. Technol. 2010;8:427–436. doi: 10.1089/adt.2009.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanford R.L., Al’Aref S.J., Andersen O.S. Statins alter lipid bilayer properties. Biophys. J. 2013;104:86A. [Google Scholar]
- 39.Mingeot-Leclercq M.P., Deleu M., Dufrêne Y.F. Atomic force microscopy of supported lipid bilayers. Nat. Protoc. 2008;3:1654–1659. doi: 10.1038/nprot.2008.149. [DOI] [PubMed] [Google Scholar]
- 40.Hutter J.L., Bechhoefer J. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 1993;64:1868–1873. [Google Scholar]
- 41.Picas L., Rico F., Scheuring S. Direct measurement of the mechanical properties of lipid phases in supported bilayers. Biophys. J. 2012;102:L01–L03. doi: 10.1016/j.bpj.2011.11.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rico F., Picas L., Scheuring S. The mechanics of membrane proteins is a signature of biological function. Soft Matter. 2013;9:7866–7873. [Google Scholar]
- 43.Dimitriadis E.K., Horkay F., Chadwick R.S. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 2002;82:2798–2810. doi: 10.1016/S0006-3495(02)75620-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picas L., Rico F., Scheuring S. Structural and mechanical heterogeneity of the erythrocyte membrane reveals hallmarks of membrane stability. ACS Nano. 2013;7:1054–1063. doi: 10.1021/nn303824j. [DOI] [PubMed] [Google Scholar]
- 45.Feigenson G.W. Phase behavior of lipid mixtures. Nat. Chem. Biol. 2006;2:560–563. doi: 10.1038/nchembio1106-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leonenko Z.V., Finot E., Cramb D.T. Investigation of temperature-induced phase transitions in DOPC and DPPC phospholipid bilayers using temperature-controlled scanning force microscopy. Biophys. J. 2004;86:3783–3793. doi: 10.1529/biophysj.103.036681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rawicz W., Olbrich K.C., Evans E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000;79:328–339. doi: 10.1016/S0006-3495(00)76295-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Das C., Sheikh K.H., Connell S.D. Nanoscale mechanical probing of supported lipid bilayers with atomic force microscopy. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2010;82:041920. doi: 10.1103/PhysRevE.82.041920. [DOI] [PubMed] [Google Scholar]
- 49.Stetter F.W.S., Hugel T. The nanomechanical properties of lipid membranes are significantly influenced by the presence of ethanol. Biophys. J. 2013;104:1049–1055. doi: 10.1016/j.bpj.2013.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steltenkamp S., Müller M.M., Janshoff A. Mechanical properties of pore-spanning lipid bilayers probed by atomic force microscopy. Biophys. J. 2006;91:217–226. doi: 10.1529/biophysj.106.081398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dieluweit S., Csiszár A., Merkel R. Mechanical properties of bare and protein-coated giant unilamellar phospholipid vesicles. A comparative study of micropipet aspiration and atomic force microscopy. Langmuir. 2010;26:11041–11049. doi: 10.1021/la1005242. [DOI] [PubMed] [Google Scholar]
- 52.Evans E., Rawicz W. Entropy-driven tension and bending elasticity in condensed-fluid membranes. Phys. Rev. Lett. 1990;64:2094–2097. doi: 10.1103/PhysRevLett.64.2094. [DOI] [PubMed] [Google Scholar]
- 53.Dimova R. Recent developments in the field of bending rigidity measurements on membranes. Adv. Colloid Interface Sci. 2014;208:225–234. doi: 10.1016/j.cis.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 54.Seghezza S., Dante S., Canale C. High resolution nanomechanical characterization of multi-domain model membranes by fast force volume. J. Mol. Recognit. 2015;28:742–750. doi: 10.1002/jmr.2490. [DOI] [PubMed] [Google Scholar]
- 55.Wallace E.J., Hooper N.M., Olmsted P.D. Effect of hydrophobic mismatch on phase behavior of lipid membranes. Biophys. J. 2006;90:4104–4118. doi: 10.1529/biophysj.105.062778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mouritsen O.G., Bloom M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984;46:141–153. doi: 10.1016/S0006-3495(84)84007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang C., Mason J.T. Structure and properties of mixed-chain phospholipid assemblies. Biochim. Biophys. Acta. 1986;864:423–470. doi: 10.1016/0304-4157(86)90005-5. [DOI] [PubMed] [Google Scholar]
- 58.Nielsen C., Goulian M., Andersen O.S. Energetics of inclusion-induced bilayer deformations. Biophys. J. 1998;74:1966–1983. doi: 10.1016/S0006-3495(98)77904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Frolov V.A.J., Chizmadzhev Y.A., Zimmerberg J. “Entropic traps” in the kinetics of phase separation in multicomponent membranes stabilize nanodomains. Biophys. J. 2006;91:189–205. doi: 10.1529/biophysj.105.068502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagle J.F., Tristram-Nagle S. Structure of lipid bilayers. Biochim. Biophys. Acta Biomembr. 2000;1469:159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schroeder R., London E., Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc. Natl. Acad. Sci. USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dietrich C., Bagatolli L.A., Gratton E. Lipid rafts reconstituted in model membranes. Biophys. J. 2001;80:1417–1428. doi: 10.1016/S0006-3495(01)76114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morandat S., El Kirat K. Membrane resistance to Triton X-100 explored by real-time atomic force microscopy. Langmuir. 2006;22:5786–5791. doi: 10.1021/la0604228. [DOI] [PubMed] [Google Scholar]
- 64.Domenech O., Francius G., Mingeot-Leclercq M.P. Interactions of oritavancin, a new lipoglycopeptide derived from vancomycin, with phospholipid bilayers: effect on membrane permeability and nanoscale lipid membrane organization. Biochim. Biophys. Acta Biomembr. 2009;1788:1832–1840. doi: 10.1016/j.bbamem.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 65.Mesquita R.M., Melo E., Vaz W.L.C. Partitioning of amphiphiles between coexisting ordered and disordered phases in two-phase lipid bilayer membranes. Biophys. J. 2000;78:3019–3025. doi: 10.1016/S0006-3495(00)76840-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baumgart T., Hunt G., Feigenson G.W. Fluorescence probe partitioning between Lo/Ld phases in lipid membranes. Biochim. Biophys. Acta. 2007;1768:2182–2194. doi: 10.1016/j.bbamem.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lundbaek J.A., Andersen O.S. Spring constants for channel-induced lipid bilayer deformations. Estimates using gramicidin channels. Biophys. J. 1999;76:889–895. doi: 10.1016/S0006-3495(99)77252-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lundbaek J.A., Koeppe R.E., Andersen O.S. Amphiphile regulation of ion channel function by changes in the bilayer spring constant. Proc. Natl. Acad. Sci. USA. 2010;107:15427–15430. doi: 10.1073/pnas.1007455107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rusinova R., Koeppe R.E., 2nd, Andersen O.S. A general mechanism for drug promiscuity: studies with amiodarone and other antiarrhythmics. J. Gen. Physiol. 2015;146:463–475. doi: 10.1085/jgp.201511470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wallace B.A., Ravikumar K. The gramicidin pore: crystal structure of a cesium complex. Science. 1988;241:182–187. doi: 10.1126/science.2455344. [DOI] [PubMed] [Google Scholar]
- 71.Khadka N.K., Cheng X., Pan J. Interactions of the anticancer drug tamoxifen with lipid membranes. Biophys. J. 2015;108:2492–2501. doi: 10.1016/j.bpj.2015.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bartlett P. Wiley-VCH; Weinheim, Germany: 2008. Bioelectrochemistry: Fundamentals, Experimental Technical and Applications. [Google Scholar]
- 73.Needham D., Nunn R.S. Elastic deformation and failure of lipid bilayer membranes containing cholesterol. Biophys. J. 1990;58:997–1009. doi: 10.1016/S0006-3495(90)82444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Björkhem-Bergman L., Lindh J.D., Bergman P. What is a relevant statin concentration in cell experiments claiming pleiotropic effects? Br. J. Clin. Pharmacol. 2011;72:164–165. doi: 10.1111/j.1365-2125.2011.03907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.