Abstract
Background
VAAM is an amino acid mixture that simulates the composition of Vespa larval saliva. VAAM enhanced physical endurance of mice and have been used by athletes as a supplementary drink before exercise. However, there is no information on the effect of VAAM on the physiology of freely moving animals. The purpose of this study was to obtain information about the VAAM-dependent regulation of liver and adipose tissue transcriptomes.
Results
Mice were orally fed a VAAM solution, an amino acid mixture mimicking casein hydrolysate (CAAM) or water under ad libitum feeding conditions for 5 days. Comparisons of the hepatic transcriptome between VAAM-, CAAM-, and water-treated groups revealed a VAAM-specific regulation of the metabolic pathway, i.e., the down-regulation of glycolysis and fatty acid oxidation and the up-regulation of polyunsaturated fatty acid synthesis and glucogenic amino acid utilization. Similar transcriptomic analyses of white and brown adipose tissues (WAT and BAT, respectively) indicated the up-regulation of phospholipid synthesis in WAT and the negative regulation of cellular processes in BAT. Because the coordinated regulation of tissue transcriptomes implied the presence of upstream signaling common to these tissues, we conducted an Ingenuity Pathways Analysis. This analysis showed that estrogenic and glucagon signals were activated in the liver and WAT and that beta-adrenergic signaling was activated in all three tissues.
Conclusions
We found that VAAM ingestion had an effect on multiple tissue transcriptomes of freely moving mice. Utilization of glycogenic amino acids may have been activated in the liver. Fatty acid conversion into phospholipid, not to triacylglycerol, may have been stimulated in adipocytes contrasting that a little effect was observed in BAT. Analysis of upstream factors revealed that multiple hormonal signals were activated in the liver, WAT, and BAT. Our data provide some clues to understanding the role of VAAM in metabolic regulation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12263-016-0534-2) contains supplementary material, which is available to authorized users.
Keywords: Gene expression, Amino acid mixture, Liver, DNA microarray, Adipose tissue
Background
The amino acid composition of food has a large impact on the metabolic homeostasis of individuals, especially when they are supplied as a major nutrient. As clearly understandable by the polyribosome model of protein synthesis, a shortage of the first-limiting amino acids readily affects the supply of proteins as the source of enzymes and other tissue-constructing materials. In this regard, essential amino acids (EAAs) are critical for the growth of the animal owing to their inability to synthesize EAAs from an organic backbone and nitrogen donor. In addition, many amino acids possess organic acid moieties that can serve as substrates for energy production (glucogenic and ketogenic amino acids).
In addition to these nutritive functions of amino acids, some dietary amino acids play roles as extracellular signals. Branched chain amino acids (BCAAs) constitute approximately 50 % of dietary EAAs and are known to activate protein anabolism via the mammalian target of rapamycin (mTOR)-dependent pathway [1, 2]. Glycine, glutamate, phenylalanine, histidine, and tryptophan act as or are metabolized to neurotransmitters and thus are able to exert a modulatory effect on the nervous system [3]. Portions of gastrointestinal epithelial cells express l-amino acid-sensing taste receptors and stimulate gastrointestinal hormone secretion in response to phenylalanine, leucine, glutamate, and tryptophan administration [4, 5]. These non-nutritive functions are attributable to a small number of amino acids but not to their whole constituent amino acids.
We have investigated the regulatory role of an amino acid mixture that simulates Vespa species (VAAM) on motor performance in animals at the dosing condition of sub-nutritional level (0.38 % of total food-derived energy/day). VAAM consists of 17 amino acids, among which glycine, tryptophan, and proline are present at an approximately twofold higher molar ratio than diet-derived amino acids (Additional file 1: Table S1). We first searched the optimum condition of VAAM administration to achieve significant enhancement of exercise performance [6]. In one study, a preceding oral administration of VAAM at the dose of 0.67 g/kg body weight improved the swimming performance of mice; this improvement was not observed with an amino acid mixture simulating the amino acid composition of bovine casein (CAAM) [7]. In addition, VAAM can lower serum lactate levels and activate fatty acid release into mice sera and into the media of cultured rat adipocytes [6, 8]. VAAM solution, as a sport supplement drink, has been in product line since 1995 and consumed by many athletes. In the human studies, the preceding administration of VAAM at the dose of one eighth of our animal studies significantly lowered the respiratory quotient during the 60 min of cycling exercise [9], and the 12 weeks of VAAM administration at the same dose as above increased maximal oxygen uptake in the older women groups [10]. These results suggest that ingested VAAM may affect the metabolic state of multiple tissues, such as liver and adipose tissues, to facilitate energy supply to the locomotor system. However, there is no information on the effect of VAAM on the physiology of resting or freely moving animals. Here we present data indicating this type of regulation in multiple tissues at the gene transcriptional level.
Results
Characterization of genes regulated by VAAM administration in murine liver
Mice under freely feeding and freely moving conditions were treated with VAAM, CAAM, or water once per day for 5 days and euthanized at 4 h after the last administration (Additional file 2: Figure S1). The energy contribution of VAAM and CAAM was 0.38 % of total food-derived energy/day (Additional file 1: Table S1). The biochemical analysis of mice sera showed no difference in terms of carbohydrates and lipids among the groups (Additional file 3: Table S2). We conducted DNA microarray analysis to investigate the physiological effect of VAAM administration. The differentially expressed genes (DEGs) were calculated by the statistic comparison between the water group and VAAM group or between the water group and CAAM group (see “Methods”). These DEGs were then set-theoretically subtracted or merged to extract DEGs specifically regulated by VAAM administration (Fig. 1). To elucidate the function of genes regulated by VAAM specifically, we applied GO analysis to the subset of the extracted genes. The group specifically up-regulated by VAAM administration was composed of 446 genes, 217 down-regulated genes, and 45 plus 15 genes inversely correlated among VAAM and CAAM, as shown in a Venn diagram (Fig. 1a, b). The GO terms attributed to these probe sets clearly indicate that the genes related to lipid metabolism are significantly enriched in these subsets (Fig. 2). It is also notable that 45 + 15 genes, which were oppositely regulated by VAAM and CAAM, respectively (Fig. 1b), include eight genes (cytochrome P450, family 7, subfamily b, polypeptide 1 (Cyp7b1), hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 5 (Hsd3b5), phosphatidylcholine transfer protein (Pctp), sterol regulatory element binding transcription factor 1 (Srebf1), aldehyde dehydrogenase family 3, subfamily A2 (Aldh3a2), fatty acid binding protein 7, brain (Fabp7), 3-hydroxy-3-methylglutaryl-Coenzyme A synthase 1 (Hmgcs1), and retinol dehydrogenase 9 (Rdh9)) related to lipid metabolism; three genes (serine hydroxymethyltransferase 1 (soluble) (Shmt), solute carrier family 46, member 1 (Slc46a1), and thiopurine methyltransferase (Tpmt)) related to one-carbon metabolism; and one gene (pyruvate kinase liver and red blood cell (Pklr)) related to glycolysis (Additional file 4: Table S3).
Fig. 1.
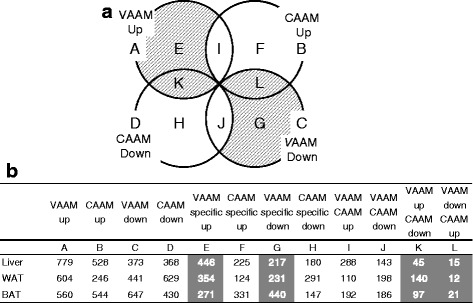
Effects of amino acid mixtures on tissue transcriptomes. a Number of genes regulated by VAAM and/or CAAM. VAAM Up or CAAM Up: up-regulated compared with the water group; VAAM Down or CAAM Down: down-regulated compared with the water group. b Number of regulated genes in the liver, WAT, and BAT
Fig. 2.
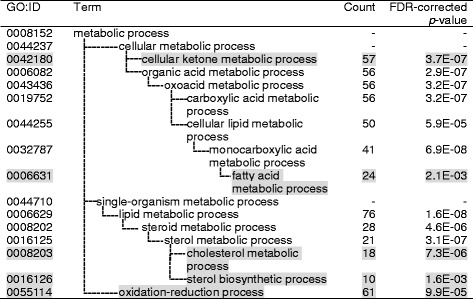
Gene ontology analysis of liver genes for which expression levels were affected by VAAM administration. 446 + 45 + 15 + 217 genes in Fig. 1b were used for analysis. GO terms located at the bottom of hierarchy are represented by shaded text
Regulation of liver metabolism by VAAM
Mapping of the enriched genes to known metabolic pathways revealed a VAAM-specific regulation of lipid and amino acid metabolism, i.e., a down-regulation of beta- and omega-oxidation (Fig. 3a), the accumulation of middle chain fatty acid (Fig. 3b), the utilization of cholesterol stock for bile acid synthesis (Fig. 3c), and the utilization of glucogenic amino acids in the TCA cycle (Additional file 5: Table S4) (Fig. 3d). In particular, the oppositely regulated genes highlight the VAAM-specific effect on metabolic pathways; e.g., Hmgcs1 and Cyp7b1 in bile acid synthesis (Fig. 3a) and Pklr and Shmt in glucogenic amino acid utilization (Fig. 3d). We confirmed expression levels of Elovl3 (elongation of very long chain fatty acids like 3), Pecr (peroxisomal trans-2-enoyl-CoA reductase), and G6pc (glucose-6-phosphatase, catalytic subunit) by quantitative PCR and found that Elovl3 and Pecr were down-regulated in the VAAM group, while G6pc was up-regulated in the VAAM group as observed in DNA microarray analysis (Additional file 6: Figure S2).
Fig. 3.
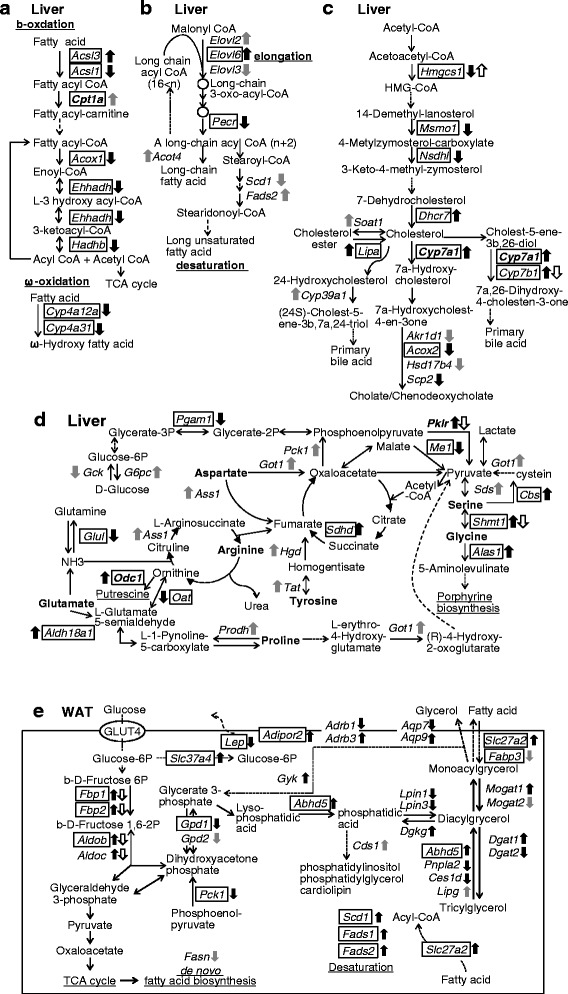
Metabolic function of VAAM-regulated genes in the liver and WAT. Gene symbols and their mode of regulation by VAAM (black arrows), by CAAM (white arrows), and by VAAM and CAAM (gray arrows) are represented in fatty acid oxidation (a), in fatty acid desaturation (b), in cholesterol metabolism (c), and in glycolysis, glucogenic amino acid metabolism, and the TCA cycle (d) in the liver. e Glucose and triacylglycerol metabolism of WAT. All gene symbols and their full names are listed in Additional file 5: Table S4
Response of adipose tissue transcriptomes to VAAM
Next, we applied a similar transcriptomic analysis to white and brown adipose tissues (WAT and BAT). In the case of WAT, the GO terms attributed to the VAAM-responsive genes (354 + 140 + 12 + 231 genes, Fig. 1a, b) were “calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules” and “monocarboxylic acid metabolism process” (Fig. 4). Approximately, one half of the genes enriched in “monocarboxylic acid metabolism process” were related to lipid synthesis (17 out of 30, Additional file 7: Table S5). Mapping these genes in metabolic pathways revealed that VAAM up-regulates several genes for phospholipid synthesis from glucose and fatty acid (Fig. 3e). Four genes for enzymes of acylglycerol synthesis were oppositely regulated: up-regulated for monoacylglycerol O-acyltransferase 1 (Mogat1) and diacylglycerol O-acyltransferase 1 (Dgat1), and down-regulated for monoacylglycerol O-acyltransferase 2 (Mogat2) and diacylglycerol O-acyltransferase 2 (Dgat2). It is also notable that solute carrier family 37 (glucose-6-phosphate transporter), member 4 (Slc37a4), which facilitates glucose-6-phosphatase activity, is up-regulated.
Fig. 4.
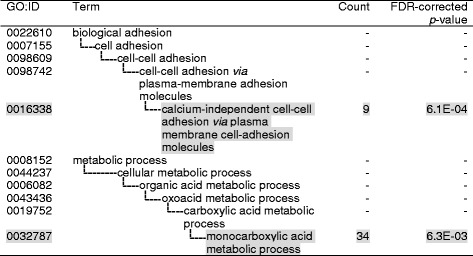
Gene ontology analysis of WAT genes for which expression levels were affected by VAAM administration. 354 + 140 + 12 + 231 genes in Fig. 1b were used for analysis. GO terms located at the bottom of hierarchy are represented by shaded text
GO analysis of VAAM-responsive genes in BAT (271 + 97 + 21 + 440 genes, Fig. 1a, b) resulted in the identification of the term “negative regulation of biological process” located at the bottom of the hierarchy (Fig. 5). These terms contain a smaller number of metabolic enzyme genes than is the case for liver and WAT, but those related to cellular signaling such as adiponectin receptor 2 (Adipor2), adenosine A1 receptor (Adora1), cyclin-dependent kinase inhibitor 1 (Cdkn1), glycogen synthase kinase 3b (Gsk3b), protein tyrosine phosphatase, non-receptor type (Ptpn), forkhead box O1 (Foxo1), insulin-like growth factor 1 (Igf1), lymphocyte protein tyrosine kinase (Lck), mitogen-activated protein kinase (Mapk), and regulator of G-protein signaling (Rgs) (Additional file 8: Table S6). This finding prompted us to search for an unknown upstream factor that may regulate the adipose tissue transcriptomes in relation to that of the liver.
Fig. 5.

Gene ontology analysis of BAT genes for which expression levels were affected by VAAM administration. 271 + 97 + 21 + 440 genes in Fig. 1b were used for analysis. GO terms located at the bottom of hierarchy are represented by shaded text
Search for upstream regulators common among liver and adipose tissues
We searched for Ingenuity Pathway Analysis (IPA) upstream regulators of VAAM-regulated genes starting with liver genes (446 + 217 genes), followed by comparison with the WAT genes (354 + 231 genes) and the BAT genes (271 + 440 genes), respectively. The liver transcriptome exhibited a total of 103 activated and 32 inhibited upstream regulators (absolute Z-scores >2.0, Table 1). Because some of these regulators have close functional relationships to each other, they were classified into the same columns (e.g., “isoproterenol”, “norepinephrine” as a beta-agonist, and “propranolol” as a beta-antagonist). Interestingly, there are obvious inter-tissue correlations of selected regulators, including “norepinephrine” (2.06 in WAT and 3.05 in BAT) and “progesterone” (Z-score of 2.50 in liver and 2.12 in WAT). Considering the functional similarity of these selected regulators, it is very possible that beta-adrenergic and TGF-related signals may be activated in these three tissues. It is also noteworthy that estrogen, glucagon, and gentamicin signals seemed to be activated in the liver and WAT and that I-kappa B, interleukins 5 and 6, insulin, PDGF, and the other signals were activated in BAT and WAT (Table 1). There was a discrepancy in the Z-score between “INS”, at −2.18 in the liver, and “Ins1”, at 2.23 in BAT. The mouse genome contains two genes for insulin (Ins1 and Ins2), each of which corresponds to “Ins1” and “INS” in the IPA database, respectively. It is possible that this discrepancy is due to functional differences between the Ins1 and Ins2 genes.
Table 1.
Correlation of IPA upstream regulators between the liver, WAT, and BAT transcriptomes
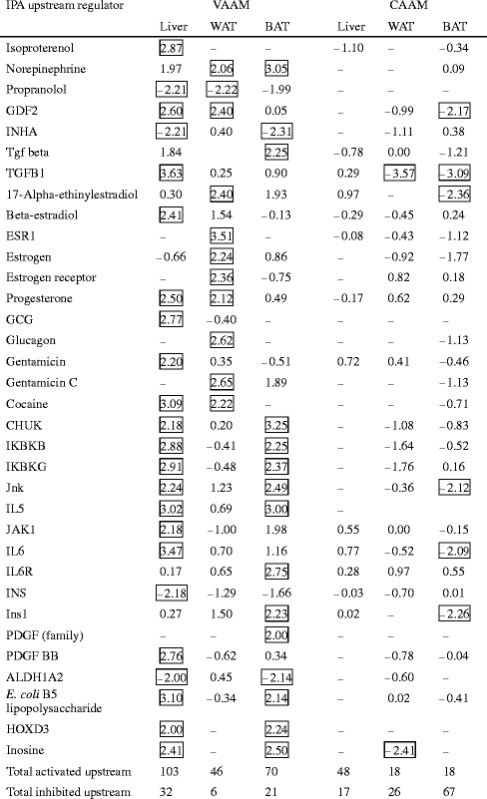
The values show the activation Z-score. The absolute Z-scores ≥2 are in squares
ALDH1A2 aldehyde dehydrogenase 1 family, member A2, CHUK conserved helix-loop-helix ubiquitous kinase, ESR1 estrogen receptor 1, GCG glucagon, GDF2 growth differentiation factor 2, HOXD3 homeobox D3, IKBKB inhibitor of kappa light polypeptide gene enhancer in B cells, kinase beta, IKBKG inhibitor of kappa light polypeptide gene enhancer in B cells, kinase gamma, IL5 interleukin 5, IL6 interleukin 6, IL6R interleukin 6 receptor, INHA inhibin, alpha, INS insulin, Ins1 insulin I, JAK1 Janus kinase 1, PDGF platelet-derived growth factor, TGFB1 transforming growth factor, beta 1
Discussion
In this study, we examined the non-nutritive effect of a specific amino acid mixture, VAAM, on the liver, WAT, and BAT transcriptomes in mice under freely moving conditions. From the gene ontology (GO) analysis of VAAM-regulated genes, it was predicted that VAAM may up-regulate sugar and lipid anabolism in these tissues. In addition, a comparative analysis of IPA upstream factors among these tissues suggested that estrogenic and glucagon signals were activated in the liver and WAT and that beta-adrenergic signaling was activated in the three tissues.
Our previous studies [6, 7] on VAAM were conducted using a mouse model under forced exercise conditions, where the glycogen storage might have been exhausted and the muscle might have continued to move with the help of a fatty acid supply from the adipose tissues. The major difference between the previous studies and the present study resides in whether or not moving conditions were present (forced swimming or free motion) and in the dosing condition (one shot or five shots). However, there are some correlations in the metabolic regulation among these studies. We observed a down-regulation of beta-oxidation and an up-regulation of glucogenic amino acid utilization in the liver (Fig. 3). The former may save the fatty acid pool in total and the latter makes it possible to supply amino acid-derived substrates to gluconeogenesis in response to exercise stimulus [11–13]. In the case of WAT, four enzyme genes for acylglycerol synthesis were regulated in a distinct manner: Mogat1 and Dgat1 were up-regulated, and Mogat2 and Dgat2 were down-regulated. Especially, the opposite regulation of Dgat1 (up) and 2 (down) should be mentioned because Dgat1 facilitates fatty acid/acylglycerol shuttling, and Dgat2 is responsible for triacylglycerol accumulation [14]. This regulation may support fatty acid release to the serum. It is also possible that the up-regulation of Slc37a4 may decrease lactate production by facilitating glucose regeneration from glucose-6-phosphate (Fig. 3e). The role of BAT in exercise performance is not clear because our experiments were performed at a moderate temperature, where no additional thermogenesis is required. However, it should be noted that the two catecholamine responsive genes, deiodinase, iodothyronine, type II (Dio2) and peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (Ppargc1a, also known as Pgc1a), are up-regulated. These genes are reported to be induced in mice and rats exposed to cold temperature [15, 16]. In the moderate temperature condition as those we adopted, this kind of regulation may result in an increased basal metabolic rate under resting conditions [12, 17, 18]. In conclusion, the VAAM-dependent regulation of multiple tissue transcriptomes seems to be a tuning mechanism that makes the metabolic state resistant to more intense exercise. It would be valuable to examine the effect of middle term (approximately 1 week) pre-administration of VAAM on exercise performance in a human interactive study.
Whether VAAM acts on these tissues directly or indirectly is an open question. From the IPA upstream factor analysis, it was predicted that the three endocrine factors adrenaline, estrogen, and glucagon seemed to stimulate these tissues. In particular, adrenaline is the most plausible factor involved in VAAM signaling because several IPA terms related to adrenergic signals were detected in the three tissues (Table 1) and its increase in the mice sera was detected in a previous study [7]. Considering that a major source of adrenaline is the hypothalamus-sympathetic nerve-adrenal medullary (HSA) axis, we think it is highly possible that VAAM controls terminal tissues via this system. This assumption raises another question as to what type of mechanism transduces VAAM signals in the HSA axis. There are two supporting information. First is that VAAM’s effect depends on the overall ratio of amino acids (Additional file 1: Table S1), and second, the maximal effect of VAAM was obtained with the pretreatment of animals 30 min before the exercise. These considerations suggest that the sensing mechanism for VAAM might have a broad spectrum to amino acids and that the mechanism may reside in the gastrointestinal tract that faces the VAAM solution but not in metabolizing tissues nor locomotor systems. Accordingly, we assume that gastrointestinal chemosensory cells are one of the candidate targets of VAAM stimulus. Further investigation is needed to elucidate the VAAM-sensing and VAAM-transducing machinery.
Conclusions
We found that VAAM ingestion had an effect on multiple tissue transcriptomes of freely moving mice. Utilization of glucogenic amino acids may have been activated in the liver. Fatty acid conversion into phospholipid, not to triacylglycerol, may have been stimulated in adipocytes contrasting that a little effect was observed in BAT. Analysis of upstream factors revealed that multiple hormonal signals were activated in the liver, WAT, and BAT. Our data provide some clues to understanding the role of VAAM in metabolic regulation.
Methods
Animals
Five-week-old male ddY mice (SLC Japan, Shizuoka, Japan) were housed in plastic cages with food (MF, Oriental yeast, Tokyo, Japan) and deionized water ad libitum in a temperature- and humidity-controlled room with a 12-h light/dark cycle (light 08:00–20:00; dark 20:00–08:00). The mice were weighed every day. All animal experimental protocols were approved by the Animal Use Committee of the Faculty of Agriculture at the University of Tokyo with approval number: P12-669.
Amino acids
Glycine and all l-amino acids were purchased from Wako Pure Chemical Industries (Osaka, Japan). The amino acid mixtures VAAM and CAAM were prepared in the ratios described by Abe et al. [6] and were analyzed for amino acid composition using a Hitachi l-8500A amino acid analyzer (Hitachi High Technologies, Tokyo) (Additional file 1: Table S1). The concentrations of VAAM and CAAM were each adjusted to 1.8 %.
Experimental procedure
The experimental schedule is shown in Additional file 2: Figure S1. The mice (n = 21) were acclimated to a laboratory environment for 1 week. They were divided randomly into three treatment groups: VAAM, CAAM, or water. Using a feeding tube, 1.8 % VAAM, 1.8 % CAAM, or water was orally administered five times once a day at 10:00AM. The dosage of VAAM, CAAM, or water was adjusted to 37.5 μL per gram of body weight as described previously [6]. On the day of last administration, the food was removed and the mice were moved to clean cages at 8:00. The last treatments began at 10:00. At 4 h after the last administration, the mice were euthanized by cervical dislocation and the blood, liver, WAT, and BAT were collected. The blood was separated to obtain the serum. Small hepatic pieces were immersed into RNAlater (Qiagen, Tokyo, Japan). The WAT and BAT were immediately frozen after extraction using liquid nitrogen. All samples were maintained at −80 °C until use. Two mice in CAAM group were excluded because of their morphological abnormality in the liver. Subsequently, the mice number of each group became n = 7 for the VAAM group, n = 5 for the CAAM group, and n = 7 for the water group.
Measurement of serum biochemical parameters
Serum glucose, non-esterified fatty acid (NEFA), and triacylglycerol were enzymatically assayed using the glucose C II test Wako, the NEFA C test Wako, and the triglyceride E test Wako (Wako Pure Chemical Industries, Osaka, Japan), respectively. Total lipids, total ketone bodies, total cholesterol, HDL cholesterol and LDL cholesterol were assayed on Nagahama Life Science (Shiga, Japan). Each of the values is represented as the mean ± standard error of the mean (SEM). Significant differences (p ≤ 0.05) between the experimental groups were assessed by Tukey-Kramer comparison.
DNA microarray assay
Total RNA was isolated from each hepatic, WAT, and BAT sample by TRIzol reagent (Invitrogen Japan, Tokyo, Japan) and purified using an RNeasy mini kit (Qiagen, Tokyo, Japan). The RNA Integrity Number (RIN) was estimated as an index of the quality of the total RNA using an Agilent 2100 Bioanalyzer (Agilent Technologies Japan, Tokyo, Japan). The values for RIN were over 8.3.
Then, amplified RNA (aRNA) was synthesized from 200 ng of purified total RNA, and biotinylated aRNA was obtained using a GeneChip® 3′IVT Express Kit (Affymetrix, Santa Clara, CA, USA). The aRNA was fragmented and hybridized to a GeneChip® Mouse Genome 430 2.0 Array (Affymetrix) for 16 h at 45 °C. The arrays were washed and stained with phycoerythrin using the GeneChip® Fluidics Station 450 (Affymetrix). The arrays were submitted to scanning on an Affymetrix GeneChip® Scanner 3000 7G (Affymetrix). The Affymetrix® GeneChip® Command Console® Software (Affymetrix) was used to make CEL files.
DNA microarray data analysis
The CEL files were quantified by the distribution free weighted method (DFW) [19] using the statistical language R (2.7.1) (http://www.r-project.org/) [20] and Bioconductor (2.2) (http://www.bioconductor.org/) [21]. Hierarchical clustering was performed using the pvclust() function in R [22]. The rank products (RP) method was used to identify differentially expressed gene probe sets of the DFW-quantified data [23]. The probe sets with a false discovery rate (FDR) <0.05 were considered to be differentially expressed between two groups. RP was performed for a comparison of the water treatment group and the VAAM or CAAM treatment groups.
Gene ontology analysis
The up- and down-regulated probe sets picked out at FDR <0.05 were functionally classified by the biological process in gene ontology (GO) with the Functional Annotation Tool of the Database for Annotation, Visualization, and Integrated Discovery (DAVID) [24, 25] and Quick GO (http://www.ebi.ac.uk/QuickGO/) [26]. EASE scores, which are modified Fisher’s exact test p values, were used to extract statistically overrepresented GO terms. GO terms with FDR-corrected p values <0.01 were regarded as significantly enriched.
Pathway analysis
The differentially expressed genes were mapped on metabolic pathways in reference to the Kyoto Encyclopedia of Genes and Genomes (www.genome.ad.jp/kegg) [27].
Upstream regulator analysis
Predicted upstream regulators from VAAM- and CAAM-specific data sets were analyzed using QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). The activation Z-score was calculated as a measure of upstream regulator analysis. An absolute Z-score ≥2 was judged as significantly activated or inhibited, while an absolute Z-score ≤1.0 could not judge a direction. Common upstream regulators that were predicted to be activated or inhibited in the liver, WAT, and BAT were selected from the all upstream regulators list.
Quantitative PCR analysis
Total RNA was prepared as described in “DNA microarray assay” section. Complementary DNA (cDNA) was synthesized with PrimeScript RT reagent kit (TaKaRa, Shiga, Japan) and subjected to PCR analysis using the following primer sets and SYBR Premix Ex Taq II kit (TaKaRa). The cDNA was amplified by 30 s at 95 °C followed by 40 cycles of incubation at 95 °C for 15 s and at 60 °C for 30 s in CFX96 real-time PCR detection system (Bio-Rad). The primers sets were as follows: Elovl3 forward; 5′-TTCTCACGCGGGTTAAAAATGG-3′, Elovl3 reverse; 5′- GAGCAACAGATAGACGACCAC-3′, G6pc forward; 5′-TTCAAGTGGATTCTGTTTGG-3′, G6pc reverse; 5′- AGATAGCAAGAGTAGAAGTGAC-3′, Pecr forward; 5′-GAAGGGATGGCATGCTGTGA-3′, Pecr reverse; 5′- TTGACAATCGACCCTCCGTG-3′, beta-actin forward; 5′-GATGTATGAAGGCTTTGGTC-3′, beta-actin reverse; 5′- TGTGCACTTTTATTGGTCTC-3′. Each cycle threshold (CT) value of an amplified fragment was compared with that of beta-actin and represented as a relative expression level. Values for VAAM or CAAM treatment group and water treatment group were examined for their significant difference using Dunnett’s test.
Abbreviations
BAT, brown adipose tissue; CAAM, casein amino acid mixture; EAAs, essential amino acids; NEFA, non-esterified fatty acid; VAAM, Vespa amino acid mixture; WAT, white adipose tissue
Acknowledgements
We thank Hiromasa Morishita for technical assistance in amino acid analysis. A part of this study was supported by KYOWA HAKKO BIO CO., LTD. This work was supported by the Council for Science, Technology and Innovation (CSTI), Cross-ministerial Strategic Innovation Promotion Program (SIP), “Technologies for creating next-generation agriculture, forestry and fisheries”, and JSPS KAKENHI Grant number 26350177.
Availability of data and materials
The data sets supporting the results of this article are available in the National Center of Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession number GSE 73266.
Authors’ contributions
FS, TA, AK, and KA conceived and designed the experiment. KKu provided the technique and materials for VAAM preparation. FS, TA, AK, YW, and KS performed animal experiment. FS, AK, AY, KKo, SA, and TK analyzed the data. FS, AY, TK, and KA wrote the paper. All authors read and approved the final manuscript.
Competing interests
Author Kumagai K is an employee of KYOWA HAKKO BIO CO., LTD. He provided the technique and materials for VAAM preparation, but the company had no control over the interpretation, the writing, or the publication of this work. Other authors declare that they have no competing interests.
Ethics approval and consent to participate
All animal experimental protocols were approved by the Animal Use Committee of the Faculty of Agriculture at the University of Tokyo with approval number: P12-669.
Additional files
Comparison of amino acid mixtures (Mol%). The amino acid mixtures VAAM and CAAM were analyzed for amino acid composition using an amino acid analyzer. (DOCX 36 kb)
Feeding schedule of amino acid mixtures. VAAM, CAAM or water was orally administered five times once a day using a feeding tube. Daily administration was performed at 10:00. On the day of last administration, the food was removed and the mice were moved to clean cages at 8:00. The last treatments began at 10:00. At 4 hours after the last administration, the mice were euthanized by cervical dislocation and blood, liver, WAT and BAT were collected. (PPTX 53 kb)
Serum biomedical indices of mice administrated amino acid mixtures. The values are represented as the mean ± standard error of the mean (SEM). (DOCX 26 kb)
The list of liver genes showing significant enrichment in the GO-terms located at the bottom of the hierarchy. VAAM-specific genes mapped on Figure 3 were represented by bold style. (XLSX 11 kb)
Genes mapped on Fig. 3. (XLSX 13 kb)
The relative hepatic mRNA expression levels of Elovl3, G6pc, and Pecr in VAAM, CAAM or Water group. Quantitative PCR analysis were performed to validate the expression of metabolic genes, Elovl3 (elongation of very long chain fatty acids like 3), Pecr (peroxisomal trans-2-enoyl-CoA reductase) and G6pc (glucose-6-phosphatase, catalytic subunit) in Water and VAAM or CAAM groups. *: p ≤ 0.05 by Dunnett’s test. (PPTX 69 kb)
The list of WAT genes showing significant enrichment in the GO-terms located at the bottom of the hierarchy. VAAM-specific genes mapped on Figure 3 were represented by bold style. (XLSX 11 kb)
The list of BAT genes showing significant enrichment in the GO-terms located at the bottom of the hierarchy. The list of BAT genes showing significant enrichment in the GO-terms located at the bottom of the hierarchy. (XLSX 12 kb)
References
- 1.Wu G. Functional amino acids in growth, reproduction, and health. Adv Nutr. 2010;1:31–7. doi: 10.3945/an.110.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattick JSA, Kamisoglu K, Ierapetritoua MG, Androulakisa IP, Berthiaume F. Branched chain amino acid supplementation impact on signaling and relevance to critical illness. Wiley Interdiscip Rev Syst Biol Med. 2013;5:449–60. doi: 10.1002/wsbm.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu G. Amino acids metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- 4.Wang JH, Inoue T, Higashiyama M, Guth PH, Engel E, Kaunitz JD, Akiba Y. Umami receptor activation increases duodenal bicarbonate secretion via glucagon-like peptide-2 release in rats. J Pharmacol Exp Ther. 2011;339:464–73. doi: 10.1124/jpet.111.184788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shirazi-Beechey SP, Daly K, Al-Rammahi M, Moran AW, Bravo D. Role of nutrient-sensing taste 1 receptor (T1R) family members in gastrointestinal chemosensing. Br J Nutr. 2014;111:S8–15. doi: 10.1017/S0007114513002286. [DOI] [PubMed] [Google Scholar]
- 6.Abe T, Takiguchi Y, Tamura M, Shimura J, Yamazaki K. Effects of Vespa amino acid mixture isolated from hornet larval saliva exercise in swimming mice of blood lactate and glucose and modified VAAM nutrients on endurance. Jpn J Phys Fitness Sports Med. 1995;44:225–38. doi: 10.7600/jspfsm1949.44.225. [DOI] [Google Scholar]
- 7.Abe T, Inamori M, Iida K, Tamura M, Takiguchi Y, Yasuda K. The activation of fatty acid metabolism by VAAM and related nutrients during endurance exercise in mice. Adv Exerc sports physiol. 1997;13:35–44. [Google Scholar]
- 8.Shinozaki F, Abe T. Synergistic effect of Vespa amino acid mixture on lipolysis in rat adipocytes. Biosci Biotechnol Biochem. 2008;72:1860–8. doi: 10.1271/bbb.80119. [DOI] [PubMed] [Google Scholar]
- 9.Demura S, Nagasawa Y, Kitabayashi T, Matsuzawa J. Effect of amino acid mixture intake on physiological response and rating of perceived exertion during cycling exercise. Percept Mot Skills. 2003;96:883–95. doi: 10.2466/pms.2003.96.3.883. [DOI] [PubMed] [Google Scholar]
- 10.Sasai H, Matsuo T, Fujita M, Saito M, Tanaka K. Effects of regular exercise combined with ingestion of vespa amino acid mixture on aerobic fitness and cardiovascular diseases risk factors in sedentary older woman: A preliminary study. Geriatr Gerontol Int. 2011;11:24–31. doi: 10.1111/j.1447-0594.2010.00630.x. [DOI] [PubMed] [Google Scholar]
- 11.Emhoff CA, Messonnier LA, Horning MA, Fattor JA, Carlson TJ, Brooks GA. Gluconeogenesis and hepatic glycogenolysis during exercise at the lactate threshold. J Appl Physiol (1985) 2013;114:297–306. doi: 10.1152/japplphysiol.01202.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haase TN, Ringholm S, Leick L, Biensø RS, Kiilerich K, Johansen S, Nielsen MM, Wojtaszewski JF, Hidalgo J, Pedersen PA, Pilegaard H. Role of PGC-1a in exercise and fasting-induced adaptations in mouse liver. Am J Physiol Regul Integr Comp Physiol. 2011;301:R1501–9. doi: 10.1152/ajpregu.00775.2010. [DOI] [PubMed] [Google Scholar]
- 13.Suh SH, Paik IY, Jacobs K. Regulation of blood glucose homeostasis during prolonged exercise. Mol Cells. 2007;23:272–9. [PubMed] [Google Scholar]
- 14.Yen CL, Stone SJ, Koliwad S, Harris C, Farese RV., Jr Thematic review series: glycerolipids. DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe M, Yamamoto T, Kakuhata R, Okada N, Kajimoto K, Yamazaki N, Kataoka M, Baba Y, Tamaki T, Shinohara Y. Synchronized changes in transcript levels of genes activating cold exposure-induced thermogenesis in brown adipose tissue of experimental animals. Biochim Biophys Acta. 2008;1777:104–12. doi: 10.1016/j.bbabio.2007.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Hondares E, Rosell M, Díaz-Delfín J, Olmos Y, Monsalve M, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor (PPARα) induces PPARγ coactivator 1 (PGC-1α) gene expression and contributes to thermogenic activation of brown fat: involvement of PRDM16. J Biol Chem. 2011;286:43112–22. doi: 10.1074/jbc.M111.252775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall JA, Ribich S, Christoffolete MA, Simovic G, Correa-Medina M, Patti ME, Bianco AC. Absence of thyroid hormone activation during development underlies a permanent defect in adaptive thermogenesis. Endocrinology. 2010;151:4573–82. doi: 10.1210/en.2010-0511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Betz MJ, Enerbäck S. Therapeutic prospects of metabolically active brown adipose tissue in humans. Front Endocrinol (Lausanne) 2011;29:86. doi: 10.3389/fendo.2011.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Z, McGee M, Liu Q, Scheuermann RH. A distribution free summarization method for Affymetrix GeneChip arrays. Bioinformatics. 2007;23:321–7. doi: 10.1093/bioinformatics/btl609. [DOI] [PubMed] [Google Scholar]
- 20.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2006. http://www.R-project.org.
- 21.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–2. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 23.Breitling R, Armengaud P, Amtmann A, Herzyk P. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 2004;573:83–92. doi: 10.1016/j.febslet.2004.07.055. [DOI] [PubMed] [Google Scholar]
- 24.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 25.Huang DW, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Current Protocols in Bioinformatics. New York: John Wiley & Sons, Inc; 2009. Extracting biological meaning from large gene lists with DAVID. [DOI] [PubMed] [Google Scholar]
- 26.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.KEGG. Kyoto Encyclopedia of Genes and Genomes. [www.genome.ad.jp/kegg]. Accessed 2 Oct 2015.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data sets supporting the results of this article are available in the National Center of Biotechnology Information (NCBI) Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through accession number GSE 73266.


