ABSTRACT
Pseudomonas aeruginosa is an important, thriving member of microbial communities of microbial bioelectrochemical systems (BES) through the production of versatile phenazine redox mediators. Pure culture experiments with a model strain revealed synergistic interactions of P. aeruginosa with fermenting microorganisms whereby the synergism was mediated through the shared fermentation product 2,3-butanediol. Our work here shows that the behavior and efficiency of P. aeruginosa in mediated current production is strongly dependent on the strain of P. aeruginosa. We compared levels of phenazine production by the previously investigated model strain P. aeruginosa PA14, the alternative model strain P. aeruginosa PAO1, and the BES isolate Pseudomonas sp. strain KRP1 with glucose and the fermentation products 2,3-butanediol and ethanol as carbon substrates. We found significant differences in substrate-dependent phenazine production and resulting anodic current generation for the three strains, with the BES isolate KRP1 being overall the best current producer and showing the highest electrochemical activity with glucose as a substrate (19 μA cm−2 with ∼150 μg ml−1 phenazine carboxylic acid as a redox mediator). Surprisingly, P. aeruginosa PAO1 showed very low phenazine production and electrochemical activity under all tested conditions.
IMPORTANCE Microbial fuel cells and other microbial bioelectrochemical systems hold great promise for environmental technologies such as wastewater treatment and bioremediation. While there is much emphasis on the development of materials and devices to realize such systems, the investigation and a deeper understanding of the underlying microbiology and ecology are lagging behind. Physiological investigations focus on microorganisms exhibiting direct electron transfer in pure culture systems. Meanwhile, mediated electron transfer with natural redox compounds produced by, for example, Pseudomonas aeruginosa might enable an entire microbial community to access a solid electrode as an alternative electron acceptor. To better understand the ecological relationships between mediator producers and mediator utilizers, we here present a comparison of the phenazine-dependent electroactivities of three Pseudomonas strains. This work forms the foundation for more complex coculture investigations of mediated electron transfer in microbial fuel cells.
INTRODUCTION
Bioelectrochemical systems (BES), including their most important variant, the microbial fuel cell (MFC), are rapidly developing and promising technologies for renewable energy production and wastewater treatment, among other applications (1, 2). The MFC technology aims at generating electrical current through extracellular transfer of electrons, which microorganisms liberate from organic substrates. Microorganisms oxidize organic compounds, and the electrons from the intracellular electron transport chains are transferred to an external electron acceptor (i.e., an anode poised at a suitable potential) (3). One of the challenges facing MFC performance is the efficiency of microbial electron transfer to an anode. The most commonly described transfer mechanisms are direct electron transfer via direct cell contact or protein nanowires and mediated electron transfer via secondary or primary metabolites (4–9). Attempts to improve the biological efficiency of MFCs have therefore focused on understanding and improving these mechanisms.
In mediated electron transfer, microorganisms utilize endogenous or exogenous soluble redox mediators that enable transmission of electrons to an external electron acceptor. In bacteria, endogenous secondary metabolites used as mediators include riboflavins in Shewanella (6), phenazines in Pseudomonas (10), and quinones in Lactococcus (11). These molecules undergo reversible oxidation and reduction and hence can be used repeatedly as electron shuttles (4). Also, the addition of natural or synthetic redox compounds to enhance electron transfer in BES has demonstrated some potential (12), and very recent work shows that the heterologous expression of natural redox mediators can enable nonelectroactive bacteria for electrode interactions (13).
Phenazines play several important roles in the physiology of Pseudomonas aeruginosa: virulence generation (14), signaling (15), iron acquisition (16), and shuttling of electrons (17, 18). Phenazines are synthesized from chorismic acid (19, 20) via the intermediate phenazine-1-carboxylic acid (PCA), which is further converted into pyocyanin (PYO), 1-hydroxyphenazine (1-HP), and phenazine-carboxyamide (PCN) (21). Since the different phenazines have different redox properties and reactivities with oxygen, they may play different roles in a P. aeruginosa biofilm, and subsequently gradients of their production exist across the biofilm structures (22).
In mixed microbial communities and biofilms, the redox mediators might be shared among different species and could be responsible for the syntrophic links between different species (17). Naturally, microbial communities build consortia that are characterized by intricate interactions, which often lead to a better utilization of resources (23, 24). Some of these interactions are synergistic, involving native redox mediator and non-redox mediator producers (25). Studies have shown that Pseudomonas aeruginosa produces phenazines that can be utilized by members of other species to transfer electrons to an external electron acceptor (10, 18). The involvement of phenazines in synergistic and syntrophic interactions among bacteria is also well documented in natural communities, where, for instance, P. aeruginosa was found to coexist and interact with Enterobacter aerogenes in marine sediments through the transfer of PYO and other metabolites (26).
A common prevalence of P. aeruginosa and E. aerogenes has also been found in a mixed microbial community of an MFC treating synthetic wastewater (27). Here, further investigations have provided the first insight into these interactions, which were especially pronounced under oxygen-limited conditions. Redox mediators from P. aeruginosa were shown to mediate extracellular electron transfer in a synergistic interaction with E. aerogenes. The Enterobacter fermentation product 2,3-butanediol (2,3-BD) was shown to enhance and influence the spectrum of phenazine production from P. aeruginosa. E. aerogenes, in turn, used the phenazines to “respire” with the electrode, resulting in increased current and biomass production. There are a number of reports confirming the possible effect of certain carbon substrates in changing and enhancing the production of phenazines by P. aeruginosa (28–31). 2,3-BD was found to enhance not only phenazine production but also other virulence factors of P. aeruginosa (32). Another recent study has shown that ethanol produced by Candida albicans can enhance the production of phenazines by P. aeruginosa in polymicrobial interactions (33). These interactions represent social networks that have evolved between members occupying certain ecological niches.
Understanding this web of interactions and gaining a deeper understanding of the physiology of mediator production and usage will help us design mixed microbial cultures that effectively self-mediate electroactivity. We also need to know whether and how the interspecies interactions, redox mediator production, and responses to metabolites are dependent on the strain of P. aeruginosa. Strains PA14 and PAO1 have been extensively used in many physiological studies. Genetically, strains PA14 and PAO1 are appreciably different, and they often show discernible physiological differences. PA14 has a slightly larger genome than PAO1 (∼6.54 and 6.26 MB, respectively), which contains several pathogenicity islands that are not present in PAO1 (34, 35). The first pure culture evaluations of P. aeruginosa in BES have been performed with strain PA14 (9, 27, 32); however, this strain differs from the ones evolved in MFC microbial communities and might show different physiological and ecological behaviors.
To elucidate the spectrum of mediator-based bioelectrochemical activity of P. aeruginosa, we here investigated the production of phenazines and the related current generation by three P. aeruginosa strains provided with different ecologically relevant carbon substrates. We examined the well-described strains PA14 and PAO1 and compared them to Pseudomonas sp. strain KRP1, which was previously isolated from a mixed microbial community of an MFC and showed 95% identity with P. aeruginosa in a 16S rRNA gene analysis (25). This evaluation forms an important foundation for future, more complex coculture investigations of mediated electron transfer in microbial fuel cells.
MATERIALS AND METHODS
P. aeruginosa strains and culture conditions.
Strains PA14 (DSMZ 19882) and PAO1 (DSMZ 19880) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). Pseudomonas sp. KRP1 was isolated from an MFC setup at the Laboratory of Microbial Ecology and Technology (LabMET), Ghent University (deposited into the Belgian Coordinated Collections of Microorganisms [BCCM] as strain number LMG 23160), and was kindly provided for this study (25). Precultures were grown overnight in Luria broth medium at 37°C and then washed in 0.9% NaCl solution before BES inoculation. For BES experiments, strains were cultured in AB medium, containing (per liter) 2.0 g (NH4)2SO4, 6.0 g Na2HPO4, 3.0 g KH2PO4, 3.0 g NaCl, 0.011 g Na2SO4, 0.2 g MgCl2, 0.010 g CaCl2, and 0.5 mg FeCl3 · 7 H2O (36). The medium was supplemented with 30 to 35 mM single or cofeed carbon and energy source (see below). The electrochemical reactors were inoculated to a starting optical density at 600 nm (OD600) of 0.05. The temperature was maintained at 37°C.
Molecular procedures.
The phenazine-null mutant of PA14 (PA14 Δphz), in which the genes phzA1 to -G1, phzS, phzM, and phzA2 to -G2 were deleted, was generated via a knockout protocol published by Martinez-Garcia and de Lorenzo (37). Primers used to generate homologous regions for the knockout crossover events and to analyze the success of the knockouts are described in Table S1 in the supplemental material. All sequence information for the genetic comparison of strains PA14 and PAO1 was obtained from the well maintained Pseudomonas Genome Database at www.pseudomonas.com. The genome sequence of the BES isolate KRP1 is not yet published. To compare the phz gene regions of KRP1 to those of PA14 and PAO1, primers (see Table S1 in the supplemental material) for the most external genes of the phz coding area were designed from the PA14 genome sequence and used to amplify the entire KRP1 phz coding area. The amplicons were sequenced by GATC, Germany. Sequence comparison for all strains was performed in Clone Manager (Scientific & Educational Software, USA).
BES setup and electrochemical procedures.
The bioelectrochemical cell was a single-chamber 500-ml glass reactor with a water jacket for temperature control (see Fig. 6b). One side port with a rubber septum was used for inoculation and sampling. Top ports were used for further installations. Microaerobic conditions were achieved through two 2-μm vent filters that allowed circulation of the room atmosphere through the reactor airspace. Oxygen-limited conditions were necessary to generate an environmental stimulus for phenazine production (27) and other virulence factors (38) but at the same time to maintain oxygen availability for the production of PYO (21).
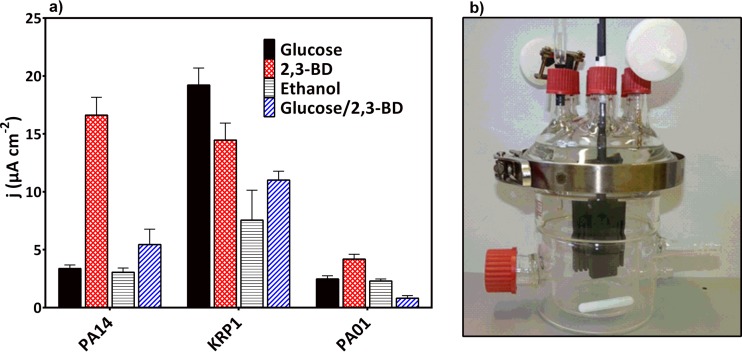
FIG 6.
(a) Observed maximum current densities for P. aeruginosa strains PA14, KRP1, and PAO1 grown in the presence of glucose, 2,3-BD, ethanol, or equimolar glucose and 2,3-BD. Values are averages from three independent biological replicates, except those for KRP1 with 2,3-BD, which are in duplicates. (b) Photograph of the bioelectrochemical setup used.
A three-electrode setup was used with a comb-like solid carbon electrode (Novotec) (surface area, 153 cm2) as the working electrode (anode), a carbon block (49 cm2) as counterelectrode (cathode), and a saturated silver/silver chloride electrode as a reference electrode (RE) (all potentials are reported versus this reference). Electrochemical tests were performed with the Ivium-n-stat potentiostat (Ivium Technologies, Eindhoven, The Netherlands). Electric current generation was measured chronoamperometrically at a set potential of 0.2 V. To determine the coulombic efficiency (CE), the collected charge was related to the charge fed with each substrate, i.e., glucose, 24 e− per molecule; 2,3-BD, 22 e− per molecule; and ethanol, 12 e− per molecule.
Cyclic voltammetry (CV) measurements were used to assess the electrochemical activity of the cultures every 23 h in a potential range of −0.5 to 0.5 V at a scan rate of 2 mV s−1. As a reference, standards of all four phenazines produced by P. aeruginosa were measured under similar aeration conditions in the same medium used for all experiments. 1-HP and PCN were not detected in the chromatographic analysis of the culture media; hence, only the CV data for the standards of PCA and PYO at a 2:1 concentration ratio (since PCA always was determined at higher concentrations than PYO) are shown (see Fig. S1 in the supplemental material). The midpeak potentials (E1/2) of PCA and PYO individually were −0.23 V and −0.18 V, respectively. When the two compounds were mixed, the derived E1/2 were −0.26 V and −0.18 V for PCA and PYO, respectively (see Fig. S1 in the supplemental material).
For the phenazine utilization assay, a slightly simpler BES setup was used. AB medium with or without glucose was used to fill a 100-ml standard laboratory bottle (Schott, Germany) with a septum screw cap through which two graphite rods (5-mm diameter, 5.5-cm submerged length; Novotec) and a reference electrode were installed. Each bottle was stirred at 100 rpm with a magnetic stir bar. Precultured and three-times-washed cells of different P. aeruginosa strains were added at equal cell density (OD600 = 1) to duplicate bottles, one supplied with 30 mM glucose and one not. The background current at an applied potential of 0.2 V was recorded for each culture for 45 min before the addition of PCA (100 μg ml−1 or 20 μg ml−1) or PYO (20 μg ml−1).
Analytical procedures.
For phenazine quantification, samples of culture supernatants were separated in a reverse-phase column (C18 Nucleodur ec column; 250-mm length, 4-mm diameter, 5-μm particle size; Macherey & Nagel, Düren, Germany) mounted on a Shimadzu Prominence ultrafast liquid chromatograph and detected using a photodiode array detector (Shimadzu SPD-M20A). The procedure for sample analysis was as described before (19), with slight modifications. Briefly, 25 mM ammonium acetate (Fluka) as solvent A and acetonitrile (Sigma-Aldrich) as solvent B were used as eluents at a flow rate of 0.35 ml min−1 and a temperature of 20°C. A linear gradient was run for 27 min as follows: 5 min at 20% acetonitrile, 10-min linear gradient ramp to 80% acetonitrile, 5 min at 80% acetonitrile, 2-min linear gradient to 20% acetonitrile, and finally 5 min at 20% acetonitrile. Phenazines were separated and detected at their characteristic wavelengths: PYO, 319 nm; PCA, 366 nm; 1-HP, 247 nm; and PCN, 247 nm. Stock solutions of phenazine standards PCA and PCN (obtained from Princeton Biomolecular) and 1-HP (obtained from TCA Europe) were made by dissolving 1,000 μg ml−1 of each phenazine in dimethyl sulfoxide (DMSO) (Sigma-Aldrich). Stock solutions of PYO (Cayman Chemical) standards were made by dissolving 2,500 μg ml−1 in 100% ethanol.
For analysis of carbon source consumption and secreted metabolites, sample supernatants were separated on an organic acid resin column (300 by 8 mm; polystyrol-divinylbenzol copolymer [PS-DVB]; CS-Chromatography) mounted on a Dionex (Sunnyvale, CA, USA) Ultimate 3000 high-pressure liquid chromatograph (HPLC). Sulfuric acid (5 mM) was used to elute the samples at a flow rate of 0.8 ml min−1 and at 60°C. A refractive index detector (RI-101; Shodex) and UV detector (Ultimate 3000 UV/visible detector; Dionex) at 210 nm were used to detect and quantify the different compounds, depending on their properties.
Biomass as cell dry weight was quantified by harvesting the entire planktonic and biofilm populations at the end of each experiment. Total samples were centrifuged, and the cell pellet was dried at 120°C for 24 h.
RESULTS
Single-substrate metabolism.
The substrates glucose, 2,3-BD, and ethanol were supplied at initial concentrations of 30 to 35 mM; by the end of the experiment, all substrate was consumed (Fig. 1). Often an initially high substrate consumption rate was observed up to day 4 or 5. The uptake rate strongly decreased for the later times of the experiment (to only 25 to 50% of initial rate) (Table 1).
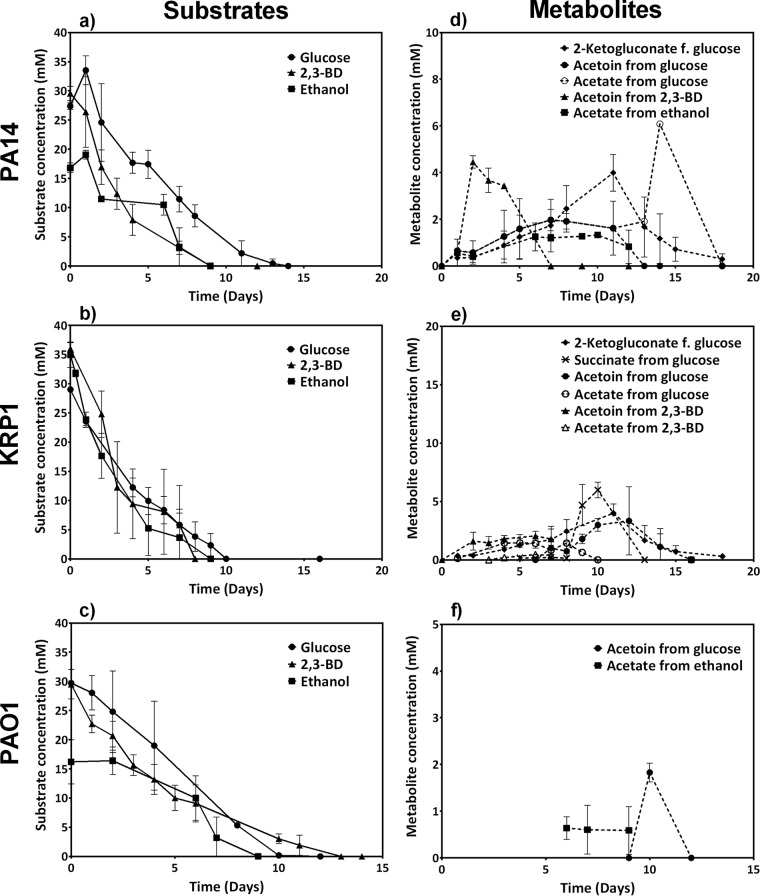
FIG 1.
Carbon substrate consumption (a, b, and c) and metabolite formation time profiles (d, e, and f) for three P. aeruginosa strains in oxygen-limited bioelectrochemical experiments. All data are means and standard deviations for three biological replicates, except those for KRP1 with 2,3-BD, which include only duplicates. The lower reported concentrations of ethanol for PAO1 (the initial concentration was also 30 mM) are due to strong evaporation during sampling and HPLC analysis.
TABLE 1.
Consumption rates for the three carbon substrates
| Strain | Substrate consumption rate (mM day−1)a |
||
|---|---|---|---|
| Glucose | 2,3-BD | Ethanol | |
| PA14 | 3.8 (days 0–5), 1.4 (days 5–11) | 5.6 (days 0–4), 1.7 (days 4–12) | 2.2 (days 0–9) |
| KRP1 | 4.1 (days 0–4), 2.0 (days 4–10) | 6.6 (days 0–4), 2.3 (days 4–8) | 5.8 (days 1–5), 1.3 (days 5–9) |
| PAO1 | 3.1 (days 0–10) | 3.9 (days 1–5), 1.3 (days 5–14) | 3.6 (days 0–3) |
Calculated from the means of the substrate concentrations over time (Fig. 1).
From the provided substrates, strains PA14 and KRP1 produced low levels of 2-ketogluconate (as a direct oxidation product from glucose only), acetate, and acetoin as primary products, which were subsequently reconsumed (Fig. 1d, e, and f). Production of acetoin from glucose may be a carbon storage strategy as well as aimed at preventing the pH effects of possible acidic products (39, 40). The direct interconversion of 2,3-BD to acetoin likely caused the increased production of acetoin during growth with 2,3-BD as the carbon source (41). Strain KRP1 also produced succinate during growth with glucose as carbon source, which was also later consumed (Fig. 1). Succinate is one product of the pyruvate fermentation pathway, which was shown to be activated for anaerobic survival of P. aeruginosa (42), especially in the presence of phenazines to discharge surplus reducing equivalents (43). This indicates that the cells were oxygen limited due to high metabolic activity (e.g., biomass production) during days 6 to 8. In contrast to the case for PA14 and KRP1, hardly any substrate degradation products or intermediates were found for strain PAO1.
Single-substrate phenazine and current generation.
Electric current generation by the three strains was recorded chronoamperometrically at a provided anode potential of 0.2 V versus the RE (Fig. 2). In parallel, we quantified phenazine production during the experiments to correlate the mediators to the electrochemical activity.
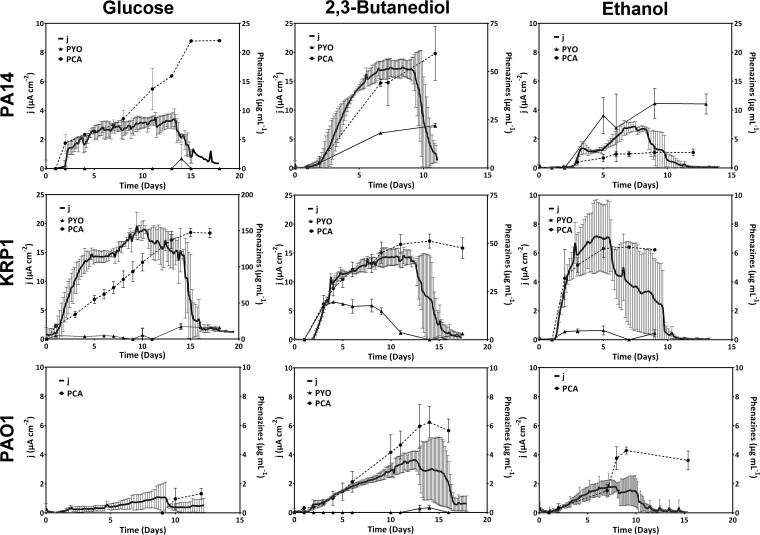
FIG 2.
Current generation and phenazine production by P. aeruginosa PA14, KRP1, and PAO1 grown with the substrates glucose, 2,3-BD, and ethanol. Data are from three biological replicates, except those for KRP1 with 2,3-BD, which are in duplicates. Note the different current density axis scaling for PA14 with 2,3-BD, KRP1 with glucose, and KRP1 with 2,3-BD.
For strain PA14, we observed a switch to production of PYO (22 μg ml−1 versus hardly any) and a 7-fold-increased production of PCA (60 μg ml−1 versus 8.5 μg ml−1) when provided with 2,3-BD compared to glucose and, consequently, a 3-fold-higher current density (17 and 5 μA cm−2 for 2,3-BD and glucose, respectively). This is comparable to an earlier report of increased maximum current density with 2,3-BD compared to glucose as a substrate (5.2 and 3.3 μA cm−2, respectively) (27).
Compared to this, KRP1 produced its highest current with glucose as a carbon source (maximum current density [jmax] = 19 μA cm−2) at very large amounts of PCA in the culture (up to 150 μg ml−1), while PYO (up to 17 μg ml−1) was produced only very late in the experiment when the current levels had already decreased (>day 11). With 2,3-BD, three-times-smaller amounts of PCA were produced, but instead considerable amounts of PYO (20 μg ml−1) were produced at the beginning of the experiment. Under this condition, a current density of 14 μA cm−2 was observed for strain KRP1. In comparison to glucose and 2,3-BD, both PA14 and KRP1 showed much lower electroactivity during growth with ethanol as the carbon source, and this is in line with the small amounts of phenazines produced (Fig. 2).
In contrast, PAO1 was barely electroactive with all substrates; the highest current was recorded with 2,3-BD (jmax = 4 μA cm−2). The low electroactivity of PAO1 concurred with low or undetectable levels of phenazines in our experiments.
The substrate-based energy yield was derived as coulombic efficiency (CE) from the integral of the individual current curves over time (Table 2). In comparison, strains PA14 and PAO1 showed low efficiency with glucose (1.8% and 0.2%, respectively), which increased during growth with 2,3-BD (4.5% and 1.5%, respectively). In contrast, KRP1 showed higher efficiency with glucose (8.9%), which decreased during growth with 2,3-BD (6.5%). With ethanol, the coulombic efficiencies were generally much lower than those with 2,3-BD.
TABLE 2.
Coulombic efficiencies of the conversion of different substrates to electric current
| P. aeruginosa strain | Coulombic efficiency (%, mean ± SD) with: |
|||
|---|---|---|---|---|
| Glucose | 2,3-BD | Ethanol | Glucose and 2,3-BD (1:1) | |
| PA14 | 1.8 ± 0.3 | 4.5 ± 0.3 | 1.1 ± 0.0 | 2.3 ± 0.4 |
| KRP1 | 8.9 ± 0.6 | 6.5 ± 0.3 | 3.4 ± 0.7 | 2.6 ± 0.4 |
| PAO1 | 0.2 ± 0.1 | 1.5 ± 0.3 | 1.1 ± 0.1 | 0.1 ± 0.0 |
Cosubstrate consumption and bioelectrochemical utilization.
To test the functional relationship of the presence of different carbon sources, equimolar glucose and 2,3-BD substrate were cofed to the three strains (at 15 mM each). PA14 showed a typical diauxic growth pattern where glucose was first consumed and depleted before 2,3-BD was taken up (Fig. 3a). The measured current profile (Fig. 3d) also reflects the two activity phases: the glucose-based current enters a stable plateau of ∼3.25 μA cm−2 around day 4.5, and the current starts to rise again to a maximum ∼4.7 μA cm−2 after day 7, when 2,3-BD is consumed as a substrate. The current increase with transition to 2,3-BD coincides with a strong increase of PCA from 0.5 μg ml−1 to 2.5 μg ml−1.
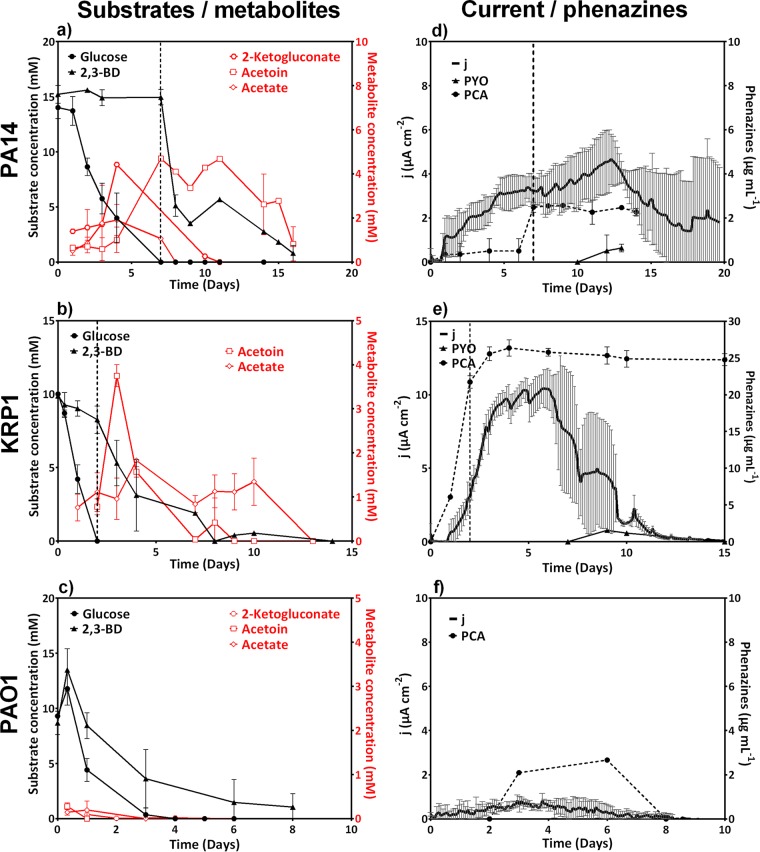
FIG 3.
Carbon substrate uptake by cultures supplied with equimolar glucose and 2,3-BD (a, b, and c) and related electric current generation and phenazine concentrations (d, e, and f) for strains PA14, KRP1, and PAO1. The vertical dashed lines in panels a, b, d, and e represent the time of substrate switch from glucose to 2,3-BD. All data are means and standard deviation for three biological replicates.
Similar to the case for PA14, HPLC analysis of carbon source consumption by KRP1 depicted a diauxic growth pattern, which was, however, not easily distinguishable in the current generation (Fig. 3b and e). Electric current increased rapidly during the initial glucose growth phase (up to ∼3.5 μA cm−2). During 2,3-BD consumption, the current further increased to over 10 μA cm−2. Thereby, strain KRP1 produced very high concentrations of PCA mainly during glucose consumption (22 μg ml−1 by day 2, which was over 80% of the total PCA production in the experiment). However, the highest metabolic activity of electron discharge to the anode took place only after glucose was consumed.
Unlike PA14 and KRP1, PAO1 consumed both the carbon sources concomitantly while producing the smallest amounts of metabolic side products (<1 mM) (Fig. 3c) and very low current densities (Fig. 3f).
The CE for the cofeed experiments was similarly low as with the single-substrate experiments.
CV analysis of redox activity.
Cyclic voltammetry (CV) measurements provide characteristic redox peak systems of different redox-active compounds, from which specific midpeak potentials (E1/2) and information on the reversibility of the redox reaction can be derived.
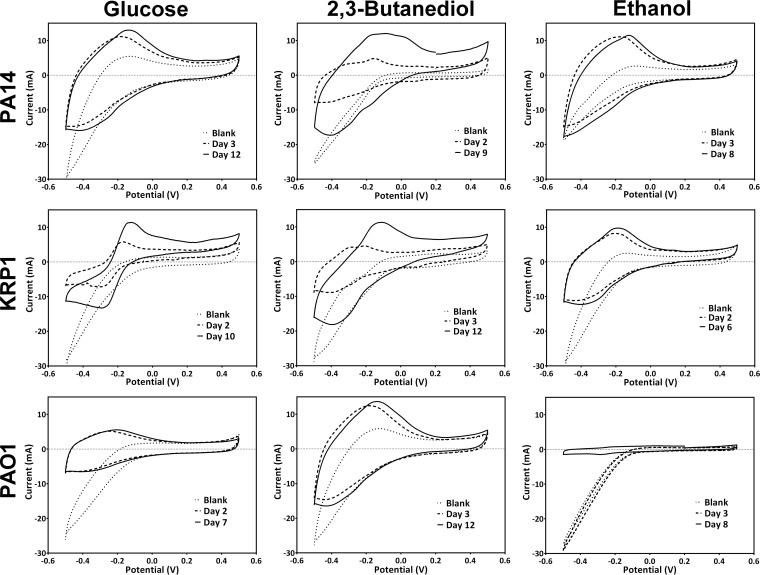
In microbial experiments, the complex cyclic voltamograms differed from those for the phenazine standards (see Fig. S1 in the supplemental material) depending on the strain, carbon source, time point of the experiment, and culture pH (Fig. 4). Here, the mixed potentials can be evaluated only in combination with a chemical analysis (via HPLC) of the medium at the respective time points (Table 3). From the daily scans, we show representative CV and HPLC data for the early phase of increasing bioelectrochemical activity and during maximum detected current production.
FIG 4.
Cyclic voltamograms for the three P. aeruginosa strains grown with the three carbon sources. Plots show a representative blank scan (immediately after inoculation), one scan during increasing electrochemical activity, and one scan during maximum electrochemical activity for one representative biological replicate for each condition.
TABLE 3.
Comparison of electrochemical and chromatographic phenazine analyses
| Strain | Carbon source | Day | pH | Eox (V)a | Ered (V)a | E1/2 (V) | Phenazinesb (μg/ml) |
|
|---|---|---|---|---|---|---|---|---|
| PYO | PCA | |||||||
| PA14 | Glucose | 3 | 6.8 | −0.20 | −0.43 | −0.31 | —c | 5.8 |
| 12 | 6.2 | −0.14 | −0.26 | −0.20 | — | 15.9 | ||
| 2,3-BD | 2 | 6.7 | −0.17 | −0.21 | −0.19 | 2.6 | 2.6 | |
| 9 | 6.7 | −0.19 | −0.42 | −0.30 | 20.0 | 51.0 | ||
| −0.10 | −0.28 | −0.19 | ||||||
| −0.01 | −0.17 | −0.09 | ||||||
| Ethanol | 3 | 7.3 | −0.19 | −0.22 | −0.20 | 3.9 | 1.0 | |
| 8 | 6.7 | −0.13 | −0.21 | −0.17 | 10.5 | 2.7 | ||
| KRP1 | Glucose | 2 | 6.7 | −0.18 | −0.28 | −0.23 | 3.6 | 29.0 |
| 10 | 6.2 | −0.14 | −0.28 | −0.21 | — | 106.3 | ||
| 2,3-BD | 3 | 6.7 | −0.30 | −0.39 | −0.34 | 16.1 | 19.8 | |
| −0.22 | −0.27 | −0.25 | ||||||
| 12 | 6.7 | −0.12 | −0.37 | −0.25 | 3.2 | 51.2 | ||
| Ethanol | 2 | 6.7 | −0.22 | −0.40 | −0.31 | 6.1 | 8.1 | |
| 6 | 6.7 | −0.19 | −0.38 | −0.28 | — | 6.4 | ||
| PAO1 | Glucose | 2 | 6.7 | −0.266 | −0.385 | −0.325 | — | — |
| 7 | 6.7 | −0.22 | −0.36 | −0.29 | — | — | ||
| 2,3-BD | 3 | 7.1 | −0.19 | −0.41 | −0.30 | — | 0.4 | |
| 12 | 6.7 | −0.15 | −0.40 | −0.27 | 4.5 | |||
| 0.00 | −0.26 | −0.13 | ||||||
| Ethanol | 3 | 6.7 | —c | — | — | — | 0.8 | |
| 8 | 6.7 | −0.21 | −0.27 | −0.24 | — | 3.8 | ||
Eox and Ered, oxidation and reduction peak potentials, respectively, derived from the representative CVs in Fig. 4.
Corresponding phenazine concentration related to the representative CV in Fig. 4, not to the averaged phenazine concentrations from Fig. 3.
—, no phenazines were detected or a redox peak was not distinguishable.
A fairly broad redox peak system was found for strain PA14 supplied with glucose, which shifted the E1/2 from −0.31 to −0.20 from day 3 to day 12 (Fig. 4). The HPLC analysis indicated only the presence of PCA at these time points (Table 3). In comparison, PA14 supplied with 2,3-BD as the carbon source showed more defined redox peak systems at day 9 (E1/2 = −0.30, −0.19, and −0.09 V), during which PYO and PCA were present in the culture (20 and 51 μg ml−1, respectively). During growth with ethanol as the carbon source, a redox peak system at ∼ E1/2 = −0.18 V was identified at days 3 and 8, corresponding to PYO as the dominating phenazine in this culture (with 4 times more PYO than PCA at both time points).
Unlike for PA14, easily distinguishable redox peaks were observed for KRP1 supplied with glucose. Notably, at day 10 KRP1 produced very large amounts of PCA (106 μg ml−1), which correlated with a prominent corresponding peak system (E1/2 = −0.21 V) (Fig. 4). Similarly defined peak systems were observed during growth with 2,3-BD, where roughly equal amounts of PYO and PCA were present in the culture on day 3 (16 versus 20 μg ml−1 for PYO and PCA, respectively). During maximum activity (day 12), PCA clearly dominated the culture, and a mixed CV with a strong peak separation was measured. Small amounts of PCA were produced by KRP1 supplied with ethanol, resulting in the observed redox peak system at E1/2 = −0.28V (Fig. 4; Table 3). Strain PAO1 produced very small amounts of phenazines, and redox peak systems for strain PAO1 were not clearly defined.
Evaluating differences in usage and production of phenazines.
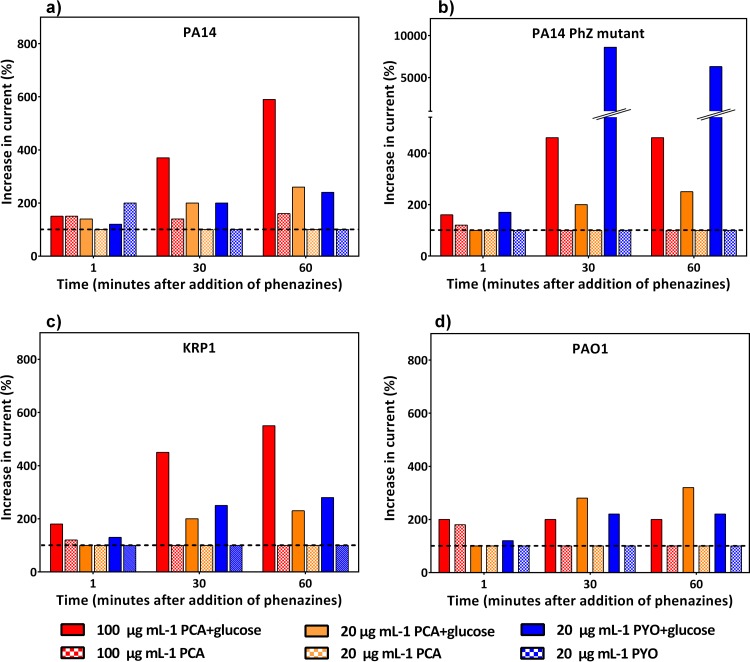
To evaluate whether the strains differ more in electrochemical behavior because of a difference in electrochemical utilization of the phenazines or because of a difference in phenazine production, we performed two additional analyses. In the first, we washed all strains thoroughly to remove endogenously produced phenazines from the cells, added them to medium with glucose (to generate metabolic activity) or without glucose (resting cells), and added defined concentrations of PCA or PYO similar to the ones found in the experiments. Figure 5 shows the relative increase in current production for different time points after the addition of the phenazines to the respective strain (current production before addition corresponds to 100%). A significant and sustained current increase was observed almost exclusively for metabolically active cells (with glucose). For all three strains, PA14, KRP1, and PAO1, the addition of 20 μg ml−1 PCA or PYO resulted in a similar current between 200 and 300% of that with the nonphenazine background. The addition of the observed elevated concentration of PCA (here 100 μg ml−1) resulted in an increase in current production capability only for strains PA14 and KRP1, while PAO1 was unresponsive to the increased levels of PCA. For this phenazine utilization assay, we also included a phenazine-null mutant of PA14. We expected this strain to behave similarly to the washed PA14 culture. However, while this was true for the addition of PCA at both tested concentrations, the addition of PYO yielded a 35-times-higher current increase with PA14 Δphz compared to the PA14 wild type.
FIG 5.
Phenazine utilization assay with defined phenazine concentrations. The bar graphs show the relative increase in current generation (at 0.2 V) with the addition of defined concentrations of phenazines to a resuspension of washed cells of strains PA14 (a), PA14 Δphz (b), KRP1 (c), and PAO1 (d). The dashed horizontal line at 100% highlights the activity of the cells before phenazine addition. Solid bars represent cultures with 30 mM glucose, and crosshatched bars represent cultures without glucose addition. Values at 1 min, 30 min, and 60 min after phenazine addition are reported.
In the second analysis, we performed a direct genome sequence comparison between the coding and regulatory regions for phenazine synthesis of the three strains. Table S2 in the supplemental material summarizes the sequence similarity for all phenazine synthesis genes, and although there is a very high sequence identity level (mostly >98%), it is clear that all genes also contain nucleotide mismatches, which could potentially result in different enzyme activities during phenazine synthesis. We also evaluated all major transcription regulatory elements for all strains and gene clusters with a focus on the upstream areas of phzA1 and phzA2 and found no considerable difference (see Fig. S2 in the supplemental material). Therefore, differences in regulation are most likely driven by a different modulation of the overlaying quorum-sensing regulatory network and not by any remarkable genetic differences between the strains' phenazine gene clusters.
DISCUSSION
Substrate preference of P. aeruginosa strains.
The carbon source preference of P. aeruginosa and the control of their utilization in nature have been extensively explored (44, 45). Here, P. aeruginosa strains PA14 and PAO1 and Pseudomonas sp. isolate KRP1 were grown in passively aerated one-chamber BES setups (Fig. 6) supplied with either glucose or the fermentation product 2,3-BD or ethanol. Our results suggest an enhanced uptake of the fermentation product 2,3-BD compared to glucose, with an ∼50% higher consumption rate for 2,3-BD with PA14 and KRP1 and an ∼25% higher consumption rate with PAO1 (Table 1). This might be linked to the dynamics of their transport rather than to carbon source prevalence. Cofeed experiments (Fig. 2), which showed a diauxic growth pattern (except for PAO1) when both glucose and 2,3-BD are present as substrates, indicated that glucose is preferred over 2,3-BD. In a cofeed experiment, it is generally expected that the catabolite repression system will favor the uptake of the preferred substrate. It is widely accepted that P. aeruginosa prefers organic acids and amino acids to glucose in a sequential hierarchy (45, 46). Thereby, the catabolism of glucose, via the Entner-Doudoroff pathway, is under the control of the catabolite repression system (44, 47). Our cofeed experiments suggest that glucose is a preferred substrate over 2,3-BD for P. aeruginosa. Indeed, the repression of transcription of the genes responsible for 2,3-BD catabolism by glucose has been experimentally confirmed in Bacillus subtilis (the model organism for the study of 2,3-BD and acetoin catabolism) (39).
Strains dictate redox mediator and electric current production.
Increased current generation has been reported before for PA14 in microbial cocultures containing 2,3-BD-producing microorganisms and in monocultures supplied with 2,3-BD as a carbon source (27). Thereby, 2,3-BD was thought to interact with the quorum-sensing regulatory system, leading to increased production of phenazines, including a switch in the phenazine spectrum toward PYO. Our experiments confirmed this behavior for strain PA14 for the substrates glucose and 2,3-BD. In the cofeed experiments with these substrates, the observed current for PA14 resembles more the current profile of glucose; the high-performance current of PA14 with 2,3-BD was not established after the transition. The produced phenazine (especially PCA) levels were much lower in this cofeed experiment than with the pure substrates. The regulatory differences in a 7-day-old culture at transition to 2,3-BD consumption might have prevented the elevated production of both PYO and PCA as observed with 2,3-BD as single substrate.
It is intriguing that the influence of this fermentation product seems to be strain dependent. Unlike PA14, KRP1 produced its highest current with glucose as a carbon source, which correlated with very large amounts of PCA and hardly any PYO. In the glucose/2,3-BD cofeed experiment, KRP1 also produced high levels of PCA, with over 80% of it in the initial consumption phase. However, the highest metabolic activity of electron discharge to the anode took place only after glucose was consumed, hinting that indeed the large amounts of PCA in this strain are responsible for enhanced current generation. During the cofeed experiments, KRP1 produced the highest current density among the three strains. This also corresponds to the high maximum concentration of PCA in the culture of KRP1 compared to PA14 and PAO1 (26.4 versus 2.6 and 2.7 μg ml−1, respectively) (Fig. 3d, e, and f). While our data confirm the switch to more PYO production with 2,3-BD in both PA14 and KRP1, the increased current production in KRP1 seems to be mediated mainly by PCA.
Thus, the influence of especially glucose on the phenazine spectrum appears to be different among the strains. As noted before, large amounts of succinate (∼6 mM) were produced during growth of KRP1 on glucose, and the reconsumption of this intermediate correlates in time with the highest observed electrochemical activity in the glucose experiments. For P. aeruginosa (strain PAO1), it was shown before that the highly preferred carbon substrate succinate suppresses PYO synthesis (and to a lesser extent also PCA synthesis) through catabolite repression via the Crc protein (48). It should be investigated whether a change in this regulatory path is responsible for the strong production of PCA and electric current in strain KRP1.
However, we cannot linearly correlate the concentration of phenazines to the amplitude of current generation. Addition of defined phenazine concentrations to washed cells (Fig. 5) indicated a similar capacity to utilize the phenazines for PA14 and KRP1, but more investigations into the roles of the different phenazines in redox cycling are required. It should also be noted that the ratio of PCA to PYO is oxygen dependent because of an oxygen-dependent monooxygenase (PhzM), which converts PCA to PYO. Thus, the instantaneous oxygen availability in our oxygen-limited experiments influenced the production of PCA versus PYO for the evaluated substrates. It is likely that the amplitude of current generation is a multivariate problem, influenced by phenazines, oxygen availability, metabolic and physiological preferences, and complex regulatory networks.
In contrast, strain PAO1 barely showed any phenazine production and electroactivity in our study. The phenazine addition experiment also indicates a lower capacity to utilize PCA as redox shuttle. These findings have important implications for the study of electrochemical activity of P. aeruginosa. In many laboratories, PAO1 is the model strain for P. aeruginosa investigations. However, our results indicate that this strain might not be the best choice for P. aeruginosa electroactivity or BES microbial ecology research.
PCA is mainly responsible for reversible redox cycling.
CV measurements together with chemical analysis can be instrumental in making inferences about the redox activity and the redox species present in a BES setup. The voltamograms observed for cultures with higher concentrations of PYO (or a higher PYO/PCA ratio) depict irreversible or weakly reversible redox peaks (e.g., with strain PA14). Instead, in experiments with KRP1 where large amounts or only PCA were detected, CVs with clearly defined oxidation and reduction peaks were observed (Fig. 4; Table 3). This could suggest that PYO was limited in redox cycling under the conditions of our BES. It has been postulated that, depending on the reactivity of the phenazines, PCA might be located deep in the biofilm, where it is involved in iron acquisition, whereas PYO occurs on the surface, where it reacts with oxygen to produce reactive oxygen species as an antibiotic against competitors (22). Further, the reversible intracellular reduction of PYO might not be efficient: an earlier report demonstrated the possibility that P. aeruginosa lacks an NADPH:PYO oxidoreductase, resulting in a limitation in redox cycling (49). Another recent report has shown that PYO can be irreversibly oxidized by the pseudomonas quinolone signal (PQS), a quorum-sensing molecule, making it unavailable for redox cycling (50). Thus, our results reflected in the light of these earlier reports support the hypothesis that PCA rather than PYO is more directly involved in redox cycling under oxygen-limited conditions. However, in our phenazine addition test, a similar relative current increase was observed for the three strains with the addition of 20 μg/ml PCA or PYO, indicating a comparable capacity of the strains to utilize fresh PCA and PYO. Effects of phenazine aging and degradation are currently under investigation. In the comparison of the PA14 Δphz mutant, which is not able to synthesize phenazines, to washed cells of strain PA14, a surprising 35-fold increase in current was observed with the addition of 20 μg/ml PYO (Fig. 5b). Currently, we are not able to explain this observation, but it points toward a suppressed PYO utilization in the strain that was already primed for phenazine production and usage. Thus, besides the high complexity of the phenazine regulation through quorum-sensing cascades on the production side, the overall utilization of available phenazines also might be strongly regulated within the cell.
Anodic electron discharge is a metabolic side reaction under oxygen limitation.
To estimate the amount of energy recovered as charge from the substrates provided, the coulombic efficiency was calculated. In general, CE was low for all strains, showing that current production is likely a metabolic side reaction for all three strains. The main contributor to this low proportion of anodic electron discharge likely was oxygen. Under our oxygen-limited conditions, the three strains showed different levels of efficiency of substrate conversion to side products (i.e., metabolic products and electric current) and biomass formation. Growth or biomass quantification during the experiments was not trivial, since varying levels of biofilm formation prevented reliable measurements of optical density or cell counts. Therefore, biomass was assessed only as total cell dry weight (planktonic plus biofilm biomass) at the end of each experiment (Table 4). Table 4 also indicates the observed tendency to form a strong biofilm on the air-liquid interface. No direct correlation between the final biomass, substrate utilization, and metabolic side products could be derived. However, it is likely that a strong biofilm formation on the surface of the reactor liquid will cause many cells to be in close proximity to the headspace oxygen to discharge electrons (e.g., with PA14 grown on glucose). Similarly, a low biofilm formation tendency in combination with a low general biomass formation might allow oxygen to be available for electron discharge throughout the reactor liquid (e.g., all strains with ethanol). However, besides this, fundamental strain differences might also largely influence CE; for example, PAO1 generally grows well under our oxygen-limited conditions without the discharge of metabolic products or electrons.
TABLE 4.
Final bioreactor biomass as cell dry weight and biofilm formation tendency for different carbon sources
| P. aeruginosa strain | Final reactor biomass (g [cell dry wt]/liter, mean ± SD)a |
|||
|---|---|---|---|---|
| Glucose | 2,3-BD | Ethanol | 1:1 Glucose:2,3-BD | |
| PA14 | 1.29 ± 0.06 (+++) | —b (+++) | 0.27 ± 0.02 (+) | 0.48 ± 0.09 (+) |
| KRP1 | 0.46 ± 0.23 (++) | 0.34 ± 0.01c (++) | 0.26 ± 0.03 (+) | 0.58 ± 0.04 (+) |
| PAO1 | — (+) | 0.43 ± 0.15 (+) | 0.37 ± 0.01 (+) | 0.74d (+) |
Biofilm formation tendency is given in parentheses as a rough estimate: +++, strong biofilm formation on reactor liquid; ++, medium biofilm formation; +, weak biofilm formation.
—, no cell dry weight data were available.
Average for two replicates.
Biomass data were available for only one replicate.
Overall, the observed current is a function of a wide array of factors that influence the amount of electrons and their efficiency of transmission to the electrode. If we consider a similar oxygen availability, the most important contributor for efficient cell-to-electrode electron transfer is the phenazine concentration. High currents correlate with high phenazine concentrations, while PAO1 produces hardly any phenazines and consequently no anodic current. The main driver for a high concentration of phenazines therefore was the identity of the Pseudomonas strain, which determined the dominating phenazine species and the influence or lack of influence of the carbon source on phenazine production. Both parameters might provide an ecological advantage in certain environments, also because the different phenazines might have different capacities for reversible redox cycling. In comparison to the influence of the strain identity and oxygen as a direct competitor for electrons, the amount of biomass seems to play a smaller role in influencing the phenazine-based anodic current in our experiments.
Conclusions.
One approach to enhance electron transfer efficiency in MFCs is to coculture microorganisms that can synergistically mediate current production by employing soluble redox mediators. Gaining more insight into the interactions that shape these electroactive communities will enable the definition of cocultures that can effectively self-mediate current generation. We here compared the electroactivities of three strains of the mediator producer P. aeruginosa to deepen our understanding of this important member of such communities. Overall, our data indicate differences in the electroactivities of the three strains which, most likely, mirror their adaptabilities to different environments. Comparing all strains and carbon sources, we conclude that the BES isolate KRP1 is the most electroactive when supplied with the three carbon sources considered (Fig. 6). Ongoing investigations of KRP1 in an ecological, i.e., coculture, context and a comparative transcriptome analysis with PA14 will shed further light into this naturally increased capability of electron discharge to an extracellular electron acceptor. Building on this work, more complex coculture investigations of mediated electron transfer in microbial fuel cells will become possible.
Supplementary Material
ACKNOWLEDGMENTS
E.M.B. designed, executed, and analyzed the experiments and drafted the primary manuscript, L.M.B. discussed the experiments and edited the manuscript, and M.A.R. conceived of the study, designed and interpreted the experiments, and prepared the final manuscript.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (DFG, AG156/1-1) to M.A.R. E.M.B was supported by a personal stipend from the Deutscher Akademischer Austauch Dienst (DAAD).
We thank Korneel Rabaey for kindly providing us with Pseudomonas sp. KRP1, Simone Schmitz for the generation of strain PA14 Δphz, and Simone Schmitz and Carola Berger for fruitful discussions.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01342-16.
REFERENCES
- 1.Rabaey KV, Verstraete W. 2005. Microbial fuel cells: novel biotechnology for energy generation. Trends Biotechnol 23:291–298. doi: 10.1016/j.tibtech.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 2.Fornero JJ, Rosenbaum M, Angenent LT. 2010. Electric power generation from municipal, food, and animal wastewaters using microbial fuel cells. Electroanalysis 22:832–843. doi: 10.1002/elan.200980011. [DOI] [Google Scholar]
- 3.Schröder U, Harnisch F, Angenent L. 2015. Microbial electrochemistry and technology: terminology and classification. Energy Environ Sci 8:513. doi: 10.1039/C4EE03359K. [DOI] [Google Scholar]
- 4.Schröder U. 2007. Anodic electron transfer mechanisms in microbial fuel cells and their energy efficiency. Phys Chem Chem Phys 9:2619–2629. doi: 10.1039/B703627M. [DOI] [PubMed] [Google Scholar]
- 5.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S, Logan B, Nealson KH, Fredrickson JK. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Nat Acad Sci U S A 103:11358–11363. doi: 10.1073/pnas.0604517103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marsili E, Baron DB, Shikhare ID, Coursolle D, Gralnick JA, Bond DR. 2008. Shewanella secretes flavins that mediate extracellular electron transfer. Proc Natl Acad Sci U S A 105:3968–3973. doi: 10.1073/pnas.0710525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrighton KC, Thrash JC, Melnyk RA, Bigi JP, Byrne-Bailey KG, Remis JP, Schichnes D, Auer M, Chang CJ, Coates JD. 2011. Evidence for direct electron transfer by a gram-positive bacterium isolated from a microbial fuel cell. Appl Environ Microbiol 77:7633–7639. doi: 10.1128/AEM.05365-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lovley DR, Ueki T, Zhang T, Malvankar NS, Shrestha PM, Flanagan KA, Aklujkar M, Butler JE, Giloteaux L, Rotaru AE, Holmes DE, Franks AE, Orellana R, Risso C, Nevin KP. 2011. Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv Microb Physiol 59:1–100. doi: 10.1016/B978-0-12-387661-4.00004-5. [DOI] [PubMed] [Google Scholar]
- 9.Venkataraman A, Rosenbaum M, Arends JBA, Halitsche R, Angenent LT. 2010. Quorum sensing regulates electric current generation of Pseudomonas aeruginosa PA14 in bioelectrochemical systems. Electrochem Commun 12:459–462. doi: 10.1016/j.elecom.2010.01.019. [DOI] [Google Scholar]
- 10.Pham TH, Boon N, Aelterman P, Clauwaert P, De Schamphelaire L, Vanhaecke L, De Maeyer K, Hofte M, Verstraete W, Rabaey K. 2008. Metabolites produced by Pseudomonas sp. enable a Gram-positive bacterium to achieve extracellular electron transfer. Appl Microbiol Biotechnol 77:1119–1129. doi: 10.1007/s00253-007-1248-6. [DOI] [PubMed] [Google Scholar]
- 11.Freguia S, Masuda M; Tsujimura S, Kano K. 2009. Lactococcus lactis catalyses electricity generation at microbial fuel cell anodes via excretion of a soluble quinone. Bioelectrochemistry 76:14–18. doi: 10.1016/j.bioelechem.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 12.Liu XW, Sun XF, Chen JJ, Huang YX, Xie JF, Li WW, Sheng GP, Zhang YY, Zhao F, Lu R, Yu HQ. 2013. Phenothiazine derivative-accelerated microbial extracellular electron transfer in bioelectrochemical system. Sci Rep 3:1616. doi: 10.1038/srep01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitz S, Nies S, Wierckx N, Blank LM, Rosenbaum MA. 2015. Engineering mediator-based electroactivity in the obligate aerobic bacterium Pseudomonas putida KT2440. Front Microbiol 6:284. doi: 10.3389/fmicb.2015.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson R, Pitt T, Taylor G, Watson D, MacDermot J, Sykes D, Roberts D, Cole P. 1987. Pyocyanin and 1-hydroxyphenazine produced by Pseudomonas aeruginosa inhibit the beating of human respiratory cilia in vitro. J Clin Invest 79:221–229. doi: 10.1172/JCI112787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich LE, Price-Whelan A, Petersen A, Whiteley M, Newman DK. 2006. The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Mol Microbiol 61:1308–1321. doi: 10.1111/j.1365-2958.2006.05306.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Wilks JC, Danhorn T, Ramos I, Croal L, Newman DK. 2011. Phenazine-1-carboxylic acid promotes bacterial biofilm development via ferrous iron acquisition. J Bacteriol 193:3606–3617. doi: 10.1128/JB.00396-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez ME, Newman DK. 2001. Extracellular electron transfer. Cell Mol Life Sci 58:1562–1571. doi: 10.1007/PL00000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rabaey K, Boon N, Höfte M, Verstraete W. 2005. Microbial phenazine production enhances electron transfer in biofuel cells. Environ Sci Technol 39:3401–3408. doi: 10.1021/es048563o. [DOI] [PubMed] [Google Scholar]
- 19.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierson LS III, Pierson EA. 2010. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol 86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mavrodi DV, Blankenfeldt W, Thomashow LS. 2006. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol 44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- 22.Wang N, Newman DK. 2008. Redox reactions of phenazine antibiotics with ferric (hydr)oxides and molecular oxygen. Environ Sci Technol 42:2380–2386. doi: 10.1021/es702290a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rinaldi A, Mecheri B, Garavaglia V, Licoccia S, Di Nardo P, Traversa E. 2008. Engineering materials and biology to boost performance of microbial fuel cells: a critical review. Energy Environ Sci 1:417. doi: 10.1039/b806498a. [DOI] [Google Scholar]
- 24.Morris BE, Henneberger R, Huber H, Moissl-Eichinger C. 2013. Microbial syntrophy: interaction for the common good. FEMS Microbiol Rev 37:384–406. doi: 10.1111/1574-6976.12019. [DOI] [PubMed] [Google Scholar]
- 25.Rabaey K, Boon N, Siciliano SD, Verhaege M, Verstraete W. 2004. Biofuel cells select for microbial consortia that self-mediate electron transfer. Appl Environ Microbiol 70:5373–5382. doi: 10.1128/AEM.70.9.5373-5382.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angell S, Bench BJ, Williams H, Watanabe CM. 2006. Pyocyanin isolated from a marine microbial population: synergistic production between two distinct bacterial species and mode of action. Chem Biol 13:1349–1359. doi: 10.1016/j.chembiol.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Venkataraman A, Rosenbaum MA, Perkins SD, Werner JJ, Angenent LT. 2011. Metabolite-based mutualism between Pseudomonas aeruginosa PA14 and Enterobacter aerogenes enhances current generation in bioelectrochemical systems. Energy Environ Sci 4:4550–4559. doi: 10.1039/c1ee01377g. [DOI] [Google Scholar]
- 28.Korgaonkar AK, Whiteley M. 2011. Pseudomonas aeruginosa enhances production of an antimicrobial in response to N-acetylglucosamine and peptidoglycan. J Bacteriol 193:909–917. doi: 10.1128/JB.01175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whiteson KL, Meinardi S, Lim YW, Schmieder R, Maughan H, Quinn R, Blake DR, Conrad D, Rohwer F. 2014. Breath gas metabolites and bacterial metagenomes from cystic fibrosis airways indicate active pH neutral 2,3-butanedione fermentation. ISME J 8:1247–1258. doi: 10.1038/ismej.2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dusane DH, Matkar P, Venugopalan VP, Kumar AR, Zinjarde SS. 2011. Cross-species induction of antimicrobial compounds, biosurfactants and quorum-sensing inhibitors in tropical marine epibiotic bacteria by pathogens and biofouling microorganisms. Curr Microbiol 62:974–980. doi: 10.1007/s00284-010-9812-1. [DOI] [PubMed] [Google Scholar]
- 31.Korgaonkar A, Trivedi U, Rumbaugh KP, Whiteley M. 2013. Community surveillance enhances Pseudomonas aeruginosa virulence during polymicrobial infection. Proc Natl Acad Sci U S A 110:1059–1064. doi: 10.1073/pnas.1214550110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkataraman A, Rosenbaum MA, Werner JJ, Winans SC, Angenent LT. 2014. Metabolite transfer with the fermentation product 2,3-butanediol enhances virulence by Pseudomonas aeruginosa. ISME J 8:1210–1220. doi: 10.1038/ismej.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S, Ha DG, Willger SD, O'Toole GA, Harwood CS, Dietrich LEP, Hogan DA. 2014. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. 2006. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mikkelsen H, Sivaneson M, Filloux A. 2011. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. Environ Microbiol 13:1666–1681. doi: 10.1111/j.1462-2920.2011.02495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark DJ, Maaløe O. 1967. DNA replication and the division cycle in Escherichia coli. J Mol Biol 23:99–112. doi: 10.1016/S0022-2836(67)80070-6. [DOI] [Google Scholar]
- 37.Martinez-Garcia E, de Lorenzo V. 2011. Engineering multiple genomic deletions in Gram-negative bacteria: analysis of the multi-resistant antibiotic profile of Pseudomonas putida KT2440. Environ Microbiol 13:2702–2716. doi: 10.1111/j.1462-2920.2011.02538.x. [DOI] [PubMed] [Google Scholar]
- 38.Sabra W, Kim EJ, Zeng A-P. 2002. Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology 148:3195–3202. doi: 10.1099/00221287-148-10-3195. [DOI] [PubMed] [Google Scholar]
- 39.Ali NO, Bignon J, Rapoport G, Debarbouille M. 2001. Regulation of the acetoin catabolic pathway is controlled by sigma L in Bacillus subtilis. J Bacteriol 183:2497–2504. doi: 10.1128/JB.183.8.2497-2504.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovacikova G, Lin W, Skorupski K. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol Microbiol 57:420–433. doi: 10.1111/j.1365-2958.2005.04700.x. [DOI] [PubMed] [Google Scholar]
- 41.Xiao Z, Xu P. 2007. Acetoin metabolism in bacteria. Crit Rev Microbiol 33:127–140. doi: 10.1080/10408410701364604. [DOI] [PubMed] [Google Scholar]
- 42.Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Glasser NR, Kern SE, Newman DK. 2014. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol 92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frimmersdorf E, Horatzek S, Pelnikevich A, Wiehlmann L, Schomburg D. 2010. How Pseudomonas aeruginosa adapts to various environments: a metabolomic approach. Environ Microbiol 12:1734–1747. doi: 10.1111/j.1462-2920.2010.02253.x. [DOI] [PubMed] [Google Scholar]
- 45.Rojo F. 2010. Carbon catabolite repression in Pseudomonas: optimizing metabolic versatility and interactions with the environment. FEMS Microbiol Rev 34:658–684. doi: 10.1111/j.1574-6976.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 46.Valentini M, Lapouge K. 2013. Catabolite repression in Pseudomonas aeruginosa PAO1 regulates the uptake of C-4-dicarboxylates depending on succinate concentration. Environ Microbiol 15:1707–1716. doi: 10.1111/1462-2920.12056. [DOI] [PubMed] [Google Scholar]
- 47.Wang HC, Stern IJ, Gilmour CM. 1959. The catabolism of glucose and gluconate in Pseudomonas species. Arch Biochem Biophys 81:489–492. doi: 10.1016/0003-9861(59)90229-2. [DOI] [PubMed] [Google Scholar]
- 48.Huang JF, Sonnleitner E, Ren B, Xu YQ, Haas D. 2012. Catabolite repression control of pyocyanin biosynthesis at an intersection of primary and secondary metabolism in Pseudomonas aeruginosa. Appl Environ Microbiol 78:5016–5020. doi: 10.1128/AEM.00026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hassett DJ, Charniga L, Bean K, Ohman DE, Cohen MS. 1992. Response of Pseudomonas aeruginosa to pyocyanin: mechanism of resistance, antioxidant defenses, and demonstration of a manganese-cofactored superoxide dismutase. Infect Immun 60:328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seviour T, Doyle LE, Lauw SJ, Hinks J, Rice SA, Nesatyy VJ, Webster RD, Kjelleberg S, Marsili E. 2015. Voltammetric profiling of redox-active metabolites expressed by Pseudomonas aeruginosa for diagnostic purposes. Chem Commun (Camb) 51:3789–3792. doi: 10.1039/C4CC08590F. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.