ABSTRACT
Treatment of human excreta and animal manure (HEAM) is key in controlling the spread of persistent enteric pathogens, such as viruses. The extent of virus inactivation during HEAM storage and treatment appears to vary with virus genome type, although the reasons for this variability are not clear. Here, we investigated the inactivation of viruses of different genome types under conditions representative of HEAM storage or mesophilic digestion. The goals were to characterize the influence of HEAM solution conditions on inactivation and to determine the potential mechanisms involved. Specifically, eight viruses representing the four viral genome types (single-stranded RNA [ssRNA], double-stranded RNA [dsRNA], single-stranded DNA [ssDNA], and double-stranded DNA [dsDNA]) were exposed to synthetic solutions with well-controlled temperature (20 to 35°C), pH (8 to 9), and ammonia (NH3) concentrations (0 to 40 mmol liter−1). DNA and dsRNA viruses were considerably more resistant than ssRNA viruses, resulting in up to 1,000-fold-longer treatment times to reach a 4-log inactivation. The apparently slower inactivation of DNA viruses was rationalized by the higher stability of DNA than that of ssRNA in HEAM. Pushing the system toward harsher pH (>9) and temperature (>35°C) conditions, such as those encountered in thermophilic digestion and alkaline treatments, led to more consistent inactivation kinetics among ssRNA and other viruses. This suggests that the dependence of inactivation on genome type disappeared in favor of protein-mediated inactivation mechanisms common to all viruses. Finally, we recommend the use of MS2 as a conservative indicator to assess the inactivation of ssRNA viruses and the stable ΦX174 or dsDNA phages as indicators for persistent viruses.
IMPORTANCE Viruses are among the most environmentally persistent pathogens. They can be present in high concentrations in human excreta and animal manure (HEAM). Therefore, appropriate treatment of HEAM is important prior to its reuse or discharge into the environment. Here, we investigated the factors that determine the persistence of viruses in HEAM, and we determined the main mechanisms that lead to their inactivation. Unlike other organisms, viruses can have four different genome types (double- or single-stranded RNA or DNA), and the viruses studied herein represent all four types. Genome type appeared to be the major determinant for persistence. Single-stranded RNA viruses are the most labile, because this genome type is susceptible to degradation in HEAM. In contrast, the other genome types are more stable; therefore, inactivation is slower and mainly driven by the degradation of viral proteins. Overall, this study allows us to better understand the behavior of viruses in HEAM.
INTRODUCTION
Human excreta and animal manure (HEAM) may pose a threat to people because they are a major reservoir for many fecally-orally transmissible pathogenic organisms (1). In addition, untreated HEAM may release chemical contaminants into the environment (e.g., macronutrients and microorganic pollutants or heavy metals). While HEAM thus represents a source of pollution, it simultaneously is a valuable product containing nutrients, e.g., nitrogen, phosphorus, and potassium, which are essential for plant growth (2, 3), and water for irrigation (4). In order to safely harvest these valuable components, adequate waste treatment is a necessity to prevent the spread of diseases and the contamination of the environment.
Many fecally-orally transmissible pathogens, such as viruses, protozoa, and helminthes, are parasitic organisms that cannot reproduce or grow outside their host. Thus, from the time of excretion from the host, the infective pathogen concentration generally declines with time (5). HEAM treatment and stabilization processes may accelerate this decline by creating harsh environmental conditions that promote pathogen inactivation. Effective treatment of HEAM can generate safe soil conditioner or fertilizer products for land application (6). A wide array of processes are available for sanitation (6, 7), ranging from storage at ambient temperature to chemical treatment with sanitizing substances (8, 9), to aerobic or anaerobic digestion, composting, and alkaline and heat treatment. For most of these processes, the temperature, pH, water content, and the exposure to sanitizing substances, such as ammonia, govern the extent of pathogen inactivation (1, 10). Yet, the individual and synergistic contributions of these parameters to pathogen inactivation remain to be systematically characterized.
Among possible sanitizing compounds, total ammonia (NH4+/NH3) is of particular interest because it may be naturally present at substantial levels in stored urine, stored fecal sludge, and anaerobically digested sludge (Table 1). Total ammonia is produced by urea and protein hydrolysis during the storage and digestion of HEAM (11, 12). In its neutral dissolved form, aqueous ammonia (NH3(aq)) is a major nitrogen source for bacteria, eukaryotic microbes, fungi, and plants. However, NH3(aq) may become harmful at elevated concentrations (13); it was found to have biocidal activity against most pathogenic microorganisms (14–18), and it was shown to be the main substance responsible for virus die-off in sludge (19). NH3(aq) can thus be considered an in situ sanitizer naturally present in HEAM.
TABLE 1.
Typical temperature, pH, and ammonia conditions in human/animal waste during storage and mesophilic anaerobic digestion
| Type of waste by treatment type | Range |
Reference(s) | |||
|---|---|---|---|---|---|
| NH4+/NH3 (mmol liter−1) | Temp (°C) | pH | {NH3(aq)} (mmol liter−1)a | ||
| Storage | |||||
| Urine | 5–475 | 4–35 | 8.2–9.8 | 2–246 | 25, 87, 104 |
| Feces (raw) | 80–247 | 20–34 | 6.8–8.3 | 0.2–32 | 27, 105 |
| Feces (+additives)b | 60–862 | 20–34 | 7.5–12.8 | 1–340 | |
| Feces + urine | 104–1,098 | 10–28 | 8.8–9.2 | 9–439 | 106 |
| Anaerobic digestionc | |||||
| Animal manure | 35–450 | 25–38 | 7.0–8.1 | 0–37 | 107–110 |
| Sewage sludge | 42–45 | 37 | 7.5–8.0 | 1–5 | 34 |
| Activated sludge | 149–191 | 37 | 7.7–7.9 | 7–13 | 111 |
Ammonia activity (evaluated with PHREEQC, as described in Materials and Methods).
Urea, wood ash, or oyster shell.
Under mesophilic conditions.
The mechanisms involved in the toxicity of NH3(aq) toward eukaryotic and prokaryotic cells are not fully understood. Several hypotheses have been put forward to explain its biocidal effect, namely, intracellular pH change, disturbance of the electrochemical gradient across the cell membrane, and inhibition of enzymatic reactions (20–23). Unlike other pathogenic microorganisms, viruses do not have a cell metabolism of their own but use the host machinery to reproduce (24). Therefore, the biocidal processes involving NH3(aq) relevant to cells do not apply to viruses. Instead, the virucidal activity of NH3(aq) likely involves modification or damage of the virus components (i.e., protein, envelope, or nucleic acid). However, while numerous studies have investigated the fate of viruses in complex matrices containing NH3(aq) (8, 17, 19, 25–37), the virucidal mechanism of action of NH3(aq) has received little scrutiny to date (38, 39).
Unlike other living organisms, viruses carry their genetic information in different forms, specifically as single-stranded (ss) or double-stranded (ds) RNA or DNA (24). Several studies have observed differences in inactivation behavior among viruses with different genome types (19, 25, 26, 30, 33–36). This suggests that the genome may be an important target of NH3(aq), and that the different genome types differ in their NH3(aq) susceptibilities. Other factors besides genome type, such as genome geometry or packaging, capsid properties, or the presence of an envelope, may additionally influence virus stability in the presence of NH3(aq), although the extent to which they influence NH3-mediated inactivation is not clear. Given that viruses are generally permeable to water and to salt ions (40, 41), however, none of these structural features should have a greatly protective effect against the action of NH3(aq) on the viral genomes.
Here, we exposed a suite of viruses to NH3(aq) in controlled laboratory solutions in order to characterize how the genome type influences the kinetics of NH3(aq)-mediated inactivation. We focused on naked viruses, which represent the majority of enteric viruses of interest. Furthermore, we put our observations in the context of literature data to elucidate the potential mechanisms of inactivation. Our working hypothesis is that the viral genome type is the major determinant for susceptibility to NH3(aq), and that the influence of other virus-specific parameters, such as genome sequence or protein structure, is minor in comparison. A total of eight viruses representing all genome types were studied: bacteriophages MS2 and GA and human echovirus and coxsackievirus (single-stranded RNA [ssRNA]), mammalian reovirus (double-stranded RNA [dsRNA]), bacteriophage ΦX174 (single-stranded DNA [ssDNA]), and bacteriophage T4 and human adenovirus (double-stranded DNA [dsDNA]). The ultimate goal of this study was to advance our understanding of the dominant mechanisms involved in virus inactivation over a range of environmental conditions relevant to HEAM storage or processing (Table 1).
MATERIALS AND METHODS
Definition of virus.
According to Bândea (42), infective viral particles outside their host should be referred to as virions. Correspondingly, in this study, we were interested in the effect of HEAM treatment and storage on virions. However, viral solutions always contain both virions and noninfective viral particles, and the processes addressed herein act on both. As such, we use the less-specific word “virus” throughout the manuscript, even if it is the virions that are of the most interest.
Viruses and cells.
Human adenovirus (HAdV) type 2 was kindly provided by Rosina Gironès (University of Barcelona, Spain). Echovirus (EV) type 11 (ATCC VR-41), coxsackievirus (CV) B5 Faulkner (ATCC VR-185), and mammalian reovirus (ReoV) type 1 (ATCC VR-230) were purchased from LGC Standards (Molsheim, France). HAdV, EV/CV, and ReoV were propagated on A549 human lung carcinoma epithelial cells, buffalo green monkey kidney (BGMK) cells, and L929 mouse fibroblasts, respectively. A549 and L929 cells were kindly provided by the Lausanne University Hospital (Switzerland) and BGMK cells by the University of Barcelona. A549 cells were cultivated in high-glucose pyruvate Dulbecco's modified Eagle's medium (DMEM; Invitrogen), and L929 and BGMK cells were cultivated in minimum essential medium (MEM; Invitrogen). Both media were supplemented with penicillin (20 U ml−1), streptomycin (20 μg ml−1) (Invitrogen), and 2 or 10% fetal bovine serum (FBS; Invitrogen), and cells were incubated at 37°C in 5% CO2 and 95% humidity. Viruses were propagated by spiking 10 μl of HAdV or CV (1010 to 1011 most probable number of cytopathogenic units [MPNCU] ml−1) or 100 μl of EV or ReoV (107 to 108 MPNCU ml−1) into 160-cm2 flasks (TPP Techno Plastic Products, Trasadingen, Switzerland) containing 95% confluent cells. The flasks were incubated until cytopathic effects were apparent. The viruses were then purified as described by Bosshard et al. (43). From each flask, 1 ml of sample containing 1010 to 1011 MPNCU ml−1 of HAdV, 109 to 1010 MPNCU ml−1 of CV, 107 to 108 MPNCU ml−1 of EV, or 107 to 108 MPNCU ml−1 of ReoV was collected and stored at 4°C as virus stocks for the experiments. New stocks were produced before each set of experiments. Virus titers were determined by most probable number analysis from 5 × 100 μl of samples on 96-well plates (Greiner Bio-One, Frickenhausen, Germany), as described by Bosshard et al. (43). Briefly, the DMEM/MEM containing 10% FBS on a 95% confluent cell monolayer was replaced by 100 μl of virus solution and augmented with 200 μl of DMEM/MEM containing 2% FBS. Cytopathogenic units could be seen after incubation times of 14, 4 to 7, and 10 days for HAdV, EV/CV, and ReoV, respectively. The detection limit for all viruses was 102 to 103 MPNCU ml−1.
Phages and bacteria.
Coliphages MS2 (DSMZ 13767) and ΦX174 (DSMZ 4497) and their host Escherichia coli (DSMZ 5695 and DSMZ 13127) were purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany). Coliphage GA was kindly provided by Joan Jofre (University of Barcelona) and was cultured in the same E. coli host as MS2. Coliphage T4 and its E. coli B1 host were kindly provided by Petr Leiman (École Polytechnique Fédérale de Lausanne, Switzerland). All phages were propagated and purified as described previously (44), except that E. coli B1 was grown in the absence of antibiotics. Stock solutions were kept in virus dilution buffer (VDB) (5 mmol liter−1 NaH2PO4, 10 mmol liter−1 NaCl [pH 7.5]) and were stored at 4°C. The same stocks were used for all the experiments. Infectivity was assessed using the double-agar-layer method and was expressed as the PFU per milliliter (45). The detection limit for all phages was 300 PFU ml−1.
Chemicals.
Sodium carbonate (Na2CO3; Fluka), sodium bicarbonate (NaHCO3; Acros), ammonium chloride (NH4Cl; Acros), potassium chloride (KCl; Acros), sodium chloride (NaCl; Acros), and sodium phosphate (NaH2PO4; Acros) were used to make the experimental solutions described below.
Experimental solutions.
Virus inactivation was assessed over a range of temperatures, pH values, and NH3(aq) activities ({NH3(aq)}). Temperature, pH, and {NH3(aq)} were chosen within an environmentally relevant range, as detailed in Table 1. pH 8 and 20°C (pH 8/20°C) represents the baseline conditions from which pH and temperature effects were assessed. For all pH/temperature (T) conditions, inactivation was quantified at approximate {NH3(aq)} of 0, 20, and 40 mmol liter−1. Phosphate carbonate buffer (PCa) was used as the NH3(aq)-free control, whereas ammonium carbonate buffer (AmCa) was used to generate solutions containing NH3(aq). The specific composition of the buffers to attain a given NH3(aq) activity was determined with PHREEQC (version 2.18.00) (46) and a database using the Pitzer approach for calculating the ion activity, as described by Decrey et al. (38). The exact buffer composition and resulting {NH3(aq)} for each experiment can be found in Table S1 in the supplemental material.
Under baseline conditions (pH 8/20°C), the AmCa solutions with {NH3(aq)} of ≈40 mmol liter−1 exhibited a high ionic strength compared to the PCa solution. Therefore, control experiments were conducted to distinguish between the effects of ionic strength and NH3(aq). These control experiments were conducted under baseline conditions in high-ionic-strength PCa (PCaH), where the ionic strength was increased by adding sodium chloride. The ionic strength was assessed by measurement of the electrical conductivity (EC) with a Cond315i conductivity meter and a TetraCon 325 probe (WTW, Weilheim, Germany) (see Table S1 in the supplemental material).
For phages MS2, ΦX174, and T4, additional experiments were conducted at pH 10 and 11/20°C in carbonate buffer, pH 11.5 and 12/20°C in phosphate buffer, and at pH 8/50°C and 60°C in PCa, to assess the influence of extreme pH and temperature, respectively. Potassium carbonate solutions (KCa) with K+ activity similar to NH4+ in AmCa at pH 9 and 20°C were used to assess the effect of monovalent cations on EV inactivation.
Experimental setup.
For MS2, GA, ΦX174, T4, and HAdV, 1 ml of a virus solution containing 107 to 1010 PFU or MPNCU ml−1 in VDB was added to airtight 116-ml glass serum flasks (Infochroma, Zug, Switzerland) containing 114 ml of experimental solution. For the lower-titer ReoV, EV, and CV, 1 ml of virus stock (106 to 1010 MPNCU ml−1) was added to airtight 16-ml glass serum flasks containing 14 ml of experimental buffer. After mixing, a 1-ml sample was taken from each flask with a sterile syringe and filtered through a 0.22-μm-pore-size filter (Millipore). The filtered samples were directly diluted in medium containing 2% FBS (HAdV, ReoV, EV, and CV) or VDB (MS2, GA, ΦX174, and T4) and were stored at 4°C for no more than 6 h prior to enumeration. Each pH, T, and {NH3(aq)} condition was tested in duplicate flasks for all organisms. Phage titers were determined in triplicate from the same flasks, whereas viruses were enumerated once per flask. At the end of each experiments, the pH was measured at experimental temperature, and the NH4+/NH3 concentration was determined by ion chromatography (ICS-3000A, IonPac CS16 column) with electrical conductivity detection (Dionex, Switzerland). From these measurements, {NH3(aq)} was calculated as described by Decrey et al. (38). In order to stop the fast inactivation at high pH, samples were mixed with HCl prior to dilution in VDB as follows: for pH 11.5, 940 μl of sample was amended with 60 μl of HCl (0.1 N), and for pH 12.0, 400 μl of sample was mixed with 600 μl HCl (0.01 N).
Data analysis.
Inactivation kinetics were determined by least-square fit of the data to a first-order model according to the following equation:
where C0 and C (in PFU or MPNCU per milliliter) are the virus concentrations at time zero and time t, respectively, and kobs is the first-order inactivation rate constant (day−1). The data of all replicates were pooled, and the 95% confidence interval of kobs was calculated from the standard error of the slope of the pooled data.
The second-order rate constant for inactivation by NH3(aq) (kNH3 [day−1 liter mol−1]) was determined by the best fit of a linear model according to the following relationship:
where {NH3(aq)} is the actual activity of NH3(aq) (listed in Table S1 in the supplemental material) and kbackground is the first-order inactivation rate constant (day−1) in the absence of NH3(aq).
Inactivation rate constants were compared by means of an analysis of covariance (ANCOVA) using the R software (47), applying a confidence threshold of 95%.
DNA electrophoresis.
Electrophoresis experiments were performed to determine the dsDNA stability in the presence of NH3(aq) at mildly alkaline pH. Due to the high detection limit of the electrophoresis instrument, it was not possible to directly measure dsDNA genomes extracted from viruses. Instead, linearized plasmid DNA was used as our model dsDNA. Specifically, a concentrated stock of plasmid pBSK 22 (2,985 bp) was linearized with SpeI (lysis step: CutSmart buffer for 1 h at 37°C enzyme deactivation, 20 min at 80°C; NEB reference no. R0133) and dephosphorylated with shrimp alkaline phosphatase (rSAP) (lysis step: CutSmart buffer for 30 min at 37°C; enzyme deactivation, 5 min at 65°C; NEB reference no. M0371). A 50-μl volume of 4 μg liter−1 plasmid solution was spiked into 10-ml serum flasks (Infochroma) containing VDB or AmCa (pH 9; {NH3(aq)}, 40 mmol liter−1) at 20°C. Samples were periodically collected and stored at −20°C until subjected to electrophoresis. Electrophoresis was carried out with an Agilent DNA high-sensitivity kit and a 2100 Bioanalyzer (Agilent Technologies, Inc.), according to the manufacturer's instructions.
RESULTS
Influence of NH3(aq) on virus inactivation kinetics.
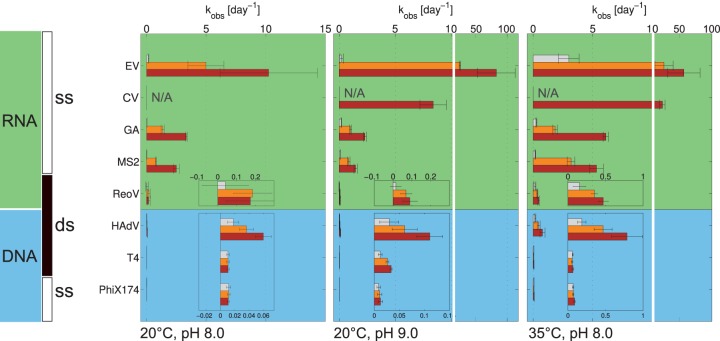
All viruses studied exhibited a loss of infectivity with time, and inactivation kinetics generally demonstrated a first-order model (see Fig. S1 and S2 in the supplemental material). Only for ReoV was poor adherence to first-order kinetics observed, in particular at pH 8/20°C, as well as at pH 9/20°C in PCa. Figure 1 shows an overview of the corresponding first-order inactivation rate constants at pH 8 and 9 and at temperatures of 20 and 35°C. For each virus, the inactivation rate constant in ammonia-free PCa is shown, along with that in solutions with {NH3(aq)} of approximately 20 and 40 mmol liter−1. In PCa, the kobs of the different viruses were all within one order of magnitude (for exact values, see Tables S2 and S3 in the supplemental material). In the presence of NH3(aq), however, noticeable differences in inactivation were observed between the different viruses, which seemed to be correlated with genome type (Fig. 1). For ssRNA viruses, the presence of NH3(aq) led to a substantially faster inactivation, resulting in a 100- to 1,000-fold greater kobs than that of viruses with other genome types.
FIG 1.
Comparison of kobs values for all viruses tested at pH 8/20°C, pH 9/20°C, and pH 8/35°C. The gray, orange, and brown bars correspond to intended {NH3(aq)} of approximately 0, 20, and 40 mmol liter−1, respectively. The measured {NH3(aq)} can be found in Table S1 in the supplemental material. The error bars depict the 95% confidence intervals associated with kobs values. N/A, not available.
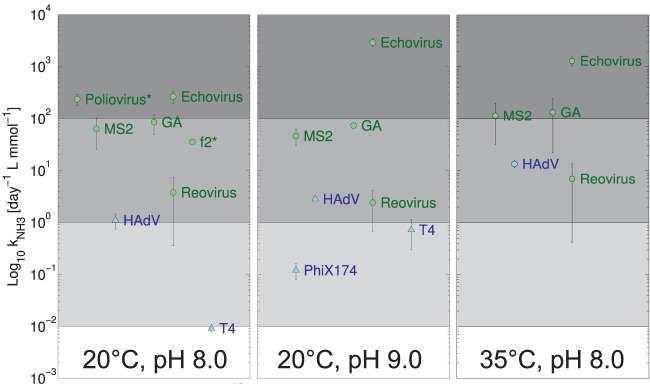
The differing susceptibilities of viruses to NH3(aq) as a function of genome type are further illustrated in Fig. 2, which shows the kNH3 for the different viruses obtained at two pH values and temperatures. For ssRNA viruses, the kNH3 was 10- to 1,000-fold higher than for viruses with other genome types under all conditions studied (for exact values of kNH3, see Table S4 in the supplemental material). The magnitude of kNH3 reflects a virus' susceptibility to inactivation by NH3(aq). The large kNH3 of ssRNA viruses thus indicates that NH3(aq) affects ssRNA viruses more drastically than other viruses. Although kNH3 was lower for DNA viruses, it was still significant for HAdV under all conditions tested and for ΦX174 and T4 at pH 9.
FIG 2.
Dependence of kNH3 on temperature and pH. At pH 8 and 20°C, additional data for f2 and poliovirus (indicated by *) were determined from the work of Burge et al. (57). The error bars depict the 95% confidence interval associated with kNH3 values. Data points associated with the inactivation of ΦX174 and T4 at pH 8.0 and temperatures of 20 and 35°C that were not significantly different from zero were omitted (see Table S4 in the supplemental material).
Differences in inactivation behavior were also observed within a single genome type, but they were small compared to the differences between genome types. For example, at 40 mM NH3(aq), the differences in inactivation rate constants between genome types varied by a factor of 5 to 8,200 between ssRNA and dsDNA viruses, whereas among dsDNA viruses, inactivation varied by a factor of 3 to 10 and among ssRNA viruses by a factor of 1 to 50. Among the four ssRNA viruses studied, MS2 and GA showed significantly different kobs (P < 0.05) under all conditions tested, although they were of the same order of magnitude and exhibited the same trends upon changes in the solution conditions (Fig. 1; see also Table S2 in the supplemental material). EV exhibited a 4-fold greater kobs than MS2 at pH 8/20°C in AmCa, and this difference was more pronounced when the temperature or pH was increased (Fig. 1). CV was also more readily inactivated than MS2 and GA, although it was more persistent than EV under all conditions tested.
Effect of other solution conditions on inactivation. (i) pH.
The effect of pH on kobs can be assessed by comparing the left and middle panels of Fig. 1. In the absence of NH3(aq), no relevant increase in the kobs of DNA viruses could be observed when raising the pH from 8 to 9. Among the RNA viruses studied, MS2 and GA exhibited an approximately 5-fold increase in kobs (see Table S2 in the supplemental material). For EV and ReoV, the scatter in the data did not allow us to conclusively establish if the pH shift affected the kobs (see Table S3 in the supplemental material). For three of the viruses studied (MS2, ΦX174, and T4), inactivation was additionally quantified in PCa at pH values higher than 9. As shown in Fig. 3, increasing the pH to 12 resulted in a large effect on kobs independent of the viral genome type.
FIG 3.
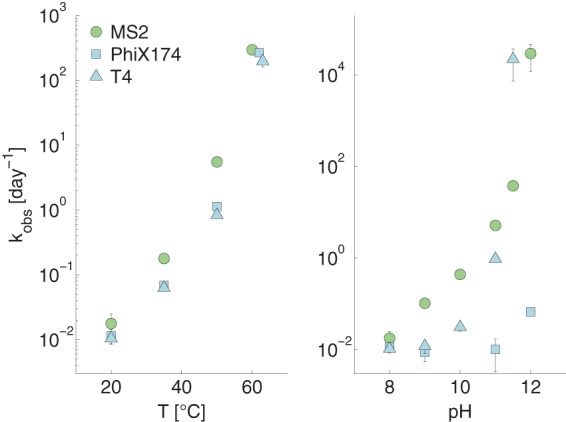
Effect of temperature and pH on kobs of MS2, T4, and ΦX174. The effect of temperature was determined at pH 8 and the effect of pH at 20°C. The buffer composition can be found in Table S1 in the supplemental material. The error bars depict the 95% confidence intervals associated with kobs values.
Furthermore, the solution pH affected the susceptibilities of some viruses to NH3(aq). Most notably, the dsDNA viruses HAdV and T4 exhibited a higher kNH3 at pH 9 than at pH 8 (Fig. 2). Similarly, the kNH3 of EV increased when raising the pH from 8 to 9. This indicates that for these viruses, kNH3 is not a simple second-order rate constant. Instead, there appears to be a synergistic effect between hydroxide and NH3(aq) that promotes the inactivation of these viruses. For the other viruses tested, however, kNH3 was independent of pH.
(ii) Temperature.
A comparison of the left and right panels in Fig. 1 and 2 illustrates the effect of temperature on kobs and kNH3, respectively. As expected, an increase in temperature from 20 to 35°C led to greater kobs and kNH3 for all viruses tested (see Tables S2 to S4 in the supplemental material). However, DNA viruses showed a greater increase in kobs (6- to 8-fold) than RNA viruses (2- to 4-fold), even though the absolute value of kobs remained low for DNA viruses at 35°C, between 0.1 and 1 day−1. Three viruses (MS2, ΦX174, and T4) were additionally tested at temperatures up to 60°C in PCa at pH 8 (Fig. 3), and they exhibited an exponential increase in kobs with increasing temperature. MS2 was more sensitive to inactivation by temperature than the two DNA phages ΦX174 and T4 up to 50°C. At 60°C, however, the gap between the kobs of the three viruses narrowed.
(iii) Ionic strength.
Increasing the ionic strength of PCa to match that of a 40 mM AmCa solution slightly enhanced the inactivation of the ssRNA viruses MS2, GA, and EV (see Tables S2 and S3 in the supplemental material). However, the effect of ionic strength on kobs of ssRNA viruses remained negligible compared to the effect of {NH3(aq)}, confirming that NH3(aq), not ionic strength, is the inactivating agent in AmCa. Among DNA viruses, an increase the ionic strength had either no effect on inactivation (ΦX174) or lowered the inactivation rate constant (HAdV and T4).
(iv) Monovalent cations.
Besides ionic strength, the specific effect of monovalent cations on the inactivation of EV was also tested, as cations, such as K+ or NH4+, were implicated in promoting genome degradation via the induction of nuclease activity of the virus capsid (48–50). At pH 9 and 20°C, no significant increase in kobs could be observed with increasing {NH3(aq)}, whereas a comparable increase in {NH3(aq)} did enhance the kobs (see Fig. S3 in the supplemental material).
Effect of NH3(aq) on genomic material.
To elucidate the observed differences in the susceptibility of ssRNA and dsDNA viruses to NH3(aq) and to pH changes (Fig. 1), the effect of these two solution parameters on genomic material was investigated. Our previous work demonstrated that the ssRNA of MS2 readily degraded over the course of 3 days in the presence of NH3(aq) and upon increasing the pH from 7.5 to 9 (38). In contrast, as shown in Fig. 4, dsDNA remained intact in solutions containing NH3(aq) at pH 9.
FIG 4.
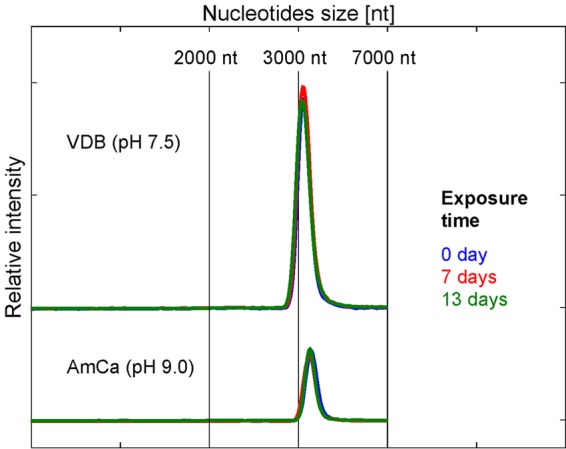
Effect of NH3(aq) on dsDNA integrity. Electrophoresis measurement of linearized plasmid pBSK 22 (2,985 bp) after 0, 7, and 13 days in VDB and AmCa ({NH3(aq)} = 40 mmol liter−1) at 20°C.
DISCUSSION
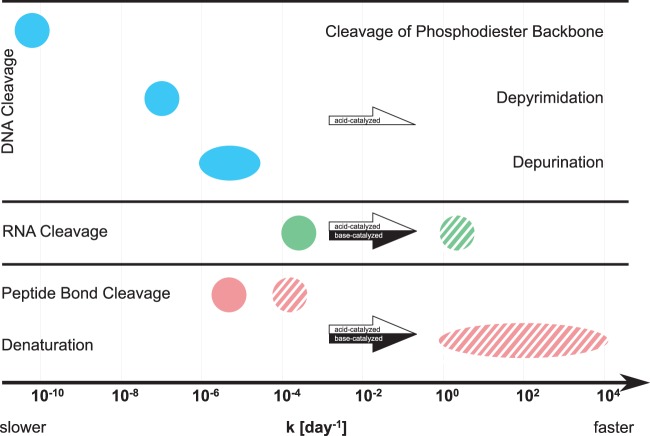
In the following, we aim to explain the source heterogeneity in virus inactivation observed in this study. Specifically, this discussion focuses on the stability of the different viral components under conditions of HEAM storage and processing and the mechanisms contributing to their degradation. For this purpose, we draw on our experimental results, as well as on genome and protein degradation rate constants reported in the literature (compiled in Table S5 in the supplemental material and summarized in Fig. 5). Unfortunately, only few such degradation rates are published for viral genomes or proteins. We therefore included data in our analysis that are not specific to viruses. We acknowledge that reported genome and protein degradation rates vary widely, because they are inherently influenced by the specific structure and composition of the individual biomolecules studied. As such, they cannot be directly applied to any given virus. Despite this variation, the reported rates allowed us to establish a relative order of importance of the different degradation processes considered and thus were useful in establishing a general framework of the processes contributing to virus inactivation in HEAM.
FIG 5.
Summary and comparison of reported DNA, RNA, and protein degradation rate constants. Data refer to degradation at pH 7/37°C (filled symbols) and at pH 11/37°C (striped symbols), and were obtained from the literature (for references and exact numbers, see Table S5 in the supplemental material). The rate for spontaneous cleavage of the phosphodiester backbone corresponds to pH 7/25°C. The arrows indicate the direction of change of the rate constants upon changes in pH conditions (white for acid catalyzed, black for base catalyzed).
Causes underlying the differences in inactivation kinetics between DNA and RNA viruses.
The heterogeneous susceptibilities of viruses with different genome types to inactivation by NH3(aq) suggest that inactivation is associated with the stability of the different genome types in the presence of NH3(aq). The observation that the variation among a single genome type was lower than the variation between genome types (e.g., ssRNA and DNA viruses) supports this hypothesis. Indeed, for ssRNA viruses, it has previously been shown that under the main range of temperatures (20 to 35°C), pH (8 to 9) and {NH3(aq)} conditions tested in this study, infectivity loss can mainly be related to genome degradation (38). In contrast, dsDNA exposed to NH3(aq) under the same conditions did not lose its integrity over the time period studied (Fig. 4). To rationalize this finding, the mechanisms involved in genome degradation by NH3(aq) must be considered.
For ssRNA phage MS2, genome degradation could be attributed to RNA cleavage via a general base-catalyzed transesterification (38), where the bases involved included NH3(aq), OH−, and any other base in solution. In this process, the presence of the 2′-hydroxyl group of ribose renders the 3′,5′-phosphodiester linkages of RNA molecules susceptible to base-catalyzed transesterification (51). This mechanism explains why ssRNA viruses are sensitive to changes in both pH and NH3(aq), since both OH− and NH3(aq) can act as the base catalyzing ssRNA transesterification. In contrast, the absence of the 2′-hydroxyl group in deoxyribose protects DNA from base-catalyzed transesterification (51). DNA cleavage can, however, spontaneously occur through the cleavage of the phosphodiester backbone, in particular at abasic sites (i.e., DNA sites lacking a purine or pyrimidine base) (52). The rate-limiting step in this process is acid catalyzed and involves the depurination, and to a lesser extent the depyrimidation, of a DNA base to form an abasic site. Except in the case of site-specific self-catalyzed depurination (53), the occurrence of which has not been studied for the DNA viruses considered herein, spontaneous depurination, and hence DNA cleavage, proceeds at an approximately 100-fold lower rate at pH 7.0 and 37°C than that with RNA cleavage (Fig. 5; see also Table S5 in the supplemental material). This lower rate of genome degradation explains why DNA viruses are more resistant to inactivation by NH3(aq) than RNA viruses under the conditions of this study (Fig. 1).
Besides genome degradation, virus inactivation can involve viral protein degradation through denaturation or peptide bond cleavage. To our knowledge, the degradation of viral proteins under typical HEAM conditions has not been studied. Furthermore, denaturation and peptide cleavage rates depend on protein composition and structure and can vary by orders of magnitude (see Table S5 in the supplemental material). Nevertheless, while not specific to viruses, reported peptide bond cleavage rates illustrate that this process is likely slower than ssRNA cleavage (Fig. 5). Similarly, around neutral pH, protein denaturation is not a relevant process, because the protein folding rate is higher than the corresponding unfolding rate (see database produced by Bogatyreva et al. [54]). As a result, and as was established in Decrey et al. (38), protein modifications do not contribute significantly to the inactivation of ssRNA viruses under typical conditions of HEAM storage.
In the case of DNA viruses, in contrast, under conditions close to those investigated herein (pH 7.0 and 37°C), reported genome cleavage rates are similar to reported protein degradation rates (Fig. 5; see also Table S5 in the supplemental material). Thus, the inactivation of DNA viruses may involve both genome and protein destabilization under neutral pH conditions. However, the exact mechanism by which mildly alkaline conditions, and in particular NH3(aq), promote DNA virus inactivation is currently not understood and warrants further investigation.
Causes underlying differences in inactivation among RNA viruses.
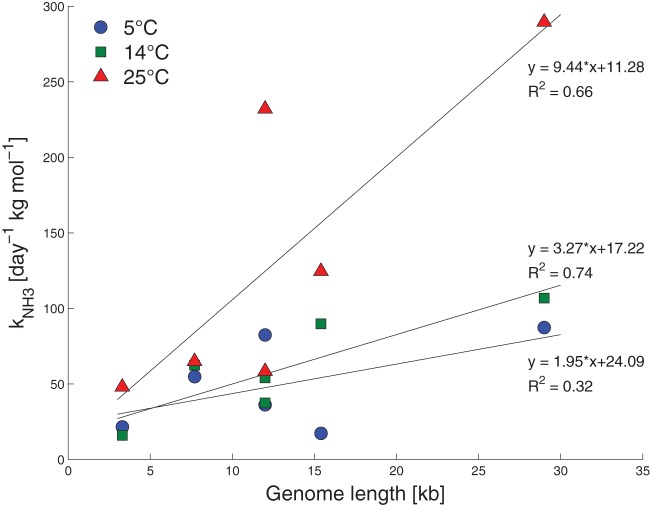
Differences in inactivation kinetics were not only observed between DNA and RNA viruses but also among RNA viruses (Fig. 1 and 2). While these differences were small compared to those between ssRNA and DNA viruses, they were nevertheless statistically significant. A likely cause for this observation is the complex structure of ssRNA. Any ssRNA forms higher-ordered structures through base pairing and tertiary interactions (55). Among these structures, single-stranded regions are more likely to adopt in-line conformations, which are susceptible to cleavage by base-catalyzed transesterification (56). In comparison, highly structured and folded regions are locked into positions that are less accessible to attack by a base and therefore less likely to be cleaved. Thus, heterogeneity among ssRNA virus toward inactivation under the conditions of this study may be explained by the differences in sequence and length of the genome, which further determine its structure. Specifically, the greater length of the EV and CV genomes (7,440 bases) compared to that of MS2 and GA (3,570 bases) may be in part responsible for their more rapid inactivation, as a longer genome implies a higher probability to form an in-line structure susceptible to cleavage. The differences in inactivation kinetics between short ssRNA genomes (MS2 and GA) and long ssRNA genomes (EV and CV) are consistent with reports by others. At pH 8/20°C, Burge et al. (57) observed similar differences between phage f2 and poliovirus (Fig. 2). Furthermore, the susceptibility of ssRNA viruses to NH3(aq) during the disinfection of hatchery waste was shown to be positively correlated with viral genome length (Fig. 6) (17). The genome length of EV, however, fails to explain why its sensitivity to NH3(aq) (and change in kNH3) increases so drastically compared to MS2, GA, and CV upon increasing the pH from 8 to 9 or upon raising the temperature from 20 to 35°C (Fig. 1 and 2). These strong increases in kNH3 imply that additional factors, possibly linked to specific capsid features, promote the virucidal action of NH3(aq) on EV. As discussed by Decrey et al. (38), monovalent cations, such as NH4+ or K+, have been postulated to enhance endonuclease activity exerted by the viral capsid, causing additional genome degradation. However, this process could be confirmed neither for MS2 (38) nor for EV, as K+ failed to promote inactivation (see Fig. S3 in the supplemental material). The reason for the great sensitivity of EV toward NH3(aq) thus remains unknown.
FIG 6.
kNH3 for ssRNA viruses as a function of the genome length at different temperatures. Values for kNH3 were calculated based on the kobs reported by Emmoth et al. (17). The following genome lengths were assumed: MS2, 3.6 kb; feline calicivirus, 7.7 kb; avian influenza virus (H7N1 and H5N3), 12.0 kb; bovine parainfluenza virus type 3, 15.4 kb; and feline coronavirus, 29.0 kb. Note that the pH was not constant for given temperature conditions, and kNH3 was determined from solutions with increasing pH (from 8.0 to 9.7). Furthermore, feline calicivirus, avian influenza virus, and bovine parainfluenza virus type 3 are enveloped viruses. Note that the concentration of NH3 was reported in moles per kilogram of treated hatchery waste.
Unlike DNA viruses, which did not exhibit significant differences in inactivation whether they were single or double stranded, the dsRNA ReoV exhibited higher resistance against NH3(aq) inactivation than ssRNA viruses. Similar trends were reported by others, including Ward and Ashley (19) and Makaya et al. (58), who also observed that ReoV was more resistant than other ssRNA viruses (polio-, coxsackie-, and echovirus) in anaerobically digested sludge, and bovine rotavirus A was more resistant than murine norovirus in stored urine, respectively. A likely explanation for the resistance of dsRNA to NH3(aq) lies in its configuration. Usher (59) suggested that the 5′-oxygen of any internal nucleotide unit of a RNA double helix is unlikely to take on an in-line conformation conducive to cleavage by base-catalyzed transesterification (56). This was later experimentally confirmed by observations of single-stranded RNA, in which folded domains with double-stranded configuration were more resistant to cleavage than single-stranded regions (60–62). Finally, Burge et al. (57) suggested that cleavage of double-stranded genomes is slower because the double strands would require two chains to rupture to be cleaved.
No relevant effect of strand conformation could be observed in the case of DNA viruses, likely because the cleavage process was very slow compared to that of RNA viruses.
Inactivation mechanisms under extreme pH and temperature conditions.
One way to enhance virus inactivation in HEAM is to push the system toward even harsher conditions of pH and temperature than those naturally encountered during storage. Under such extreme conditions, the inactivation kinetics of ssRNA and DNA viruses no longer exhibited major differences, with the notable exception of ΦX174 at pH 12 (Fig. 3). It was also shown that in lime-stabilized biosolids at pH 11.5 to 12.0 and temperatures of 4 and 28°C, differences in inactivation were rather low for MS2, reovirus, and hepatitis A (29) and for MS2, rotavirus, and adenovirus (63). A shift to more-alkaline conditions enhances RNA and peptide bond cleavage, as well as protein denaturation, which are base-catalyzed processes (64–69), but not the rate-limiting step of DNA cleavage, depurination, which is an acid-catalyzed process (70) (Fig. 5). Thus, when increasing the pH, the rate of protein degradation, in particular denaturation, will be similar to or higher than that of RNA cleavage and much higher than that of DNA cleavage (Fig. 5). Therefore, RNA and DNA virus inactivation should shift from only genome- and genome/protein-mediated inactivation, respectively, to mostly protein-mediated inactivation under more-alkaline conditions. We thus hypothesize that protein denaturation is the main mechanism responsible for the inactivation of all viruses under highly alkaline pH conditions.
While reported genome and protein degradation rates for high temperature were not available, several studies present evidence for protein denaturation as the main inactivation mechanism at temperatures characteristic of thermophilic HEAM digestion. Aitken et al. (71) observed a high activation energy characteristic of protein denaturation for poliovirus inactivation in biosolids under thermophilic anaerobic conditions (49 to 53°C). Nuanualsuwan and Cliver (72) reported that the primary target of poliovirus, hepatitis A virus, and feline calicivirus inactivation at 72°C was the capsid, and that inactivation occurred by conformational change of the viral proteins. Similarly, Romero-Maraccini et al. (73) proposed that rotavirus inactivation at 57°C was linked to a disruption of protein-mediated steps of the early life cycle. Finally, the virucidal mode of action of high temperature and pH may also involve a denaturation of the viral capsid proteins that allows for the release of the genome from the capsid. Genome release was found to occur at a pH between 8 and 9 for an ssRNA insect virus (74) and at a temperature of 45 to 70°C at pH 7.0 for dsDNA phage λ and HK97 (75, 76) and ssRNA poliovirus, bovine enterovirus, and equine rhinitis A virus (77, 78). Specifically, Duda et al. (75) reported a rate constant of around 102 day−1 for the release of the HK97 genome from capsid at 65°C, which is the same order of magnitude than the kobs for phages at 60°C in this study (Fig. 3; see also Table S2 in the supplemental material).
Whether protein degradation causes inactivation through loss of host attachment, genome release, or a combination of these viral functions remains unclear and is likely virus dependent. However, it can be concluded that inactivation under extreme conditions appears to be protein related and therefore independent of genome type. While our data as well as those of other studies (34, 79) thus showed a reduction in the heterogeneity among viruses under extreme conditions, other works have demonstrated that some heterogeneity in virus inactivation kinetics is conserved even under high-pH and -temperature conditions (29, 57, 63, 80, 81). Very resistant viruses include phage λ(82) and ΦX174 (this study) for high pH and parvovirus and Salmonella phage 28B for high temperature (30, 83–86). The reasons for these resistant behaviors remain unknown.
Implication of this study for virus inactivation in HEAM.
This work suggests that the heterogeneity in virus inactivation kinetics in HEAM is correlated with genome type in neutral to mildly alkaline conditions and at temperatures below 50°C. Under more-extreme conditions of pH and temperature, however, inactivation is associated with a loss of protein structure and stability. Thus, for the subset of viruses studied herein, virus genome type is of less importance during alkaline stabilization or thermophilic digestion, whereas it is a key parameter in determining virus fate during HEAM storage or mesophilic digestion. While our results remain to be confirmed for additional viruses, we suggest, based on the basic genome and protein degradation mechanisms considered herein, that DNA and dsRNA viruses, e.g., adenovirus and rotavirus, respectively, are the most persistent during storage or mesophilic digestion. Consistent with this notion, a recent survey of urine storage tanks in Durban (South Africa) revealed that JC polyomavirus (dsDNA), adenovirus, and rotavirus were frequently detected, whereas ssRNA virus, such as norovirus, hepatitis A virus, astrovirus, and enterovirus, were only rarely found (87).
The kinetic trends observed in this study were generally consistent with studies using real matrices (19, 26, 30, 34–36, 58), although some works reported contradictory results (25, 27, 32, 37). This may be due to the high complexity of real matrices compared to the well-controlled solutions used in this study. Real matrices, like sludge or urine, however, may contain additional inactivating factors than the ones considered herein. For example, the presence of metal ions may enhance inactivation, especially in the case of ssRNA virus, by accelerating RNA base-catalyzed transesterification (66). Detergents were also shown to reduce and increase the inactivation of enterovirus and reovirus, respectively (88). In the case of sludge, viruses may adsorb to the solid fraction and thereby be protected from the inactivating agents present in the bulk (89). Furthermore, the water content of sludge can be an important parameter, as it determines the mobility and concentration of inactivating or protective agents, and water evaporation itself can lead to virus inactivation during the dewatering process (90–92). Finally, microbial or related enzyme activity may also enhance inactivation during sludge digestion (93–96).
The conclusions of this study were drawn from experiments with naked viruses, which constitute the main fraction of enteric viruses. However, enveloped viruses, such as coronaviruses, can also be excreted in HEAM (97), and they are expected to behave differently due to the instability of the lipid bilayer under alkaline conditions. It was observed that phospholipids were cleaved at a rate of ∼1 × 10−2 day−1 at 40°C and pH 9 (98), which is close to the RNA cleavage rate constant for similar conditions (1.6 × 10−2 day−1 at pH 9 and 37°C) (65). Consequently, Elving et al. (99) showed that MS2 was more persistent than the enveloped dsRNA phage Φ6 and the enveloped ssRNA influenza A virus during composting of manure at 35 to 55°C. Similarly, Ye et al. determined that, independent of genome type, enveloped viruses (ssRNA murine hepatitis virus and dsRNA phage Φ6) were inactivated at higher rates than naked viruses (MS2 and dsDNA phage T3) in wastewater (100). In contrast, Emmoth et al. (17) did not observe a significant influence of the envelope among ssRNA virus inactivated in manure at 4 to 25°C.
Concerns about fecal-oral transmission of viruses responsible for recent outbreaks, such as coronavirus (severe acute respiratory syndrome [SARS]) (101, 102) or filovirus (Ebola virus) (103), may be partially quelled from the results of this study. The facts that they are ssRNA viruses and possess an envelope make them very sensitive to inactivation in human excreta during simple treatment, such as storage or mesophilic digestion, if mildly alkaline conditions with substantial amount of NH3(aq) are reached.
Finally, based on the results of this study and work by others referenced herein, we can issue recommendations regarding the use of indicators for the inactivation of actual human viruses in HEAM. Consistent with the suggestion by Emmoth et al. (17), we propose the use of MS2 as a conservative indicator to assess the inactivation of ssRNA viruses in HEAM treated under mildly alkaline conditions (pH 7.0 to 9.5) and at moderate temperatures (4 to 35°C). The very stable ΦX174 or dsDNA phages (T phages, PRD1, or Salmonella phage 28B) can be used as indicators for persistent viruses. Under higher pH and temperature conditions, however, the use of very resistant viruses, such as ΦX174 or parvovirus, may overestimate treatment requirements.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Swiss National Science Foundation (grant 200021_146829/1) and by EPFL discretionary funding. S.K. was supported by the JSPS Institutional Program for Young Researcher Overseas Visits. This study was conducted within the framework of the Valorization of Urine Nutrients in Africa (VUNA) project.
We thank Simon Meister for experimental assistance and Kai Udert for valuable input.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01106-16.
REFERENCES
- 1.WHO. 2006. Guidelines for the safe use of wastewater, excreta and greywater, vol 4 Excreta and greywater use in agriculture. World Health Organization, Geneva, Switzerland: http://www.who.int/water_sanitation_health/publications/gsuweg4/en/. [Google Scholar]
- 2.Langergraber G, Muellegger E. 2005. Ecological sanitation—a way to solve global sanitation problems? Environ Int 31:433–444. doi: 10.1016/j.envint.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 3.Winker M, Vinneras B, Muskolus A, Arnold U, Clemens J. 2009. Fertiliser products from new sanitation systems: their potential values and risks. Bioresour Technol 100:4090–4096. doi: 10.1016/j.biortech.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 4.Keraita B, Jimenez B, Drechsel P. 2008. Extent and implications of agricultural reuse of untreated, partly treated and diluted wastewater in developing countries. CAB Rev Perspect Agric Vet Sci Nutr Nat Resour 3:15. [Google Scholar]
- 5.Feachem RG, Bradley DJ, Garelick H, Mara DD. 1983. Sanitation and disease: health aspects of excreta and wastewater management. World Bank studies in water supply and sanitation. John Wiley & Sons, Chichester, United Kingdom: http://www-wds.worldbank.org/external/default/WDSContentServer/WDSP/IB/1999/12/23/000178830_98101911180473/Rendered/PDF/multi0page.pdf. [Google Scholar]
- 6.Arthurson V. 2008. Proper sanitization of sewage sludge: a critical issue for a sustainable society. Appl Environ Microbiol 74:5267–5275. doi: 10.1128/AEM.00438-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maurer M, Pronk W, Larsen TA. 2006. Treatment processes for source-separated urine. Water Res 40:3151–3166. doi: 10.1016/j.watres.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 8.Vinnerås B, Holmqvist A, Bagge E, Albihn A, Jonsson H. 2003. The potential for disinfection of separated faecal matter by urea and by peracetic acid for hygienic nutrient recycling. Bioresour Technol 89:155–161. doi: 10.1016/S0960-8524(03)00044-0. [DOI] [PubMed] [Google Scholar]
- 9.Vinnerås B, Hedenkvist M, Nordin A, Wilhelmson A. 2009. Peepoo bag: self-sanitising single use biodegradable toilet. Water Sci Technol 59:1743–1749. doi: 10.2166/wst.2009.184. [DOI] [PubMed] [Google Scholar]
- 10.Strande L, Ronteltap M, Brdjanovic D (ed). 2014. Faecal sludge management: systems approach implementation and operation. IWA Publishing, London, United Kingdom: http://www.unesco-ihe.org/sites/default/files/fsm_book_lr.pdf. [Google Scholar]
- 11.Chen Y, Cheng JJ, Creamer KS. 2008. Inhibition of anaerobic digestion process: a review. Bioresour Technol 99:4044–4064. doi: 10.1016/j.biortech.2007.01.057. [DOI] [PubMed] [Google Scholar]
- 12.Udert KM, Larsen TA, Gujer W. 2006. Fate of major compounds in source-separated urine. Water Sci Technol 54:413–420. doi: 10.2166/wst.2006.921. [DOI] [PubMed] [Google Scholar]
- 13.von Wiren N, Merrick M. 2004. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Top Curr Genet 9:95–120. doi: 10.1007/b95775. [DOI] [Google Scholar]
- 14.Jenkins MB, Bowman DD, Ghiorse WC. 1998. Inactivation of Cryptosporidium parvum oocysts by ammonia. Appl Environ Microbiol 64:784–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pecson BM, Nelson KL. 2005. Inactivation of Ascaris suum eggs by ammonia. Environ Sci Technol 39:7909–7914. doi: 10.1021/es050659a. [DOI] [PubMed] [Google Scholar]
- 16.Himathongkham S, Riemann H, Bahari S, Nuanualsuwan S, Kass P, Cliver DO. 2000. Survival of Salmonella Typhimurium and Escherichia coli O157:H7 in poultry manure and manure slurry at sublethal temperatures. Avian Dis 44:853. doi: 10.2307/1593057. [DOI] [PubMed] [Google Scholar]
- 17.Emmoth E, Ottoson J, Albihn A, Belak S, Vinneras B. 2011. Ammonia disinfection of hatchery waste for elimination of single-stranded RNA viruses. Appl Environ Microbiol 77:3960–3966. doi: 10.1128/AEM.02990-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fidjeland J, Nordin A, Pecson BM, Nelson KL, Vinnerås B. 2015. Modeling the inactivation of Ascaris eggs as a function of ammonia concentration and temperature. Water Res 83:153–160. doi: 10.1016/j.watres.2015.06.030. [DOI] [PubMed] [Google Scholar]
- 19.Ward RL, Ashley CS. 1977. Identification of virucidal agent in wastewater sludge. Appl Environ Microbiol 33:860–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinelle K, Häggström L. 1993. Mechanisms of ammonia and ammonium ion toxicity in animal cells: transport across cell membranes. J Biotechnol 30:339–350. doi: 10.1016/0168-1656(93)90148-G. [DOI] [PubMed] [Google Scholar]
- 21.Sprott G, Patel G. 1986. Ammonia toxicity in pure culture of methanogenic bacteria. Syst Appl Microbiol 7:358–363. doi: 10.1016/S0723-2020(86)80034-0. [DOI] [Google Scholar]
- 22.Kadam PC, Boone DR. 1996. Influence of pH on ammonia accumulation and toxicity in halophilic, methylotrophic methanogens. Appl Environ Microbiol 62:4486–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lay J-J, Li Y-Y, Noike T. 1998. The influence of pH and ammonia concentration on the methane production in high-solids digestion processes. Water Environ Res 70:1075–1082. doi: 10.2175/106143098X123426. [DOI] [Google Scholar]
- 24.Fields BN, Knipe DM, Howley PM. 2007. Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 25.Vinnerås B, Nordin A, Niwagaba C, Nyberg K. 2008. Inactivation of bacteria and viruses in human urine depending on temperature and dilution rate. Water Res 42:4067–4074. doi: 10.1016/j.watres.2008.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Höglund C, Ashbolt N, Stenström TA, Svensson L. 2002. Viral persistence in source-separated human urine. Adv Environ Res 6:265–275. doi: 10.1016/S1093-0191(01)00057-0. [DOI] [Google Scholar]
- 27.Magri ME, Philippi LS, Vinnerås B. 2013. Inactivation of pathogens in feces by desiccation and urea treatment for application in urine-diverting dry toilets. Appl Environ Microbiol 79:2156–2163. doi: 10.1128/AEM.03920-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandran A, Pradhan SK, Heinonen-Tanski H. 2009. Survival of enteric bacteria and coliphage MS2 in pure human urine. J Appl Microbiol 107:1651–1657. doi: 10.1111/j.1365-2672.2009.04353.x. [DOI] [PubMed] [Google Scholar]
- 29.Katz BD, Margolin AB. 2007. Inactivation of hepatitis A HM-175/18f, reovirus T1 Lang and MS2 during alkaline stabilization of human biosolids. J Appl Microbiol 103:2225–2233. doi: 10.1111/j.1365-2672.2007.03463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lund B, Jensen VF, Have P, Ahring B. 1996. Inactivation of virus during anaerobic digestion of manure in laboratory scale biogas reactors. Antonie Van Leeuwenhoek Int 69:25–31. doi: 10.1007/BF00641608. [DOI] [PubMed] [Google Scholar]
- 31.Ottoson JR, Schnürer A, Vinnerås B. 2008. In situ ammonia production as a sanitation agent during anaerobic digestion at mesophilic temperature. Lett Appl Microbiol 46:325–330. doi: 10.1111/j.1472-765X.2007.02317.x. [DOI] [PubMed] [Google Scholar]
- 32.Pesaro F, Sorg I, Metzler A. 1995. In situ inactivation of animal viruses and a coliphage in nonaerated liquid and semiliquid animal wastes. Appl Environ Microbiol 61:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guzmán C, Jofre J, Montemayor M, Lucena F. 2007. Occurrence and levels of indicators and selected pathogens in different sludges and biosolids. J Appl Microbiol 103:2420–2429. doi: 10.1111/j.1365-2672.2007.03487.x. [DOI] [PubMed] [Google Scholar]
- 34.Astals S, Venegas C, Peces M, Jofre J, Lucena F, Mata-Alvarez J. 2012. Balancing hygienization and anaerobic digestion of raw sewage sludge. Water Res 46:6218–6227. doi: 10.1016/j.watres.2012.07.035. [DOI] [PubMed] [Google Scholar]
- 35.Yen-Phi VT, Rechenburg A, Vinneras B, Clemens J, Kistemann T. 2010. Pathogens in septage in Vietnam. Sci Total Environ 408:2050–2053. doi: 10.1016/j.scitotenv.2010.01.030. [DOI] [PubMed] [Google Scholar]
- 36.Nordin A, Niwagaba C, Jönsson H, Vinnerås B. 2013. Pathogen and indicator inactivation in source-separated human urine heated by the sun. J Water Sanit Hyg Dev 3:181. doi: 10.2166/washdev.2013.174. [DOI] [Google Scholar]
- 37.Magri ME, Fidjeland J, Jönsson H, Albihn A, Vinnerås B. 2015. Inactivation of adenovirus, reovirus and bacteriophages in fecal sludge by pH and ammonia. Sci Total Environ 520:213–221. doi: 10.1016/j.scitotenv.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 38.Decrey L, Kazama S, Udert KM, Kohn T. 2014. Ammonia as an in situ sanitizer: inactivation kinetics and mechanisms of the ssRNA virus MS2 by NH3. Environ Sci Technol 49:1060–1067. doi: 10.1021/es5044529. [DOI] [PubMed] [Google Scholar]
- 39.Ward RL. 1978. Mechanism of poliovirus inactivation by ammonia. J Virol 26:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roos WH, Ivanovska IL, Evilevitch A, Wuite GJL. 2007. Viral capsids: mechanical characteristics, genome packaging and delivery mechanisms. Cell Mol Life Sci 64:1484–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larsson DSD, Liljas L, van der Spoel D. 2012. Virus capsid dissolution studied by microsecond molecular dynamics simulations. PLoS Comput Biol 8:e1002502. doi: 10.1371/journal.pcbi.1002502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bândea CI. 1983. A new theory on the origin and the nature of viruses. J Theor Biol 105:591–602. doi: 10.1016/0022-5193(83)90221-7. [DOI] [PubMed] [Google Scholar]
- 43.Bosshard F, Armand F, Hamelin R, Kohn T. 2013. Mechanisms of human adenovirus inactivation by sunlight and UVC light as examined by quantitative PCR and quantitative proteomics. Appl Environ Microbiol 79:1325–1332. doi: 10.1128/AEM.03457-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pecson BM, Martin LV, Kohn T. 2009. Quantitative PCR for determining the infectivity of bacteriophage MS2 upon inactivation by heat, UV-B radiation, and singlet oxygen: advantages and limitations of an enzymatic treatment to reduce false-positive results. Appl Environ Microbiol 75:5544–5554. doi: 10.1128/AEM.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohn T, Nelson KL. 2007. Sunlight-mediated inactivation of MS2 coliphage via exogenous oxygen produced by sensitizers in natural waters. Environ Sci Technol 41:192–197. doi: 10.1021/es061716i. [DOI] [PubMed] [Google Scholar]
- 46.Parkhurst DL, Appelo CAJ. 2013. PHREEQC (version 3)—a computer program for speciation, batch-reaction, one-dimensional transport, and inverse geochemical calculations. US Geological Survey, Reston, VA: http://wwwbrr.cr.usgs.gov/projects/GWC_coupled/phreeqc/phreeqc3-html/phreeqc3.htm. [Google Scholar]
- 47.The R Core Team. 2013. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 48.Newman JF, Piatti PG, Gorman BM, Burrage TG, Ryan MD, Flint M, Brown F. 1994. Foot-and-mouth disease virus particles contain replicase protein 3D. Proc Natl Acad Sci U S A 91:733–737. doi: 10.1073/pnas.91.2.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newman J, Brown F. 1997. Foot-and-mouth disease virus and poliovirus particles contain proteins of the replication complex. J Virol 71:7657–7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scodeller EA, Lebendiker MA, Dubra MS, Crespo OA, Basarab O, La Torre JL, Vasquez C. 1984. Inactivation of foot-and-mouth disease virus vaccine strains by activation of virus-associated endonuclease. J Gen Virol 65:1567–1573. [DOI] [PubMed] [Google Scholar]
- 51.Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 52.Gates KS. 2009. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem Res Toxicol 22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amosova O, Coulter R, Fresco JR. 2006. Self-catalyzed site-specific depurination of guanine residues within gene sequences. Proc Natl Acad Sci U S A 103:4392–4397. doi: 10.1073/pnas.0508499103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogatyreva NS, Osypov AA, Ivankov DN. 2009. KineticDB: a database of protein folding kinetics. Nucleic Acids Res 37:D342–D346. doi: 10.1093/nar/gkn696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weeks KM. 2010. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 20:295–304. doi: 10.1016/j.sbi.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soukup GA, Breaker RR. 1999. Relationship between internucleotide linkage geometry and the stability of RNA. RNA 5:1308–1325. doi: 10.1017/S1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burge WD, Cramer WN, Kawata K. 1983. Effect of heat on virus inactivation by ammonia. Appl Environ Microbiol 46:446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Makaya JM, Kaplon J, Fremy C, Barro N, Aho S, Pothier P, Belliot G, Traoré AS. 2014. Norovirus and rotavirus survival in urine collected from a public ecological sanitation system in Ouagadougou, Burkina Faso. Food Environ Virol 7:41–48. [DOI] [PubMed] [Google Scholar]
- 59.Usher DA. 1972. RNA double helix and the evolution of the 3′,5′ linkage. Nature 235:207–208. [DOI] [PubMed] [Google Scholar]
- 60.Ciesolka J, Lorenz S, Erdmann VA. 1992. Different conformational forms of Escherichia coli and rat liver 5S rRNA revealed by Pb(II)-induced hydrolysis. Eur J Biochem 204:583–589. doi: 10.1111/j.1432-1033.1992.tb16671.x. [DOI] [PubMed] [Google Scholar]
- 61.Reynolds MA, Beck TA, Say PB, Schwartz DA, Dwyer BP, Daily WJ, Vaghefi MM, Metzler MD, Klem RE, Arnold LJ. 1996. Antisense oligonucleotides containing an internal, non-nucleotide-based linker promote site-specific cleavage of RNA. Nucleic Acids Res 24:760–765. doi: 10.1093/nar/24.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Podyminogin MA, VIassov VV, Giegé R. 1993. Synthetic RNA-cleaving molecules mimicking ribonuclease A active center. Design and cleavage of tRNA transcripts. Nucleic Acids Res 21:5950–5956. doi: 10.1093/nar/21.25.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hansen JJ, Warden PS, Margolin AB. 2007. Inactivation of adenovirus type 5, rotavirus WA and male specific coliphage (MS2) in biosolids by lime stabilization. Int J Environ Res Public Health 4:61–67. doi: 10.3390/ijerph2007010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smith RM, Hansen DE. 1998. The pH-rate profile for the hydrolysis of a peptide bond. J Am Chem Soc 120:8910–8913. doi: 10.1021/ja9804565. [DOI] [Google Scholar]
- 65.Li Y, Breaker RR. 1999. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc 121:5364–5372. doi: 10.1021/ja990592p. [DOI] [Google Scholar]
- 66.Corona-Martínez DO, Gomez-Tagle P, Yatsimirsky AK. 2012. Electrophilic assistance to the cleavage of an RNA model phosphodiester via specific and general base-catalyzed mechanisms. J Org Chem 77:9110–9119. doi: 10.1021/jo301649u. [DOI] [PubMed] [Google Scholar]
- 67.Xia F, Bronowska AK, Cheng S, Gräter F. 2011. Base-catalyzed peptide hydrolysis is insensitive to mechanical stress. J Phys Chem B 115:10126–10132. doi: 10.1021/jp202162r. [DOI] [PubMed] [Google Scholar]
- 68.Levy M, Benaglia AE. 1950. The influence of temperature and pH upon the rate of denaturation of ricin. J Biol Chem 186:829–847. [PubMed] [Google Scholar]
- 69.Grinberg VY, Burova TV, Haertlé T, Tolstoguzov VB. 2000. Interpretation of DSC data on protein denaturation complicated by kinetic and irreversible effects. J Biotechnol 79:269–280. doi: 10.1016/S0168-1656(00)00243-1. [DOI] [PubMed] [Google Scholar]
- 70.An R, Jia Y, Wan B, Zhang Y, Dong P, Li J, Liang X. 2014. Non-enzymatic depurination of nucleic acids: factors and mechanisms. PLoS One 9:e115950. doi: 10.1371/journal.pone.0115950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aitken MD, Sobsey MD, Blauth KE, Shehee M, Crunk PL, Walters GW. 2005. Inactivation of Ascaris suum and poliovirus in biosolids under thermophilic anaerobic digestion conditions. Environ Sci Technol 39:5804–5809. doi: 10.1021/es048004h. [DOI] [PubMed] [Google Scholar]
- 72.Nuanualsuwan S, Cliver DO. 2003. Capsid functions of inactivated human picornaviruses and feline calicivirus. Appl Environ Microbiol 69:350–357. doi: 10.1128/AEM.69.1.350-357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Romero-Maraccini OC, Shisler JL, Nguyen TH. 2015. Solar and temperature treatments affect the ability of human rotavirus Wa to bind to host cells and synthesize viral RNA. Appl Environ Microbiol 81:4090–4097. doi: 10.1128/AEM.00027-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Snijder J, Uetrecht C, Rose RJ, Sanchez-Eugenia R, Marti GA, Agirre J, Guérin DMA, Wuite GJL, Heck AJR, Roos WH. 2013. Probing the biophysical interplay between a viral genome and its capsid. Nat Chem 5:502–509. doi: 10.1038/nchem.1627. [DOI] [PubMed] [Google Scholar]
- 75.Duda RL, Ross PD, Cheng N, Firek BA, Hendrix RW, Conway JF, Steven AC. 2009. Structure and energetics of encapsidated DNA in bacteriophage HK97 studied by scanning calorimetry and cryo-electron microscopy. J Mol Biol 391:471–483. doi: 10.1016/j.jmb.2009.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qiu X. 2012. Heat induced capsid disassembly and DNA release of bacteriophage λ. PLoS One 7:e39793. doi: 10.1371/journal.pone.0039793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Larkin EP, Fassolitis AC. 1979. Viral heat resistance and infectious ribonucleic acid. Appl Environ Microbiol 38:650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walter TS, Ren J, Tuthill TJ, Rowlands DJ, Stuart DI, Fry EE. 2012. A plate-based high-throughput assay for virus stability and vaccine formulation. J Virol Methods 185:166–170. doi: 10.1016/j.jviromet.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tuladhar E, Bouwknegt M, Zwietering MH, Koopmans M, Duizer E. 2012. Thermal stability of structurally different viruses with proven or potential relevance to food safety. J Appl Microbiol 112:1050–1057. doi: 10.1111/j.1365-2672.2012.05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bertrand I, Schijven JF, Sánchez G, Wyn-Jones P, Ottoson J, Morin T, Muscillo M, Verani M, Nasser A, de Roda Husman AM, Myrmel M, Sellwood J, Cook N, Gantzer C. 2012. The impact of temperature on the inactivation of enteric viruses in food and water: a review. J Appl Microbiol 112:1059–1074. doi: 10.1111/j.1365-2672.2012.05267.x. [DOI] [PubMed] [Google Scholar]
- 81.Feng YY, Ong SL, Hu JY, Tan XL, Ng WJ. 2003. Effects of pH and temperature on the survival of coliphages MS2 and Qβ. J Ind Microbiol Biotechnol 30:549–552. doi: 10.1007/s10295-003-0080-y. [DOI] [PubMed] [Google Scholar]
- 82.Jepson CD, March JB. 2004. Bacteriophage lambda is a highly stable DNA vaccine delivery vehicle. Vaccine 22:2413–2419. doi: 10.1016/j.vaccine.2003.11.065. [DOI] [PubMed] [Google Scholar]
- 83.Spillmann SK, Traub F, Schwyzer M, Wyler R. 1987. Inactivation of animal viruses during sewage sludge treatment. Appl Environ Microbiol 53:2077–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sahlström L, Bagge E, Emmoth E, Holmqvist A, Danielsson-Tham ML, Albihn A. 2008. A laboratory study of survival of selected microorganisms after heat treatment of biowaste used in biogas plants. Bioresour Technol 99:7859–7865. doi: 10.1016/j.biortech.2007.09.071. [DOI] [PubMed] [Google Scholar]
- 85.Haas B, Ahl R, Böhm R, Strauch D. 1995. Inactivation of viruses in liquid manure. Rev Sci Tech 14:435–445. [DOI] [PubMed] [Google Scholar]
- 86.Elving J, Vinnerås B, Albihn A, Ottoson JR. 2014. Thermal treatment for pathogen inactivation as a risk mitigation strategy for safe recycling of organic waste in agriculture. J Environ Sci Health B 49:679–689. doi: 10.1080/03601234.2014.922783. [DOI] [PubMed] [Google Scholar]
- 87.Bischel HN, Özel Duygan BD, Strande L, McArdell CS, Udert KM, Kohn T. 2015. Pathogens and pharmaceuticals in source-separated urine in eThekwini, South Africa. Water Res 85:57–65. doi: 10.1016/j.watres.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 88.Ward RL, Ashley CS. 1978. Identification of detergents as components of wastewater sludge that modify the thermal stability of reovirus and enteroviruses. Appl Environ Microbiol 36:889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Templeton MR, Andrews RC, Hofmann R. Particle-associated viruses in water: impacts on disinfection processes. Crit Rev Environ Sci Technol 38:137–164. [Google Scholar]
- 90.Straub TM, Pepper IL, Gerba CP. 1992. Persistence of viruses in desert soils amended with anaerobically digested sewage sludge. Appl Environ Microbiol 58:636–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ward R, Ashley C. 1977. Inactivation of enteric viruses in wastewater sludge through dewatering by evaporation. Appl Environ Microbiol 34:564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ward R, Ashley C. 1978. Heat inactivation of enteric viruses in dewatered wastewater sludge. Appl Environ Microbiol 36:898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Knowlton DR, Ward RL. 1987. Characterization of virucidal agents in activated sludge. Appl Environ Microbiol 53:621–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cliver DO, Herrmann JE. 1972. Proteolytic and microbial inactivation of enteroviruses. Water Res 6:797–805. doi: 10.1016/0043-1354(72)90032-2. [DOI] [Google Scholar]
- 95.Deng MY, Cliver DO. 1995. Antiviral effects of bacteria isolated from manure. Microb Ecol 30:43–54. [DOI] [PubMed] [Google Scholar]
- 96.Ward RL. 1982. Evidence that microorganisms cause inactivation of viruses inactivated sludge. Appl Environ Microbiol 43:1221–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wigginton KR, Ye Y, Ellenberg RM. 2015. Emerging investigators series: the source and fate of pandemic viruses in the urban water cycle. Environ Sci Water Res Technol 1:735–746. doi: 10.1039/C5EW00125K. [DOI] [Google Scholar]
- 98.Grit M, Crommelin DJ. 1993. Chemical stability of liposomes: implications for their physical stability. Chem Phys Lipids 64:3–18. doi: 10.1016/0009-3084(93)90053-6. [DOI] [PubMed] [Google Scholar]
- 99.Elving J, Emmoth E, Albihn A, Vinnerås B, Ottoson J. 2012. Composting for avian influenza virus elimination. Appl Environ Microbiol 78:3280–3285. doi: 10.1128/AEM.07947-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ye Y, Ellenberg RM, Graham KE, Wigginton KR. 2016. Survivability, partitioning, and recovery of enveloped viruses in untreated municipal wastewater. Environ Sci Technol 50:5077–5085. doi: 10.1021/acs.est.6b00876. [DOI] [PubMed] [Google Scholar]
- 101.Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RAM, Berger A, Burguière A-M, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra J-C, Müller S, Rickerts V, Stürmer M, Vieth S, Klenk H-D, Osterhaus ADME, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348:1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 102.Peiris JSM, Chu CM, Cheng VCC, Chan KS, Hung IFN, Poon LLM, Law KI, Tang BSF, Hon TYW, Chan CS, Chan KH, Ng JSC, Zheng BJ, Ng WL, Lai RWM, Guan Y, Yuen KY, HKU/UCH SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bibby K, Casson LW, Stachler E, Haas CN. 2015. Ebola virus persistence in the environment: state of the knowledge and research needs. Environ Sci Technol Lett 2:2–6. doi: 10.1021/ez5003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kraft L. 2010. Final sampling report for products from double-chamber UDDTs (faeces and urine). EU-SIDA GTZ EcoSan promotion project. Deutsche Gesellschaft für Technische Zusammenarbeit (GTZ) GmbH, Eschborn, Germany. http://www.susana.org/_resources/documents/default/2-1026-en-eu-sida-gtz-ecosan-promotion-project-final-report-2010.pdf.
- 105.Nordin A, Nyberg K, Vinneras B. 2009. Inactivation of Ascaris eggs in source-separated urine and feces by ammonia at ambient temperatures. Appl Environ Microbiol 75:662–667. doi: 10.1128/AEM.01250-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fidjeland J, Magri ME, Jönsson H, Albihn A, Vinnerås B. 2013. The potential for self-sanitisation of faecal sludge by intrinsic ammonia. Water Res 47:6014–6023. doi: 10.1016/j.watres.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 107.Hafner SD, Bisogni JJ Jr, Jewell WJ. 2006. Measurement of un-ionized ammonia in complex mixtures. Environ Sci Technol 40:1597–1602. doi: 10.1021/es051638j. [DOI] [PubMed] [Google Scholar]
- 108.Hafner SD, Bisogni JJ Jr. 2009. Modeling of ammonia speciation in anaerobic digesters. Water Res 43:4105–4114. doi: 10.1016/j.watres.2009.05.044. [DOI] [PubMed] [Google Scholar]
- 109.Garcia ML, Angenent LT. 2009. Interaction between temperature and ammonia in mesophilic digesters for animal waste treatment. Water Res 43:2373–2382. doi: 10.1016/j.watres.2009.02.036. [DOI] [PubMed] [Google Scholar]
- 110.Niu Q, Qiao W, Qiang H, Hojo T, Li Y-Y. 2013. Mesophilic methane fermentation of chicken manure at a wide range of ammonia concentration: stability, inhibition and recovery. Bioresour Technol 137:358–367. doi: 10.1016/j.biortech.2013.03.080. [DOI] [PubMed] [Google Scholar]
- 111.Bolzonella D, Cavinato C, Fatone F, Pavan P, Cecchi F. 2012. High rate mesophilic, thermophilic, and temperature phased anaerobic digestion of waste activated sludge: a pilot scale study. Waste Manag 32:1196–1201. doi: 10.1016/j.wasman.2012.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.