Abstract
Metastasis is responsible for most cancer-related deaths, and, among common tumor types, melanoma is one with great potential to metastasize. Here we study the contribution of epigenetic changes to the dissemination process by analyzing the changes that occur at the DNA methylation level between primary cancer cells and metastases. We found a hypomethylation event that reactivates a cryptic transcript of the Rab GTPase activating protein TBC1D16 (TBC1D16-47 kDa; referred to hereafter as TBC1D16-47KD) to be a characteristic feature of the metastatic cascade. This short isoform of TBC1D16 exacerbates melanoma growth and metastasis both in vitro and in vivo. By combining immunoprecipitation and mass spectrometry, we identified RAB5C as a new TBC1D16 target and showed that it regulates EGFR in melanoma cells. We also found that epigenetic reactivation of TBC1D16-47KD is associated with poor clinical outcome in melanoma, while conferring greater sensitivity to BRAF and MEK inhibitors.
Metastases account for 90% of cancer-associated deaths1, so it is critical to understand the mechanisms that mediate this process. The plastic nature of the epigenetic landscape of tumoral cells2–4 contributes to cancer progression and metastasis. In this regard, transcriptional silencing associated with promoter CpG island hypermethylation5–8 has been linked to tumor invasion. However, we know little about the epigenetic events involved in melanoma metastasis9,10, a deficit that is of particular concern because cutaneous melanoma has a growing incidence and mortality11. In the clinical setting, it is necessary to find molecular biomarkers for predicting whether metastatic disease will develop or for determining prognosis; in addition, it is necessary to determine histopathological features such as Breslow stage or the presence of ulceration12,13. Despite the introduction of small-molecule drugs that target kinases, such as BRAF and MEK inhibitors, and immunological treatments14–16, little improvement in the survival of individuals with disseminated disease has been achieved. Thus, there is a need to discover melanoma markers that are able to identify subjects with the most pro-metastatic forms of the disease. Most importantly, it would be optimal to use knowledge of molecular mechanisms of melanoma metastasis to identify therapeutic targets. By using epigenomic approaches, combined with functional in vitro and in vivo data and clinical cohorts of melanoma, we show how the DNA hypomethylation-associated reactivation of the Rab GTPase activating protein TBC1D16 isoform may be such a candidate.
Results
Identification of DNA methylation loci associated with metastasis
To identify DNA methylation-associated changes involved in the formation of metastatic melanoma, we analyzed cell lines derived from the primary tumor and the lymph node metastasis of the same individual. We first selected the primary melanoma cell line IGR39 and its paired metastasis cell line IGR37 (ref. 17). We obtained the DNA methylation profile of each sample using the Infinium HumanMethylation450 (450K) microarray18. We wanted to find genes that participate directly in metastasis, so we also hybridized additional pairs of primary and metastasis cell lines from two other malignancies: colon (SW480 (primary) and SW620 (metastasis)) and breast (MDA-MB-468PT (primary) and MDA-MB-468LN (metastasis)).
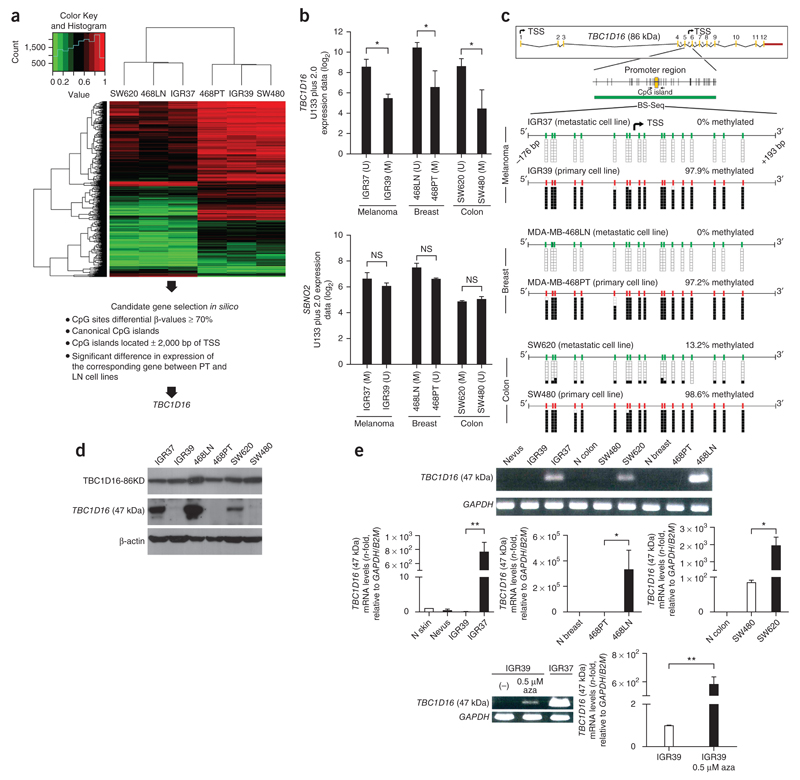
We analyzed 482,422 CpGs in the three paired cancer cell lines, in which we explored the probes with low dispersion (s.d. < 0.1) within the primary and metastatic groups (to identify common CpG methylation events involved in the metastasis in the three tumor types) and high dispersion between the groups (to identify common CpG sites that discriminated primary from metastases sites for all three tumor types). Using this approach, we obtained 2,620 CpG probes that distinguished between cell lines derived from primary or metastatic sites independently of the tumor type (Fig. 1a and Supplementary Data 1). To identify candidate genes with differential methylation, we imposed stringent criteria: we considered only CpG sites with a ≥70% change in CpG methylation level; the CpG had to be in a CpG island (differential methylation outside CpG islands is shown in Supplementary Data 2); the CpG had to be within ± 2,000 bp of the transcription start site; and the differential CpG methylation between primary and metastases had to occur in the three tumor types. Under these conditions, we identified only two candidate genes: TBC1 domain family member 16 (TBC1D16)19,20, which loses CpG methylation in the metastatic cell lines compared with the primary cancer cells; and strawberry notch homolog 2 (SBNO2)21, which gains CpG methylation. We then merged the DNA methylation data with the transcriptional profiles obtained from an Affymetrix GeneChip Human Genome U133 Plus 2.0 microarray hybridization (melanoma) or data available from Gene Expression Omnibus (breast and colon). For SBNO2 there was no correlation between methylation and expression (Fig. 1b). However, TBC1D16 was hypermethylated and downregulated in the primary cancer cell lines and overexpressed and unmethylated in the paired corresponding metastatic cells (Fig. 1b). Thus, the DNA demethylation-associated re-expression of TBC1D16 in the metastatic cells became our main focus of interest.
Figure 1.
DNA hypomethylation-associated transcriptional activation of a TBC1D16 cryptic isoform in metastatic cancer cells. (a) DNA methylation heatmap clustering of 2,620 CpG sites showing DNA methylation events that distinguish primary (IGR39, MDA-MB-468PT and SW480) or metastatic-derived cell lines (IGR37, MDA-MB-468LN and SW620). (b) Representation of the correlation between DNA methylation microarray data and gene expression microarray values for TBC1D16 and SBNO2. Expression is represented as the average intensity of three probes for TBC1D16 and two probes for SBNO2. (c) Genomic structure of TBC1D16. Transcription start sites (TSSs) of the isoforms are denoted by small black arrows. Locations of CpG islands (green bars) and designed bisulfite sequencing primers (black horizontal arrows) are shown. Bisulfite genomic sequencing was carried out in eight individual clones. The presence of a methylated or unmethylated cytosine is indicated by a black or white square, respectively. (d) Protein expression levels for the TBC1D16-47KD and TBC1D16-86KD isoforms analyzed by western blot in the paired primary/metastasis cancer cell lines. (e) Conventional (top) and quantitative RT-PCR (middle) analyses of TBC1D16 (47 kDa) expression in paired primary and metastasis cancer cell lines and normal tissues. Bottom, reactivation of the TBC1D16 (47 kDa) transcript upon use of the DNA demethylating agent 5-aza-2′-deoxycytidine (aza). NS, nonsignificant; *P < 0.05; **P < 0.01, using Student’s t-test. Error bars show means ± s.d.
TBC1D16-47KD epigenetic activation in metastatic melanoma
The loss of methylation from the metastatic cells occurred in the CpGs located around the transcription start site of two short isoforms of TBC1D16 (TBC1D16-45KD and TBC1D16-47KD) (Fig. 1c), whereas the CpGs surrounding the long isoform (TBC1D16-86KD) remained unmethylated in all six cell lines (Supplementary Fig. 1). Western blot analyses confirmed that TBC1D16 demethylation was associated with reactivation of TBC1D16-47KD expression (the short isoform recognized by the only available antibody), whereas TBC1D16-86KD expression remained unaltered in the three paired cancer cell lines (Fig. 1d). RT-PCR assays designed to recognize TBC1D16-47KD (Fig. 1e) and TBC1D16-45KD (Supplementary Fig. 2a,b) confirmed the described pattern and a DNA demethylating agent restored their expression, without affecting TBC1D16-86KD (Supplementary Fig. 3a). We focused our subsequent studies on TBC1D16-47KD because it is readily detected and shares high homology with the other short isoform (TBC1D16-45KD, 98% homologous), which behaves in a similar fashion in all performed assays (Supplementary Fig. 2)
We established a mechanistic link between TBC1D16-47KD hypomethylation and transcript reactivation by studying the microphthalmia-associated transcription factor (MITF), a master regulator of melanocyte development and a recognized oncogene14. The TBC1D16-47KD CpG island contains a MITF E-box binding site (Supplementary Fig. 4a). Chromatin immunoprecipitation revealed that MITF binds to the unmethylated CpG island in IGR37 cells, but not to the methylated sequence of IGR39 (Supplementary Fig. 4b). Reporter assays demonstrated that the E-box of TBC1D16-47KD was required for its regulation by MITF (Supplementary Fig. 4c). Importantly, depletion of MITF mediated by small interfering RNA in unmethylated cells downregulated TBC1D16-47KD, as well as TBC1D16-86KD, which contains two E-boxes in its promoter (Supplementary Fig. 4d).
We found TBC1D16-47KD methylation in the normal counterparts of the analyzed malignancies (Supplementary Fig. 5a) and absence or minimal levels of the transcript (Fig. 1e). Data mining of TCGA 450K DNA methylation data confirmed hypermethylation of these CpGs in all normal samples, with two exceptions: central nervous system and embryonic stem cells (Supplementary Fig. 5b). Bisulfite genomic sequencing confirmed the unmethylated status in gray and white brain matter and we detected TBC1D16-47KD expression (Supplementary Fig. 5c,d). Thus, TBC1D16-47KD probably has a physiological role in the embryo and the neuronal lineage, whereas it remains densely methylated and silenced in the remaining tissues.
TBC1D16-47KD promotes melanoma progression
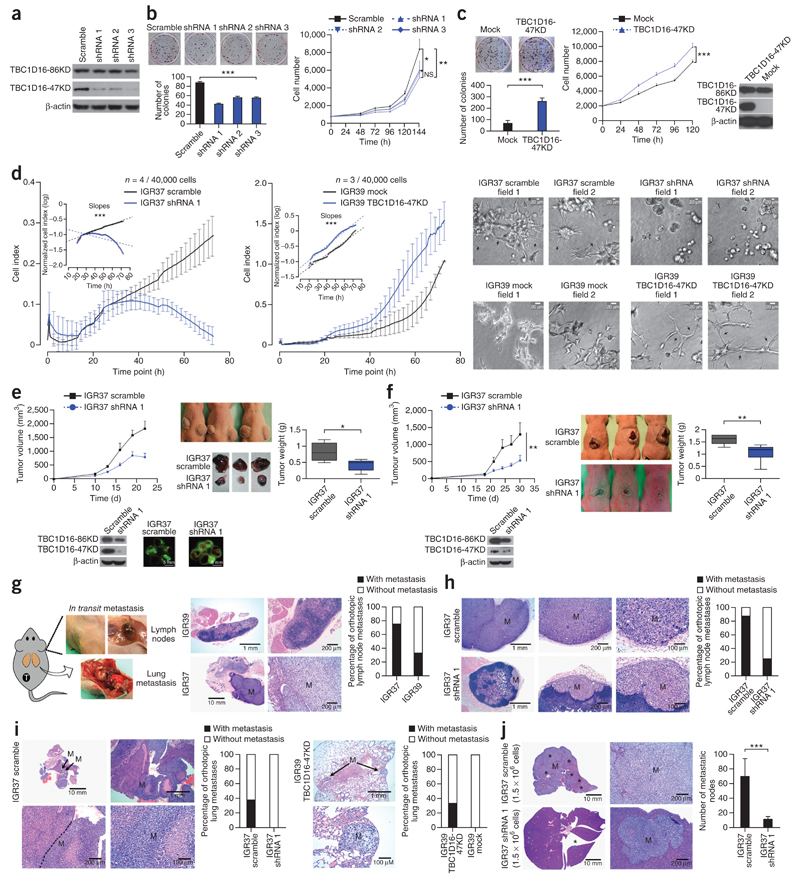
We next pursued cellular approaches to demonstrate that the epigenetic reactivation of this transcript contributed to melanoma progression. We created stable knockdowns of TBC1D16-47KD by the short hairpin RNA (shRNA) method in TBC1D16-expressing and unmethylated melanoma metastasis-derived cells (IGR37). Upon shRNA-mediated depletion of TBC1D16-47KD from IGR37 cells (Fig. 2a and Supplementary Fig. 6a), we observed lower cell proliferation (Fig. 2b). We obtained similar results when we knocked-down TBC1D16-47KD in the metastatic breast and colon cells (Supplementary Fig. 6b). Conversely, ectopic expression of TBC1D16-47KD in IGR39 increased cellular proliferation (Fig. 2c). TBC1D16-47KD transfection did not affect TBC1D16-86KD expression (Supplementary Fig. 3b). shRNA-mediated depletion of TBC1D16-47KD from IGR37 reduced invasion capacity according to XCELLigence real-time cell analysis22 (Fig. 2d). Conversely, TBC1D16-47KD transfection in IGR39 increased invasion capacity (Fig. 2d).
Figure 2.
TBC1D16-47KD enhances melanoma progression in vitro and in vivo. (a) Western blot of TBC1D16-47KD shRNA interference in IGR37. (b) Colony formation and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assays in TBC1D16-47KD shRNA-depleted IGR37 cells. Values derived from two experiments with 12 replicates (mean ± s.d.); significance according to 10,000-permutation t-test with Holm correction. (c) Cellular proliferation upon TBC1D16-47KD ectopic expression in IGR39. (d) Invasion capacity upon shRNA-mediated depletion in IGR37 and TBC1D16-47KD transfection in IGR39 cells. (e) Top, tumor volume in subcutaneous implanted shRNA TBC1D16-47KD-depleted IGR37. Bottom, TBC1D16-47KD and TBC1D16-86KD protein levels (22 d; n = 5 for group). Right, illustrative samples obtained at the end point and mean ± s.d. weights. (f) Left, orthotopic volumes from TBC1D16-47KD shRNA-depleted IGR37 cells. Bottom, TBC1D16-47KD and TBC1D16-86KD protein levels (30 d, n = 8 for group). Right, illustrative endpoint tumor samples and mean ± s.d. tumor weights. (g) Left, model of orthotopic melanoma implantation for studying metastasis. Right, H&E staining of lymph nodes showing metastases (M). Percentage of metastatic lymph nodes in orthotopics derived from IGR37 (eight mice) and IGR39 (six mice). (h) H&E staining. Percentage of metastatic lymph nodes upon TBC1D16-47KD shRNA depletion in IGR37-derived orthotopics (eight mice). (i) H&E staining of lungs showing metastases. Percentage of metastases upon TBC1D16-47KD depletion in IGR37-derived orthotopics (eight mice) (left) and in IGR39 (eight mice) tumors with TBC1D16-47KD ectopic expression (right). (j) Number of liver metastases (black stars) upon direct spleen injection of TBC1D16-47KD shRNA-depleted IGR37 cells (ten mice). NS, nonsignificant; *P < 0.05; **P < 0.01, ***P < 0.001; Student’s t-test. Error bars show means ± s.d.
We translated these results to an in vivo setting in athymic nude mice. Tumors originating from TBC1D16-47KD shRNA-downregulated IGR37 cells had smaller volumes and weighed less (Fig. 2e) than those derived from scrambled-shRNA cells. TBC1D16-47KD orthotopic-depleted tumors were also smaller and lighter than the scrambled shRNA-derived tumors (Fig. 2f). Breast and colon cancer cells showed similar results (Supplementary Fig. 6c). TBC1D16-86KD expression was not affected (Supplementary Fig. 6c). We also studied the potential contribution of TBC1D16-47KD to metastasis. After confirming that IGR37 cells had higher metastatic potential than IGR39 (Fig. 2g), we studied the effect of TBC1D16-47KD depletion in IGR37. We found that although 88% scrambled-orthotopic tumors were able to metastasize to the lymph nodes, only 25% of the TBC1D16-47KD orthotopic-depleted tumors had this ability (Fisher’s exact test, P = 0.04) (Fig. 2h). With respect to blood dissemination, none of the eight orthotopic TBC1D16-47KD shRNA-downregulated tumors yielded distant metastasis, whereas there were lung metastases in 38% of the scrambled-orthotopic tumors (Fig. 2i). Conversely, ectopic expression of TBC1D16-47KD in IGR39 increased its metastatic potential (Fig. 2i). We also used an additional model of metastasis: direct spleen injection. We observed that scrambled-shRNA IGR37 cells induced a higher formation of liver metastases than TBC1D16-47KD shRNA-depleted IGR37 cells (Student’s t-test, P < 0.001) (Fig. 2j).
We extended our analysis to an additional paired primary and metastasis melanoma model derived from the same individual: WM115 (primary) and WM266.4 (metastasis). We observed hypomethylation-associated reactivation of TBC1D16-47KD in WM266.4 cells, whereas WM115 showed methylation and silencing (Supplementary Fig. 7a). TBC1D16-47KD ectopic expression in WM115 increased melanoma growth and progression (Supplementary Fig. 7b). Conversely, TBC1D16-47KD depletion by shRNA from WM266.4 cells had the opposite effects (Supplementary Fig. 7c). We also used two additional melanoma cell lines: WM793, derived from a primary melanoma, in which we found methylated and repressed TBC1D16-47KD; and SK-MEL-28, derived from a melanoma metastasis with an unmethylated and expressed TBC1D16-47KD (Supplementary Fig. 8a). TBC1D16-47KD transfection in WM793 increased growth and progression (Supplementary Fig. 8b). Conversely, TBC1D16-47KD shRNA-mediated depletion in SK-MEL-28 had the opposite effects (Supplementary Fig. 8c).
TBC1D16-47KD regulates Rab GTPases and EGFR activation
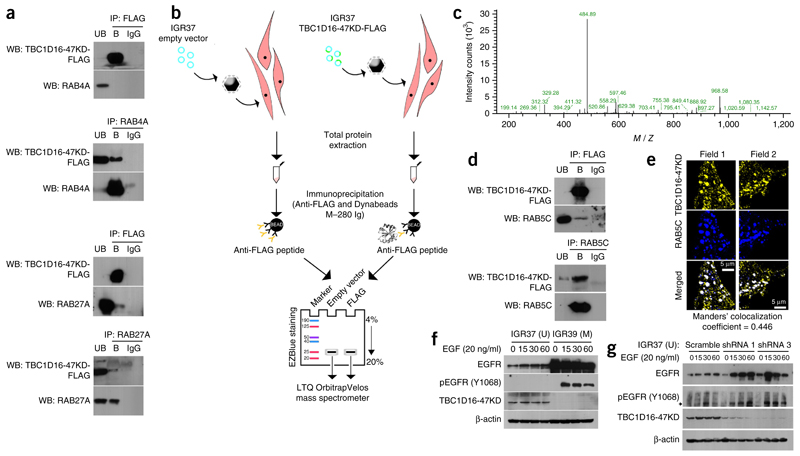
TBC1D16 is a member of the family of proteins containing the Tre2/Bub2/Cdc16 (TBC) domain, which are associated with cell signaling and intracellular receptor trafficking19,20. These TBC domains form the catalytic core of many GTPase-activating proteins that regulate Rab GTPases23. Here, using co-immunoprecipitation, we have confirmed the described direct binding of TBC1D16-47KD with RAB27A and RAB4A19,20 (Fig. 3a).
Figure 3.
TBC1D16-47KD regulates Rab GTPases and EGFR activation in melanoma cells. (a) Co-immunoprecipitation experiments show the interaction of TBC1D16-47KD with the Rab GTPases, RAB4A and RAB27A. Immunoprecipitation (IP) and western blotting (WB) were carried out using anti-FLAG and anti-RAB antibodies. Normal mouse IgG was used as a negative control. UB, unbound fraction; B, bound fraction. (b) Strategy used to identify TBC1D16-47KD novel interacting RABs based on MS analysis comparing the protein immunoprecipitates obtained in IGR37 cells from the empty vector–transfected condition versus the TBC1D16-47KD-FLAG–transfected cells. (c) Data from a representative RAB5C peptide identified by MS. (d) Co-immunoprecipitation experiments confirmed the interaction between TBC1D16-47KD and RAB5C. Immunoprecipitation and western blotting were performed using anti-FLAG and anti-RAB5C. UB, unbound fraction; B, bound fraction. (e) Examples of multicolored immunofluorescence images show colocalization of the TBC1D16-47KD and RAB5C proteins. (f) Western blot analyses indicate that IGR39 (methylated and silenced for TBC1D16-47KD) had large amounts of EGFR and its phosphorylated Y1068 form, whereas IGR37 cells (unmethylated and expressing TBC1D16-47KD) had minimal amounts of EGFR and is hypophosphorylated. Protein levels were determined at 0, 15, 30 and 60 min after EGF stimulation. (g) Western blot analyses show that shRNA-mediated downregulation of TBC1D16-47KD induces higher levels of EGFR and phospho-EGFR compared with control cells.
To identify targets involved in melanoma by TBC1D16-47KD epigenetic reactivation, we performed mass spectrometry (MS) to compare the immunoprecipitates obtained from empty vector and TBC1D16-FLAG-(47KD)-transfected IGR37 cells (Fig. 3b). We identified 39 proteins in the TBC1D16-47KD-immunoprecipitated sample (Supplementary Table 1). The first hit was TBC1D16-47KD itself, showing that the designed MS workflow was efficient. Notably, we identified a new RAB binding partner for TBC1D16-47KD: RAB5C (Fig. 3c). Co-immunoprecipitation confirmed the direct binding of TBC1D16-47KD to RAB5C (Fig. 3d), and immunofluorescence demonstrated the colocalization of both proteins (Fig. 3e).
The reported role of TBC1D16 in enhancing EGF-stimulated EGFR degradation mediated by RAB4A20 and the action of RAB5C in the EGFR downstream pathway24 prompted us to examine whether TBC1D16-47KD exerted an EGFR regulator effect mediated by RAB5C. First, we observed that TBC1D16-47KD enhanced RAB5C GTPase activity, and thus counteracted RAB5C function, in a concentration-dependent manner (Supplementary Fig. 9a). In this regard, RAB5C shRNA-mediated depletion in IGR37 cells had the opposite effect to TBC1D16-47KD depletion: enhanced growth and invasion capacity (Supplementary Fig. 9b) and reduced EGFR expression (Supplementary Fig. 9c). Second, we overexpressed RAB5C in IGR37 cells and found that it increased membrane–associated EGFR (Supplementary Fig. 9d), as it occurs with RAB4A20. Third, we found that IGR39 (TBC1D16-47KD methylated) had a large amount of EGFR and its phosphorylated Y1068 form, whereas IGR37 cells (TBC1D16-47KD unmethylated) had minimal amounts of EGFR and a hypophosphorylated status (Fig. 3f). We observed the same pattern in the paired breast-colon primary-metastasis cells (Supplementary Fig. 9e). Finally, we proceeded with IGR37 serum starvation followed by EGF stimulation. Downregulation of TBC1D16-47KD induced higher levels of EGFR and phospho-EGFR than in control cells (Fig. 3g). We obtained similar results upon TBC1D16-47KD depletion in SK-MEL-28 (another unmethylated melanoma cell line) (Supplementary Fig. 8d). Conversely, TBC1D16-47KD ectopic expression in IGR39 cells decreased EGFR (Supplementary Fig. 9f). We observed identical findings upon TBC1D16-47KD transfection in WM793, another methylated and silenced melanoma cell line, (Supplementary Fig. 8d). Importantly, we validated these findings in the unmethylated metastasis-derived WM266.6 cell line, which showed low levels of EGFR, whereas we observed high levels in the WM115 methylated cells (Supplementary Fig. 7d). Transfection of TBC1D16-47KD in WM115 induced EGFR downregulation (Supplementary Fig. 7e).
These results suggest that the epigenetically reactivated TBC1D16-47KD transcript, acting through RABs, is able to reduce EGFR activity in metastatic cells.
TBC1D16-47KD increases sensitivity to BRAF and MEK inhibitors
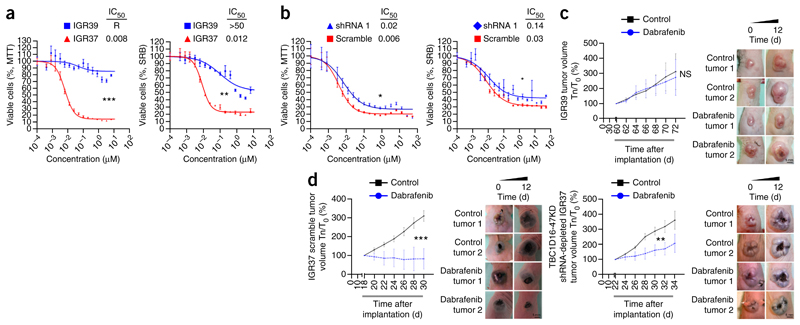
The observed EGFR downregulation and its hypophosphorylated status upon TBC1D16-47KD DNA hypomethylation-mediated re-expression prompted us to determine whether it affects drug sensitivity. BRAF inhibitors have clinical benefits in melanomas with mutant BRAF14,15, and although resistance to these drugs emerges, it can be mediated through pathways involving EGFR25,26 and PI3K/AKT27,28 activation. Thus, we examined whether epigenetic reactivation of TBC1D16-47KD conferred distinct responses to BRAF inhibitors and also to MEK inhibitors, other targetable proteins in that pathway.
In our original model of the paired primary and metastasis melanoma cells, IGR39 and IGR37, both share the same BRAF activating mutation (V600E) and, thus, it is thought they share a high drug sensitivity. However, IGR37 cells, with a less active EGFR, were more sensitive to the BRAF inhibitor dabrafenib than IGR39 cells, with highly active EGFR (Fig. 4a). Furthermore, TBC1D16-47KD shRNA depletion in IGR37 cells induced higher resistance to dabrafenib (Fig. 4b). Orthotopic tumors from IGR39 (Fig. 4c) were resistant to the BRAF inhibitor. Tumors from TBC1D16-47KD scrambled IGR37 cells were more responsive to the BRAF inhibitor than IGR37 TBC1D16-47KD shRNA-depleted tumors (Fig. 4d).
Figure 4.
Epigenetic reactivation of TBC1D16-47KD predicts response to BRAF inhibition. (a) MTT and sulforhodamine B (SRB) assays for IGR37 (TBC1D16-47KD unmethylated and expressed) and IGR39 (TBC1D16-47KD methylated and repressed) cells upon the use of the BRAF inhibitor dabrafenib. Corresponding half-maximal inhibitory concentration (IC50) values are also shown. R = resistance to drug inhibition. Values are the mean ± s.e.m (n = 3 for each group). (b) MTT and SRB assays in TBC1D16-47KD shRNA-downregulated IGR37 cells upon the use of dabrafenib. Values are the mean ± s.e.m (n = 3 for each group). (c) Volume of TBC1D16-47KD methylated orthotopic tumors derived from IGR39 upon daily BRAF inhibitor (30 mg/kg) or vehicle treatment. Values are the mean ± s.d. (n = 5 for each group.) Right, representative images of the studied melanoma grafts. Tn/T0 (%), tumor size on day “n” in relation to day 0 of drug treatment. (d) Left, volume of TBC1D16-47KD unmethylated orthotopic tumors derived from scrambled shRNA IGR37 upon daily treatment with BRAF inhibitor (30 mg/kg) (n = 9) or vehicle (n = 5). Right, TBC1D16-47KD shRNA-depleted IGR37-derived orthotopic tumors (n = 7) are less responsive to the BRAF inhibitor than the shRNA scrambled IGR37-derived tumors. Tn/T0 (%), tumor size on day “n” in relation to day 0 of drug treatment. Values are the mean ± s.d. (n = 7 for each group). Significance was determined from a 10,000-permutation t-test with Holm correction of the difference between growth or volume curves. NS, nonsignificant; *P < 0.05; **P < 0.01, ***P < 0.001. Representative images of the studied melanoma grafts are shown.
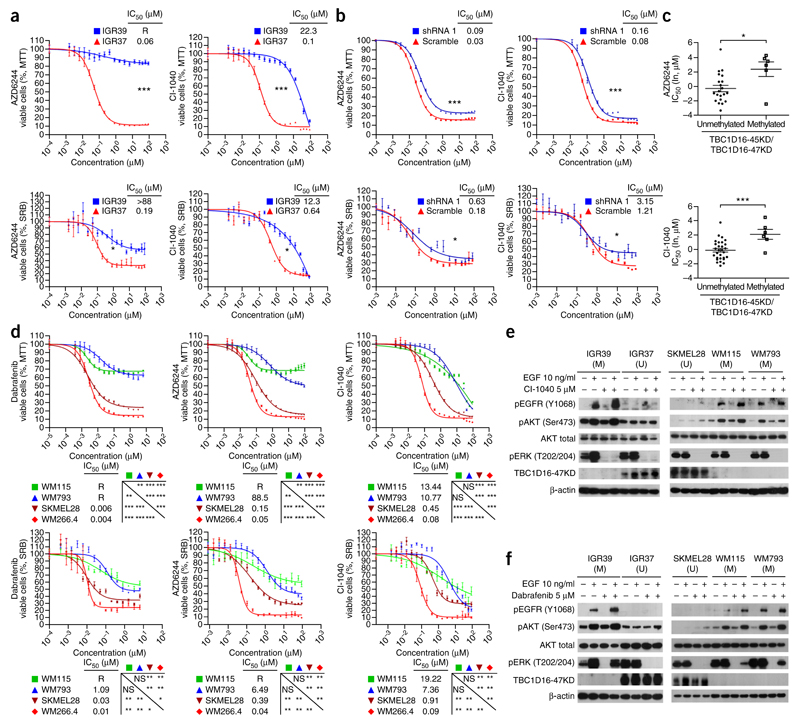
We found the same pattern for MEK inhibitors: IGR37 epigenetically reactivated by TBC1D16-47KD was more sensitive to AZD6244 (selumetinib) and CI-1040 than TBC1D16-47KD methylated IGR39 (Fig. 5a). Downregulation of TBC1D16-47KD in IGR37 increased resistance to MEK inhibitors (Fig. 5b). We extended these observations by comparing the TBC1D16-47KD methylation status in 36 additional melanoma cell lines (Supplementary Data 3) with the corresponding sensitivity to MEK inhibitors29. Therein, we observed that TBC1D16-47KD unmethylated status was also associated with sensitivity to AZD6244 and CI-1040 (Student’s t-test, P = 0.0110 and P = 0.0015, respectively) (Fig. 5c).
Figure 5.
Epigenetic reactivation of TBC1D16-47KD predicts response to BRAF and MEK inhibitors by targeting two signaling pathways. (a) MTT and SRB assays in IGR37 and IGR39 cells upon treatment with MEK inhibitors (AZD6244 and CI-1040). Corresponding half-maximal inhibitory concentration (IC50) values are also shown. R = resistance to drug inhibition. (b) shRNA-mediated downregulation of TBC1D16-47KD induced higher resistance to MEK inhibitors compared with the shRNA-scrambled control cells. (c) TBC1D16-47KD CpG island unmethylated status is associated with enhanced sensitivity to MEK inhibitors in the Sanger panel of melanoma cell lines. y axis, drug IC50 values (natural log, μM); x axis, DNA methylation status. Values are means ± s.e.m. (d) MTT and SRB assays in the WM115 (methylated), WM793 (methylated), SK-MEL-28 (unmethylated) and WM266.4 (unmethylated) cell lines upon the use of BRAF or MEK inhibitors. (e) Western blot analyses of the RAS/BRAF/MEK/ERK and PI3K/AKT signaling pathways in TBC1D16-47KD unmethylated (U) or methylated (M) melanoma cells upon using the MEK inhibitor CI-1040. (f) Western blot analyses of the RAS/BRAF/MEK/ERK and PI3K/AKT signaling pathways in TBC1D16-47KD unmethylated (U) or methylated (M) melanoma cells upon using the BRAF inhibitor dabrafenib. Significance was determined from a 10,000-permutation t-test with Holm correction of the difference between growth curves. NS, nonsignificant; *P < 0.05; **P < 0.01, ***P < 0.001. Error bars show means ± s.e.m.
We complemented these findings using the two described melanoma cell lines that also harbor a BRAF mutation: WM793 (primary, TBC1D16-47KD methylated) and SK-MEL-28 (metastasis TBC1D16-47KD unmethylated) (Supplementary Fig. 8). SK-MEL-28 cells showed low levels of EGFR, whereas we saw high levels in WM793 cells (Supplementary Fig. 8d). Thus, the SK-MEL-28 cells were more sensitive to the BRAF and MEK inhibitors than WM793 cells (Fig. 5d). Furthermore, shRNA-depletion of TBC1D16-47KD in SK-MEL-28 cells triggered EGFR upregulation and increased resistance to BRAF and MEK inhibitors, whereas TBC1D16-47KD transfection in WM793 decreased EGFR levels (Supplementary Fig. 8d) and increased drug sensitivity (Supplementary Fig. 8e). We found the same patterns in the paired breast-colon primary-metastasis cells (Supplementary Fig. 10a). Bearing in mind that both long and short TBC1D16 isoforms have the ability to stimulate Rab4A GTPase activity20, we carried out TBC1D16-86KD knockdown to show that depletion was also associated with resistance to BRAF and MEK inhibitors (Supplementary Fig. 3c).
It is possible that the high levels of phosphorylated EGFR in TBC1D16-47KD methylated cells cause, upon treatment with BRAF and MEK inhibitors, a shift in cellular proliferation that depends on the PI3K/AKT pathway, which cannot be blocked by the described drugs27,28. Conversely, the TBC1D16-47KD unmethylated cells, with their low associated phospho-EGFR levels, cannot properly activate this alternative growth pathway to survive and are more sensitive to the BRAF and MEK inhibitors. We addressed this hypothesis by studying ERK (representative of the RAS/BRAF/MEK/ERK pathway) and AKT (representative of the PI3K/AKT pathway) activation. We observed that the MEK inhibitor blocked phosphorylation of ERK independently of TBC1D16-47KD methylation and expression status (Fig. 5e). However, the BRAF inhibitor only blocked ERK phosphorylation in the context of TBC1D16-47KD-unmethylated cells, such as IGR37 and SK-MEL-28 (Fig. 5f). Thus, there is an initial distinct sensitivity to these drugs targeting the RAS/BRAF/MEK/ERK pathway at different levels that is associated with TBC1D16-47KD methylation status. However, in addition, melanoma cells with a methylated TBC1D16-47KD show drug-resistance mechanisms that involve the PI3K/AKT pathway. After MEK and BRAF inhibitor treatment, we found that high levels of phospho-EGFR, associated with hypermethylated TBC1D16-47KD (IGR39, WM115 and WM793), were able to hyperphosphorylate AKT to escape from growth inhibition (Fig. 5e,f). However, this was not the case for the hypophosphorylated EGFR in TBC1D16-47KD-unmethylated cells (IGR37 and SK-MEL-28), which were unable to phosphorylate AKT efficiently upon administration of the MEK or BRAF inhibitor (Fig. 5e,f). We obtained similar results using non-serum-depleted medium (Supplementary Fig. 10b).
These findings suggest that TBC1D16-47KD epigenetic reactivation renders melanoma cells more sensitive to BRAF and MEK inhibitors, probably through the inability to shift to an EGFR-driven PI3K/AKT pathway. Notably, for those TBC1D16-47KD methylated melanomas that are more resistant to these drugs, the addition of an EGFR inhibitor such as Iressa (gefinitib) increased the antitumoral effect of BRAF and MEK inhibitors (Supplementary Fig. 11a). This effect is probably observed because we are blocking the survival escape route mediated by EGFR-led PI3K/AKT signaling (Supplementary Fig. 11b). This combination therapy might warrant further study in BRAF and MEK drug-resistant melanomas.
TBC1D16-47KD demethylation predicts poor clinical outcome
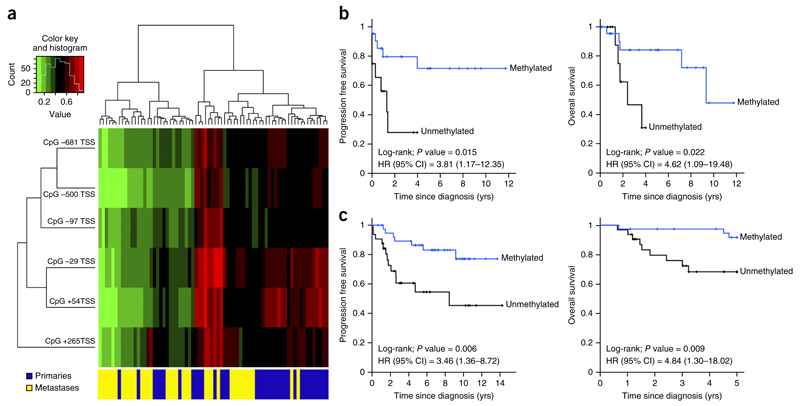
The analyses of a collection of human melanomas showed a TBC1D16-47KD unmethylated CpG island status in 41% (30 of 73) of subjects. Hypomethylation was associated with transcript overexpression (Supplementary Fig. 12a). We observed enrichment of TBC1D16-47KD hypomethylation in more melanoma metastases (61%, 22 of 36) compared with primary melanomas (21%, 8 of 37) (Fisher’s exact test, P = 0.0008). We found a similar degree of TBC1D16-47KD hypomethylation among the The Cancer Genome Atlas (TCGA) metastatic melanomas (https://tcga-data.nci.nih.gov/tcga/) (54%, 144 of 267 cases). TCGA data confirmed the enrichment of TBC1D16-47KD hypomethylation in distant melanoma metastases compared with non-disseminated primary melanomas (Mann-Whitney test, P = 0.015) (Supplementary Fig. 12b). Furthermore, TCGA RNA-sequencing data showed that TBC1D16-47KD hypomethylation was associated with transcript overexpression (Spearman correlation test, rho, −0.44, P < 0.00001) (Supplementary Fig. 12c). The study of an additional set of 14 paired patients’ primary and metastatic melanomas confirmed the enrichment of TBC1D16-47KD hypomethylation in the metastatic samples compared with the respective primary melanomas (Wilcoxon signed-rank test, P = 0.043). Paired metastases of ten colorectal tumors were also enriched for TBC1D16-47KD hypomethylation compared with their corresponding primary tumors (Wilcoxon signed-rank test, P = 0.014). TCGA data set highlighted the functional relevance of TBC1D16-47KD hypomethylation by showing that, in the context of widespread blocks of DNA hypomethylation in cancer30–32, there was not an association between our identified demethylation event and a global DNA methylation loss within this region (r2 = 0.006) (Supplementary Fig. 12d). Normal melanocytes (n = 10) were methylated at TBC1D16-47KD. Unsupervised clustering analysis showed enrichment of the TBC1D16-47KD hypomethylated sites in our metastasis samples (Fig. 6a).
Figure 6.
TBC1D16-47KD promoter demethylation is an independent prognostic factor for poor clinical outcome in melanoma patients. (a) Unsupervised clustering analysis using the CpGs located in the TBC1D16-47KD promoter shows enrichment of the hypomethylated sites in the metastatic melanoma samples. Green, unmethylated; red, methylated. (b) Kaplan-Meier analysis of PFS and OS in the melanoma discovery cohort with respect to TBC1D16-47KD methylation status. Significance of the log-rank test is shown. Results of the univariate Cox regression analysis are represented by the HR and 95% CI. Twelve unmethylated and 22 methylated cases had a mean PFS of 1.6 and 8.8 years (yrs), respectively. 12 unmethylated and 24 methylated cases had a mean time OS of 2.8 and 9 years, respectively. (c) Kaplan-Meier analysis of PFS and OS among the melanoma validation cohort with respect to TBC1D16-47KD methylation status. Significance of the log-rank test is shown. Results of the univariate Cox regression analysis are represented by the HR and 95% CI. Thirty-two unmethylated and 39 methylated cases had a mean PFS of 8.1 and 11.6 years, respectively. Thirty-two unmethylated and 39 methylated cases had a mean OS of 4.0 and 4.9 years, respectively.
We also wondered whether TBC1D16-47KD hypomethylation conferred any prognostic value. TBC1D16-47KD hypomethylation was associated with shorter progression-free survival (PFS) (log-rank; P = 0.015; hazard ratio (HR) = 3.81, 95% CI = 1.17–12.35) and lower overall survival (OS) (log-rank; P = 0.022; HR = 4.62, 95% confidence interval (CI) = 1.09–19.48) (Fig. 6b). TBC1D16-47KD hypomethylation was also associated with lower OS in the TCGA cohort (log-rank; P = 0.013; HR = 1.96; 95% CI 1.14–3.36) (Supplementary Fig. 12e). PFS data are not available in TCGA. In a validation cohort of 71 individuals with melanoma, TBC1D16-47KD demethylation was associated with metastatic melanomas (Fisher’s test; P = 0.032), shorter PFS (log-rank; P = 0.006; HR = 3.46, 95% CI = 1.36–8.72) and lower OS (log-rank; P = 0.009; HR = 4.84, 95% CI = 1.30–18.02) (Fig. 6c). TBC1D16-47KD hypomethylation predicted poor outcome across the different Breslow and ulceration statuses (Supplementary Fig. 12f). In this regard, TBC1D16-47KD hypomethylation was an independent predictor of poor PFS and OS in the multivariate analysis (Supplementary Table 2).
Given our findings that melanoma cells with TBC1D16-47KD hypomethylation were more sensitive to BRAF and MEK inhibitors, we wondered whether the same effect occurred in clinical samples. We studied the metastatic melanoma of a cohort of 33 subjects who were carriers of the BRAF V600E mutation and treated with either of the BRAF inhibitors, dabrafenib or vemurafenib. We observed a TBC1D16-47KD-unmethylated status in 33% (n = 11) of the melanomas (Supplementary Table 2). The TBC1D16-47KD unmethylated status was associated with complete clinical response to the BRAF inhibitor, whereas partial response, stable disease or progression occurred in individuals with TBC1D16-47KD methylation (Fisher’s exact test, P = 0.008) (Supplementary Table 2). Finally, we wondered whether TBC1D16-47KD methylation increased at the time of clinical progression during BRAF and MEK inhibitor treatment. We obtained paired melanoma samples from five individuals at the time of disease presentation and at relapse after therapy failure. We observed that initial drug response occurred in TBC1D16-47KD unmethylated samples, but when the melanoma relapsed,40% (2 of 5) of these resistant samples showed TBC1D16-47KD hypermethylation. Thus, the clinical data supported the experimental observations that TBC1D16-47KD unmethylated status increases sensitivity to BRAF inhibitors.
Discussion
Melanoma is one of the most aggressive and therapy-resistant malignancies1. Many mechanisms are involved in melanoma progression, but dynamic epigenetic changes are likely to be critical contributors. Here we took an approach to identify DNA methylation events that were associated with metastasis by minimizing the effect of the genetic background, using paired primary and metastasis samples from the same individual. Using this strategy, we identified a hypomethylation event in TBC1D16 in the pro-metastatic melanoma samples. Because TBC1D16 participates in intracellular receptor recycling19,20, our findings reinforce the concept that melanoma is dependent on a high volume of vesicle trafficking19,33,34.
The contribution of TBC1D16-47KD epigenetic reactivation to melanoma progression can provide additional opportunities for therapeutic intervention related to EGFR targeting and the effects in RAS/BRAF/MEK/ERK and PI3K/AKT pathways. Many melanomas carry BRAF activating mutations that render these patients more sensitive to treatment with BRAF inhibitors35, whereas wild-type BRAF melanomas are treated by standard chemotherapy, with only 10–20% of these individuals responding to treatment36. In addition to immunomodulatory treatments37, new strategies are being considered based on the knowledge that the BRAF protein modulates the MEK signaling cascade downstream of EGFR. In this context, alternative treatments using EGFR inhibitors, MEK inhibitors alone and various combinations with BRAF inhibitors are underway38,39. However, biomarkers to predict whether a patient will respond to these new drugs are needed. Here, we have shown that individuals with melanoma with TBC1D16-47KD hypomethylation have a poor prognosis, but at the same time, their tumors are ‘addicted’ to BRAF and MEK signaling and could greatly benefit from treatment with drugs that target this pathway.
Our findings highlight the contribution of epigenetics to the generation of metastasis and to the prediction of drug response. The example provided here of how the loss of DNA methylation status of an internal promoter of TBC1D16 unleashes melanoma progression and generates sensitivity to BRAF and MEK inhibitors may be helpful in clinical trials that seek to improve the outcome of melanoma patients.
Online Methods
Human cancer cells and tissues
Human paired primary and metastasis cancer cell lines derived from melanoma IGR39/IGR37 and WM115/WM266.4 were obtained from the Leibniz Institute DSMZ-German Collection of Microorganisms (Braunschweig, Germany) and the American Type Culture Collection (ATCC), respectively. The paired primary and metastasis cancer cell lines derived from colon (SW480 and SW620) were obtained from the ATCC. The breast cancer pair MDA-MB-468PT and MDA-MB-468LN was kindly provided by Anne Chambers (London Regional Cancer Program, London, Ontario, Canada). The primary melanoma cell line WM793 and the metastasis melanoma cell line SK-MEL-28 were obtained from the Coriell Institute for Medical Research and the ATCC, respectively. All cell lines were characterized by short tandem repeat analysis (STR) profiling (LGS Standards SLU) within 6 months after receipt and tested on a monthly basis for mycoplasma contamination. DNA from 36 additional melanoma cell lines, with previously known IC50 values for MEK inhibitors29, was obtained from the Wellcome Trust Sanger Institute (Hinxton, Cambridge, UK). To study the association of DNA methylation with clinical progression and OS of melanoma patients, we used fresh-frozen melanoma samples supplied by the Katholieke Universiteit Leuven (discovery cohort). As an independent validation cohort, we used primary melanomas diagnosed with Clark stage I-V and Breslow index provided by Lund University (Sweden). An additional set of 14 paired patients’ primary and metastatic melanomas was also included. To analyze the effects of DNA methylation with clinical response to first-line treatments in melanoma patients, we studied a cohort of 33 melanoma patients treated with BRAF inhibitors. An additional five paired melanoma samples (pre- and post-BRAF inhibitor therapy) were also obtained. Overall, we studied 196 melanoma patients with a gender distribution of 51% male and 49% female and a mean age of 67 ± 12 years. None of the human individual samples were excluded before analysis. Tissue collection and the study protocol were approved by the ethical committees of the FP7/2007-2013 “Target-Melanoma: Molecular Dissection of Melanoma Progression: An Integrated Pan-European Approach” consortium (#PIAPP-GA-2009-230614), the Germans Trias i Pujol University Hospital Biobank (BB14007), the Vall d’Hebron University Hospital Biobank (RE-017-VO3) and the Manchester Cancer Research Centre Biobank (12_JOBR_01 and 13_RIMA_01). Full informed consent was obtained from all subjects. DNA methylation data in melanoma were collected from the TCGA Data Portal (https://tcga-data.nci.nih.gov/tcga/). Patient samples were reversibly encoded for blinding and only the investigator responsible for each collection was able to decodify the cases. Sample size has been chosen considering the balance between clinical feasibility and study power, taking incidence of melanoma cases into account in the different stages. To ensure the statistic power of the results, we assumed as significant a two-tailed P value under 0.05. We followed the REporting recommendations for tumor MARKer prognostic studies (REMARK) reporting guidelines for tumor marker prognostic studies40. PFS and OS were determined using Kaplan-Meier plots and log-rank tests. Statistical analysis was performed by using SPSS for Windows.
DNA methylation arrays
We analyzed whole-genome DNA methylation in the three paired tumor cell lines, 10 nevi, 37 primary melanomas, and 36 melanoma metastases samples using the Illumina Infinium HumanMethylation450Beadchips as described18,41. DNA was extracted from cell lines and tissues by the phenol: chloroform method. All DNA samples were assessed for integrity, quantity and purity by electrophoresis in a 1.3% agarose gel, PicoGreen quantification and NanoDrop measurement. All samples were randomly distributed into 96-well plates. We performed bisulfite conversion of 500 ng of genomic DNA using an EZ DNA methylation kit (Zymo Research). Bisulfite converted DNA (200 ng) was used for hybridization on the HumanMethylation450 BeadChip (Illumina). Raw fluorescence intensity values were normalized with Illumina Genome Studio software (V2011.1) using ‘control normalization’ with background correction. Normalized intensities were then used to calculate DNA methylation levels (beta values). Likewise, data points with statistically low power (as reported by detection values of P > 0.01) were designated as NA and excluded from the analysis. Genotyping probes present on the chip and DNA methylation probes overlapping with known single-nucleotide polymorphisms (SNPs) were also removed. Probes were considered to be in a promoter CpG island if they were located within a CpG island (UCSC database) and <2,000 bp away from a transcription start site (outside chromosome X). We only considered CpG sites with a ≥70% change in CpG methylation level between primary and metastases sites, and the differential CpG methylation primary and metastasis had to occur in the three tumor types studied.
Bisulfite sequencing and pyrosequencing
For primer sequences please refer to Supplementary Table 3. We used the EZ DNA Methylation Gold kit (Zymo Research, Orange, CA, USA) for genomic DNA bisulfite conversion. For bisulfite sequencing, a minimum of eight single clones was interrogated for each sample and the methylation frequency was calculated in each case, as described42. DNA methylation in clinical samples was studied by pyrosequencing on bisulfite-treated DNA extracted from formalin-fixed paraffin-embedded (FFPE) samples. Pyrosequencing reactions and quantification of DNA methylation were performed in a PyroMark Q96 System version 2.0.6 (Qiagen) including appropriate controls as described7. Specific primers were designed using the MethylExpress program (Applied Biosystems) for bisulfite sequencing and PyroMark Assay Design Software (Qiagen, version 2.0.01.15) for pyrosequencing to examine the methylation status of particular CG sites covering the candidate genes promoter regions.
Expression analysis
Total RNA samples of paired melanoma cell lines IGR39 and IGR37 were extracted and sent to the IRB Functional Genomic Core External Service (Barcelona). Once samples had been reversed-transcribed and labeled, they were hybridized onto Affymetrix GeneChip Human Genome U133 Plus 2.0 arrays. Hybridization, washing, staining and scanning were performed using Affymetrix GeneChip system instruments and protocols. For breast and colon cancer cell lines, expression data were obtained from the Gene Expression Omnibus (GSE11683, GSE10843 and GSE57083). For qRT–PCR experiments, total RNA was extracted using Trizol reagent and retrotranscribed using the ThermoScript RT-PCR system (Invitrogen). The reaction was carried out following the methods for use of SYBR Green (Applied Biosystems), and B2M, GAPDH and ACTB were used as housekeeping genes to enable normalization. For primer sequences please refer to Supplementary Table 3. We performed reactivation treatments with the demethylating agent 5-aza-2′-deoxycytidine (AZA; Sigma) at 0.5 μM for 72 h. For immunoblotting assays, we extracted total protein using RIPA (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA and EGTA, 1% NP40, 0.5% of sodium deoxycholate, 0.1% of SDS, and protease and phosphatase inhibitors from Roche). All the antibodies used in this study are described in Supplementary Table 3.
Short hairpin interference and ectopic expression assays
For shRNA and cloning sequences please refer to Supplementary Table 3.Three and four different shRNAs were designed over the TBC1D16-45KD/47KD and 86KD mRNA, respectively, to target melanoma, breast and colon TBC1D16-expressing cells. shRNA against the MSS2 yeast protein (not present in mammals) was used as scrambled. All annealed shRNA oligos were ligated into pLVX-shRNA2 plasmid, purchased from Clontech, using BamH1 and EcoR1 restriction sites. Each shRNA-encoding plasmid (10 μg) was mixed with 7.5 μg of ps-PAX2 and 2.5 μg of PMD2.G plasmid in 1 ml JetPRIME buffer and 50 μl of JetPRIME (Polyplus-transfection S.A., Illkirch, France). After 10 min of RT incubation, the transfection mix was added drop-wise to a 10-cm dish containing 10 ml of DMEM and 293T cells at 80% confluence. After 48 h, viral supernatant was recovered and 0.45-μm filtered. After 48–72 h, cells were checked for infection efficiency. For transduction efficiencies lower than 95%, cells grown and green positive cells were isolated by flow cytometry. After cell sorting, 10–20 clones were isolated by limiting dilution, checked for silencing, and finally 5–10 of the best clones were mixed as a pool to perform the in vitro and in vivo experiments to avoid the cloning bias-effect. If the infection efficiency was higher than 95%, we worked with the cells as a pool. We also engineered the construct corresponding to TBC1D16-47KD ORF so that the three target regions of the three shRNAs used have been mutated to render the mRNA insensitive to shRNA-mediated depletion. To construct the RNAi-insensitive TBC1D16 overexpressing construct, we used a three-step PCR-based strategy in which mutant sites at third codon-positions were introduced by the PCR primers. The mutations introduced are synonymous changes and encode the same TBC1D16 wild-type protein. Both the wild-type cDNA (shRNA-sensitive) and the mutant cDNA (shRNA-insensitive) were tested in parallel in MDA-MB-468LN cells stably expressing each shRNA. Additionally, we carried out cotransfection experiments in MDA-MB-468LN and IGR37 cell lines in which the wild-type cDNA was fused to a FLAG tag (pLVX-ZsGreen1, Clontech) and the mutated cDNA was fused to a HA tag (pLVX-tdTomato, Clontech). For ectopic overexpression experiments, TBC1D16-47KD cDNA was amplified from IGR37 using specific primers with end adaptors containing EcoRI and NotI sequences and a Kozak sequence. The PCR product was verified by Sanger-sequencing and ligated into pLVX-ZsGreen1 plasmid from Clontech using EcoRI and NotI restriction sites. The 293T transfections and cell infections (IGR39, WM793 and WM115) were performed following the same procedures as for shRNA constructs. TBC1D16-45KD cDNA was obtained using TBC1D16-47KD cDNA as a template following a three-step PCR based strategy.
In vitro proliferation and colony formation
Cell proliferation was determined by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. The proliferation of the cells indicated in the figures were quantified for 6 d, staining the cells with MTT for 3 h and blocking the reaction adding lysis buffer (HCl 20 mM, acetic acid 2.5%, SDS 20%, dimethylformamide 50%, pH 4.7). Measurements were done at 560 nm after overnight incubation at 37 °C. Colony-formation assay was performed by seeding 300 cells onto 12-well plates and maintaining them on DMEM medium. After 2 weeks, cells were MTT-stained and fixed, and colonies were quantified using Fiji software.
Mass spectrometry analysis and validation
Once we had infected the IGR37 cells with TBC1D16-47KD-FLAG (DYKDDDDK)-expressing vector, we performed the immunoprecipitation as follows: 2 mg of total protein extract was recovered in 1 ml of RIPA (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM EDTA and EGTA, 1% NP40, 0.5% of sodium deoxycholate, 0.1% of SDS, and protease and phosphatase inhibitors from Roche) and put on ice. After cleaning the membrane residues, overnight precleaning was done with magnetic beads (Dynabeads M-280 sheep anti-mouse IgG, Invitrogen) at 4 °C. Anti-FLAG antibody (30 μg) from Sigma (F1804) was incubated with 300 μl of magnetic beads overnight at 4 °C in BSA 0.5%. Beads were removed from the protein solution, and magnetic beads with anti-FLAG antibody were recovered. Following bead cleaning, eluted proteins were reduced with β-mercaptoethanol at 70 °C for 10 min. Samples were loaded and run on a gradient NuPAGE Novex 4–15% Bis-Tris Polyacrylamide Gel (Life Technology). Bands were identified incubating the gel overnight with EZBlue Gel Staining Reagent (G1041, Sigma). Single bands were excised and sent to the external proteomics service of the Barcelona Parc Scientific (Spain). Mass spectrometry was performed in a NanoAcquity (Waters) HPLC coupled to an LTQ OrbitrapVelos mass spectrometer (Thermo Scientific). Data analysis was carried out in the LTQ OrbitrapVelos. Peptide masses were measured in the Orbitrap at a resolution of 60,000 (m/z: 300–1700). Most abundant peptides were selected from each MS scan and fragmented in the linear ion trap with helium as the collision gas. All results were filtered so only proteins identified with high confidence peptides (FDR ≤ 0.01) and with at least three peptides were included. Finally, results were filtered by Homo sapiens. For co-immunoprecipitation validation, IGR37 TBC1D16-NH2-FLAG cells were grown at 70% confluence. Cells were detached using a scraper. Total protein extract (1 mg) was prepared in RIPA. Samples were precleared using Dynabeads M-280 Sheep Anti-Mouse IgG (Invitrogen), overnight at 4 °C in rotation. Antibodies against the candidate proteins were pre-incubated with beads overnight at 4 °C in rotation. For colocalization analysis, the cells were cultured directly on poly-L-lysine coverslips and fixed with 4% paraformaldehyde for 30 min. Cells were permeabilized with 0.1% Triton X-100 for 10 min and blocked with 2% blocking reagent (Roche) for 1 hour. The immunostainings were performed for 16 h at 4 °C. The coverslips were then mounted on glass slides with Mowiol. Multicolor immunofluorescence images were captured, and the images were analyzed with the plugin Intensity Correlation Analysis (ImageJ) to calculate the Manders’ R overlap colocalization coefficient.
GAP assays
FLAG-TBC1D16-47KD protein was purified from HEK293T cells. Recombinant RAB5C GTPase (100 pmol) (Origene, TP310698) was loaded with [γ–32P]GTP for 10 min at 25 °C and desalted on PD-SpinTrapTM G-25 columns (GE Healthcare, # 28-9180-04) to remove free nucleotide. GAP assay was then conducted in 1x assay buffer (20 mM HEPES, KOH pH 7.6, 50 mM KCl, 5 mM MgCl2, 0.1 mg/ml BSA) containing 0.3 μM RAB5C-GTP and increasing concentrations of TBC1D16-47KD at 25 °C. Aliquots were removed at various times and quenched by vortexing the reaction mixture with 700 μl of a suspension of 70 mg/ml charcoal in 20% (vol/vol) ethanol, 0.2 M HCl and 0.1 M KH2PO4. The quenched samples were spun to pellet the charcoal and 32Pi in the supernatant was analyzed by counting Cerenkov radiation in a scintillation counter (Beckman Coulter).
EGFR time course assay
Parental cell lines, IGR37 scramble and IGR37 shRNA-1 and shRNA-3 were grown to 80% confluence and then serum-starved overnight, incubating at 37 °C. During the last 4 h, 25 μg/ml cycloheximide was added to the medium to prevent protein synthesis. Cells were placed on ice for 30 min, 20 ng/ml of human EGF was added, and cells were kept on ice for an additional 30 min. Subsequently, cells in six-well plates were moved to a 37 °C incubator for the indicated times. The cells were then washed three times with cold PBS and lysed in ice-cold RIPA buffer.
xCELLigence real-time cell invasion assay
The invasion assays were performed in CIM-16 plates with 8-μm-pore membranes (ACEA Biosciences). Wells were coated with 30 μl of 5% Matrigel (BD Bioscience) and allowed to gel at 37 °C and 5% CO2 for 4 h. After 4 h, the lower chamber wells were filled with 160 μl of 10% FBS containing medium and the top chamber wells with 40 μl of serum-free medium. The two portions were assembled together and allowed to equilibrate for 1 h at 37 °C and 5% CO2. Cells were incubated for 16 h in 0.05% FBS media; for seeding, the cells were rinsed with PBS, trypsinized and resuspended in serum-free medium. A total of 4 × 104 cells and wells for IGR39, IGR37 and SKMEL28 and 12 × 104 for WM266.4, WM793 and WM115 were seeded onto the top chamber of CIM-16 plates and placed into the xCELLigence system for data collection after background measurement. The xCELLigence software was set to collect impedance data every 15 min during 72 h. The cell index represents the capacity for cell invasion, whereas the slope of the curve can be related to the invasion velocity.
Cell viability assays
Cells were seeded in either 96-well microplates at ~15% confluence in medium with 10% FBS, penicillin and streptomycin. The optimal cell number for each was determined to ensure that each one was in growth phase at the end of the assay. After overnight incubation, cells were treated with 18 concentrations of each compound (twofold dilution series) and then returned to the incubator for assay after 72 h. The media of treatments involving lentiviral depletions or overexpressions was always supplemented with EGF according to the EGFR modulation induced by TBC1D16-47KD protein. Cells were plated and incubated overnight with serum-free medium before treatment. The day after, drugs were dispensed at different concentrations and after 1 h, EGF was added to a final concentration of 10 ng/ml. After 72 h of incubation at 37 °C, cell viability was determined by the MTT and the sulforhodamine B (SRB) assays.
Preparation of protein lysates for western blot analysis of MEK and BRAF inhibition
Cells were treated with the MEK inhibitor CI-1040 or BRAF inhibitor dabrafenib in 10% FBS supplemented medium or in low-serum conditions. For the serum-depleted assay, cells were washed in medium containing 0.1% FBS (FBS) and incubated in this medium for 48 h. Drugs (5 μM) or DMSO (0.05%) were added directly to this medium, and 45 min later, EGF was added at a final dose of 10 ng/ml. Cells were harvested after 30 min of EGF stimulation. Protein lysates were prepared from cells at 80% confluence. Total protein was extracted adding Laemmli buffer 1× directly to the plates after washing the cells twice with cold PBS. The lysates were processed using a Branson Ultrasonics 25/450 Sonifier and boiled at 100 °C for 5 min. Protein concentrations were determined by Lowry procedures and western blot was performed using NuPAGE Novex 4–12% Bis-Tris gels and standard techniques.
Mouse studies
Mice tumor growth and metastasis experiments were performed as described43. Tumor cells lines IGR37 scramble and IGR37 shRNA-1 (1 × 106 cells diluted in 200 μl of Matrigel BD) were grafted subcutaneously in anesthetized athymic nu/nu mice (Harlan Laboratories), each cell type in one side of the mice back (n = 10). Tumor volumes were estimated each three days from two-dimensional caliper measurements using the equation V = (π/6) × L × W2, where V = volume (mm3), L = length (mm), and W = width (mm), and reported as volume mean ± s.d. for each mouse group. Tumor weights were obtained after death and small pieces of tumor were stored to confirm the transfection stability and TBC1D16-47KD silencing. After subcutaneous assay, additional subcutaneous tumors were used for the orthotopic model. Once the tumor had grown to 600–800 mm3, it was cut into 3 × 3 mm3 pieces and maintained in DMEM supplemented medium with 10% FBS and penicillin/streptomycin. Those fragments with macroscopically low or absent levels of necrotic areas were selected for orthotopic implantation. A cutaneous back shave biopsy of normal skin was done by removing a rhomboid area of around 6 mm2 (4 × 3 mm), and then a small fragment of solid tumor tissue was anchored with a prolene 7.0 suture to the edge of the skin, fixing the tumor at four anchored sites coinciding with each corner. To improve the implantation process, the anchored-skin tumor was protected making a skin fold that was fixed with three points with 5.0 silk sutures or two surgery staples. Thus, eight animals were implanted for each cell line tumor derived, and the orthotopic tumor was allowed to grow for several weeks. Mice were weighed, and the tumor width (W) and length (L) measured every 3 d. Tumor volume was estimated according to the formula V = π/6 × L × W2. The animals were maintained in a sterile environment; their cages, food and bedding were sterilized by autoclaving. All experiments with mice were approved by the Institutional Animal Care and Use Committee. Following the orthotopic growth assay, for subgroups of eight IGR37 scramble and eight IGR37 shRNA-1 mice, the tumor was surgery removed by an excisional surgery and the skin closed by 5.0 silk sutures suture, and mice kept alive for up to 3 months. Mice were monitored over this period for the presence of transit metastases following back and axillary lymph node affection. Finally, after the mice were sacrificed, the lung, liver and brain were examined by the pathologist for the presence of distal metastases. For each tumor, small fragments of 2–5 mm3 obtained in the previous pre-experiment were reimplanted orthotopically by a cutaneous shave biopsy, in the normal skin of 20 athymic nu/nu mice aged 4–5 weeks. These tumors in addition to those derived from IGR39 were then used in tumor drug response assays. When tumors reached a size of 200–400 mm3 mice, 15 mice harboring homogeneous skin tumors were randomized into two groups and treated with dabrafenib or vehicle alone (DMSO) by daily oral gavage of 0.5% hydroxypropylmethylcellulose (Sigma-Aldrich), 0.2% Tween 80 in pH 8.0 distilled water at 30 mg/kg for 12 d. For the metastasis model without the influence of tumor burden, 1.5 × 106 IGR37 and WM115 cells were injected into the spleen of 20 mice (10 mice for each condition). To avoid local tumor growth, the spleens were removed 48 h after cell injection. Mice were sacrificed when first symptomatologies that suggested liver and/or lung dysfunction appeared. Hepatic and lung metastases were examined macroscopically and microscopically following H&E stain tissue staining. The total number of metastatic nodes from several randomly chosen areas of each liver was counted for the analysis. Additional tumor growth assay were performed in melanoma, breast and colon cancer models. All animal specimens used for the experiments were male, except for the breast cancer cell lines MDA-MB-468PT and MDA-MB-468LN, which were female. None of the mice samples were excluded before analysis. Mice samples were reversibly encoded for blinding. All animal experiments were approved by the IDIBELL Ethical Committee (no. AAALAC-3880) and performed in accordance with guidelines stated in The International Guiding Principles for Biomedical Research involving Animals, developed by the Council for International Organizations of Medical Sciences (CIOMS)44.
Supplementary Material
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
Acknowledgments
We thank the patients and their families. The research leading to these results has received funding from the European Community’s Seventh Framework Programme FP7/2007-2013 under grant agreement no. PIAPP-GA-2009-230614–Target-Melanoma project (F.P., J.O., W.M.G., M.E.), the Worldwide Cancer Research grant reference no. 15-0354 (M.E.), the European Research Council Advanced grant no. 268626–EPINORC project (M.E.), the Ministerio de Ciencia e Innovacion grant numbers SAF2011-22803 (M.E.) and FIS PI13-01339 (A. Villanueva), the CRUK Manchester Institute (C5759/A12328 to R.M.), the Wellcome Trust (100282/Z/12/Z to R.M.), the Cellex Foundation (M.E.) and the Health and Science Departments of the Catalan Government Generalitat de Catalunya 2005-SGR00727 (A. Villanueva) and 2014-SGR 633 (M.E.). M.V. was supported by a Formacion de Profesorado Universitario fellowship from the Spanish Ministry of Education. We thank the staff of the Animal Core Facility of Bellvitge Biomedical Research Institute for mouse care and maintenance. M.E. is an Institucio Catalana de Recerca i Estudis Avançats Research Professor.
Footnotes
Accession codes. Gene Expression Omnibus: Coordinates have been deposited with accession code GSE46522, DNA methylation and expression microarray data.
Author Contributions
M.V. and M.E. conceived the study and wrote the manuscript. M.V. performed most experiments with the help of H.J.F., P.L.-S., F.J.C., S.G., C.M., J.L., A.P., H.H. and S.M. A.Vidal and A. Villanueva, together with M.M.-I., performed the mouse studies. A.M.-C., M.R.G., J.L.M., M.T.F.-F., E.E., E.M.-C., R.B.-E., A.B., F.P., J.v.d.O., W.M.G., D.T.F., K.T.F., U.M., P.L. and R.M. analyzed the clinical outcome and drug response data and provided conceptual input. All authors discussed the results and commented on the manuscript.
Competing Financial Interests
The authors declare no competing financial interests.
Reprints and permissions information is available online at http://www.nature.com/reprints/index.html.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–692. doi: 10.1038/nrg3270. [DOI] [PubMed] [Google Scholar]
- 4.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang F, et al. Breast cancer methylomes establish an epigenomic foundation for metastasis. Sci Transl Med. 2011;3:75ra25. doi: 10.1126/scitranslmed.3001875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cunha S, et al. The RON receptor tyrosine kinase promotes metastasis by triggering MBD4-dependent DNA methylation reprogramming. Cell Reports. 2014;6:141–154. doi: 10.1016/j.celrep.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carmona FJ, et al. A comprehensive DNA methylation profile of epithelial-to-mesenchymal transition. Cancer Res. 2014;74:5608–5619. doi: 10.1158/0008-5472.CAN-13-3659. [DOI] [PubMed] [Google Scholar]
- 8.Lujambio A, et al. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci USA. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harbst K, et al. Multiple metastases from cutaneous malignant melanoma patients may display heterogeneous genomic and epigenomic patterns. Melanoma Res. 2010;20:381–391. [PubMed] [Google Scholar]
- 10.Marzese DM, et al. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum Mol Genet. 2014;23:226–238. doi: 10.1093/hmg/ddt420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacKie RM, Hauschild A, Eggermont AM. Epidemiology of invasive cutaneous melanoma. Ann Oncol. 2009;20(suppl 6):vi1–vi7. doi: 10.1093/annonc/mdp252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Villanueva MT. Skin cancer: in melanoma ulceration, size matters. Nat Rev Clin Oncol. 2012;9:370. doi: 10.1038/nrclinonc.2012.90. [DOI] [PubMed] [Google Scholar]
- 13.Mandalà M, Massi D. Tissue prognostic biomarkers in primary cutaneous melanoma. Virchows Arch. 2014;464:265–281. doi: 10.1007/s00428-013-1526-x. [DOI] [PubMed] [Google Scholar]
- 14.Tsao H, Chin L, Garraway LA, Fisher DE. Melanoma: from mutations to medicine. Genes Dev. 2012;26:1131–1155. doi: 10.1101/gad.191999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman HL, et al. The Society for Immunotherapy of Cancer consensus statement on tumour immunotherapy for the treatment of cutaneous melanoma. Nat Rev Clin Oncol. 2013;10:588–598. doi: 10.1038/nrclinonc.2013.153. [DOI] [PubMed] [Google Scholar]
- 16.Kaufman HL. Melanoma as a model for precision medicine in oncology. Lancet Oncol. 2014;15:251–253. doi: 10.1016/S1470-2045(14)70059-2. [DOI] [PubMed] [Google Scholar]
- 17.Weinreb A, Travo P. Discrimination between human melanoma cell lines by fluorescence anisotropy. Eur J Cancer Clin Oncol. 1984;20:673–677. doi: 10.1016/0277-5379(84)90015-4. [DOI] [PubMed] [Google Scholar]
- 18.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 19.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goueli BS, Powell MB, Finger EC, Pfeffer SR. TBC1D16 is a Rab4A GTPase activating protein that regulates receptor recycling and EGF receptor signaling. Proc Natl Acad Sci USA. 2012;109:15787–15792. doi: 10.1073/pnas.1204540109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Kasmi KC, et al. Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signalling pathway. J Immunol. 2007;179:7215–7219. doi: 10.4049/jimmunol.179.11.7215. [DOI] [PubMed] [Google Scholar]
- 22.Eisenberg MC, et al. Mechanistic modeling of the effects of myoferlin on tumor cell invasion. Proc Natl Acad Sci USA. 2011;108:20078–20083. doi: 10.1073/pnas.1116327108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frasa MA, Koessmeier KT, Ahmadian MR, Braga VM. Illuminating the functional and structural repertoire of human TBC/RABGAPs. Nat Rev Mol Cell Biol. 2012;13:67–73. doi: 10.1038/nrm3267. [DOI] [PubMed] [Google Scholar]
- 24.Onodera Y, et al. Rab5c promotes AMAP1–PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J Cell Biol. 2012;197:983–996. doi: 10.1083/jcb.201201065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corcoran RB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov. 2012;2:227–235. doi: 10.1158/2159-8290.CD-11-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prahallad A, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature. 2012;483:100–103. doi: 10.1038/nature10868. [DOI] [PubMed] [Google Scholar]
- 27.Yoon YK, et al. Combination of EGFR and MEK1/2 inhibitor shows synergistic effects by suppressing EGFR/HER3-dependent AKT activation in human gastric cancer cells. Mol Cancer Ther. 2009;8:2526–2536. doi: 10.1158/1535-7163.MCT-09-0300. [DOI] [PubMed] [Google Scholar]
- 28.Mirzoeva OK, et al. Basal subtype and MAPK/ERK kinase (MEK)-phosphoinositide 3-kinase feedback signaling determine susceptibility of breast cancer cells to MEK inhibition. Cancer Res. 2009;69:565–572. doi: 10.1158/0008-5472.CAN-08-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garnett MJ, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–775. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hon GC, et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2012;22:246–258. doi: 10.1101/gr.125872.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bert SA, et al. Regional activation of the cancer genome by long-range epigenetic remodeling. Cancer Cell. 2013;23:9–22. doi: 10.1016/j.ccr.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Kedlaya R, et al. Interactions between GIPC-APPL and GIPC-TRP1 regulate melanosomal protein trafficking and melanogenesis in human melanocytes. Arch Biochem Biophys. 2011;508:227–233. doi: 10.1016/j.abb.2011.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang ZM, et al. Targeting protein-trafficking pathways alters melanoma treatment sensitivity. Proc Natl Acad Sci USA. 2012;109:553–558. doi: 10.1073/pnas.1118366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gray-Schopfer V, Wellbrock C, Marais R. Melanoma biology and new targeted therapy. Nature. 2007;445:851–857. doi: 10.1038/nature05661. [DOI] [PubMed] [Google Scholar]
- 36.Middleton MR, et al. Randomized phase III study of temozolomide versus dacarbazine in the treatment of patients with advanced metastatic malignant melanoma. J Clin Oncol. 2000;18:158–166. doi: 10.1200/JCO.2000.18.1.158. [DOI] [PubMed] [Google Scholar]
- 37.Eggermont AM, Spatz A, Robert C. Cutaneous melanoma. Lancet. 2014;383:816–827. doi: 10.1016/S0140-6736(13)60802-8. [DOI] [PubMed] [Google Scholar]
- 38.Catalanotti F, et al. Phase II trial of MEK inhibitor selumetinib (AZD6244, ARRY-142886) in patients with BRAFV600E/K-mutated melanoma. Clin Cancer Res. 2013;19:2257–2264. doi: 10.1158/1078-0432.CCR-12-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Girotti MR, et al. Inhibiting EGF receptor or SRC family kinase signaling overcomes BRAF inhibitor resistance in melanoma. Cancer Discov. 2013;3:158–167. doi: 10.1158/2159-8290.CD-12-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altman DG, et al. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci USA. 2012;109:10522–10527. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moutinho C, et al. Epigenetic inactivation of the BRCA1 interactor SRBC and resistance to oxaliplatin in colorectal cancer. J Natl Cancer Inst. 2014;106:djt322. doi: 10.1093/jnci/djt322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez-Serra P, et al. A DERL3-associated defect in the degradation of SLC2A1 mediates the Warburg effect. Nat Commun. 2014;5:3608. doi: 10.1038/ncomms4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard-Jones N. A CIOMS ethical code for animal experimentation. WHO Chron. 1985;39:51–56. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.