Abstract
Mass spectrometry (MS) is the main technology used in proteomics approaches. However, on average 75% of spectra analysed in an MS experiment remain unidentified. We propose to use spectrum clustering at a large-scale to shed a light on these unidentified spectra. PRoteomics IDEntifications database (PRIDE) Archive is one of the largest MS proteomics public data repositories worldwide. By clustering all tandem MS spectra publicly available in PRIDE Archive, coming from hundreds of datasets, we were able to consistently characterize three distinct groups of spectra: 1) incorrectly identified spectra, 2) spectra correctly identified but below the set scoring threshold, and 3) truly unidentified spectra. Using a multitude of complementary analysis approaches, we were able to identify less than 20% of the consistently unidentified spectra. The complete spectrum clustering results are available through the new version of the PRIDE Cluster resource (http://www.ebi.ac.uk/pride/cluster). This resource is intended, among other aims, to encourage and simplify further investigation into these unidentified spectra.
“Untargeted” or “discovery” proteomics approaches have become a key instrument in systems biology to unravel a sample’s underlying biological functions. The most common approach in the field is shotgun proteomics. Proteins are digested using a protease and the resulting peptides identified using tandem mass spectrometry (MS/MS). Thereby, most proteins present in the sample can theoretically be identified and quantified1. However, on average, around three-quarters of spectra measured in an MS/MS experiment remain unidentified (Supplementary Figure 1). When investigating these spectra, many seem to be of high quality and likely originate from peptides2. Sequence database-based search engines3–5 rely on theoretical spectra based on a user-defined protein sequence database and a set of protein post-translational modifications (PTMs). Therefore, unidentified peptides are most likely either low signal-to-noise events, not present in the used sequence database (e.g. sequence variants), or contain unexpected PTMs.
Several alternative approaches exist that improve the rate of identified spectra such as de-novo sequencing6, sequence tagging-based approaches7, precursor mass-tolerant sequence database searches2 and spectral library search engines8. Additionally, open modification searches rely on a sequence database or spectral library but allow for mass shifts to occur9. All of these approaches share two disadvantages which currently prevent their general use: they are computationally expensive and tend to return ambiguous results where more than one resulting peptide sequence can be derived from the same spectrum at equal probabilities.
We propose to use spectrum clustering at a large-scale to shed light on unidentified spectra. The likelihood that sequence variants and peptides with unexpected PTMs have been observed grows with increasing numbers of datasets coming from different origins and experimental settings. Thus, unidentified spectra can be systematically studied by exploiting the continuously growing number of MS/MS datasets available. More concretely, we use our approach to identify three distinct sets of spectra: 1) incorrectly identified spectra, 2) spectra correctly identified but below the set scoring threshold, and 3) truly unidentified spectra.
The PRIDE Archive database10 is one of the largest public proteomics MS data repositories worldwide and part of the ProteomeXchange Consortium of proteomics resources11. In 2013, we introduced a spectrum clustering algorithm that accurately grouped all identified spectra in PRIDE Archive at the time12. This allowed us to identify reliable peptide spectrum matches (PSMs) within the submitted heterogeneous data. However, the amount of data in PRIDE Archive has grown exponentially in recent years.
We first report the development of a novel spectrum clustering algorithm that is highly scalable and can cluster large amounts of unidentified spectra without incurring a high degree of false positive matching. We show that we can accurately discriminate all three subsets of spectra mentioned above. Most interestingly, for the very first time we are able to recognize millions of spectra that are commonly observed in hundreds of proteomics experiments but consistently remain unidentified.
These complete data are now publicly available as part of the redeveloped PRIDE Cluster resource. As an example, we show how more expensive computational methods can be used to identify the true origin of many of these spectra. Using a multitude of complementary analysis approaches we were able to explain roughly 20% of the originally unidentified spectra. We hope that this new resource encourages the development of new methodologies to unravel these now characterised unidentified spectra.
Results
Probabilistic Spectrum Comparison to Accurately Cluster MS/MS Spectra
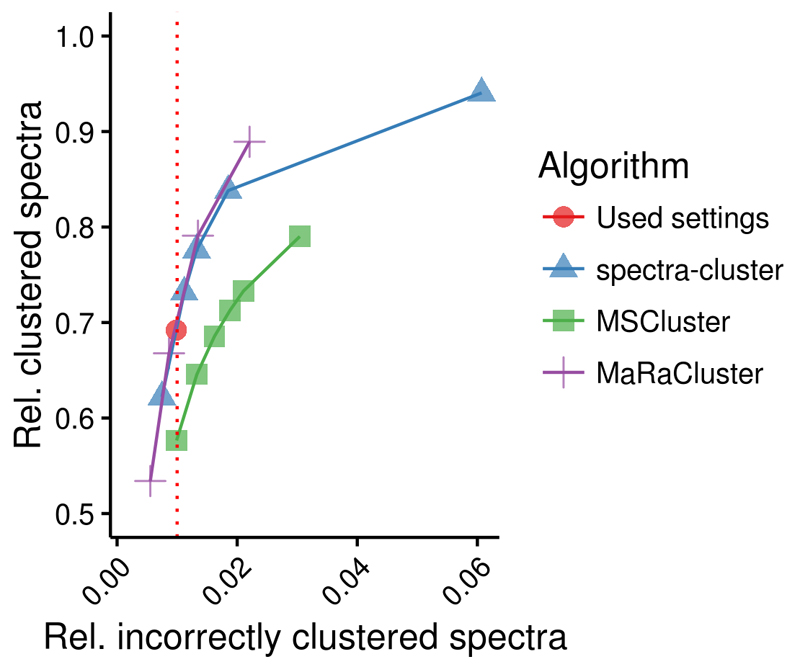
The new spectra-cluster algorithm was specifically developed using the Apache Hadoop framework13, 14 to reach two main goals: 1) to increase spectrum clustering accuracy and 2) to be scalable to handle the exponential data increase in PRIDE Archive. To increase spectrum clustering accuracy, based on the proportion of incorrectly clustered spectra, we developed a novel method to assess the similarity between two spectra: instead of the commonly used normalized dot product we employed a probabilistic scoring approach similar to that of the spectrum library search engine Pepitome15 (Online Methods). To evaluate the accuracy of the algorithm we first reanalysed 209 human datasets from PRIDE Archive (Supplementary Table 1) resulting in 10 million reliably identified spectra using SpectraST at a 1% peptide FDR (Online Methods). Based on this comprehensive test dataset we concluded that the new spectra-cluster algorithm is considerably more accurate while being able to process a large and heterogeneous dataset (Figure 1, Supplementary Note 1). Additionally, it is robust when handling chimeric spectra and shows a stable accuracy with an increasing cluster size (Supplementary Note 1).
Figure 1.
Accuracy of the spectra-cluster algorithm compared to the MSCluster36 and MaRaCluster37 algorithms. The three algorithms were tested with a test dataset built from 209 human datasets from PRIDE Archive (Online Methods, Supplementary Table 1). Clustering sensitivity (y-axis) was assessed based on the number of clustered spectra (shown as relative to the total number of spectra in the test dataset). Clustering specificity (x-axis) was assessed based on the proportion of spectra that were not identified as the most common peptide in a cluster. Only clusters with at least five spectra were taken into consideration (Supplementary Note 1).
We then clustered all identified and unidentified spectra from all publicly available “complete” datasets16 in PRIDE Archive by April 2015 (256 million spectra, 190 million unidentified and 66 million identified spectra). First, unidentified spectra were filtered using the de-novo search engine PepNovo’s peptide filtering function, that estimates if a MS/MS spectrum represents a peptide6 (Online Methods). This reduced the number of unidentified spectra by 42% (from 190 million to 111 million). The remaining unidentified and identified spectra were clustered, resulting in 28 million clusters – approximately a six-fold reduction in the initial number of spectra. This analysis took five days on a 30-node Hadoop cluster using 340 CPU cores (Online Methods).
Validating Spectrum Clustering Accuracy
The original PRIDE Cluster algorithm was developed to identify reliable PSMs in the heterogeneous data submitted to PRIDE Archive12. Then, we found that if at least 70% of the spectra within a cluster were identified as the same peptide, these identifications could be regarded as reliable. To validate the accuracy of the new spectra-cluster algorithm we repeated this analysis with the same test dataset mentioned above - considerably larger than the one we previously used12 (Supplementary Note 2). We again found that if at least 70% of spectra in a cluster were identified as the same peptide sequence, these identifications could be considered reliable. This indicates that the clustering accuracy in the new algorithm remained stable despite the increased amount of data. Reliable clusters were therefore defined as clusters with at least three identified spectra where at least 70% of the spectra were identified as the same peptide (Supplementary Note 2).
The Updated PRIDE Cluster Resource
The redeveloped PRIDE Cluster resource provides full access to these spectrum clustering results and links them with the originally submitted data in PRIDE Archive (Supplementary Note 3). The clustering process will be repeated and the data updated at least once a year. Data can be accessed using a RESTful API (Application Programming Interface, Supplementary Note 3) and a web interface (http://www.ebi.ac.uk/pride/cluster, Supplementary Note 4). Additionally, spectral libraries for 16 species are made available (http://www.ebi.ac.uk/pride/cluster/#/libraries). Two additional filters were used to generate these libraries from reliable clusters: 1) spectra must not be dominated by a single peak, and 2) at least 20% of the total ion current must be explained through b- and y-ions. The PRIDE Cluster derived human spectral library showed comparable accuracy to the human one from NIST (National Institute of Standards and Technology, Supplementary Note 5).
Additionally, the complete raw spectrum clustering results are made available for further re-analysis. The freely available open-source clustering-file-reader Java API can be used to parse the raw result files (https://github.com/spectra-cluster/clustering-file-reader). Furthermore, consensus spectra of selected subsets (e.g. large unidentified spectrum clusters for multiple species) are available as MGF (Mascot Generic Files) and mzML files (http://www.ebi.ac.uk/pride/cluster/#/results).
The complete source code of the PRIDE Cluster project is available as open source software under the permissive Apache 2.0 license at https://github.com/spectra-cluster (clustering algorithm) and https://github.com/PRIDE-Cluster (web application). Additionally, we have created a stand-alone Java application of the spectra-cluster algorithm, the spectra-cluster-cli (https://github.com/spectra-cluster/spectra-cluster-cli), which can be run on any standard computer (Windows, Mac OS or Linux).
The presented release of the PRIDE Cluster resource (version 2015-04) contains 2.6 million reliable spectrum clusters containing 37 million identified and 9 million unidentified spectra. The relatively low number of validated originally submitted identifications (56%) is a result of the rigorous thresholds required in this highly heterogeneous dataset. The vast majority of validated clusters stem from human experiments (990,853), followed by mouse (313,247), Arabidopsis thaliana (268,281), and rat (84,970). These correspond to 222,777, 103,834, 70,119, and 37,552 distinct peptides for human, mouse, A. thaliana and rat, respectively. While the two other major repositories for MS/MS data, PeptideAtlas17 and the Global Proteome Machine database (GPMDB)18 hold MS-based evidence for most of these peptides, 50,319 human, 25,101 mouse, 14,671 A. thaliana and 6,873 rat peptides are only found in the PRIDE Cluster dataset (Supplementary Figure 2). Additionally, the list of reliable peptides identifies 870 human proteins with at least two unique peptides that have at least nine amino acids (see guidelines of the Human Proteome Project19) annotated without “experimental evidence at protein level” in the human UniProtKB/SwissProt database (release 2016-03, Supplementary Figure 3).
Identifying Common Incorrect Peptide Identifications
We observed 75,310 clusters (containing 2.2 million identified and 3.4 million unidentified spectra) with at least 10 identified spectra of which less than 50% were identified as the same peptide in the original data submitted to PRIDE Archive. Therefore, these spectra were either incorrectly clustered or represent peptides which are prone to be incorrectly identified.
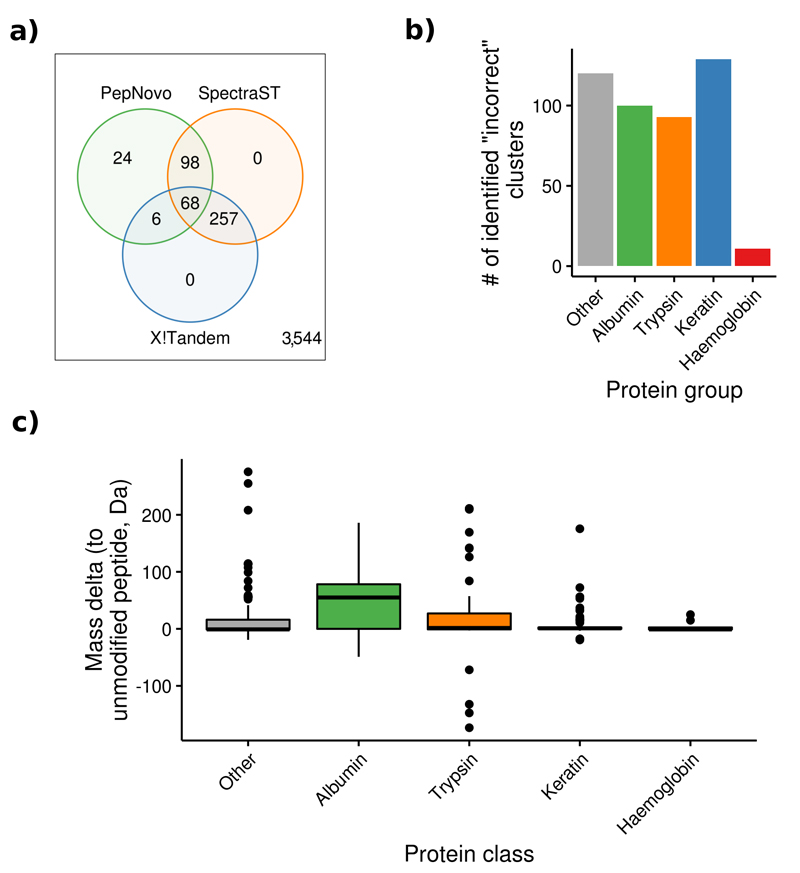
We reprocessed the originally submitted spectra of the 3,997 large clusters containing at least 100 spectra where at least one spectrum came from a human experiment (corresponding to 555,339 identified and 3.2 million unidentified spectra, Online Methods, Supplementary Figure 4). A spectrum cluster was accepted as identified if: 1) at least two of the approaches identified the majority of spectra (more than 50% of spectra in a cluster) as the same peptide, or 2) if PepNovo derived a sequence from the majority of spectra that matched a known common contaminant or proteins that are commonly found in proteomics experiments.
We were able to identify 453 clusters (11%, Figure 2a). Overall, 74% of these peptides originated from keratins, trypsin, albumin and haemoglobin (Figure 2b). Albumin peptides often contained PTMs, which could explain the reason behind the originally missed identifications. Keratin, haemoglobin and trypsin peptides, however, were mostly unmodified (Figure 2c). Originally incorrectly identified keratin peptides were mostly found in non-human experiments. In these cases, the originally used search databases most likely did not contain contaminants, which prevented the search engine from providing the correct peptide assignments for these spectra.
Figure 2.
Overview of the results of the analysis to highlight commonly found incorrect peptide identifications. (a) Overall, 424 (11%) human clusters were identified using X!Tandem, SpectraST and PepNovo. (b) The vast majority of identified peptides originated from keratin, albumin, trypsin, and haemoglobin. (c) Albumin peptides were modified more often than peptides from any other protein (center line marks the median, edges the first and third quartile, whiskers extend to +/-1.58 times the inter-quartile ratio divided by the square root of the number of observations, single points denote measurements outside this range).
Inferring Identifications for Originally Unidentified Spectra through Spectrum Clustering
In our analysis, 9.1 million originally unidentified spectra were matched to reliably identified spectra (included in reliable clusters). These additional identifications included peptides containing biologically relevant PTMs such as phosphorylation: 49,263 reliable clusters (containing 560,000 identified and 130,000 unidentified spectra) contained phosphorylated peptides. These clusters originated from 81 phosphoproteomics studies (where enrichment for phosphopeptides was performed) and 145 non-phosphorylation studies (where no enrichment was performed, Supplementary Table 2). To validate these clusters, we additionally analysed the consensus spectra of those clusters containing spectra from human experiments (28,821 clusters) using SpectraST and a human phosphopeptide spectral library20 (Online Methods). Overall, 8,417 (34%) of the spectra were identified at a 1% peptide FDR, of which 8,089 (96%) were identified as the most common peptide sequence in the cluster.
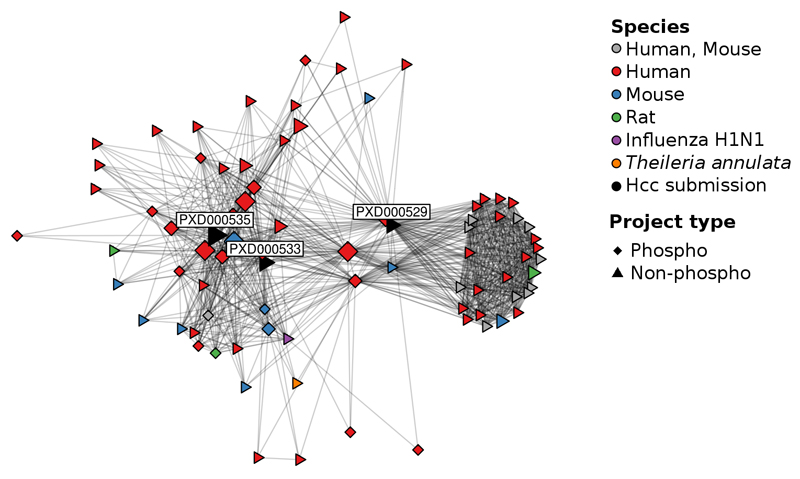
One example is a study included in the chromosome-centric Human Proteome Project (HPP, Chromosomes 1, 8 and 20, datasets PXD000529, PXD000533 and PXD000535)21. The researchers analysed the hepatocellular carcinoma cell lines Hep3B and MHCC97H and, since they did not perform any phospho-enrichment, the modification was not taken into consideration during the original analysis. However, 1,859 originally unidentified spectra were clustered with reliable identifications of phosphorylated peptides. Most of these reliably identified spectra came from five phosphorylation studies (Figure 3): PXD00031422, a study on human lung cancer, PXD00094823, a study on breast cancer, PRD00071124, a study on data extraction techniques, PRD00011825, a study on human leukocytes, and PXD00018526, a study on kinase substrates in leukaemia cells. The 1,859 originally unidentified spectra corresponded to 290 distinct peptides (coming from 344 PSMs) and to 222 proteins (Supplementary Table 3), which primarily regulate translation or are involved in RNA processing and DNA repair (Online Methods, data not shown).
Figure 3.
Identified spectra from a diverse range of datasets, including spectra from experiments in other species, led to newly identified phosphorylated peptides in the Chromosome-Centric HPP datasets (PXD000529, PXD000533 and PXD000535). Connections between datasets are based on the shared spectra within a cluster, only taking clusters of phosphorylated peptides into consideration.
A second example is a non-phosphorylation study performed by Menschaert et al. using mouse embryonic stem cells (PXD000124)27. Here, among others, a phosphorylation study on human leukaemia cells (PXD000185)26 and on human breast cancer cells (PXD000472)28 led to potential additional phosphopeptide identifications. Albeit only 82 unidentified spectra were clustered with phosphorylated peptides (Supplementary Table 3), this example illustrates that additional identifications are possible across different species. A more peculiar but highly plausible example is that a phosphorylation study on Plasmodium falciparum (PXD000070)29 led to phosphopeptide identifications in a non-phosphorylation study characterising the erythrocyte membrane human proteome (PRD000092, Supplementary Table 3)30. Since Plasmodium falciparium develops in human erythrocytes, it is likely that both studies contained peptides from human erythrocytes as well.
Analysing Clusters Containing Only Unidentified Spectra
A total of 19 million clusters (corresponding to 105 million spectra) contained only unidentified spectra. Out of these, 41,155 clusters contained more than 100 spectra (corresponding to 12.1 million spectra, 12%) indicating that these may represent highly abundant molecules that have been detected (but not identified) across many different experimental settings.
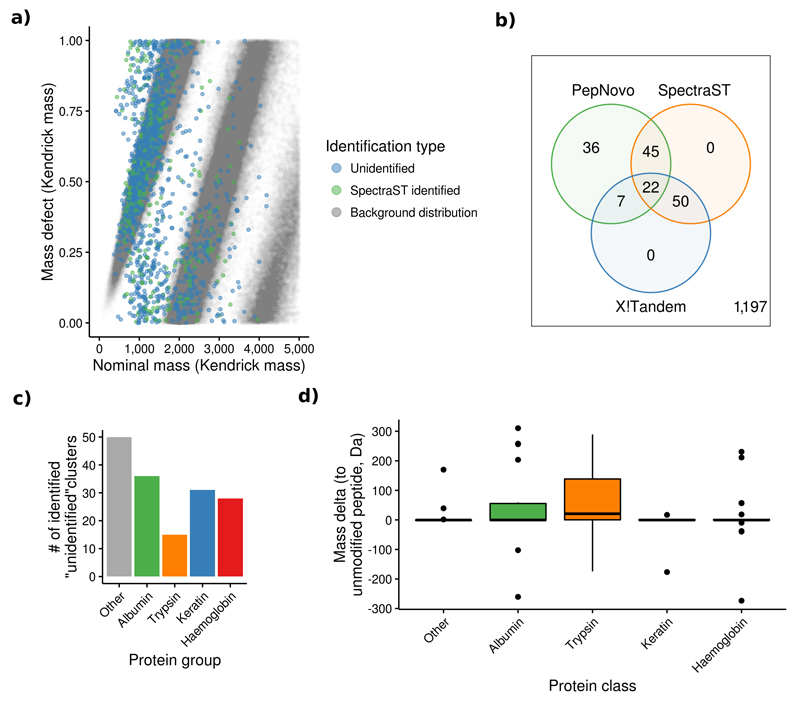
A mass defect analysis31 of all 22,344 large human clusters (taking only high-resolution spectra from Orbitrap instruments into consideration) indicated that the vast majority of these unidentified spectra originated from peptides (Figure 4a). An additional search against a metabolite database using OpenMS32 did not result in any reliable identifications (Online Methods). Finally, a search using MSPLIT33 did not reliably identify any chimeric spectra (Online Methods, Supplementary Figure 5).
Figure 4.
Overview of the results of the analysis of clusters containing only unidentified spectra. (a) The mass defect analysis showed that ~80% of the unidentified human spectra have a similar distribution as the background one created from all in silico digested tryptic peptides in UniProtKB/SwissProt. The remaining 20% of spectra not included within this distribution may be explained by the fact that only unmodified, fully tryptic peptides were considered for this distribution. (b) 160 (12%) of the large unidentified human clusters were identified using SpectraST, X!Tandem and PepNovo. (c) More than 50% of these identifications were peptides coming from albumin, trypsin, keratin and haemoglobin. (d) Only trypsin peptides were commonly modified (e.g. dimethylated, center line marks the median, edges the first and third quartile, whiskers extend to +/-1.58 times the inter-quartile ratio divided by the square root of the number of observations, single points denote measurements outside this range).
We reprocessed the consensus spectra of large unidentified clusters containing spectra from human (more than 100 spectra), mouse (more than 10 spectra), and A. thaliana (more than 10 spectra) experiments. These amounted to 22,344 unidentified clusters for human (7 million spectra), 131,353 unidentified clusters for mouse (5 million spectra), and 8,055 unidentified clusters for A. thaliana (294,079 spectra). The consensus spectra were searched with SpectraST using its open modification search function with a precursor tolerance of 500 m/z units against the corresponding NIST spectral library (for human and mouse) or, in the case of A. thaliana, the PRIDE Cluster spectral library (version 2015-04).
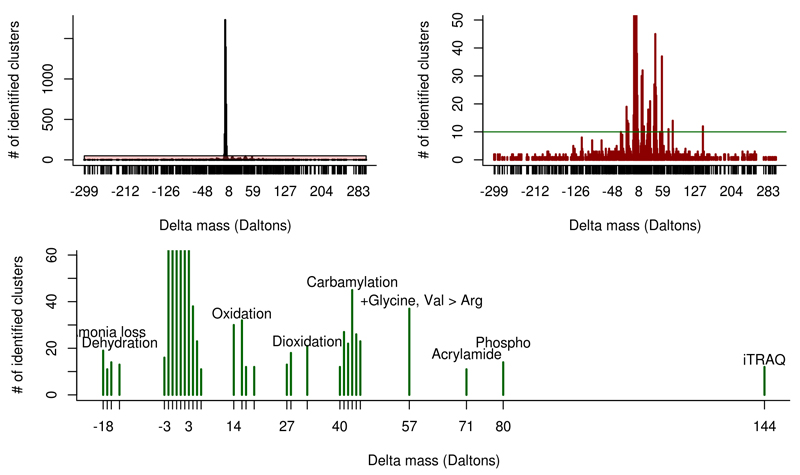
We were able to identify 5,560 human (25%), 16,439 mouse (13%), and 250 A. thaliana (3%) consensus spectra at a 1% peptide FDR (Figure 5, Supplementary Figures 6 and 7). The vast majority of newly identified peptides were detected with a delta mass between -2 and +4 Da, similar to previous findings2 (Supplementary Table 4).
Figure 5.
Summary of results for the analysis of human clusters containing only unidentified spectra. The vast majority of delta masses observed in the open modification search were between -2 and +4 Da (top left panel). After adjusting the y-axis it becomes apparent that several other delta masses were observed at high frequency (top right panel). When limiting these delta masses to only masses that were observed at least for ten different clusters, the vast majority of delta masses could be mapped to known PTMs as well as to one potential amino acid substitution (lower panel). For the complete list of the found delta masses see Supplementary Table 4.
We then reprocessed the originally submitted spectra of the 1,357 human clusters with at least 1,000 spectra, 835 mouse clusters with at least 500 spectra and 576 A. thaliana clusters with at least 50 spectra using the above mentioned pipeline (Supplementary Figure 4). In total, 160 (12%) human, 122 (15%) mouse, and 50 (9%) A. thaliana clusters were identified. Most human clusters were identified as peptides corresponding to trypsin, albumin, haemoglobin, and keratin (Figure 4b-d). The identifications of the mouse and A. thaliana clusters were more heterogeneous but could not be related to any additional protein subgroups (Supplementary Figures 8 and 9). In all three species, trypsin peptides often contained PTMs such as methylation and dimethylation, which can be artificially introduced to prevent self-digestion34 (Figure 5c, Supplementary Figures 8c, 9c). Interestingly, 242 additional human clusters had high-score PepNovo results that could not be matched to the used human sequence database.
Discussion
We studied three distinct subsets of mass spectra using a spectrum clustering approach: 1) incorrectly identified spectra, 2) spectra correctly identified but that fall below the set scoring threshold, and 3) truly unidentified spectra. This enabled us to highlight spectra consistently unidentified across thousands of experiments available in PRIDE Archive and assign identifications to 9 million originally unidentified spectra. Additionally, inadequate originally used search engine settings such as missed PTMs can be alleviated. The fact that many of these incorrect identifications are caused by known contaminants will be used as basis for a future service in PRIDE Archive: we plan that submitters will be automatically warned if their datasets contain a high proportion of such potentially incorrect identifications.
However, these data must be seen as an initial first step. We are unaware of any method to aptly quantify the FDR for the shown “rescued”, inferred identifications. Nevertheless, this information can be used to re-analyse the dataset of interest taking highlighted missed PTMs or missing sequences into account. Thereby, spectrum clustering may act as an unbiased assessment of the used search strategy.
The large amount of seemingly “good” unidentified spectra in MS/MS based experiments is currently a core interest in the field and many of these unidentified spectra seem to be originating from modified peptides2. We are now able to accurately target and in some cases identify spectra that are observed across a multitude of experiments but remain unidentified. The majority of mass shifts that were observed could be linked to known common PTMs. The accuracy of this analysis is limited though as we only analysed consensus spectra (see the mass deltas in Supplementary Table 4). As these spectra were generated from both high- and low-resolution data, the resulting mass accuracy is insufficient to accurately analyse the observed mass shifts. Still, it is intriguing that we were only able to identify less than 20% of these data and still cannot explain a large number of observed mass deltas.
The available raw clustering results represent a spectral archive of the public data in PRIDE Archive and can be seen as a compressed data storage mechanism. This has been proposed as an ideal way to make such huge amounts of data available for reanalysis35 with two main challenges to overcome: (i) to enable the transfer of GBs of data across the Internet; and (ii) to improve the spectrum clustering algorithms to handle the growth in data while maintaining accuracy. The first issue has been overcome by the availability of faster file transfer protocols like Aspera (http://asperasoft.com/), routinely used by PRIDE Archive submitters. The second issue has now been tackled with the new spectra-cluster algorithm.
Our analysis clearly only touches the tip of the iceberg. Deriving novel biological knowledge from these potential novel identifications goes far beyond the capabilities of a single research group. Creating a sensible subset of spectra to start an in-depth analysis of unidentified spectra has been very challenging until now. Therefore, we provide these ready-to-use collections of commonly unidentified spectra representing the source data of the shown analyses. To our knowledge, this is the first time that the clustered version of a complete proteomics repository is made available. Thereby, the described three fractions of spectra can be readily recognized and investigated at a repository scale.
Online Methods
Test dataset and assessment of clustering accuracy
To validate the spectrum clustering algorithm, 209 human datasets from PRIDE Archive were reprocessed (Supplementary Table 1). The submitted identified spectra were searched using SpectraST8 (version 5.0) with a precursor tolerance of 3 m/z units, ignoring the original charge states, and enabling the calculation of p-values. All other settings were left at their default values. As spectral library, we used a combination of the NIST human Orbitrap (November 2014) and iontrap (May 2014) libraries, and the global proteome machine’s (GPM) common Repository of Adventitious Proteins’ (cRAP) (downloaded on July 2014). The libraries were combined and decoy spectra appended using SpectraST. Roughly 10 million spectra were identified at a 1% peptide FDR.
The identified spectra were clustered using the spectra-cluster algorithm and the MSCluster36 and MaRaCluster37 algorithms as benchmark. Sensitivity was assessed based on the proportion of clustered spectra (spectra clustered with at least another spectrum). Specificity was assessed based on the proportion of spectra identified as a different peptide than the most common peptide identification reported in the cluster. The MSCluster algorithm was run using the default settings, which are optimized for heterogeneous, low-resolution data. The MaRaCluster algorithm was set to a precursor tolerance of 1,000 ppm. All other parameters were left at the default setting.
Probabilistic Spectrum Comparison
A probabilistic method was developed to assess the similarity between two spectra. The approach is based on the scoring function used in Pepitome15 (Supplementary Note 6). First, precursor peaks are removed from the MS/MS spectrum and peak picking is performed keeping the 70 highest peaks per spectrum (Supplementary Note 7). Next, the probability that the number of matched peaks occurred by random is modelled using a hypergeometric distribution. The probability that the rank distribution of matched peaks occurred by chance is assessed using the Kendall’s Tau correlation. For both tests point probabilities are calculated only, instead of cumulative probabilities. This sacrifices mathematical accuracy for improved speed, which is essential for the clustering process of large amounts of spectra. The two probabilities are then combined using Fisher’s method38 and reported as the negative logarithm. This comparison is only performed using the peaks that explain 50% of the total ion current (of the pre-filtered spectrum) or at least the 25 highest peaks. The consensus spectrum is then built using all peaks.
“MapReduce” Adaptation of the MSCluster Algorithm
The algorithm’s logic follows the MSCluster approach developed by Frank et al.36, adapted to the requirements of the “MapReduce” programming model, using our probabilistic spectrum comparison method instead of the normalized dot product (Supplementary Note 7). First, as described above, peak filtering is performed in a pure mapping job. Next, five successive rounds of clustering are performed with decreasing similarity thresholds to reach a final accuracy of 99%. Spectra are mapped into bins depending on the precursor’s m/z value. In the initial round, a bin width of 0.2 m/z units is used. Subsequent rounds are repeated using a bin width of 4 m/z units. Finally, the five rounds of clustering are repeated offsetting the bins by half of the used bin width. Thereby, the overlapping regions of the previous bins are processed together. In the first and second round, only spectra that share one of their five highest peaks are compared. In subsequent rounds, only spectra (or the corresponding clusters) that were among the previously 30 highest scoring matches are compared.
Similarly to the MSCluster algorithm an empirically derived cumulative distribution function is used to adapt the clustering threshold based on the number of comparisons (Supplementary Note 8). This prevents a decreased clustering accuracy caused by the multiple testing problem when processing very large datasets. The cumulative distribution function was derived by comparing randomly selected spectra from the test dataset (> 10 billion comparisons). Spectra were considered different if they were originally identified as different peptides and had a precursor mass difference of at least 4 m/z units. Thereby, the proportion of incorrectly matched spectra at given similarity scores could be estimated.
Code Availability
The complete source code of the PRIDE Cluster project is available as open source software under the permissive Apache 2.0 license at https://github.com/spectra-cluster (clustering algorithm, Hadoop implementation, stand-alone implementation) and https://github.com/PRIDE-Cluster (web application). A stand-alone Java application of the spectra-cluster algorithm, the spectra-cluster-cli is available at https://github.com/spectra-cluster/spectra-cluster-cli.
Identifying consensus spectra from reliable phosphopeptide spectral clusters
Consensus spectra of reliable human clusters representing phosphopeptides were searched using SpectraST (version 5.0)8 against the human phospho library from PeptideAtlas17, 20 (version 2013-07-15). The precursor tolerance was set to 3.0 m/z units, the spectra’s charge states were ignored and the calculation of p-values enabled and used for peptide FDR filtering at 1% FDR.
Identifying incorrectly identified and unidentified spectra
Consensus spectra or originally submitted spectra were processed using SpectraST8 (version 5.0), X!Tandem5 (version Sledgehammer, 2013.09.01.1), and PepNovo6 (release 20101117).
For SpectraST, the spectral library was either the combined human spectral library used for the test dataset or the cRAP spectral library alone. Decoy spectra were appended using SpectraST. All SpectraST searches were performed using a precursor mass tolerance of 500 m/z units, the open modification search option ignoring the spectra’s charge states and enabled the calculation of p-values which were used for FDR filtering (Supplementary Figure 4).
For X!Tandem, either the cRAP sequence database alone (downloaded on July 2014) or the concatenation of cRAP and UniProt’s human proteome (2014-07) were used. The precursor tolerance was set to 3 m/z units, fragment tolerance to 0.4 m/z units and the refinement mode disabled. Carbamidomethylation was set as fixed modification and oxidation of M and N-terminal acetylation as variable modifications. If the search was performed only against the cRAP library the following additional variable modifications were taken into consideration: Formylation on S and K, deamidation on N and Q, carboxylation on K, and N-terminal methylation. All other settings were the default ones.
PepNovo was set to use the “CID_IT_TRYP” fragmentation model. Allowed protein modifications were defined as follows: carbamidomethylation, oxidation on M, phosphorylation on S, T, and Y, acetylation on T, S, Y, and N-terminal, methylation on C, H, K, N, Q, R, and N-terminal, and formylation on K, S, and T. The ten highest scoring solutions were taken into consideration. In addition the best scoring as well as the most common solution across all spectra in one cluster were considered for the analysis.
MSPLIT33 (version 1.0) was used to identify spectra originating from more than one peptide. Precursor tolerance was set to 3.0 Da, results were filtered at 1% peptide FDR using the application’s spectrumMatchClassify.pl script and the above-mentioned spectral library used for the search.
The pathway overrepresentation analysis was performed using the PANTHER39 Overrepresentation Test (release 20150430, PANTHER version 10.0 released 2015-05-15) with ‘Homo sapiens (all genes)’ as reference list and ‘PANTHER GO-Slim Biological Process’ as annotation data set. P-values were adjusted using a Bonferroni correction.
The mass defect analysis was performed on re-calculated consensus spectra taking only spectra from Orbitrap instruments into consideration. Then, the nominal and fractional masses of the clusters’ precursor ions were plotted as previously described31. The background distribution was created based on an in silico tryptic digest of the UniProtKB/SwissProt database (release 2013-09). These high-resolution consensus spectra were additionally searched against MassBank using its SOAP (Simple Object Access Protocol) API (performed on September 2015).
Supplementary Material
Acknowledgements
This work was supported by the Vienna Science and Technology Fund (WWTF) (grant LS11-045), the Wellcome Trust (grant WT101477MA to H.H. and J.A.V.), the BBSRC (‘PROCESS’ grant BB/K01997X/1 to H.H. and J.A.V, ‘Quantitative Proteomics’ grant BB/I00095X/1 to H.H.), the Deutsche Forschungsgemeinschaft (grant SFB685/B1 to O.K.), and the BMBF (grant 01ZX1301F to O.K.). We would like to acknowledge the attendants to the Midwinter Proteomics Bioinformatics Seminar 2015 at Semmering (Austria), and the Bioinformatics Hub at the HUPO conference 2015 at Vancouver (Canada), who provided valuable feedback on the data analysis. Finally, we want to acknowledge M. The and L. Käll for their support during the benchmarking of their MaRaCluster algorithm.
Abbreviations
- API
Application Programming Interface
- cRAP
common Repository of Adventitious Proteins
- GPM
Global Proteome Machine
- HPP
Human Proteome Project
- PRIDE
PRoteomics IDEntifications (database)
- MGF
Mascot Generic Files
- MS
Mass Spectrometry
- NIST
National Institute of Standards and Technology
- PSMs
Peptide Spectrum Matches
- PTMs
Post-Translational Modifications
- PANTHER
Protein ANalysis THrough Evolutionary Relationships
- SOAP
Simple Object Access Protocol
- MS/MS
tandem Mass Spectrometry
Footnotes
Author Contributions
J.G. developed the clustering algorithm, ran the experiments and performed the data analysis. D.L.T. contributed to the development of the probabilistic scoring approach. Y.P.R. contributed to the data analysis. J.G. and R.W. developed the Java APIs for the spectrum clustering analysis pipeline. S.L., R.W. and J.G. developed the Hadoop implementation. J.A.D., N.d.T., Y.P.R. and R.W. created the web interface and the API of the PRIDE Cluster resource. M.R., M.W. and O.K. performed the metabolite search. J.G., R.W., H.H. and J.A.V. supervised the project. J.G. and J.A.V. wrote the manuscript, with contributions from the rest of the authors.
Competing Financial Interest
The authors declare no competing financial interests.
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Chick JM, et al. A mass-tolerant database search identifies a large proportion of unassigned spectra in shotgun proteomics as modified peptides. Nature biotechnology. 2015;33:743–749. doi: 10.1038/nbt.3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eng JK, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 4.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 6.Frank A, Pevzner P. PepNovo: de novo peptide sequencing via probabilistic network modeling. Analytical chemistry. 2005;77:964–973. doi: 10.1021/ac048788h. [DOI] [PubMed] [Google Scholar]
- 7.Tabb DL, Ma ZQ, Martin DB, Ham AJ, Chambers MC. DirecTag: accurate sequence tags from peptide MS/MS through statistical scoring. Journal of proteome research. 2008;7:3838–3846. doi: 10.1021/pr800154p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam H, et al. Development and validation of a spectral library searching method for peptide identification from MS/MS. Proteomics. 2007;7:655–667. doi: 10.1002/pmic.200600625. [DOI] [PubMed] [Google Scholar]
- 9.Ma CW, Lam H. Hunting for unexpected post-translational modifications by spectral library searching with tier-wise scoring. Journal of proteome research. 2014;13:2262–2271. doi: 10.1021/pr401006g. [DOI] [PubMed] [Google Scholar]
- 10.Vizcaino JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic acids research. 2016;44:D447–D556. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vizcaino JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature biotechnology. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griss J, Foster JM, Hermjakob H, Vizcaino JA. PRIDE Cluster: building a consensus of proteomics data. Nature methods. 2013;10:95–96. doi: 10.1038/nmeth.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao Q, et al. Design and development of a medical big data processing system based on Hadoop. Journal of medical systems. 2015;39:23. doi: 10.1007/s10916-015-0220-8. [DOI] [PubMed] [Google Scholar]
- 14.Hodor P, Chawla A, Clark A, Neal L. cl-dash: rapid configuration and deployment of Hadoop clusters for bioinformatics research in the cloud. Bioinformatics. 2015;32:301–303. doi: 10.1093/bioinformatics/btv553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dasari S, et al. Pepitome: evaluating improved spectral library search for identification complementarity and quality assessment. Journal of proteome research. 2012;11:1686–1695. doi: 10.1021/pr200874e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ternent T, et al. How to submit MS proteomics data to ProteomeXchange via the PRIDE database. Proteomics. 2014;14:2233–2241. doi: 10.1002/pmic.201400120. [DOI] [PubMed] [Google Scholar]
- 17.Desiere F, et al. The PeptideAtlas project. Nucleic acids research. 2006;34:D655–658. doi: 10.1093/nar/gkj040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. Journal of proteome research. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 19.Omenn GS, et al. Metrics for the Human Proteome Project 2015: Progress on the Human Proteome and Guidelines for High-Confidence Protein Identification. Journal of proteome research. 2015;14:3452–3460. doi: 10.1021/acs.jproteome.5b00499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu Y, Lam H. Expanding tandem mass spectral libraries of phosphorylated peptides: advances and applications. Journal of proteome research. 2013;12:5971–5977. doi: 10.1021/pr4007443. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Chromosome-8-coded proteome of Chinese Chromosome Proteome Data set (CCPD) 2.0 with partial immunohistochemical verifications. Journal of proteome research. 2014;13:126–136. doi: 10.1021/pr400902u. [DOI] [PubMed] [Google Scholar]
- 22.Tsai CF, et al. Sequential phosphoproteomic enrichment through complementary metal-directed immobilized metal ion affinity chromatography. Analytical chemistry. 2014;86:685–693. doi: 10.1021/ac4031175. [DOI] [PubMed] [Google Scholar]
- 23.Ye X, Li L. Macroporous reversed-phase separation of proteins combined with reversed-phase separation of phosphopeptides and tandem mass spectrometry for profiling the phosphoproteome of MDA-MB-231 cells. Electrophoresis. 2014;35:3479–3486. doi: 10.1002/elps.201300586. [DOI] [PubMed] [Google Scholar]
- 24.Mancuso F, Bunkenborg J, Wierer M, Molina H. Data extraction from proteomics raw data: an evaluation of nine tandem MS tools using a large Orbitrap data set. Journal of proteomics. 2012;75:5293–5303. doi: 10.1016/j.jprot.2012.06.012. [DOI] [PubMed] [Google Scholar]
- 25.Raijmakers R, Kraiczek K, de Jong AP, Mohammed S, Heck AJ. Exploring the human leukocyte phosphoproteome using a microfluidic reversed-phase-TiO2-reversed-phase high-performance liquid chromatography phosphochip coupled to a quadrupole time-of-flight mass spectrometer. Analytical chemistry. 2010;82:824–832. doi: 10.1021/ac901764g. [DOI] [PubMed] [Google Scholar]
- 26.Casado P, et al. Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Science signaling. 2013;6:rs6. doi: 10.1126/scisignal.2003573. [DOI] [PubMed] [Google Scholar]
- 27.Menschaert G, et al. Deep proteome coverage based on ribosome profiling aids mass spectrometry-based protein and peptide discovery and provides evidence of alternative translation products and near-cognate translation initiation events. Molecular & cellular proteomics : MCP. 2013;12:1780–1790. doi: 10.1074/mcp.M113.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Casado P, Bilanges B, Rajeeve V, Vanhaesebroeck B, Cutillas PR. Environmental stress affects the activity of metabolic and growth factor signaling networks and induces autophagy markers in MCF7 breast cancer cells. Molecular & cellular proteomics : MCP. 2014;13:836–848. doi: 10.1074/mcp.M113.034751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Collins MO, Wright JC, Jones M, Rayner JC, Choudhary JS. Confident and sensitive phosphoproteomics using combinations of collision induced dissociation and electron transfer dissociation. Journal of proteomics. 2014;103:1–14. doi: 10.1016/j.jprot.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Gestel RA, et al. Quantitative erythrocyte membrane proteome analysis with Blue-native/SDS PAGE. Journal of proteomics. 2010;73:456–465. doi: 10.1016/j.jprot.2009.08.010. [DOI] [PubMed] [Google Scholar]
- 31.Sleno L. The use of mass defect in modern mass spectrometry. Journal of mass spectrometry : JMS. 2012;47:226–236. doi: 10.1002/jms.2953. [DOI] [PubMed] [Google Scholar]
- 32.Sturm M, et al. OpenMS - an open-source software framework for mass spectrometry. BMC bioinformatics. 2008;9:163. doi: 10.1186/1471-2105-9-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang J, Perez-Santiago J, Katz JE, Mallick P, Bandeira N. Peptide identification from mixture tandem mass spectra. Molecular & cellular proteomics : MCP. 2010;9:1476–1485. doi: 10.1074/mcp.M000136-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schittmayer M, Fritz K, Liesinger L, Griss J, Birner-Gruenberger R. Cleaning out the Litterbox of Proteomic Scientists’ Favorite Pet: Optimized Data Analysis Avoiding Trypsin Artifacts. Journal of proteome research. 2016;15:1222–1229. doi: 10.1021/acs.jproteome.5b01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lam H. Spectral archives: a vision for future proteomics data repositories. Nature methods. 2011;8:546–548. doi: 10.1038/nmeth.1633. [DOI] [PubMed] [Google Scholar]
- 36.Frank AM, et al. Spectral archives: extending spectral libraries to analyze both identified and unidentified spectra. Nature methods. 2011;8:587–591. doi: 10.1038/nmeth.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.The M, Kall L. MaRaCluster: A Fragment Rarity Metric for Clustering Fragment Spectra in Shotgun Proteomics. Journal of proteome research. 2016;15:713–720. doi: 10.1021/acs.jproteome.5b00749. [DOI] [PubMed] [Google Scholar]
- 38.Mosteller F, Fisher RA. Questions and Answers. The American Statistician. 1948;2:30. [Google Scholar]
- 39.Mi H, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic acids research. 2013;41:D377–386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.