Significance
Although DNA damage is a well-known cause of disease, its connection, if any, to mistranslation is unclear. Here, we used an editing-defective tRNA synthetase to show how an editing defect in zebrafish leads to severe consequences during development and to a shortened overall life span of adult fish. These effects were p53-dependent and reflect a strong connection of mistranslation to the DNA damage response. In addition, deep sequencing of genomic DNA revealed that expression of the editing-defective tRNA synthetase was mutagenic. We suggest that the p53-dependent DNA damage response links translational and genomic fidelity to promote vertebrate homeostasis.
Keywords: mistranslation, genomic fidelity, shortened lifespan, morphological changes, genomic mutations
Abstract
Brain and heart pathologies are caused by editing defects of transfer RNA (tRNA) synthetases, which preserve genetic code fidelity by removing incorrect amino acids misattached to tRNAs. To extend understanding of the broader impact of synthetase editing reactions on organismal homeostasis, and based on effects in bacteria ostensibly from small amounts of mistranslation of components of the replication apparatus, we investigated the sensitivity to editing of the vertebrate genome. We show here that in zebrafish embryos, transient overexpression of editing-defective valyl-tRNA synthetase (ValRSED) activated DNA break-responsive H2AX and p53-responsive downstream proteins, such as cyclin-dependent kinase (CDK) inhibitor p21, which promotes cell-cycle arrest at DNA damage checkpoints, and Gadd45 and p53R2, with pivotal roles in DNA repair. In contrast, the response of these proteins to expression of ValRSED was abolished in p53-deficient fish. The p53-activated downstream signaling events correlated with suppression of abnormal morphological changes caused by the editing defect and, in adults, reversed a shortened life span (followed for 2 y). Conversely, with normal editing activities, p53-deficient fish have a normal life span and few morphological changes. Whole-fish deep sequencing showed genomic mutations associated with the editing defect. We suggest that the sensitivity of p53 to expression of an editing-defective tRNA synthetase has a critical role in promoting genome integrity and organismal homeostasis.
In the first step of protein synthesis, amino acids are attached to their cognate tRNAs in reactions catalyzed by aminoacyl-tRNA synthetases (1, 2). Mistakes of aminoacylation occur with a frequency of less than 1% (3, 4), and if not corrected, will result in the insertion of a mischarged amino acid into a growing polypeptide chain at the wrong codon (5–7). This sort of error is usually corrected by a universal editing mechanism in tRNA synthetases, which hydrolytically removes the mischarged amino acid from the tRNA before the wrong amino acid is inserted into a nascent protein (8–14).
Serious pathologies and cell death result from rare errors in protein synthesis (mistranslation). For example, a mild editing defect of an alanyl-tRNA synthetase (AlaRS) in the mouse was shown to cause the death of Purkinje cells and result in neurodegeneration (5). More recently, cardioproteinopathy was also shown to have connections to mistranslation (15). Although the cellular and physiological importance of editing has been established, the extent of how editing defects impact major cellular mechanisms is only beginning to be clarified. Because assaults on the genome by environmental factors lead to somatic cell mutations that cause diseases in humans, we were especially interested in the role editing might play in protecting vertebrates against DNA damage. Earlier work in bacteria was supportive of such a connection (16–18), and later work established that the occasional misincorporation of the wrong amino acids into enzymes of the DNA replication and repair apparatus was mutagenic by virtue of error-prone repair of DNA damage (19) (Fig. 1A).
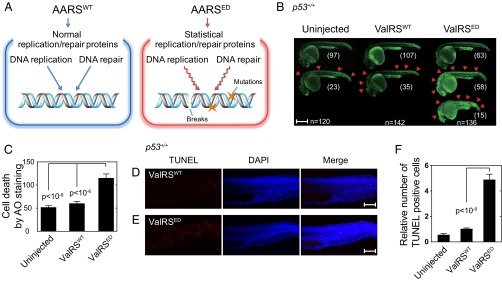
Fig. 1.
ED ValRS causes cell death and DNA damage in zebrafish. (A) The ED tRNA synthetase is proposed to generate statistical replication and repair proteins that lead to increased DNA breaks. (B) AO staining at 1 dpf of zebrafish uninjected (n = 120), or injected with either ValRSWT mRNA (n = 142) or ValRSED mRNA (n = 136). The red arrows show points of cell death. Zebrafish are aligned vertically based on cell death severity and morphology; normal (Top), increased cell death (Middle), and increased cell death with abnormal morphology (Bottom). (Scale bar, 500 μm.) (C) Quantification of AO staining to determine average number of cell deaths per fish. Bars represent mean ± SEM. Images of TUNEL staining on fish injected with either (D) ValRSWT or (E) ValRSED mRNA. TUNEL staining, DAPI staining, and TUNEL merged with DAPI, are shown. (Scale bars, 100 μm.) (F) Quantification of the average number of TUNEL+ cells per fish (periderm or basal epidermal cells on the trunk). Independent areas [from the trunk region starting at the rostral start point of the yolk extension (the distal end of yolk) and extending through the end of the caudal fin] were selected from individual animals that had been injected with mRNA and were used for the quantification. Quantifications (Uninjected n = 13, ValRSWT n = 16, ValRSED n = 23) are shown in the graph. Bars represent mean ± SEM.
Here, we investigated whether this phenomenon could extend to vertebrates. Because coupling of mistranslation and genome fidelity would have obvious implications for organismal homeostasis and disease, we chose the zebrafish, a popular model organism (with many of the same genes and disorders as mammals) (20, 21) as a system to explore the linkage between translation and genome fidelity. Using editing-defective (ED) valyl-tRNA synthetase (ValRS), we demonstrated that, in response to mistranslation, the DNA damage response (DDR) is activated, as shown by phosphorylation of H2AX and up-regulation of p53 downstream genes. In addition, ED ValRS shortened the lifespan of p53-deficient zebrafish, and whole-fish genomic DNA sequencing showed genomic mutations were associated with the editing defect. Thus, p53 surveillance of mistranslation links translational and genomic fidelity to sustain vertebrate homeostasis.
ED ValRS Causes Cell Death in Zebrafish
For a mistranslational system, we used an ED tRNA synthetase, which has impaired ability to correct an error of aminoacylation of its cognate tRNA. For this purpose, ValRS, which by editing eliminates the occasional production of Thr-tRNAVal (22), was chosen. Replacement of a universally conserved T535 (which is T539 in zebrafish ValRS) with a T535P substitution yields an enzyme that is completely deficient in editing and retains full aminoacylation activity (6). This mutant was designated as ValRSED, where ED denotes “editing-defective.” As shown by direct analysis of mammalian cells, ValRSED (encoded as a stable transfectant) occasionally inserts threonine at the positions of valine codons (6).
Because prior work showed that animals have a profound sensitivity to even mild defects in editing, the creation of T539P knockin alleles was not of use for these studies. In the mouse with AlaRS, an ED knockin was embryonic-lethal; a heterozygous knockin had extreme pathologies, such as cardioproteinopathy (15). Moreover, even a homozygous mild editing defect (about twofold) resulted in neurotoxicity and ataxia (5). Thus, a knockin of T539P ValRS in the fish would almost certainly be embryonic-lethal. In addition, we doubted that tissue-specific transgenic constructions would be useful. A liver-specific transgenic, for example, would not likely develop a liver and survive. Instead, as part of our studies, we turned to the use of transgenics generated postfertilization, and selected for survivors. These survivors are a mosaic of expression profiles distributed in different tissues of the animal, and had great utility for our studies.
We deliberately used fish that retained their wild alleles for the editing domain, so as to maintain a background level of the editing function. Then, we either introduced mRNAs that transiently expressed an ED synthetase in trans in the embryo or, in separate studies, we created a mosaic population of transgenic fish that retained the WT alleles of the editing function. By maintaining a strong background of the editing function in all studies, we were able to “tease out” the effects of a competing ED synthetase on the p53-mediated DDR.
To start, we injected synthetic mRNAs encoding ValRSWT and ValRSED into 120–142 randomly chosen zebrafish embryos. [Except where noted, all data reported below were carried out with zebrafish (zf) ValRS constructs. Because of the extensive prior work (6), many of our first experiments were done with murine (m) ValRS (SI Appendix, Fig. S1). In all of these experiments, the results closely mirrored what was observed with the zfValRS constructs.] Using the green color from Acridine orange (AO) staining at 1 d postfertilization (dpf), areas of apparent cell death within the live embryos (Fig. 1B, red arrows) were observed in 35 of 142 of embryos injected with ValRSWT mRNA. In significant contrast (P < 10−6) (Fig. 1C), 73 of 136 live embryos injected with ValRSED mRNA had regions of severe cell death and 15 of 136 of them exhibited dramatic morphological changes as well. (We observed no comparable morphological changes in the embryos injected with ValRSWT mRNA.) To further check and confirm specificity for cell death, we performed the TUNEL assay at 1 dpf, with embryos injected with either ValRSWT or ValRSED mRNA (Fig. 1 D–F). Quantification of the TUNEL+ cells (n = 13–23) confirmed the higher cell death frequency of the ValRSED-injected fish compared with the ValRSWT-injected fish (P < 10−8). In addition, this assay, which is specific for measuring DNA breaks, shows a much greater change in the frequency of ValRSED-induced DNA breaks compared with the change in cell death measured by AO staining. Thus, the increase in production of breaks goes well beyond what would be predicted from increased cell death alone. This observation supports the conclusion that there is a proliferation of ValRSED-induced breaks that is not directly associated with cell death. This line of thinking led us subsequently to investigate ValRSED-induced mutations in the genome (see below).
Pursuant to more rigorously addressing the question of artifacts from injection or overexpression variability, we investigated levels of both ValRSWT and ValRSED in normal p53+/+ fish (SI Appendix, Fig. S2). These data show that expression of ValRSED is less than or equal to that of ValRSWT. However, with three different doses of the two ValRS’s, ValRSED consistently gave much more cell death (SI Appendix, Fig. S3). This difference was even seen at the lowest dose, whereas the uninjected and ValRSWT mRNA injected zebrafish were the same (SI Appendix, Fig. S3). (In experiments described below, unless otherwise noted, we then used this “lowest dose.”) These results came from three independent experiments, using about 59–136 fish in each of seven groups. These additional data further support that the effects of ValRSED are not a result of microinjection/overexpression artifacts.
ED ValRS Activates DNA Damage-Responsive H2AX
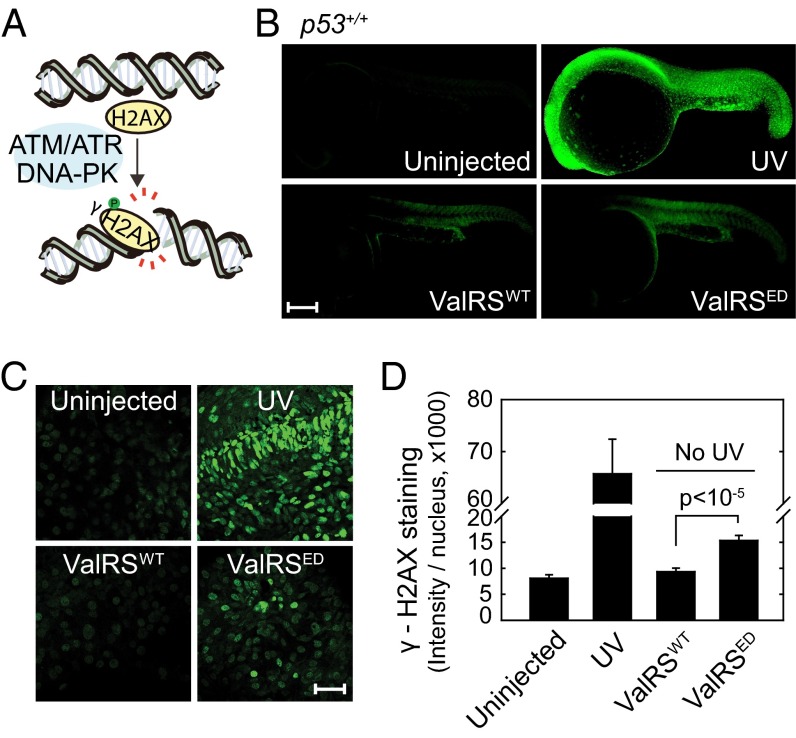
To more sharply focus on DNA breaks per se, we looked at the H2AX protein that, in response to DNA damage, is rapidly phosphorylated at S139 by ATM (Ataxia telangiectasia mutated), ATR (Ataxia telangiectasia and Rad3 related), and DNA-PK (DNA-dependent protein kinase), among others (Fig. 2A) (23, 24). For this analysis, we used UV-treated zebrafish embryos as a positive control. The activation of H2AX to give γ-H2AX was determined by microscopy and quantitation of staining with a specific antibody (α-γ-H2AX). With fish injected with ValRSED mRNA, even in the absence of UV-irradiation, the amount of γ-H2AX was increased (Fig. 2B). With an enlarged view of γ-H2AX staining, strong green signals were found in the UV control and in ValRSED mRNA injected fish, but not in the uninjected or ValRSWT mRNA-injected fish (Fig. 2C). The γ-H2AX staining was quantified and is plotted in Fig. 2D. Whereas the uninjected and ValRSWT mRNA-injected samples showed background levels of γ-H2AX staining, the ValRSED mRNA-injected fish showed significantly elevated γ-H2AX levels (P < 10−5). Thus, the editing defect by itself leads to DNA breaks. These results further support the idea that suppression of miscoding is protective against DNA damage.
Fig. 2.
Response of H2AX to an ED ValRS. (A) Cartoon of H2AX phosphorylation. (B) γ-H2AX staining in uninjected (n = 60), ValRSWT mRNA injected (n = 65), ValRSED mRNA injected (n = 79), or UV treated zebrafish (n = 30) at 1 dpf. (Scale bar, 250 μm.) (C) Enlarged pictures of γ-H2AX staining in the nuclei of different samples of zebrafish. (Scale bar, 40 μm.) (D) Quantification of γ-H2AX staining. Bars represent mean ± SEM.
ED ValRS Activates the p53 Pathway
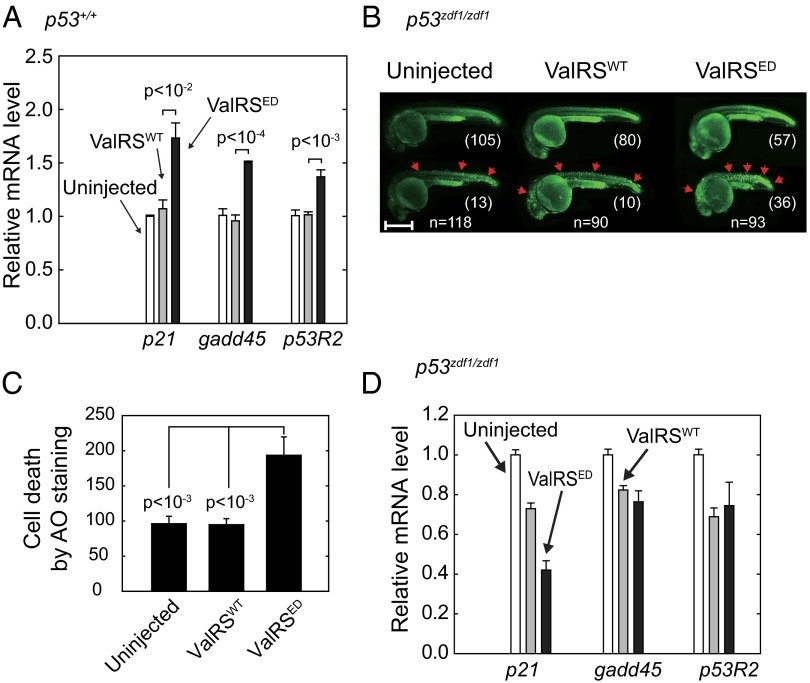
To verify the expected DDR in the subject animals (Fig. 2), we monitored downstream markers of p53 activation for our analysis. These markers included cyclin-dependent kinase (CDK) inhibitor p21, which promotes cell-cycle arrest at DNA damage checkpoints (25), and Gadd45 (26) and p53R2 (27), which have pivotal roles in DNA repair. Strikingly, although embryos injected with mRNA encoding ValRSWT showed little change in expression of these p53-downstream target genes, p21, gadd45, and p53R2 were all significantly up-regulated in embryos that harbored ValRSED (all P < 10−2) (Fig. 3A). (Because of the inevitable presence of cells not harboring ValRSED in the samples, we speculate that the level of activation of the downstream markers may be underestimated.) We also checked Mdm2, an E3 ubiquitin ligase, which negatively regulates the presence of p53 proteins to suppress p53-induced apoptosis (28). Neither injected ValRSWT nor ValRSED mRNA affected mdm2 expression (SI Appendix, Fig. S4A). As expected from the cell-death–associated AO staining seen in Fig. 1 B and C, we observed that ValRSED mRNA injection caused increased expression of the apoptosis-inducing bax (29) (SI Appendix, Fig. S4A). These results demonstrated a strong sensitivity of the induction of the DDR to miscoding in zebrafish.
Fig. 3.
ED ValRS activates p53 in zebrafish. (A) RT-PCR results of p53 downstream markers p21, gadd45, and p53R2 in ValRSWT or ValRSED mRNA-injected zebrafish. The bar graphs show the mean values ± SEM after normalization to the β-actin level. (B) Uninjected (n = 118), ValRSWT mRNA-injected (n = 90), or ValRSED mRNA-injected (n = 93) p53zdf1/zdf1 zebrafish were stained with AO at 1 dpf. The red arrows show points of cell death. Zebrafish are aligned based on cell death severity. (Scale bar, 500 μm.) (C) Quantification of AO staining. Bars represent mean ± SEM. (D) RT-PCR results of p53 downstream markers p21, gadd45, and p53R2 in p53zdf1/zdf1 zebrafish injected with either ValRSWT or ValRSED mRNA. The bar graphs show the mean values ± SEM after normalization to the β-actin level.
In addition to using ValRSWT as a control, we created an ALGA substitution of the highly conserved HLGH motif important for ValRS aminoacylation activity. This mutant (ValRSAD) is aminoacylation-defective, but retains the editing activity encoded by a separate site for editing (30). [Here the mouse ValRS construct was used. In a separate analysis, we found that cell death in surviving fish, as monitored by AO staining, was similar for fertilized eggs injected with WT and ED constructs of either zfValRS mRNA or mValRS mRNA (SI Appendix, Fig. S1 A–D). In addition, the mRNA levels of the p53 downstream target genes were also similar for both the zfValRS mRNA- and mValRS mRNA-injected fish (SI Appendix, Fig. S1 E and F). This similarity is consistent with the high conservation of the two ValRS sequences.] We found that 5 of 24 embryos injected with mValRSAD mRNA had localized regions of cell death (SI Appendix, Fig. S5 A and B), similar to those injected with ValRSWT mRNA (10 of 56) (SI Appendix, Fig. S1 A and B). However, the quantification of the AO staining suggested that there was a statistically significant difference in cell death between the mValRSWT and the mValRSAD mRNA-injected zebrafish (P < 10−2) (SI Appendix, Fig. S5B). This observation could be a manifestation of prior results showing that active-site mutations can impede bacterial cell growth because the inactive synthetase retains its tRNA binding site and soaks up the cognate tRNA in abortive complexes (31).
Next, we examined expression of the three p53 DDR downstream target genes: p21, gadd45, and p53R2. Expression of these genes did not differ from those injected with ValRSWT mRNA (SI Appendix, Fig. S5C). [Although expression of mdm2 was not altered significantly by injection of ValRSAD mRNA, the apoptosis marker bax was elevated (P < 10−2), and thus consistent with the somewhat higher cell death associated with ValRSAD-expressing fish (SI Appendix, Fig. S5 A and B).] These results show that the DDR is not directly sensitive to the site for aminoacylation.
In p53-Deficient Fish, ED ValRS Leads to Increased Abnormal Morphology
To further investigate the connection of miscoding to the p53-directed DDR, we performed mRNA injections into embryos that were p53-deficient (p53zdf1/zdf1). Here again, the expression levels of ValRSWT and ValRSED were similar (SI Appendix, Fig. S6). Additionally, as in the p53+/+ background, with three different doses of the two ValRS’s, ValRSED consistently gave much more cell death (SI Appendix, Fig. S7). In these experiments, the p53-deficiency had little or no morphology effect on either uninjected embryos or embryos injected with mRNA for ValRSWT (Fig. 3B). With ValRSED mRNA injections into p53+/+ fish, we observed cell death in zebrafish with both normal and abnormal morphology (Fig. 1 B and C). However, in the p53zdf1/zdf1 fish injected with ValRSED mRNA, cell death was only observed in zebrafish with abnormal morphology (Fig. 3 B and C). Thus, 36 of 93 of the surviving embryos injected with ValRSED mRNA showed many localized regions of AO staining and had simultaneously undergone morphological changes, whereas the remaining 57 fish had neither. (The limited cell death without morphological changes in the ValRSED-expressing p53+/+ fish of Fig. 1 B and C is consistent with the protective role of p53 against more extensive damage to the whole organism.) As expected, because of the absence of p53 activity, there was not a statistically significant difference between ValRSWT and ValRSED mRNA-injected embryos in the expression of any of the p53 downstream markers p21, gadd45, p53R2, mdm2, and bax (Fig. 3D and SI Appendix, Fig. S4B). It is worth noting that, with both the p53+/+ and p53zdf1/zdf1 fish, the protein levels of WT and mutant p53 were unaffected by the expression of ValRSWT or ValRSED (SI Appendix, Figs. S8 and S9). [The p53 +/zdf1 heterozygote behaves like the p53+/+ fish for the activation of p53 downstream targets and tumor predisposition after γ-irradiation (32). For that reason, we did not feel justified to pursue confirmatory work showing that p53 heterozygotes would behave the same as the p53 WT fish in our system.]
ED ValRS Shortens the Short- and Long-Term Survival of Zebrafish
Accumulation of DNA damage can lead to mutations through the error-prone DNA repair machinery and ostensibly shorten the lifespan (33, 34). To investigate this question, we redirected our efforts to p53zdf1/zdf1 transgenic fish that expressed ValRSED. We inserted the cDNA encoding ValRSWT and ValRSED into the pT2-mCherry vector, which fuses the coding sequence of the mCherry vector to the coding sequence for the C terminus of the ValRS’s. (We designate these constructs as pT2-ValRSWT and pT2-ValRSED, respectively.) The resulting vector constructions were then injected into one-cell stage p53zdf1/zdf1 embryos. After 2 dpf (when the fish appear developed), we sorted for mCherry+ fish under a fluorescent microscope and harvested samples for Western blot analysis to confirm the fusion protein of the expected size was indeed produced (SI Appendix, Fig. S10).
Next, we investigated both the short- and long-term survival of fish harboring WT and ED alleles of ValRS. For the short-term analysis, ValRSWT or ValRSED mRNAs were injected into separate fertilized p53+/+ or p53zdf1/zdf1 eggs. Deaths of embryos were monitored during 6–24 h postfertilization (hpf). Both p53+/+ and p53zdf1/zdf1 fish injected with ValRSED mRNA showed increased embryo death (approximately 13–35% increase compared with WT) compared with the uninjected and ValRSWT mRNA-injected fish (which were similar) (SI Appendix, Fig. S11).
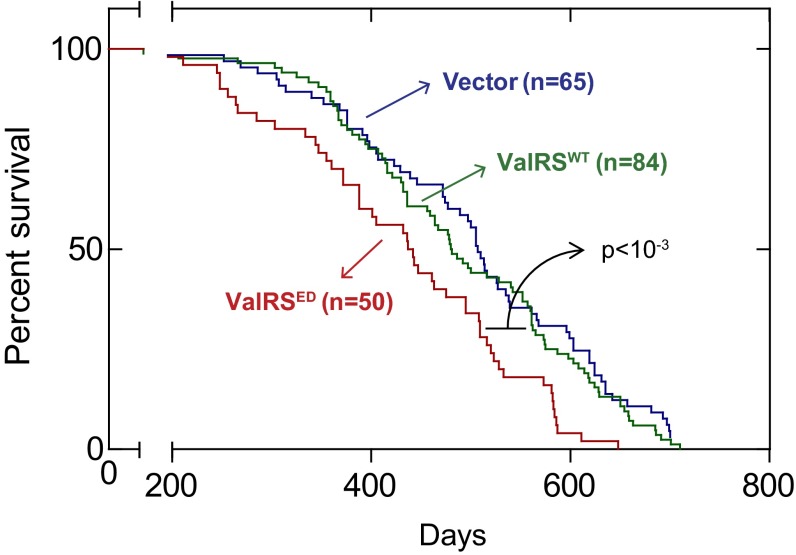
For the analysis of long-term survival, we injected p53+/+ and p53zdf1/zdf1 fertilized eggs, separately, with either the pT2-vector alone or the pT2-ValRSWT or pT2-ValRSED construct. For this work we injected pT2-ValRS constructs into the embryos that were created from siblings' group mating (a group mating of a few males and females in the same generation). This protocol established six separate groups of fish for long-term survival analysis. After development, and depending on the experiment, mCherry+ fish were selected and maintained over a period of almost 2 y. The long-term survival rates of these fish were then compared. We observed that, in the p53zdf1/zdf1 background, the long-term survival of fish harboring the pT2-vector alone and the pT2-ValRSWT construct were not significantly different. In contrast, fish harboring the pT2-ValRSED construct had sharply reduced survival rates (P < 10−3) (Fig. 4). Consistent with the analysis shown above of the p53-dependent effects of expression of ValRSED on the DDR, the long-term survival of fish harboring the pT2-vector alone, and of those harboring either the pT2-ValRSWT or pT2-ValRSED construct, were not different in a statistically significant way in the p53+/+ background (SI Appendix, Fig. S12). Thus, the p53-dependent response to DNA insults caused by mistranslation preserves the lifespan in the fish.
Fig. 4.
ED ValRS decreases long-term survival rate in p53zdf1/zdf1zebrafish. The survival rates of transgenic p53zdf1/zdf1 zebrafish expressing ValRS–mCherry fusions from either the pT2 vector alone (mCherry alone, blue), pT2-ValRSWT (green), or pT2-ValRSED (red) were recorded and compared.
Increased Genomic Variants Associate with the Presence of an ED tRNA synthetase
To confirm that fish harboring ValRSED had accumulated mutations, we selected from the siblings’ cohort, one zebrafish at 2 dpf expressing a ValRSWT fusion and another expressing a ValRSED fusion. Genomic DNA was extracted from each fish for sequencing analysis on an Illumina HiSeq2000 system. Given the nature of the phenomenon in question, likely leading to pronounced genomic mosaicism between tissues (especially in ValRSED fish), we recognized that a relatively high depth of coverage was needed to achieve a strong enough signal for genotyping. To gain sufficient coverage, we concentrated on sequencing a portion of the genome to great depth. Pursuant to this objective, we digested the genomic DNA samples with the DraI restriction enzyme (digests at TTTAAA hexanucleotide sequences). The digested samples were run out on an agarose gel and visualized by ethidium bromide staining under a UV lamp, and we excised a dense band around 1,250 bps that was common to the WT and ED samples and used it to create a DNA library for sequencing.
Three replicates of each sample library were independently sequenced, resulting in a total of 542,811,425 sequencing reads of 50–100 bases. Our goal was to examine the difference between the nonreference alleles (variants) found in the genomes of the ValRSED- versus the ValRSWT-expressing fish. Additionally, aware of experiments in bacteria showing that overexpression of a WT tRNA synthetase can lead to promiscuous, uncorrected misacylations [in this case attachment of a cognate amino acid to a noncognate tRNA (35)], we anticipated that zebrafish overexpressing ValRSWT could harbor additional variants as well.
We aligned our sequences to the zebrafish genome (v9, build 68) and processed according to established best practices for the Genome Analysis Toolkit (36). SNP- and InDel-variants were distributed across all 25 autosomes. We rejected those variants having a quality log-odds score below −0.21, the score at which 99% of known variants from the database of SNPs were recovered, and kept only those where replicates within the same fish (either ValRSWT- or ValRSED-expressing) were reliably genotyped to not be in disagreement. As expected, the discovered variants were overwhelmingly (87.8%) identical between the ValRSWT- and ValRSED-expressing fish. Those variants represent inherent differences between the fish used in these studies and the reference fish. Of the remaining 228,520 variants (those not shared between phenotypes), a notable aggregation of 67.3% more variants were specific to only the ValRSED-expressing fish, revealing that the ValRSED phenotype led to genomic instability outpacing that of the ValRSWT phenotype even in these young fish with relatively few cellular cycles compared with an adult (SI Appendix, Fig. S13). Variants from this set that were reliably determined to be reference alleles in ValRSWT-expressing fish and nonreference alleles in ValRSED-expressing fish (Fig. 5A; for estimated loci P value of 0.0095, see SI Appendix, SI Materials and Methods) were analyzed and loaded into the University of California, Santa Cruz Variant Annotation Integrator for further analysis.
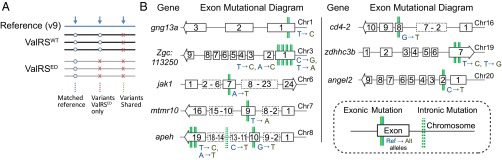
Fig. 5.
Examples of variants in genomes of ValRSED- and ValRSWT-expressing zebrafish, with the replicate filtration strategy used. (A) Three replicate libraries of ValRSWT- or ValRSED-injected p53zdf1/zdf1 zebrafish genomic DNA were sequenced, mapped, processed, and genotyped. The resultant nonreference alleles (variants) that could be found in all three replicates for each sample had a high degree of overlap, and these variants—and many of those specific to the ValRSWT-expressing fish—most likely represent intrinsic differences between the zebrafish strain used in these studies and that of the reference. [In addition, because overexpression of a WT tRNA synthetase, at least in bacteria, can lead to mistranslation (35) and presumably mutagenesis (19), we did not attempt to interpret further the variants specific to the WT fish.] Overall, the sequence match between the WT and reference was greater than 98.5%. The discovered variants were 96% SNPs and 4% InDels. (B) Examples located in specific genes on eight different chromosomes are SNPs that were reliably called as variants in the ValRSED fish and as reference alleles in the ValRSWT fish.
Fig. 5B displays examples of these variants in specific genes distributed across eight chromosomes. These examples include variant alleles (specific to the fish harboring an editing defect) in coding exons, introns, and 5′- and 3′-untranslated regions. Thus, the DNA damage associated with miscoding is correlated with mutations widely distributed throughout the genome. No specific mutational signature was readily observed in either the direct mutational scenarios (A→T, T→G, and so forth) or in any di-nucleotide scenarios (CC→CT, CC→CG, and so forth). More particulars are cast into a visual display in SI Appendix, Fig. S13.
Discussion
The zebrafish system used here seems to be especially robust for detection of p53-sensitive DNA damage and therefore could be useful as a “tester system” analogous to the Ames test for carcinogens (37). With this system, a powerful genome-protective effect of an editing mechanism that suppresses miscoding was demonstrated. Even in the p53zdf1/zdf1 background, the WT editing activity was sufficiently robust to suppress miscoding so that fish embryo morphology and a normal life span were maintained. However, in that same p53zdf1/zdf1 background, the miscoding-defect we introduced was transdominant, because all fish still harbored the diploid WT alleles for the native tRNA synthetase. This observation highlights the sensitivity of the genome to miscoding.
The results also independently reveal a critical protective effect of the p53-directed surveillance of miscoding. Despite miscoding, the morphological changes in fish embryos and lifespan decrease of adult fish were much less altered in the p53+/+ background. (Because these changes were marginal, we did not attempt to investigate genome sequence alterations.) Thus, in circumstances where p53 surveillance of miscoding is compromised, mechanisms like editing that suppress miscoding are particularly important, because even mild miscoding could, for example, contribute to the “mutator phenotype” seen in disease connected to p53 defects (38–40).
Methods
AO Staining for Detecting Cell Death in Zebrafish Embryos.
The zebrafish (Danio rerio, strain: AB/Tu; sex, N/D; age, 0–2.5 y old) used in our study were maintained at 28.5 °C under continuous water flow and filtration with automatic control for a 14-h/10-h light/dark cycle. The night before injection, male and female fish were placed in a 1-L tank containing a fish mating cage with an inner mesh and divider. Zebrafish embryos were obtained from natural spawning by removing the divider and stimulating with light. The embryos were kept at 28.5 °C before and after microinjection.
Synthetic mRNAs (made with mMESSAGE mMACHINE kit, Ambion) were injected into the one-cell stage embryos at a dosage of 250–1,000 pg per embryo (sample size was determined by standards of the field of three independent experiments of 30–50 embryos each). For the injection of pT2 vector constructs, the dosage of DNA was ∼1 ng per embryo together with ∼250 pg transposase-encoding mRNA. At 1 dpf, dead embryos were removed and all live embryos were used for experiments. For AO staining, live zebrafish embryos were dechorionated in pronase (2.0 mg/mL in egg water, 5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM magnesium sulfate) for 3–5 min and rinsed five times in egg water at 1 dpf. Embryos were then incubated in 10 μg/mL AO (Sigma A-6014) in egg water for 15 min at 28.5 °C, followed by five quick rinses. Embryos were anesthetized in 160 μg/mL tricaine (3-aminobenzoic acid ethyl ester; Sigma A-5040), mounted laterally on the glass slide, and photographed with a Nikon fluorescent microscope (AZ100) equipped with a Nikon CCD camera (Qimaging Retiga 2000R). All of the experiments involving zebrafish had been conducted according to the guidelines established by the Institutional Animal Care and Use Committee (IACUC) at The Scripps Research Institute, IACUC approval number 09–0009. Results shown are from three independent experiments.
Other Procedures.
Additional procedures are described in SI Appendix, SI Materials and Methods and Table S1.
Supplementary Material
Acknowledgments
We thank Profs. Susan Ackerman (Jackson Laboratories, Bar Harbor), Arnold Levine (Institute for Advance Studies, Princeton University), Xiaohua Wu (The Scripps Research Institute), and Leonard Zon (Children’s Hospital, Harvard Medical School, and Harvard University) for helpful comments on this work. This work was supported by National Cancer Institute Grant CA92577; NIH NIAAA Grants AA007456 and AA013525; aTyr Pharma; and by a fellowship from the National Foundation for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608139113/-/DCSupplemental.
References
- 1.Carter CW., Jr Cognition, mechanism, and evolutionary relationships in aminoacyl-tRNA synthetases. Annu Rev Biochem. 1993;62:715–748. doi: 10.1146/annurev.bi.62.070193.003435. [DOI] [PubMed] [Google Scholar]
- 2.Giegé R. The early history of tRNA recognition by aminoacyl-tRNA synthetases. J Biosci. 2006;31(4):477–488. doi: 10.1007/BF02705187. [DOI] [PubMed] [Google Scholar]
- 3.Loftfield RB. The frequency of errors in protein biosynthesis. Biochem J. 1963;89:82–92. doi: 10.1042/bj0890082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loftfield RB, Vanderjagt D. The frequency of errors in protein biosynthesis. Biochem J. 1972;128(5):1353–1356. doi: 10.1042/bj1281353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JW, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature. 2006;443(7107):50–55. doi: 10.1038/nature05096. [DOI] [PubMed] [Google Scholar]
- 6.Nangle LA, Motta CM, Schimmel P. Global effects of mistranslation from an editing defect in mammalian cells. Chem Biol. 2006;13(10):1091–1100. doi: 10.1016/j.chembiol.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Netzer N, et al. Innate immune and chemically triggered oxidative stress modifies translational fidelity. Nature. 2009;462(7272):522–526. doi: 10.1038/nature08576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin AN, Berg P. Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J Biol Chem. 1966;241(4):839–845. [PubMed] [Google Scholar]
- 9.Eldred EW, Schimmel PR. Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J Biol Chem. 1972;247(9):2961–2964. [PubMed] [Google Scholar]
- 10.Martinis SA, Boniecki MT. The balance between pre- and post-transfer editing in tRNA synthetases. FEBS Lett. 2010;584(2):455–459. doi: 10.1016/j.febslet.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadavalli SS, Ibba M. Quality control in aminoacyl-tRNA synthesis its role in translational fidelity. Adv Protein Chem Struct Biol. 2012;86:1–43. doi: 10.1016/B978-0-12-386497-0.00001-3. [DOI] [PubMed] [Google Scholar]
- 12.Kumar S, Das M, Hadad CM, Musier-Forsyth K. Substrate and enzyme functional groups contribute to translational quality control by bacterial prolyl-tRNA synthetase. J Phys Chem B. 2012;116(23):6991–6999. doi: 10.1021/jp300845h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreier AA, Schimmel PR. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11(9):1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- 14.Beuning PJ, Musier-Forsyth K. Hydrolytic editing by a class II aminoacyl-tRNA synthetase. Proc Natl Acad Sci USA. 2000;97(16):8916–8920. doi: 10.1073/pnas.97.16.8916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y, et al. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci USA. 2014;111(49):17570–17575. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy HS, Humayun MZ. Escherichia coli cells expressing a mutant glyV (glycine tRNA) gene have a UVM-constitutive phenotype: Implications for mechanisms underlying the mutA or mutC mutator effect. J Bacteriol. 1997;179(23):7507–7514. doi: 10.1128/jb.179.23.7507-7514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorazi R, Lingutla JJ, Humayun MZ. Expression of mutant alanine tRNAs increases spontaneous mutagenesis in Escherichia coli. Mol Microbiol. 2002;44(1):131–141. doi: 10.1046/j.1365-2958.2002.02847.x. [DOI] [PubMed] [Google Scholar]
- 18.Balashov S, Humayun MZ. Mistranslation induced by streptomycin provokes a RecABC/RuvABC-dependent mutator phenotype in Escherichia coli cells. J Mol Biol. 2002;315(4):513–527. doi: 10.1006/jmbi.2001.5273. [DOI] [PubMed] [Google Scholar]
- 19.Bacher JM, Schimmel P. An editing-defective aminoacyl-tRNA synthetase is mutagenic in aging bacteria via the SOS response. Proc Natl Acad Sci USA. 2007;104(6):1907–1912. doi: 10.1073/pnas.0610835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dooley K, Zon LI. Zebrafish: A model system for the study of human disease. Curr Opin Genet Dev. 2000;10(3):252–256. doi: 10.1016/s0959-437x(00)00074-5. [DOI] [PubMed] [Google Scholar]
- 21.Amsterdam A, Hopkins N. Mutagenesis strategies in zebrafish for identifying genes involved in development and disease. Trends Genet. 2006;22(9):473–478. doi: 10.1016/j.tig.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 22.Fersht AR, Kaethner MM. Enzyme hyperspecificity. Rejection of threonine by the valyl-tRNA synthetase by misacylation and hydrolytic editing. Biochemistry. 1976;15(15):3342–3346. doi: 10.1021/bi00660a026. [DOI] [PubMed] [Google Scholar]
- 23.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 24.Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 2001;276(45):42462–42467. doi: 10.1074/jbc.C100466200. [DOI] [PubMed] [Google Scholar]
- 25.Bunz F, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 26.Maeda T, et al. GADD45 regulates G2/M arrest, DNA repair, and cell death in keratinocytes following ultraviolet exposure. J Invest Dermatol. 2002;119(1):22–26. doi: 10.1046/j.1523-1747.2002.01781.x. [DOI] [PubMed] [Google Scholar]
- 27.Tanaka H, et al. A ribonucleotide reductase gene involved in a p53-dependent cell-cycle checkpoint for DNA damage. Nature. 2000;404(6773):42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 28.Buschmann T, Fuchs SY, Lee CG, Pan ZQ, Ronai Z. SUMO-1 modification of Mdm2 prevents its self-ubiquitination and increases Mdm2 ability to ubiquitinate p53. Cell. 2000;101(7):753–762. doi: 10.1016/s0092-8674(00)80887-9. [DOI] [PubMed] [Google Scholar]
- 29.Yin C, Knudson CM, Korsmeyer SJ, Van Dyke T. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature. 1997;385(6617):637–640. doi: 10.1038/385637a0. [DOI] [PubMed] [Google Scholar]
- 30.Lin L, Schimmel P. Mutational analysis suggests the same design for editing activities of two tRNA synthetases. Biochemistry. 1996;35(17):5596–5601. doi: 10.1021/bi960011y. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt E, Schimmel P. Dominant lethality by expression of a catalytically inactive class I tRNA synthetase. Proc Natl Acad Sci USA. 1993;90(15):6919–6923. doi: 10.1073/pnas.90.15.6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berghmans S, et al. tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci USA. 2005;102(2):407–412. doi: 10.1073/pnas.0406252102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vogel H, Lim DS, Karsenty G, Finegold M, Hasty P. Deletion of Ku86 causes early onset of senescence in mice. Proc Natl Acad Sci USA. 1999;96(19):10770–10775. doi: 10.1073/pnas.96.19.10770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vijg J, Suh Y. Genome instability and aging. Annu Rev Physiol. 2013;75:645–668. doi: 10.1146/annurev-physiol-030212-183715. [DOI] [PubMed] [Google Scholar]
- 35.Swanson R, et al. Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science. 1988;242(4885):1548–1551. doi: 10.1126/science.3144042. [DOI] [PubMed] [Google Scholar]
- 36.Van der Auwera GA, et al. From FastQ data to high-confidence variant calls: The Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ames BN, Lee FD, Durston WE. An improved bacterial test system for the detection and classification of mutagens and carcinogens. Proc Natl Acad Sci USA. 1973;70(3):782–786. doi: 10.1073/pnas.70.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havre PA, Yuan J, Hedrick L, Cho KR, Glazer PM. p53 inactivation by HPV16 E6 results in increased mutagenesis in human cells. Cancer Res. 1995;55(19):4420–4424. [PubMed] [Google Scholar]
- 39.Bishop AJ, et al. Atm-, p53-, and Gadd45a-deficient mice show an increased frequency of homologous recombination at different stages during development. Cancer Res. 2003;63(17):5335–5343. [PubMed] [Google Scholar]
- 40.Feng Z, Hu W, Rajagopal G, Levine AJ. The tumor suppressor p53: Cancer and aging. Cell Cycle. 2008;7(7):842–847. doi: 10.4161/cc.7.7.5657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.