Abstract
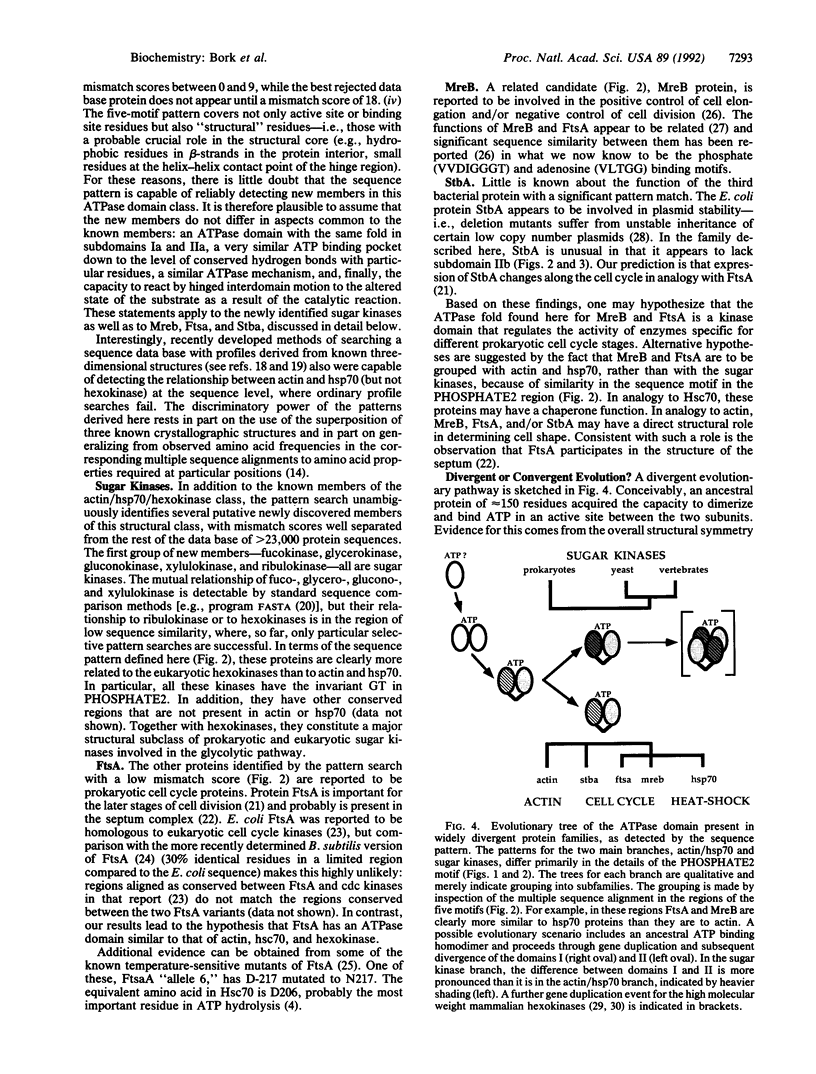
The functionally diverse actin, hexokinase, and hsp70 protein families have in common an ATPase domain of known three-dimensional structure. Optimal superposition of the three structures and alignment of many sequences in each of the three families has revealed a set of common conserved residues, distributed in five sequence motifs, which are involved in ATP binding and in a putative interdomain hinge. From the multiple sequence alignment in these motifs a pattern of amino acid properties required at each position is defined. The discriminatory power of the pattern is in part due to the use of several known three-dimensional structures and many sequences and in part to the "property" method of generalizing from observed amino acid frequencies to amino acid fitness at each sequence position. A sequence data base search with the pattern significantly matches sugar kinases, such as fuco-, glucono-, xylulo-, ribulo-, and glycerokinase, as well as the prokaryotic cell cycle proteins MreB, FtsA, and StbA. These are predicted to have subdomains with the same tertiary structure as the ATPase subdomains Ia and IIa of hexokinase, actin, and Hsc70, a very similar ATP binding pocket, and the capacity for interdomain hinge motion accompanying functional state changes. A common evolutionary origin for all of the proteins in this class is proposed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Agranovsky A. A., Boyko V. P., Karasev A. V., Koonin E. V., Dolja V. V. Putative 65 kDa protein of beet yellows closterovirus is a homologue of HSP70 heat shock proteins. J Mol Biol. 1991 Feb 20;217(4):603–610. doi: 10.1016/0022-2836(91)90517-a. [DOI] [PubMed] [Google Scholar]
- Anderson C. M., Stenkamp R. E., Steitz T. A. Sequencing a protein by x-ray crystallography. II. Refinement of yeast hexokinase B co-ordinates and sequence at 2.1 A resolution. J Mol Biol. 1978 Jul 25;123(1):15–33. doi: 10.1016/0022-2836(78)90374-1. [DOI] [PubMed] [Google Scholar]
- Arora K. K., Filburn C. R., Pedersen P. L. Glucose phosphorylation. Site-directed mutations which impair the catalytic function of hexokinase. J Biol Chem. 1991 Mar 25;266(9):5359–5362. [PubMed] [Google Scholar]
- Bairoch A., Boeckmann B. The SWISS-PROT protein sequence data bank. Nucleic Acids Res. 1991 Apr 25;19 (Suppl):2247–2249. doi: 10.1093/nar/19.suppl.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. Nucleotide sequence and insertional inactivation of a Bacillus subtilis gene that affects cell division, sporulation, and temperature sensitivity. J Bacteriol. 1989 Dec;171(12):6821–6834. doi: 10.1128/jb.171.12.6821-6834.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett W. S., Jr, Steitz T. A. Glucose-induced conformational change in yeast hexokinase. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4848–4852. doi: 10.1073/pnas.75.10.4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Grunwald C. Recognition of different nucleotide-binding sites in primary structures using a property-pattern approach. Eur J Biochem. 1990 Jul 31;191(2):347–358. doi: 10.1111/j.1432-1033.1990.tb19129.x. [DOI] [PubMed] [Google Scholar]
- Bowie J. U., Lüthy R., Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991 Jul 12;253(5016):164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- Doi M., Wachi M., Ishino F., Tomioka S., Ito M., Sakagami Y., Suzuki A., Matsuhashi M. Determinations of the DNA sequence of the mreB gene and of the gene products of the mre region that function in formation of the rod shape of Escherichia coli cells. J Bacteriol. 1988 Oct;170(10):4619–4624. doi: 10.1128/jb.170.10.4619-4624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie W. D., Begg K. J., Lutkenhaus J. F., Salmond G. P., Martinez-Salas E., Vincente M. Role of the ftsA gene product in control of Escherichia coli cell division. J Bacteriol. 1979 Nov;140(2):388–394. doi: 10.1128/jb.140.2.388-394.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle R. F. Stein and Moore Award address. Reconstructing history with amino acid sequences. Protein Sci. 1992 Feb;1(2):191–200. doi: 10.1002/pro.5560010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Easterby J. S. The polypeptide chain molecular weight of a mammalian hexokinase. FEBS Lett. 1971 Oct 15;18(1):23–26. doi: 10.1016/0014-5793(71)80397-6. [DOI] [PubMed] [Google Scholar]
- Flaherty K. M., DeLuca-Flaherty C., McKay D. B. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990 Aug 16;346(6285):623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Flaherty K. M., McKay D. B., Kabsch W., Holmes K. C. Similarity of the three-dimensional structures of actin and the ATPase fragment of a 70-kDa heat shock cognate protein. Proc Natl Acad Sci U S A. 1991 Jun 1;88(11):5041–5045. doi: 10.1073/pnas.88.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., Sambrook J. Protein folding in the cell. Nature. 1992 Jan 2;355(6355):33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- Holm L., Sander C. Database algorithm for generating protein backbone and side-chain co-ordinates from a C alpha trace application to model building and detection of co-ordinate errors. J Mol Biol. 1991 Mar 5;218(1):183–194. doi: 10.1016/0022-2836(91)90883-8. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Mannherz H. G., Suck D., Pai E. F., Holmes K. C. Atomic structure of the actin:DNase I complex. Nature. 1990 Sep 6;347(6288):37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- Keim P., Heinrikson R. L., Fitch W. M. An examination of the expected degree of sequence similarity that might arise in proteins that have converged to similar conformational states. The impact of such expectations on the search for homology between the structurally similar domains of rhodanese. J Mol Biol. 1981 Sep 5;151(1):179–197. doi: 10.1016/0022-2836(81)90227-8. [DOI] [PubMed] [Google Scholar]
- Lasters I., Wodak S. J., Alard P., van Cutsem E. Structural principles of parallel beta-barrels in proteins. Proc Natl Acad Sci U S A. 1988 May;85(10):3338–3342. doi: 10.1073/pnas.85.10.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton R. J. Hexokinases and glucokinases. Biochem Soc Trans. 1990 Apr;18(2):180–183. doi: 10.1042/bst0180180. [DOI] [PubMed] [Google Scholar]
- Pla J., Dopazo A., Vicente M. The native form of FtsA, a septal protein of Escherichia coli, is located in the cytoplasmic membrane. J Bacteriol. 1990 Sep;172(9):5097–5102. doi: 10.1128/jb.172.9.5097-5102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D. Actin. Curr Opin Cell Biol. 1990 Feb;2(1):33–40. doi: 10.1016/s0955-0674(05)80028-6. [DOI] [PubMed] [Google Scholar]
- Robinson A. C., Begg K. J., Sweeney J., Condie A., Donachie W. D. Mapping and characterization of mutants of the Escherichia coli cell division gene, ftsA. Mol Microbiol. 1988 Sep;2(5):581–588. doi: 10.1111/j.1365-2958.1988.tb00066.x. [DOI] [PubMed] [Google Scholar]
- Robinson A. C., Collins J. F., Donachie W. D. Prokaryotic and eukaryotic cell-cycle proteins. 1987 Aug 27-Sep 2Nature. 328(6133):766–766. doi: 10.1038/328766a0. [DOI] [PubMed] [Google Scholar]
- Sander C., Schneider R. Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins. 1991;9(1):56–68. doi: 10.1002/prot.340090107. [DOI] [PubMed] [Google Scholar]
- Schwab D. A., Wilson J. E. Complete amino acid sequence of rat brain hexokinase, deduced from the cloned cDNA, and proposed structure of a mammalian hexokinase. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2563–2567. doi: 10.1073/pnas.86.8.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz T. A., Shoham M., Bennett W. S., Jr Structural dynamics of yeast hexokinase during catalysis. Philos Trans R Soc Lond B Biol Sci. 1981 Jun 26;293(1063):43–52. doi: 10.1098/rstb.1981.0058. [DOI] [PubMed] [Google Scholar]
- Tabuchi A., Min Y. N., Kim C. K., Fan Y. L., Womble D. D., Rownd R. H. Genetic organization and nucleotide sequence of the stability locus of IncFII plasmid NR1. J Mol Biol. 1988 Aug 5;202(3):511–525. doi: 10.1016/0022-2836(88)90282-3. [DOI] [PubMed] [Google Scholar]
- Vriend G., Sander C. Detection of common three-dimensional substructures in proteins. Proteins. 1991;11(1):52–58. doi: 10.1002/prot.340110107. [DOI] [PubMed] [Google Scholar]
- Wachi M., Matsuhashi M. Negative control of cell division by mreB, a gene that functions in determining the rod shape of Escherichia coli cells. J Bacteriol. 1989 Jun;171(6):3123–3127. doi: 10.1128/jb.171.6.3123-3127.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]






