Abstract
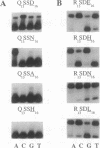
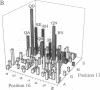
Zinc finger proteins of the Cys2-His2 type consist of tandem arrays of domains, where each domain appears to contact three adjacent base pairs of DNA through three key residues. We have designed and prepared a series of variants of the central zinc finger within the DNA binding domain of Sp1 by using information from an analysis of a large data base of zinc finger protein sequences. Through systematic variations at two of the three contact positions (underlined), relatively specific recognition of sequences of the form 5'-GGGGN(G or T)GGG-3' has been achieved. These results provide the basis for rules that may develop into a code that will allow the design of zinc finger proteins with preselected DNA site specificity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berg J. M. Proposed structure for the zinc-binding domains from transcription factor IIIA and related proteins. Proc Natl Acad Sci U S A. 1988 Jan;85(1):99–102. doi: 10.1073/pnas.85.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg J. M. Zinc finger domains: hypotheses and current knowledge. Annu Rev Biophys Biophys Chem. 1990;19:405–421. doi: 10.1146/annurev.bb.19.060190.002201. [DOI] [PubMed] [Google Scholar]
- Desjarlais, Berg J. M. Redesigning the DNA-binding specificity of a zinc finger protein: a data base-guided approach. Proteins. 1992 Jul;13(3):272–272. doi: 10.1002/prot.340130309. [DOI] [PubMed] [Google Scholar]
- Desjarlais J. R., Berg J. M. Redesigning the DNA-binding specificity of a zinc finger protein: a data base-guided approach. Proteins. 1992 Feb;12(2):101–104. doi: 10.1002/prot.340120202. [DOI] [PubMed] [Google Scholar]
- Eisen A., Taylor W. E., Blumberg H., Young E. T. The yeast regulatory protein ADR1 binds in a zinc-dependent manner to the upstream activating sequence of ADH2. Mol Cell Biol. 1988 Oct;8(10):4552–4556. doi: 10.1128/mcb.8.10.4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R. M., Hollenberg S. M. Zinc fingers: gilt by association. Cell. 1988 Jan 15;52(1):1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Gibson T. J., Postma J. P., Brown R. S., Argos P. A model for the tertiary structure of the 28 residue DNA-binding motif ('zinc finger') common to many eukaryotic transcriptional regulatory proteins. Protein Eng. 1988 Sep;2(3):209–218. doi: 10.1093/protein/2.3.209. [DOI] [PubMed] [Google Scholar]
- Klevit R. E. Recognition of DNA by Cys2,His2 zinc fingers. Science. 1991 Sep 20;253(5026):1367–1393. doi: 10.1126/science.1896847. [DOI] [PubMed] [Google Scholar]
- Lehming N., Sartorius J., Kisters-Woike B., von Wilcken-Bergmann B., Müller-Hill B. Mutant lac repressors with new specificities hint at rules for protein--DNA recognition. EMBO J. 1990 Mar;9(3):615–621. doi: 10.1002/j.1460-2075.1990.tb08153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letovsky J., Dynan W. S. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989 Apr 11;17(7):2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Protein-DNA interaction. No code for recognition. Nature. 1988 Sep 22;335(6188):294–295. doi: 10.1038/335294a0. [DOI] [PubMed] [Google Scholar]
- Nardelli J., Gibson T. J., Vesque C., Charnay P. Base sequence discrimination by zinc-finger DNA-binding domains. Nature. 1991 Jan 10;349(6305):175–178. doi: 10.1038/349175a0. [DOI] [PubMed] [Google Scholar]
- Pavletich N. P., Pabo C. O. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991 May 10;252(5007):809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Rhodes D., Klug A. An underlying repeat in some transcriptional control sequences corresponding to half a double helical turn of DNA. Cell. 1986 Jul 4;46(1):123–132. doi: 10.1016/0092-8674(86)90866-4. [DOI] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Perry-O'Keefe H., Melton D. A. Xfin: an embryonic gene encoding a multifingered protein in Xenopus. EMBO J. 1987 Oct;6(10):3065–3070. doi: 10.1002/j.1460-2075.1987.tb02613.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thukral S. K., Morrison M. L., Young E. T. Alanine scanning site-directed mutagenesis of the zinc fingers of transcription factor ADR1: residues that contact DNA and that transactivate. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9188–9192. doi: 10.1073/pnas.88.20.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953 Apr 25;171(4356):737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]