Abstract
Nucleotide pyrophosphatase/phosphodiesterases (NPPs) are widely distributed N-glycosylated enzymes that catalyze the hydrolytic breakdown of numerous nucleotides and nucleotide sugars. In many plant species, NPPs are encoded by a small multigene family, which in rice are referred to NPP1–NPP6. Although recent investigations showed that N-glycosylated NPP1 is transported from the endoplasmic reticulum (ER)–Golgi system to the chloroplast through the secretory pathway in rice cells, information on N-glycan composition and subcellular localization of other NPPs is still lacking. Computer-assisted analyses of the amino acid sequences deduced from different Oryza sativa NPP-encoding cDNAs predicted all NPPs to be secretory glycoproteins. Confocal fluorescence microscopy observation of cells expressing NPP2 and NPP6 fused with green fluorescent protein (GFP) revealed that NPP2 and NPP6 are plastidial proteins. Plastid targeting of NPP2–GFP and NPP6–GFP was prevented by brefeldin A and by the expression of ARF1(Q71L), a dominant negative mutant of ADP-ribosylation factor 1 that arrests the ER to Golgi traffic, indicating that NPP2 and NPP6 are transported from the ER–Golgi to the plastidial compartment. Confocal laser scanning microscopy and high-pressure frozen/freeze-substituted electron microscopy analyses of transgenic rice cells ectopically expressing the trans-Golgi marker sialyltransferase fused with GFP showed the occurrence of contact of Golgi-derived membrane vesicles with cargo and subsequent absorption into plastids. Sensitive and high-throughput glycoblotting/mass spectrometric analyses showed that complex-type and paucimannosidic-type glycans with fucose and xylose residues occupy approximately 80% of total glycans of NPP1, NPP2 and NPP6. The overall data strongly indicate that the trans-Golgi compartments participate in the Golgi to plastid trafficking and targeting mechanism of NPPs.
Keywords: Glycoprotein, Golgi, Membrane traffic, N-glycome, Oryza sativa, Plastid
Introduction
Nucleotide pyrophosphatase/phosphodiesterases (NPPs) are N-glycosylated enzymes occurring in both mono- and dicotyledonous plants that catalyze the hydrolytic breakdown of the pyrophosphate and phosphodiester bonds of numerous nucleotides and nucleotide sugars (Rodríguez-López et al. 2000, Nanjo et al. 2006). Six different NPP-encoding genes (NPP1–NPP6) have been identified in the rice genome (Kaneko et al. 2014). Rice NPP amino acid sequences are highly similar to those of metallophosphatases from yellow lupin (Olczak and Olczak 2002) and purple acid phosphatases 1, 24 and 27 from Arabidopsis (Zhu et al. 2005), indicating that NPPs belong to a large family of structurally related nucleotide hydrolases (Kaneko et al. 2014). Changes in NPP1 expression in genetically engineered rice plants are accompanied by changes in shoot growth and starch content, especially under high CO2 concentration and high temperature conditions, indicating that NPP1 is involved in the control of plant growth and reserve carbohydrate accumulation in rice (Kaneko et al. 2014).
In both mammalian and plant cells, the conjugation of oligosaccharide chains to asparagine residues of newly synthesized proteins occurs by means of a highly conserved biosynthetic pathway (Lerouge et al. 1998, Helenius and Abei 2001, Saint-Jore-Dupas et al. 2006). N-Glycosylation is initiated in the endoplasmic reticulum (ER) by the transfer of Glc3Man9GlcNAc2 glycans to aspargine residues in the sequence Asn-X-Ser/Thr of the nascent polypeptide (Kelleher and Gilmore 2006). In the ER, glucosidase and mannosidase convert Glc3Man9GlcNAc2 to Man8GlcNAc2 (Tremblay and Herscovics 1999, Boisson et al. 2001, Saint-Jore-Dupas et al. 2006, Soussillane et al. 2009), and glycoproteins bearing the trimmed glycan are exported to the Golgi apparatus. The Man8GlcNAc2 glycan is then trimmed to Man5GlcNAc2 by cis-Golgi-resident mannosidase I (Nebenführ et al. 1999, Liebminger et al. 2009, Kajiura et al. 2010), and further modified to complex and hybrid glycan chains by Golgi mannosidase II and glycosyltransferases (Kornfeld and Kornfeld 1985). Structures of mature complex-type oligosaccharide chains are diverse between plants and animals. Plant complex-type N-glycans are generally smaller than those of animals, and contain unique terminal sugar residues, such as, β1,2-xylose and an ⟨1,3-fucose core (Takahashi et al. 1986, Hayashi et al. 1990, Ogawa et al. 1996, Fitchette-Lainé et al. 1997, Melo et al. 1997, Olczak and Watorek 2000). N-Acetylglucosaminyltransferase I, ⟨-mannosidase II and N-acetylglucosaminyltransferase II occur in the cis-Golgi to medial-Golgi (Strasser et al. 1999a, Strasser et al. 1999b, Strasser et al. 2006), and a series of plant-specific glycosyltransferases, such as β1,2-xylosyltransferase, ⟨1,3-fucosyltransferase, ⟨1,4-fucosyltransferase and β1,3-galactosyltransferase, occur in the medial-Golgi to trans-Golgi compartments (Strasser et al. 2000, Wilson et al. 2001, Léonard et al. 2002, Bondili et al. 2006, Saint-Jore-Dupas et al. 2006, Strasser et al. 2007) that are involved in processing and maturation of N-glycans.
The general pathway of protein import from the cytosol into plastids is mediated by the Toc/Tic translocon complex (Inaba et al. 2005, Inoue and Akita 2008, Kessler and Schnell 2009). However, several reports have provided evidence supporting the occurrence of traffic of proteins and glycoproteins to the plastid via the endoplasmic reticulum (ER)–Golgi system (Chen et al. 2004, Asatsuma et al. 2005, Villarejo et al. 2005, Nanjo et al. 2006, Kitajima et al. 2009, Burén et al. 2011). Thus, Samuelsson and colleagues (Villarejo et al. 2005, Burén et al. 2011) showed that the glycosylated carbonic anhydrase 1 (CAH1) is transported and localized to the chloroplasts of Arabidopsis cells. Immunochemical analyses using anti-⟨1,3-fucose and anti-β1,2-xylose antibodies revealed that this protein possesses N-linked oligosaccharide chains with fucose and xylose residues (Villarejo et al. 2005). Because β1,2-xylosyltransferase- and ⟨1,3-fucosyltransferase-mediated glycosylation of N-linked oligosaccharide chains occurs exclusively in the late compartments of the Golgi apparatus (Saint-Jore-Dupas et al. 2006), Villarejo et al. (2005) proposed that Arabidopsis CAH1 must traverse the Golgi apparatus prior to chloroplast targeting. Brefeldin A (BFA), a drug that inhibits ER to Golgi traffic by inactivating ADP-ribosylation factor 1 (ARF1) GTPase recruitment to Golgi membranes, reversibly prevented targeting of CAH1 to the chloroplast (Villarejo et al. 2005). Similarly, studies carried out by Nanjo et al. (2006) revealed that rice NPP1 bearing N-linked oligosaccharide chains is transported to the plastid along a BFA-sensitive vesicular transport pathway. Burén et al. (2011) provided evidence that N-glycosylation is important in the folding and trafficking of some glycoproteins from the secretory pathway to the chloroplast. Asatsuma et al. (2005) showed that rice ⟨-amylase isoform I-1 (AmyI-1), a well-characterized secretory glycoprotein, has a plastidial localization. Furthermore, studies using three-dimensional time-lapse imaging and electron microscopy of high-pressure frozen/freeze-substituted cells demonstrated that contact and subsequent absorption of Golgi-derived membrane vesicles with cargo by plastids is involved in the Golgi to plastid traffic of AmyI-1 (Kitajima et al. 2009).
To obtain insights into the mechanisms involved in the traffic of NPPs from the ER–Golgi system to the plastid, in this work we characterized the N-glycan composition of several plastidial NPPs employing a highly sensitive and high-throughput ‘glycoblotting’ method for oligosaccharide purification/derivatization (Nishimura et al. 2005, Furukawa et al. 2008, Amano et al. 2010). These studies revealed that NPP1, NPP2 and NPP6 contain complex-type and paucimannosidic-type oligosaccharide chains with β1,2-xylose and/or core ⟨1,3-fucose residue(s), providing strong evidence that the trans-Golgi compartments participate in the Golgi to plastid trafficking and targeting mechanism of NPP in rice.
Results and Discussion
NPP2 and NPP6 have a plastidial localization
Computer-assisted analyses using the SignalP (http://www.cbs.dtu.dk/services/SignalP/) and PSORT (http://psort.ims.u-tokyo.ac.jp/) algorithms revealed that all the deduced NPP amino acid sequences have cleavable hydrophobic N-terminal extensions that act as potential signal peptides to the ER and putative N-glycosylation sites (Fig. 1). NPP1, NPP2 and NPP6 form homo-oligomeric structures composed of 70, 71 or 74kDa polypeptides, respectively (Kaneko et al. 2014). Edman sequence analyses of the purified enzymes revealed that the N-terminal amino acid residues of mature NPP1, NPP2 and NPP6 are Gly52, Ala41 and Ala55, respectively (Kaneko et al. 2014). These amino acid residues do not match the predicted cleavable sites of the signal sequence of the proteins (Fig. 1), which can be ascribed to additional proteolytic processing occurring in different subcellular compartments during the maturation process of the NPPs (Kaneko et al. 2011). To determine the subcellular localization of NPP2 and NPP6, we produced rice cells stably transformed with either NPP2–GFP (green fluorescent protein) or NPP6–GFP and subjected them to confocal microscopy analysis. As controls, we used GFP- and NPP1–GFP-expressing cells. As shown in Fig. 2, the fluorescence distribution patterns of NPP1–GFP, NPP2–GFP and NPP6–GFP largely matched those of Chl autofluorescence. Quantitative analyses showed that approximately 73–82% of the fluorescence emitted by each NPP–GFP form was localized in the chloroplasts of transformed cells (Fig. 2Q). We further characterized the localization of NPP2 and NPP6 using onion epidermal cells transiently expressing NPP–GFP. NPP2–GFP and NPP6–GFP were simultaneously expressed with a plastid marker, the plastid transit peptide of starch granule-bound starch synthase I, Waxy, fused to red fluorescent protein from Discosoma sp. (WxTP–DsRed, Kitajima et al. 2009). As shown in Fig. 3D–F and G–I, a a large portion of NPP2–GFP and NPP6–GFP fluorescence overlapped with the plastids visualized by WxTP–DsRed, respectively, providing further evidence that NPP2 and NPP6 have a plastidial localization.
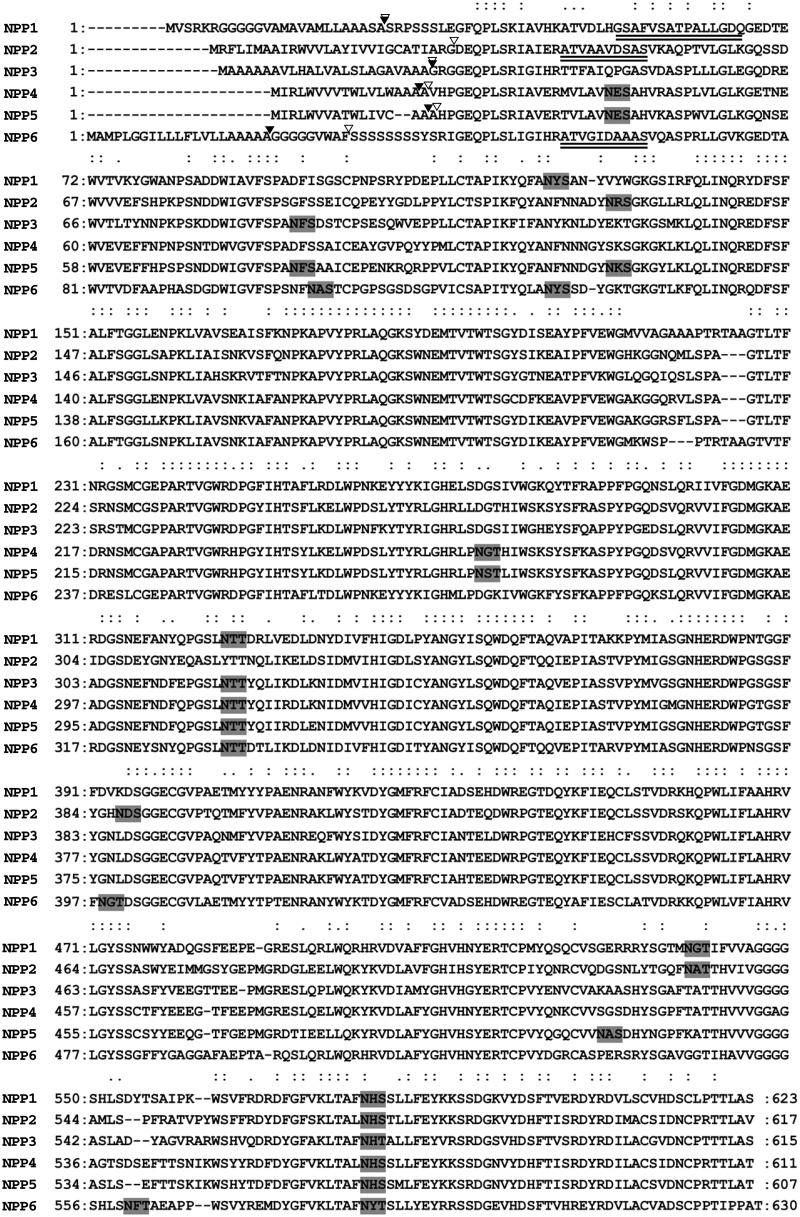
Fig. 1.
Amino acid sequence alignments of Oryza sativa NPPs. Double dots represent completely conserved amino acid residues in all NPPs, and single dots represent conserved amino acid residues among NPP1, NPP2 and NPP6. Predicted N-glycosylation sites are boxed. Double underline, N-terminal sequence in mature protein; filled triangle, cleavage site of signal peptide (PSORT prediction). Open triangle, cleavage site of signal peptide (SignalP prediction). All NPP precursor proteins possess a potential signal peptide and several N-glycosylation sites.
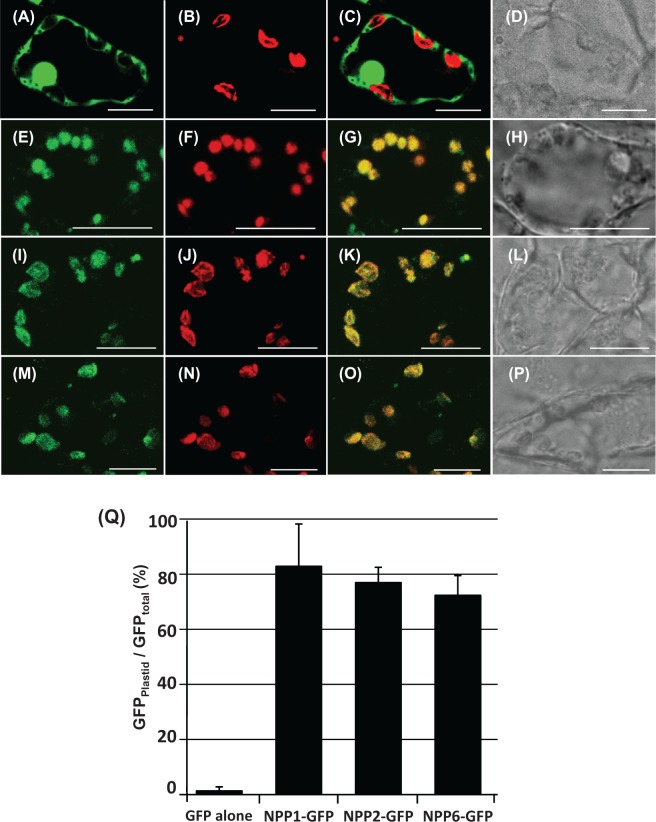
Fig. 2.
Fluorescence images in transgenic rice cells expressing either GFP, NPP1–GFP, NPP2–GFP or NPP6–GFP. The stable transformant cells were sectioned with a vibratome to 25μm thickness, and immediately observed by means of confocal laser scanning microscopy. (A–D) Rice cells expressing GFP. (E–H) Rice cells expressing NPP1–GFP. (I–L) Rice cells expressing NPP2–GFP. (M–P) Rice cells expressing NPP6–GFP. (A), (E), (I) and (M) GFP fluorescence; (B), (F), (J) and (N) Chl autofluorescence; (C), (G), (K) and (O) GFP and Chl autofluorescence merged; (D), (H), (L) and (P) Nomarski images. Scale bars represent 10μm. (Q) Statistical evaluation of the plastidial localization of NPP–GFPs. Ratios of the fluorescence intensity of GFP in the plastidial area to GFP in the whole cell (GFPplastid/GFPtotal) (%) were determined. Values show the means ± SD (n = 3). NPP1–GFP, NPP2–GFP and NPP3–GFP were mostly localized in the chloroplasts of rice cells.
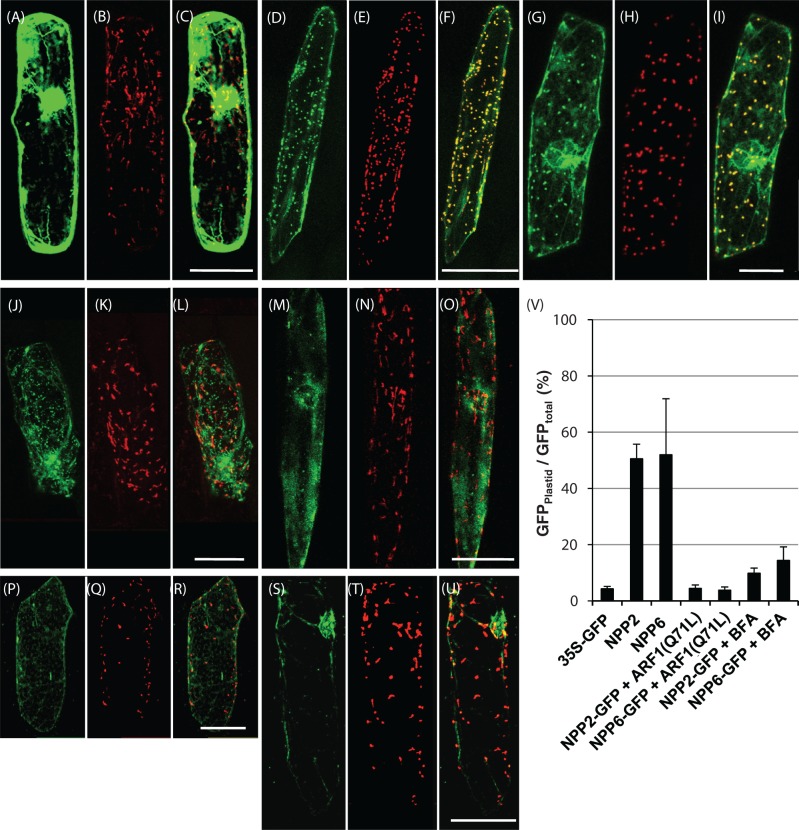
Fig. 3.
Expression and localization of NPP2–GFP and NPP6–GFP in onion epidermal cells. (A–C) Control experiments. Double labeling with GFP and WxTP–DsRed was performed in onion cells. (D–F) Onion cells expressing NPP2–GFP and WxTP–DsRed. (G–I) Onion cells expressing NPP6–GFP and WxTP–DsRed. (J–O) Effect of Arabidopsis ARF1(Q71L) on the localization of NPP2–GFP (J–L) and NPP6–GFP (M–O). (P–U) Effect of 90μM BFA on the localization of NPP2–GFP (P–R) and NPP6–GFP (S–U). (A), (D), (G), (J), (M), (P) and (S) GFP, green; (B), (E), (H), (K), (N), (Q) and (T) WxTP–DsRed, red; (C), (F), (I), (L), (O), (R) and (U) GFP and DsRed merged. (A–U) Projections of stacks of 20–30 images per cell, acquired from the top to the middle of the cell, every 1.5μm. Scale bars = 100μm. (V) Statistical evaluation of the plastidial localization of NPP2–GFP and NPP6–GFP. Ratios of the fluorescence intensity of GFP in the plastidial area to GFP in the whole cell (GFPplastid/GFPtotal) (%) were determined. Values are represented as mean ± SD (n = 4). NPP2–GFP and NPP6–GFP were evidently localized in the plastids visualized with WxTP–DsRed, and the plastid targeting of NPP2–GFP and NPP6–GFP was significantly prevented in cells expressing ARF1 mutants or BFA treatments.
Membrane trafficking from the ER to the Golgi apparatus is essential for the plastid targeting of NPP2 and NPP6
ARF1 is essential for membrane trafficking between the ER and the Golgi apparatus in plant cells. Expression of dominant negative mutants of ARF1 such as ARF1(Q71L) inhibits the ER to Golgi traffic (Takeuchi et al. 2002). To explore the possible traffic of NPP2 and NPP6 from the ER to the Golgi, we investigated the effect of ARF1(Q71L) expression on the distribution of NPP2–GFP and NPP6–GFP in onion epidermal cells. These analyses revealed that the transport of NPP2–GFP and NPP6–GFP into plastids is largely prevented by ARF1(Q71L) (Fig. 3J–O). A statistical analysis indicated that the transport activities of GFP-labeled NPPs into plastids are reduced to <10% in ARF1(Q71L)-expressing cells (Fig. 3V). We further examined the effect of BFA on NPP2–GFP and NPP6–GFP targeting in onion epidermal cells. As shown in Fig. 3P–V, BFA (90μM) prevented NPP2–GFP and NPP6–GFP targeting into the plastid in onion cells. A resemblance between abnormal distributions (a network structure with puncta) of NPP2–GFP could be observed in both ARF1(Q71L)-expressing and BFA-treated cells (Fig. 3J, P). Together with our previous studies showing that NPP1 is targeted to plastids of rice cells in a BFA-sensitive manner (Nanjo et al. 2006), these data strongly indicated that membrane trafficking from the ER to the Golgi apparatus is essential for the plastid targeting of NPP1, NPP2 and NPP6.
Small membrane vesicles derived from the trans-Golgi participate in the traffic of NPP1 to the chloroplast
To gain insight into the mechanism(s) of Golgi to plastid traffic of NPPs, we carried out confocal laser scanning microscopy and high-pressure frozen/freeze-substituted electron microscopy analyses using cultured rice cells ectopically expressing the trans-Golgi marker sialyltransferase fused with GFP (ST–GFP) without or with a constitutively high expression of NPP1. As shown in Fig. 4C and D, when NPP1 was ectopically expressed (which would activate Golgi to plastid traffic in rice cells), significant merging of GFP and Chl autofluorescence could be detected both on the surface and inside the chloroplast. Such incorporation of ST–GFP-labeled membrane vesicles into the chloroplasts appeared to be much less frequent in cells with wild-type NPP1 expression levels (Fig. 4A, B; Supplementary Fig. S1). High-pressure frozen/freeze-substituted electron microscopy analyses revealed that plastids of NPP1-overexpressing cells frequently contained small membrane vesicles whose morphology resembled that of Golgi vesicles (Fig. 4F). The presence of such vesicles within the chloroplast was less frequent in rice cells expressing wild-type levels of NPP1 (Fig. 4E). Immunocytochemical analyses using anti-GFP antibodies showed that a significant portion of ST–GFP was detected in the interior of plastids of NPP1-overexpressing cells (Fig. 4H) in addition to the endomembrane system (Fig. 4G; Supplementary Fig. S2). Notably, membrane-anchored ST–GFP could be observed within the plastid (Fig. 4H). Some immuno-gold particles were deposited on the membrane-free area in the plastids (Fig. 4H), which can be ascribed to degradation of the imported Golgi vesicles. The overall data would thus indicate that small membrane vesicles derived from the trans-Golgi are involved in the mechanisms of NPP1 targeting to plastids.
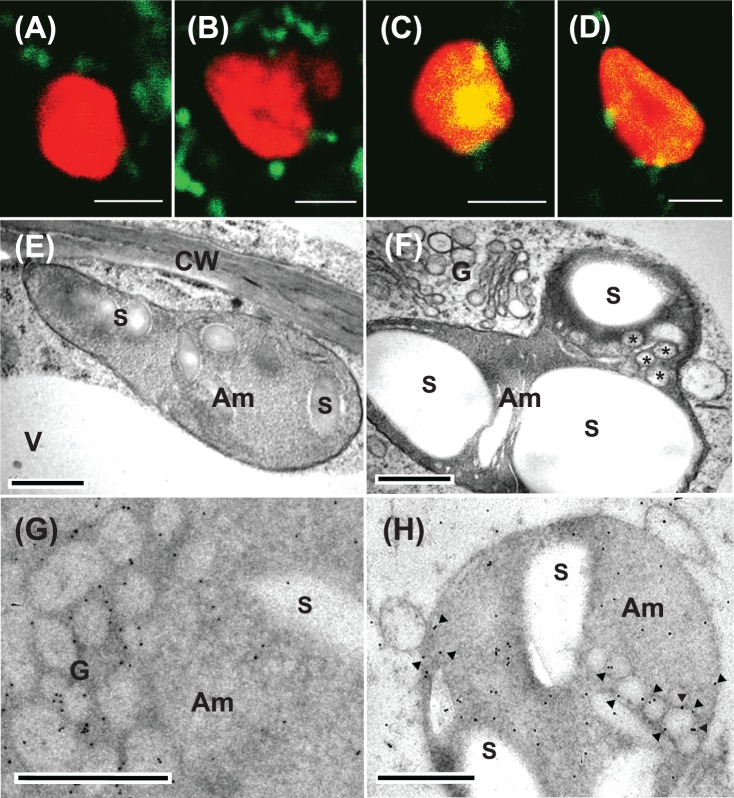
Fig. 4.
Confocal imaging and electron microscopy characterization of trans-Golgi to plastid communication in rice cells expressing ST–GFP without or with a constitutively high expression of NPP1. (A–D) The stable transformant rice cells were sectioned with a vibratome to 25μm thickness, and immediately observed by means of confocal laser scanning microscopy. (A and B) Rice cells expressing ST–GFP without NPP1 overexpression. (C and D) Rice cells expressing ST–GFP with NPP1. GFP (green) and Chl autofluorescence (red) merged. Some Golgi marker ST–GFP complexes (green blobs) were tightly adhered to the surface of chloroplasts. The change in color from red to yellow in the interior of the plastid is due to the uptake of ST–GFP. Scale bars represent 2μm. (E–H) The rice cells cultured in MS medium containing 3% sucrose were rapidly frozen in a high-pressure freezer and subjected to electron microscopic studies. Am, amyloplasts; CW, cell wall; G, Golgi; V, vacuole; S, starch. (E and F) Morphological observations of the cells expressing ST–GFP without and with NPP1 overexpression, respectively. Asterisks represent small membrane vesicles observed in the plastid. (G and H) Immunocytochemical observations with anti-GFP antibodies of the cells expressing ST–GFP with NPP1 overexpression. (G) shows remakable labeling of immunogold particles in the Golgi vesicle area. Arrowheads in (H) show immunogold particles deposited on membrane vesicles inside plastids. Scale bars represent 0.5μm.
We can only speculate as to the mechanism(s) of uptake of small membrane vesicles derived from the trans-Golgi. If microautophagy-like invaginations were involved in this mechanism, vesicles bounded by the plastid envelope membranes in addition to small invaginations should be frequently detected in the stroma of plastids. However, we never observed vesicles bounded by three membranes in the plastids. Therefore, it is conceivable that vesicles traverse the plastid envelope membranes by mechanism(s) other than by microautophagy-like invagination into the organelle. Once in the chloroplast, it is likely that the vesicles are digested by enzymes occurring in the stroma, allowing the liberation of the cargo glycoproteins. It is obvious that further investigations will be necessary to verify this hypothesis.
N-Glycomic characterization of plastidial NPPs
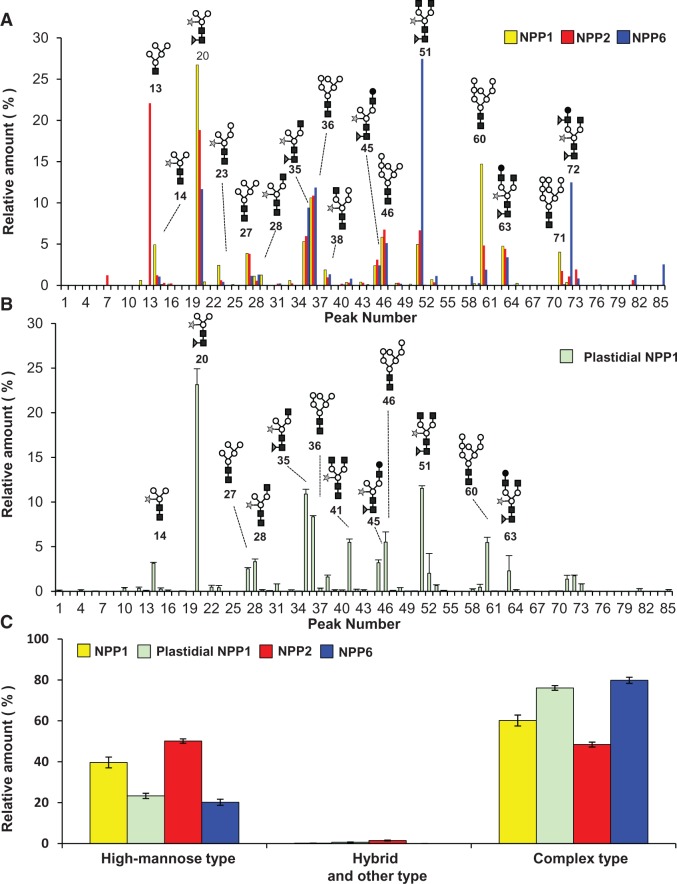
Maturation of the N-linked oligosaccharide chain is a well-characterized event occurring in the late compartments of the Golgi apparatus, involving the conjugation of terminal sugar(s) such as xylose and fucose (Strasser et al. 2000, Wilson et al. 2001, Léonard et al. 2002, Bondili et al. 2006, Saint-Jore-Dupas et al. 2006, Strasser et al. 2007). Therefore, compositional analysis of N-glycans of plastidial glycoproteins should constitute a powerful tool to investigate the possible involvement of the trans-Golgi compartment in the maturation and traffic of glycoproteins to the plastid. We thus purified NPP1 from chloroplasts isolated from shoots of transgenic UNP1 rice plants and carried out N-glycomic analyses of this protein. As shown in Supplementary Figs. S3 and S4, intact chloroplast-enriched fractions contained negligible contamination by cytosol, mitochondria, Golgi and tonoplast membranes. NPP-specific activity in the chloroplast preparation was approximately 9-fold higher than in crude leaf extracts (Table 1), which further confirms the presence of NPPs in the plastidial compartment. Subsequent characterization of the N-glycome of the purified chloroplastic NPP1 revealed the occurrence of at least 66 different glycans registered in the KEGG Glycan (Hashimoto et al. 2006), GlycoMod Tool (Cooper et al. 2001a) and GlycoSuite (Cooper et al. 2001b) databases. Man3GlcNAc2Fuc1Xyl1 (peak 20, 23.1%), Man3GlcNAc4Fuc1Xyl1 (peak 51, 11.5%), Man3GlcNAc3Fuc1Xyl1 (peak 35, 10.9%), Man6GlcNAc2 (peak 36, 8.3%) and Man7GlcNAc2 (peak 46, 5.5%) were detected as the top five glycan chains in the chloroplastic NPP1 (Fig. 5B; Table 2; Supplementary Fig. S5). It is noteworthy that, consistent with the idea that NPP1 is transported into the chloroplast after modifying the oligosaccharide chains in the late compartment of the Golgi apparatus, 76.0% of the plastidial NPP1 glycoforms corresponded to complex-type oligosaccharide chains modified with fucose and xylose residues, whereas the high mannose-type forms accounted for 23.3% of the total glycoforms (Table 2).
Table 1.
Purification of chloroplastic NPP1 from leaves of the UNP1 line
| Step | Total volume (ml) | Total protein (mg) | Total activity (Ua) | Specific activity (U mg–1 protein) | Purification (fold) |
|---|---|---|---|---|---|
| Leaf extract | 400 | 183.76 | 170.68 | 0.93 | – |
| Isolated chloroplast | 10 | 1.27 | 10.86 | 8.58 | 9.24 |
| Con A–Sepharose 4B | 8.5 | 0.39 | 12.31 | 31.96 | 34.41 |
| Microcon YM-100 | 0.1 | 0.02 | 2.67 | 107.52 | 115.76 |
aμmol min–1.
Fig. 5.
N-Glycoforms of NPP1, NPP2, NPP6 and plastidial NPP1 in rice cells. N-Glycan chains released from NPP1, NPP2 and NPP6 were captured by the glycoblotting method and subjected to MALDI-TOF-MS with labeling reagents. Details of mass values and contents for the detected N-glycans are shown in Tables 2–5. Compositional annotation and prediction of absolute structures for these N-glycans were carried out by the GlycoMod Tool and GlycoSuite websites. (A) Comparison of N-glycoforms of NPP1, NPP2 and NPP6 purified from the whole cells of transgenic rice lines, UNP1, UNP2 and UNP6, respectively. Values show the averages of triplicate experiments. (B) N-Glycoforms of NPP1 purified from the leaf chloroplasts of the UNP1 line. Values show the averages of duplicate experiments. Filled squares, N-acetylglucosamine; open circles, mannose; stars, xylose; filled triangles, fucose; filled circles, galactose. (C) Differential N-glycan complexities of NPP1, NPP2, NPP6 and plastidial NPP1. Glycans were grouped into high mannose type (Glc3Man9GlcNAc2 to Man5GlcNAc2), hybrid and other type (including intermediates between GlcNAc2Man3GlcNAc2 and Man3GlcNAc2), and complex type (including Man3GlcNAc2 conjugated with xylose and/or fucose).
Table 2.
Glycoforms detected in rice plastidial NPP1
| Peak no.a | m/zb | Δc | Estimated composition | Relative contents (%)d |
|---|---|---|---|---|
| 1 | 688.9915 | −0.268 | HexNAc2 Pent2 | 0.108 ± 0.033 |
| 4 | 749.0722 | −0.209 | Hex2 HexNAc2 | 0.132 ± 0.056 |
| 5 | 780.0588 | −0.217 | Hex1 HexNAc1 Pent3 | 0.007 ± 0.005 |
| 6 | 851.0808 | −0.232 | Hex1 HexNAc2 Pent2 | 0.037 ± 0.003 |
| 9 | 895.1576 | −0.181 | Hex2 HexNAc2 dHex1 | 0.006 ± 0.001 |
| 10 | 911.149 | −0.185 | Hex3 HexNAc2 | 0.359 ± 0.075 |
| 12 | 1,027.204 | −0.178 | Hex2 HexNAc2 dHex1 Pent1 | 0.338 ± 0.143 |
| 13 | 1,032.178 | −0.182 | Hex5 HexNAc1 | 0.088 ± 0.034 |
| 14 | 1,043.203 | −0.174 | Hex3 HexNAc2 Pent1 | 3.144 ± 0.109 |
| 15 | 1,057.220 | −0.171 | Hex3 HexNAc2 dHex1 | 0.249 ± 0.139 |
| 16 | 1,073.216 | −0.171 | Hex4 HexNAc2 | 0.137 ± 0.013 |
| 18 | 1,114.242 | −0.172 | Hex3HexNAc3 | 0.061 ± 0.000 |
| 19 | 1,175.243 | −0.176 | Hex3 HexNAc2 Pent2 | 0.011 ± 0.006 |
| 20 | 1,189.274 | −0.160 | Hex3 HexNAc2 dHex1 Pent1 | 23.146 ± 1.786 |
| 22 | 1,203.290 | −0.159 | Hex3 HexNAc2 dHex2 | 0.464 ± 0.215 |
| 23 | 1,205.274 | −0.155 | Hex4 HexNAc2 Pent1 | 0.406 ± 0.245 |
| 24 | 1,219.304 | −0.141 | Hex4 HexNAc2 dHex1 | 0.010 ± 0.006 |
| 27 | 1,235.288 | −0.152 | Hex5 HexNAc2 | 2.519 ± 0.135 |
| 28 | 1,246.306 | −0.150 | Hex3 HexNAc3 Pent1 | 3.321 ± 0.305 |
| 29 | 1,260.319 | −0.153 | Hex3 HexNAc3 dHex1 | 0.148 ± 0.059 |
| 30 | 1,276.311 | −0.155 | Hex4 HexNAc3 | 0.063 ± 0.026 |
| 31 | 1,351.343 | −0.144 | Hex4 HexNAc2 dHex1 Pent1 | 0.828 ± 0.011 |
| 33 | 1,367.343 | −0.138 | Hex5 HexNAc2 Pent1 | 0.161 ± 0.009 |
| 34 | 1,381.356 | −0.142 | Hex5 HexNAc2 dHex1 | 0.030 ± 0.007 |
| 35 | 1,392.371 | −0.143 | Hex3 HexNAc3 dHex1 Pent1 | 10.904 ± 0.531 |
| 36 | 1,397.347 | −0.146 | Hex6 HexNAc2 | 8.345 ± 0.129 |
| 37 | 1,406.389 | −0.140 | Hex3 HexNAc3 dHex2 | 0.331 ± 0.043 |
| 38 | 1,408.377 | −0.132 | Hex4 HexNAc3 Pent1 | 1.611 ± 0.218 |
| 39 | 1,422.396 | −0.128 | Hex4 HexNAc3 dHex1 | 0.138 ± 0.046 |
| 40 | 1,438.388 | −0.131 | Hex5 HexNAc3 | 0.151 ± 0.013 |
| 41 | 1,449.402 | −0.133 | Hex3 HexNAc4 Pent1 | 5.496 ± 0.372 |
| 42 | 1,463.431 | −0.120 | Hex3 HexNAc4 dHex1 | 0.222 ± 0.018 |
| 43 | 1,513.414 | −0.125 | Hex5 HexNAc2 dHex1 Pent1 | 0.143 ± 0.044 |
| 45 | 1,554.439 | −0.127 | Hex4 HexNAc3 dHex1 Pent1 | 3.191 ± 0.327 |
| 46 | 1,559.417 | −0.128 | Hex7 HexNAc2 | 5.511 ± 1.141 |
| 47 | 1,568.455 | −0.126 | Hex4 HexNAc3 dHex2 | 0.108 ± 0.002 |
| 48 | 1,570.446 | −0.116 | Hex5 HexNAc3 Pent1 | 0.415 ± 0.005 |
| 49 | 1,581.453 | −0.125 | Hex3 HexNAc4 Pent2 | 0.036 ± 0.014 |
| 50 | 1,584.458 | −0.119 | Hex5 HexNAc3 dHex1 | 0.059 ± 0.001 |
| 51 | 1,595.470 | −0.122 | Hex3 HexNAc4 dHex1 Pent1 | 11.533 ± 0.292 |
| 52 | 1,609.496 | −0.113 | Hex3 HexNAc4 dHex2 | 2.017 ± 2.223 |
| 53 | 1,611.481 | −0.107 | Hex4 HexNAc4 Pent1 | 0.651 ± 0.087 |
| 54 | 1,625.495 | −0.109 | Hex4 HexNAc4 dHex1 | 0.106 ± 0.000 |
| 56 | 1,656.547 | −0.051 | Hex3 HexNAc3 dHex1 Pent3 | 0.004 ± 0.000 |
| 57 | 1,666.579 | −0.051 | Hex3 HexNAc5 dHex1 | 0.005 ± 0.005 |
| 58 | 1,700.517 | −0.107 | Hex4 HexNAc3 dHex2 Pent1 | 0.229 ± 0.059 |
| 59 | 1,716.518 | −0.101 | Hex5 HexNAc3 dHex1 Pent1 | 0.458 ± 0.335 |
| 60 | 1,721.490 | −0.108 | Hex8 HexNAc2 | 5.471 ± 0.583 |
| 61 | 1,730.529 | −0.105 | Hex5 HexNAc3 dHex2 | 0.045 ± 0.007 |
| 62 | 1,732.518 | −0.096 | Hex6 HexNAc3 Pent1 | 0.025 ± 0.015 |
| 63 | 1,757.543 | −0.103 | Hex4 HexNAc4 dHex1 Pent1 | 2.282 ± 1.720 |
| 64 | 1,771.565 | −0.096 | Hex4 HexNAc4 dHex2 | 0.151 ± 0.042 |
| 66 | 1,787.561 | −0.096 | Hex5 HexNAc4 dHex1 | 0.025 ± 0.001 |
| 68 | 1,846.610 | −0.072 | Hex4 HexNAc3 dHex3 Pent1 | 0.002 ± 0.001 |
| 69 | 1,862.593 | −0.084 | Hex5 HexNAc3 dHex2 Pent1 | 0.054 ± 0.005 |
| 70 | 1,878.6 | −0.072 | Hex6 HexNAc3 dHex1 Pent1 | 0.031 ± 0.024 |
| 71 | 1,883.565 | −0.086 | Hex9 HexNAc2 | 1.370 ± 0.428 |
| 72 | 1,903.616 | −0.088 | Hex4 HexNAc4 dHex2 Pent1 | 1.738 ± 0.086 |
| 73 | 1,919.620 | −0.079 | Hex5 HexNAc4 dHex1 Pent1 | 0.815 ± 0.035 |
| 74 | 1,933.639 | −0.075 | Hex5 HexNAc4 dHex2 | 0.038 ± 0.005 |
| 78 | 1,962.665 | −0.065 | Hex3 HexNAc3 dHex4 Pent2 | 0.008 ± 0.004 |
| 79 | 1,964.611 | −0.098 | Hex4 HexNAc3 dHex2 Pent3 | 0.012 ± 0.002 |
| 80 | 2,045.653 | −0.050 | Hex10 HexNAc2 | 0.005 ± 0.001 |
| 81 | 2,065.692 | −0.064 | Hex5 HexNAc4 dHex2 Pent1 | 0.045 ± 0.012 |
| 82 | 2,079.713 | −0.059 | Hex5 HexNAc4 dHex3 | 0.025 ± 0.000 |
| 83 | 2,081.756 | 0.003 | Hex6 HexNAc4 dHex1 Pent1 | 2.282 ± 0.006 |
| 85 | 2,211.765 | −0.050 | Hex5 HexNAc4 dHex3 Pent1 | 0.151 ± 0.027 |
Compositional annotations for the peaks detected in MALDI-TOF MS were achieved using KEGG Glycan and GlycoSuite online databases.
Hex, hexose; HexNAc, N-acetylhexosamine; dHex, deoxyhexose; Pent, pentose.
aPeak number indicated in Fig. 5.
bMass-to-charge ratio.
cDifference from the theoretical mass value.
dValues show the averages of duplicate experiments.
We also identified some paucimannosidic-type glycan chains (Lerouge et al. 1998, Bardor et al. 1999) such as Man3GlcNAc2Fuc1Xyl1 and Man2GlcNAc2Fuc1Xyl1 (peaks 20 and 12, respectively; Table 2; Fig. 5B) in the N-glycoforms of the chloroplastic NPP1. Lerouge et al. (1998) argued that paucimannosidic-type N-glycans resulting from the elimination of terminal residues from complex-type N-glycans in the post-Golgi compartments are typical plant vacuole type N-glycans. However, it was later shown that chloroplast and amyloplast proteomes contain exo- and endoglycosidases that can process the oligosaccharide chains of plastidial glycoproteins (Andon et al. 2002, Tanaka et al. 2004, Sun et al. 2009) (http://gene64.dna.affrc.go.jp/RPD/main_en.html; http://ppdb.tc.cornell.edu/subproteome.aspx). We thus infer that these paucimannosidic-type glycans might be produced from the complex-type glycans within the plastids. Furthermore, we detected many novel N-linked, deoxyhexose(s)-containing oligosaccharides in the plastidial NPP1 (peaks 22, 24, 34, 42, 47, 50, 52, 54, 57, 64, 66, 74, 81 and 82 in Table 2) that have not been previously registered in databases as plant N-glycans.
We performed structural evaluation of N-linked oligosaccharide chains of NPP1, NPP2 and NPP6, purified from the UNP1, UNP2 and UNP6 lines, respectively. Similar to the choloroplastic NPP1, the N-glycome of NPP1, NPP2 and NPP6 included complex-type and paucimannosidic-type glycans that contained no acidic sugars, such as sulfated, carboxylated and phosphorylated sugars (Tables 3–5; Fig. 5). The ratios of modified complex-type vs. total glycans of NPP1, NPP2 and NPP6 were estimated to be 60, 48 and 80%, respectively (Fig. 5C). In the glycoforms of NPP1, NPP2 and NPP6, Man3GlcNAc2Fuc1Xyl1 (peak 20, 26.73% of total glycans), Man5GlcNAc1 (peak 13, 22.08% of total glycans) and Man3GlcNAc4Fuc1Xyl1 (peak 51, 27.45% of total glycans) were the most abundant glycan chains, respectively (Tables 3–5). The content of the complex-type oligosaccharide chains conjugated to whole-cell NPP1 was slightly lower than that of the plastidial NPP1 (Fig. 5C). This can be ascribed to the fact that approximately 20% of NPP1 was distributed in extra-chloroplastic compartments including the ER network of the cells (cf. Fig. 2Q).
Table 3.
Glycoforms detected in rice NPP1
| Peak no.a | m/zb | Δc | Estimated composition | Relative contents (%)d |
|---|---|---|---|---|
| 2 | 708.259 | 0.003 | Hex3 HexNAc1 | 0.00 ± 0.00e |
| 3 | 733.201 | −0.086 | Hex1 HexNAc2 dHex1 | 0.00 ± 0.00e |
| 4 | 749.196 | −0.085 | Hex2 HexNAc2 | 0.00 ± 0.00e |
| 7 | 870.222 | −0.086 | Hex4 HexNAc1 | 0.02 ± 0.01 |
| 8 | 881.238 | −0.086 | Hex2 HexNAc2 Pent1 | 0.01 ± 0.00 |
| 10 | 911.249 | −0.085 | Hex3 HexNAc2 | 0.02 ± 0.01 |
| 11 | 952.276 | −0.085 | Hex2 HexNAc3 | 0.01 ± 0.02 |
| 12 | 1,027.296 | −0.086 | Hex2 HexNAc2 dHex1 Pent1 | 0.60 ± 0.21 |
| 13 | 1,032.275 | −0.085 | Hex5 HexNAc1 | 0.00 ± 0.00e |
| 14 | 1,043.291 | −0.086 | Hex3 HexNAc2 Pent1 | 4.91 ± 0.57 |
| 15 | 1,057.307 | −0.085 | Hex3 HexNAc2 dHex1 | 0.11 ± 0.00 |
| 16 | 1,073.302 | −0.085 | Hex4 HexNAc2 | 0.16 ± 0.01 |
| 17 | 1,084.318 | −0.288 | Hex2 HexNAc3 Pent1 | 0.00 ± 0.00e |
| 18 | 1,114.328 | −0.086 | Hex3 HexNAc3 | 0.01 ± 0.00 |
| 20 | 1,189.349 | −0.085 | Hex3 HexNAc2 dHex1 Pent1 | 26.73 ± 3.99 |
| 21 | 1,194.328 | −0.921 | Hex6 HexNAc1 | 0.42 ± 0.10 |
| 23 | 1,205.344 | −0.085 | Hex4 HexNAc2 Pent1 | 2.44 ± 0.37 |
| 25 | 1,230.376 | −0.922 | Hex2 HexNAc3 dHex1 Pent1 | 0.13 ± 0.09 |
| 27 | 1,235.355 | −0.085 | Hex5 HexNAc2 | 3.88 ± 1.16 |
| 28 | 1,246.371 | −0.085 | Hex3 HexNAc3 Pent1 | 1.11 ± 0.33 |
| 29 | 1,260.386 | −0.086 | Hex3 HexNAc3 dHex1 | 1.26 ± 0.54 |
| 32 | 1,356.381 | −0.922 | Hex7 HexNAc1 | 0.01 ± 0.01 |
| 33 | 1,367.397 | −0.085 | Hex5 HexNAc2 Pent1 | 0.58 ± 0.56 |
| 35 | 1,392.429 | −0.085 | Hex3 HexNAc3 dHex1 Pent1 | 5.33 ± 3.95 |
| 36 | 1,397.408 | −0.085 | Hex6 HexNAc2 | 10.62 ± 0.17 |
| 38 | 1,408.423 | −0.086 | Hex4 HexNAc3 Pent1 | 1.87 ± 0.27 |
| 39 | 1,422.439 | −0.085 | Hex4 HexNAc3 dHex1 | 0.02 ± 0.01 |
| 41 | 1,449.45 | −0.085 | Hex3 HexNAc4 Pent1 | 0.34 ± 0.24 |
| 43 | 1,513.455 | −0.085 | Hex5 HexNAc2 dHex1 Pent1 | 0.39 ± 0.31 |
| 44 | 1,518.434 | −0.922 | Hex8 HexNAc1 | 0.09 ± 0.14 |
| 45 | 1,554.481 | −0.086 | Hex4 HexNAc3 dHex1 Pent1 | 2.40 ± 1.83 |
| 46 | 1,559.46 | −0.085 | Hex7 HexNAc2 | 5.85 ± 0.79 |
| 48 | 1,570.476 | −0.086 | Hex5 HexNAc3 Pent1 | 0.26 ± 0.10 |
| 50 | 1,584.492 | −0.085 | Hex5 HexNAc3 dHex1 | 0.14 ± 0.21 |
| 51 | 1,595.508 | −0.085 | Hex3 HexNAc4 dHex1 Pent1 | 4.96 ± 3.78 |
| 52 | 1,609.524 | −0.085 | Hex3 HexNAc4 dHex2 | 0.05 ± 0.04 |
| 53 | 1,611.503 | −0.085 | Hex4 HexNAc4 Pent1 | 0.71 ± 0.18 |
| 58 | 1,700.539 | −0.086 | Hex4 HexNAc3 dHex2 Pent1 | 0.00 ± 0.00e |
| 59 | 1,716.534 | −0.085 | Hex5 HexNAc3 dHex1 Pent1 | 0.23 ± 0.05 |
| 60 | 1,721.513 | −0.085 | Hex8 HexNAc2 | 14.71 ± 3.20 |
| 62 | 1,732.529 | −0.085 | Hex6 HexNAc3 Pent1 | 0.02 ± 0.01 |
| 63 | 1,757.561 | −0.085 | Hex4 HexNAc4 dHex1 Pent1 | 4.75 ± 0.93 |
| 65 | 1,773.556 | −0.085 | Hex5 HexNAc4 Pent1 | 0.26 ± 0.07 |
| 66 | 1,787.571 | −0.086 | Hex5 HexNAc4 dHex1 | 0.00 ± 0.00e |
| 69 | 1,862.592 | −0.085 | Hex5 HexNAc3 dHex2 Pent1 | 0.00 ± 0.00e |
| 70 | 1,878.587 | −0.085 | Hex6 HexNAc3 dHex1 Pent1 | 0.01 ± 0.00 |
| 71 | 1,883.566 | −0.085 | Hex9 HexNAc2 | 4.03 ± 0.89 |
| 72 | 1,903.619 | −0.085 | Hex4 HexNAc4 dHex2 Pent1 | 0.35 ± 0.10 |
| 73 | 1,919.614 | −0.085 | Hex5 HexNAc4 dHex1 Pent1 | 0.00 ± 0.00e |
| 74 | 1,933.629 | −0.085 | Hex5 HexNAc4 dHex2 | 0.02 ± 0.02 |
| 80 | 2,045.619 | −0.085 | Hex10 HexNAc2 | 0.03 ± 0.02 |
| 81 | 2,065.672 | −0.085 | Hex5 HexNAc4 dHex2 Pent1 | 0.06 ± 0.05 |
| 82 | 2,079.687 | −0.085 | Hex5 HexNAc4 dHex3 | 0.06 ± 0.09 |
| 84 | 2,179.714 | −0.086 | Hex5 HexNAc6 Pent1 | 0.00 ± 0.00e |
| 85 | 2,211.729 | −0.086 | Hex5 HexNAc4 dHex3 Pent1 | 0.00 ± 0.00e |
Compositional annotations for the peaks detected in MALDI-TOF MS were achieved using KEGG Glycan and GlycoSuite online databases.
Hex, hexose; HexNAc, N-acetylhexosamine; dHex, deoxyhexose; Pent, pentose.
aPeak number indicated in Fig. 5.
bMass-to-charge ratio.
cDifference from the theoretical mass value.
dValues are represented as mean ± SD (n = 3).
eGlycan peaks are detected in this analysis.
Table 4.
Glycoforms detected in rice NPP2
| Peak no.a | m/zb | Δc | Estimated composition | Relative contents (%)d |
|---|---|---|---|---|
| 2 | 707.6797 | −0.575 | Hex3 HexNAc1 | 0.00 ± 0.01 |
| 7 | 869.7067 | −0.601 | Hex4 HexNAc1 | 1.21 ± 0.15 |
| 8 | 880.7287 | −0.595 | Hex2 HexNAc2 Pent1 | 0.00 ± 0.00e |
| 9 | 894.7418 | −0.597 | Hex2 HexNAc2 dHex1 | 0.00 ± 0.00e |
| 10 | 910.7386 | −0.595 | Hex3 HexNAc2 | 0.05 ± 0.01 |
| 12 | 1,026.7538 | −0.628 | Hex2 HexNAc2 dHex1 Pent1 | 0.00 ± 0.01 |
| 13 | 1,032.3988 | 0.038 | Hex5 HexNAc1 | 22.08 ± 1.02 |
| 14 | 1,042.7447 | −0.632 | Hex3 HexNAc2 Pent1 | 1.22 ± 0.22 |
| 15 | 1,056.7657 | −0.626 | Hex3 HexNAc2 dHex1 | 0.30 ± 0.04 |
| 16 | 1,072.7539 | −0.633 | Hex4 HexNAc2 | 0.23 ± 0.05 |
| 18 | 1,113.7991 | −0.615 | Hex3 HexNAc3 | 0.01 ± 0.00 |
| 20 | 1,188.7929 | −0.641 | Hex3 HexNAc2 dHex1 Pent1 | 18.84 ± 0.16 |
| 23 | 1,204.7875 | −0.641 | Hex4 HexNAc2 Pent1 | 0.62 ± 0.05 |
| 27 | 1,234.7847 | −0.655 | Hex5 HexNAc2 | 3.83 ± 0.24 |
| 28 | 1,244.8355 | −1.620 | Hex3 HexNAc3 Pent1 | 0.55 ± 0.06 |
| 29 | 1,260.8385 | 0.366 | Hex3 HexNAc3 dHex1 | 0.01 ± 0.00 |
| 30 | 1,275.8167 | −0.649 | Hex4 HexNAc3 | 0.03 ± 0.01 |
| 31 | 1,350.8126 | −0.674 | Hex4 HexNAc2 dHex1 Pent1 | 0.20 ± 0.01 |
| 33 | 1,366.7937 | −0.688 | Hex5 HexNAc2 Pent1 | 0.24 ± 0.02 |
| 35 | 1,391.8347 | −0.679 | Hex3 HexNAc3 dHex1 Pent1 | 5.97 ± 0.24 |
| 36 | 1,396.8037 | −0.696 | Hex6 HexNAc2 | 10.88 ± 0.02 |
| 38 | 1,407.8360 | −0.673 | Hex4 HexNAc3 Pent1 | 0.96 ± 0.04 |
| 40 | 1,437.8385 | −0.680 | Hex5 HexNAc3 | 0.19 ± 0.02 |
| 41 | 1,447.9069 | −1.628 | Hex3 HexNAc4 Pent1 | 0.33 ± 0.01 |
| 43 | 1,512.8520 | −0.688 | Hex5 HexNAc2 dHex1 Pent1 | 0.32 ± 0.01 |
| 45 | 1,553.8680 | −0.699 | Hex4 HexNAc3 dHex1 Pent1 | 3.11 ± 0.16 |
| 46 | 1,558.8304 | −0.714 | Hex7 HexNAc2 | 6.75 ± 0.06 |
| 48 | 1,569.8841 | −0.678 | Hex5 HexNAc3 Pent1 | 0.32 ± 0.02 |
| 51 | 1,594.8909 | −0.702 | Hex3 HexNAc4 dHex1 Pent1 | 6.67 ± 0.42 |
| 53 | 1,609.9986 | −1.593 | Hex4 HexNAc4 Pent1 | 0.36 ± 0.01 |
| 55 | 1,650.9748 | −1.640 | Hex3 HexNAc5 Pent1 | 0.00 ± 0.00e |
| 60 | 1,720.8731 | −0.725 | Hex8 HexNAc2 | 4.82 ± 0.70 |
| 63 | 1,756.9414 | −0.704 | Hex4 HexNAc4 dHex1 Pent1 | 4.43 ± 0.30 |
| 67 | 1,813.0323 | 0.343 | Hex3 HexNAc5 dHex2 | 0.00 ± 0.00e |
| 71 | 1,882.9253 | −0.725 | Hex9 HexNAc2 | 1.75 ± 0.03 |
| 72 | 1,903.0037 | −0.700 | Hex4 HexNAc4 dHex2 Pent1 | 1.08 ± 0.02 |
| 73 | 1,918.9961 | −0.703 | Hex5 HexNAc4 dHex1 Pent1 | 1.92 ± 0.13 |
| 81 | 2,065.0560 | −0.701 | Hex5 HexNAc4 dHex2 Pent1 | 0.65 ± 0.00 |
| 85 | 2,211.1471 | −0.668 | Hex5 HexNAc4 dHex3 Pent1 | 0.07 ± 0.16 |
Compositional annotations for the peaks detected in MALDI-TOF MS were achieved using KEGG Glycan and GlycoSuite online databases.
Hex, hexose; HexNAc, N-acetylhexosamine; dHex, deoxyhexose; Pent, pentose.
aPeak number indicated in Fig. 5.
bMass-to-charge ratio.
cDifference from the theoretical mass value.
dValues are represented as mean ± SD (n = 3).
eGlycan peaks are detected in this analysis.
Table 5.
Glycoforms detected in rice NPP6
| Peak no.a | m/zb | Δc | Estimated composition | Relative contents (%)d |
|---|---|---|---|---|
| 8 | 880.7558 | −0.568 | Hex2 HexNAc2 Pent1 | 0.00 ± 0.00e |
| 9 | 894.7836 | −0.555 | Hex2 HexNAc2 dHex1 | 0.00 ± 0.00e |
| 10 | 910.753 | −0.581 | Hex3 HexNAc2 | 0.00 ± 0.00e |
| 12 | 1,026.8222 | −0.560 | Hex2 HexNAc2 dHex1 Pent1 | 0.02 ± 0.00 |
| 14 | 1,042.805 | −0.572 | Hex3 HexNAc2 Pent1 | 1.05 ± 0.16 |
| 15 | 1,056.806 | −0.586 | Hex3 HexNAc2 dHex1 | 0.00 ± 0.00e |
| 16 | 1,072.7896 | −0.601 | Hex4 HexNAc2 | 0.02 ± 0.01 |
| 20 | 1,188.8735 | −0.560 | Hex3 HexNAc2 dHex1 Pent1 | 11.66 ± 0.75 |
| 23 | 1,204.8407 | −0.588 | Hex4 HexNAc2 Pent1 | 0.43 ± 0.02 |
| 26 | 1,230.8615 | 0.397 | Hex2 HexNAc3 Pent1 dHex1 | 0.04 ± 0.01 |
| 27 | 1,234.8493 | −0.590 | Hex5 HexNAc2 | 1.16 ± 0.19 |
| 28 | 1,244.9241 | −1.532 | Hex3 HexNAc3 Pent1 | 1.30 ± 0.22 |
| 29 | 1,261.8621 | 1.389 | Hex3 HexNAc3 dHex1 | 0.01 ± 0.00 |
| 31 | 1,350.8699 | −0.617 | Hex4 HexNAc2 dHex1 Pent1 | 0.24 ± 0.04 |
| 33 | 1,366.8419 | −0.640 | Hex5 HexNAc2 Pent1 | 0.03 ± 0.01 |
| 35 | 1,391.9241 | −0.590 | Hex3 HexNAc3 dHex1 Pent1 | 9.42 ± 0.32 |
| 36 | 1,396.8932 | −0.600 | Hex6 HexNAc2 | 11.85 ± 0.72 |
| 38 | 1,407.9079 | −0.601 | Hex4 HexNAc3 Pent1 | 1.35 ± 0.17 |
| 40 | 1,438.9019 | 0.382 | Hex5 HexNAc3 | 0.10 ± 0.02 |
| 41 | 1,447.9816 | −1.553 | Hex3 HexNAc4 Pent1 | 0.82 ± 0.17 |
| 43 | 1,512.9161 | −0.624 | Hex5 HexNAc2 dHex1 Pent1 | 0.01 ± 0.00 |
| 45 | 1,553.9508 | −0.616 | Hex4 HexNAc3 dHex1 Pent1 | 2.42 ± 0.12 |
| 46 | 1,558.9192 | −0.626 | Hex7 HexNAc2 | 5.12 ± 0.42 |
| 48 | 1,569.939 | −0.623 | Hex5 HexNAc3 Pent1 | 0.19 ± 0.05 |
| 51 | 1,594.9819 | −0.611 | Hex3 HexNAc4 dHex1 Pent1 | 27.45 ± 1.32 |
| 53 | 1,610.0227 | −1.569 | Hex4 HexNAc4 Pent1 | 1.13 ± 0.06 |
| 55 | 1,651.0562 | −1.559 | Hex3 HexNAc5 Pent1 | 0.00 ± 0.00e |
| 58 | 1,699.9968 | −0.628 | Hex4 HexNAc3 dHex2 Pent1 | 1.10 ± 0.10 |
| 59 | 1,715.9949 | −0.624 | Hex5 HexNAc3 dHex1 Pent1 | 0.27 ± 0.05 |
| 60 | 1,720.9711 | −0.627 | Hex8 HexNAc2 | 1.91 ± 0.23 |
| 63 | 1,757.0222 | −0.624 | Hex4 HexNAc4 dHex1 Pent1 | 3.41 ± 0.21 |
| 65 | 1,773.0269 | −0.614 | Hex5 HexNAc4 Pent1 | 0.08 ± 0.02 |
| 67 | 1,813.0896 | 0.401 | Hex3 HexNAc3 dHex2 | 0.00 ± 0.00e |
| 69 | 1,862.0539 | −0.623 | Hex5 HexNAc3 dHex2 Pent1 | 0.05 ± 0.01 |
| 71 | 1,883.0133 | −0.638 | Hex9 HexNAc2 | 0.15 ± 0.05 |
| 72 | 1,903.0693 | −0.635 | Hex4 HexNAc4 dHex2 Pent1 | 12.48 ± 0.67 |
| 73 | 1,919.0714 | −0.627 | Hex5 HexNAc4 dHex1 Pent1 | 0.83 ± 0.10 |
| 75 | 1,935.1201 | −0.574 | Hex6 HexNAc4 Pent1 | 0.02 ± 0.02 |
| 76 | 1,945.1061 | 0.374 | Hex3 HexNAc5 dHex2 Pent1 | 0.10 ± 0.03 |
| 77 | 1,959.1654 | 0.419 | Hex3 HexNAc5 dHex3 | 0.00 ± 0.00e |
| 81 | 2,065.1188 | −0.638 | Hex5 HexNAc4 dHex2 Pent1 | 1.26 ± 0.19 |
| 85 | 2,211.1646 | −0.650 | Hex5 HexNAc4 dHex3 Pent1 | 2.54 ± 0.30 |
Compositional annotations for the peaks detected in MALDI-TOF MS were achieved using KEGG Glycan and GlycoSuite online databases.
Hex, hexose; HexNAc, N-acetylhexosamine; dHex, deoxyhexose; Pent, pentose.
aPeak number indicated in Fig. 5.
bMass-to-charge ratio.
cDifference from the theoretical mass value.
dValues are represented as mean ± SD (n = 3).
eGlycan peaks are detected in this analysis.
Some oligosaccharides without a chitobiose core (GlcNAc-GlcNAc) were detected in the NPP family glycoproteins, particularly in NPP2. This type of oligosaccharide chain was also observed in mammalian N-glycans prepared by PNGase F, and their contents were shown to be changed in different cell types (Amano et al. 2010). It seems that glycopeptidase A with a faint contaminant of endo-®β-N-acetylglucosaminidase further cleaved the chitobiose core of the oligosaccharide chain.
A measurable amount of high mannose-type glycan chains was detected in the plastidial NPPs (Table 2, Fig. 5). We consider three possible interpretations for this result.
Significant contamination by microsomal membranes including the ER occurs during chloroplast preparation. However, as shown in Supplementary Fig. S6, the chloroplast preparations were virtually free from contamination by microsomal membrane glycoproteins recognized by Concanavalin A (Con A).
A protein traffic pathway exists from the ER to the plastid. In this respect we must emphasize that, similar to previous reports showing a physical association between ER tubules and chloroplasts (Andersson et al. 2007, Schattat et al. 2011), we also observed that the ER tubules cling tightly to the plastids of onion epidermal cells (cf. fig. 3D in Kitajima et al. 2009). It is thus tempting to speculate that a translocator complex for importing NPP glycoproteins from the ER lumen to the chloroplast stroma occurs in the ER–chloroplast interacting surfaces.
Incomplete maturation of glycans occurs in the late compartment of the Golgi apparatus. In this respect, it is worth noting that rice Amyl-1 possesses only one N-glycosylation site (Mitsui and Itoh 1997, Ochiai et al. 2014) with high mannose-type that accounts for 37.3% of total glycans conjugated to Amyl-1 (Hayashi et al. 1990), suggesting incomplete maturation of N-glycans along the secretory pathway.
Additional and concluding remarks
Various glycoproteins including Arabidopsis CAH1 (Burén et al. 2011), Oryza sativa Amyl-1 (Kitajima et al. 2009) and NPP1 (Nanjo et al. 2006) have been reported to occur in the plastids of higher plant cells. Immunodetection with anti-⟨1,3-fucose and anti-β1,2-xylose antibodies, and analyses of glycan susceptibility to endoglycosidase H (which specifically digests high mannose-type oligosaccharide chains) strongly suggested that chloroplastic glycoproteins bear complex-type glycan chains (Villarejo et al. 2005). However, the precise N-glycome in the plastidial glycoproteins of plant cells remained to be clarified. In the present study, we characterized the N-glycome of rice plastidial NPP1, NPP2 and NPP6 by employing rapid, sensitive and high-throughput ‘glycoblotting’ for N-glycan purification/derivatization, followed by mass spectrometry (Nishimura et al. 2005, Furukawa et al. 2008, Amano et al. 2010). We found that the N-glycoforms in NPP1, NPP2 and NPP6 contain at least 78 chains, including deoxyhexose(s)-containing oligosaccharides that have hitherto not been deposited in databases as plant N-glycans (Tables 2–5). Importantly, the complex-type and paucimannosidic-type glycans with fucose and xylose residues occupied approximately 80% of total glycans in plastidial NPP (Fig. 5; Table 2), strongly suggesting that the trans-Golgi to plastid traffic pathway is the main route for plastid targeting of NPP in rice cells.
Unlike NPP1, NPP2 and NPP6, NPP3 is largely distributed in extra-chloroplastic compartments, probably in the endomembrane system of rice cells (Kaneko et al. 2011). The degree of amino acid sequence similarity between NPP1 and NPP2, NPP3 and NPP6 is 62.2, 61.1 and 71.6%, respectively (Fig. 1). The examination of plastid localization of a series of C-terminally truncated NPP1 molecules fused with GFP suggested that the peptide region from amino acid residues 308 to 478 is important for targeting of NPP1 into plastids in rice cells (Kaneko et al. 2011). Amino acid sequence alignment between the region from 308 to 478 of NPP1 and the corresponding region of NPP2, NPP3 and NPP6 revealed that the negatively charged residue Asp336 in NPP1 (Asp329 in NPP2, Asp342 in NPP6) is substituted for the positively charged residue Lys328 in NPP3, and the hydrophobic residue Ala416 in NPP1 (Ala409 in NPP2, Ala422 in NPP6) is substituted for the negatively charged residue Glu408 in NPP3 (Fig. 1). Therefore, it is highly likely that these amino acid residues are major determinants of the subcellular localization of NPPs.
The structure of N-linked glycans in mature glycoproteins varies among glycans bound to proteins, and among cell types, tissues and species (Kornfeld and Kornfeld 1985, Helenius and Aebi 2001, Hirose et al. 2011). In mammalian cells, variation in the cellular N-glycome allows the modulation of dynamic cellular mechanisms such as cell–cell adhesion, cell activation and malignant alterations (Varki 1993, Haltiwanger et al. 2004, Guérardel et al. 2006, Ohtsubo and Marth 2006). It should be stressed that the different patterns of N-glycosylation of the external proteins of human and simian immunodeficiency viruses, influenza virus and hepatitis C viruses are crucially important for viral infectivity, antigen conformation and the immune response (Zhang et al. 2004). The conjugation of N-glycans to plant glycoprotein is certainly necessary for maintaining a fully functional protein conformation (Helenius and Aebi 2001, Burén et al. 2011). Nevertheless, the physiological importance of glycan modification of glycoproteins in plants remains obscure. Defects in N-glycosylation modification exhibit no visible phenotypes in plants growing under normal conditions (von Schaewen et al. 1993, Strasser et al. 2004, Strasser et al. 2006, Strasser et al. 2007). On the other hand, it has been reported that plant N-glycans have biological significance beyond facilitating protein folding in the ER, and that phenotypes of mutants defective in N-glycosylation and N-glycan maturation are extremely sensitive to environmental conditions (Kang et al. 2008). The highly diverse sialic acid-containing glycans are key determinants in a range of molecular recognition events (Schauer 2009); however, the presence of negatively charged sugar residues has hitherto not been reported in plant glycoproteins. ⟨(1,3)-Fucose linked to the proximal N-acetylglucosamine of the core oligosaccharide is uniquely expressed in plant glycoproteins (Takahashi et al. 1986, Hayashi et al. 1990 Ogawa et al. 1996, Melo et al. 1997, Fitchette-Lainé et al. 1997, Lerouge et al. 1998, Olczak and Watorek 2000); therefore, the variation in the ⟨(1,3)-fucosylation pattern should be plant specific. The present N-glycome analyses revealed differential fucosylation of glycan chains of rice NPP1, NPP2 and NPP6: the contents of the oligosaccharide chains with fucose residues in NPP1, NPP2 and NPP6 were calculated to be 47.6–62.3, 43.6 and 73.3%, respectively (Tables 3–5). It is possible that variations in fucosylation reflect the different protein structures and functions of NPPs, though more investigations will be necessary to confirm (or refute) this hypothesis.
Materials and Methods
Plant materials
The rice variety used in this study was Oryza sativa L. cv. Nipponbare. To obtain cultured cells, rice seeds were sterilized in a 1% sodium hypochloride solution for 15 min and rinsed well in sterile water. Rice calli derived from the embryo portions of seeds were cultured according to the procedure of Kaneko et al. (2014).
Plasmid constructs
Plasmids used in this study and references describing how they were constructed are listed in Supplementary Table S1. Complete rice NPP cDNAs were cloned from rice shoot cDNA libraries as described previously (Nanjo et al. 2006, Kaneko et al. 2014). Complete NPP1, NPP2 and NPP6 cDNAs were cloned into pBluescript SK(–) (Stratagene) to create pOsNPP1, pOsNPP2 and pOsNPP6, respectively. To produce NPP1-, NPP2- and NPP6-overexpressing rice cultured cells, BamHI–KpnI PCR fragments were amplified from pOsNPP1, pOsNPP2 and pOsNPP6, and cloned into the BamHI and KpnI sites of the p2K-1+ plant expression vector (Christensen et al. 1992, Miki et al. 2004, Nanjo et al. 2006), resulting in p2K-Ubi-OsNPP1, p2K-Ubi-OsNPP2 and p2K-Ubi-OsNPP6 used for genetic transformation. For NPP subcellular localization studies, we PCR-amplified NPP1, NPP2 and NPP6 from pOsNPP1, pOsNPP2 and pOsNPP6 with specific primer sets and then digested the PCR product with BamHI. The resulting fragments were then ligated to BamHI-digested pZH2B-35S-GFP (Hajdukiewicz et al. 1994, Asatsuma et al. 2005) to create the pZH2B-35S-OsNPP1-GFP, pZH2B-35S-OsNPP2-GFP and pZH2B-35S-OsNPP6-GFP expression vectors. For labeling and visualizing the trans-Golgi in rice cells, the BamHI-digested fragment of pST-mRFP (Matsuura-Tokita et al. 2006) was inserted into the BamHI site as pGFP to produce pST-GFP, and then the HindIII- and SacI-digested fragment of pST-GFP was inserted into the same site as pZH2B to create pZH2B-35S-ST-GFP.
Genetic transformation
The above expression vectors were incorporated into competent cells of Agrobacterium tumefaciens strain EHA101 (Hood et al. 1986). Subsequent Agrobacterium-mediated transformation and regeneration of rice plants were performed as described by Hiei et al. (1994). Cultured rice cells were grown in hygromycin selective medium for 2 weeks and then transferred to a redifferentiative medium for 1 month. The following transgenic rice lines were established: UNP1, UNP2 and UNP6 transformed with p2K-Ubi-OsNPP1, p2K-Ubi-OsNPP2 and p2K-Ubi-OsNPP6, respectively; SNP1–GFP, SNP2–GFP and SNP6–GFP transformed with pZH2B-35S-OsNPP1-GFP, pZH2B-35S-OsNPP2-GFP and pZH2B-35S-OsNPP6-GFP, respectively; and ST–GFP transformed with pZH2B-35S-ST-GFP. In order to generate a transgenic rice line (UNP1/ST-GFP) expressing ST–GFP with a constitutively high expression of NPP1, the cells derived from UNP1 seeds were transformed with pZH2B-35S-ST-GFP, and subjected to the redifferentiation processes.
To introduce plasmid DNA into onion epidermal cells, the particle bombardment method was adopted using a helium-driven particle accelerator (PDS-1000/He; Bio-Rad) with all basic adjustments set according to the manufacturer’s recommendations. A 3 μg aliquot of plasmid DNA [pGFP, pWxTP-DsRed, pZH2B-35S-OsNPP2-GFP and -OsNPP6-GFP, and pMT121-ARF1 Q71L (Takeuchi et al. 2002)] in 10 μl of pure water (18.2 MΩ cm–1 at 25°C) were mixed with 10 μl of a 60 mg ml–1 gold particle (diameter 1.0 μm) solution, 10 μl of 2.5 mM CaCl2 and 4 μl of 0.1 M spermidine, and incubated for 30 min at room temperature. Gold particles coated with plasmid DNA were rinsed with cold ethanol and then gently suspended in 10 μl of ethanol. The gold particles were bombarded twice into onion cells using a particle delivery system with 1,100 p.s.i. rupture discs. The bombarded onion epidermal cells were cultured without or with 90 μM BFA at 25°C in darkness.
Confocal laser scanning microscopy
A confocal laser scanning microscope (FV1000- and FV300-BX-61; Olympus) was used for imaging GFP, DsRed and Chl autofluorescence in rice and onion cells (Kitajima et al. 2009). For quantitative analysis, the fluorescence intensity in plastids and in whole cells was determined using Lumina Vision imaging software (Mitani Corporation). The background was always set at the maximum value of fluorescence intensity for the area in which no structural image was present. The area visualized with either Chl autofluorescence or WxTP–DsRed was defined as the plastidial area. For analysis of the whole rice cell, each individual image from the top to the bottom of the cell, 10–15 frames every 0.7–1.4 μm, was evaluated. In the case of onion cells, each image from the top to the middle of the cell, 20–30 frames every 1.5–2 μm, was evaluated. To evaluate the plastid-targeting abilities of GFP-labeled proteins, we determined the ratio of the fluorescent intensity of GFP in the plastidial area to that in the whole cell (GFPplastid/GFPtotal).
Electron microscopy and immunocytochemical analyses
Transgenic rice cells expressing ST–GFP without or with a constitutively high expression of NPP1 cultured in Murashige and Skoog (MS) medium containing 3% sucrose at 28°C in darkness were immediately placed on a flat specimen carrier and frozen in a high-pressure freezer (EM-PACT; Leica Microsystems). The frozen samples were fixed in anhydrous acetone containing 2% osmic acid (OsO4) for 3–4 d at –80°C for morphological observation or fixed with anhydrous acetone containing 1% glutaraldehyde for 3–4 d at –80°C for immunocytochemistry. Tubes containing the frozen samples were warmed at 3°C h–1 to a temperature of –20°C, and at 1°C h–1 from –20°C to 4°C, and kept for 2 h at 4°C using an automatic freeze substitution system (EM-AFS; Leica Microsystems). The samples were then washed with 100% acetone and embedded in epoxy resin Epon 812 (Shell Chemicals). The fixed samples for immunocytochemistry were embedded in LR White resin (London Resin). Ultrathin sections for morphological and immunocytochemical observations were mounted on 300 mesh and 200 mesh Ni grids, respectively. Immunocytochemical detection of ST–GFP was performed according to Toyooka et al. (2006, 2009). Sections on Ni grids were treated with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 30 min at room temperature. They were then incubated with an affinity-purified polyclonal antibody toward GFP (Abcam) in PBS (1 : 1,000). After washing with PBS, the sections were further incubated with 10 nm colloidal gold particles conjugated to Protein A (Sigma), washed with PBS and rinsed in distilled water. Sections for morphological observations were finally stained with 5% aqueous uranyl acetate for 15 min, while sections for immunochemistry were stained with 5% aqueous uranyl acetate for 10 min and 5% lead citrate for 5 min sequentially, and examined using a transmission electron microscope (H-7650; Hitachi) at 80 kV. Images were acquired using a Gatan DualView camera and Digital Micrograph software or transmission electron microscopy films.
Isolation of chloroplasts
UNP1 seeds were germinated for 2 weeks at 28°C in the dark, and then further incubated for 3 d at 28°C in the light (20,000 lux). UNP1 leaves were homogenized with an equal volume of an isolation buffer consisting of 50 mM HEPES-KOH pH 7.5, 0.33 M sorbitol, 5 mM MgCl2, 5 mM MnCl2, 5 mM EDTA and 50 mM sodium ascorbate. The shoot homogenates were passed through four layers of gauze and then four layers of Miracloth (Merck). The filtrate was layered onto an 80% (v/v) Percoll (Sigma) cushion containing 50 mM HEPES-KOH pH 7.5, 0.33 M sorbitol, 5 mM MgCl2, 5 mM MnCl2 and 5 mM EDTA, and centrifuged at 2,000×g for 4 min at 4°C. The crude chloroplasts on the Percoll surface were collected, and diluted with more than twice the volume of the isolation buffer and then layered onto a discontinuous density gradient consisting of 40% and 80% Percoll solutions. The gradient was centrifuged at 4,000×g for 10 min at 4°C. Intact chloroplasts enriched around the 40/80% Percoll interface were collected, and Percoll gradient centrifugation was carried out again. Intact chloroplasts were collected, diluted with five times the volume of the isolation buffer, and centrifuged at 2,000×g for 4 min at 4°C, followed by protein extraction. The shoot extracts were also subjected to differential centrifugation at 8,000×g for 10 min, at 20,000×g for 10 min and at 100,000×g for 1 h using a himac CP80MX and P70AT rotor type (Hitachi), respectively. All of the above steps were carried out at 4°C.
Immuno- and lectin blots
An aliquot of a sample was subjected to SDS–PAGE, followed by the electrical transfer of proteins separated in SDS–polyarcrylamide gels to a polyvinylidene difluoride (PVDF) membrane (Amersham™ Hybond™, GE Healthcare). In immunoblots, antisera were diluted to 1 : 1,000–1 : 5,000 and peroxidase-conjugated anti-rabbit IgG to 1 μg ml–1, respectively. The blotted PVDF sheets were soaked with 1% (w/v) skim milk in PBST (PBS with 0.05% Tween-20) for 12 h at 4°C, washed with PBST for 15 min three times, and incubated with the primary antibody in PBST for 1 h at room temperature. The sheets were then incubated with the second antibody for 1 h at room temperature in the same way as for the primary antibody. The reacted protein bands were visualized with Amersham ECL prime (GE Healthcare). Anti-NPP1 (Nanjo et al. 2006), anti-RbcL (chloroplast marker) (Nishimura and Akazawa 1974) and anti-UGP (cytosol marker) (Kimura et al. 1992) antibodies were prepared as described previously. Anti-Arf1 (Golgi marker), anti-v-ATPase (tonoplast membrane marker) and anti-COX2 (mitochondria marker) were purchased from Funakoshi Co. For lectin blots, the blotted nitrocellulose sheets were soaked with PBST for 15 min three times and incubated with 0.4 μg ml–1 Con A–peroxidase in PBST for 12 h at 4°C. The reacted protein bands were visualized with Amersham ECL prime.
NPP activity assay
NPP activity was assayed as described in Rodríguez-López et al. (2000) and Kaneko et al. (2014). Protein content was measured using the Bradford method, employing Bio-Rad prepared reagent (Bio-Rad). One unit (U) is defined as the amount of enzyme that catalyzes the production of 1 μmol of product per min.
Purification of NPPs
Chloroplastic NPP1 was purified from chloroplasts isolated from UNP1 leaves. A 500 g aliquot of rice leaves was homogenized with an equal volume of 50 mM HEPES-KOH (pH 7.5), 0.3 M mannitol, 1 mM MgCl2 and 2 mM EDTA, and subjected to chloroplast separation employing Percoll density gradient centrifugation. The chloroplast-enriched fraction (10 ml) was resuspended in 5 vols. of 10 mM Tris–HCl (pH 8.8) and 1% (v/v) Triton X-100, and centrifuged at 20,000×g for 10 min. The resulting supernatants were applied to a Con A–Sepharose 4B column (ø 1.0×8 cm, Pharmacia) equilibrated with 40 mM Tris–HCl (pH 7.4), 0.5 M NaCl, 1 mM MnCl2 and 1 mM CaCl2, and eluted with 25 ml of 0.5 M ⟨-methyl-d-mannopyranoside in 10 mM Tris–HCl (pH 7.4). Finally, the enzyme preparation was desalted by ultrafiltration on a Microcon YM-100 (Amicon). Purification procedures for NPP1, NPP2 and NPP6 expressed in the transgenic rice cells (UNP1, UNP2 and UNP6, respectively) were described previously (Kaneko et al. 2014).
Amino acid sequencing and mass spectrometric analyses
NPP protein bands separated by SDS–PAGE were blotted onto a PVDFmembrane (Hybond-P, Amersham), excised and subjected to N-terminal sequencing analysis with a Shimadzu PPSQ-21 amino acid sequencer (Nanjo et al. 2006).
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) for protein identification was performed on an DiNA-A (KYA Tech), interfaced to a LTQ Orbitrap XL (Thermo Fisher Scientific). Ionization voltage was set to 1.8 kV and the capillary transfer temperature was set at 200°C. A 5 μl aliquot (∼1 μg) of in-gel digested tryptic peptide solution was loaded onto a trapping column packed with HiQ sil C18, 0.5×1 mm (KYA Tech) at a flow rate of 10 μl min–1 in 2% (v/v) acetonitrile containing 0.1% formic acid. Peptides were eluted into HiQ sil C18W-3 (0.1×50 mm, KYA Tech) at a flow rate of 300 nl min–1. Peptides were separated using a mobile phase gradient of solvent A and B: 0–50% solvent B in 25 min, 50–100% solvent B in 30 min, 100 % solvent B for 40 min, and 0% solvent B for 45 min. Solvent A was 2% acetonitrile containing 0.1% formic acid; solvent B was 80% acetonitrile containing 0.1% formic acid. LC-MS/MS data were acquired in data-dependent acquisition (DDA) mode controlled by Xcalibur 2.0 software (Thermo Fisher Scientific). A typical DDA cycle consisted of an MS scan within m/z 350–1,600 performed under the target mass resolution of 60,000 followed by MS/MS fragmentation of the three most intense precursor ions under the normalized collision energy of 35% in the linear trap. Singly charged ions were excluded from MS/MS experiments, and m/z values of fragmented precursor ions were dynamically excluded for a further 60 s. Protein identification was carried out by Proteome Discoverer version 1.3 and SEQUEST search engine (Thermo Fisher Scientific) using the O. sativa database (63,539 entries) of Uniprot (http://www.uniprot.org). The identification of peptide was performed with the following parameters: enzyme, trypsin; missed cleavages, 2; MS tolerance, 10 p.p.m.; MS/MS tolerance, 0.8 Da; static modification, carbamidomethylation; dynamic modification, oxidation (H, M, W). False discovery rates for peptide identification were <5.0%.
N-Glycomics of NPPs
Analysis of N-glycans of NPPs was performed employing the glycoblotting procedure described in earlier studies (Nishimura et al. 2005, Amano et al. 2010). Release of N-glycans from each preparation of NPP isozyme protein was carried out as follows. The protein samples were added to cold acetone (1 : 4) to precipitate proteinaceous materials, and the precipitates were collected by centrifuging at 12,000×g for 15 min at 4°C, followed by serial washing with acetonitrile. The resulting precipitates were dissolved in 50 μl of 80 mM ammonium bicarbonate containing 0.2% 1-propanesulfonic acid 2-hydroxyl-3-myristamide and incubated at 60°C for 10 min. The solubilized proteinaceous materials were reduced by 10 mM 1,4-dithiothreitol at 60°C for 30 min, followed by alkylation with 20 mM iodoacetamide by incubation in the dark at room temperature for 30 min. The mixture was then treated with 400 U of proteinase K (Sigma-Aldrich) at 37°C overnight, followed by heat inactivation of the enzyme at 90°C for 10 min. After cooling to room temperature, N-glycans of glycopeptides were released from the digested samples by incubation with 250 μU of glycopeptidase A (Seikagaku Corp.) at 37°C overnight. The sample mixture was then dried using a SpeedVac and stored at –20°C until use.
Glycoblotting of the sample mixtures containing N-glycans by means of BlotGlyco H beads was performed according to the procedure described previously (Furukawa et al. 2008). Aliquots of BlotGlyco H beads (500 μl) (10 mg ml–1 suspension, Sumitomo Bakelite Co.) were placed in the wells of a MultiScreen Solvinert filter plate (Millipore). Glycopeptidase A-digested samples were dissolved in 20 μl of water and applied to the well, followed by the addition of 180 μl of 2% (v/v) acetic acid in acetonitrile. The plate was incubated at 80°C for 45 min to capture total glycans in sample mixtures specifically onto beads via stable hydrazone bonds. The plate was washed with 200 μl of 2 M guanidine-HCl in ammonium bicarbonate, followed by washing with the same volume of water and 1% triethylamine in methanol. Each washing step was performed twice. Unreacted hydrazide functional groups on beads were capped by incubation with 10% acetic anhydride in methanol for 30 min at room temperature. The solution was then removed by vacuum and the beads serially washed twice with 200 μl of 10 mM HCl, methanol and dioxane, respectively. On-bead methyl esterification of carboxyl groups in sialic acids was carried out by incubation to dryness with 150 mM 3-methyl-1-p-tolyltriazene in dioxane at 60°C. This took 90 min in a conventional oven. The beads were then serially washed in 200 μl of dioxane, water, methanol and water. The glycans blotted on the beads were subjected to the transiminization reaction with N⟨-[(aminooxy)acetyl]tryptophanylarginine methyl ester (aoWR) (Nishimura et al. 2005, Furukawa et al. 2008) for 45 min at 80°C. WR-tagged glycans were eluted by adding 100 μl of water, and then purified using a Mass PREP™ HILIC mElution Plate (Waters) according to the manufacturer’s instructions. The purified N-glycans were concentrated 10-fold by SpeedVac, followed by direct dissolution in 2,5-dihydroxybenzoic acid (10 mg ml–1 in 30% acetonitrile) and were crystallized. The analytes were then subjected to matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF-MS) analysis using an Ultraflex time-of-flight mass spectrometer III (Bruker Daltonics) in a reflector, in positive-ion mode, typically summing 1,000 shots. The detected N-glycan peaks in MALDI-TOF-MS spectra were picked using the FlexAnalysis ver. 3 (Bruker Daltonics) software in independently performed experiments in plastidial NPP1, NPP2 and NPP6, respectively. We determined the difference (Δstd) from the theoretical mass value of an internal standard (M+H 831.33533 m/z) in each measurement. The identification of glycan was manually performed with the following parameters: Δ value of glycan <2 Da, Δstd + 0.3 Da > Δ > Δstd – 0.3 Da. The intensity of the isotopic peak of each glycan was normalized to 15 pmol of internal standard (A2 amide glycan) for each status. The structures of the glycans were speculated using the GlycoMod Tool (http://br.expasy.org/tools/glycomod/) and GlycoSuite website (https://glycosuite.proteomesystems.com/glycosuite/glycodb).
Accession numbers
Rice NPP genes whose complete cDNAs were isolated were: NPP1 (AB100451, AK072408), NPP2 (AB196673), NPP3 (AK101976), NPP4 (AK073512), NPP5 (AK121432) and NPP6 (AK102346).
Supplementary data
Supplementary data are available at PCP online.
Funding
This research was supported by the Japan Society for the Promotion of Sciences [KAKENHI Grants-in-Aid for Scientific Research (A) (15H02486) to T.M.]; the Comisión Interministerial de Ciencia y Tecnología and Fondo Europeo de Desarrollo Regional (Spain) [grants BIO2010-18239 and BIO2013-49125-C2-1-P]; the Government of Navarra [grant IIQ14067.RI1].
Supplementary Material
Acknowledgments
We thank Dr. Aya Kitajima, Ai Yanagida, Tsutomu Koshu, and Yukiho Umezawa (Niigata University) for their valued contributions to the confocal laser scanning microscopy and the production of rice mutants, and Mayuko Sato and Takako Kawai (RIKEN Center for Sustainable Resource Science) for their valued contributions to the EM study.
Glossary
Abbreviations
- AmyI-1
α-amylase isoform I-1
- ARF1
ADP-ribosylation factor 1
- BFA
brefeldin A
- CAH1
carbonic anhydrase 1
- Con A
Concanavalin A
- ER
endoplasmic reticulum
- GFP
green fluorescent protein
- NPP
nucleotide pyrophosphatase/phosphodiesterase
- PBS
phosphate-buffered saline
- PBST
phosphate-buffered saline with 0.05% Tween-20
- PVDF
polyvinylidene difluoride
- ST
sialyltransferase
- WxTP
waxy transit peptide
Disclosures
The authors have no conflicts of interest to declare.
References
- Amano M., Yamaguchi M., Takegawa Y., Yamashita T., Terashima M., Furukawa J.-I., et al. (2010) Threshold in stage-specific embryonic glycotypes uncovered by a full portrait of dynamic N-glycan expression during cell differentiation. Mol. Cell. Proteomics 9: 523–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M.X., Goksör M., Sandelius A.S. (2007) Optical manipulation reveals strong attracting forces at membrane contact sites between endoplasmic reticulum and chloroplasts. J. Biol. Chem. 282: 1170–1174. [DOI] [PubMed] [Google Scholar]
- Andon N.L., Hollingworth S., Koller A., Greenland A.J., Yates J.R., 3rd, Haynes P.A. (2002) Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics 2: 1156–1168. [DOI] [PubMed] [Google Scholar]
- Asatsuma S., Sawada C., Itoh K., Okito M., Kitajima A., Mitsui T. (2005) Involvement of α-amylase I-1 in starch degradation in rice chloroplasts. Plant Cell Physiol. 46: 858–869. [DOI] [PubMed] [Google Scholar]
- Bardor M., Faye L., Lerouge P. (1999) Analysis of the N-glycosylation of recombinant glycoproteins produced in transgenic plants. Trends Plant Sci. 4: 376–380. [DOI] [PubMed] [Google Scholar]
- Boisson M., Gomord V., Audran C., Berger N., Dubreucq B., Granier F., et al. (2001) Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20: 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondili J.S., Castilho A., Mach L., Glössl J., Steinkellner H., Altmann F., et al. (2006) Molecular cloning and heterologous expression of α1,2-xylosyltransferase and core α1,3-fucosyltransferase from maize. Phytochemistry 67: 2215–2224. [DOI] [PubMed] [Google Scholar]
- Burén S., Ortega-Villasante C., Blanco-Rivero A., Martínez-Bernardini A., Shutova T., Shevela D., et al. (2011) Importance of post-translational modifications for functionality of a chloroplast-localized carbonic anhydrase (CAH1) in Arabidopsis thaliana. PLoS One 6: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper C.A., Gasteiger E., Packer N.H. (2001a) GlycoMod—a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1: 340–349. [DOI] [PubMed] [Google Scholar]
- Cooper C.A., Harrison M.J., Wilkins M.R., Packer N.H. (2001b) GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.H., Huang L.F., Li H.M., Chen Y.R., Yu S.M. (2004) Signal peptide-dependent targeting of a rice α-amylase and cargo proteins to plastids and extracellular compartments of plant cells. Plant Physiol. 135: 1367–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen A.H., Sharrock R.A., Quail P.H. (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol. Biol. 18: 675–689. [DOI] [PubMed] [Google Scholar]
- Fitchette-Lainé A.-C., Gomord V., Cabanes M., Michalski J.-C., Saint Macary M., Foucher B., et al. (1997) N-glycans harboring the Lewis a epitope are expressed at the surface of plant cells. Plant J. 12: 1411–1417. [DOI] [PubMed] [Google Scholar]
- Furukawa J., Shinohara Y., Kuramoto H., Miura Y., Shimaoka H., Kurogochi M., et al. (2008) Comprehensive approach to structural and functional glycomics based on chemoselective glycoblotting and sequential tag conversion. Anal. Chem. 80: 1094–1101. [DOI] [PubMed] [Google Scholar]
- Guérardel Y., Chang L.Y., Maes E., Huang C.J., Khoo K.H. (2006) Glycomic survey mapping of zebrafish identifies unique sialylation pattern. Glycobiology 16: 244–257. [DOI] [PubMed] [Google Scholar]
- Haltiwanger R.S., Lowe J.B. (2004) Role of glycosylation in development. Annu. Rev. Biochem. 73: 491–537. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994. [DOI] [PubMed] [Google Scholar]
- Hashimoto K., Goto S., Kawano S., Aoki-Kinoshita K.F., Ueda N., Hamajima M., et al. (2006) KEGG as a glycome informatics resource. Glycobiology 16: 63R–70R. [DOI] [PubMed] [Google Scholar]
- Hayashi M., Tsuru A., Mitsui T., Takahashi N., Hanzawa H., Arata Y., et al. (1990) Structure and biosynthesis of the xylose-containing carbohydrate moiety of rice α-amylase. Eur. J. Biochem. 191: 287–295. [DOI] [PubMed] [Google Scholar]
- Helenius A., Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291: 2364–2369. [DOI] [PubMed] [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hirose K., Amano M., Hashimoto R., Lee Y.C., Nishimura S.-I. (2011) Insight into glycan diversity and evolutionary lineage based on comparative avio-N-glycomics and sialic acid analysis of 88 egg whites of Galloanserae. Biochemistry 50: 4757–4774. [DOI] [PubMed] [Google Scholar]
- Hood E.E., Helmer G.L., Fraley R.T., Chilton M.D. (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J. Bacteriol. 168: 1291–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba T., Alvarez-Huerta M., Li M., Bauer J., Ewers C., Kessler F., et al. (2005) Arabidopsis tic110 is essential for the assembly and function of the protein import machinery of plastids. Plant Cell 17: 1482–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue H., Akita M. (2008) Three sets of translocation intermediates are formed during the early stage of protein import into chloroplasts. J. Biol. Chem. 283: 7491–7502. [DOI] [PubMed] [Google Scholar]
- Kajiura H., Koiwa H., Nakazawa Y., Okazawa A., Kobayashi A., Seki T., et al. (2010) Two Arabidopsis thaliana Golgi α-mannosidase I enzymes are responsible for plant N-glycan maturation. Glycobiology 20: 235–247. [DOI] [PubMed] [Google Scholar]
- Kaneko K., Inomata T., Masui T., Koshu T., Umezawa Y., Itoh K., et al. (2014) Nucleotide pyrophosphatase/phosphodiesterase 1 exerts a negative effect on starch accumulation and growth in rice seedlings under high temperature and CO2 concentration conditions. Plant Cell Physiol. 55: 320–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko K., Yamada C., Yanagida A., Koshu T., Umezawa Y., Itoh K., et al. (2011) Differential localizations and functions of rice nucleotide pyrophosphatase/phosphodiesterase isozymes 1 and 3. Plant Biotechnol. 28: 69–76. [Google Scholar]
- Kang J.S., Frank J., Kang C.H., Kajiura H., Vikram M., Ueda A., et al. (2008) Salt tolerance of Arabidopsis thaliana requires maturation of N-glycosylated proteins in the Golgi apparatus. Proc. Natl. Acad. Sci. USA 105: 5933–5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher D.J., Gilmore R. (2006). An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R. [DOI] [PubMed] [Google Scholar]
- Kessler F., Schnell D. (2009) Chloroplast biogenesis: diversity and regulation of the protein import apparatus. Curr. Opin. Cell Biol. 21: 494–500. [DOI] [PubMed] [Google Scholar]
- Kimura S., Mitsui T., Matsuoka T., Igaue I. (1992) Purification, characterization and localization of rice UDP-glucose pyrophosphorylase. Plant Physiol. Biochem. 30: 683–693. [Google Scholar]
- Kitajima A., Asatsuma S., Okada H., Hamada Y., Kaneko K., Nanjo Y., et al. (2009) The rice α-amylase glycoprotein is targeted from the Golgi apparatus through the secretory pathway to the plastids. Plant Cell 21: 2844–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R., Kornfeld S. (1985) Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54: 631–664. [DOI] [PubMed] [Google Scholar]
- Léonard R., Costa G., Darrambide E., Lhernould S., Fleurat-Lessard P., Carlué M., et al. (2002) The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active α1,4-fucosyltransferase gene. Glycobiology 12: 299–306. [DOI] [PubMed] [Google Scholar]
- Lerouge P., Cabanes-Macheteau M., Rayon C., Fischette-Lainé A.C., Gomord V., Faye L. (1998) N-glycoprotein biosynthesis in plants: recent developments and future trends. Plant Mol. Biol. 38: 31–48. [PubMed] [Google Scholar]
- Liebminger E., Hüttner S., Vavra U., Fischl R., Schoberer J., Grass J., et al. (2009) Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21: 3850–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura-Tokita K., Takeuchi M., Ichihara A., Mikuriya K., Nakano A. (2006) Live imaging of yeast Golgi cisternal maturation. Nature 441: 1007–1010. [DOI] [PubMed] [Google Scholar]
- Melo N.S., Nimtz M., Conradt H.S., Fevereiro P.S., Costa J. (1997) Identification of the human Lewis (a) carbohydrate motif in a secretory peroxidase from a plant cell suspension culture (Vaccinium myrtillus L.). FEBS Lett. 415: 186–191. [DOI] [PubMed] [Google Scholar]
- Miki D., Shimamoto K. (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 45: 490–495. [DOI] [PubMed] [Google Scholar]
- Mitsui T., Itoh K. (1997) The α-amylase multigene family. Trends Plant Sci. 2: 255–261. [Google Scholar]
- Nanjo Y., Oka H., Ikarashi N., Kaneko K., Kitajima A., Mitsui T., et al. (2006) Rice plastidial N-glycosylated nucleotide pyrophosphatase/phosphodiesterase is transported from the ER–Golgi to the chloroplast through the secretory pathway. Plant Cell 18: 2582–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebenführ A., Gallagher L.A., Dunahay T.G., Frohlick J.A., Mazurkiewicz A.M., Meehl J.B., et al. (1999) Stop-and-go movements of plant Golgi stacks are mediated by the acto-myosin system. Plant Physiol. 121: 1127–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M., Akazawa T. (1974) Studies on spinach ribulosebisphosphate carboxylase. Carboxylase and oxygenase reaction examined by immunochemical methods. Biochemistry 13: 2277–2281. [DOI] [PubMed] [Google Scholar]
- Nishimura S.-I., Niikura K., Kurogochi M., Matsushita T., Fumoto M., Hinou H., et al. (2005) High-throughput protein glycomics: combined use of chemoselective glycoblotting and MALDI-TOF/TOF mass spectrometry. Angew. Chem. Int. Ed. 44: 91–96. [DOI] [PubMed] [Google Scholar]
- Ochial A., Sugai H., Harada K., Tanaka S., Ishiyama Y., Ito K., Tanaka T., Uchiumi T., Taniguchi M., Mitsui T. (2014) Crystal structure of α-amylase from Oryza sativa: Molecular insights into enzyme activity and thermostability. Biosci. Biotechnol. Biochem. 78: 989–997. [DOI] [PubMed] [Google Scholar]
- Ogawa H., Hijikata A., Amano M., Kojima K., Fukushima H., Ishizuka I., et al. (1996) Structures and contribution to the antigenicity of oligosaccharides of Japanese cedar (Cryptomeria japonica) pollen allergen Cry j I: relationship between the structures and antigenic epitopes of plant N-linked complex-type glycans. Glycoconj. J. 13: 555–566. [DOI] [PubMed] [Google Scholar]
- Ohtsubo K., Marth J.D. (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126: 855–867. [DOI] [PubMed] [Google Scholar]
- Olczak M., Olczak T. (2002) Diphosphonucleotide phosphatase/phosphodiesterase from yellow lupin (Lupinus luteus L.) belongs to a novel group of specific metallophosphatases. FEBS Lett. 519: 159–163. [DOI] [PubMed] [Google Scholar]
- Olczak M., Watorek W. (2000) Structural analysis of N-glycans from yellow lupin (Lupinus luteus) seed diphosphonucleotide phosphatase/phosphodiesterase. Biochim. Biophys. Acta 1523: 236–245. [DOI] [PubMed] [Google Scholar]
- Rodríguez-López M., Baroja-Fernández E., Zandueta-Criado A., Pozueta-Romero J. (2000) Adenosine diphosphate glucose pyrophosphatase: a plastidial phosphodiesterase that prevents starch biosynthesis. Proc. Natl. Acad. Sci. USA 97: 8705–8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Jore-Dupas C., Nebenführ A., Boulaflous A., Follet-Gueye M.L., Plasson C., Hawes C., et al. (2006) Plant N-glycan processing enzymes employ different targeting mechanisms for their spatial arrangement along the secretory pathway. Plant Cell 18: 3182–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattat M., Barton K., Baudisch B., Klösgen R.B., Mathur J. (2011) Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 155: 1667–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer R. (2009) Sialic acids as regulators of molecular and cellular interactions. Curr. Opin. Struct. Biol. 19: 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussillane P., D’Alessio C., Paccalet T., Fitchette A.-C., Parodi A.J., Williamson R., et al. (2009) N-glycan trimming by glucosidase II is essential for Arabidopsis development. Glycoconj. J. 26: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R., Altmann F., Mach L., Glössl J., Steinkellner H. (2004) Generation of Arabidopsis thaliana plants with complex N-glycans lacking β1,2-linked xylose and core α1,3-linked fucose. FEBS Lett. 561: 132–136. [DOI] [PubMed] [Google Scholar]
- Strasser R., Bondili J.S., Vavra U., Schoberer J., Svoboda B., Glössl J., et al. (2007) A unique β1,3-galactosyltransferase is indispensable for the biosynthesis of N-glycans containing Lewis a structures in Arabidopsis thaliana. Plant Cell 19: 2278–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R., Mucha J., Mach L., Altmann F., Wilson I.B.H., Glössl J., et al. (2000) Molecular cloning and functional expression of β1,2-xylosyltransferase cDNA from Arabidopsis thaliana. FEBS Lett. 472: 105–108. [DOI] [PubMed] [Google Scholar]
- Strasser R., Mucha J., Schwihla H., Altmann F., Glössl J., Steinkellner H. (1999a) Molecular cloning and characterization of cDNA coding for β1,2N-acetylglucosaminyltransferase I (GlcNAc-TI) from Nicotiana tabacum. Glycobiology 9: 779–785. [DOI] [PubMed] [Google Scholar]
- Strasser R., Schoberer J., Jin C., Glössl J., Mach L., Steinkellner H. (2006) Molecular cloning and characterization of Arabidopsis thaliana Golgi α-mannosidase II, a key enzyme in the formation of complex N-glycans in plants. Plant J. 45: 789–803. [DOI] [PubMed] [Google Scholar]
- Strasser R., Steinkellner H., Borén M., Altmann F., Mach L., Glössl J., et al. (1999b) Molecular cloning of cDNA encoding N-acetylglucosaminyltransferase II from Arabidopsis thaliana. Glycoconj. J. 16: 787–791. [DOI] [PubMed] [Google Scholar]
- Sun Q., Zybailov B., Majeran W., Friso G., Olinares P.D.B., van Wijk K.J. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 37: D969–D974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Hotta T., Ishihara H., Mori M., Tejima S., Bligny R., et al. (1986) Xylose-containing common structural unit in N-linked oligosaccharides of laccase from sycamore cells. Biochemistry 25: 388–395. [Google Scholar]
- Takeuchi M., Ueda T., Yahara N., Nakano A. (2002) Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31: 499–515. [DOI] [PubMed] [Google Scholar]
- Tanaka N., Fujita M., Handa H., Murayama S., Uemura M., Kawamura Y., et al. (2004) Proteomics of the rice cell: systematic identification of the protein populations in subcellular compartments. Mol. Genet. Genomics 271: 566–576. [DOI] [PubMed] [Google Scholar]
- Tremblay L.O., Herscovics A. (1999) Cloning and expression of a specific human α1,2-mannosidase that trims Man9GlcNAc2 to Man8GlcNAc2 isomer B during N-glycan biosynthesis. Glycobiology 9: 1073–1078. [DOI] [PubMed] [Google Scholar]
- Toyooka K., Goto Y., Asatsuma S., Koizumi M., Mitsui T., Matsuoka K. (2009) A mobile secretory vesicle cluster involved in mass transport from the Golgi to plant cell exterior. Plant Cell 21: 1212–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyooka K., Moriyasu Y., Goto Y., Takeuchi M., Fukuda H., Matsuoka K. (2006) Protein aggregates are transported to vacuoles by a macroautophagic mechanism in nutrient-starved plant cells. Autophagy 2: 96–106. [DOI] [PubMed] [Google Scholar]
- Varki A. (1993) Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3: 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarejo A., Burén S., Larsson S., Déjardin A., Monné M., Rudhe C., et al. (2005) Evidence for a protein transported through the secretory pathway en route to the higher plant chloroplast. Nat. Cell Biol. 7: 1224–1231. [DOI] [PubMed] [Google Scholar]
- von Schaewen A., Sturm A., O’Neill J., Chrispeels M.J. (1993) Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102: 1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson I.B.H., Rendić D., Freilinger A., Dumić J., Altmann F., Mucha J., et al. (2001) Cloning and expression of cDNAs encoding α1,3-fucosyltransferase homologues from Arabidopsis thaliana. Biochim. Biophys. Acta 1527: 88–96. [DOI] [PubMed] [Google Scholar]
- Zhang M., Gaschen B., Blay W., Foley B., Haigwood N., Kuiken C., et al. (2004) Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14: 1229–1246. [DOI] [PubMed] [Google Scholar]
- Zhu H., Qian W., Lu X., Li D., Liu X., Liu K., et al. (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 59: 581–594. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.