Abstract
Replanting disease is a major factor limiting the artificial cultivation of the traditional Chinese medicinal herb Salvia miltiorrhiza. At present, little information is available regarding the role of miRNAs in response to replanting disease. In this study, two small RNA libraries obtained from first-year (FPR) and second-year plant (SPR) roots were subjected to a high-throughput sequencing method. Bioinformatics analysis revealed that 110 known and 7 novel miRNAs were annotated in the roots of S. miltiorrhiza. Moreover, 39 known and 2 novel miRNAs were identified and validated for differential expression in FPR compared with SPR. Thirty-one of these miRNAs were further analyzed by qRT-PCR, which revealed that 5 miRNAs negatively regulated the expression levels of 7 target genes involved in root development or stress responses. This study not only provides novel insights into the miRNA content of S. miltiorrhiza in response to replanting disease but also demonstrates that 5 miRNAs may be involved in these responses. Interactions among the differentially expressed miRNAs with their targets may form an important component of the molecular basis of replanting disease in S. miltiorrhiza.
Introduction
Salvia miltiorrhiza Bunge is a very popular traditional Chinese medicinal plant. The economic importance of S. miltiorrhiza results from the medicinal activity of extracts of its tuberous roots, which include tanshinones and salvianolic acid, and the herb has been used extensively to treat a variety of conditions, especially cardiovascular and cerebrovascular diseases [1–3]. Unfortunately, while several studies have described how to improve the major medicinal bioactive constituents of S. miltiorrhiza [4–8], little attention has been paid to S. miltiorrhiza yields and quality.
The cultivation area of S. miltiorrhiza in China has expanded over the years because of increased demands for the plant in the domestic and international markets. Environmental conditions, such as soil, climate, and altitude, play an important role in S. miltiorrhiza cultivation. Land suitable for planting S. miltiorrhiza is extremely limited. Consequently, replanting disease has become a major factor limiting the artificial cultivation of S. miltiorrhiza [9–11].
Replanting disease, also known as the continuous monoculturing problem, has been observed to induce sharps drop in the growth, development, yield, and quality of plants grown in a field where the same species had been grown over the past several years even under normal cultivation and management measures. This syndrome has been observed in approximately 70% of all medicinal plants with roots or rhizomes used as traditional medicine, including Panax ginseng [12], Rhizomacoptidis [13], and P. notoginseng [14]. Replanting disease presents serious influences on the sustainable development of medicinal plants and is a major issue that must be addressed in the industry.
Previous studies have suggested that changes in the soil properties, microbial population, and allelopathic autotoxicity promote replanting disease [15–16]. However, the molecular basis of the sensitivity of a species to its own exudate remains unknown. Yang et al. [13] recently studied the responses of genes of Rehmannia glutinosa to replanting disease during continuous cropping and obtained a global perspective of differentially transcribed genes and miRNAs in the plant.
Increasing lines of evidence suggest that miRNAs play an important role in plant stress, and that the expression profiles of most miRNAs involved in plant growth and development are significantly altered during environmental stress [17]. Given that replanting disease represents a form of stress, miRNAs may be involved in the replanting disease of S. miltiorrhiza.
We hypothesize that miRNA activity may underlie at least some of the problems associated with the continuous cropping of S. miltiorrhiza. To gain novel insights into the miRNA content of S. miltiorrhiza in replanting disease, we employed a high throughput parallel sequencing platform (Solexa sequencing) to generate two small libraries of the first-year (FPR) and second-year (SPR) plant roots of the herb. We applied this technology to achieve comparative profiles of miRNAs of the two libraries with the aim of identifying the miRNAs expressed differentially in FPR and SPR. We also identified and confirmed the responses of novel miRNAs to replanting disease using RT-PCR. The expression of miRNAs expressed differentially was confirmed using qRT-PCR, and the target genes were elucidated. In this study, a total of 110 known miRNAs were identified, and 7 novel miRNAs were discovered. Among these miRNAs,41 were differentially expressed between the two libraries. The targets of the differentially expressed miRNAs were also analyzed, and 7 targets from 5 miRNAs, including transcriptional regulation factors, proteases, and disease-resistance proteins, were eventually verified. The results suggest that the obtained miRNAs and their targets may play an important role in gene pathway responses to replanting disease.
Materials and Methods
The experimental materials were planted at the GAP planting base of S. miltiorrhiza (Shangluo, Shanxi Province, China); this base belongs to Tasly R&D Institute, Tasly Holdings Group Co. Ltd., Tianjin, China. We obtained permission to conduct the study at this site and performed sample collection after authorization by Yan Liu, the official in charge of the GAP planting base. No endangered or protected species were involved in this study.
Plants material and RNA extraction
The S. miltiorrhiza cultivar ‘violet flower S. miltiorrhiza’ was collected from the GAP planting base. The first-year crop was grown in a field where S. miltiorrhiza had not been planted for more than 5 years. A second group of plants was grown in a field where the same cultivar had been grown in the previous year. For convenience, we named members of the former group as first-year plants and members of the latter group as second-year plants. Root samples were obtained from five independent plants at the tuberous root expansion stage (Aug. 10, 2015), and their RNA content was extracted using an improved CTAB-LiCl method [18].
Identification of novel miRNA in S. miltiorrhiza
Raw read sequences in the two libraries were combined into one small RNA library for novel miRNA prediction, and all reads matching the tRNA, rRNA, or conserved miRNA sequences with two mismatches were removed using Bowtie [19]. The remaining reads were mapped to the assembled transcript sequences (downloaded from http://bi.sky.zstu.edu.cn/Bio111/DsTRD/home.php) [20] using Bowtie with perfect matching [19]. With one end anchored 20 bp away from the mapped small RNA location, 120–360 bp sequences, each with an extension of 20 bp covering the small RNA region, were collected using a Perl script. Secondary structures of each sequence were predicted using RNAfold from the Vienna package (version 1.8.2) [21]. Under conditions similar to those proposed by Thakur et al. [22], a stem-loop structure with ≤ 3 gaps involving ≤ 8 bases at the small RNA location was considered a candidate miRNA precursor.
Identification of replant-responsive miRNAs
The frequency of the miRNAs from two libraries was normalized to 1 million by total clean reads of miRNAs (RPM) in each sample. If the normalized read count of a given miRNA was zero, the expression value was modified to 0.01 for further analysis. The fold change between the SPR and FPR libraries was calculated as follows: fold change = log2(SPR/FPR). miRNAs with fold changes > 1 or < −1 and with P≤ 0.001 were respectively considered to be upregulated or downregulated in response to replanting disease. The P-value was calculated according to previously established methods [23].
Prediction and validation of miRNA target genes
For identifying the miRNA targets, the FASTA files of S. miltiorrhiza transcriptome sequences were downloaded from the website http://bi.sky.zstu.edu.cn/DsTRD/downloads.php?codingRNAfa=codingRNA.fa. Following this, the online psRNAtarget program was first employed for target identification (http://plantgrn.noble.org/psRNA-Target/?function=3). Then degradome data of S. miltiorrhiza was downloaded from NCBI SRA database (accession number: SRR1557864) [24]. After removing the adaptor [24], clean reads were furtherly used for detecting the cleaved targets of miRNAs using CleaveLand pipeline [25].
Quantitative RT-PCR analysis
Total RNA was treated with RNase-free DNase I (TaKaRa, Dalian, China) to remove genomic DNA. Forward primers for 5 selected miRNAs were designed based on the sequence of the miRNAs and are listed in S1 Table. The reverse transcription reaction was performed with the One Step PrimeScript miRNA cDNA Synthesis Kit (TaKaRa, Dalian, China) according to the manufacturer's protocol.
SYBR Green PCR was performed following the manufacturer’s instructions (Takara, Japan). In brief, 2 μl of cDNA template was added to 10 μl of 2× SYBR Green PCR master mix (Takara), 1 μM each primer, and ddH2O to a final volume of 20 μl. The reactions were amplified for 1min at 95°C, followed by 40 cycles of 95°C for 5 s and 60°C for 34 s. All reactions were performed in triplicate, and the controls (no template and no RT) were included for each gene. The threshold cycle (CT) values were automatically determined by the instrument. The fold-changes were calculated using 2 –Delta DeltaCt method, where DeltaDeltaCt = (Cttarget − Ctinner)treatment − (Cttarget − Ctinner)control[26].
Results
Deep sequencing of sRNAs in S.miltiorrhiza root
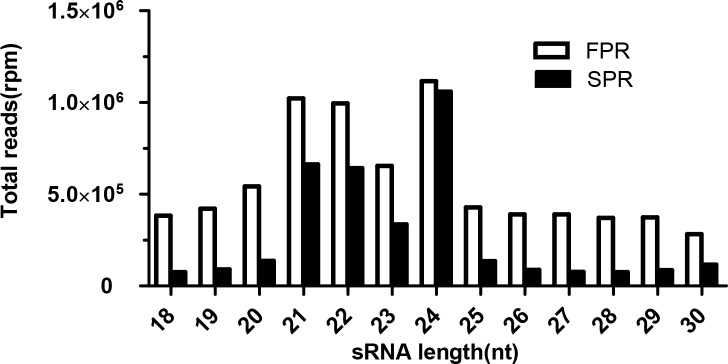
Two sRNA libraries were constructed from the root tissues of FPR and SPR to identify miRNAs responding to replanting disease in S. miltiorrhiza roots. The sequencing results were submitted to the GenBank SRA database (accession numbers: SRR3056582 and SRR3056449). A total of 14.73 million raw reads were generated from the two sRNA libraries. Low-quality tags and adaptor contaminations were removed to obtain 7,374,092 (representing 2,050,638 unique sequences) and 3,581,868 (representing 1,512,866 unique sequences) clean reads from the FPR and SPR libraries, respectively; these reads ranged from 18 nt to 30 nt in both groups. Sequences of about 21 and 24 nt in length were dominant in both libraries (Fig 1). This result is consistent with those presented in previous studies on other plant genera, such as Arabidopsis [27], Oryza [28], Medicago [29], and Populus [30].
Fig 1. sRNA length distributions in S. miltiorrhiza FPR and SPR.
Identification of known miRNAs in S. miltiorrhiza
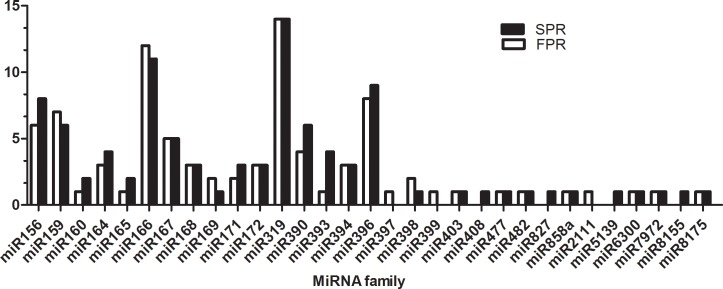
Unique sRNA sequences were mapped to miRBase 21.0 with perfect matches using Perl script [31]. A total of 110 unique sequences belonging to 31 conserved miRNA families were identified in the FPR and SPR libraries (S2 Table). Among these sequences, 87miRNAs belonging to 27 families (except for miR408, miRNA827, miR5139 and miR8155) were detected in FPR, while 96 miRNAs belonging to 28 families (except for miR397, miR399 and miR2111) were detected in SPR. Between the two libraries, the more abundant sequences were found in the miR156, miR396, miR 319, and miR166 families. The miR319 family, which was composed of 14 members, dominated the sequences (Fig 2).
Fig 2. Number of known miRNA families in the FPR and SPR libraries.
Identification and validation of novel miRNAs in S. miltiorrhiza
sRNA reads that were homologous to known miRNAs and other non-coding sRNAs (Rfam 10) were excluded, and the secondary structures of the precursors of the remaining 19–24 nt sRNAs were analyzed using RNAfold software to determine novel miRNAs in S. miltiorrhiza. Precursors with canonical stem–loop structures were further analyzed using more stringent filters to ensure that they satisfied the criteria established by the research community [32, 33]. Thirty-two miRNAs candidates derived from 71 loci satisfied the screening criteria. The 32 putative miRNA precursors were then used to extract miRNA*s, which are considered strong evidence of DICER-LIKE-1 (DCL1)-derived products. Thirteen of the 32 miRNA candidates contained miRNA star (miRNA*) sequences identified from the same libraries; the remaining 19 candidates did not contain any miRNA* sequences (S3 Table). The 13 candidates with miRNA* sequences were considered to be novel S. miltiorrhiza miRNAs, whereas the19 remaining candidates without miRNA* sequences were considered potential S. miltiorrhiza miRNAs. The stem-loop structures and miRNA* sequences of the13 novel miRNAs are shown in S1 Fig.
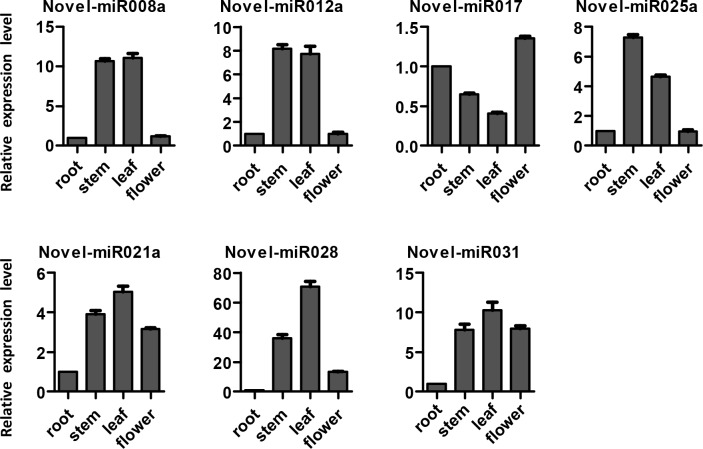
RT-PCR was performed to validate the 13 novel miRNAs and determine their expression patterns in the root, stem, leaf, and flower of S. miltiorrhiza. The electrophoresis results indicated that the 7 novel miRNAs all showed amplification in the root, stem, leaf, and flower of S. miltiorrhiza (S2 Fig). Seven novel miRNAs were analyzed for tissue-specificity by qRT-PCR, and the results showed obvious tissue specificity. The expressions of all of the novel miRNA tissues, except that of miR017, were higher in the stem and leaf than in the root and flower. MiR021a, miR028, and miR031 expression levels were highest in the leaf, while miR025a expression levels were highest in the stem. The expression levels of miR031 in the root were lower by about 6 times than those in the stem, leaf, and flower of S. miltiorrhiza. miRNA017 gene expression was lower in the stem and leaf than in the root and flower; the highest expression of this gene was observed in the flower (Fig 3).The 7 novel miRNAs miR001a, miR008a, miR012a, miR017, miR021a, miR028 and miR031 were renamed as smi-miR35829, smi-miR35830, smi-miR35831, smi-miR35832, smi-miR35833, smi-miR35834 and smi-miR35835, respectively.
Fig 3. Expression levels of 7 novel miRNAs in the root, stem, leaf, and flower of S.miltiorrhiza.
Differential expression identification of known and novel miRNAs involved in response to replanting disease
To identify the miRNAs involved in the response of S. miltiorrhiza to replanting disease, differential expressions in the two libraries were estimated from the read counts determined through high-throughput sequencing. We selected RPMs that exceeded 1 and presented a P value of less than 0.001 from both libraries. miRNAs that met this condition and exhibited a log2 (SPR RPM/FPR RPM) fold-change higher than 1 were designated as upregulated, whereas those with a log2 (SPR RPM/FPR RPM) fold-change of less than −1 were designated as downregulated (S2 and S3 Tables).
The read counts of 110 known unique sequences from both libraries were retrieved to determine miRNAs responding to replanting disease. A total of 39 known miRNAs from 15 families and 2 novel miRNAs were differentially expressed in response to replanting disease (Table 1). Among these differentially expressed miRNAs, 38 were upregulated and only 3 were downregulated in the SPR library compared with those in the FPR library. Among these 41 sequences, the miR166, miR319 and miR396 families, which possessed 8, 5 and 5 members, respectively, were in a dominant position. Only pab-miR160a-like was absent from the FPR library.
Table 1. Differentially expressed miRNAs in the FPR and SPR libraries of S. miltiorrhiza.
| miR_name | Mature Sequence(5'-3') | FPR_RPM | SPR_RPM | log2(SP/FP) | Pvalue |
|---|---|---|---|---|---|
| smi-miR156a-1 | TGACAGAAGAGAGTGAGCAC | 8.54 | 24.85 | 1.540938 | 6E-11 |
| smi-miR156a-2 | TGACAGAAGAGAGTGAGCAC | 8.54 | 24.85 | 1.540938 | 6E-11 |
| smi-miR156a-3 | TGACAGAAGAGAGTGAGCAC | 8.54 | 24.85 | 1.540938 | 6E-11 |
| smi-miR159a | TTTGGATTGAAGGGAGCTCTA | 691.88 | 2714.23 | 1.971949 | 0 |
| aly-miR159b-3p-like | TTTGGATTGAAGGGAGCTCTT | 0.81 | 58.91 | 6.184447 | 3.05E-93 |
| pab-miR160a-like | TGCCTGGCTCCCTGTATGCCA | 0.01 | 2.79 | 8.124121 | 9.12E-06 |
| smi-miR164a-1 | TGGAGAAGCAGGGCACGTGCA | 12.61 | 112.51 | 3.157413 | 8.1E-110 |
| smi-miR164a-2 | TGGAGAAGCAGGGCACGTGCA | 12.61 | 112.51 | 3.157413 | 8.1E-110 |
| ath-miR165a-3p-like | TCGGACCAGGCTTCATCCCCC | 2.31 | 6.7 | 1.536268 | 0.000636 |
| smi-miR166a-3p-1 | TCGGACCAGGCTTCATTCCCC | 20432.21 | 47641.34 | 1.221369 | 0 |
| smi-miR166a-3p-2 | TCGGACCAGGCTTCATTCCCC | 20432.21 | 47641.34 | 1.221369 | 0 |
| smi-miR166a-3p-3 | TCGGACCAGGCTTCATTCCCC | 20432.21 | 47641.34 | 1.221369 | 0 |
| smi-miR166a-3p-4 | TCGGACCAGGCTTCATTCCCC | 20432.21 | 47641.34 | 1.221369 | 0 |
| smi-miR166a-5p-4 | GGAATGTTGTTTGGCTCGAGG | 10.85 | 4.19 | -1.37267 | 0.000252 |
| osa-miR166e-3p-like | TCGAACCAGGCTTCATTCCCC | 24.55 | 115.86 | 2.238588 | 8.35E-76 |
| smi-miR166h-3p | TCTCGGACCAGGCTTCATTCC | 495.25 | 1758.58 | 1.828182 | 0 |
| smi-miR166a-5p | GGAATGTTGTCTGGCTCGAGG | 99.81 | 33.5 | -1.57502 | 6.67E-36 |
| smi-miR167b-5p | TGAAGCTGCCAGCATGATCTG | 3.93 | 34.62 | 3.139005 | 8.75E-35 |
| smi-miR168a-5p-1 | TCGCTTGGTGCAGGTCGGGAA | 39.06 | 200.73 | 2.361492 | 4.3E-139 |
| smi-miR168a-5p-2 | TCGCTTGGTGCAGGTCGGGAA | 39.06 | 200.73 | 2.361492 | 4.3E-139 |
| smi-miR171a-3p | TGAGCCGAACCAATATCACTC | 200.57 | 456.19 | 1.185529 | 1.8E-114 |
| smi-miR319a-3p-1 | TTGGACTGAAGGGAGCTCCC | 166.39 | 424.36 | 1.35072 | 4.1E-131 |
| smi-miR319a-3p-2 | TTGGACTGAAGGGAGCTCCC | 166.39 | 424.36 | 1.35072 | 4.1E-131 |
| smi-miR319a-3p-3 | TTGGACTGAAGGGAGCTCCC | 166.39 | 424.36 | 1.35072 | 4.1E-131 |
| smi-miR319a-3p-4 | TTGGACTGAAGGGAGCTCCC | 166.39 | 424.36 | 1.35072 | 4.1E-131 |
| smi-miR319a-3p-5 | TTGGACTGAAGGGAGCTCCC | 166.39 | 424.36 | 1.35072 | 4.1E-131 |
| smi-miR390a-5p-1 | AAGCTCAGGAGGGATAGCGCC | 3.25 | 8.1 | 1.317482 | 0.000892 |
| smi-miR390a-5p-2 | AAGCTCAGGAGGGATAGCGCC | 3.25 | 8.1 | 1.317482 | 0.000892 |
| smi-miR390a-5p-3 | AAGCTCAGGAGGGATAGCGCC | 3.25 | 8.1 | 1.317482 | 0.000892 |
| smi-miR394a-5p-1 | TTGGCATTCTGTCCACCTCC | 7.87 | 16.75 | 1.089726 | 4.31E-05 |
| smi-miR394a-5p-2 | TTGGCATTCTGTCCACCTCC | 7.87 | 16.75 | 1.089726 | 4.31E-05 |
| smi-miR394a-5p-3 | TTGGCATTCTGTCCACCTCC | 7.87 | 16.75 | 1.089726 | 4.31E-05 |
| smi-miR396b-5p-1 | TTCCACAGCTTTCTTGAACTT | 62.79 | 317.15 | 2.336559 | 1.3E-215 |
| smi-miR396b-5p-2 | TTCCACAGCTTTCTTGAACTT | 62.79 | 317.15 | 2.336559 | 1.3E-215 |
| smi-miR396a-5p | TTCCACAGCTTTCTTGAACTG | 1408.85 | 6879.65 | 2.287817 | 0 |
| smi-miR396c-5p | TTCCACAGCTTTCTTGAACTA | 143.07 | 1340.36 | 3.227827 | 0 |
| gma-miR396e-like | TTCCACAGCTTTCTTGAACTGT | 0.54 | 3.35 | 2.63313 | 0.000549 |
| har-miR403a-like | TTAGATTCACGCACAAACTCG | 58.99 | 227.26 | 1.945801 | 1.2E-121 |
| smi-miR482a | TTTCCAACTCCACCCATTCCTA | 24.55 | 222.79 | 3.18189 | 2.7E-217 |
| smi-miR35832 | GGTGCAATGGGCGAACGCCGAGG | 50.85 | 15.36 | -1.72707 | 2.13E-21 |
| smi-miR35835 | TTTCCAATGCCGCCCATACCGA | 327.09 | 1715.59 | 2.390945 | 0 |
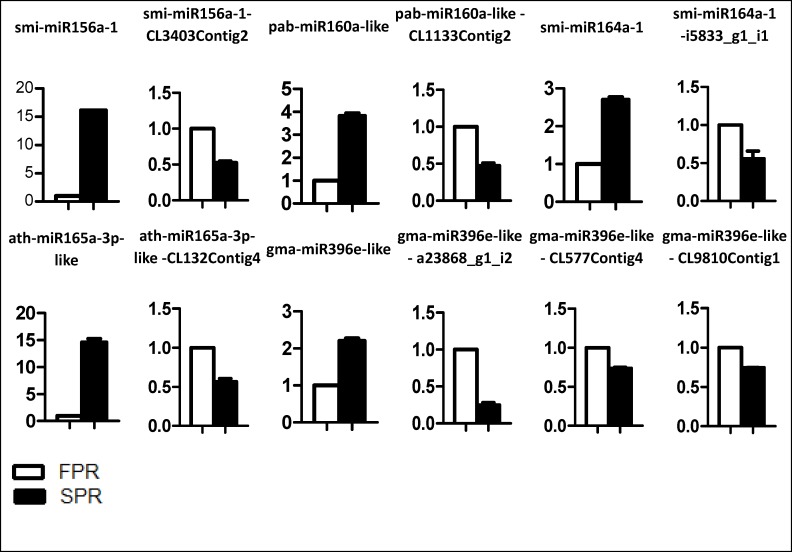
The expression levels of the differentially expressed miRNAs were reanalyzed using qRT-PCR. The 5 miRNAs (smi-miR319a-3p-1, smi-miR319a-3p-2, smi-miR319a-3p-3, smi-miR319a-3p-4 and smi-miR319a-3p-5) could not be amplified. The abundances estimated by sequencing and qRT-PCR analysis were consistent for31 of the 36 miRNAs (the exceptions were smi-miR166a-5p-4, smi-miR394a-5p-1, smi-miR394a-5p-2, smi-miR394a-5p-3 and smi-miR35832 (Fig 4 and S3 Fig). However, the fold-changes obtained from the qRT-PCR data were much higher than those estimated from the high-throughput sequencing data, presumably because of differences in the sensitivity and specificity of the experimental approaches.
Fig 4. Verification of differential expression miRNAs and their targets by qRT-PCR in FPR and SPR.
Analysis and validation of target genes
The psRNAtarget program was used to predict the targets of 31 replant-responsive miRNAs, only 41 CDSs were targeted by 12 miRNA families (S4 Table). CleaveLand pipeline was performed for the second screening of the targets (S4 Fig). The results showed that 20 CDSs were targeted by 10 miRNA families except for miR403 (Table 2). The expression profiles of these 20 CDSs were furtherly determined using qRT-PCR. The result showed that PCR failed to amplify three targets containing g7803_g1_i2, CL765Contig5 and CL2Contig108. Among remaining 18 target cDNAs, SPL13 (CL3403Contig2), ARF18-like (CL1133Contig2), NAC100-like (i5833_g1_i1), athb-15-like (CL132Contig4) and three members of the growth-regulating factor family (a23868_g1_i2, CL577Contig4 and CL9810Contig1), which were targeted by smi-miR156a-1, pab-miR160a-like, smi-miR164a-1, ath-miR165a-3p-like and gma-miR396e-like, respectively, were significantly downregulated in SPR, and exhibited a negative relationship to the expression of the 5 miRNAs (Fig 4). However, GRF4-like (CL1008Contig3), disease resistance protein rpm1-like (CL3Contig201) and hypothetical protein MIMGU_mgv1a023991mg(CL2Contig108), which were targeted by gma-miR396e-like, smi-miR482a and novel miRNA smi-miR35835, respectively, were significantly upregulated, and exhibited a consistent expression pattern with that of these miRNAs (S3B Fig). In addition, no significant differential expression in SPR was observed in the remaining 8target cDNAs (S3A Fig). Therefore, the results strongly suggested that the smi-miR156a-1, pab-miR160a-like, smi-miR164a-1, ath-miR165a-3p-like and gma-miR396e-like may be involved in the responses to replanting disease in S. miltiorrhiza.
Table 2. Identified targets of differential expression miRNAs in S. miltiorrhiza by degradome sequencing.
| miR_name | Target_Acc. | Length | Target_Desc. |
|---|---|---|---|
| smi-miR156a-1 | CL1799Contig3 | 2055 | squamosa promoter-binding-like 12-like, SPL12L |
| smi-miR156a-1 | CL3403Contig2 | 1281 | squamosa promoter-binding-like 13, SPL13 |
| smi-miR159a | CL542Contig9 | 1294 | transcription factor gamyb-like isoform x1 |
| pab-miR160a-like | g7803_g1_i2 | 2690 | auxin response factor 18-like, ARF18-like |
| pab-miR160a-like | CL1133Contig2 | 2925 | auxin response factor 18-like, ARF18-like |
| pab-miR160a-like | CL16970Contig1 | 1429 | auxin response factor 18-like, ARF18-like |
| smi-miR164a-1 | i5833_g1_i1 | 906 | NAC domain-containing protein 100-like, NAC100 |
| ath-miR165a-3p-like | CL132Contig4 | 2174 | homeobox-leucine zipper protein athb-15-like |
| smi-miR167b-5p | a28228_g1_i3 | 933 | mitochondrial substrate carrier family protein b-like |
| smi-miR167b-5p | k12130_g1_i1 | 1378 | mitochondrial substrate carrier family protein b-like |
| smi-miR167b-5p | CL1585Contig3 | 1545 | mitochondrial substrate carrier family protein b-like |
| smi-miR168a-5p-1 | CL600Contig1 | 3706 | argonaute 1 |
| gma-miR396e-like | a23868_g1_i2 | 772 | growth-regulating factor 3-like, GRF3-like |
| gma-miR396e-like | e14536_g1_i4 | 1078 | growth-regulating factor 4-like, GRF4-like |
| gma-miR396e-like | CL1008Contig3 | 1155 | growth-regulating factor 4-like, GRF4-like |
| gma-miR396e-like | CL577Contig4 | 2862 | growth-regulating factor 4-like, GRF4-like |
| gma-miR396e-like | CL1003Contig3 | 3662 | 6-phosphofructokinase 3-like |
| gma-miR396e-like | CL9810Contig1 | 1625 | growth-regulating factor 2-like, GRF2-like |
| smi-miR482a | CL3Contig201 | 422 | disease resistance protein rpm1-like |
| smi-miR35835 | CL2Contig108 | 1770 | hypothetical protein MIMGU_mgv1a023991mg |
Discussion
Identification of S. miltiorrhiza miRNAs by high-throughput sequencing
Identification of miRNAs in medicinal plants, such as R. glutinosa [13], Panax ginseng [34], and Pinellia pedatisecta [35], using high-throughput sequencing or miRNA arrays has previously been reported. High-throughput sequencing is particularly useful for identifying involved abiotic and biotic stress responses [36, 37]. In this study, miRNA libraries constructed from the FPR and SPR were used to identify novel and replant-responsive miRNAs in S. miltiorrhiza, and their mechanisms of action were subsequently investigated. Among the miRNAs identified using high-throughput sequencing, 65% of those already known were expressed at low levels (less than 10 raw reads; S1 Table). This finding suggests that high-throughput sequencing is a powerful tool for identifying poorly expressed miRNAs in plants. The miR160, miR165/166, and miR396 families, which are believed to target auxin response factor (ARF), homeodomain leucine zipper (HD-ZIP III), and growth-regulating factor (GRF) transcription factors, respectively, were abundantly represented in both libraries. By comparing the expression levels of all members of a miRNA family, dominant members, such as miR166 and miR396, may be found. These dominant members may perform key regulatory roles in response to stress. Some family members, such as Smi-miR166a/b/c/e/f/g/h/I in the miR166 family, exhibited comparable expression levels, thereby indicating that several members of a family may exert synergistic effects in the relevant regulatory network.
With the discovery of miRNAs as ubiquitous regulators of plant growth and development, including nearly all biological, metabolic, and stress processes, researchers have focused on miRNAs in S. miltiorrhiza. However, given that the complete genome of S. miltiorrhiza is currently unavailable, the discovery of miRNAs from S. miltiorrhiza is relatively limited.
Xu et al. [24] studied tissue-specific miRNAs in S.miltiorrhiza and identified 164 miRNAs after redundancy elimination; of these miRNAs, 28 were known, 22 were novel, and another 114 miRNAs were considered homologous with known miRNAs. Some of these 114 miRNAs were 1–4 bases to the left or right of known miRNA sequences or differed from known miRNA sequences in terms of one base. In this study, we identified 117 miRNAs, 110 of which belonged to 31 known families and 7 of which were novel miRNAs. While twenty-three known miRNAs were consistently identified between the present study and Xu et al.’s research [24] (S5 Table), no novel miRNA were identified between both works, likely because assembled transcriptome sequences were used as reference RNAs for the miRNA precursors analysis in our study whereas ESTs were used in Xu et al.’s work.
Medicinal plant miRNAs responding to replanting
Yang et al. [13] identified 16 miRNAs from 13 miRNA families, including 2 novel miRNAs, responding to R. glutinosa replanting disease. In this study, we identified 31 miRNAs from 14 miRNA families, including 1 novel miRNA responding to S. miltiorrhiza replanting disease. In addition, three replant-responsive miRNAs, namely sim-miR156a and sim- miR160a, were identified both in S. miltiorrhiza and R. glutinosa (S6 Table), suggesting that these 2 miRNAs might have important role in respons to replanting disease.
Potential targets of replanting disease-associated miRNAs
Several studies have shown that miR165 [38], miR396 [39], miR164 [40], miR160 [41], and miR156 [42] are related to the growth and development of plant root. MiR160 is a major regulator of root growth and gravitropism, negatively regulating the target auxin response factors 10 (ARF10), ARF16, and ARF17 [42]. Transgenic plants overexpressing the miR160c gene, which downregulates the expression levels of ARF10 and ARF16 mRNAs, show short roots lacking gravity-sensing capability as well as enlarged tumor-like apices [43]. Overexpression ARF17, another target of miR160, has been observed to lead to the reduction of primary root length in A. thaliana [44]. MiR164 negatively regulates lateral root development by targeting NAC1 [45, 46]. Transgenic Arabidopsis overexpressing ZmNAC1 leads to increased numbers of lateral roots in comparison with wild-type plants, and miR164 negatively regulates ZmNAC1 [47]. Overexpression of miR166/165 downregulates target HD-ZIP IIIs and promotes root growth [38]. MiR396 targets seven growth-regulating factor (GRF) genes to regulate root development [48]. Thus, miR396a knock-down lines develop longer roots than wild-type plants, whereas miR396a and miR396b over-expressing lines and plants overexpressing miR396-resistant AtGRF1 or AtGRF3 exhibit shorter roots [39,48]. MiR156 regulates lateral roots by targeting SPL3, SPL9, and SPL10 in Arabidopsis [42].
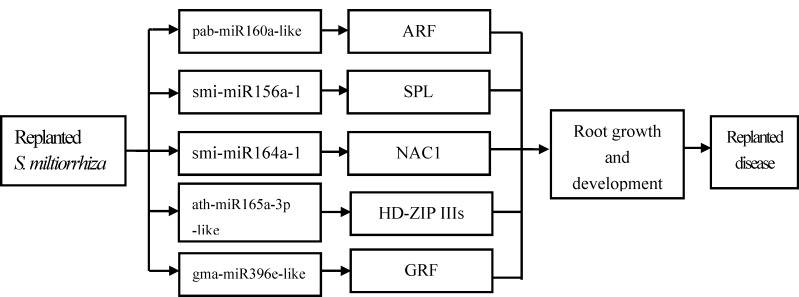
In our studies, smi-miR156a-1, pab-miR160a-like, smi-miR164a-1, ath-miR165a-3p-like, and gma-miR396e-like were upregulated in SPR and negatively regulated the targets of SPL13, ARF18-like, NAC100-like, homeobox-leucine zipper protein athb-14-like, and GRF3-like, respectively. Combining with the phenotype of poor growth in second-year crop, our results revealed that replanting disease upregulates the expressions of smi-miR156a-1, pab-miR160a-like, sim-miR164a-1, ath-miR165a-3p-like, and gma-miR396e-like. Correspondingly, their target genes including SPL13, ARF18-like, NAC100-like, athb-14-like and GRF3-like which were related to root growth were downregulated and then might be presented a growth inhibition (Fig 5).
Fig 5. Proposed regulatory mechanism of miRNAs in replanted S. miltiorrhiza.
Conclusion
Our study provides a global perspective of differential miRNA expression in S. miltiorrhiza in response to replanting disease for the first time. We identified 110 known and 7 novel miRNAs, of which 5 known miRNAs were expressed differentially in the first- and second-year crops. Further investigation revealed that the target genes of differentially expressed miRNAs were related to root growth and development. Combining with the phenotype of poor growth in second-year crop, our results revealed that replanting disease upregulates the expressions of smi-miR156a-1, pab-miR160a-like, smi-miR164a-1, ath-miR165a-3p-like, and gma-miR396e-like. Correspondingly, their target genes including SPL13, ARF18-like, NAC100-like, athb-14-like and GRF3-like which were related to root growth were downregulated and then might be presented a growth inhibition.
Supporting Information
(PDF)
(PDF)
A) The miRNAs were significantly upregulated and the targets were no significant differential expression in SPR; B) The miRNAs and the targets were both significantly upregulated in SPR;C) Differential expression miRNAs without matching targets.
(PDF)
The red dot indicate the cleavage sites.
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability
All data files are available from the NCBI SRA (BioProject:PRJNA307038, accession numbers SRR3056582, SRR3056449).
Funding Statement
This study was supported by the National Natural Science Foundation of China (no. 81373908 and no. 81403033), ZL; and the Natural Science Foundation of Zhejiang Province (no. LQ15H280009), HZ.
References
- 1.Xu ZC, Peters RJ, Weirather J, Luo HM, Liao BS, Zhang X, et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis.Plant J. 2015; 82(6): 951–961. 10.1111/tpj.12865 [DOI] [PubMed] [Google Scholar]
- 2.Kai GY, Hao XL, Cui LJ, Ni XL, Zekria D, Wu JY. Metabolic engineering and biotechnological approaches for production of bioactive diterpene tanshinones in Salvia miltiorrhiza. Biotechnol Adv. 2014; pii: S0734-9750(14)00150-5. [DOI] [PubMed] [Google Scholar]
- 3.Zhang S, Ma P, Yang D, Li W, Liang Z, Liu Y, et al. Cloning and characterization of a putative R2R3 MYB transcriptional repressor of the rosmarinic acid biosynthetic pathway from Salvia miltiorrhiza. Plos One. 2013; 8(9): e73259 10.1371/journal.pone.0073259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao S, Zhang J, Tan R, Yang L, Zheng X. Enhancing diterpenoid concentration in Salvia miltiorrhiza hairy roots through pathway engineering with maize C1 transcription factor. J Exp Bot. 2015; pii: erv418. [DOI] [PubMed] [Google Scholar]
- 5.Xing BC, Yang DF,Guo WL, Liang ZS, Yan XJ, Zhu YH, et al. Ag+ as a more effective elicitor for production of tanshinones than phenolic acids in Salvia miltiorrhiza hairy roots. Molecules 2014; 20(1):309–324. 10.3390/molecules20010309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo HM, Zhu YJ, Song JY, Xu LJ, Sun C, Zhang X, et al. Transcriptional data mining of Salvia miltiorrhiza in response to methyl jasmonate to examine the mechanism of bioactive compound biosynthesis and regulation. Physiol Plant. 2014; 152(2): 241–255. 10.1111/ppl.12193 [DOI] [PubMed] [Google Scholar]
- 7.Gao W, Sun HX, Xiao H, Cui G, Hillwig ML, Jackson A, et al. Combining metabolomics and transcriptomics to characterize tanshinone biosynthesis in Salvia miltiorrhiza. BMC Genomics. 2014; 15:73 10.1186/1471-2164-15-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo J, Zhou YJ, Hillwig ML, Shen Y, Yang L, Wang Y, et al. CYP76AH1 catalyzes turnover of miltiradiene in tanshinones biosynthesis and enables heterologous production of ferruginol in yeasts. Proc Natl Acad Sci U S A. 2013; 110(29): 12108–12113. 10.1073/pnas.1218061110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W, Wei YY, Liu DH, Zhou J, Wang X, Li FS, et al. Biological and Phenological Properties of Salvia miltiorrhiza f.alba for Anti-continuous Cropping. Journal of Chinese Medicinal Materials. 2015; 38(1): 5–7, Chinese. [PubMed] [Google Scholar]
- 10.Yan ZY, Wang H, He B, Liu M, Wang GZ, Chen X, et al. Study on controlling measurement for continuous cropping obstacle in Traditional Chinese Medicinal plants by microecological research model. Pharmacy and Clinics of Chinese Materia Medica. 2012; 3(2): 5–9. Chinese. [Google Scholar]
- 11.Zhang CL, Sun Q, Ye Q. Obstacle effect of continuous cropping on Salvia miltiorrhiza growth. Acta Bot. Boreal. Occident. Sin. 2005; 25 (5): 1029–1034. Chinese. [Google Scholar]
- 12.Xiao C P, Yang LM, Ma FM. Effects of growing time on Panax ginseng rhizosphere soil microbial activity and biomass. Zhongguo Zhong Yao Za Zhi. 2014; 39(24): 4740–4747. Chinese. [PubMed] [Google Scholar]
- 13.Yang YH, Chen XJ, Chen JY, Xu HX, Li J, Zhang ZY. Differential miRNA expression in Rehmannia glutinosa plants subjected to continuous cropping. BMC Plant Biology. 2011; 11: 53 10.1186/1471-2229-11-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang M, Zhang X, Xu Y, Mei X, Jiang B, Liao J, et al. Autotoxic ginsenosides in the rhizosphere contribute to the replant failure of Panax notoginseng. PloS One. 2015; 10(2): e0118555 10.1371/journal.pone.0118555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogweno J O, Yu JQ. Autotoxic potential in soil sickness: A re-examination. Allelopathy Journal. 2006; 18: 93–101. [Google Scholar]
- 16.Wu LJ, Zhao YH, Guan YM, Peng SF. A review on studies of the reason and control methods of succession cropping obstacle of Panax ginseng C.A. Mey. Special Wild Economic Animal Plant Research. 2008; 2: 68–72. [Google Scholar]
- 17.Sunkar R, Li YF. Jagadeeswaran G: Functions of microRNAs in plant stress Responses. Trends in Plant Science. 2012; 17(4): 196–203. 10.1016/j.tplants.2012.01.010 [DOI] [PubMed] [Google Scholar]
- 18.Gambino G, Perrone I, Gribaudo I. A Rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal. 2008; 19(6): 520–525. 10.1002/pca.1078 [DOI] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memoryefficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao Y, Wei J, Wu F, Zhang H, Yang D, Liang Z, et al. DsTRD: Danshen Transcriptional Resource Database. PloS ONE, 2016; 11(2):e0149747 10.1371/journal.pone.0149747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofacker IL.Vienna RNA secondary structure server. Nucleic Acids Res. 2003; 31: 3429–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thakur V, Wanchana S, Xu M, Bruskiewich R, Quick WP, Mosig A, et al. Characterization of statistical features for plant microRNA prediction. BMC Genomics. 2011; 12: 108 10.1186/1471-2164-12-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000; 16: 953–959. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Jiang Q, Ma X, Ying Q, Shen B, Qian Y, et al. Deep Sequencing Identifies Tissue-Specific MicroRNAs and Their Target Genes Involving in the Biosynthesis of Tanshinones in Salvia miltiorrhiza. PLoS ONE, 2014; 9(11): e111679 10.1371/journal.pone.0111679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 2009; 25:130–131 10.1093/bioinformatics/btn604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-△△CT Method. Methods. 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 27.Hsieh LC, Lin SI, Shih AC, Chen JW, Lin WY, Tseng CY, et al. Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiology. 2009; 151: 2120–2132. 10.1104/pp.109.147280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones-Rhoades MW, Bartel DP, Bartel B.MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006; 57: 19–53. [DOI] [PubMed] [Google Scholar]
- 29.Lelandais-Brière C, Naya L, Sallet E, Calenge F, Frugier F, Hartmann C, et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009; 21; 2780–2796. 10.1105/tpc.109.068130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen L, Ren YY, Zhang YY, Xu JC, Sun FS, Zhang ZY, et al. Genome-wide identification and expression analysis of heat-responsive and novel microRNAs in Populus tomentosa. Gene. 2012; 504: 160–165. 10.1016/j.gene.2012.05.034 [DOI] [PubMed] [Google Scholar]
- 31.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences,targets and gene nomenclature. Nucleic Acids Res. 2006; (Database issue): D140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, et al. Criteria for annotation of plant microRNAs. Plant Cell. 2008; 20:3186–3190. 10.1105/tpc.108.064311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thakur V, Wanchana S, Xu M, Bruskiewich R, Quick WP, Mosig A, et al. Characterization of statistical features for plant microRNA prediction. BMC Genomics. 2011;12: 108 10.1186/1471-2164-12-108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu B, Wang M, Ma Y, Yuan L, Lu S.High-throughput sequencing and characterization of the small RNA transcriptome reveal features of novel and conserved microRNAs in Panax ginseng. Plos One.2012;7(9): e44385 10.1371/journal.pone.0044385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong M, Yang DF, Lang QL, Zhou W, Xu SW, Xu T. Microarray and degradome sequencing reveal microRNA differential expression profiles and their targets in Pinellia pedatisecta. Plos One. 2013; 8(9): e75978 10.1371/journal.pone.0075978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin W, Wu F. Identification and characterization of cucumber microRNAs in response to Pseudoperonospora cubensis infection. Gene, 2015; 569(2): 225–232. 10.1016/j.gene.2015.05.064 [DOI] [PubMed] [Google Scholar]
- 37.Jin W, Wu F. Characterization of miRNAs associated with Botrytis cinerea infection of tomato leaves. BMC Plant Biology. 2015; 15:1; 10.1186/s12870-014-0410-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh A, Singh S, Panigrahi KC, Reski R, Sarkar AK.Blanced activity of microRNA166/165 and its target transcripts from the class III homeodomain-leucine zipper family regulates root growth in Arabidopsis thaliana. Plant Cell Rep. 2014; 33: 945–953. 10.1007/s00299-014-1573-z [DOI] [PubMed] [Google Scholar]
- 39.Hewezi T, Maier TR, Nettleton D, Baum TJ. The Arabidopsis microRNA396-GRF1/GRF3 regulatory module acts as a developmental regulator in the reprogramming of root cells during cyst nematode infection. Plant Physiol. 2012; 159: 321–335. 10.1104/pp.112.193649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li J, Bi FC, Yin J, Wu JX, Rong C, Wu JL, et al. An Arabidopsis neutral ceramidasemutantncer1 accumulateshydroxyceramidesandissensitiveto oxidative stress. Frontiers in Plant Science. 2015; 6: 460 10.3389/fpls.2015.00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Turner M, Nizampatnam NR, Baron M, Coppin S, Damodaran S, Adhikari S, et al. Ectopic expression of miR160 results in auxin hypersensitivity, cytokinin hyposensitivity, and inhibition of symbiotic nodule development in soybean. Plant Physiol. 2013;162: 2042–2055. 10.1104/pp.113.220699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu N, Niu QW, Ng KH, Chua NH. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant Journal. 2015; 83: 673–685. 10.1111/tpj.12919 [DOI] [PubMed] [Google Scholar]
- 43.Wang J W, Wang LJ, Mao Y B, Cai WJ, Xue HW, Chen XY. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005; 17: 2204–2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mallory AC, Bartel DP, Bartel B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell. 2005; 17: 1360–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev. 2000; 14(23): 3024–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo H S, Xie Q, Fei J F, Chua N H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to down-regulate auxin signals for Arabidopsis lateral root development. Plant Cell. 2005; 17: 1376–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Guo GH, Guo WW, Guo GG, Tong D, Ni ZF, et al. miRNA164-directed cleavage of ZmNAC1 confers lateral root development in maize (Zea mays L.) BMC Plant Biol. 2012; 12: 220 10.1186/1471-2229-12-220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao ML, Bian HW, Zha YL, Li FY, Sun YZ, Bai B, et al. MiR396a-mediated basic helix-loophelix transcription factor bHLH74 repression acts as a regulator for root growth in Arabidopsis seedlings. Plant Cell Physiol. 2014; 6: 1343–1353. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
A) The miRNAs were significantly upregulated and the targets were no significant differential expression in SPR; B) The miRNAs and the targets were both significantly upregulated in SPR;C) Differential expression miRNAs without matching targets.
(PDF)
The red dot indicate the cleavage sites.
(PPTX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
All data files are available from the NCBI SRA (BioProject:PRJNA307038, accession numbers SRR3056582, SRR3056449).