Abstract
Purpose
Castration therapy in advanced prostate cancer eventually fails and leads to development of castration resistant prostate cancer (CRPC) which has no cure. Characteristic features of CRPC can be increased androgen receptor (AR) expression and altered transcriptional output. We investigated expression of Nuclear Receptor Corepressor 1 (NCOR1) in human prostate and prostate cancer and the role of NCOR1 in response to antiandrogens.
Experimental Design
NCOR1 protein levels were compared between matched normal prostate and prostate cancer in 409 patient samples. NCOR1 knockdown was used to investigate its effect on bicalutamide response in androgen-dependent prostate cancer cell lines and transcriptional changes associated with loss of NCOR1. NCOR1 transcriptional signature was also examined in prostate cancer gene expression datasets.
Results
NCOR1 protein was detected in cytoplasm and nuclei of secretory epithelial cells in normal prostate. Both cytoplasmic and nuclear NCOR1 protein levels were lower in prostate cancer than in normal prostate. Prostate cancer metastases show significant decrease in NCOR1 transcriptional output. Inhibition of LNCaP cellular proliferation by bicalutamide requires NCOR1. NCOR1 regulated genes suppress cellular proliferation and mediate bicalutamide resistance. In mouse, NCOR1 is required for bicalutamide dependent regulation of a subset of the AR target genes.
Conclusions
In summary, we demonstrated that NCOR1 function declines with prostate cancer progression. Reduction in NCOR1 levels causes bicalutamide resistance in LNCaP cells and compromises response to bicalutamide in mouse prostate in vivo.
Keywords: Prostate cancer, NCOR1
Introduction
Metastatic prostate cancer (PCa) is treated with androgen ablation. While it is initially successful, the resistance to the treatment almost invariably develops. Recurrent castration resistant prostate cancer (CRPC) is the major cause of mortality of prostate cancer patients and better understanding of its biology is required for the development of novel therapies. Multiple studies show that the common mechanisms in PCa progression are an increase in the androgen receptor (AR) expression and a reprograming of AR transcriptional output (1–3); both are accelerated by androgen ablation. AR is a transcription factor that binds a broad range of androgen response elements (ARE) in regulatory regions of genes and intergenic loci. It assembles promoter-specific complexes composed of coactivators, corepressors, and general transcription factors and regulates gene expression (4). Inhibition of AR transcriptional activity eventually leads to an activation of pro-survival Akt signaling both in prostate cancer patients and AR-dependent cell lines, caused largely by downregulation of PHLPP (5) and INPP4B (6). Castration triggers increased expression of AR coactivators, such as TIF2 and CBP that stimulate AR activity at low androgen levels (7). Another adaptation of PCa to castration can be an increased synthesis and/or retention of androgens in tumor cells (8, 9).
NCOR1 and its close homolog, NCOR2, were first discovered as corepressors of unliganded thyroid receptor (TR). NCOR1 has both nuclear and cytoplasmic fractions that have distinct cellular functions. Nuclear – cytoplasmic shuttling is regulated in part by NCOR1 phosphorylation. In HeLa cells, NCOR1 is distributed between cytoplasm and the nucleus; PKA activation causes NCOR1 phosphorylation and its complete nuclear translocation (10). In thyroid tumor cells, cytoplasmic NCOR1 interacts with PI3K regulatory subunit p85a and downregulates PI3K signaling (11). In the nucleus, NCOR1 binds various transcription factors and modulates their activity. Both NCOR1 and NCOR2 are recruited to PSA promoter in an AR-dependent manner (12) and can inhibit both agonist and partial antagonist-dependent AR activity (13). In addition to common activities, these paralogs have unique functions: NCOR2 is required for DNA repair (14), while NCOR1 regulates mitochondrial activity in mouse (15, 16). Recent report suggests that prostate cancer response to castration therapies is dependent on functional AR-NCOR1 complexes. The ubiquitin ligase SIAH2 that clears promoter associated AR-NCOR1 complexes for degradation modulates castration response (1).
In this manuscript we show that response of prostate cancer cell lines LNCaP and LAPC4 to the AR antagonist bicalutamide (casodex) requires expression of NCOR1. We demonstrate that NCOR1 modulates bicalutamide-dependent transcriptome. We show that the loss of NCOR1 changes expression of a number of genes strongly implicated in prostate cancer progression. In agreement with previous reports, we discovered the AR and SIAH2 gene expression signatures among NCOR1 regulated genes (1, 13). We show that the NCOR1 protein levels are significantly lower in prostate cancer than in normal prostate with the corresponding increase in activated Akt. In advanced prostate cancer, NCOR1 signatures are significantly diminished in multiple cohorts. Our data indicate that NCOR1 is required for optimal bicalutamide response in mouse prostate and loss of its function is associated with prostate cancer progression.
Materials and Methods
Cell culture and reagents
LNCaP, LAPC4, PC3, VCaP, DU145, RWPE, PNT1A, and HeLa cell lines were purchased from ATCC collection and maintained in the recommended media. LNCaPAR-V7/pLenti was described previously (17). Media was purchased from Life Technologies (Grand Island, NY). FBS and steroid depleted charcoal stripped serum (css) were purchased from Sigma (St. Louis, MO). Bicalutamide (Casodex) and MDV3100 (enzalutamide) were purchased from Selleckchem (Houston, TX).
siRNA transfections
siRNAs were transfected using Lipofectamine 2000 (Life Technologies) or electroporated in R buffer (Lonza, Allendale, NJ) with the Nucleofector Device (Lonza, Allendale, NJ). TARP1 was downregulated using siRNAs s226672 and s54578 (Life Technologies). NCOR1 was targeted with s201 and s203 (Life Technologies) and on-target SMARTpool L-003518-00-0050 (Thermo Fisher Scientific, Fair Lawn, NJ). CHEK1 specific siRNAs were s504 and s503 (Life Technologies). TARP was downregulated using s226672 and s54578 (Life Technologies). NCOR2, UGT2B15, and UGT2B17 were downregulated using SMARTpools (Thermo Fisher Scientific) and prevalidated siRNA from Life Technologies (s18467). Control noncoding siRNAs were purchased from Life Technologies and Thermo Fisher Scientific.
Western Blotting
For AR, total Akt, pAkt (S473), actin and tubulin, 30 μg of protein were resolved on SDS PAGE and transferred to nitrocellulose membrane. For NCOR1 and NCOR2 50 μg of protein were similarly processed. Membranes were blocked with 2% milk in TBST and incubated with 1:1000 dilution of NCOR1 (Bethyl Laboratories, Montgomery, TX), 1:1000 dilution of NCOR2, 1:1000 of total Akt (Cell Signaling Technology, Danvers, MA), 1:1000 pAkt (S473), 1:2000 dilution of N20 AR (Santa Cruz Biotechnology, Dallas, TX), 1:2000 tubulin (Millipore, Billerica, MA), 1:1000 actin (Sigma) primary antibodies overnight at 4°C. Membranes were washed and incubated with HRP conjugated secondary antibodies (Promega, Madison, WI) for 1 hour and signals were captured on a Gel Logic 2000 imaging system with Carestream Molecular Imaging software (Carestream, Rochester, NY).
CHIP Assay was performed as previously described (6) using AR antibody N-20 (Santa Cruz Biotechnology) and NCOR1 antibody (Bethyl Laboratories).
DNA synthesis and proliferation assays
The [3H]thymidine incorporation was performed exactly as previously described (7). Cellular proliferation and motility was compared using Roche xCelligence RTCA (Roche Diagnostics, Indianapolis, IN) as previously described (6, 18). Cellular impedance in these assays is proportionate to the number of cells covering the E-plate and CIM-plate membrane.
Cell Motility assays were performed using xCelligence RTCA (Roche Diagnostics) as described before (18) and Cellomics Celll Motility Kit as recommended by manufacturer (Thermo Fisher Scientific).
Gene expression array analysis
To determine NCOR1 regulated genes, 2x106 LNCaP cells were transfected with 800 pmol of either noncoding control or NCOR1 SMARTpool siRNA (Thermo Fisher Scientific) using R Buffer (Lonza) and Nucleofector Device (Lonza) exactly as recommended by the manufacturer. After 24 hours later cells were treated with either ethanol or 1μM bicalutamide for 48 hours and RNA was purified using TRIzol reagent (Life Technologies). For DHT regulated gene expression, LNCaP cells were plated in RPMI medium supplemented with 10% css. 24 hours later cells were treated with 10 nM DHT or ethanol vehicle. Cells were harvested 24 or 48 hours after treatment. RNAs were used for expression analysis with Affymetrix 133A 2.0 Arrays at Genomic & RNA Profiling Core, Baylor College of Medicine. Gene expression was marked as changed if the difference was statistically significant (p<0.01) and the change was twofold or more. Gene expression data was deposited into GEO repository, series numbers GSE60721 and GSE60722.
Gene Set Enrichment Analysis (GSEA) was performed using JAVA program (http://www.broadinstitute.org/gsea) as previously described (19). The AR gene signature was generated from genes upregulated in LNCaP after 48 hour DHT treatment (p<0.01) (Supplemental Table 3). The SIAH2 gene signature was generated by extracting genes changed more than twofold (p<0.01) in LNCaP cell following SIAH2 knockdown (1) (Supplemental Table 4).
RNA extraction and quantitative PCR analysis
RNA was extracted using TRI reagent (Thermo Fisher Scientific) as recommended by the manufacturer. RNA was used to prepare cDNA using Verso cDNA expression kit (Thermo Fisher Scientific). Primers and probes are listed in Supplemental Table 5.
Animal Studies
The mice were maintained under standard conditions at Florida International University (FIU) animal facilities. All procedures were reviewed and approved by the Institutional Animal Care and Use Committee at FIU and conducted in accordance with the National Academy of Science Guide for Care and Use of Laboratory Animals. Mice with Ncor1 floxed allele (Ncor1fl) (16) were kindly provided by Dr. Johan Auwerx (Switzerland); B6.Cg-Tg(Pbsn-cre)4Prb/Nci transgenic mice with Cre recombinase gene driven by a derivative of the rat prostate-specific probasin (Pbsn) promoter (20) were obtained from the NCI Mouse Repository (Bethesda, MD). The mice were intercrossed to obtain wild-type Ncor1fl/fl males and Ncor1fl/fl, Pbsn-cre males with conditional inactivation of Ncor1 gene in prostate epithelium. Genotyping of the animals was conducted as described in original publication (20). Ten month-old wild-type and mutant males were treated by a single oral gavage with either oil alone or bicalutamide in sesame oil at 50 mg/kg. Each of four groups contained 7–8 males. Forty eight hours later mice were euthanized and ventral, lateral, and dorsal lobes of the prostate were dissected. Total RNA was extracted with a Trizol reagent (Life Technilogies). cDNA was synthesized using a Verso cDNA kit (Thermo Fisher Scientific). qRT-PCR was performed using primers and probes listed in Supplemental Table 5.
Immunohistochemistry and tissue microarray analysis
Tissue microarray analysis was described previously (21). Briefly, sections were deparaffinized and antigen retrieval performed in 10 mM TRIS-HCl buffer (pH 8). Microarrays were stained using NCOR1 antibody (Millipore) and counterstained with hematoxylin. Automated slide scanner was used to digitize the staining and slides were scored as previously described (22). Immunostaining and analysis for pAkt(S473) was described previously (23).
Association of a NCOR1 silencing signature with metastatic progression
A gene signature of NCOR1 silencing were inferred (t-test, p<0.05, fold change exceeding 1.5x) using the R statistical system. The association of the NCOR1 silencing transcriptome footprint with metastatic progression was evaluated using three PC patient cohorts containing both primary and metastatic prostate cancer: Taylor et al (2) (GSE21034), Varambally et al (24) (GSE3325), and Cai et al (25) (GSE32269). For each gene in NCOR1 silencing transcriptomic response and for each PC specimen, we first computed the z-score for its expression within the cohort, as described previously (2), and next computed the sum z-score for each specimen. Specifically, the z-scores of genes repressed in the NCOR1 silencing signature were subtracted from the z-scores of genes induced in the NCOR1 silencing signature, resulting in a corresponding NCOR1 silencing activity score for each specimen. We next evaluated the difference between the distribution of activity scores in primary prostate cancer patients and metastatic cancer patients using the t-test (p<0.05) as implemented in the R statistical system.
Statistical Methods
Comparison of NCOR1 levels between tissue types was done on paired observations, i.e. patients without matching tumor and normal tissue measurements were excluded from analysis. Wilcoxon Signed Ranks test was used to evaluate the differences due to skewedness of the data. The association of cytoplasmic NCOR1 with pAkt (S473) levels was evaluated using the Spearman Correlation. All analyses were performed using the SPSS 15.0 software package (SPSS for Windows, Version 15.0. Chicago, SPSS Inc.).
Comparisons of mean levels of expression of specific mRNAs and CI indices were done using independent samples Student’s t-test. P-values of <0.05 were considered statistically significant.
Results
NCOR1 expression modulates response to bicalutamide in androgen dependent prostate cancer cell lines
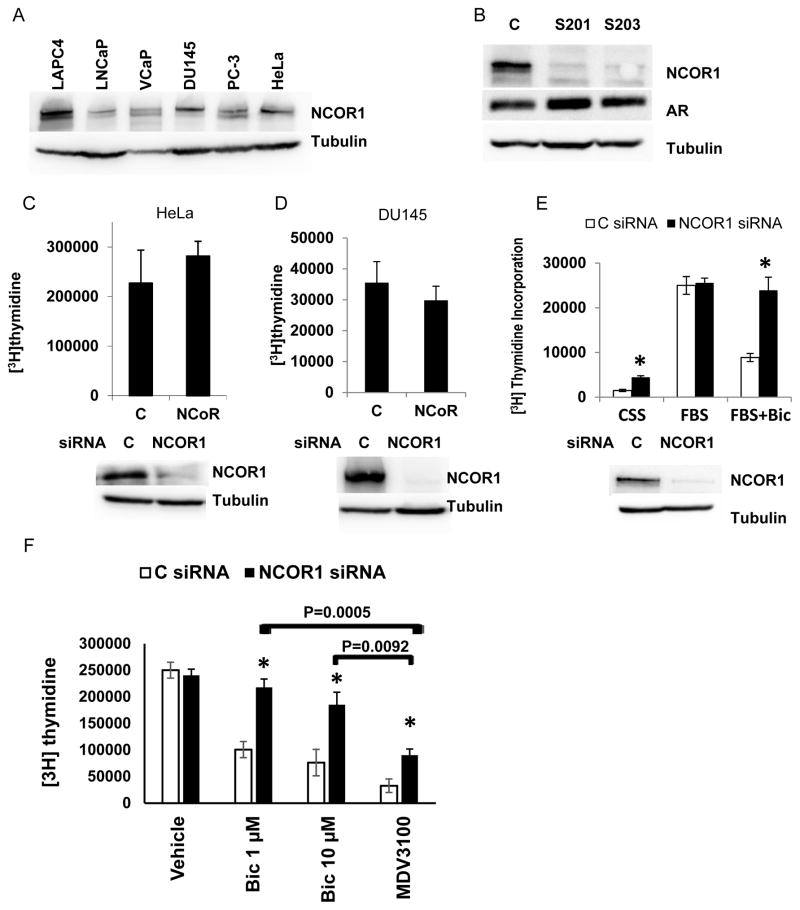
Clearance of AR/NCOR1 complexes from a subset of promoters by SIAH2 interferes with androgen ablation (1). We asked whether direct NCOR1 depletion would similarly alter cellular responses to androgen ablation. First, we compared NCOR1 expression in several AR positive and AR negative cell lines. As shown in Figure 1A and Supplementary Figure 1A, all cell lines expressed NCOR1 protein with somewhat higher expression in VCaP and LAPC4 prostate cancer cell lines. Next, we evaluated whether loss of NCOR1 altered AR protein levels in androgen responsive cell lines, LNCaP and LAPC4. No change of AR expression was observed following NCOR1 loss in both LNCaP (Figure 1B) and LAPC4 cells (Supplemental Figures 1B and 1C). In AR negative cell lines, HeLa and DU145, knockdown of NCOR1 did not affect proliferation of cell lines grown in complete media (Figure 1C and 1D). Depletion of NCOR1 in LNCaP cells increased proliferation in medium supplemented with css (Figure 1E) but similar to HeLa and DU145 had no effect on cells growth in complete medium. A striking increase in proliferation following NCOR1 loss was observed in LNCaP cells treated with bicalutamide (Figure 1E). We next tested whether NCOR1 mediates response to pure AR antagonist MDV3100 (enzalutamide). Both bicalutamide and MDV3100 at 10 μM similarly suppressed DNA synthesis in LNCaP cells transfected with control siRNA (no statistically significant difference). However, MDV3100 inhibited much better then bicalutamide after NCOR1 knockdown (Figure 1F). NCOR1 knockdown stimulated proliferation of LAPC4 cells under all treatment conditions (Supplemental Figure 1D). Opposite to NCOR1, NCOR2 knockdown did not stimulate LNCaP cell growth under any conditions (Supplemental Figures 1E).
Figure 1. NCOR1 suppresses proliferation of LNCaP cells in css supplemented medium and in the presence of bicalutamide.
A. Fifty microgram of protein extracted from LAPC4, LNCaP, VCaP, DU145, PC-3, and HeLa cells were resolved on SDS PAGE and analyzed for NCOR1 and tubulin expression. B. LNCaP cells were transfected with control (C) or two independent NCOR1 specific siRNAs (s201, s203). Cells were harvested 48 hours later and analyzed for NCOR1, AR, and tubulin expression by Western blot analysis. C. HeLa cells were transfected with either control or NCOR1 specific siRNA and cells were grown for 48 hours in complete medium. DNA synthesis was compared by measuring rates of [3H] thymidine incorporation. Cells transfected in parallel were analyzed for levels of NCOR1 by western blotting. D. DU145 cells were transfected and analyzed exactly as in C. E. LNCaP cells were transfected with control or NCOR1 specific siRNA and grown for 24 hours in medium supplemented with css or FBS serum as indicated. Cells were then treated for 24 hours with either vehicle (ethanol) or 1 μM bicalutamide and [3H] thymidine incorporation measured. Cells transfected in parallel and treated with vehicle were analyzed for NCOR1 and tubulin expression by Western blot analysis. F. LNCaP cells were transfected with either Control or NCOR1 specific siRNA and 24 hours later treated with either vehicle (ethanol), 1 μM bicalutamide, 10 μM bicalutamide, or 10 μM MDV3100 for additional 24 hours. In panels B through F experiments were repeated at least three times with two different NCOR1 specific siRNAs. Each point in DNA synthesis assay was done in triplicates. Average and standard deviations are shown. Unless the exact value is shown, * denotes differences between control and NCOR1 siRNA transfected cell with p < 0.05
Bicalutamide also reduced LNCaP motility and NCOR1 knockdown reversed this effect (Supplementary Figure 2). LNCaP remained non-invasive after NCOR1 knockdown (not shown). In PC-3 and DU145 cell lines which express NCOR1 but not AR, NCOR1 knockdown did not alter cell motility and invasion (not shown).
Loss of NCOR1 alters bicalutamide regulated gene expression profile
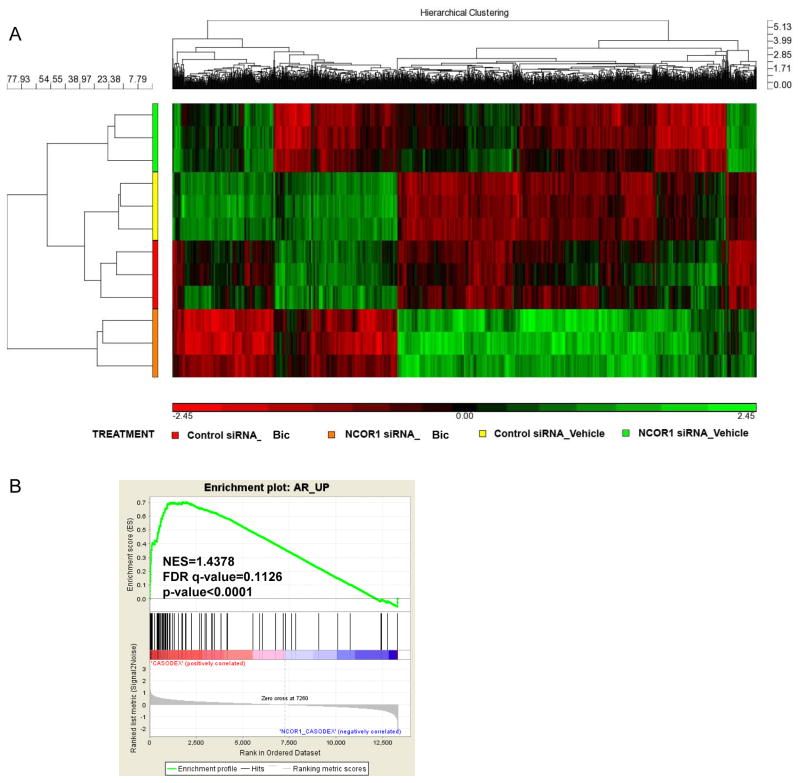
To understand how NCOR1 mediates bicalutamide resistance, we explored changes in gene expression caused by bicalutamide treatment and knockdown of NCOR1 using Affymetrix microarray. LNCaP cells were transfected with either control or NCOR1 specific siRNAs and treated with either ethanol or 1 μM bicalutamide generating 4 groups: Control siRNA_Vehicle (Control siRNA transfected, treated with ethanol), Control siRNA_Bic (Control siRNA transfected, treated with bicalutamide), NCOR1 siRNA_Vehicle (NCOR1 siRNA transfected, treated with ethanol), and NCOR1 siRNA_Bic (NCOR1 siRNA transfected, treated with bicalutamide) (Figure 2A). Gene expression analysis revealed pathways significantly affected by NCOR1 loss with or without bicalutamide treatment (Supplemental Table 1); we observed significant alteration in metabolic, cell signaling, and prostate cancer-associated pathways (Supplemental Table 1). We next examined whether NCOR1 regulated AR transcriptional activity using GSEA analysis. To determine DHT regulated genes, we treated LNCaP cell line with DHT or vehicle and performed microarray analysis. DHT treatment caused significant changes in genes associated with cell cycle, prostate cancer, and biosynthesis of steroids (Supplemental Table 2). A list of genes that was significantly increased by DHT treatment (p<0.01) (Supplemental Table 3) was used to analyze changes in gene expression in Control siRNA_Bic and NCOR1 siRNA_Bic pair (Figure 2B). As expected, AR signature was significantly represented among bicalutamide regulated genes with or without NCOR1 expression (Supplemental Figures 3A and 3B). As seen from Figure 2B, the AR signature was represented among NCOR1 regulated genes (NES=1.4378, FDR q-value=0.1126, p<0.0001). Using a published list of SIAH2 regulated genes in LNCaP cells (1), we created the SIAH2 signature comprised of top SIAH2 up and down regulated genes (Supplemental Table 4). As shown in Supplemental Figure 3C, SIAH2 signature is present among NCOR1 regulated genes in bicalutamide treated LNCaP cells (NES=1.2258, FDR q-value=0.2101, p=0.046).
Figure 2. Modulation of gene expression by NCOR1 and bicalutamide.
A. LNCaP cells were transfected with either noncoding control or NCOR1 siRNA and treated with either ethanol or 1μM bicalutamide for 48 hours. Gene expression was evaluated using Affymetrix 133A 2.0 Arrays and differentially expressed genes were clustered. B. GSEA of the AR target gene signature among NCOR1 regulated genes in bicalutamide treated LNCaP cells (Control siRNA_Bic and NCOR1 siRNA_Bic pair).
NCOR1 regulated genes modulate cellular proliferation and response to castration and bicalutamide treatment
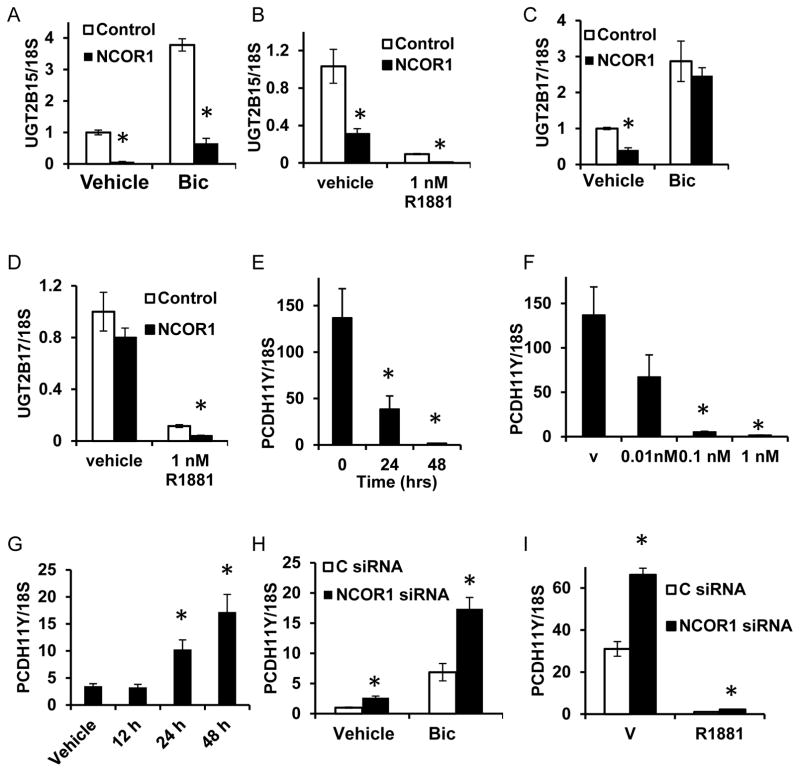
UDP glucuronosyltransferases 2B17 and 2B15 are expressed in luminal epithelium of human prostate and in LNCaP cells (26). They stimulate androgen removal and are implicated in PCa (27). UGT2B15 and UGT2B17 are directly repressed by AR in an agonist-dependent manner (28). Loss of NCOR1 caused decrease in UGT2B15 expression under all treatment conditions (Figures 3A and 3B). Expression of UGT2B17 declined in parallel with NCOR1 loss only in FBS or in css and R1881 supplemented media suggesting that agonist bound AR is needed for this regulation (Figures 3C and 3D). Interestingly, the UGT2B17 and UGT2B15 expression was significantly lower in C4-2 cells, an LNCaP derived cell line that proliferates and produces PSA in steroid depleted medium (Supplemental Figure 4). Consistent with previous reports (28), concomitant knockdown of both UGT2B15 and UGT2B17 resulted in increased AR transcriptional activity (Supplemental Figure 5).
Figure 3. NCOR1 regulates AR repressed genes.
A. LNCaP cells were transfected with either control or NCOR1 specific siRNAs. Cells were plated in medium supplemented with FBS and treated with ethanol (vehicle) or 1μM bicalutamide for 48 hours. RNA was analyzed for UGT2B15 expression. B. Cells were transfected as in (A), plated in css supplemented medium and treated with ethanol vehicle or 1 nM R1881. RNA was analyzed for UGT2B15 expression. C. RNAs form 3A were analyzed for UGT2B17 expression. D. RNAs from 3B were analyzed for expression of UGT2B17. E. LNCaP cells were grown in medium supplemented with css, treated with vehicle or 1nM R1881, and harvested at indicated time points. RNA was extracted and analyzed for expression of PCDH11Y. F. LNCaP cells grown css supplemented medium and treated with vehicle or indicated concentrations of R1881 for 48 hours, RNA extracted, and analyzed for PCDH11Y expression. G. LNCaP cells were grown in FBS supplemented medium and treated with 1 μM bicalutamide and RNA extracted at indicated time points. PCDH11Y expression was analyzed. H. LNCaP cells transfected and treated as in (A) were analyzed for PCDH11Y expression. I. LNCaP cells transfected and treated as in (B) were analyzed for PCDH11Y expression. Every point is an average of three biological replicates ± standard deviation; all experiments were done at least three times and a representative experiment shown. In panels A-D, H, I at least 3 independent NCOR1 siRNAs were used. * indicates difference with corresponding control with p<0.05.
The most upregulated gene upon NCOR1 depletion was protocadherin 11 Y-linked (PCDH11Y). Previous reports showed that PCDH11Y overexpression allowed LNCaP cells proliferate in medium supplemented with css and made them resistant to TPA and serum starvation induced apoptosis (29–31). We showed that PCDH11Y was suppressed by androgens in LNCaP cells at androgen concentrations as low as 0.1 nM (Figure 3E and 3F). Androgen depletion, bicalutamide treatment, and NCOR1 knockdown stimulated PCDH11Y expression (Figures 3G, 3H, and 3I).
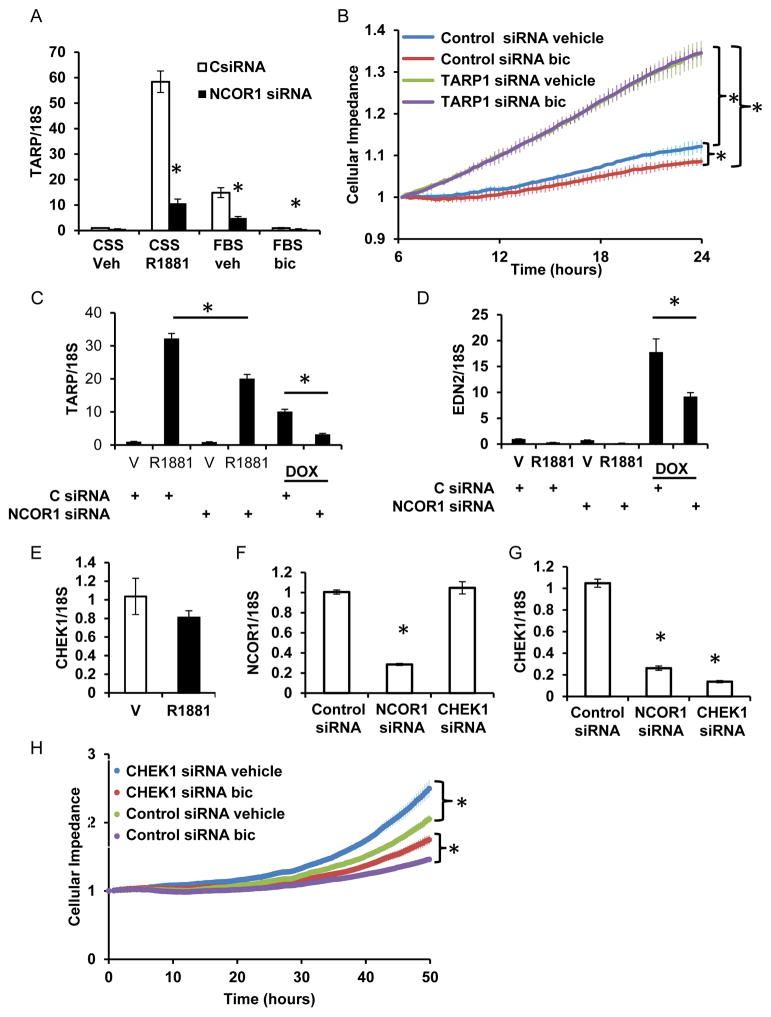
Following NCOR1 knockdown, expression of T-Cell Receptor γ Chain Alternative Reading Frame Protein (TARP) was significantly reduced. As previously reported, we show that TARP expression is induced by R1881 (32) almost 60 fold and suppressed by bicalutamide in medium with FBS (Figure 4A). Intriguingly, TARP expression was consistently lower in medium supplemented with FBS than css plus R1881, suggesting additional modes of regulation by signaling pathways (Figure 4A). NCOR1 knockdown significantly reduced TARP expression both in medium supplemented with css plus R1881 and with FBS. TARP is regulated similarly by NCOR1 and AR in LAPC4 cells (Supplemental Figure 6A). As seen from Figure 4B, TARP depletion increased LNCaP cell proliferation. Moreover, loss of TARP expression reduced LNCaP cells sensitivity to bicalutamide treatment (Figure 4B).
Figure 4. NCOR1 is required for optimal expression of TARP and CHEK1.
A. LNCaP cells transfected with either control or NCOR1 specific siRNAs were placed in medium supplemented with either css or FBS. LNCaP cells grown in css supplemented medium were treated with ethanol vehicle or 1 nM R1881. Cells grown in FBS supplemented medium were treated with ethanol vehicle or 1 μM bicalutamide. RNA was extracted and levels of TARP mRNA compared by qRT PCR. Level of TARP expression in cells grown in css supplemented medium transfected with control siRNA and treated with vehicle was assigned a value of 1 and all other values adjusted accordingly. B. Cells transfected as in A were plated into E-plate of xCelligence analyzer at 10,000 cells per well 12 hours after transfection and treated with vehicle or 1 μM bicalutamide. Cellular impedance was measured every 30 min and average and standard error calculated for each time point. Statistical significance was calculated for the last time point. C-D. Parental LNCaP and LNCaPAR-V7/pLenti cells were transfected with control or NCOR1 siRNA and grown in css supplemented medium. Cells were treated with either vehicle, 1 nM R1881 or doxycycline (Dox) and RNA evaluated for TARP (C) and EDN2 (D) expression. E. LNCaP cells were treated with 1 nM R1881 for 24 hours and RNA extracted and examined for CHEK1 expression. F-G. LNCaP cells grown in complete medium were transfected with either control, CHEK1, or NCOR1 siRNAs and expression of NCOR1 (F) and CHEK1 (G) compared 48 hours after transfection. H. LNCaP cells were transfected with CHEK1 or control siRNA. Twelve hours later 10,000 cells were plated into each well of E-plate of xCELLigence analyzer. Ethanol vehicle or 1 μM bicalutamide were added and changes in cellular impedance measured every 30 min during 50 hours. An average and standard error from 4 biological replicates were calculated for each time point. The statistical significance was calculated for the last cellular impedance measurement between cells transfected with control and NCOR1 siRNAs. * signifies p<0.05. C.
We next tested whether the AR splice variant V7 can induce TARP expression and whether NCOR1 is required for TARP optimal induction using previously described LNCaPAR-V7/pLenti cell line inducibly expressing AR-V7 (17). As seen from Figure 4C, doxycycline induction of AR-V7 expression increased TARP expression levels and NCOR1 knockdown reduced AR and AR-V7 dependent induction of TARP. To analyze whether NCOR1 regulates AR-V7 specific transcription, we measured EDN2 gene expression. As previously reported (17), EDN2 was induced specifically by AR-V7 and optimal induction required NCOR1 expression (Figure 4D).
NCOR1 knockdown resulted in significant loss of expression of checkpoint kinase 1 (CHEK1). CHEK1 kinase was not regulated by androgens in our LNCaP and LAPC4 cell lines (Figure 4E and Supplemental Figure 6B) and no recruitment sites were reported within 30 Kb upstream or downstream of its gene locus (3). Depletion of NCOR1 (Figure 4F) significantly reduced CHEK1 expression in LNCaP cells (Figure 4G). In LAPC4 cells, NCOR1 depletion reduced CHEK1 expression in css (Supplemental Figure 6B). As seen from the Figure 4H, loss of CHEK1 in LNCaP cells stimulated proliferation but did not abolish responsiveness to bicalutamide.
We next tested whether NCOR1 modulates AR activity by changing its recruitment to PSA and INPP4B promoters, genes whose expression is regulated by both AR and NCOR1 (6, 12). We observed NCOR1 recruitment to PSA and INPP4B promoters (6, 12). However, NCOR1 knockdown did not significantly altered AR recruitment to these loci (Supplementary Figure 7).
NCOR1 mediates bicalutamide response in mouse prostate
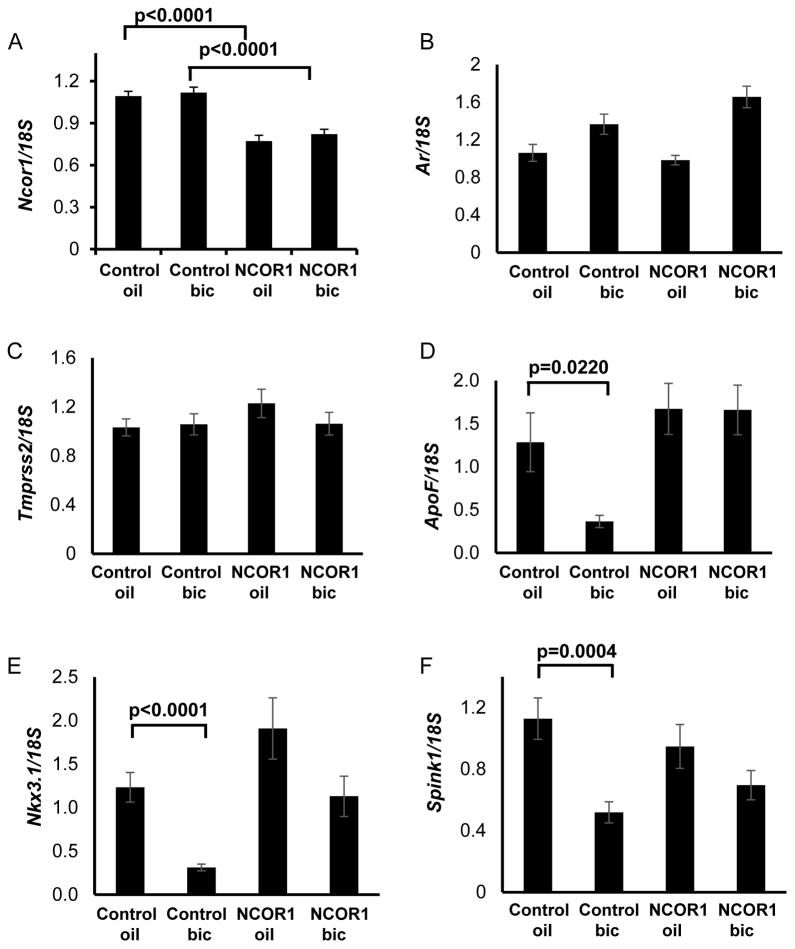
To test whether NCOR1 is required for response to bicalutamide in mouse prostate, we obtained Ncor1fl/fl, Prbn-cre male mice with conditional deletion of Ncor1 gene in prostate epithelium and control Ncor1fl/fl littermate males with functional Ncor1 gene. Mutant males did not display any overt abnormality: total body weight and the weights of testes or seminal vesicles did not differ between two genotypes. Males with both genotypes were fertile and had normal prostates. Levels of Ncor1 expression evaluated by qRT-PCR were significantly lower in males with conditional deletion of Ncor1 (Figure 5A), whereas Ar expression did not change when compared to the control group (Figure 5B). After 48 hour treatment with bicalutamide at 50 mg/kg, no change was detected in Tmprss2 gene expression (Figure 5C), confirming hypothesis that SIAH2/NCOR1/AR complexes do not regulate its expression in mouse prostate (1). Another three SIAH2/AR target genes in mouse prostate are ApoF, Nkx3.1, and Spink1 (1). In control animals, expression of AR target gene, ApoF, was reduced after bicalutamide treatment; downregulation of Ncor1 in mutant prostates abolished bicalutamide-dependent repression of ApoF. In control animals, we observed statistically significant downregulation of Nkx3.1 and Spink1 expression after treatment with bicalutamide. In mutant prostates, changes in expression of these genes were not statistically significant. Total reduction in Ncor1 was modest in prostate, which contains multiple cell types, but there is substantial change in regulation of epithelial specific genes. Thus we observed that the loss of Ncor1 compromised response to bicalutamide for a subset of AR target genes (Figure 5D–5F).
Figure 5. NCOR1 loss in mouse dampens response to bicalutamide of some AR regulated genes.
A. Ncor1fl/fl and Ncor1fl/fl, Pbsn-cre mice were treated with single oral gavage of either sesame oil or 50 mg/kg bicalutamide in sesame oil. Ventral, lateral, and dorsal prostate lobes were dissected 48 hours later and RNA analyzed for Ncor1 by qRT-PCR. B. RNA from A was analyzed by qRT-PCR for Ar (B), Tmprss2 (C), ApoF (D), Nkx3.1 (E), Spink1 (F). Values were normalized for 18S. Significance was evaluated using t-test and all statistically significant differences are marked on bar graphs with corresponding p values.
NCOR1 protein levels decline with prostate cancer progression in prostate cancer patients
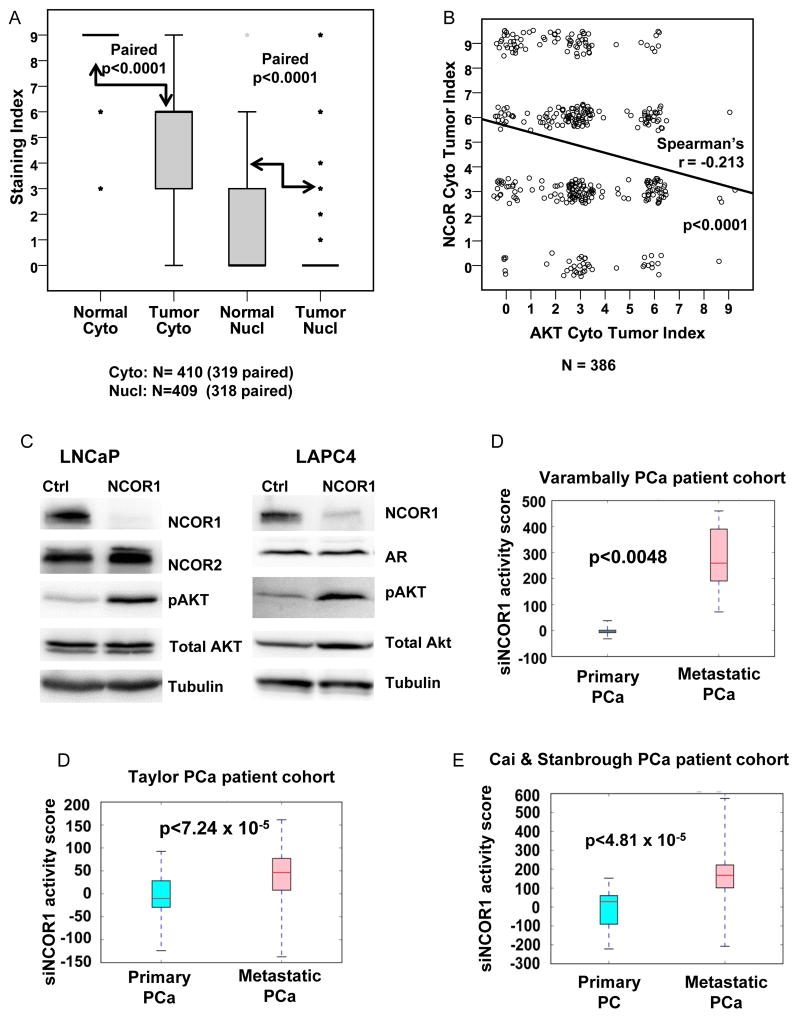
We next analyzed NCOR1 protein levels in normal prostate and prostate cancer samples from radical prostatectomy specimens using tissue microarrays. Consistent with previous observations, strong staining was detected both in cytoplasm and nuclei of prostate luminal epithelium (Supplemental Figure 8). In primary prostate tumors, both cytoplasmic and nuclear levels of NCOR1 were significantly reduced (Figure 6A). We correlated previously reported pAkt levels determined using the same array (23) with NCOR1 levels. In agreement with the observation that NCOR1 inhibits Akt pathway in thyroid cancer cells (11), we detected statistically significant negative correlation between levels of pAkt(S473) and NCOR1 proteins in prostate cancer tissues (Figure 6B). Consistent with this finding, NCOR1 knockdown in LNCaP and LAPC4 cells increased levels of pAkt(S473) (Figure 6C). On the other hand, NCOR1 knockdown did not change NCOR2 protein levels in LNCaP cells (Figure 6C). In addition, no correlation between NCOR1 and NCOR2 was detected in prostate cancer patients using Grasso (primary and CRPC) (33), Stanborough (primary and metastatic) (34) and Taylor (2) Cohorts (Supplementary Table 7).
Figure 6. NCOR1 expression and transcriptional signature in prostate cancer.
A. Tissue microarrays constructed from tissues obtained from radical prostatectomy were analyzed for NCOR1 expression in nuclear and cytoplasmic compartments of luminal epithelium of benign prostate, and in tumor tissue. Wilcoxon Signed-Ranks test was used to compare levels of NCOR1 in these compartments in normal and tumor cells and to calculate p values for each comparison. B. Cytoplasmic staining with phospho-S473 was correlated with NCOR1 staining using a scatter plot with jitter and Spearman’s correlation coefficient calculated. C. LNCaP and LAPC4 cells were transfected with either control or NCOR1 specific siRNA. Cells were maintained in FBS supplemented medium for 48 hours, harvested, and analyzed for NCOR1, NCOR2, AR, total Akt, pAkt (Ser473), and tubulin expression. D-F. NCOR1 silencing signatures primary and metastatic prostate cancer tissues in Taylor et al (GSE21034), Varambally et al (GSE3325), and Cai et al (GSE32269) cohorts.
To determine whether NCOR1 regulated genes change with prostate cancer progression we inferred transcriptome signature of NCOR1 silencing. Using previously employed methodology (2), we evaluated the NCOR1 silencing signature activity in three cohorts containing both primary and metastatic prostate cancer patients: Taylor et al (2), Varambally et al (24), and Cai et al (25). In each cohort, the metastatic patients exhibited significantly higher activity scores for the NCOR1 silencing gene signature (p<0.005 for each cohort), as presented in Figures 6D–F, indicating a diminished activity of NCOR1 in metastatic prostate cancer.
Discussion
NCOR1 is a steroid receptor coregulatory protein, which was first described as a repressor of steroid receptor activity (13, 35). Unique to AR, NCOR1 was shown to repress both agonist- and antagonist-dependent transcriptional activity on androgen response element driven reporters (13). We investigated whether NCOR1 modulates endogenous AR transcriptional activity and plays a role in response to bicalutamide treatment. Using independently derived androgen-dependent cell lines LNCaP and LAPC4 we determined that NCOR1 suppresses cellular proliferation. Interestingly, in the LNCaP cells, which express moderate levels of NCOR1, the loss of NCOR1 stimulated proliferation in both css supplemented medium and in full medium treated with bicalutamide. In the high NCOR1-expressing LAPC4 cells, depletion increased proliferation under all conditions (Supplemental Figure 1D). Importantly, loss of NCOR1 did not alter AR protein levels in either cell line suggesting NCOR1-dependent changes in AR activity rather than AR level. Depletion of NCOR2 had the opposite effect on proliferation of LNCaP cells (Supplemental Figures 1E) confirming previous reports of nonredundant roles for these corepressors. Bicalutamide and the pure AR inhibitor MDV3100 suppressed LNCaP proliferation to a similar degree. However, in the absence of NCOR1 bicalutamide was much less effective than MDV3100 in inhibiting proliferation (Fig 1E,F). This difference may be due to the different mechanisms of action of the two compounds. Bicalutamide allows some AR DNA binding and recruitment of protein complexes that include NCOR1. Depletion of NCOR1 reduces the inhibitory complexes. In contrast MDV3100 bound AR does not bind to the DNA and thus the elimination of NCOR1 should have little or no effect on AR dependent activity under these conditions whereas AR independent actions would be retained. Recently, MDV3100 regulated changes in gene expression have been reported for LNCaP (36). A comparison of MDV3100 transcriptional regulation (GSE44905 and GSE44924) in LNCaP and bicalutamide dependent changes in our experiments showed that approximately 20% of the genes were regulated by both ligands.
To identify NCOR1 regulated genes and cellular pathways that mediate resistance to bicalutamide, we performed whole genome gene expression analysis in LNCaP cells. As seen from Figure 2A, NCOR1 regulates both bicalutamide-dependent and independent transcription. In a previous report, loss of the ubiquitin ligase, SIAH2, reduced LNCaP proliferation, growth of colonies in soft agar, and sphere formation both in the presence and absence of androgens (1). This was presumably mediated by an increase in NCOR1-AR complexes on selected promoters. Indeed, the DHT-upregulated gene signature was highly significantly represented among NCOR1 target genes suggesting a functional interaction between AR and NCOR1. This conclusion is supported by previous reports of physical and functional AR-NCOR1 interactions in the presence of androgens (1, 13) and bicalutamide (12). Significantly, the SIAH2 target gene signature was also significantly represented among NCOR1 regulated genes, confirming the finding that SIAH2 targets NCOR1-AR complexes for degradation in the presence of both androgen and bicalutamide (1).
Depletion of NCOR1 in control and bicalutamide treated LNCaP cells led to changes in expression of genes that play significant roles in PCa progression. The most downregulated gene was NCOR1, confirming successful knockdown. Many, but not all of the genes identified are also androgen receptor regulated. The second most downregulated gene was an androgen induced tumor suppressor, INPP4B, encoding inositol polyphosphate 4-phosphatase, type II. We have previously reported concordant induction of INPP4B by AR and NCOR1 (6). Consistent with this, we found that NCOR1 protein levels decline with PCa progression. We and others reported that INPP4B levels decrease with PCa progression both in primary and especially in metastatic PCa (2, 6) consistent with the positive correlation between NCOR1 and INPP4B expression observed in prostate cancer cell lines (6). NCOR1 loss also caused a significant induction of the PCDH11Y transcript, an androgen repressed target. Androgen deprivation increases PCDH11Y expression in LNCaP cells. High levels of endogenously or exogenously expressed PCDH11Y stimulate androgen-independent LNCaP cell growth (29), confer resistance to apoptosis (31), and activate oncogenic Wnt signaling (30). PCDH11Y mRNA in normal prostates, primary prostate tumors, and untreated PCa is low but increases significantly in CRPC (29), consistent with a negative correlation between NCOR1 and PCDH11Y observed in LNCaP cells. AR-repressed genes, UGT2B15 and UGT2B17, cause DHT conjugation and excretion (28, 37) and are associated with prostate cancer risk (38, 39). We show that NCOR1 is required for optimal UGT2B15 expression under all conditions, while for UGT2B17 expression of NCOR1 was required only in the presence of androgens. Both increased and decreased expression of these enzymes have been reported in prostate cancer (34, 40) potentially due to using samples from prostate tumors at different stages and treatments. It is also possible that levels of UGT2B15 and UGT2B17 enzymes decline only in a subset of prostate cancers as a mechanism to adapt to castration therapy by raising intracellular levels of androgens.
One of the most strongly downregulated transcripts whenNCOR1 is depleted was TARP mRNA, a mitochondrial protein expressed specifically in normal prostate, prostate cancer, and breast cancer. At least four distinct protein coding TARP variants are produced from this locus in response to androgens (32, 41). Splice variant, NM_001003799.1, changed expression with NCOR1 knockdown. TARP was strongly induced by androgens and by NCOR1 expression in LNCaP and LAPC4 cells. We found that TARP depletion significantly increased LNCaP cellular proliferation (Figure 4B) and contributed to bicalutamide resistance. Our finding that AR-V7 also upregulates TARP and that NCOR1 is required for optimal induction suggests that in prostate cancers driven by various AR splice variants, NCOR1 is an important coregulator. This is confirmed by our observation, that the induction of the AR-V7 specific target gene EDN2 requires NCOR1 expression (Figure 5G and 5H). In an earlier report, exogenous expression of TARP increased cellular proliferation in an AR-negative PCa cell line, by increasing CAV1, AGREG, and CXCL1 and suppressing IL-1β expression (42). In LNCaP cells, levels of IL-1β, AREG, and CAV1 are low to undetectable, while loss of CXCL1 causes the opposite effect, increasing cell proliferation and anchorage independent growth (43). TARP may also affect AR signaling contributing to differences in its effect between AR-negative and AR-positive cells.
As shown in Figure 4, NCOR1 also regulates CHEK1, and this regulation is androgen-independent. CHEK1 is required for cell cycle arrest in response to DNA damage and may also regulate cell cycle (44). A decline in CHEK1 expression was reported in prostate cancer (45) and compound loss of CHEK1 and PTEN triggers progression from high grade prostatic intraepithelial neoplasia to invasive prostate carcinoma (46). CHEK1 knockdown increased LNCaP cellular proliferation but bicalutamide was still able to suppress proliferation (Figure 4H). It is possible that in the NCOR1 negative prostate tumors a decline in CHEK1 might contribute to more aggressive proliferative phenotype.
Additional confirmation of a role for NCOR1 in bicalutamide action was obtained using mutant mice with conditional prostate-specific Ncor1 gene deletion. We chose 48 hour time point for bicalutamide treatment because it would inhibit AR action without significant cell death in prostate epithelial compartment. Importantly, Ncor1 expression is not exclusive to luminal epithelium of mouse prostate, the site of Pbsn-cre expression. Similar to what we saw in LNCaP and LAPC4 cells, NCOR1 deletion in mouse prostate did not reduce AR mRNA levels. However, some of the AR target genes that are highly expressed in prostate epithelium (1, 47) lost their response to bicalutamide in mutant mice. In agreement with the previous report that Siah2 regulates gene expression by removing NCOR1 complexes from a sub-set of genes (1), Siah2 regulated AR target genes, ApoF, Nkx3.1, and Spink1, lost their ability to respond to bicalutamide treatment in Ncor1 knockout mouse prostates, while Tmprss2 expression did not change.
NCOR1 has emerged as an important regulatory protein in prostate and other cancers. Analysis of changes in signaling pathways with PCa progression by Taylor et al. showed that NCOR1 is lost in 8% of primary and 16% metastatic prostate tumors due to deletions, loss of expression and/or mutations (2, 33, 48). A comparison of high grade untreated localized prostate cancers and lethal metastatic CRPC showed a high level of NCOR1 somatic mutations and copy number alterations in CRPC while none were detected in untreated tumors (33). Additional evidence for NCOR1’s contribution to the development of resistance to anti-hormonal treatments was found in breast cancer. In ERα-positive breast tumors, low NCOR1 expression is an independent predictor of tamoxifen resistance (49). Loss of NCOR1 at the protein level also correlated with acquired tamoxifen resistance in mouse models of breast cancer (50) and with breast cancer recurrence in human patients (51). Tamborero et al. identified NCOR1 as a high confidence cancer driver gene using three unbiased screening techniques: MuSIC-SMG, OncodriveFM, and Active Driver (48). We have compared NCOR1 protein expression in normal prostate and prostate cancer in over 400 paired tissues (Figure 6). NCOR1 was detected mostly in the prostate secretory epithelium (Supplemental Figure 8). Consistent with previous reports we observed NCOR1 staining in both the cytoplasm and the nuclei of the cells (10, 11) and levels significantly decline in both compartments in malignant cells (Figure 6A). Significantly, the NCOR1-regulated transcriptional signature declines in advanced prostate cancer in multiple cohorts (Figures 6D–F). Similar to thyroid cancer cells (11), we observe a highly significant negative correlation between cytoplasmic levels of NCOR1 and levels of pAkt staining in PCa tissues (Figure 6B). These data suggest that a decline in NCOR1 protein level occurs with PCa progression and may contribute to patients’ response to antiandrogen treatments.
Our data illustrate the importance of NCOR1 in prostate response to bicalutamide in two cell based and in Ncor1-deficient mice in vivo. Together with the observation that NCOR1 levels decline during prostate cancer progression in men, we demonstrate that NCOR1 is involved in development of resistance to bicalutamide treatment and provide an additional mechanism to explain the better response to MDV3100.
Supplementary Material
Translational Relevance.
Castration therapies are the standard-of-care treatment for men with advanced prostate cancer. The short and long term morbidities of this treatment are substantial. In addition, time lost for ineffective treatment can lead to the progression of the disease. With the advances of personalized medicine, it will be possible to more precisely characterize the tumor type for individual patients. Thus markers that would predict therapeutic response are important. We examined the role of androgen receptor coregulator NCOR1 in response to antiandrogen treatments. In multiple data sets, NCOR1 mutations and loss of expression has been reported and, in this manuscript, we show decline in NCOR1 protein and transcriptional output in prostate cancer tissues. We present data that suggests that patients with the loss of NCOR1 function due to mutation or loss of protein expression may be predictive of resistance to castration therapy in AR expressing tumors.
Footnotes
Conflicts of interest: None
Financial support: This work was supported by R15 CA179287-01A1 (IA)
References
- 1.Qi J, Tripathi M, Mishra R, Sahgal N, Fazil L, Ettinger S, et al. The e3 ubiquitin ligase siah2 contributes to castration-resistant prostate cancer by regulation of androgen receptor transcriptional activity. Cancer Cell. 2013;23:332–46. doi: 10.1016/j.ccr.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agoulnik IU, Weigel NL. Androgen receptor coactivators and prostate cancer. Adv Exp Med Biol. 2008;617:245–55. doi: 10.1007/978-0-387-69080-3_23. [DOI] [PubMed] [Google Scholar]
- 5.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hodgson MC, Shao LJ, Frolov A, Li R, Peterson LE, Ayala G, et al. Decreased Expression and Androgen Regulation of the Tumor Suppressor Gene INPP4B in Prostate Cancer. Cancer Res. 2011;71:572–82. doi: 10.1158/0008-5472.CAN-10-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agoulnik IU, Vaid A, Nakka M, Alvarado M, Bingman WE, 3rd, Erdem H, et al. Androgens modulate expression of transcription intermediary factor 2, an androgen receptor coactivator whose expression level correlates with early biochemical recurrence in prostate cancer. Cancer Res. 2006;66:10594–602. doi: 10.1158/0008-5472.CAN-06-1023. [DOI] [PubMed] [Google Scholar]
- 8.Locke JA, Guns ES, Lubik AA, Adomat HH, Hendy SC, Wood CA, et al. Androgen levels increase by intratumoral de novo steroidogenesis during progression of castration-resistant prostate cancer. Cancer Res. 2008;68:6407–15. doi: 10.1158/0008-5472.CAN-07-5997. [DOI] [PubMed] [Google Scholar]
- 9.Mohler JL, Gregory CW, Ford OH, 3rd, Kim D, Weaver CM, Petrusz P, et al. The androgen axis in recurrent prostate cancer. Clin Cancer Res. 2004;10:440–8. doi: 10.1158/1078-0432.ccr-1146-03. [DOI] [PubMed] [Google Scholar]
- 10.Choi HK, Yoo JY, Jeong MH, Park SY, Shin DM, Jang SW, et al. Protein Kinase A phosphorylates NCoR to enhance its nuclear translocation and repressive function in human prostate cancer cells. J Cell Physiol. 2013;228:1159–65. doi: 10.1002/jcp.24269. [DOI] [PubMed] [Google Scholar]
- 11.Furuya F, Guigon CJ, Zhao L, Lu C, Hanover JA, Cheng SY. Nuclear receptor corepressor is a novel regulator of phosphatidylinositol 3-kinase signaling. Mol Cell Biol. 2007;27:6116–26. doi: 10.1128/MCB.00900-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol Cell. 2002;9:601–10. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- 13.Agoulnik IU, Krause WC, Bingman WE, 3rd, Rahman HT, Amrikachi M, Ayala GE, et al. Repressors of androgen and progesterone receptor action. The Journal of biological chemistry. 2003;278:31136–48. doi: 10.1074/jbc.M305153200. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Palmer C, Alenghat T, Li Y, Kao G, Lazar MA. The corepressor silencing mediator for retinoid and thyroid hormone receptor facilitates cellular recovery from DNA double-strand breaks. Cancer Res. 2006;66:9316–22. doi: 10.1158/0008-5472.CAN-06-1902. [DOI] [PubMed] [Google Scholar]
- 15.Catic A, Suh CY, Hill CT, Daheron L, Henkel T, Orford KW, et al. Genome-wide Map of Nuclear Protein Degradation Shows NCoR1 Turnover as a Key to Mitochondrial Gene Regulation. Cell. 2013;155:1380–95. doi: 10.1016/j.cell.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamamoto H, Williams EG, Mouchiroud L, Canto C, Fan W, Downes M, et al. NCoR1 is a conserved physiological modulator of muscle mass and oxidative function. Cell. 2011;147:827–39. doi: 10.1016/j.cell.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krause WC, Shafi AA, Nakka M, Weigel NL. Androgen receptor and its splice variant, AR-V7, differentially regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int J Biochem Cell Biol. 2014;54:49–59. doi: 10.1016/j.biocel.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodgson MC, Deryugina EI, Suarez E, Lopez SM, Lin D, Xue H, et al. INPP4B suppresses prostate cancer cell invasion. Cell communication and signaling : CCS. 2014;12:61. doi: 10.1186/s12964-014-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu X, Wu J, Huang J, Powell WC, Zhang J, Matusik RJ, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–9. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 21.Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003;63:6244–51. [PubMed] [Google Scholar]
- 22.Ding Y, He D, Florentin D, Frolov A, Hilsenbeck S, Ittmann M, et al. Semaphorin 4F as a critical regulator of neuroepithelial interactions and a biomarker of aggressive prostate cancer. Clin Cancer Res. 2013;19:6101–11. doi: 10.1158/1078-0432.CCR-12-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ayala G, Thompson T, Yang G, Frolov A, Li R, Scardino P, et al. High levels of phosphorylated form of Akt-1 in prostate cancer and non-neoplastic prostate tissues are strong predictors of biochemical recurrence. Clin Cancer Res. 2004;10:6572–8. doi: 10.1158/1078-0432.CCR-04-0477. [DOI] [PubMed] [Google Scholar]
- 24.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 25.Cai C, Wang H, He HH, Chen S, He L, Ma F, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. The Journal of clinical investigation. 2013;123:1109–22. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chouinard S, Pelletier G, Belanger A, Barbier O. Cellular specific expression of the androgen-conjugating enzymes UGT2B15 and UGT2B17 in the human prostate epithelium. Endocr Res. 2004;30:717–25. doi: 10.1081/erc-200044014. [DOI] [PubMed] [Google Scholar]
- 27.Grosse L, Paquet S, Caron P, Fazli L, Rennie PS, Belanger A, et al. Androgen glucuronidation: an unexpected target for androgen deprivation therapy, with prognosis and diagnostic implications. Cancer Res. 2013;73:6963–71. doi: 10.1158/0008-5472.CAN-13-1462. [DOI] [PubMed] [Google Scholar]
- 28.Bao BY, Chuang BF, Wang Q, Sartor O, Balk SP, Brown M, et al. Androgen receptor mediates the expression of UDP-glucuronosyltransferase 2 B15 and B17 genes. Prostate. 2008;68:839–48. doi: 10.1002/pros.20749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Terry S, Queires L, Gil-Diez-de-Medina S, Chen MW, de la Taille A, Allory Y, et al. Protocadherin-PC promotes androgen-independent prostate cancer cell growth. Prostate. 2006;66:1100–13. doi: 10.1002/pros.20446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Chen MW, Terry S, Vacherot F, Chopin DK, Bemis DL, et al. A human- and male-specific protocadherin that acts through the wnt signaling pathway to induce neuroendocrine transdifferentiation of prostate cancer cells. Cancer Res. 2005;65:5263–71. doi: 10.1158/0008-5472.CAN-05-0162. [DOI] [PubMed] [Google Scholar]
- 31.Chen MW, Vacherot F, De La Taille A, Gil-Diez-De-Medina S, Shen R, Friedman RA, et al. The emergence of protocadherin-PC expression during the acquisition of apoptosis-resistance by prostate cancer cells. Oncogene. 2002;21:7861–71. doi: 10.1038/sj.onc.1205991. [DOI] [PubMed] [Google Scholar]
- 32.Cheng WS, Giandomenico V, Pastan I, Essand M. Characterization of the androgen-regulated prostate-specific T cell receptor gamma-chain alternate reading frame protein (TARP) promoter. Endocrinology. 2003;144:3433–40. doi: 10.1210/en.2003-0121. [DOI] [PubMed] [Google Scholar]
- 33.Grasso CS, Wu YM, Robinson DR, Cao X, Dhanasekaran SM, Khan AP, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–43. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanbrough M, Bubley GJ, Ross K, Golub TR, Rubin MA, Penning TM, et al. Increased expression of genes converting adrenal androgens to testosterone in androgen-independent prostate cancer. Cancer Res. 2006;66:2815–25. doi: 10.1158/0008-5472.CAN-05-4000. [DOI] [PubMed] [Google Scholar]
- 35.Smith CL, Nawaz Z, O’Malley BW. Coactivator and corepressor regulation of the agonist/antagonist activity of the mixed antiestrogen, 4-hydroxytamoxifen. Mol Endocrinol. 1997;11:657–66. doi: 10.1210/mend.11.6.0009. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z, Lan X, Thomas-Ahner JM, Wu D, Liu X, Ye Z, et al. Agonist and antagonist switch DNA motifs recognized by human androgen receptor in prostate cancer. EMBO J. 2015;34:502–16. doi: 10.15252/embj.201490306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chouinard S, Barbier O, Belanger A. UDP-glucuronosyltransferase 2B15 (UGT2B15) and UGT2B17 enzymes are major determinants of the androgen response in prostate cancer LNCaP cells. The Journal of biological chemistry. 2007;282:33466–74. doi: 10.1074/jbc.M703370200. [DOI] [PubMed] [Google Scholar]
- 38.Grant DJ, Hoyo C, Oliver SD, Gerber L, Shuler K, Calloway E, et al. Association of uridine diphosphate-glucuronosyltransferase 2B gene variants with serum glucuronide levels and prostate cancer risk. Genet Test Mol Biomarkers. 2013;17:3–9. doi: 10.1089/gtmb.2012.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karypidis AH, Olsson M, Andersson SO, Rane A, Ekstrom L. Deletion polymorphism of the UGT2B17 gene is associated with increased risk for prostate cancer and correlated to gene expression in the prostate. Pharmacogenomics J. 2008;8:147–51. doi: 10.1038/sj.tpj.6500449. [DOI] [PubMed] [Google Scholar]
- 40.Paquet S, Fazli L, Grosse L, Verreault M, Tetu B, Rennie PS, et al. Differential expression of the androgen-conjugating UGT2B15 and UGT2B17 enzymes in prostate tumor cells during cancer progression. J Clin Endocrinol Metab. 2012;97:E428–32. doi: 10.1210/jc.2011-2064. [DOI] [PubMed] [Google Scholar]
- 41.Wolfgang CD, Essand M, Vincent JJ, Lee B, Pastan I. TARP: a nuclear protein expressed in prostate and breast cancer cells derived from an alternate reading frame of the T cell receptor gamma chain locus. Proc Natl Acad Sci U S A. 2000;97:9437–42. doi: 10.1073/pnas.160270597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolfgang CD, Essand M, Lee B, Pastan I. T-cell receptor gamma chain alternate reading frame protein (TARP) expression in prostate cancer cells leads to an increased growth rate and induction of caveolins and amphiregulin. Cancer Res. 2001;61:8122–6. [PubMed] [Google Scholar]
- 43.Liu XF, Xiang L, Zhang Y, Becker KG, Bera TK, Pastan I. CAPC negatively regulates NF-kappaB activation and suppresses tumor growth and metastasis. Oncogene. 2012;31:1673–82. doi: 10.1038/onc.2011.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, et al. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- 45.Kilpinen S, Ojala K, Kallioniemi O. Analysis of kinase gene expression patterns across 5681 human tissue samples reveals functional genomic taxonomy of the kinome. PLoS One. 2010;5:e15068. doi: 10.1371/journal.pone.0015068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lunardi A, Varmeh S, Chen M, Taulli R, Guarnerio J, Ala U, et al. Suppression of CHK1 by ETS Family Members Promotes DNA Damage Response Bypass and Tumorigenesis. Cancer Discov. 2015;5:550–63. doi: 10.1158/2159-8290.CD-13-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma C, Yoshioka M, Boivin A, Gan L, Takase Y, Labrie F, et al. Atlas of dihydrotestosterone actions on the transcriptome of prostate in vivo. Prostate. 2009;69:293–316. doi: 10.1002/pros.20883. [DOI] [PubMed] [Google Scholar]
- 48.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Kandoth C, Reimand J, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Girault I, Lerebours F, Amarir S, Tozlu S, Tubiana-Hulin M, Lidereau R, et al. Expression analysis of estrogen receptor alpha coregulators in breast carcinoma: evidence that NCOR1 expression is predictive of the response to tamoxifen. Clin Cancer Res. 2003;9:1259–66. [PubMed] [Google Scholar]
- 50.Lavinsky RM, Jepsen K, Heinzel T, Torchia J, Mullen TM, Schiff R, et al. Diverse signaling pathways modulate nuclear receptor recruitment of N-CoR and SMRT complexes. Proc Natl Acad Sci U S A. 1998;95:2920–5. doi: 10.1073/pnas.95.6.2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kurebayashi J, Otsuki T, Kunisue H, Tanaka K, Yamamoto S, Sonoo H. Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res. 2000;6:512–8. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.