Abstract
Prologue ‘As the study of natural science advances, the language of scientific description may be greatly simplified and abridged. This has already been done by Linneaus and may be carried still further by other invention. The descriptions of natural orders and genera may be reduced to short definitions, and employment of signs, somewhat in the manner of algebra, instead of long descriptions. It is more easy to conceive this, than it is to conceive with what facility, and in how short a time, a knowledge of all the objects of natural history may ultimately be acquired; and that which is now considered learning and science, and confined to a few specially devoted to it, may at length be universally possessed in every civilized country and in every rank of life’. J. C. Louden 1829. Magazine of natural history, vol. 1.
This article is part of the themed issue ‘From DNA barcodes to biomes’.
Keywords: DNA barcoding, species, genomics, biodiversity
1. Introduction
For more than two centuries, biodiversity science has focused on the inventory of species, on probing their relationships and on clarifying the factors responsible for their diversification. The sheer diversity of life, the fact that millions of species of multi-cellular organisms await description, is a serious barrier to scientific progress. Moreover, morphological approaches cannot enable the census of these species in a timely or affordable fashion; the cost of describing five million animal species has been estimated at $250 billion and as requiring six centuries [1]. Eleven years ago, Savolainen et al. [2] considered the possibility that DNA barcoding might allow the encyclopedia of life to be written in decades rather than a millennium. The present issue considers progress towards this goal and provides a glimpse of the ways in which DNA barcoding is transforming biodiversity science. The 16 articles included in this issue derive from plenary presentations at the 6th International Barcode of Life Conference held in August 2015. When coupled with the conference abstracts [3], it is clear that DNA barcoding is contributing to rapid scientific progress on diverse fronts. This outcome might not have been predicted just a decade ago.
When the Natural History Museum in London hosted the 1st International Barcode of Life Conference in 2005, it anticipated a lively discussion with an uncertain outcome. Some researchers involved in large-scale biodiversity inventories viewed DNA barcoding as a breakthrough [4–6], but endorsements from other segments of the community were restrained. Because prior genetic approaches [7–9] had modest impact on their workflows, some taxonomists anticipated that DNA barcoding would also have limited influence [10]. Others [11] highlighted the risks in basing taxonomic decisions on sequence variation in a single gene, noting the potential complexities introduced by paraphyly and polyphyly [12] and by the possible discordances between gene trees and species trees [13]. These concerns could only be addressed by examining the efficacy of DNA barcoding in varied taxonomic assemblages in diverse environments. About five million specimens have now been analysed, and these results indicate that DNA barcodes can discriminate most species.
The balance of this introductory article considers the factors that were important in mobilizing a DNA barcode research community, and the issues that needed consideration in construction of the reference library. It also addresses the effectiveness of DNA barcoding as a tool for specimen identification and species discovery before examining the scientific impacts of work in this field and future prospects.
2. Community mobilization
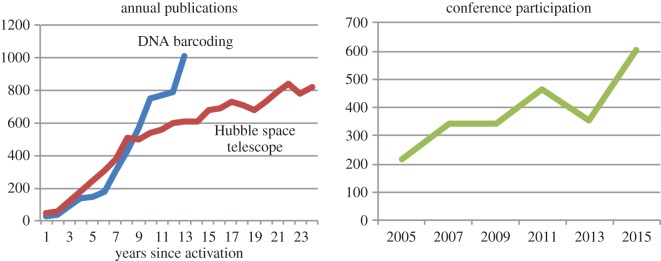
More than 1000 publications on DNA barcoding appeared in 2015, a count higher than that for many other major scientific programmes (figure 1). The growth in interest and global involvements in this field [14] are further signalled by the increasing participation in the International Barcode of Life Conferences (figure 1); 600 researchers from 55 nations joined the latest meeting.
Figure 1.
Metrics showing the growth of the DNA barcode research community through time as measured by the yearly number of publications on DNA barcoding and by the number of participants in the International Barcode of Life Conferences. Data on publication activity by the Hubble Space Telescope research community are presented for comparison.
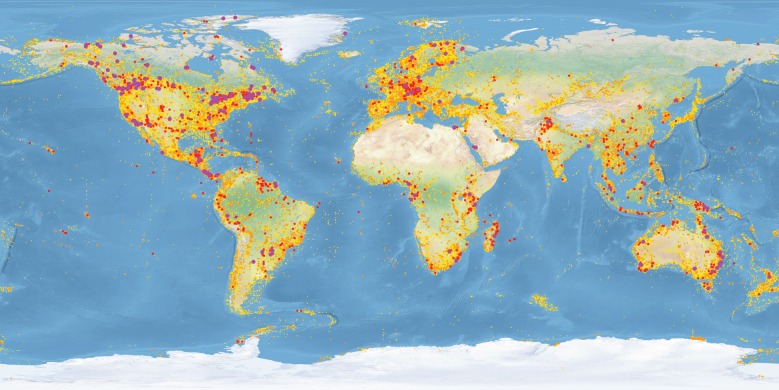
DNA barcoding has rapidly become the largest research collaboration in biodiversity science. What provoked this? The establishment of the Consortium for the Barcode of Life (CBOL) in 2004 was a key development. It galvanized the community and quickly organized meetings to advance understanding of DNA barcoding, including the International Conferences in London (2005), Taipei (2007), Mexico City (2009) and Adelaide (2011). CBOL also worked with researchers to achieve consensus on the best DNA barcode marker(s) for each eukaryote kingdom [15–18]. While these activities were critical, there was also a great need to clarify the efficacy of DNA barcoding. The Gordon and Betty Moore Foundation sponsored the first large-scale evaluations [19,20], and the flow of data was reinforced with activation of the Canadian Barcode of Life Network in 2005 [21]. By 2007, it was recognized that the development of a global DNA barcode reference library required a broad alliance, stimulating plans for iBOL, the International Barcode of Life project (www.iBOL.org), which aimed to deliver barcode records for 500 000 species within 5 years of activation. Because substantial resources (more than $100 million) were required, and plans called for research nodes in 25 nations, it took 3 years before fundraising and network development were sufficiently advanced for its activation. National barcode networks were ultimately established in 11 countries (Argentina, Austria, Brazil, Canada, China, Finland, Germany, Mexico, Norway, South Africa and Switzerland), but they emerged asynchronously; those in Argentina and Mexico launched in 2008 and 2009, while the Austrian and Norwegian networks began work in 2014. Researchers in other countries (e.g. Costa Rica, France, Kenya, The Netherlands, UK and USA) made major contributions without a formal network. Despite this organizational fluidity and varied activation dates, iBOL met its target for species coverage in August 2015 (figure 2).
Figure 2.
Heat map of the five million DNA barcode records in July 2016. Purple circles >1000 records, red >100 records, orange >10 records, yellow 1–10 records.
Although CBOL, iBOL and the national networks stimulated the rapid rise of DNA barcoding, these grant-funded entities had finite lifespans. As a result, the research community needed to assume certain activities initiated by CBOL, such as the International Barcode of Life Conferences, and responsibility for their organization transitioned to countries with lead roles in iBOL (China, 2013; Canada, 2015; South Africa, 2017). Looking to the future, there will be an ongoing need for a research consortium to ensure that barcode coverage is extended efficiently and to aid the acquisition of the funds required for this purpose.
3. Constructing the DNA barcode reference library
Although DNA barcoding is conceptually simple, the assembly and curation of sequence information from one or more standard gene regions across millions of species is challenging. Over the past decade, improved laboratory protocols have simplified barcode acquisition [22–24]. As a result, five million specimens were analysed by July 2016, providing coverage for some 60 000 plant and 450 000 animal species, although many of the later taxa were undescribed. As these totals likely represent no more than 20 and 5% of the species in these kingdoms, much work remains. However, achieving the level of barcode coverage required for an effective identification system [25] is a realistic goal for the biotas of Europe and North America by 2025 [26]. Completion of the global library might require the analysis of 100 million specimens, presuming a target of 10× coverage per species, but it could be completed in a few decades with adequate resources. Because achieving a well-parametrized global library will require specimens, sequence analysis and data management, the rest of this section considers these matters in more detail.
(a). Sourcing specimens
The most expensive component in DNA barcode analysis is specimen acquisition. Obtaining sets of many thousands of voucher specimens with expert annotation requires enormous effort. Viewed from this perspective, natural history collections are a valuable legacy, especially herbaria as barcode recovery is high, even from specimens a century old [27]. Some animal groups are challenging, particularly those preserved in formaldehyde [28], but others are more tractable [24]. Because of their greater sensitivity, high-throughput sequencers (HTS) allow barcode recovery from specimens recalcitrant to Sanger analysis [29]. Their use to obtain barcodes from type specimens is particularly important as the resulting data serve to create a searchable index of specimens linked to binomials, facilitating the correct application of names and the resolution of synonymies [30–32]. While the analysis of museum specimens will extend barcode coverage for named species, new collections will be required for groups that have seen little taxonomic attention and for under-collected regions of the planet. However, as evidenced over the past decade, the biodiversity science community has a strong capacity to make collections. In considering the task ahead, it is important to emphasize that a highly effective identification system is achieved long before the last species is analysed because most surveys encounter common, widely distributed taxa rather than those that are either very rare or narrow endemics. Moreover, when one of the latter taxa is encountered, its presence is ordinarily signalled by its assignment to a new barcode cluster, provoking referral of the specimens to a taxonomist working on the group, allowing its subsequent inclusion in the barcode reference library.
(b). Acquiring sequences
Presuming access to specimens, their barcode sequences must be recovered. As polymerase chain reaction (PCR) is employed to amplify the barcode region from genomic DNA, analysis is disrupted if amplicons are generated from pseudogenes [33] or from bacterial and fungal endosymbionts [34]. Pseudogenes have proven an infrequent problem because they are typically shorter [35] and are present in lower copy numbers than the barcode regions targeted for analysis. Sequences from bacterial endosymbionts are encountered more commonly [36], but they are easily excluded during data validation. Sequences from fungal endosymbionts can fail to be recognized, especially when the barcode is a DNA region, such as the internal transcribed spacers (ITS) of nuclear ribosomal DNA, which cannot be aligned across phyla [37], but spurious records will be excised as valid entries are acquired for each species.
Aside from problems introduced by the recovery of non-target DNA, library construction is also impeded if PCR fails to generate an amplicon, a situation that arises because no primer set is truly universal. This is particularly pertinent to maturase K (matK), one of the two core barcode markers for vascular plants, as existing primer sets have high failure rates [38]. Recovery of cytochrome c oxidase subunit I (COI) from animals is more reliable, although each primer set targets a particular constellation of species (e.g. fishes, insects). While these primer sets are effective for their designated group, they occasionally fail, especially in fast-evolving lineages [39]. Although amplification success could be raised by adopting a more conserved gene region, this would reduce taxonomic resolution [40]. Moreover, the difficulty in recovery of COI amplicons has been exaggerated by in silico predictions of primer binding [41]. In practice, primer sets employed for animals have strong performance with, for example, a single primer set generating sequences from 88% of specimens in 579 insect families [39]. This result and those from similar studies on other groups of animals indicate that amplification failure is too infrequent to justify the shift to a more conserved gene region. However, there is a need for further work on primer design to conquer problems in certain groups, especially some marine taxa.
Presuming amplicon recovery, sequence characterization is the next step in the analytical chain. The barcode standard currently requires bidirectional Sanger analysis of each amplicon, an approach that generates a high fidelity, full-length read. In practice, unidirectional analysis delivers reads that meet key elements of the standard, suggesting the possibility of relaxing the requirement for bidirectional coverage. Aside from considering this adjustment, the barcode standard needs to be revisited in light of the very different attributes of the sequence records generated by HTS. It is certain that the volume of data generated by these platforms will rapidly expand because they enable the barcode characterization of bulk DNA extracts, a key advance for environmental monitoring [42–49]. A shift to HTS for barcode recovery from single specimens might also reduce costs for library construction [50,51], but substantial work will be needed to optimize data quality and bioinformatics protocols [52]. Certainly, for the foreseeable future, the barcode standard needs enough flexibility to recognize the validity of records generated by different sequencing platforms so long as they satisfy the requirements for sequence quality, length and verifiability.
(c). Data management
The early development of BOLD, the Barcode of Life Data System, has been critical for the storage, validation and analysis of DNA barcode records generated via Sanger sequencing [53]. Because it couples specimen and sequence information, this platform plays an increasingly important role as data volumes expand. Moreover, BOLD is gaining the capabilities needed to support large-scale biodiversity analyses. For example, its Barcode Index Number (BIN) system automates the delineation of molecular operational taxonomic units [54] as proxies for animal species and embeds each new BIN in a persistent registry [55]. Work is also underway to allow BOLD to automatically position new BINs in the Linnaean hierarchy by exploiting taxonomic information linked to barcode records from known species. There will be a need for sustained vigilance to ensure that specimens providing barcode records have reliable taxonomic assignments. While major errors are easily recognized, misidentifications of closely allied species require careful examination to recognize and correct [56,57]. The development of a barcode library for known species is aided by ongoing efforts to create a registry of valid species names [58], but ‘dark taxa’, those only known from their DNA barcode sequences, will represent an increasingly important challenge [32]. Although it is ultimately desirable to have all specimens with a sequence, a name and associated data, the fact that BINs provide a stable framework for subsequent annotation and data enrichment is a major breakthrough for tackling poorly known mega-diverse groups [39]. Aside from the well-recognized challenges in the storage and analysis of the data generated by HTS, studies enabled by this technology will undoubtedly illuminate massive numbers of dark taxa.
(d). Data sharing and release
The traditional model has been to ‘publish and then release data’. However, wider cultural scientific changes focusing on building infrastructure and access to big-data have driven a shift to rapid data release and sharing. DNA barcoding has tracked this change, transforming from a series of large-connected research projects into a community movement using BOLD as a project management system and as a central repository of searchable barcode sequences.
4. DNA barcodes for specimen identification and species discovery
DNA barcoding is advancing biodiversity science by enabling the automated identification of specimens belonging to known species and by facilitating the recognition of new species [59]. Its capacity to deliver these insights depends upon the reliability with which sequence variation in each barcode region discriminates species. Within the animal kingdom, there is generally a gap between intraspecific and interspecific variation in COI sequences, so barcoding is highly effective. The situation in plants is more challenging; barcode divergences at ribulose-biphosphate carboxylase (rbcL) and matK are often so low that closely allied species cannot be discriminated [52,60]. Studies on fungi [61] and the many lineages of Protista also indicate cases of variable discriminatory power.
(a). Animals
DNA barcodes typically discriminate about 95% of known species; cases of compromised resolution involve sister taxa, often species that hybridize [19,20,62,63]. In the many taxa where geographical variation in barcode sequences is small [64], a few records per species are sufficient to create an effective identification system. However, the analysis of more specimens is advantageous because it often reveals discordances that indicate misidentifications or cryptic taxa [65], and it also provides insights into the extent of geographical variation in barcode sequences [66,67]. There are two animal phyla in which COI often fails to deliver species-level resolution, sponges [68,69] and some benthic cnidarians [70], apparently because of their slowed rates of mitochondrial evolution. Barcoding also fails to distinguish a small fraction of species in other groups, typically sister taxa or those whose status is uncertain [71,72]. Conversely, barcode analysis frequently exposes deep ‘intraspecific’ variation, situations that often represent overlooked species as evidenced by covariation with ecological or morphological traits [73–75]. However, some cases have other explanations; they seem linked to the merger of phylogeographic isolates [76], to rate acceleration [77] or to doubly uniparental inheritance [78]. There remains a need to clarify patterns of DNA barcode sequence variation by examining selected nuclear loci or through genome-wide approaches such as RAD sequencing [79] to extend understanding of factors explaining the origins and maintenance of these cases of deep mitochondrial divergence.
(b). Plants
DNA barcoding confronts the challenge that many plant species are exposed to hybridization and introgression, while others have arisen via polyploidy in a near-instantaneous fashion. Moreover, the evolutionary rates of their mitochondrial and plastid genomes are far slower than those in animals, creating a further barrier to species resolution. Given these factors, it is unsurprising that the designation of barcode markers for plants has been challenging. Although it was recognized that they would often not deliver species-level resolution, two plastid markers (matK and rbcL) were selected as the core barcodes for plants [16], supplemented with ancillary markers such as trnH–psbA, a plastid inter-genic spacer, and the ITS of nuclear ribosomal DNA [80,81]. Researchers focusing on highly degraded DNAs have also used a small plastid region from the trnL intron [82]. Collectively, DNA barcoding has been deployed widely for discriminating plant species or species groups [83–85]. The quest for improved barcode resolution in plants is ongoing [52]. The benefits of complete plastid genome sequencing have been noted by several authors [86–88] although this will not solve identification failures arising from plastid introgression, such as those presumed in Salix [89]. Ultimately, further substantial gains in plant species discrimination will depend on cost-effective, standardized and scalable approaches for accessing data from multiple unlinked nuclear markers [38,52,88].
(c). Fungi
ITS is the standard DNA barcode marker for fungi and has been widely adopted and used by mycologists [18,61]. The use of sequence data for species discovery and identification is particularly important in this kingdom, because so many fungal species are both undescribed and unculturable [90]. The recovery of ITS barcode sequences is sometimes compromised by intra-individual heterogeneity, reflecting its multi-copy nature [91], and alignment ambiguities can make it difficult to establish if the recovered sequence derives from the target species or a symbiont. As a consequence, there has been a search for secondary markers. COI has shown strong resolution in some groups [92], but its utility is constrained because the introns prevalent in fungal mitochondrial genomes often disrupt its PCR amplification from genomic DNA [93]. This fact has provoked studies on diverse nuclear gene regions, such as large and small subunit ribosomal DNA [94], but no secondary marker has gained broad adoption. As with plants, efforts are shifting towards the incorporation of wider genomic coverage into barcoding workflows, creating a challenge to balance between the need for increased resolution with the requirement for a cost-effective, highly scalable assay. Another key issue for fungi is the growing divide between identified taxa and sequences, driven by the rapid growth of ‘sequences without names’ produced from metabarcoding studies, and also the need to increase the proportion of newly described species that have barcode sequences generated from type material. This parallels the dark taxa challenges for other highly diverse groups [32,39] and further sequencing of fungal types coupled with community agreement on linking sequence-only records to a naming system is a high priority [61].
(d). Protists
Work on protists is in the early stages, but 18S RNA has been adopted as the core barcode marker [17] with full recognition that this gene region evolves too slowly to provide species resolution in most cases [40]. However, because primers for 18S are effective across diverse phyla, they can provide the sequence information needed for a ‘rough’ taxonomic placement that can be followed by the analysis of secondary barcodes to obtain species-level resolution. The selection of secondary markers for varied protistan lineages is underway, and some core markers, such as COI and rbcL, have demonstrated utility [95,96]. However, it is certain that both the selection and testing of the efficacy of barcode regions will be challenging given the extreme diversity of protistan lineages [97].
5. Impacts of DNA barcoding
Although motivated by the goal of accelerating the inventory of biodiversity and making taxonomic information more accessible, DNA barcoding is providing opportunities for important investigations in other fields of enquiry [98,99]. The balance of this section briefly considers some of the research areas aided by its advance.
(a). Probing species
DNA barcoding is shifting taxonomic workflows in two ways. Firstly, it is providing an increasingly effective identification ‘service’ for groups with a well-parametrized barcode reference library. Secondly, it is accelerating taxonomic progress by aiding the recognition of species and by facilitating the connection of their life stages [100] and sexes [101], associations that are often challenging without barcode data. For example, more than half of all genera of phorid flies are only known from one sex [102], creating high risk for synonymy. DNA barcoding also has a strong role in species discovery, especially in little-studied groups, because it can rapidly screen collections for presumptive species, which can then be targeted for taxonomic study [103]. DNA barcodes are additionally being employed to streamline and expedite species descriptions [104]. In fact, in hyperdiverse groups, the BIN registry may be the terminal taxonomic system, one allowing the assembly of morphological, ecological and distributional data for the members of each barcode cluster.
(b). Probing species assemblages
DNA barcoding is a powerful tool for advancing knowledge of species interactions and distributions [98,99]. It is often the sole way to clarify dietary preferences in taxa where direct observation of feeding behaviour is impossible [105,106]. It can also provide new details on host–parasitoid interactions [107], on pollination syndromes [108,109] and on symbiotic associations [110,111]. Aside from revealing interactions, DNA barcoding allows the assessment of biodiversity on scales [112] and in settings where this would otherwise be impossible [113]. By exploiting its capacity to improve species recognition and to reveal their interactions, DNA barcoding is also providing new details on food web structure [114–117]. Finally, DNA barcodes have been retrieved from ancient DNA, delivering insights into the evolution and ecology of extinct organisms [118].
(c). Probing evolution
Although DNA barcoding was initiated to empower taxonomy, the assembly of sequence information for a particular gene region across diverse taxa creates a resource useful in evolutionary contexts [119]. For example, patterns of sequence variation in the barcode region are an effective sentinel for shifts in the nucleotide composition of mitochondrial genomes [120] and provide a rich source of data for investigating molecular evolutionary rates [121,122]. Because species coverage is so comprehensive, DNA barcoding can also make useful contributions to phylogenetic studies [123]. Other potential applications await exploration. For example, expansion of each barcode record to include the entire sequence for COI or rbcL would deliver an unrivalled database for studying the evolutionary trajectories of these key proteins. It is important to emphasize the mutualism between DNA barcoding and studies which aim for deeper genomic characterization. For example, barcode analysis played an important role in verifying identifications for specimens analysed in the 1KITE initiative [124] because transcriptomic analysis required specimens to be processed while alive, often making morphological identification impossible. Aside from this role in validating identifications, the DNA extracts resulting from barcode analysis represent a resource for a future when sequencing costs have declined enough to allow the assembly of a whole-genome sequence for every species.
(d). Applying DNA barcodes
Because it facilitates specimen identifications, DNA barcoding has gained adoption in diverse applied contexts. It is, for example, now widely used to identify agricultural and forestry pests and pathogens [61,125,126], to detect invasive species [127] and for environmental impact assessments [43]. It has also become the standard method for suppressing marketplace fraud [128] and for deterring trade in endangered wildlife [129]. In addition, it is gaining use in forensic contexts [130], and in preventing illegal timber harvest [131]. Finally, DNA barcoding has proven a superb vehicle for exposing students to the practice of science [132].
6. What next?
Given past progress, what goals might the DNA barcode community set for the next quarter century? The assembly of a DNA barcode reference library for all multi-cellular species will effectively write the encyclopedia of life. Moreover, by coupling the automation of specimen identifications with the power of HTS to screen massive numbers of individuals, barcoding will enable a future in which reading life is routine. A global network of stations provisioned with sequencers, computational hardware and autonomous samplers [133] could track the shifting spectra of species in space and time, an Internet of living things, a world in which organisms act as transducers of biosphere change.
By completing the registry of all species by 2040, biodiversity science would deliver the foundation needed to track and forecast biotic change. Although this advance is within reach, it will require biodiversity science to join those disciplines that regard mega-science as everyday business. New structures, new alliances, and new leaders will be required to propel this transition. There is a critical need for action. More than half of all biodiversity hotspots have lost 90% of their vegetation [134], and the remnant patches are impacted by climate change. In fact, the least disturbed hotspot, the California Floristic Province, recently experienced its most severe drought in 1500 years [135]. Habitat fragmentation is also increasing; 70% of global forests lie within 1 km of a road [136]. These changes have lowered species abundances [137] and have increased extinction rates [138]. Because a sixth of all multi-cellular species may be extinct by the end of this century [139], there is an urgent need to complete the inventory of life and to use this information to track shifts in species abundances and distributions. Without interventions enabled by such knowledge, it is certain that endless forms most beautiful and most wonderful [140] will be lost. This prospect is surely a call to arms.
Acknowledgements
We are grateful to Sarah Adamowicz, Karl Kjer, John La Salle, Scott Miller and Dirk Steinke for helpful comments on earlier versions of this article. We also thank Suzanne Bateson, Sujeevan Ratnasingham and Dirk Steinke for their aid in generating the figures. We are very thankful to Ann McCain Evans and Chris Evans for their generosity in defraying the Open Access charges for this special issue.
Authors' contributions
P.D.N.H., P.M.H. and M.H. wrote the manuscript.
Competing interests
The authors declare no conflict of interests.
Funding
P.D.N.H. gratefully acknowledges the support of the Canada Research Chairs Program, the Canada Foundation for Innovation, NSERC and the Ontario Ministry of Research and Innovation. P.M.H. acknowledges funding from the Scottish Government's Rural and Environment Science and Analytical Services Division. M.H. acknowledges funding from Environment and Climate Change Canada.
References
- 1.Carbayo F, Marques AC. 2011. The costs of describing the entire animal kingdom. Trends Ecol. Evol. 25, 154–155. ( 10.1016/j.tree.2011.01.004) [DOI] [PubMed] [Google Scholar]
- 2.Savolainen V, Cowan RS, Vogler AP, Roderick GK, Lane R. 2005. On writing the encyclopaedia of life. Phil. Trans. R. Soc. B 360, 1805–1811. ( 10.1098/rstb.2005.1730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamowicz SJ. 2015. International barcode of life: evolution of a global research community. Genome 58, 151–162. ( 10.1139/gen-2015-0094) [DOI] [PubMed] [Google Scholar]
- 4.Janzen DH, Hajibabaei M, Burns JM, Hallwachs W, Remigio E, Hebert PDN. 2005. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B 360, 1835–1845. ( 10.1098/rstb.2005.1715) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller SE. 2007. DNA barcoding and the renaissance of taxonomy. Proc. Natl Acad. Sci. USA 104, 4775–4776. ( 10.1073/pnas.0700466104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janzen DH, et al. 2009. Integration of DNA barcoding into an ongoing inventory of complex tropical biodiversity. Mol. Ecol. Res. 9(Suppl 1), 1–26. ( 10.1111/j.1755-0998.2009.02628.x) [DOI] [PubMed] [Google Scholar]
- 7.Oxford GS, Rollinson D. 1983. Protein polymorphism: adaptive and taxonomic significance. Systematics Association. Special volume 24. London, UK: Academic Press. [Google Scholar]
- 8.Petitpierre E. 1996. Molecular cytogenetics and taxonomy of insects with particular reference to Coleoptera. Int. J. Insect Morph. Embryol. 25, 115–134. ( 10.1016/0020-7322(95)00024-0) [DOI] [Google Scholar]
- 9.Frey JE, Pfunder M. 2006. Molecular techniques for identification of quarantine insects and mites: the potential of microarrays. In Molecular diagnostics: current technology and applications (eds Roa JR, Fleming CC, Moore J), ch. 6, pp. 141–163. Wymondham, Norfolk, UK: Horizon Bioscience. [Google Scholar]
- 10.Will KN, Mishler BD, Wheeler QD. 2005. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 54, 844–851. ( 10.1080/10635150500354878) [DOI] [PubMed] [Google Scholar]
- 11.Rubinoff D, Cameron S, Will K. 2006. A genomic perspective on the shortcomings of mitochondrial DNA ‘barcoding’ identification. J. Heredity 97, 581–594. ( 10.1093/jhered/esl036) [DOI] [PubMed] [Google Scholar]
- 12.Funk DJ, Omland KE. 2003. Species-level paraphyly and polyphyly: frequency, causes and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Syst. 34, 397–423. ( 10.1146/annurev.ecolsys.34.011802.132421) [DOI] [Google Scholar]
- 13.Degnan JH, Rosenberg NA. 2009. Gene tree discordance, phylogenetic inference and the multispecies coalescent. Trends Ecol. Evol. 24, 332–340. ( 10.1016/j.tree.2009.01.009) [DOI] [PubMed] [Google Scholar]
- 14.Adamowicz SJ, Steinke D. 2015. Increased global participation in genetics research through DNA barcoding. Genome 58, 519–526. ( 10.1139/gen-2015-0130) [DOI] [PubMed] [Google Scholar]
- 15.Hebert PDN, Cywinska A, Ball SL, deWaard JR. 2003. Biological identifications through DNA barcodes. Proc. R. Soc. Lond B 270, 313–321. ( 10.1098/rspb.2002.2218) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollingsworth PM, et al. 2009. A DNA barcode for land plants. Proc. Natl Acad. Sci. USA, 106, 12 794–12 797. ( 10.1073/pnas.0905845106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pawlowski J, et al. 2012. CBOL protist working group: barcoding eukaryotic richness beyond the animal, plant, and fungal kingdoms. PLOS Biol. 17, e1001419 ( 10.1371/journal.pbio.1001419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schoch CL, et al. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl Acad. Sci. USA 109, 6241–6246. ( 10.1073/pnas.1117018109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. 2005. DNA barcoding Australia's fish species. Phil. Trans. R. Soc. B 360, 1847–1857. ( 10.1098/rstb.2005.1716) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PDN. 2006. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl Acad. Sci. USA 103, 968–971. ( 10.1073/pnas.0510466103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golding GB, Hanner RH, Hebert PDN. 2009. Preface. Mol. Ecol. Res. 9(Supp l. 1), iv–vi. ( 10.1111/j.1755-0998.2009.02654.x) [DOI] [PubMed] [Google Scholar]
- 22.Ivanova NV, deWaard JR, Hebert PDN. 2006. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol. Ecol. Notes 6, 998–1002. ( 10.1111/j.1471-8286.2006.01428.x) [DOI] [Google Scholar]
- 23.deWaard JR, Ivanova NV, Hajibabaei M, Hebert PDN. 2007. Assembling DNA barcodes: analytical protocols. In Methods in molecular biology: environmental genetics (ed. Martin C.), pp. 275–293. Totowa, NJ: Humana Press. [DOI] [PubMed] [Google Scholar]
- 24.Hebert PDN, deWaard JR, Zakahov EV, Prosser SWJ, Sones JE, McKeown JTA, Mantle DB, La Salle J. 2013. A DNA ‘Barcode Blitz’: rapid digitization and sequencing of a natural history collection. PLoS ONE 8, e68535 ( 10.1371/journal.pone.0068535) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ekrem T, Willasen E, Stur E. 2007. A comprehensive DNA sequence library is essential for identification with DNA barcodes. Mol. Phylogen. Evol. 43, 530–542. ( 10.1016/j.ympev.2006.11.021) [DOI] [PubMed] [Google Scholar]
- 26.Geiger MF, et al. In press. How to tackle the molecular species inventory for an industrialized nation – lessons from the first phase of the German Barcode of Life initiative GBOL (2012–2015). Genome 59 ( 10.1139/gen-2015-0185) [DOI] [PubMed] [Google Scholar]
- 27.Xu C, et al. 2015. Accelerating plant DNA barcode reference library construction using herbarium specimens: improved experimental techniques. Mol. Ecol. Res. 15, 1366–1374. ( 10.1111/1755-0998.12413) [DOI] [PubMed] [Google Scholar]
- 28.Hykin SM, Bi K, McGuire JA. 2015. Fixing formalin: a method to recover genomic-scale DNA sequence data from formalin fixed museum specimens using high-throughput sequencing. PLoS ONE 10, e0141579 ( 10.1371/journal.pone.0141579) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prosser S, deWaard JR, Miller SE, Hebert PDN. 2016. DNA barcodes from century-old type specimens using next generation sequencing. Mol. Ecol. Res. 16, 487–497. ( 10.1111/1755-0998.12474) [DOI] [PubMed] [Google Scholar]
- 30.Hausmann A, Miller SE, Holloway JD, deWaard JR, Pollock D, Prosser SWJ, Hebert PDN. In press. Calibrating the taxonomy of a megadiverse insect family: 3000 DNA barcodes from geometrid type specimens (Lepidoptera: Geometridae). Genome 59 ( 10.1139/gen-2015-0197) [DOI] [PubMed] [Google Scholar]
- 31.La Salle J, Williams KJ, Moritz C. 2016. Biodiversity analysis in the digital era. Phil. Trans. R. Soc. B 371, 20150337 ( 10.1098/rstb.2015.0337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Page RDM. 2016. DNA barcoding and taxonomy: dark taxa and dark texts. Phil. Trans. R. Soc. B 371, 20150334 ( 10.1098/rstb.2015.0334) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song H, Buhay JE, Whiting MF, Crandall KE. 2008. Many species in one: DNA barcoding overestimates the number of species when pseudogenes are amplified. Proc. Natl Acad. Sci. USA 105, 13 486–13 491. ( 10.1073/pnas.0803076105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gibson CM, Hunter MS. 2010. Extraordinarily widespread and fantastically complex: comparative biology of endosymbiotic bacterial and fungal mutualists of insects. Ecol. Lett. 13, 223–234. ( 10.1111/j.1461-0248.2009.01416.x) [DOI] [PubMed] [Google Scholar]
- 35.Pamilo P, Viljakainen L, Vahavainen A. 2007. Exceptionally high density of NUMTs in the honeybee genome. Mol. Biol. Evol. 24, 1340–1346. ( 10.1093/molbev/msm055) [DOI] [PubMed] [Google Scholar]
- 36.Smith MA, et al. 2012. Wolbachia and DNA barcoding insects: patterns, potential, and problems. PLoS ONE 7, e36514 ( 10.1371/journal.pone.0036514) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang W, Wendel JF, Clark LG. 1997. Bamboozled again! Inadvertent isolation of fungal DNA sequences from bamboos (Poeaceae: Bambusoideae). Mol. Phylogen. Evol. 8, 205–217. ( 10.1006/mpev.1997.0422) [DOI] [PubMed] [Google Scholar]
- 38.Hollingsworth PM, Graham SW, Little DP. 2011. Choosing and using a plant DNA barcode. PLoS ONE 6, e19254 ( 10.1371/journal.pone.0019254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hebert PDN, Ratnasingham S, Zakharov EV, Telfer AC, Levesque-Beaudin V, Milton MA, Pedersen S, Jannetta P, deWaard JR. 2016. Counting animal species with DNA barcodes: Canadian insects. Phil. Trans. R. Soc. B 371, 20150333 ( 10.1098/rstb.2015.0333) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang CQ, Leasi F, Obertegger U, Kieneke A, Barraclough TG, Fontaneto D. 2012. The widely used small subunit 18S rDNA molecule greatly underestimates true diversity in biodiversity surveys of the meiofauna. Proc. Natl Acad. Sci. USA 109, 16 208–16 212. ( 10.1073/pnas.1209160109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deagle BE, Jarman SN, Coissac E, Pompanon F, Taberlet P. 2014. DNA metabarcoding and the cytochrome c oxidase subunit I marker: not a perfect match. Biol. Lett. 10, 20140562 ( 10.1098/rsbl.2014.0562) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hajibabaei M, Shokralla S, Zhou X, Singer GA, Baird DJ. 2011. Environmental barcoding: a next-generation sequencing approach for biomonitoring applications using river benthos. PLoS ONE 6, e17497 ( 10.1371/journal.pone.0017497) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hajibabaei M, Baird DJ, Fahner NA, Beiko R, Golding GB. 2016. A new way to contemplate Darwin's tangled bank: how DNA barcodes are reconnecting biodiversity science and biomonitoring. Phil. Trans. R. Soc. B 371, 20150330 ( 10.1098/rstb.2015.0330) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E. 2012. Towards next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. ( 10.1111/j.1365-294X.2012.05470.x) [DOI] [PubMed] [Google Scholar]
- 45.Yu DW, Ji Y, Emerson BC, Wang X, Ye C, Yang C, Ding Z. 2012. Biodiversity soup: metabarcoding of arthropods for rapid biodiversity assessment and biomonitoring. Methods Ecol. Evol. 3, 613–623. ( 10.1111/j.2041-210X.2012.00198.x) [DOI] [Google Scholar]
- 46.Gibson JF, Shokralla S, Curry C, Baird DJ, Monk WA, King I, Hajibabaei M. 2015. Large-scale biomonitoring of remote and threatened ecosystems via high-throughput sequencing. PLoS ONE 10, e0138432 ( 10.1371/journal.pone.0138432) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lejzerowicz F, Esling P, Pillet L, Wilding TA, Black KD, Pawlowski J. 2015. High-throughput sequencing and morphology perform equally well for benthic monitoring of marine ecosystems. Sci. Rep. 5, 13932 ( 10.1038/srep13932) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leray M, Knowlton N. 2015. DNA barcoding and metabarcoding of standardized samples reveal patterns of marine benthic diversity. Proc. Natl Acad. Sci. USA 112, 2076–2081. ( 10.1073/pnas.1424997112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leray M, Knowlton N. 2016. Censusing marine eukaryotic diversity in the twenty-first century. Phil. Trans. R. Soc. B 371, 20150331 ( 10.1098/rstb.2015.0331) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shokralla S, Gibson JF, Nikbakht H, Janzen DH, Hallwachs W, Hajibabaei M. 2014. Next-generation DNA barcoding: using next-generation sequencing to enhance and accelerate DNA barcode capture from single specimens. Mol. Ecol. Res. 14, 892–901. ( 10.1111/1755-0998.12236) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shokralla S, Porter TM, Gibson JF, Dobosz R, Janzen DH, Hallwachs W, Golding GB, Hajibabaei M. 2015. Massively parallel multiplex DNA sequencing for specimen identification using an Illumina MiSeq platform. Sci. Rep. 5, 9687 ( 10.1038/srep09687) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hollingsworth PM, Li D-Z, van der Bank M, Twyford AD. 2016. Telling plant species apart with DNA: from barcodes to genomes. Phil. Trans. R. Soc. B 371, 20150338 ( 10.1098/rstb.2015.0338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ratnasingham S, Hebert PDN. 2007. BOLD: the Barcode of Life Data System. Mol. Ecol. Notes 7, 355–364. ( 10.1111/j.1471-8286.2007.01678.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blaxter M. 2016. Imagining Sisyphus happy: DNA barcoding and the unnamed majority. Phil. Trans. R. Soc. B 371, 20150329 ( 10.1098/rstb.2015.0329) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ratnasingham S, Hebert PDN. 2013. A DNA-based registry for all animal species: the Barcode Index Number (BIN) system. PLoS ONE 8, e66213 ( 10.1371/journal.pone.0066213) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nilsson RH, Ryberg M, Kristiansson E, Abarenkov K, Larsson K-H, Koljalg U. 2006. Taxonomic reliability of DNA sequences in public sequence databases: a fungal perspective. PLoS ONE 1, e59 ( 10.1371/journal.pone.0000059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen Y-Y, Chen X, Murphy RW. 2013. Assessing DNA barcoding as a tool for species identification and data quality control. PLoS ONE 8, e57125 ( 10.1371/journal.pone.0057125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roskov Y, et al. (eds). 2016. Species 2000 & ITIS catalogue of life, 2016 annual checklist. Leiden, The Netherlands: Naturalis Species 2000, Naturalis. http://www.catalogueoflife.org/annual-checklist/2016.
- 59.Mallo D, Posada D. 2016. Multilocus inference of species trees and DNA barcoding. Phil. Trans. R. Soc. B 371, 20150335 ( 10.1098/rstb.2015.0335) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cowan RS, Chase MW, Kress WJ, Savolainen V. 2006. 300,000 species to identify: problems, progress and prospects in the DNA barcoding of land plants. Taxon 55, 611–616. ( 10.2307/25065638) [DOI] [Google Scholar]
- 61.Yahr R, Schoch CL, Dentinger BTM. 2016. Scaling up discovery of hidden diversity in fungi: impacts of barcoding approaches. Phil. Trans. R. Soc. B 371, 20150336 ( 10.1098/rstb.2015.0336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hebert PDN, DeWaard JR, Landry JF. 2010. DNA barcodes for 1/1000 of the animal kingdom. Biol. Lett. 6, 359–362. ( 10.1098/rsbl.2009.0848) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pentinsaari M, Hebert PDN, Mutanen M. 2014. Barcoding beetles: a regional survey of 1872 species reveals high identification success and unusually deep interspecific divergences. PLoS ONE 9, e108651 ( 10.1371/journal.pone.0108651) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huemer P, Mutanen M, Sefc KM, Hebert PDN. 2014. Testing DNA barcode performance in 1000 species of European Lepidoptera: large geographic distances have small genetic impacts. PLoS ONE 9, e115774 ( 10.1371/journal.pone.0115774) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mutanen M, et al. In press. Species-level para- and polyphyly in DNA barcode gene trees: strong operational bias in European Lepidoptera. Syst. Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergsten J, et al. 2012. The effect of geographical scale of sampling on DNA barcoding. Syst. Biol. 61, 851–869. ( 10.1093/sysbio/sys037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mutanan M, Hausmann A, Landry JF, Hebert PDN, deWaard J, Huemer P. 2012. Allopatry as a Gordian knot for taxonomists: patterns of DNA barcode divergence in arctic-alpine Lepidoptera. PLoS ONE 7, e47214 ( 10.1371/journal.pone.0047214) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vargas S, et al. 2012. Barcoding sponges: an overview based on comprehensive sampling. PLoS ONE 7, e39345 ( 10.1371/journal.pone.0039345) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeBiasse MB, Neilsen BJ, Hellberg ME. 2014. Evaluating summary statistics used to test for incomplete lineage sorting: mito-nuclear discordance in the reef sponge Callispongia vaginalis. Mol. Ecol. 23, 225–238. ( 10.1111/mec.12584) [DOI] [PubMed] [Google Scholar]
- 70.McFadden CS, Benayahu Y, Pante E, Thoma JN, Nevarez PA, France SC. 2011. Limitations of mitochondrial gene barcoding in Octocorallia. Mol. Ecol. Res. 11, 19–31. ( 10.1111/j.1755-0998.2010.02875.x) [DOI] [PubMed] [Google Scholar]
- 71.Kerr KCR, Stoeckle MY, Dove CJ, Weigt LA, Francis CM, Hebert PDN. 2007. Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. Notes 7, 535–543. ( 10.1111/j.1471-8286.2007.01670.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hausmann A, Haszprunar G, Hebert PDN. 2011. DNA barcoding the geometrid fauna of Bavaria (Lepidoptera): successes, surprises and questions. PLoS ONE 6, e17134 ( 10.1371/journal.pone.0017134) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hebert PDN, Penton EH, Burns J, Janzen DH, Hallwachs W. 2004. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly, Astraptes fulgerator. Proc. Natl Acad. Sci. USA, 101, 14 812–14 817. ( 10.1073/pnas.0406166101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith MA, Woodley NE, Janzen DH, Hallwachs W, Hebert PDN. 2006. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae). Proc. Natl Acad. Sci. USA 103, 3657–3662. ( 10.1073/pnas.0511318103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Smith MA, et al. 2008. Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proc. Natl Acad. Sci. USA, 105, 12 359–12 364. ( 10.1073/pnas.0805319105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kvie KS, Hogner S, Aarvik L, Lifjeld JT, Johnsen A. 2013. Deep sympatric mtDNA divergence in the autumnal moth (Epirrita autumnata). Ecol. Evol. 3, 126–144. ( 10.1002/ece3.434) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pinceel J, Jordeans K, Backeljau T. 2005. Extreme mtDNA divergences in a terrestrial slug (Gastropoda, Pulmonata, Arionidae): accelerated evolution, allopatric divergence and secondary contact. J. Evol. Biol. 18, 264–280. ( 10.1111/j.1420-9101.2005.00932.x) [DOI] [PubMed] [Google Scholar]
- 78.Theologidis I, Fodeliankias S, Gaspar MB, Zouros E. 2010. Doubly uniparental inheritance (DUI) of mitochondrial DNA in Donax trunculus (Bivalvia: Donacidae) and the problem of its sporadic detection in Bivalvia. Evolution 62, 959–970. ( 10.1111/j.1558-5646.2008.00329.x) [DOI] [PubMed] [Google Scholar]
- 79.Davey JW, Blaxter ML. 2011. RADSeq: next generation population genetics. Brief. Funct. Genomics 9, 416–423. ( 10.1093/bfgp/elq031) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. 2005. Use of DNA barcodes to identify flowering plants. Proc. Natl Acad. Sci. USA 102, 8369–8374. ( 10.1073/pnas.0503123102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li DZ, et al. 2011. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl Acad. Sci. USA 108, 19 641–19 646. ( 10.1073/pnas.1104551108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Taberlet P, et al. 2007. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res. 35, e14 ( 10.1093/nar/gkl938) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, Sanjur O, Bermingham E. 2009. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc. Natl Acad. Sci. USA 106, 18 621–18 626. ( 10.1073/pnas.0909820106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Vere N, et al. 2012. DNA barcoding the native flowering plants and conifers of Wales. PLoS ONE 7, e37945 ( 10.1371/journal.pone.0037945) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kuzmina ML, Johnson KL, Barron HR, Hebert PDN. 2012. Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecol. 12, 25 ( 10.1186/1472-6785-12-25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kane NC, Cronk Q. 2008. Botany without borders: barcoding in focus. Mol. Ecol. 17, 5175–5176. ( 10.1111/j.1365-294X.2008.03972.x) [DOI] [PubMed] [Google Scholar]
- 87.Li X, Yang Y, Henry RJ, Rossetto M, Wang Y, Chen S. 2015. Plant DNA barcoding: from gene to genome. Biol. Rev. 90, 157–166. ( 10.1111/brv.12104) [DOI] [PubMed] [Google Scholar]
- 88.Coissac E, Hollingsworth PM, Lavergne S, Taberlet P. 2016. From barcodes to genomes: extending the concept of DNA barcoding. Mol. Ecol. 25, 1423–1428. ( 10.1111/mec.13549) [DOI] [PubMed] [Google Scholar]
- 89.Percy DM, et al. 2014. Understanding the spectacular failure of DNA barcoding in willows (Salix): does this result from a trans-specific selective sweep? Mol. Ecol. 23, 4737–4746. ( 10.1111/mec.12837) [DOI] [PubMed] [Google Scholar]
- 90.Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA. 2007. The current status of species recognition and identification in Aspergillus. Stud. Mycol. 59, 1–10. ( 10.3114/sim.2007.59.01) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kiss L. 2012. Limits of nuclear ribosomal DNA internal transcribed spacer (ITS) sequences as species barcodes for Fungi. Proc. Natl Acad. Sci. USA 109, E1811 ( 10.1073/pnas.1207143109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Seifert KA, Samson RA, deWaard JR, Houbraken J, Levesque C, Moncalvo JM, Louis-Seize G, Hebert PDN. 2007. Prospects for fungus identification using CO1 DNA barcodes, with Penicillium as a test case. Proc. Natl Acad. Sci. USA 104, 3901–3906. ( 10.1073/pnas.0611691104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ferandon C, Moukha S, Callac P, Benedetto JP, Castroviejo M, Barroso G. 2010. The Agaricus bisporus cox1 gene: the longest mitochondrial gene and the largest reservoir of mitochondrial group I introns. PLoS ONE 5, e14048 ( 10.1371/journal.pone.0014048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stockinger H, Kruger M, Schusler A. 2010. DNA barcoding of arbuscular mycorrhizal fungi. New Phytol. 187, 461–474. ( 10.1111/j.1469-8137.2010.03262.x) [DOI] [PubMed] [Google Scholar]
- 95.Le Gall L, Saunders GW. 2010. DNA barcoding is a powerful tool to uncover algal diversity: a case study of the Phyllophoraceae (Gigartinales, Rhodophyta) in the Canadian flora. J Phycol. 46, 374–389. ( 10.1111/j.1529-8817.2010.00807.x) [DOI] [Google Scholar]
- 96.Leliaert F, Verbruggen H, Vanormelingen P, Steen F, Lopez-Bautista JM, Zuccarello GC, de Clerck O. 2014. DNA-based species delimitation in algae. Eur. J. Phycol. 49, 179–196. ( 10.1080/09670262.2014.904524) [DOI] [Google Scholar]
- 97.Adl SM, et al. 2012. The revised classification of eukaryotes. J. Eukaryot. Microbiol. 59, 429–514. ( 10.1111/j.1550-7408.2012.00644.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Joly S, et al. 2014. Ecology in the age of DNA barcoding: the resource, the promise and the challenges ahead. Mol. Ecol. Res. 14, 221–230. ( 10.1111/1755-0998.12173) [DOI] [PubMed] [Google Scholar]
- 99.Kress WJ, Garcia-Robledo C, Uriarte M, Erickson DL. 2015. DNA barcodes for ecology, evolution and conservation. Trends Ecol. Evol. 30, 25–35. ( 10.1016/j.tree.2014.10.008) [DOI] [PubMed] [Google Scholar]
- 100.Steinke D, Connell A, Hebert PDN. In press. Linking adults and immatures of South African marine fishes. Genome 59 ( 10.1139/gen-2015-0212) [DOI] [PubMed] [Google Scholar]
- 101.Ekrem T, Stur E, Hebert PDN. 2010. Females do count: documenting Chironomidae (Diptera) species diversity using DNA barcoding. Organ. Divers. Evol. 10, 397–408. ( 10.1007/s13127-010-0034-y) [DOI] [Google Scholar]
- 102.Disney RHL. 1994. Scuttle flies: the Phoridae. London, UK: Chapman & Hall. [Google Scholar]
- 103.Kekkonen M, Hebert PDN. 2014. DNA barcode-based delineation of putative species: efficient start for taxonomic workflows. Mol. Ecol. Res. 14, 706–715. ( 10.1111/1755-0998.12233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Riedel A, Sagata K, Suhardjono YR, Tanzler R, Balke M. 2013. Integrative taxonomy on the fast track - towards more sustainability in biodiversity research. Front. Zool. 10, 15 ( 10.1186/1742-9994-10-15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Clare EL, Fraser EE, Braid HE, Fenton MB, Hebert PDN. 2009. Species on the menu of a generalist predator, the eastern red bat (Lasiurus borealis): using a molecular approach to detect arthropod prey. Mol. Ecol. 18, 2532–2542. ( 10.1111/j.1365-294X.2009.04184.x) [DOI] [PubMed] [Google Scholar]
- 106.Valdez-Moreno M, Gomez-Lozano R, del Carmen Garcia Rivas M. 2012. Monitoring an alien invasion: DNA barcoding and the identification of lionfish and their prey on coral reefs of the Mexican Caribbean. PLoS ONE 7, e36636 ( 10.1371/journal.pone.0036636) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rougerie R, Smith MA, Fernandez-Triana J, Lopez-Vaamonde C, Ratnasingham S, Hebert PDN. 2010. Molecular analysis of parasitoid linkages (MAPL): gut contents of adult parasitoids reveal larval hosts. Mol. Ecol. 20, 179–186. ( 10.1111/j.1365-294X.2010.04918.x) [DOI] [PubMed] [Google Scholar]
- 108.Clare EL, Schiestl FP, Leitch AR, Chittka L. 2013. The promise of genomics in the study of plant-pollinator interactions. Genome Biol. 14, 207 ( 10.1186/gb-2013-14-6-207) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bell KL, de Vere N, Keller A, Richardson RT, Gous A, Burgess KS, Brosi BJ. In press. Pollen DNA barcoding: current applications and future prospects. Genome 59 ( 10.1139/gen-2015-02000) [DOI] [PubMed] [Google Scholar]
- 110.Baker CCM, Bittleston LS, Sanders JG, Pierce NE. 2016. Dissecting host-associated communities with DNA barcodes. Phil. Trans. R. Soc. B 371, 20150328 ( 10.1098/rstb.2015.0328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.McLean AHC, Parker BJ, Hrček J, Henry LM, Godfray HCJ. 2016. Insect symbionts in food webs. Phil. Trans. R. Soc. B 371, 20150325 ( 10.1098/rstb.2015.0325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miller SE, Hausmann A, Hallwachs W, Janzen DH. 2016. Advancing taxonomy and bioinventories with DNA barcodes. Phil. Trans. R. Soc. B 371, 20150339 ( 10.1098/rstb.2015.0339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Brehm G, Hebert PDN, Colwell RK, Adams M-O, Bodner F, Friedemann K, Mockel L, Fiedler K. 2016. Turning up the heat on a hotspot: DNA barcodes reveal 80% more species of geometrid moths along an Andean elevational gradient. PLoS ONE 11, e0150327 ( 10.1371/journal.pone.0150327) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kaartinen R, Stone GN, Hearn J, Lohse K, Roslin T. 2010. Revealing secret liaisons: DNA barcoding changes our understanding of food webs. Ecol. Entomol. 35, 623–638. ( 10.1111/j.1365-2311.2010.01224.x) [DOI] [Google Scholar]
- 115.Smith MA, Eveleigh EE, McCann KS, Merilo MT, McCarthy PC, Van Rooyen KI. 2011. Barcoding a quantified food web: crypsis, concepts, ecology and hypotheses. PLoS ONE 6, e14424 ( 10.1371/journal.pone.0014424) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wirta HK, Hebert PDN, Kaartinen R, Prosser SW, Varkonyi G, Roslin T. 2014. Complementary molecular information changes our perception of food web structure. Proc. Natl Acad. Sci. USA 111, 1885–1890. ( 10.1073/pnas.1316990111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roslin T, Majaneva S. In press. The use of DNA barcodes in food web construction – terrestrial and aquatic ecologists unite! Genome 59 ( 10.1139/gen-2015-0229) [DOI] [PubMed] [Google Scholar]
- 118.Speller C, et al. 2016. Barcoding the largest animals on Earth: ongoing challenges and molecular solutions in the taxonomic identification of ancient cetaceans. Phil. Trans. R. Soc. B 371, 20150332 ( 10.1098/rstb.2015.0332) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hajibabaei M, Singer GAC, Hebert PDN, Hickey DA. 2007. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 23, 167–172. ( 10.1016/j.tig.2007.02.001) [DOI] [PubMed] [Google Scholar]
- 120.Min XJ, Hickey DA. 2007. DNA barcodes provide a quick preview of mitochondrial genome composition. PLoS ONE 2, e325 ( 10.1371/journal.pone.0000325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mitterboeck TF, Adamowicz SJ. 2013. Flight loss linked to faster molecular evolution in insects. Proc. R. Soc. B 280, 20131128 ( 10.1098/rspb.2013.1128) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Young MR, Hebert PDN. 2015. Patterns of protein evolution in cytochrome c oxidase 1 (COI) from the class Arachnida. PLoS ONE 10, e0138167 ( 10.1371/journal.pone.0138167) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou X, et al. 2016. The Trichoptera barcode initiative: a strategy for generating a species-level Tree of Life. Phil. Trans. R. Soc. B 371, 20160025 ( 10.1098/rstb.2016.0025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. ( 10.1126/science.1257570) [DOI] [PubMed] [Google Scholar]
- 125.Floyd R, Lima J, deWaard J, Humble L, Hanner R. 2010. Common goals: policy implications of DNA barcoding as a protocol for identification of arthropod pests. Biol. Invasions 12, 2947–2954. ( 10.1007/s10530-010-9709-8) [DOI] [Google Scholar]
- 126.Frewin A, Scott-Dupree C, Hanner RH. 2013. DNA barcoding for plant protection: applications and summary of available data for arthropod pests. CAB Rev. 8, 1–13. ( 10.1079/PAVSNNR20138018) [DOI] [Google Scholar]
- 127.Briski E, Cristescu ME, Bailey SA, MacIsaac HJ. 2011. Use of DNA barcoding to detect invertebrate invasive species from diapausing eggs. Biol. Invas. 13, 1325–1340. ( 10.1007/s10530-010-9892-7) [DOI] [Google Scholar]
- 128.Handy SM, et al. 2011. A single-laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance. J. AOAC Int. 94, 201–210. [PubMed] [Google Scholar]
- 129.Yan D, Luo JY, Han YM, Peng C, Dong XP, Gen SL, Sun LG, Xiao XH. 2013. Forensic DNA barcoding and bio-response studies of animal horn products used in traditional medicine. PLoS ONE 8, e55854 ( 10.1371/journal.pone.0055854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rolo EA, Oliviera EA, Dourado CG, Farinha A, Rebelo MT, Dias D. 2013. Identification of sarcosaprophagous Diptera species through DNA barcoding in wildlife forensics. Forensic Sci. Int. 223, 160–164. ( 10.1016/j.forsciint.2013.02.038) [DOI] [PubMed] [Google Scholar]
- 131.Nielsen LR, Kjaer ED. 2008. Tracing timber from forest to consumer with DNA markers. Copenhagen, Denmark: Danish Ministry of the Environment, Forest and Nature Agency. [Google Scholar]
- 132.Henter HJ, Imondi R, James K, Spencer D, Steinke D. 2016. DNA barcoding in diverse educational settings: five case studies. Phil. Trans. R. Soc. B 371, 20150340 ( 10.1098/rstb.2015.0340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Herfort L, et al. 2016. Use of continuous, real-time observations and model simulations to achieve autonomous, adaptive sampling of microbial processes with a robotic sampler. Limnol. Ocean. Methods 14, 50–67. ( 10.1002/lom3.10069) [DOI] [Google Scholar]
- 134.Sloan S, Jenkins CN, Joppa LN, Gaveau DLA, Laurance WF. 2014. Remaining natural vegetation in the global biodiversity hotspots. Biol. Conserv. 177, 12–24. ( 10.1016/j.biocon.2014.05.027) [DOI] [Google Scholar]
- 135.Griffin D, Anchukaitis KJ. 2014. How unusual is the 2012–2014 California drought? Geophys. Res. Lett. 41, 9017–9023. ( 10.1002/2014GL062433) [DOI] [Google Scholar]
- 136.Haddad NM, et al. 2015. Habitat fragmentation and its lasting impact on the Earth's ecosystems. Sci. Adv. 1, e15000052 ( 10.1126/sciadv.1500052) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.McLellan R, Ivengar L, Jeffries B, Oerlemans N (eds). 2014. Living Planet 2014 Report Summary. Gland, Switzerland: World Wildlife Fund. [Google Scholar]
- 138.Ceballos G, Ehrlich PR, Barnovsky AD, Garcia A, Pringle RM, Palmer TM. 2015. Accelerated modern human-induced species losses: entering the sixth mass extinction. Sci. Adv. 1, e1400253 ( 10.1126/sciadv.1400253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Urban MC. 2015. Accelerating extinction risk from climate change. Science 348, 571–573. ( 10.1126/science.aaa4984) [DOI] [PubMed] [Google Scholar]
- 140.Darwin C. 1859. On the origin of species. London, UK: John Murray. [Google Scholar]