Abstract
Background:
The Philippines is home to some ethnomedicinal Apocynaceae that has been used to cure common ailments. They are perceived to be safe, but misidentification can lead to substitution and adulteration. Morphological characters are primarily utilized to identify these species but a new method utilizing molecular characters called DNA barcoding has emerged. In this study, the efficiency of matK, rbcL, trnH-psbA, and trnL-F to molecularly authenticate selected Apocynaceae species were tested.
Materials and Methods:
Genomic DNA from silica-dried leaf samples were isolated and used as a template for generating DNA barcodes. Pair-wise sequence divergence using Kimura-2-Parameter was used to analyze inter-specific and intraspecific variations among the barcodes, whereas basic local alignment search tool (BLAST) and neighbor-joining (NJ) analyses were employed to examine discrimination success.
Results:
The results show that matK is the best barcode for Apocynaceae as it has the highest amplification and sequencing success together with rbcL while having high inter-specific and low intra-specific divergence relative to the other candidate barcodes. Furthermore, matK provided the highest discrimination both in BLAST and NJ analyses.
Conclusion:
This study proposes the use of matK as the principal barcode for Apocynaceae.
SUMMARY
Both matK and rbcL have higher universality compared to trnH-psbA and trnL-F
matK has relatively high inter-specific divergence and very minimal intra-specific divergence
matK is the best barcode to molecularly authenticate Apocynaceae with either trnH-psbA or trnL-F as supplements.
Abbreviations used: K2P: Kimura-2-parameter, BLAST: Basic local alignment search tool, NJ: Neighbor-joining.
Keywords: Apocynaceae, DNA barcoding, ethnomedicinal, molecular authentication
INTRODUCTION
The people in Philippine provinces are known to utilize the native plants of their area as logical and practical sources of medicine that provide symptomatic relief for common ailments.[1] Among these plant sources are the members of the family Apocynaceae (sub-family Rauvolfioideae) such as Alstonia macrophylla Wall. and G. Don and Alstonia scholaris (L.) R. Br. which are traditionally used as an emmenagogue, anti-choleric, and vulnerary and the endemic Voacanga globosa (Blanco) Merr. for cancer and tuberculosis.[2,3] Medicinal plants are being marketed today as herbal medicine or natural health products. They are often perceived to be safe but adulterated, counterfeit and low quality products pose serious safety threats to consumers[4,5] as well as to the existing markets. Incorrect identification using primarily morphological characters of many plants has resulted in adulteration and substitution of plant products that compromise their therapeutic value.[6,7,8] Morphological characterization remains the cornerstone of taxonomic diagnosis in plants. Unfortunately, relying solely on morphology has some considerable limitations.
The evolution of molecular biology gave rise to a new approach based on nucleotide sequence diversities among species called DNA barcoding.[9,10] Short standardized segment of the genome serving as a pattern “barcode’’ has been proposed as a technology that offers to expedite accurate species identification.[11,12] The consortium for the barcode of the life (CBOL) plant working group has suggested matK and rbcL as the core barcode regions for plants.[13]
DNA barcodes are increasingly recognized for their ability to authenticate medicinal plants.[14,15,16,17] In this study, we evaluated the efficiency of four candidate DNA barcodes-matK, rbcL, trnH-psbA, and trnL-F (cpDNA) for molecular authentication of selected Philippine ethnomedicinal Apocynaceae species namely A. macrophylla, A. scholaris, V. globosa, Allamanda cathartica (L.,) Tabernaemontana pandacaqui Poir., Catharanthus roseus (L.) G. Don., and Thevetia peruviana (Pers.) K. Schum. The candidate barcodes were assessed using the criteria set by CBOL stating that an ideal barcode should be routinely retrievable with a single primer pair, be amenable to bidirectional sequencing with little requirement for manual editing of sequence traces, and provide maximal discrimination among species.[13,18,19]
MATERIALS AND METHODS
Sample collection and preservation
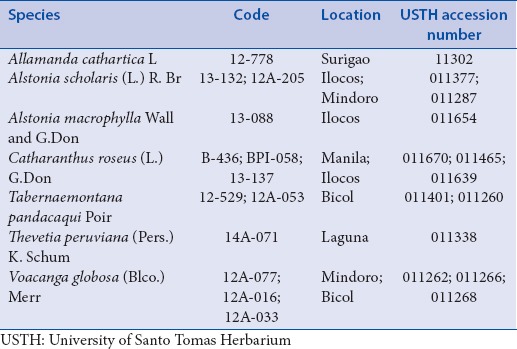
Field collections were conducted in different Philippine provinces. Leaf samples for each specimen were stored in re-sealable packs with silica beads. All specimens were provided with herbarium vouchers, currently stored in the University of Santo Tomas Herbarium [Table 1].
Table 1.
List of Apocynaceae species used in the study with their University of Santo Tomas Herbarium accession numbers
Generation of DNA barcodes
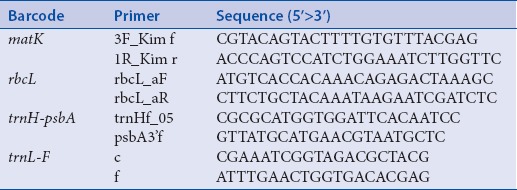
Total genomic DNA was extracted from silica gel-dried leaf tissues following the protocols of DNeasy Plant Minikit (Qiagen, Germany). The universal primer pairs for matK,[19] rbcL,[20] trnH-psbA,[18] and trnL-F[21] were amplified using Kapa Taq PCR Kit (Kapa, USA) in Biometra® T-Gradient thermocycler [Table 2]. The polymerase chain reaction (PCR) cocktail for all markers contained (25 μL reaction): 17.35 μL water, 2.5 μL ×10 buffer, 1.0 μL 25 mM MgCl2, 2 μL 2 mM dNTP, 1.0 μL of 10 μM forward and reverse primers, 0.15 μL Taq DNA polymerase, and 0.5 μL DNA template. PCR conditions were set as follows: Initial denaturation of 90s at 97°C, followed by 35 cycles of 30s 95°C; 20s 50°C (matK and rbcL)/55°C (trnH-psbA)/20s 72°C (trnL-F); 60s 72°C; finishing with 72°C for 10 min.[22]
Table 2.
Universal primers used in this study
The amplicons were resolved in agarose gel electrophoresis and specific DNA fragments were purified using the QIA-quick PCR Purification Kit (Qiagen, Germany). Purified DNA was sent to Macrogen Inc. Seoul, South Korea for bidirectional DNA sequencing. All DNA sequences were assembled and edited using CodonCode Aligner v.4.1.1 (CodonCode Co., USA).
Sequence analyses
The sequences of candidate DNA regions were automatically and manually aligned in SeaView v.4.[23] Pair-wise sequence divergence was calculated using Kimura-2-Parameter (K2P)[24] model in MEGA 6.0[25] to evaluate the mean intra-specific divergence, coalescent depth, mean inter-specific distance, and the minimum inter-specific distance.[14,26] The Wilcoxon tests for inter- and intra-specific divergences were conducted using SPSS 15.0 software (SPSS Inc., Chicago, USA). Basic local alignment search tool (BLAST, NCBI-Gen Bank) analysis of acquired sequences was conducted as previously described.[14] Neighbor-joining (NJ) method using K2P distances (1000 bootstrap replicates) were conducted to test phylogenetic relationships in MEGA 6.0.
RESULTS
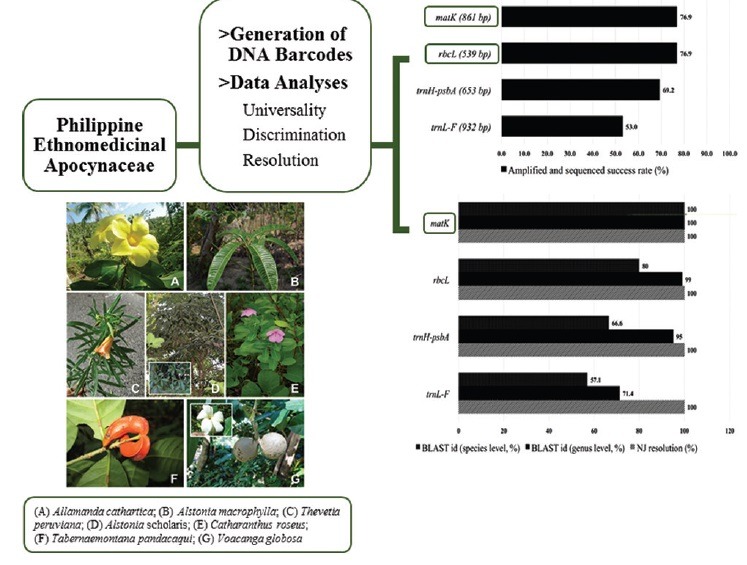
A total of 13 Apocynaceae specimens representing seven species (A. macrophylla, A. scholaris, V. globosa, A. cathartica, T. pandacaqui, C. roseus, and T. peruviana) were obtained. Four candidate barcodes (matK, rbcL, trnH-psbA, trnL-F) were used to investigate the feasibility of DNA barcoding for molecular authentication. The 36 new sequences [Appendix 1] obtained from all markers and 14 sequences from Gen Bank were combined for the succeeding analyses. The Gen Bank sequences ensure each species has, at least, a duplicate sequence for intraspecific computation. A previous study by Mahadani et al.[15] also looked into the application of DNA barcoding in ethnomedicinal Apocynaceae but they only included two markers for the analyses. This issue is addressed in our study by contrasting the viability of the four candidate barcodes. For amplification and sequencing success, both matK and rbcL have 76.9% while trnH-psb A and trnL-F have 69.2% and 53.0%, respectively [Figure 1].
Figure 1.
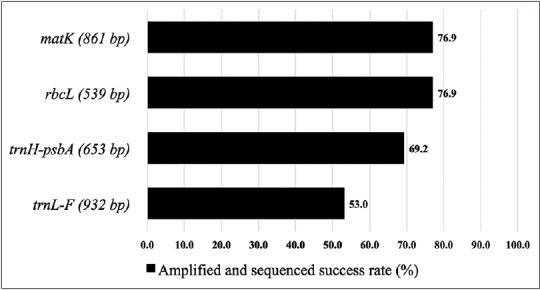
Efficiency of polymerase chain reaction amplification and sequencing success of the four candidate barcodes for the selected Apocynaceae. Numbers in parentheses are aligned lengths
To test the sequence divergence of each barcode, pair wise divergence was calculated using K2P. Table 3 summarizes the results having trnH-psbA as the barcode with the highest inter-specific divergence and rbcL with the lowest with 0.327 ± 0.090 and 0.028 ± 0.007, respectively. For the intra-specific divergences, trnH-psbA also has highest value with and 0.049 ± 0.046 while matK has the lowest the lowest with 0.002 ± 0.002. All barcodes were able to identify more than 50% of the specimens at species level using BLAST and gave good resolutions in NJ [Figures 2 and 3].
Table 3.
Inter- and intra-specific divergences of the four barcodes for selected Apocynaceae inferred using kimura-2-parameter distances
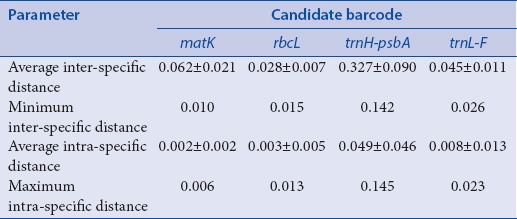
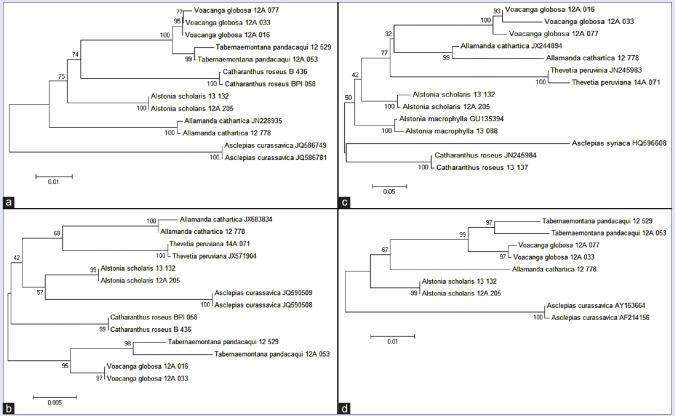
Figure 2.
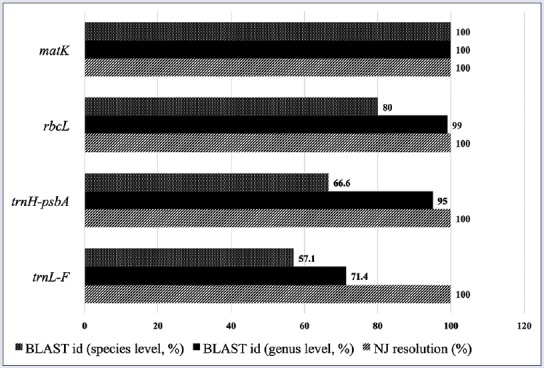
Rate of correct identification of the four barcodes using basic local alignment search tool and neighbor-joining for the selected Apocynaceae
Figure 3.
Neighbor-Joining tree of (a) matK, (b) rbcL, (c) trnH-psbA, and (d) trnL-F sequences inferred using Kimura-2-Parameter distances. All barcodes have successfully resolved each species but only matK and trnL-F provided proper polarity with Asclepia curassavica as the out group
DISCUSSION
Universality of the four candidate barcodes
For a barcode to be considered universal, it must be tractable across a wide range of species. Ideally, it should be relatively short in length to facilitate easy DNA extraction, amplification, and sequencing.[18] Both matK and rbcL have higher success rate thus, higher universality compared to trnH-psbA and trnL-F. These are after several attempts to amplify using pure and diluted (1/10 and 1/100) DNA extracts.
Inter-versus intra-specific divergence and barcoding gap analyses
Two parameters (average inter-specific distance and the minimum inter-specific distance) were employed to characterize inter-specific divergence, and two others (average intra-specific distance and maximum intra-specific distance) were used to indicate intra-specific variation. By comparison on the inter-specific divergences of four candidate DNA regions (matK, rbcL, trnH-psbA, and trnL-F), trnH-psbA has the highest average inter-specific distance followed by trnL-F and rbcL, respectively. Even with its relatively short length (approximately 450 bp), trnH-psbA is considered as the most variable plastid region in angiosperms and is easily amplified across a broad range of land plants with the potential to discriminate among the largest number of plant species for barcoding purposes.[16,18,27]
The Wilcoxon signed rank test among the markers confirms the significant difference between each pair. The same pattern for intra-specific variation is observed. Specifically, intra-specific variation of trnH-psbA is higher than those of matK, trnL-F, and rbcL, but there exists no significant difference between those of the latter three candidate DNA regions [Tables 3 and 4]. These results point out that matK is better than the rest of the candidate barcodes since it has relatively high inter-specific divergence and very minimal intraspecific divergence compared to trnH-psbA. Using the Wilcoxon two-sample test, there are significant differences between the inter- and intra-specific divergences of the candidate DNA regions, with their inter-specific divergences significantly higher than their related intra-specific variations [P < 0.05, Table 5]. In consequence, all of them have a potential to discriminate one species from another.
Table 4.
Wilcoxon signed rank tests for inter- and intra-specific divergences among the four barcodes
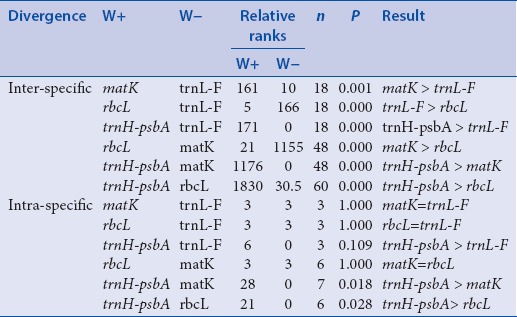
Table 5.
Wilcoxon two-sample test for inter- and intra-specific divergences of the four barcodes

Since the sample size could barely represent a portion of the entire Apocynaceae, DNA barcoding gap analysis is only restricted to the lack of overlap (“gap”) between minimum inter- and maximum intra-specific divergences, or presence thereof.[28] Based on this, only trnH-psbA has an overlap although the gaps (i.e., difference between minimum inter- and maximum intra-specific divergences) of the other three barcodes are only <0.005 [Table 3].
Efficiency of the four cpDNA markers for resolving identity
One of the main purposes of DNA barcoding is to identify unknown species by matching a particular barcode sequence to available confirmed sequences. This is addressed here using BLAST and NJ method. Correct matches in BLAST and resolutions in NJ (i.e., monophyly of conspecific specimens) are considered as positive results. As shown in Figure 2, matK has the highest rates of correct identification in BLAST, both at the genus and species level. It is followed by rbcL, trnH-psbA, and trnL-F, respectively. The limited available published sequences in the Gen Bank, especially for the endemic species V. globosa, played a big factor. It cannot however, deny the fact that matK has a superior ability for authentication compared to other candidate barcodes.[13]
This finding corroborates with the NJ resolution results [Figure 3]. All the candidate barcodes are successful in resolving the monophyly of all con-specific specimens. However, only matK and trnL-F provide the expected polarity in the NJ trees which show Asclepias curassavica L. as the out-group. This observation is in agreement with the findings of Mahadani et al.[15] Hollingsworth et al.[19] stated that for barcoding purposes, rbcL meets all the criteria by being easy to amplify, sequence, and align in most land plants. Regrettably, it only has modest discriminatory power, evident in our results. Further, no other 2-marker or multi-marker plastid barcode gave better species resolution than the rbcL + matK combination. In this study, matK is the best barcode to molecularly authenticate Apocynaceae with either trnH-psbA or trnL-F as supplements.
CONCLUSION
In total, our results support the claim that DNA barcoding in general can provide fast and reliable species identification, especially for the economically important ethnomedicinal plants. In the case of the Apocynaceae species, matK is the best barcode for molecular authentication as it gives high universality, discriminatory ability, and can resolve phylogenetic placement accurately. The authors propose the application of matK as the primary DNA barcode for Apocynaceae with trnH-psbA, and trnL-F as supplementary barcodes.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
ABOUT AUTHOR

Vincent Louie D. Cabelin
Vincent Louie D. Cabelin, is an instructor at Notre Dame of Dadiangas University. Dr. Cabelin is currently completing his PhD in Biological Science at the Graduate School (GS) of University of Santo Tomas (UST).
Acknowledgments
The authors would like to thank the Department of Science and Technology Science (DOST)-Philippine Council for Health Research and Development, the Research Center for the Natural and Applied Sciences and Thomasian Angiosperm Phylogeny and Barcoding Group (TAPBG) of UST, and DOST-Science Educational Institute under the Accelerated S and T Human Resource Development Program.
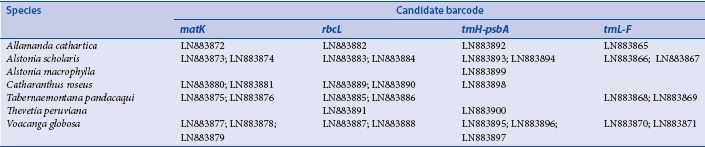
Appendix 1: EMBL accession numbers of the sequences generated in this study
REFERENCES
- 1.Maramba NPC, Saludez JD, Sia IC, Alegre OY, Solis-de Asis GA, Bagnaes LB, et al. Quezon City, Philippines: Katha Publishing Co, Inc; 1982. Guidebook on the proper use of medicinal plants. [Google Scholar]
- 2.Hendrian R. Voacanga globosa (Blanco) Merr. In: van Valkenburg JLCH, Bunyapraphatsara, editors. Plant Resources of South-East Asia No. 12(2): Medicinal and poisonous plants 2. Leiden, The Netherlands: Backhuys Publisher; 2001. pp. 584–5. [Google Scholar]
- 3.Macabeo AP, Vidar WS, Chen X, Decker M, Heilmann J, Wan B, et al. Mycobacterium tuberculosis and cholinesterase inhibitors from Voacanga globosa. Eur J Med Chem. 2011;46:3118–23. doi: 10.1016/j.ejmech.2011.04.025. [DOI] [PubMed] [Google Scholar]
- 4.Mine Y, Young D. Regulation of natural health products in Canada. Food Sci Technol Res. 2009;15:459–68. [Google Scholar]
- 5.Geneva: World Health Organization; 2008. [Last accessed on 2014 Mar 21]. World Health Organization. Traditional Medicine, Fact Sheet No. 134. Available from: http://www.who.int/mediacentre/factsheets/fs134/en/ [Google Scholar]
- 6.Bilia AR. NMR as integrative or alternative analytical tool for the quality control of herbal drugs and their preparations. Planta Med. 2013;79:OP18. [Google Scholar]
- 7.Roy A, Mallick A, Kaur A. Adulteration and substitution in Indian medicinal plants. IJPRBS. 2013;2:208–18. [Google Scholar]
- 8.Viljoen AM, Vermaak I. Standardization and quality control of herbal medicinal products – Does vibrational spectroscopy offer the solution? Planta Med. 2013;79:OP20. [Google Scholar]
- 9.Hsieh HM, Chiang HL, Tsai LC, Lai SY, Huang NE, Linacre A, et al. Cytochrome b gene for species identification of the conservation animals. Forensic Sci Int. 2001;122:7–18. doi: 10.1016/s0379-0738(01)00403-0. [DOI] [PubMed] [Google Scholar]
- 10.Parson W, Pegoraro K, Niederstätter H, Föger M, Steinlechner M. Species identification by means of the cytochrome b gene. Int J Legal Med. 2000;114:23–8. doi: 10.1007/s004140000134. [DOI] [PubMed] [Google Scholar]
- 11.Hebert PD, Cywinska A, Ball SL, deWaard JR. Biological identifications through DNA barcodes. Proc Biol Sci. 2003;270:313–21. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waugh J. DNA barcoding in animal species: Progress, potential and pitfalls. Bioessays. 2007;29:188–97. doi: 10.1002/bies.20529. [DOI] [PubMed] [Google Scholar]
- 13.Hollingsworth PM, Graham SW, Little DP. Choosing and using a plant DNA barcode. PLoS One. 2011;6:e19254. doi: 10.1371/journal.pone.0019254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Yao H, Han J, Liu C, Song J, Shi L, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One. 2010;5:e8613. doi: 10.1371/journal.pone.0008613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahadani P, Sharma GD, Ghosh SK. Identification of ethnomedicinal plants (Rauvolfioideae: Apocynaceae) through DNA barcoding from Northeast India. Pharmacogn Mag. 2013;9:255–63. doi: 10.4103/0973-1296.113284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song J, Yao H, Li Y, Li X, Lin Y, Liu C, et al. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. J Ethnopharmacol. 2009;124:434–9. doi: 10.1016/j.jep.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, Wang J, Xia T, Zhou S. DNA barcoding: Species delimitation in tree peonies. Sci China C Life Sci. 2009;52:568–78. doi: 10.1007/s11427-009-0069-5. [DOI] [PubMed] [Google Scholar]
- 18.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proc Natl Acad Sci U S A. 2005;102:8369–74. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, van der Bank M, et al. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009;106:12794–7. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kress WJ, Erickson DL. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS One. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taberlet P, Gielly L, Pautou G, Bouvet J. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol Biol. 1991;17:1105–9. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Au KY, Lam H, Cheng L, Jiang RW, But PP, et al. Identification of Baiying (Herba Solani Lyrati) commodity and its toxic substitute Xungufeng (Herba Aristolochiae Mollissimae) using DNA barcoding and chemical profiling techniques. Food Chem. 2012;135:1653–8. doi: 10.1016/j.foodchem.2012.06.049. [DOI] [PubMed] [Google Scholar]
- 23.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–4. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 24.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yao H, Song J, Liu C, Luo K, Han J, Li Y, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS One. 2010:5. doi: 10.1371/journal.pone.0013102. pii: E13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao H, Song JY, Ma XY, Liu C, Li Y, Xu HX, et al. Identification of Dendrobium species by a candidate DNA barcode sequence: The chloroplast psbA-trnH intergenic region. Planta Med. 2009;75:667–9. doi: 10.1055/s-0029-1185385. [DOI] [PubMed] [Google Scholar]
- 28.Meyer CP, Paulay G. DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol. 2005;3:e422. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]